An opportunistic bacterial infection preceded by weather of unusually high humidity and temperature caused mass death of saigas.
Abstract
In 2015, more than 200,000 saiga antelopes died in 3 weeks in central Kazakhstan. The proximate cause of death is confirmed as hemorrhagic septicemia caused by the bacterium Pasteurella multocida type B, based on multiple strands of evidence. Statistical modeling suggests that there was unusually high relative humidity and temperature in the days leading up to the mortality event; temperature and humidity anomalies were also observed in two previous similar events in the same region. The modeled influence of environmental covariates is consistent with known drivers of hemorrhagic septicemia. Given the saiga population’s vulnerability to mass mortality and the likely exacerbation of climate-related and environmental stressors in the future, management of risks to population viability such as poaching and viral livestock disease is urgently needed, as well as robust ongoing veterinary surveillance. A multidisciplinary approach is needed to research mass mortality events under rapid environmental change.
INTRODUCTION
In May 2015, researchers witnessed a catastrophic disease event causing the death of some 200,000 saiga antelopes (Saiga tatarica tatarica) in calving aggregations, across a landscape stretching for several hundreds of kilometers near the Torgai lowland, central Kazakhstan (Fig. 1, fig. S1, and the Supplementary Materials). The syndrome attracted global media attention and considerable speculation as to its cause. By June, adult numbers were reduced to around 15% of the pre-calving numbers of this population, representing a loss of around 62% of the global population of this critically endangered species (1).
Fig. 1. 2015 MME sites showing dates of onset and the location of the two sites studied in detail in the field.
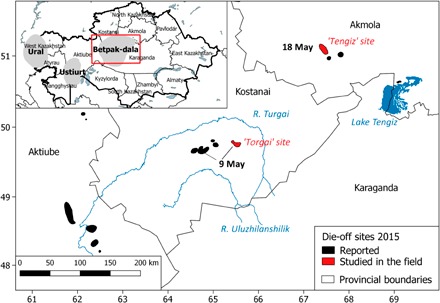
Inset shows the location of the three saiga populations within Kazakhstan (x and y units are longitude and latitude, respectively).
Mass mortality events (MMEs) of animals are demographic catastrophes that can simultaneously affect all life stages and rapidly remove a substantial proportion of a population over a short period of time relative to the generation time of the organism. They are often associated with multiple stressors including highly transmissible infections and exposure to toxins or extreme weather, alone or in combination (2). There is considerable speculation as to the increasing influence of ecological perturbations from human activities on MMEs (3, 4). Understanding and predicting the response of opportunistic microbes to changing environmental conditions, and of hosts to growing numbers of stressors, require a much greater emphasis on investigating the ecological context of MMEs than hitherto has been the case (5). This calls for a more multidisciplinary, or One Health, approach (6).
This paper reports one of the first investigations of a major MME using this approach, providing a strong empirical example of the critical role played by environmental factors in MMEs. We establish the primary cause of death in the MME in May 2015 affecting the Betpak-dala saiga population in central Kazakhstan and investigate potential causative and predisposing factors using contemporaneous and historical data analysis and insights from the literature. On the basis of our findings, we make recommendations for future management and research for this species and MMEs in general.
MATERIALS AND METHODS
Research approach at the time of the mortality event
Given the unexpected nature of the disease event, there was no prior experimental design for the study. A research team was in the field for normal monitoring of calving, led by S.Z. As animals started to die, researchers S.W. and S.Z. took observations and samples using the best available methods. An emergency response team including R.A.K. and M.O. attended within 1 week of the first observations of mortality. They carried out field postmortems and sampling according to an outbreak investigation protocol that had previously been agreed upon based on Food and Agriculture Organisation of the United Nations best practice for emergency response to disease outbreaks, customized for saiga antelopes (7).
Because mortality and morbidity appeared to be 100%, for practical reasons, we did not attempt random sampling for epidemiological analysis and no healthy animals remained for comparison. Unaffected animals and survivors of the MME (approximately 30,000 individuals, based on post-MME expeditions and aerial surveys) were either geographically far removed at the time (for example, older male groups occur to the north at this time of the year) (8) or cryptically located away from aggregations, in small herds widely distributed or peripheral to the calving areas. Because of the vast landscape, neither the precise location nor the behavior of these surviving groups could be determined.
The selection of carcasses for necropsy was not based on any randomization, given that the syndrome was so universal and consistent in character. It affected a proportion of the aggregations each day with a few initial cases observed, but soon affected thousands daily until the end of the die-off, which stopped abruptly when all were dead; there was hardly any tailing off of cases. Saiga herds during calving move up to approximately 2 km each day, and the necropsy simply followed these movements, with a number of full postmortem examinations (PMEs) completed on each day. It was not possible to catch healthy saigas for comparison of clinical pathology with dead and dying animals, because the capture technique requires specialized teams and no unaffected saigas were within easy range of the teams, which were situated hundreds of kilometers to the north. Unaffected female saigas were not observed over the period of mortality.
Samples were sent to laboratories in Kazakhstan and overseas for analysis, following normal best practice. Once the formal scientific research collaboration was established, to ensure that the research was carried out in a systematic and structured manner, the characteristics of the outbreak were listed by the research team. A set of non–mutually exclusive hypotheses for the proximate cause of the outbreak was then developed, consistent with these characteristics. These hypotheses were then explored, and refined or discarded, using a combination of review of the English and Russian language literatures (including texts available only in Russia or Kazakhstan), laboratory analyses, expert consultation with field biologists who were present at the time of previous outbreaks, consultation of archived field notes and maps, follow-up field expeditions, and statistical analysis using a range of data sets of environmental covariates.
Clinical and pathological diagnosis
Animals were observed alive from a distance. When moribund, they were observed closely and physically examined. After death, a complete postmortem examination (PME) and sampling were performed on selected cases including adults and calves. Before death, 26 saigas from Torgai and 4 from Tengiz (Fig. 1) were blood-sampled from the jugular vein using a sterile technique, and aliquots of blood were stored whole for parasitological examination and placed in clot tubes for removal of serum. For microscopy, thin blood smears were prepared fresh, fixed with 96% ethanol, and stained using the Romanovsky-Giemsa method. Given the long distances to the laboratory and the likelihood of a delay of more than a week before samples could be processed, biological material was stored in cool boxes or frozen in liquid nitrogen, and representative tissues were cut and placed in buffered formal saline. Samples from dead saigas included feces, gut contents, tissues, and any observed external or internal parasites. For bacteriology and virology, samples from liver, spleen, lungs, kidney, blood, and milk were stored frozen; 26 samples from Torgai and 6 samples from Tengiz were processed. Whole brain and rumen and gut content were taken for plant and toxin studies, as were feces and blood for parasite examination. Biological samples were taken to the Research Institute for Biological Safety Problems in Gvardeiskiy, Kazakhstan, for processing. Fresh tissues underwent polymerase chain reaction (PCR) analysis for the presence of DNA/RNA of viral or bacterial pathogens, including microscopy of tissue imprint smears stained by Gram and Giemsa, isolation of bacterial cultures on nutritional medium, microscopy of cultivated bacteria and identification by biochemical methods, and attempted isolation of viruses in cell cultures and identification through electron microscopy. Full details of the methods used are discussed in the subsequent sections.
Polymerase chain reaction
RNA was extracted from samples and tested for foot-and-mouth disease virus (FDMV), bluetongue virus, and peste des petits ruminants virus (PPRV) using the QIAamp Viral RNA Mini Kit (Qiagen). DNA was extracted from samples and tested for viruses (sheep pox, Aujeszky, and Visna-Maedi) and protozoa (Theileria, Babesia, Anaplasma, and Coxiella) using the DNeasy blood and tissue mini kit (Qiagen) following the manufacturer’s protocols. Detection of virus RNA/DNA of peste des petits ruminants (PPR), foot-and-mouth disease (FMD), bluetongue, and sheep pox was carried out using Real-Time RT-PCR Target Specific Reagents (Tetracore) (9–12). The PCR was performed following the manufacturer’s protocols. Amplification and analysis of PCR products were performed using LightCycler 2.0 (Roche). Detection of bacterial DNA of Listeria monocytogenes was carried out by PCR using the kit “Lister” (AmpliSens), again following the manufacturer’s protocols. Detection of DNA or antigens of Pasteurella, Mycoplasma, Clostridium toxins, Coxiella, Akabane, malignant catarrhal fever (MCF) and Aujeszky viruses, Theileria, Babesia, and Anaplasma was performed by PCR. PCR was completed using the primers and conditions described in the literature, as listed in table S1.
Virus isolation and identification
Two types of cell culture were used for virus isolation: primary lamb kidney cells (LKCs) and Vero cells (African green monkey kidney cells). A total of 96 tissue samples from saiga (lungs, spleen, and lymph nodes) were inoculated onto primary LKC and Vero cells. The control flask was inoculated with sterile phosphate-buffered saline only. The inoculated cell cultures were observed twice daily (morning and evening) under an inverted microscope for the appearance of cytopathic effects (CPEs). A sample was considered negative for PPRV if no CPEs were observed within 10 days after inoculation. In the case of cells where there were no CPEs, the inoculated cultures were subjected to freezing and thawing three times to disrupt the intact cells (blind passages), and 1 ml of this suspension was added to a new 25-cm2 flask of a confluent monolayer of cells. A sample was considered negative if no CPEs were observed after three blind passages. The virus isolates identified by characteristic CPEs observed on LKC and Vero cells and the presence or absence of virus were confirmed by testing the cell culture supernatant using PCR and by electron microscopy.
Electron microscopy
Samples of tissues (lungs, spleen, and lymph nodes) from all the dead saigas were examined by electron microscopy. Preparations for electron microscopy were prepared by negative staining with 2% phosphotungstic acid at pH 6.8 to 7.0. Virus was isolated from the tissue material by ultracentrifugation at 35,000 rpm for 45 min from the clarified homogenate. Virus adsorption to mesh procedures used the sputtered carbon substrate formvarovoy added to potentially virus-containing fluid (drop) placed on the well of the Teflon plate. Adsorption time was 10 min. After sample adsorption, the mesh contrasted with a drop of phosphotungstic acid for 5 min, and after drying, the sample was examined under a JEM-100 CX II electron microscope (JEOL) at an acceleration voltage of 80 kV and increased by 80,000 to 100,000.
Isolation and identification of bacterial isolates
Smears from inner organs and from broth and agar cultures were stained using Romanovsky-Giemsa, Gram, and Loeffler methylene blue (13). Isolation of Pasteurella was undertaken by cultivation of pathological material samples in meat-peptone broth, Hottinger digest broth, and plain agar (IPA) with 5% sterile horse serum. To obtain pure cultures of a single bacterial colony type from blood agar and MacConkey agar only, bacterial colonies (with the following characteristics: rod, coccobacilli, rough or smooth, Gram negative, with or with no growth on MacConkey, and presence or absence of hemolysis on blood agar) were subjected to subculturing. Then, each bacterial colony was characterized, and further bacterial identification was carried out using a series of primary and secondary biochemical tests, following standard procedures (14). Identification of the isolated Pasteurella culture was conducted according to biochemical indicators including saccharolytic and proteolytic enzyme excretion, formation of catalase, acetylmethylcarbinol, ammonia, hydrogen sulfide, indole, lysis of red blood cells and cattle bile, and nitrate reduction (13). Typing of the isolates was conducted with the use of PCR analysis applying capsular-specific primers for determining capsular groups A, B, D, E, and F (15).
FTA analysis
Flinders Technology Associates (FTA) cards were taken from 12 dead or dying saigas at the Tengiz site for rapid dispatch to international reference laboratories at the Pirbright Institute (PI) and the Friedrich-Loeffler Institute (FLI) for examination, including use of genomic tests [next-generation sequencing (NGS)].
At PI, the following procedures were followed. Samples of dried blood spots (approximately 2 cm in diameter) were received from 12 animals, with two spots provided per animal. These samples were taken postmortem to eliminate the possibility of laboratory contamination. RNA was extracted by taking a 1-cm-diameter disc of blood spot and incubating in 200 μl of RLT lysis buffer (with 1% β-mercaptoethanol) (Qiagen) before incubating at 56°C for 15 min. Each sample was processed individually, with decontamination periods of at least 30 min between the handling of each sample. After the incubation, the supernatant was then decanted off and tested for the presence of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using a previously published qualitative real-time reverse transcription (RT)–PCR assay for porcine GAPDH (16). These results provided an indication of the suitability of recovered/extracted nucleic acid within the sample for further pathogen testing by real-time RT-PCR and NGS. Testing was performed with diagnostic real-time RT-PCR assays used by both vesicular disease and non-vesicular disease reference laboratories at PI (VDRL and NVDRL, respectively). Assays used were specific for FMDV (9), bluetongue virus (10), PPRV (11), capripox (12), and epizootic hemorrhagic disease virus. In each case, 5 μl of extracted RNA was tested in duplicate using established protocols within both VDRL and NVDRL. Assays were run on an ABI 7500 Fast [with the exception of the FMDV assay, which was run on an MX3005 (Agilent)]. All assay runs were validated to standards described within in-house standard operating procedures (SOPs).
At FLI, the following procedures were followed. Eleven samples of dried blood spots on FTA cards were received. For whole-genome sequencing, RNA was extracted and was used as a template for double-stranded cDNA synthesis (Roche). The Covaris M220 ultrasonicator was used for DNA fragmentation of 0.5 to 1 μg of double-stranded DNA to an average size of about 500 base pairs (bp). For library preparation of the fragmented DNA, a GeneRead DNA Library L Core Kit and GeneRead Adapter I Set A 12-plex (both from Qiagen) were used followed by manual size selection using AMPure XP magnetic beads (Beckmann Coulter). The quality of the library was checked on a Bioanalyzer 2100 (Agilent Technologies) using a high-sensitivity DNA chip and corresponding reagents. Quantity was determined by quantitative PCR with the Ion Library TaqMan Quantitation Kit (Life Technologies). After emulsion PCR with a OneTouch 2 device and Ion PGM Hi-Q OT2 reagents (both from Life Technologies), sequencing was performed on an Ion Torrent Personal Genome Machine (PGM) using an Ion PGM Hi-Q Seq Kit (Life Technologies) for 400-bp reads. Raw sequence data were first analyzed using Reliable Information Extracted from Metagenomics Sequential Datasets (RIEMS) (17) and additionally assembled using the Genome Sequencer software suite (v3.0, Roche).
Histopathology
A set of fixed tissues was sent from the Torgai site to the Department of Pathobiology at the Royal Veterinary College for histopathological diagnosis; the remaining tissues from Tengiz and some from Torgai were processed at Research Institute for Biological Safety Problems, Kazakhstan (RIBSP) for histology, and some fresh tissue were processed for electron microscopy. Wax blocks were later sent to the Royal Veterinary College and further processed including rewaxing, slide preparation, and histological examination. A complete set of histological slides was sent to an experienced wildlife and comparative pathologist at the Wakefield School of Medicine Department of Pathology/Comparative Medicine Institute in North Carolina for review.
FISH analysis
A set of fixed tissues from several different saigas was sent to the University of Bristol, School of Veterinary Science, for examination of the distribution of bacterial populations using fluorescence in situ hydridization (FISH). Paraffin-embedded, slide-mounted tissue samples were dewaxed in Histoclear for 10 min, followed by 3-min hydrations in 100, 90, and 70% ethanol and finally rinsed in water for 3 min. Slides were hybridized with a single probe for either P. multocida (nonspecific, binding all serogroups) or Clostridium (1 μl of probe per sample) with a FISH buffer (0.9 M NaCl, 0.02 M tris-HCl, 0.01% SDS, and 25% formamide) in a thermos cycler at 95°C for 10 min and at 46°C for an hour. Slides were washed, dried, and mounted under a coverslip with Vectashield (Vector Laboratories). Fluorescing bacteria were then examined in 10 fields of view in each tissue under ×400 total magnification.
Mineral composition analysis
A subset of saiga from Tengiz was sampled for tissue mineral analysis to evaluate possible micronutrient deficiencies. Fresh liver was collected, frozen, and tested at RIBSP for tissue copper, molybdenum, cobalt, zinc, and iron using standard methods for an inductively coupled plasma mass spectrometry (ICP-MS) analysis as follows: 0.2 to 0.3 g of the sample was weighed (Shimadzu AY 220 scale) in a Teflon cuvette. The cuvette with the sample was placed in a Teflon autoclave and 5 ml of nitric acid and 1 ml of hydrogen peroxide were added, and after termination of the intensive reaction, the autoclave was sealed. The autoclaves were placed into the microwave system SpeedWave Four Technology (Berghof Products) and decomposed using the program recommended by the manufacturer for the decomposition of biological samples. At the end of the program, the autoclaves were cooled to room temperature and depressurized, and the resulting solution was transferred to a 50.0-ml volumetric flask, filled up to the mark with water, and thoroughly mixed. Afterward, metals were determined in the samples through ICP-MS. Along with the samples analyzed, a “blank” solution was prepared without any sample material, which was treated with all stages of sample preparation and analysis.
The samples were analyzed for Fe, Co, Cu, Zn, and Mo using ICP-MS in the Agilent 7500a mass spectrometer. Standard solutions for ISP-MS were used to calibrate the mass spectrometer and obtain calibration characteristics. The calibration solutions were prepared by successively diluting standard solutions with 2.5% HNO3.
Data acquisition and processing for modeling
The outcome variable
The aim of the statistical analysis was to predict the probability of an MME (cases) versus a normal calving event (controls). Our case sites were those corresponding to MMEs in 1988, 1981, and 2015. Locations and dates of onset in 2015 were recorded at two sites by R.A.K. and S.Z. Information for other sites came from rangers and other key informants in the field (fig. S1). In the case of the 1988 and 1981 events, information was obtained from literature and maps produced at the time (1981, Institute of Zoology and Department of Reserves and Hunting; 1988, Institute of Zoology and Betpak-dala State Hunting Organisation). Exact calving locations were provided by the Association for the Conservation of Biodiversity of Kazakhstan (ACBK) for the period 2008 to 2016 from field expeditions and telemetry data from collared animals. Other, more approximate, calving sites were recorded from descriptions available in the annual official saiga aerial and ground monitoring reports by the Institute of Zoology and Hunting Agency of Kazakhstan (18). Of the resulting 214 sites, the climate data used in this analysis were available for 135 sites [29 of which included estimated dates of onset (20 cases; 9 controls) and 106 of which lacked dates (4 cases; 202 controls)].
Predictor variables
Our “zero hour” is the start of calving or, for MME, the day of the index case, and all predictors were aggregated over specified periods up to these dates. MMEs in saigas appear from the literature to be strongly associated with calving (8). We used the onset of calving, rather than peak calving, to focus on environmental conditions before the onset of disease, rather than when it may already have taken hold.
Field observations from 2015 suggest that, at an individual level, the course of the disease from the first visible signs of depression to death occurred in the course of a few hours. In addition, the MME at each calving site occurred over the course of a number of days. These factors are strongly suggestive of a relatively recent common risk factor. Evidence from the literature suggests incubation periods for hemorrhagic septicaemia (HS) lasting from 12 hours to several days (19). These factors suggest that external triggers may have been important anytime from several days to approximately 12 hours before the onset of clinical signs in the index case. Uncertainties in onset dates suggest that it is better to err on the side of longer periods, and so, predictor variables were generated for 3, 5, and 10 days before onset.
Given the large geographic scale of the study, range of variables to be covered, long time series coverage, and high resolution, the daily European Research Area (ERA)–Interim reanalysis data set produced at the European Centre for Medium-Range Weather Forecasts was selected for this analysis (20). Daily aggregates (means, maxima, minima, or totals) were generated from raw 3 or 6 hourly data for temperature, dew-point temperature, soil moisture, snow depth, maximum wind gust, and precipitation. Because of uncertainties in precipitation data from this source, interpolated gauge-based products from the Global Precipitation Climatology Centre (GPCC), which are independent of ERA data, were also used, although these were available only from 1988 onward (21). For investigation of long-term anomalies at single sites, an additional lower-resolution reanalysis data set from the National Centre for Environmental Prediction (NCEP) was also used because it covers the longer time period from 1948 to 2016 (22).
Normalized difference vegetation index (NDVI) anomaly data based on Systeme Probatoire pour l’Observation de la Terre (SPOT) and PROBA-V data going back to 1998 were supplied by F.H. and J.R. at the Université catholique de Louvain–LifeWatch Wallonia-Brussels (WB) project (23). Snow anomaly data—unusual values at the date of observation compared to mean probability of snow presence—were used to look for unusual variation in winter (for example, winter length, severity, and sudden cold shocks) and likely to be related to spring NDVI patterns derived from filtered MODIS data (24). These data sets cover periods from 1998 and 2000 to 2016, respectively, and thus include only 2015 MME sites and a set of controls. An earlier data set of NDVI anomalies derived from Global Inventory Modeling and Mapping Studies data (25) was provided by the LifeWatch project team for the period 1982 to 1997, covering the 1988 MME.
Data extraction and processing
Values of climate metrics at saiga MME and calving sites were extracted from the daily rasters using a polygon shapefile for sites available from 2008 onward (which represented estimates of the actual size and shape of the aggregations). For the older sites, identified as point data from the literature, means were extracted using buffers of 6 km, corresponding to the median polygon size for those sites, which were available in that format.
From the extracted daily climate data, various aggregate metrics (totals, maxima, minima, or means of daily variables, depending on the metric in question) were produced over 3, 5, and 10 days before onset of MME or calving. In the case of precipitation, in addition to production of sum totals over the days before onset, the number of days with nonzero precipitation was counted, because the duration of wet weather may be more important than the actual total rainfall in millimeters. Metrics of temperature variability in the days to onset included daily temperature variation (difference between daily maximum and minimum temperatures) and overall temperature variation (difference between minimum daily temperatures) over 3, 5, and 10 days before onset or calving.
The ERA-interim daily snow depth variable was used to calculate the date of snow melt by finding the last three consecutive days of snow cover of the season. Variables measuring the length of snow cover (total days of snow cover from 1 January to last snow melt) and days between end of last snowmelt and calving/onset were then produced. The extracted 7-day LifeWatch-WB snow and NDVI anomaly data were interpolated to daily values. These were then used to calculate aggregates over various periods before the date of onset or calving and for fixed periods of the year (for example, snow anomaly for the last week of March and NDVI anomaly in the last week of April).
The process described above depended on knowing dates for onset of calving and MME. However, calving dates were missing for many sites. In these cases, an onset date of 9 May was used, because this is the average date of start of calving for those sites for which dates were available. For sites having onset dates, metrics were aggregated both based on actual dates and using an onset date of 9 May for comparison.
The resulting database listed 135 sites, with climate metrics aggregated in different ways and over different multiday periods up to 9 May, with a second set of these same variables aggregated using actual onset dates where these were available. In addition, the database held information on calving/MME, latitude, and longitude for each site.
Statistical analysis
Univariate analysis
We first conducted visual plotting (box plots) of all candidate variables at case and control sites to select those that appeared to show relationships with the outcome variable. Because of the structure of the data, the results of t tests to compare means would have been unreliable. Bayesian approaches were therefore used to compare means, taking into account the effect of year using the BRMS package for R (26). Last, we conducted simple t tests for differences in means between MME and calving sites in 1988 alone, the only year for which both types of site were available.
Multivariate analysis
From results of the univariate analysis, we selected a subset of eight variables, which both appeared to present differences between cases and controls and represented different types of climatic phenomena. Principal components analysis (PCA) was then applied to these variables, resulting in the reduction of the data set into two major dimensions. The eight selected climate variables and the PCA dimensions were both used to build multivariate models, again using hierarchical Bayesian approaches with year as a random variable. Latitude was also included in these models to account for spatial autocorrelation.
In the case of models using “raw” climate variables, all predictors were standardized using z scores. This enabled direct comparisons of coefficients and use of a single weakly informative prior, which assumed a normal distribution of coefficients with a mean of 0 and an SD of 3. Initially, selected variables were included in the model. Those with the smallest coefficients and having confidence intervals crossing zero were removed one by one, and the model was rerun each time until only significant variables were left. At each step, the WAIC (Watanabe-Akaike, or widely applicable, information criterion) was generated and used to compare the models (27). In the case of models using PCA results, dimensions 1 and 2, capturing the majority of variation, and latitude were entered into models and insignificant variables were removed. The same prior was also used here.
Because conditions were known to be different between MME years, models were run using subsets of the data set (i) removing MME sites from 1981 and (ii) removing MME sites from 1981 and 1988. These models had convergence problems or did not produce significant results. Thus, in the end, only the full data set including all data for 1981, 1988, and 2015 was used. Taking this full data set, models were run using climate variables aggregated over various periods to 9 May for all sites, as well as for the “real date” data set in which they were aggregated to the actual onset date (where available) or 9 May (if not). In the latter case, only those MME sites having estimated real onset dates were used, and thus, only at controls were dates set to 9 May.
Climate anomalies at single sites
In addition to the modeling of MME probability at case and control sites, we also explored long-term climatic variation at single die-off sites, to understand the extent to which climatic conditions at these sites were exceptional in MME years, focusing on those variables that seemed important from the modeling. This was conducted using ERA data aggregated over the first and second halves of May and NCEP data for the month of May.
RESULTS
Description of the 2015 MME
In each of the two sites that were intensively monitored (Tengiz and Torgai; 8000 and 62,000 animals, respectively; Fig. 1), the die-off lasted approximately 9 days. Morbidity and mortality were estimated at 100%, although it is possible that a few animals survived. The affected aggregations contained pregnant and recently calved females, their calves, and some young males.
Carcass distribution in the main areas of mortality showed a remarkably even spread of animals, with a spacing of approximately 30 to 50 m between individual adults, consistent with becoming ill while grazing and then dying on the spot (fig. S2A). Individual animals’ behavior changed from apparent normality (able to run and graze) to suffering from general depression, lethargy, and hindlimb weakness and ataxia while grazing or standing. Sicker animals were recumbent, with some dyspnea and grunting, frothy fluid or saliva forming around the mouth, and terminal diarrhea, frequently with blood and urine staining of the soil (fig. S2B). Most died within a few hours of onset of clinical signs.
No other wildlife species were seen ill or dead in large numbers, nor were there significant excess livestock deaths or human disease in the vicinity over this time period. This was confirmed during follow-up expeditions to villages and farms in the region in summer 2015 and spring 2016. The temporal and spatial distribution of the outbreak suggested that horizontal transmission of a contagion was unlikely to have been the cause. Calves were observed to suckle until and even after maternal death (fig. S2C), and then to die. Delayed death of calves was consistent with disease transmission from the mother, rather than horizontal transmission of an infectious agent through saiga aggregations, which would have affected the calves before or at the same time as their mothers.
A number of saigas were blood-sampled while moribund, and a full PME was performed on 33 animals (24 adult females and 9 calves; 4 male, 5 female) with an even split in necropsies between the two monitored sites; microbiological sampling included 32 saigas (26 from Torgai and 6 from Tengiz). PMEs were performed as soon after death as possible. The gross pathology was similar in animals from both geographical sites. Animals were generally in very good body condition, with 76% having abundant mesenteric/omental fat (fig. S2D), which, in small ruminants, correlates significantly with live weight and body condition (28). In the 10 adult animals necropsied immediately after death, all organs were diffusely congested, and there was bloodstained fluid in serous cavities (fig. S2D), generalized hemorrhages, ecchymoses and petechiae in the subcutis (fig S2E), hemorrhages on multiple serosal surfaces [extensively within the sub-endocardium of the heart (fig. S2F) and within lungs], lymph nodes (fig. S2G), and skeletal musculature. On occasion, there was severe edema and congestion of the lung (fig. S2H). Many cases showed catarrh, congestion, and intense reddening of the intestinal mucosa, with multifocal subserosal hemorrhages and congested vasculature (fig. S2I).
The preliminary diagnosis made by experienced attending veterinarians (R.A.K. and M.O.) based on the clinical presentation and necropsy was peracute HS, as described in the literature (29). This diagnosis was supported by a pure profuse culture of P. multocida serotype B from 32 microbiologically sampled saiga, which included fresh blood, milk, and tissues taken from lung, liver, spleen, and kidney, tested in the research laboratory of Kazakhstan’s Research Institute for Biological Safety Problems, with similar findings to other in-country reference laboratories. Microscopic examination of multiple tissue samples examined at the Royal Veterinary College found necrosuppurative hepatitis, splenitis, and lymphadenitis with intralesional and intravascular Gram-negative bacteria (Fig. 2 and the Supplementary Materials). In lungs, there were perivascular and intra-alveolar hemorrhage and edema. These findings are consistent with a diagnosis of HS caused by P. multocida (29).
Fig. 2. Photomicrographs showing multifocal, acute degenerative tissue changes with the light blue staining indicating Gram-negative bacteria consistent with the pure isolation of P. multocida.
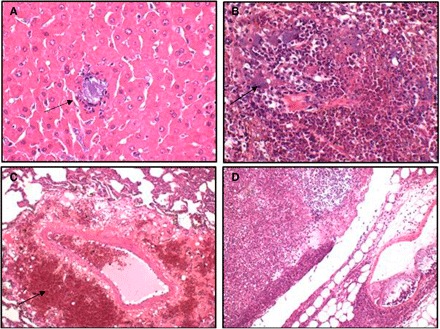
(A) Liver: necrosuppurative hepatitis, random with intralesional bacteria (arrow). (B) Spleen: splenitis, necrotizing with bacterial emboli. (C) Lung, perivascular hemorrhage (arrow) and edema. (D) Lymph node: lymphadenitis, necrosuppurative.
Extensive PCR testing of tissue samples from the same 32 individuals sampled for microbiology for the presence of viruses in several laboratories found one positive for sheep pox and no evidence for a range of other viruses (table S1 and the Supplementary Materials). In three saigas sampled in Tengiz, lung tissues showed evidence of intracytoplasmic bronchial epithelial inclusions, which were suggestive of Paramyxoviridae; however, ascribing a specific virus is highly speculative, and to date, this finding is inconclusive. Microscopy of the blood smears from sick saigas revealed Hemosporidia at various stages of development; this was confirmed by PCR. Of 23 tested blood samples, DNA of the theileriosis pathogen (Theileria annulata) was identified in 11, with negative results for anaplasmosis and babesiosis.
FISH for three animals was consistent with a breach of the mucosal immune barrier and systemic invasion of P. multocida, rather than a general invasion of mucosal bacteria into the body, which is found in dying or recently dead animals (fig. S3). The immediacy of the PME made this investigation possible and relevant.
The proximate cause of death in this MME was therefore a massive peracute HS. However, given the extraordinary mortality and extent, this was not considered a sufficient explanation. To account for such rapid, concurrent mortality of distant aggregations, latent infection with P. multocida would have to be common across the entire saiga population. Some prior influence seemed likely, priming the majority of individuals for a breakdown of immunity to opportunistic bacteria or increasing bacterial growth and/or virulence.
Exploration of potential underlying cofactors
To provide a more comprehensive explanation for such an extraordinary phenomenon, a multidisciplinary approach was used; possible hypotheses for the observed events were developed from the literature and field observations and were systematically tested (table S4). These hypotheses focused on factors that trigger pathogen invasion in the short term, although longer-term influences on P. multocida carriage and host susceptibility might also be important in the disease dynamics. Given that causation might be multifactorial, competing hypotheses were not mutually exclusive.
The existence of P. multocida as a commensal organism in healthy saigas (30), near-simultaneous onset of clinical signs within and between large groups of animals, and our understanding of HS from the literature (31) all suggest a possible environmental trigger. Saiga calving aggregations in this population have been monitored regularly since the 1970s. A review of the literature and consultation with saiga field biologists identified two previous large-scale mortality events, which could possibly be ascribed to HS, in 1981 and 1988 (table S2 and the Supplementary Materials). Although pathological evidence of this syndrome was not reported at the time, the clinical signs and morbidity were similar to those in 2015, especially in 1988 (fig. S2J), and P. multocida of the same phylogeny and pathogenic potential was isolated in each case (table S2). In 2015, no disease-free sites were observed; in 1981, although many animals survived, their locations are unknown, whereas in 1988, locations of unaffected and affected sites were documented. Using these three cases, the impact of environmental covariates on the probability of an MME versus a normal calving event was explored using statistical modeling.
The data set represents a typical case of “rare events,” having small sample sizes, a strong imbalance in the number of case and controls, and autocorrelation issues. First, MME and non-MME site data were spatially and temporally correlated; that is, there were many locations from the same year, and locations of MME sites were clustered. Second, there was perfect separation between MME and non-MME sites for certain environmental variables; that is, MME sites had nonoverlapping values of certain variables with non-MME sites. Last, there was pseudo-replication in the data set; that is, locations were not independent because many were from the same year, but the number of levels of year compared to the overall size of the data set precluded its use as a random variable in standard mixed models. To resolve these issues, we used Bayesian generalized linear hierarchical models, which enabled us to incorporate a random effect structure for year (32).
On the basis of our hypotheses and evidence concerning triggers of pasteurellosis from the literature, we tested for any differences between various measures of temperature, precipitation, wind speed, atmospheric moisture, soil moisture, and vegetation greenness (using the NDVI) in 5 to 10 days before die-off, and both NDVI and snow cover in the preceding weeks, using univariate tests (fig. S4 and table S5). A subset of these variables with good explanatory power was then used in the regression models, both as raw variables and combined as dimensions within a PCA (Fig. 3 and fig. S5).
Fig. 3. Kernel density map of the first two components of a PCA of selected climatic variables in the 10 days to disease onset (or 9 May for controls for which exact date is not available), showing separation of die-off sites from other sites.
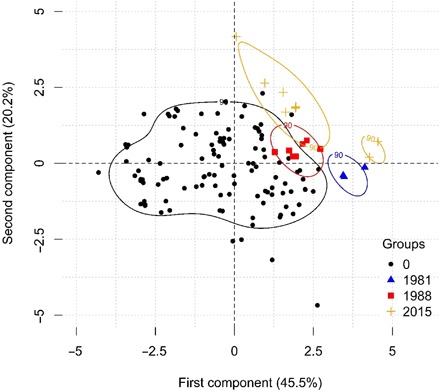
See fig. S5 for indications of the relative contribution of each climate variable to these two components.
In all analyses, average maximum daily relative humidity, average minimum daily temperature, and average maximum daily dew-point temperature showed the strongest positive relationships with the probability of a die-off; in particular, die-off sites were unusually humid, and in 2015, they were unusually warm. Of the Bayesian multivariate models, the strongest associations emerged for the 10-day aggregates of the days preceding the die-off, whether natural climate variables or PCA dimensions were used (see the Supplementary Materials). The final model contained only temperature and humidity metrics. There was a clear threshold association with humidity, with most die-offs occurring at a mean average maximum daily relative humidity of more than 80% (Fig. 4A); mean daily relative humidity was 84.6 ± 1.7% at die-off sites and 71.2 ± 1.7% at control sites. There was a weak positive association between minimum temperature and die-off events (Fig. 4B and the Supplementary Materials).
Fig. 4. Probability of die-off related to selected environmental variables in the 10 day period to onset, 1979 to 2015.
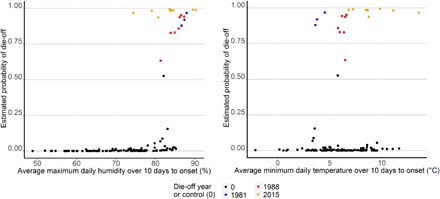
(A) Fitted values for the probability of a die-off event against mean maximum daily relative humidity in the previous 10 days, with the three die-off years in different colors. (B) Fitted values for the probability of a die-off event against mean minimum daily temperature in the previous 10 days, with the three MME years in different colors.
Long-term climate anomaly data at the Kostanai cluster of sites (affected in all three MME years) support these findings (table S5 and the Supplementary Materials). At this site, atmospheric moisture in May 2015 not only was extreme but also exhibited the highest recorded values of two long-term time series for certain metrics (covering 1948 to 2016 and 1979 to 2016). In 1981, in the second half of May, humidity levels were above the 95th percentile, precipitation was the highest of the 1979 to 2016 time series, and minimum temperature was unusually low for the time of year. Conditions at 1988 MME sites appear less extreme than in the other two MME years; however, in that year, the animals were in very poor condition following a harsh winter and so may have been more vulnerable. Average minimum daily temperature was significantly higher at the seven case sites than at the four control sites in 1988 (cases, 6.2°C; controls, 4.0°C; t = −4.32; P = 0.03), as was average daily maximum humidity (cases, 85%; controls, 82%; t = −3.623; P = 0.006). In contrast, although total precipitation was overall very high in 1988, it did not differ significantly between control and case locations.
DISCUSSION
MMEs in the context of saiga life history
Given the precedents from other MMEs in the literature, particularly regarding HS events, our expectation was that certain combinations of climatic factors might be associated with the 2015 die-off, but would be unlikely to fully explain it (table S4). We found strong, but not complete, differentiation between climatic conditions in sites with and without MMEs (Figs. 3 and 4). HS caused by P. multocida has been linked to humidity and high temperatures in other studies and species (33–35), but the total numbers of dead animals, geographic scale, and mortality rate in other recorded outbreaks do not even approach the levels seen in saigas (table S2). Therefore, additional factors must also be at play.
The saiga antelope appears particularly vulnerable to MMEs, especially around calving, having suffered from several such episodes in the recorded past (table S2 and the Supplementary Materials), although we found no record of MMEs in folklore or pre-Soviet literature. Apart from the 2015 event, recent years have seen high levels of disease-induced mortality in other saiga populations; in 2010, around two-thirds of a calving aggregation of 20,000 in the Ural population died of a peracute respiratory disease with bloat, and this was followed in 2011 by a smaller die-off of several hundreds of saiga with similar clinical signs, immediately after calving, grazing in the exact same location as the earlier MME, suggesting a site-specific cause. Pasteurellosis was implicated based on clinical signs and bacteriology, but very little pathology was undertaken, putting some doubt on the diagnosis (36). The endemic Mongolian saiga subspecies is currently experiencing high levels of mortality from PPR (37, 38), caused by a morbillivirus of the Paramyxoviridae family. P. multocida was concurrently isolated from tissues, but in this case, P. multocida is not implicated in the primary pathology.
This propensity for MMEs may be a result of the species’ life history strategy of very high investment in reproduction in a harsh continental climate. The species has the largest recorded mass at birth as a proportion of maternal mass of any ungulate (39), producing precocious calves and enabling large-scale seasonal migrations tracking greenery across thousands of kilometers (40). High mortality as a result of hard winters (dzhuts), where ice forms over snow and thus restricting access to pasture, and summer droughts is well documented, and the species has a history of rapid recovery from low numbers (8). However, high levels of poaching since the 1990s have depleted populations, holding out the possibility that an MME could reduce numbers below the level at which recovery is possible (41).
FUTURE RESEARCH NEEDS
The mechanisms by which climatic factors triggered bacterial invasion in the saiga MMEs remain unknown. A trigger factor, such as stress or infection with a paramyxovirus, is usually invoked in P. multocida outbreaks (42, 43), but there is no serological evidence for previous viral infection other than Akabane, in the population that died in 2015 based on samples collected opportunistically from 2012 to 2014 and tested for bluetongue virus, PPRV, and Akabane and Schmallenberg viruses (44). Despite the negative results to date, there is still therefore the possibility of an undiagnosed predisposing infection, microbiome perturbation, or environmental change that reduced the resistance of a proportion of the individuals, enabling bacterial opportunism. In the light of the emerging threat from PPR, and the presence of Theileria in approximately 40% of the sampled saigas (see the Supplementary Materials), interactions between saigas and livestock in a changing social and ecological environment also require further study.
The fact that Pasteurella strains of near-homologous genotype are found in both apparently healthy and sick populations of saigas throughout their range and over decades suggests that latent infection with endemic strains occurs, which are all potentially virulent. Studies of bacterial virulence in controlled environments could cast light on the mechanisms by which these strains become virulent, or new strains emerge.
Recent history suggests that the saiga must be managed with the high probability of future MMEs in mind; this requires investment in preventative measures where possible (for example, livestock vaccination for PPR), strong antipoaching actions so that populations are large and resilient enough to withstand large-scale mortality, and a sustainable landscape-level approach to coexistence between saigas, livestock, and people to enable its migratory lifestyle to continue.
The fact that P. multocida infection in saigas, as in bovines (31), appears strongly linked to high humidity and temperature is of concern going forward, given that a climate change–induced increase in temperature is projected for the region over the short to medium term (45, 46) and evidence for recent change is strong (47, 48). Predicted precipitation trends are uncertain and unreliable (46), although some models do suggest possible increases over the saiga range, particularly in winter (45, 47). Although recent observations do not yet reflect these predictions with any certainty (48), there is some evidence to suggest increased spring precipitation in northern Kazakhstan since 1980 in comparison with the preceding 30-year period (49), leading to increased air humidity. Predicted increases in extreme rainfall events (45) and abrupt variation in primary productivity (50) also suggest that saigas might have to adapt to increased climatic variability in the future, affecting resource acquisition and migration patterns (18) as well as disease risks.
CONCLUSIONS
This study illustrates the power of a multidisciplinary, collaborative approach to investigating MMEs, the importance of understanding the contextual factors as well as the proximate cause, and taking a historical, evidence-based approach to investigations. A narrow focus on etiology of MMEs could fail to elucidate causation, where this involves multifactorial interactions between pathogens, hosts, and environment. The scale and nature of this event also point to the need for ongoing scientific and veterinary monitoring of wildlife populations and the need to be prepared for rapid and rigorous responses to disease outbreaks when they occur; in the absence of a research presence at the time of disease onset, little could have been said about the epidemiology of the 2015 event.
Supplementary Material
Acknowledgments
This work was undertaken with the approval of the Royal Veterinary College Ethical Review Board (URN 2015 1435) and under the authority of the Biosafety Institute of the Ministry of Science and Education and the Ministry of Agriculture, Committee for Forestry and Wildlife, Ministry of Agriculture, Republic of Kazakhstan. This work did not involve any experimental use of animals, only routine clinical examination and sampling on moribund animals in extremis during the MME. Funding: This study was made possible through various funding mechanisms, the main one being the National Environment Research Council of the United Kingdom urgency research grant (NE/N007646/1); the team also received substantial support financially or logistically from the following agencies, which are gratefully acknowledged: the ACBK, the Flora and Fauna International, the People’s Trust for Endangered Species and the Saiga Conservation Alliance, the Royal Society for the Protection of Birds, and the Frankfurt Zoological Society. We thank D. King and G. Freimanis of the PI, S. Feroudini, and the FLI for the laboratory support on advanced diagnostics. We also thank the Convention on Migratory Species of the United Nations and the Wildlife Conservation Network for their support and assistance. We acknowledge cooperation from I. Sytnyk (former Vice Director) and S. Tyulegenov (Director) and staff from the National Reference Veterinary Centre. At the University of Bristol, we thank B. Stuijfzand and W. Browne for advice on statistical approaches and E. Hector and T. Cogan for pathogen visualization in tissues. We also thank A. Lushchekina and Y. Grachev for provision of key literature and insights and A. Salemgareyev, S. Kopeyev, E. Shalgynbaev, and S. Inkarbekov for their participation in the fieldwork. We thank N. Kock (Wake Forest School of Medicine Winston-Salem) for review of the histopathology. Remote sensing image processing was supported by the Fédération Wallonie-Bruxelles in the frame of LifeWatch ERIC. Author contributions: R.A.K. led all elements of the study and carried out clinical observations, field postmortems, and epidemiological assessment. M.O. led the laboratory testing and carried out field postmortems. S.R. carried out the review of historical evidence and statistical analyses. S.Z. led the fieldwork component. N.J.S. carried out statistical analyses. W.B. and E.R.M. contributed to the evidence review and statistical analyses. A.K., R.R., and Z.O. carried out microbiological analyses. S.K. participated in the fieldwork. H.M.M. carried out histopathological analyses. S.W. carried out field investigations. F.H. and J.R. produced all NDVI and snow anomaly data. E.J.M.-G. led the evidence review, statistical components, and paper writing. R.A.K., M.O., S.R., S.Z., N.S., W.B., E.R.M., H.M.M., and E.J.M.-G. wrote the paper. All authors contributed to hypothesis development and testing. Conflicts of interest: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors and data presented in this paper are archived at NERC’s Environmental Information Data Centre (accession number pending).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/1/eaao2314/DC1
Supplementary Text
fig. S1. Locations of the three P. multocida–associated disease events (1981, 1988, and 2015) in the Betpak-dala population and control sites at which calving proceeded without a die-off since 1979, used in the climate analysis (x and y units are longitude and latitude).
fig. S2. Photographs depicting the die-off of saiga and their clinical signs and pathology.
fig. S3. Fluorescence in situ hydridization (FISH) photomicrographs showing position of fluorescing P. multocida bacteria.
fig. S4. Box plots for cases (die-off sites) and controls (no die-off) using the full data set, with real dates of onset if available or 9 May if not, showing the relationship between cases/controls and selected climate metrics aggregated over 10-day periods before onset.
fig. S5. Contributions of variables to components 1 and 2 of the PCA using the full data set, with real dates of onset if available or 9 May if not.
table S1. PCR primers used for microorganism detection.
table S2. A summary of the largest MMEs in saiga attributed to Pasteurellaceae-related syndromes.
table S3. Results of an ICP-MS analysis of livers (w/w) of three dead saigas from May 2015 (Tengiz group).
table S4. Assuming a diagnosis of HS as a primary cause of death, this table is a list of possible stressors, which may cause suppressed immunity in the host or increased virulence and invasion of a commensal parasite such as P. multocida, subsequent septicemia in infected hosts, and/or enhanced transmission.
table S5. Comparison of means for cases and controls using the full data set (all MME years); real dates of onset or 9 May; and climate metrics aggregated over the 10 days to onset.
table S6. Long-term climate anomaly data at Kostanai sites (ERA data using actual die-off site, NCEP data using pixel, covering entire Torgai area).
REFERENCES AND NOTES
- 1.H. Nicholls, Mysterious die-off sparks race to save Saiga antelope. Nature (2015); doi:10.1038/nature.2015.17675.
- 2.Fey S. B., Siepielski A. M., Nusslé S., Cervantes-Yoshida K., Hwan J. L., Huber E. R., Fey M. J., Catenazzi A., Carlson S. M., Recent shifts in the occurrence, cause, and magnitude of animal mass mortality events. Proc. Natl. Acad. Sci. U.S.A. 112, 1083–1088 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harvell C. D., Kim K., Burkholder J. M., Colwell R. R., Epstein P. R., Grimes D. J., Hofmann E. E., Lipp E. K., Osterhaus A. D. M. E., Overstreet R. M., Porter J. W., Smith G. W., Vasta G. R., Emerging marine diseases—Climate links and anthropogenic factors. Science 285, 1505–1510 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Smith K. F., Sax D. F., Lafferty K. D., Evidence for the role of infectious disease in species extinction and endangerment. Conserv. Biol. 20, 1349–1357 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Tompkins D. M., Carver S., Jones M. E., Krkošek M., Skerratt L. F., Emerging infectious diseases of wildlife: A critical perspective. Trends Parasitol. 31, 149–159 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Travis D. A., Sriramarao P., Cardona C., Steer C. J., Kennedy S., Sreevatsan S., Murtaugh M. P., One medicine one science: A framework for exploring challenges at the intersection of animals, humans, and the environment. Ann. N.Y. Acad. Sci. 1334, 26–44 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.CMS, “Standard Operating Procedures for detecting and reacting to incidents of health risks for and die-offs in Saiga antelopes and other wildlife in Kazakhstan” (Convention on the Conservation of Migratory Species of Wild Animals, 2017).
- 8.Bekenov A. B., Grachev Iu. A., Milner-Gulland E. J., The ecology and management of the Saiga antelope in Kazakhstan. Mammal Rev. 28, 1–52 (1998). [Google Scholar]
- 9.Callahan J. D., Brown F., Osorio F. A., Sur J. H., Kramer E., Long G. W., Lubroth J., Ellis S. J., Shoulars K. S., Gaffney K. L., Rock D. L., Nelson W. M., Use of a portable real-time reverse transcriptase-polymerase chain reaction assay for rapid detection of foot-and-mouth disease virus. J. Am. Vet. Med. Assoc. 220, 1636–1642 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Hofmann M., Griot C., Chaignat V., Perler L., Thür B., Blauzungenkrankheit erreicht die Schweiz. Schweiz. Arch. Tierheilkd. 150, 49–56 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Batten C. A., Banyard A. C., King D. P., Henstock M. R., Edwards L., Sanders A., Buczkowski H., Oura C. C. L., Barrett T., A real time RT-PCR assay for the specific detection of Peste des petits ruminants virus. J. Virol. Methods 171, 401–404 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Bowden T. R., Babiuk S. L., Parkyn G. R., Copps J. S., Boyle D. B., Capripoxvirus tissue tropism and shedding: A quantitative study in experimentally infected sheep and goats. Virology 371, 380–393 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.OIE, Hemorrhagic septicemia, in Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2016 (World Organization for Animal Health, 2016), vol. 2, chap. 2.4.11. [Google Scholar]
- 14.P. J. Quinn, B. K. Markey, M. E. Carter, W. J. Donnelly, F. C. Leonard, Veterinary Microbiology and Microbial Disease (Blackwell Science, 2002), pp. 137–143. [Google Scholar]
- 15.Townsend K. M., Boyce J. D., Chung J. Y., Frost A. J., Adler B., Genetic organization of Pasteurella multocida cap loci and development of a multiplex capsular PCR typing system. J. Clin. Microbiol. 39, 924–929 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King D. P., Burman A., Gold S., Shaw A. E., Jackson T., Ferris N. P., Integrin sub-unit expression in cell cultures used for the diagnosis of foot-and-mouth disease. Vet. Immunol. Immunopathol. 140, 259–265 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Scheuch M., Höper D., Beer M., RIEMS: A software pipeline for sensitive and comprehensive taxonomic classification of reads from metagenomics datasets. BMC Bioinformatics 16, 69 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh N. J., Grachev I. A., Bekenov A. B., Milner-Gulland E. J., Saiga antelope calving site selection is increasingly driven by human disturbance. Biol. Conserv. 143, 1770–1779 (2010). [Google Scholar]
- 19.S. S. Bastianello, M. M. Henton, Hemorrhagic septicemia, in Infectious Diseases of Livestock, J. A. W. Coetzer, R. C. Tustin, Eds. (Oxford Univ. Press, 1994), vol. 3, chap. 164, 1690 pp. [Google Scholar]
- 20.Dee D. P., Uppala S. M., Simmons A. J., Berrisford P., Poli P., Kobayashi S., Andrae U., Balmaseda M. A., Balsamo G., Bauer P., Bechtold P., Beljaars A. C. M., van de Berg L., Bidlot J., Bormann N., Delsol C., Dragani R., Fuentes M., Geer A. J., Haimberger L., Healy S. B., Hersbach H., Hólm E. V., Isaksen L., Kållberg P., Köhler M., Matricardi M., McNally A. P., Monge-Sanz B. M., Morcrette J.-J., Park B.-K., Peubey C., de Rosnay P., Tavolato C., Thépaut J.-N., Vitart F., The ERA-Interim reanalysis: Configuration and performance of the data assimilation system. Q. J. R. Meteorol. Soc. 137, 553–597 (2011). [Google Scholar]
- 21.Schamm K., Ziese M., Becker A., Finger P., Meyer-Christoffer A., Schneider U., Schröder M., Stender P., Global gridded precipitation over land: A description of the new GPCC first guess daily product. Earth Syst. Sci. Data 6, 49–60 (2014). [Google Scholar]
- 22.Kalnay E., Kanamitsu M., Kistler R., Collins W., Deaven D., Gandin L., Iredell M., Saha S., White G., Woollen J., Zhu Y., Leetmaa A., Reynolds R., Chelliah M., Ebisuzaki W., Higgins W., Janowiak J., Mo K. C., Ropelewski C., Wang J., Jenne R., Joseph D., The NCEP/NCAR 40-year reanalysis project. Bull. Am. Meteorol. Soc. 77, 437–471 (1996). [Google Scholar]
- 23.J. Radoux, C. Rousseau, C. Lamarche, T. De Maet, P. Defourny, Seasonal metrics and anomaly detection based on SPOT-VEGETATION archive in Europe, paper presented at the International Geoscience and Remote Sensing Symposium, Milano, Italy, 26 to 31 July 2015. [Google Scholar]
- 24.C. Rousseau, J. Radoux, C. Lamarche, T. De Maet, P. Defourny, Snow cover anomalies from 2000 to 2014 and highlight of two exceptional years over Europe, paper presented at the International Geoscience and Remote Sensing Symposium, Milano, Italy, 26 to 31 July 2015. [Google Scholar]
- 25.Pinzon J. E., Tucker C. J., A non-stationary 1981–2012 AVHRR NDVI3g time series. Remote Sens. 6, 6929–6960 (2014). [Google Scholar]
- 26.P. C. Buerkner, brms: Bayesian Regression Models Using Stan. R package version 0.9.0, (2016); https://CRAN.R-project.org/package=brms.
- 27.Gelman A., Hwang J., Vehtari A., Understanding predictive information criteria for Bayesian models. Stat. Comput. 24, 997–1016 (2014). [Google Scholar]
- 28.Sezenler T., Özder M., Yildirir M., Ceyhan A., Yüksel M. A., The relationship between body weight and body condition score in some indigenous sheep breeds in Turkey. J. Anim. Plant Sci. 21, 443–447 (2011). [Google Scholar]
- 29.J. A. W. Coetzer, R. C. Tustin, Infectious Diseases of Livestock (Oxford Univ. Press, ed. 2, 2004). [Google Scholar]
- 30.F. Sanchez-Monge, “Calving status and commensalism of Pasteurella multocida in the surviving Betpak-dala saiga population in May 2016, after a mass mortality event in May 2015,” thesis, Royal Veterinary College, University of London (2016). [Google Scholar]
- 31.De Alwis M. C. L., Haemorrhagic septicaemia—A general review. Br. Vet. J. 148, 99–112 (1992). [DOI] [PubMed] [Google Scholar]
- 32.Millar R. B., Anderson M. J., Remedies for pseudoreplication. Fish. Res. 70, 397–407 (2004). [Google Scholar]
- 33.Dutta J., Rathore B. S., Mullick S. G., Singh R., Sharma G. C., Epidemiological studies on occurrence of haemorrhagic septicaemia in India. Indian Vet. J. 67, 893–899 (1990). [Google Scholar]
- 34.V. G. Lunitsyn, A. S. Donchenko, S. I. Ognev, N. A. Frolov, Pantovoe Olenevodstvo i Bolezni Olenei: Deer Farming and Disease (Barnaul, 2007). [Google Scholar]
- 35.Gao X., Xiao J., Qin H., Cao Z., Wang H., Impact of meteorological factors on the prevalence of porcine pasteurellosis in southcentral of Mainland China. Prev. Vet. Med. 125, 75–81 (2016). [DOI] [PubMed] [Google Scholar]
- 36.R. Kock, “Final report of mission by Prof Richard Kock RVC to Kazakhstan September 13th to October 1st 2011 and related analysis” (Royal Veterinary College, 2011).
- 37.Shatar M., Khanui B., Purevtseren D., Khishgee B., Loitsch A., Unger H., Settypalli T. B. K., Cattoli G., Damdinjav B., Dundon W. G., First genetic characterization of peste des petits ruminants virus from Mongolia. Arch. Virol. 162, 3157–3160 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Food and Agriculture Organization of the United Nations, Crisis Management Centre for Animal Health Mongolia. Investigation of peste des petits ruminants (PPR) among wild animals and its potential impact on the current PPR situation in livestock. Mission Report 20th January – 1st February 2017. FAO, Rome (2017).
- 39.Kühl A., Mysterud A., Erdnenov G. I., Lushchekina A. A., Grachev I. A., Bekenov A. B., Milner-Gulland E. J., The ‘big spenders’ of the steppe: Sex-specific maternal allocation and twinning in the saiga antelope. Proc. Biol. Sci. 274, 1293–1299 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh N. J., Grachev I. A., Bekenov A. B., Milner-Gulland E. J., Tracking greenery across a latitudinal gradient in central Asia – The migration of the saiga antelope. Divers. Distrib. 16, 663–675 (2010). [Google Scholar]
- 41.Milner-Gulland E. J., Kholodova M. V., Bekenov A., Bukreeva O. M., Grachev I. A., Amgalan L., Lushchekina A. A., Dramatic declines in saiga antelope populations. Oryx 35, 340–345 (2001). [Google Scholar]
- 42.Murray G. M., O’Neill R. G., More S. J., McElroy M. C., Earley B., Cassidy J. P., Evolving views on bovine respiratory disease: An appraisal of selected key pathogens—Part 1. Vet. J. 217, 95–102 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor J. D., Fulton R. W., Lehenbauer T. W., Step D. L., Confer A. W., The epidemiology of bovine respiratory disease: What is the evidence for preventive measures? Can. Vet. J. 51, 1351–1359 (2010). [PMC free article] [PubMed] [Google Scholar]
- 44.Orynbayev M. B., Beauvais W., Sansyzbay A. R., Rystaeva R. A., Sultankulova K. T., Kerimbaev A. A., Kospanova M. N., Kock R. A., Seroprevalence of infectious diseases in saiga antelope (Saiga tatarica tatarica) in Kazakhstan 2012–2014. Prev. Vet. Med. 127, 100–104 (2016). [DOI] [PubMed] [Google Scholar]
- 45.V. Kattsov, V. Govorkova, V. Meleshko, T. Pavlova, I. Shkolnik, Climate Change Projections and Impacts in Russian Federation and Central Asia States (North Eurasia Climate Centre, 2008); http://neacc.meteoinfo.ru/research/20-research.
- 46.Lioubimtseva E., Henebry G. M., Climate and environmental change in arid Central Asia: Impacts, vulnerability and adaptations. J. Arid Environ. 73, 963–977 (2009). [Google Scholar]
- 47.IPCC, Climate Change 2014: Impacts, Adaptation, and Vulnerability. Part B: Regional Aspects. Contribution of Working Group II to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change (Cambridge Univ. Press, 2014). [Google Scholar]
- 48.Salnikov V., Turulina G., Polyakova S., Petrova Y., Skakova A., Climate change in Kazakhstan during the past 70 years. Quatern. Int. 358, 77–82 (2015). [Google Scholar]
- 49.Turco M., Palazzi E., von Hardenberg J., Provenzale A., Observed climate change hotspots. Geophys. Res. Lett. 42, 3521–3528 (2015). [Google Scholar]
- 50.de Jong R., Verbesselt J., Schaepman M. E., de Bruin S., Trend changes in global greening and browning: Contribution of short-term trends to longer-term change. Glob. Chang. Biol. 18, 642–655 (2012). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/1/eaao2314/DC1
Supplementary Text
fig. S1. Locations of the three P. multocida–associated disease events (1981, 1988, and 2015) in the Betpak-dala population and control sites at which calving proceeded without a die-off since 1979, used in the climate analysis (x and y units are longitude and latitude).
fig. S2. Photographs depicting the die-off of saiga and their clinical signs and pathology.
fig. S3. Fluorescence in situ hydridization (FISH) photomicrographs showing position of fluorescing P. multocida bacteria.
fig. S4. Box plots for cases (die-off sites) and controls (no die-off) using the full data set, with real dates of onset if available or 9 May if not, showing the relationship between cases/controls and selected climate metrics aggregated over 10-day periods before onset.
fig. S5. Contributions of variables to components 1 and 2 of the PCA using the full data set, with real dates of onset if available or 9 May if not.
table S1. PCR primers used for microorganism detection.
table S2. A summary of the largest MMEs in saiga attributed to Pasteurellaceae-related syndromes.
table S3. Results of an ICP-MS analysis of livers (w/w) of three dead saigas from May 2015 (Tengiz group).
table S4. Assuming a diagnosis of HS as a primary cause of death, this table is a list of possible stressors, which may cause suppressed immunity in the host or increased virulence and invasion of a commensal parasite such as P. multocida, subsequent septicemia in infected hosts, and/or enhanced transmission.
table S5. Comparison of means for cases and controls using the full data set (all MME years); real dates of onset or 9 May; and climate metrics aggregated over the 10 days to onset.
table S6. Long-term climate anomaly data at Kostanai sites (ERA data using actual die-off site, NCEP data using pixel, covering entire Torgai area).


