Abstract
Background
Glioblastoma (GBM) is almost invariably fatal due to failure of standard therapy. The relapse of GBM following surgery, radiation, and systemic temozolomide (TMZ) is attributed to the ability of glioma stem cells (GSCs) to survive, evolve, and repopulate the tumor mass, events on which therapy exerts a poorly understood influence.
Methods
Here we explore the molecular and cellular evolution of TMZ resistance as it emerges in vivo (xenograft models) in a series of human GSCs with either proneural (PN) or mesenchymal (MES) molecular characteristics.
Results
We observed that the initial response of GSC-initiated intracranial xenografts to TMZ is eventually replaced by refractory growth pattern. Individual tumors derived from the same isogenic GSC line expressed divergent and complex profiles of TMZ resistance markers, with a minor representation of O6-methylguanine DNA methyltransferase (MGMT) upregulation. In several independent TMZ-resistant tumors originating from MES GSCs we observed a consistent diminution of mesenchymal features, which persisted in cell culture and correlated with increased expression of Nestin, decline in transglutaminase 2 and sensitivity to radiation. The corresponding mRNA expression profiles reflective of TMZ resistance and stem cell phenotype were recapitulated in the transcriptome of exosome-like extracellular vesicles (EVs) released by GSCs into the culture medium.
Conclusions
Intrinsic changes in the tumor-initiating cell compartment may include loss of subtype characteristics and reciprocal alterations in sensitivity to chemo- and radiation therapy. These observations suggest that exploiting therapy-induced changes in the GSC phenotype and alternating cycles of therapy may be explored to improve GBM outcomes.
Keywords: exosomes, glioma stem cells, molecular subtypes, radiation, temozolomide resistance
Importance of the study
We report several unsuspected contributions of GSCs to TMZ resistance in GBM. First, diverse, complex, and consequential programs of TMZ resistance (beyond upregulation of MGMT) exhibited by GSCs in vivo may curtail and bypass attempts to target individual candidate drug-resistance pathways. Second, molecular GSC subtypes may differ in their TMZ-resistant profiles and evolve on therapy. As this plasticity may necessitate real-time, noninvasive disease monitoring, such opportunities may exist in mRNA profiling of tumour-derived EVs. Importantly, TMZ resistance of MES GSCs unexpectedly led to their increased sensitivity to radiation, suggesting a potential benefit of alternating chemoradiation therapy protocols as observed in a recent clinical study.
Glioblastoma (GBM) represents the most frequent and almost uniformly fatal class of World Health Organization grade IV primary astrocytic brain tumors, associated with median survival of only 12–15 months.1 This dismal prospect has remained essentially unchanged since the introduction of the intensive trimodal backbone therapy protocol, which combines surgical resection followed by concurrent temozolomide (TMZ) chemoradiation and adjuvant courses of oral TMZ.2 Whether this sequence is optimal from the biological standpoint remains unclear.3
The efficacy of TMZ is predicated on the ability of the spontaneously generated active metabolite (methyldiazonium cation) to donate methyl groups to at least 3 acceptor sites on purine residues. Such modification of cellular DNA, most notably on O6-guanine, results in replication errors and cytotoxicity.4 These effects can be offset in normal cells by the ubiquitously expressed DNA repair enzyme O6-methylguanine DNA methyltransferase (MGMT), which removes the cytotoxic O6-methylguanine DNA adducts. MGMT expression is selectively downregulated in approximately 45% of GBM cases due to promoter methylation, a feature associated with some improvement in therapeutic effects and prognosis. However, patients vary in the degree of drug responsiveness and eventually relapse.2,4
While the expression of MGMT in a subset of GBM tumors is often viewed as the predominant cause of TMZ resistance,5 along with other biological effects,6 other plausible causes of treatment failure have also been studied. Those include reduced activity of mismatch repair (MMR) genes, activation of base excision repair (BER), including poly(ADP-ribose) polymerase 1 (PARP-1), and deregulation of other pathways, such as those involving excision repair cross-complement 1 and 2, tumor protein 53/murine double minute 2, drug efflux molecules (ATP-binding cassette [ABC]B1, ABCG2, ABCC3), and other effectors, as summarized by Messaoudi and colleagues4 and in Supplementary Table S1.
These emerging findings continue to motivate the search for the corresponding TMZ-sensitizing agents4 and drug-resistance biomarkers,7 including within the GBM secretome and tumor derived extracellular vesicles (EVs).8 EVs are heterogeneous, vesicular, membrane-bound structures of varying sizes (50 nm to over 2 μm) emitted by all cells, from which they carry a wide repertoire of molecular cargo (lipids, proteins, and nucleic acids, including RNA). Thus, EVs represent unique assemblies of informative macromolecules that can be noninvasively sampled from biofluids and analyzed for signatures of cancer cell properties and states.8–11
What complicates the implementation of these emerging opportunities is the biological complexity of GBM. This includes regional, genetic, and phenotypic heterogeneity of cancer cells12–14 and inter-individual diversity of GBM subtypes.15 In this regard, The Cancer Genome Atlas distinguishes proneural (PN), neural, classical, and mesenchymal (MES) forms of GBM at diagnosis.16 However, the underlying disease plasticity often results in phenotypic shifts, such as from PN to MES transition upon therapy and relapse.17
Underneath this molecular mosaic lies the increasingly recognized functional hierarchy of GBM cells comprising the minority subsets of brain tumor initiating cells, also referred to as glioma stem cells (GSCs), amidst their more abundant progeny with a more restricted growth potential.18–20 While their origin remains controversial, GSCs are believed to carry unique molecular signatures, such as the expression of cluster of differentiation (CD)133 antigen, as well as Nestin, oligodendrocyte transcription factor (OLIG2), and sex determining region Y-box 2 (Sox2). They are also viewed as the key cellular cause of treatment failure in GBM due to their intrinsic drug and radiation resistance and their ability to repopulate the tumor mass.20 Indeed, GSCs represent the central therapeutic target in GBM necessitating a better understanding of their diversity,21–23 as exemplified by the discovery of molecular GSC subtypes.24 Those include PN-like GSCs, positive for CD133 and CD15, which form compact spheres in cell culture and exhibit a less aggressive and relatively radiation-sensitive phenotype. In contrast, MES-like GSCs are positive for aldehyde dehydrogenase 1 family member A3, CD44, and epidermal growth factor receptor (EGFR); form loose structures in neurosphere cultures; exhibit highly aggressive phenotypes in vivo; and are essentially radiation resistant.25,26 Whether distinct subtypes of GSCs simply persist within tumor masses (remnants) or evolve under the influence of therapy remains poorly understood.
Here we studied human PN and MES GSCs in vivo and under the influence of TMZ chemotherapy. While both PN and MES GSCs gave rise to aggressive tumors that initially responded to TMZ, this treatment ultimately failed, leading to disease relapse. A series of secondary GSCs (SGSCs) isolated in sphere cultures from individual recurrent tumors expressed non-uniform profiles of TMZ resistance markers, of which the upregulation of MGMT occurred in a minority of cases. Transcripts linked to TMZ resistance were readily detectable in the cargo of EVs isolated from the respective GSC cultures. Interestingly, TMZ-resistant SGSCs exhibited a common diminution of mesenchymal characteristics and an increase in radiation sensitivity. These observations suggest that a complex reprogramming of GSCs under the influence of TMZ may lead to a phenotype detectable and more vulnerable to radiotherapy.
Material and Methods
Cell Culture and Treatments
Cells were cultured according to the provisions of the RIMUHC Biohazard Safety Certificate. All cell lines were established by the coauthor (I.N.) as glioblastoma stem cells (GSCs) within 5 years of their initial publication in 2013.24 The sphere cultures representing either mesenchymal (MES83, MES1123) or proneural GSC subtype (PN157, PN528)24,27–29 were cultured (as neurospheres) in Dulbecco’s modified Eagle’s medium/F12 supplemented with B27 (2%), Glutamax, epidermal growth factor (20 ng/mL), basic fibroblast growth factor (20 ng/mL), penicillin/streptomycin (Life Technologies), and heparin 5 μg/mL (Stem Cell Technologies). In some experiments cells were treated with TMZ (S1237, Selleckchem) or O6-benzylguanine (B2292, Sigma-Aldrich), dissolved in dimethyl sulfoxide, and used at indicated concentrations. Radiation treatment of GSC cultures was performed using a Faxitron irradiator followed by 11-day culture and cell viability assessment. The viability and growth assays were conducted either using MTS reagent or by automatic count of trypan blue excluding cells (see Supplementary Methods for details).
Tumor Models
To generate orthotopic tumors 1 × 104 GSCs were stereotactically injected into brains of NSG mice (Charles River Labs) in a total volume of 2 μL. Subcutaneous tumors were initiated by injection of 1 × 106 SGSCs resuspended in 200 μL of Matrigel (BD Biosciences) into the flanks of NSG mice. Bioluminescent imaging was carried out following the injection of D-Luciferin (15 μg/mL; PerkinElmer) under an IVIS 200 scanner (PerkinElmer) as described.30 Tumor dissociation was performed to isolate SGSCs and establish them in culture. Collagenase/dispase digestion and growth conditions are detailed in the Supplementary Methods. All procedures involving animals were performed in accordance with the guidelines of the Canadian Council of Animal Care and the Animal Utilization Protocols, approved by the Institutional Animal Care Committee at the McGill University Health Centre Research Institute and McGill University.
Molecular Analysis
As described earlier,31 protein lysates were subjected to electrophoresis, and western blots were probed with primary antibodies: rabbit anti-Notch1 (Cell Signaling, 4380), rabbit anti-Nestin (Abcam, ab105389), goat anti-Sox2 (Santa Cruz Biotechnology, sc17320), rabbit anti‒microtubule-associated protein 2 (MAP2) (Millipore, AB5622), rabbit anti‒transglutaminase 2 (TGM2) (Cell Signaling, 3557), rabbit anti-RAD50 (Cell Signaling, 3427), rabbit anti-EGFR (Cell Signaling, 4267S), mouse anti-keratin18 (KRT18) (Cell Signaling, 4548), mouse anti-MGMT (Millipore, AB16200), and mouse anti‒β-actin (Sigma). Appropriate horseradish peroxidase‒conjugated secondary antibodies (Dako/Agilent Technologies) were used to visualize the protein bands following enhanced chemiluminescence detection (GE Healthcare). For mass spectrometry,32 cell pellets were subjected to denaturation, reduction, and alkylation, followed by trypsin digestion as described elsewhere.33 Analysis of mRNA was conducted using customized 96-well RT2 Profiler PCR Arrays (SABiosciences/Qiagen).
Preparation of Extracellular Vesicles
EVs were obtained by ultracentrifugation as described earlier.31,34 Briefly, cell culture supernatants were centrifuged for 10 min at 400g, and 30 min at 10000g to remove cells and debris. The liquid fraction was then centrifuged for 1 h at 100000g to pellet EVs, which were then washed extensively in phosphate buffered saline. Nanoparticle tracking analysis was performed using the NS500 system (NanoSight).35
Statistical Analysis
All experiments were reproduced at least twice with similar results and presented as numbers of replicates (N) and mean value of replicates ± SD. Statistical significance was evaluated with GraphPad Prism software. A log-rank (Mantel–Cox) test was used for survival studies in mice, whereas statistical analysis on other assays was performed using 2-tailed unpaired t-tests. Differences were considered significant for P < 0.05 (see Supplementary Methods for experimental detail).
Results
Phenotypic Heterogeneity of Glioma Stem Cells
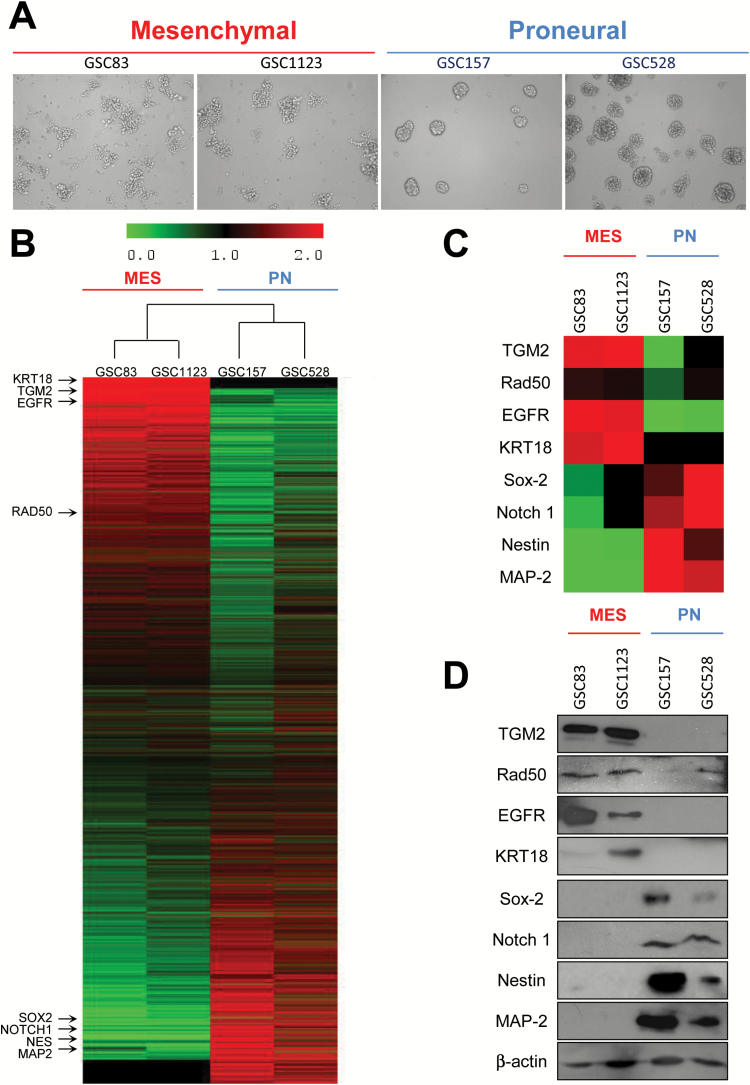
In our hands, 4 different previously characterized neurosphere (tumor sphere)-forming GBM isolates clearly shared either MES (GSC83 and GSC1123) or PN (GSC157 and GSC528) phenotypes, including morphology, protein expression profiles, and distribution of stem cell markers (Fig. 1). For example, PN-like GSCs consistently grew as compact tumor spheres in culture, whereas their MES-like GSC counterparts formed loose aggregates with well-defined individual cells and minimal cell-cell contacts (Fig. 1A). Similarly, PN-like GSCs share common protein profiles, as do MES-like cells (Fig. 1B). PN GSCs shared higher levels of Sox2, Notch1, Nestin, and MAP2, while both MES GSCs tested expressed low levels of these proteins, and instead were positive for TGM2, RAD50, EGFR, and KRT18 proteins, as revealed by mass spectrometry (Fig. 1C) or immunoblotting (Fig. 1D). Thus, protein expression profiles support the earlier identification of PN- and MES-like GSC subtypes24 and highlight the heterogeneity of GSC populations.
Fig. 1.
Phenotypic and molecular heterogeneity of glioma stem cells (GSCs). (A) Morphology of mesenchymal (MES) and proneural (PN) GSCs in sphere culture—phase contrast microscopy, under 10x objective. (B) Proteomes of MES and PN cell lines. The main validated subtype-specific proteins are indicated (arrows). (C) Differentially expressed proteins extracted from the proteome of MES and PN GSCs. (D) Western blot validation of differentially expressed marker proteins in MES and PN GSC lines.
Heterogeneous Responses of GSC-Initiated Xenografts to Temozolomide
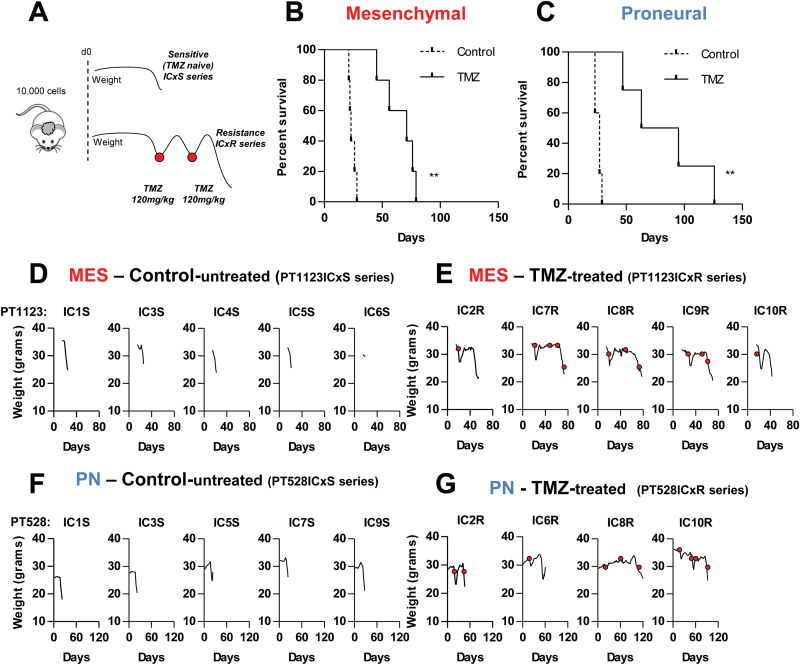
Limited therapeutic gains in GBM23,36,37 come with little understanding of the changes occurring within GSC populations during treatment and relapse.21,25,38 To explore these questions in more depth we compared tumor growth and TMZ responses of intracranial GBM xenografts initiated by injection of PN-like (528) or MES-like (1123) GSCs in immunodeficient mice (Fig. 2). The drug scheduling protocol was predicated on our observation that clinically apparent disease progression could be reliably detected in individual mice by a rapid onset of weight loss. This event could be repeatedly reversed by rounds of single-dose TMZ therapy (120 mg/kg) until tumors became unresponsive (Fig. 2A). The corresponding GBM-like lesions in the brain were verified using bioluminescence and autopsy (Supplementary Figure S1, data not shown). Under these conditions, both PN- and MES-like GSCs initiated aggressive disease that reached the humane endpoint (surrogate for survival) in less than 50 days. The maximal progression-free survival achieved under TMZ treatment for the MES GSC-initiated tumors was approximately 60 days, and this value reached 107 days in the PN GSC-initiated disease (Fig. 2B, C; Supplementary Figure S1A), but these differences were not statistically significant and all tumors eventually became TMZ unresponsive. We also observed heterogeneous responses among individual tumors, even upon injection of the same GSC line into several different mice, as documented by weight loss (Fig. 2D–G) and bioluminescent imaging (Supplementary Figure S1B, C). Thus, GSC-initiated tumors invariably progress in vivo from a TMZ-sensitive to a TMZ-resistant state.
Fig. 2.
Differential responses of GSC-initiated intracranial glioblastoma xenografts to TMZ. (A) Experimental design—tumor regression was induced by initial dose of TMZ (120 mg/kg), which was repeated at relapse of individual tumors (red dots) until response was no longer observed. (B, C) Survival curves of mice harboring xenografts initiated by either MES-1123 (B) or PN-528 (C) GSCs. Cells were injected at 1 × 104 cells per inoculum into the brains of NSG mice. Untreated control (discontinuous line; N = 5) or treated with TMZ (continuous line; N = 5 for MES, N = 4 for PN) are shown. (D–G) Heterogeneity in the natural history of the disease in individual mice harboring GSC-initiated GBMs. Clinical response to progression in individual mice was measured by weight changes. Numerical designations in panels D–G indicate the respective individual xenografts with either TMZ sensitive (S) or resistant (R) phenotype (see text). **P < 0.01.
In Vivo Acquired Drug Resistance Results in Formation of Secondary GSCs with Intrinsically Reduced Responsiveness to TMZ
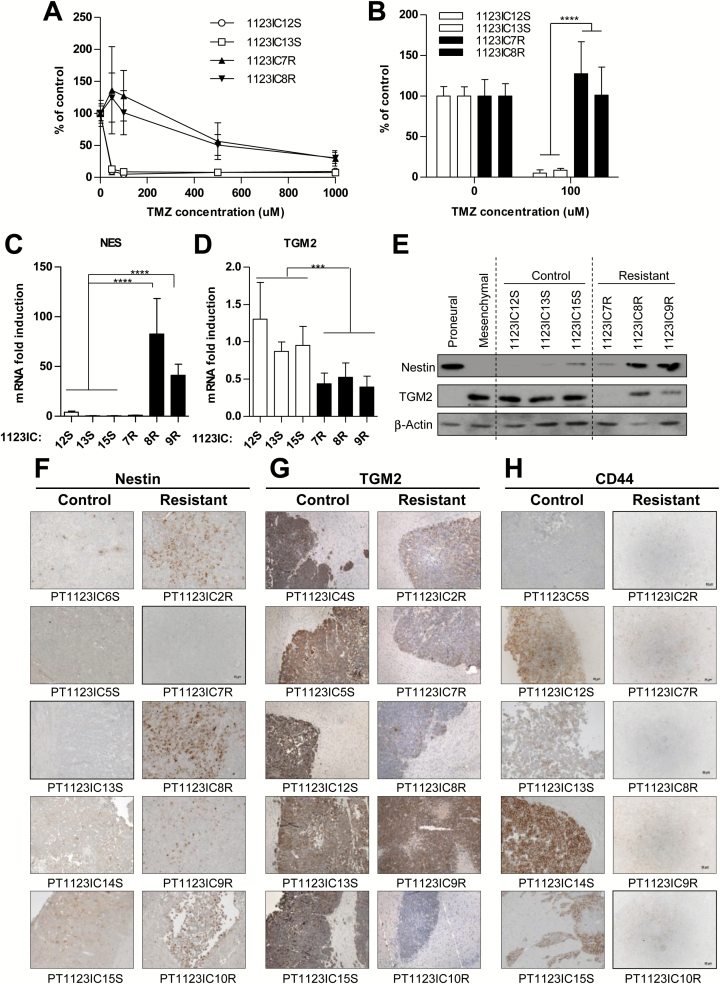
It is presently unknown whether GBM relapse and drug resistance can be accurately predicted from the properties and evolution of the related GSCs. To explore this question we focused on the MES-like GSC 1123 model due to the rapid loss of TMZ responsiveness by tumors initiated by these cells in vivo (Supplementary Figure S2) and the reported intrinsic resistance of MES GSCs to radiation.24 Thus, untreated TMZ-sensitive (S) and TMZ-resistant (R) intracranial (IC) GSC 1123-derived tumors were enzymatically dissociated and several GSC-like sphere forming cultures were established from individual xenografts (1123IC12S, 1123IC13S, 1123IC15S series and 1123IC7R, 1123IC8R, 1123IC9R series, respectively; Supplementary Figure S2). The TMZ-resistant phenotype was maintained by SGSCs in culture in the presence of up to 100 μM TMZ, while the treatment-naïve SGSCs were highly drug sensitive (Fig. 3A, B). In vivo these SGSC lines efficiently initiated aggressive tumor growth upon subcutaneous re-inoculation (1 × 106 cells per inoculum) into NSG mice, whereupon they faithfully recapitulated the patterns of TMZ (120 mg/kg) responsiveness/resistance observed in the case of original GSC xenografts (Supplementary Figure S3). For example, the established tumors initiated by TMZ-naïve SGSC cell lines (1123SC12 or 1123SC15) exhibited a near complete, albeit transient, regression upon exposure to a single dose of TMZ, a treatment which was ineffective against tumors initiated by SGSC (1123IC7R and 1123IC8R) lines derived from tumors that had previously acquired resistance to TMZ in vivo (Supplementary Figure S3). Collectively, these observations indicate that in vivo exposure of GSCs to TMZ leads to the rise of SGSCs with cell-autonomous drug-resistant phenotypes.
Fig. 3.
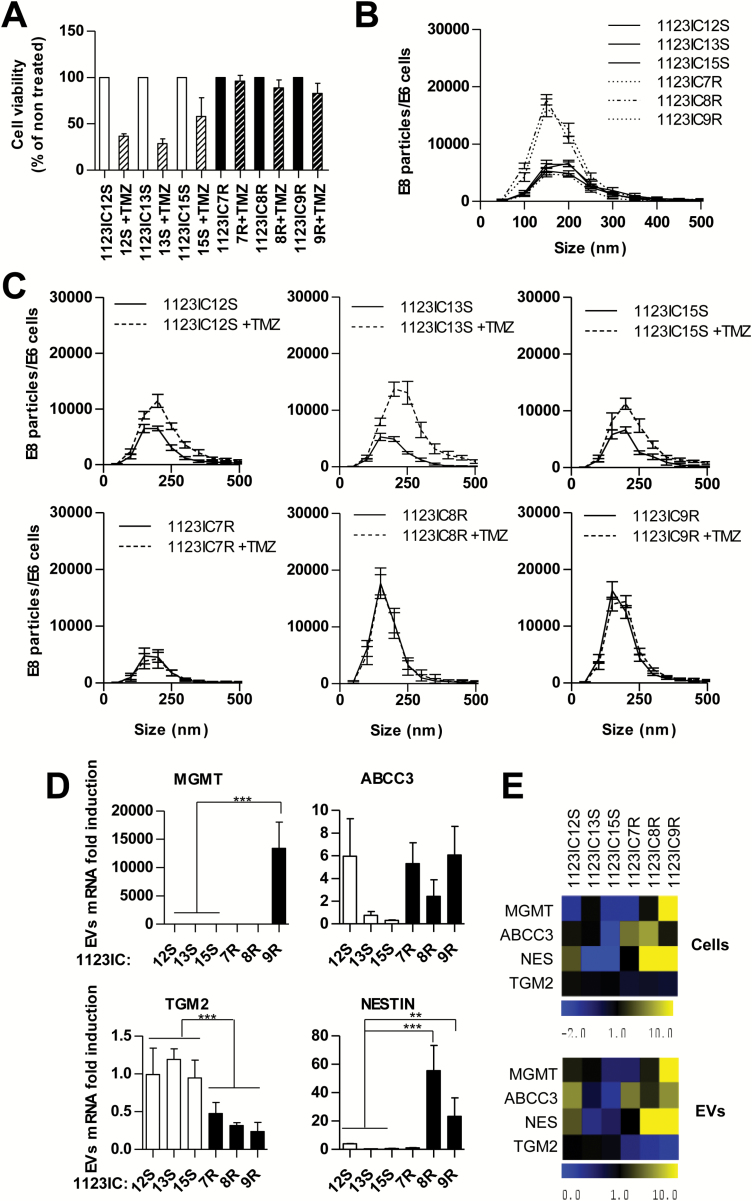
Secondary GSCs represent a stable and cell intrinsic TMZ resistance phenotype, coupled with a loss of mesenchymal characteristics. (A, B) TMZ resistance of GSCs is maintained in sphere culture (MTS viability assay, see text). MTS assay after TMZ treatment in 2 SGSC lines (1123IC8R, 1123IC7R) from TMZ-unresponsive tumors and 2 SGSC lines from control tumors (1123IC12S, 1123IC13S) (N = 3; 100% = no TMZ). ****P < 0.0001. (C–E) TGM2 downregulation and Nestin induction in TMZ resistant phenotype were observed in vitro after isolation of SGSCs from xenograft tumors, at the mRNA level tested by qPCR (C, D) (1 = average mRNA expression in TMZ-sensitive cell lines), and at the protein level by western blot (E). ****P < 0.0001; **P < 0.01 (N = 3). (F–H) Immunostaining of mesenchymal control (PT1123ICxS) and TMZ-treated (PT1123ICxR) intracranial tumors confirmed the variability in protein expression, a trend toward a decrease in mesenchymal markers TGM2 and CD44, and an upregulation of Nestin expression after acquisition of resistance to TMZ (objective 20x).
Loss of Mesenchymal Features Parallels the Acquisition of Resistance of GSCs to Temozolomide
Interestingly, 4 out of 5 SGSCs resistant to TMZ exhibited a significant increase in the expression of Nestin (mesenchymal SGSCs [Fig. 3C and E]; proneural SGSCs [Supplementary Figure S7]). This is surprising in the case of SGSCs derived from GSCs with MES phenotype, which unlike PN-type GSCs are usually Nestin-low/negative (Fig. 1C, D). Conversely, TGM2, a marker of mesenchymal GSCs,39 was expressed highly in tumor spheres of MES-like 1123 GSCs (but not in PN GSCs), but downregulated in all TMZ-resistant derivatives of this cell line at both mRNA and protein levels (Fig. 3D, E). Also tumor tissues of treatment-naïve GSC 1123-derived primary xenografts (PT1123ICxS tumors) were negative for Nestin, positive for TGM2, and, with some exceptions, positive for CD44, while their isogenic TMZ-resistant counterparts (PT1123ICxR tumors) exhibited the opposite staining pattern, no longer consistent with the MES-like phenotype (Fig. 3F, H). These changes were maintained in secondary tumors (ST1123IC12-15 vs ST1123IC7-9R; Supplementary Figure S4A). No consistent increase in PN markers (MAP2, Sox2, OLIG2) was observed (Supplementary Figure S4B, C), suggesting no obvious mesenchymal-to-proneural transition. Some of the TMZ-resistant GSC variants also exhibited reduced invasiveness at the tumor margin (Supplementary Figure S5A, B). These observations suggest that the exposure to TMZ in vivo is associated with reprogramming of the GSC phenotypes, as exemplified by their loss of MES-like features.
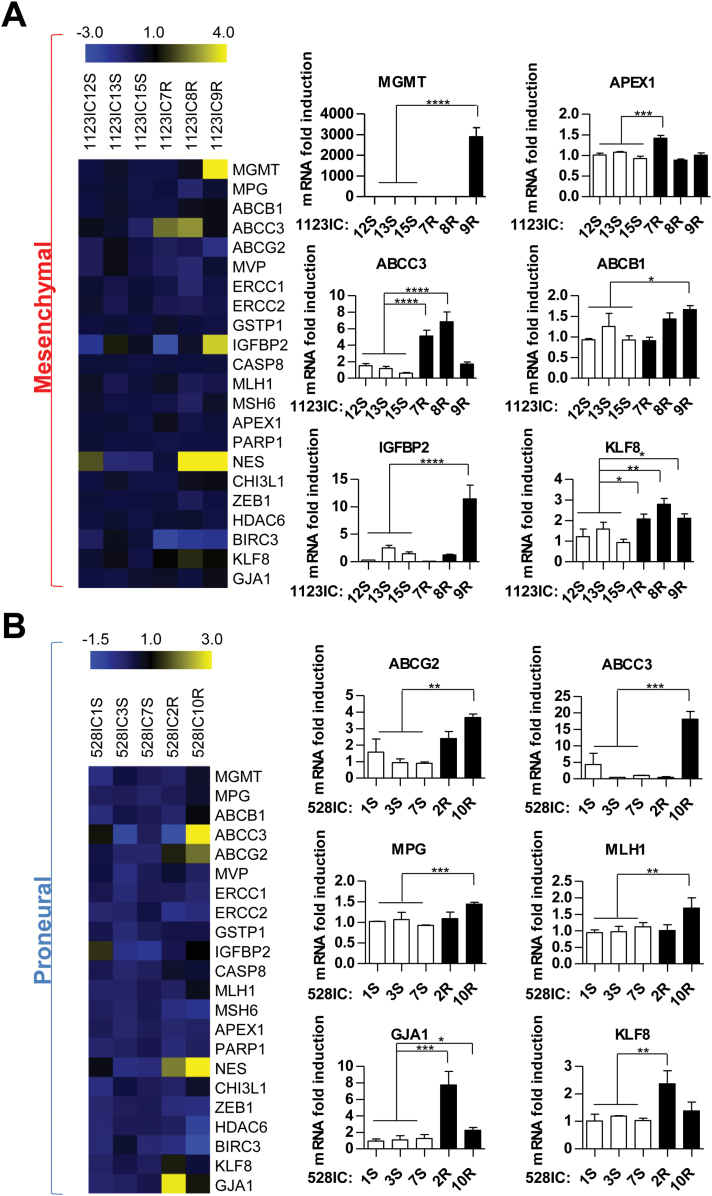
Diverse Expression of Genes Implicated in Temozolomide Resistance Among Secondary GSCs
To explore the possible markers associated with drug refractory SGSCs, we chose to assess their expression of genes implicated in TMZ resistance (Supplementary Table S1; Fig. 4; Supplementary Figures S6–S10). Here, we included MGMT, genes involved in MMR, nucleotide excision repair, BER, and other DNA repair pathways, ABC transporters, as well as regulators of cellular growth, differentiation, and stemness (Supplementary Tables S1–S24,8). The quantitative (q)PCR analysis of these respective transcripts revealed an extensive heterogeneity (Fig. 4A, B). For example, among 3 independent TMZ-resistant derivatives of a single MES-like GSC line (1123), only one SGSC (1123IC9R) exhibited robust upregulation of MGMT. This finding was verified at the protein level (Supplementary Figure S6A) and its functional significance was documented by a selective, but partial, rescue of TMZ toxicity upon addition of O6-benzylguanine, a potent inhibitor of MGMT4 (Supplementary Figure S6B). This effect was not observed in the case of 2 related MGMT-negative and TMZ-resistant cell lines (1123IC7R and 1123IC8R; Supplementary Figure S6B). Interestingly, MGMT-positive 1123IC9R cells also upregulated insulin-like growth factor binding protein 2 (IGFBP2) and ABCB1 transcripts, relative to TMZ-naïve SGSCs (Fig. 4A). At the same time, the remaining 2 SGSCs derived from GSC 1123 cells expressed different combinations of putative TMZ resistance markers, such as apurinic/apyrimidinic endodeoxyribonuclease 1 (APEX1) (1123IC7R), ABCC3 (1123IC7R and 1123IC8R), ABCB1 (1123IC8R and 1123IC9R), and Krüppel-like factor (KLF)8 (1123IC8R) (Fig. 4A; Supplementary Figures S7 and S8, Supplementary Table S2). These MES cell lines harbored no isocitrate dehydrogenase 1 mutations but differed in overall mutational loads depending on the MGMT status (Supplementary Table S4). Similarly, the SGSCs derived from relapsed tumors initiated by PN-like GSCs exhibited a diversity of drug resistance genes distinguishable from those observed for the MES (1123) GSC series (ABCG2, ABCC3, mutant L homolog 1 [MLH1], gap junction alpha-1 protein [GJA1]), and without MGMT upregulation (Fig. 4B). These profiles also diverged among genetically related PN-like SGSC lines (Fig. 4B; Supplementary Figure S7, Supplementary Table S2). All 3 TMZ-resistant cell lines derived from GSC 1123 and one of 2 related to PN-like GSC 528 cells were found to exhibit downregulation of BIRC3 (baculoviral inhibitors of apoptosis proteins repeat containing 3; Supplementary Figure S7). Overall, these experiments suggest that repertoires of genes associated with the onset of TMZ resistance are complex, subtype-related, and divergent (difficult to predict) even among clonally related SGSCs.
Fig. 4 .
Heterogeneous expression of genes associated with TMZ resistance among secondary GSCs isolated from individual mice following chemotherapy in vivo. Profiles of mRNA among clonally related SGSCs isolated from PN (528) or MES (1123) brain tumors: control (S), TMZ resistant (R) (as in Fig. 2). Markers for which differences were significant are shown as bar graphs (right panel), in MES (A) and PN (B) cell lines (1 = average mRNA expression in TMZ-sensitive cell lines). The heatmap (left panel) represents the global profile of all markers of resistance tested. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05 (N = 3).
Representation of Molecular Markers Associated with TMZ Resistance in the Transcriptome of GSC-Derived Extracellular Vesicles
While TMZ-related complex changes in the GSC phenotype during GBM relapse are presently difficult to molecularly assess, or oppose, their detection may be of paramount importance. In this regard monitoring changes in the GBM molecular makeup by the detection of EV-associated RNA and protein markers represents a promising avenue.8,9 To explore this possibility in the case of GSCs, we first isolated EVs from culture media by ultracentrifugation31 and performed nanoparticle tracking analysis to compare their abundance, size distribution, and TMZ-induced changes (Fig. 5A–C). Indeed, all GSCs produced readily detectable EVs, mostly near the size range of exosomes (150–200 nm). In the case of 2 out of 3 TMZ-resistant cell lines (GSC 1123IC8R and 1123IC9R), this EV emission was markedly (3-fold) greater than for their drug-sensitive counterparts (1123IC12S, 1123IC13S, and 1123IC15S; Fig. 5B). Interestingly, the latter cell lines produced increased numbers of EVs in the presence of TMZ and in parallel to the apparent drug-induced cell death (Fig. 5A). TMZ-related changes in vesiculation were not observed in the case of 3 different drug-resistant GSC variants (1123IC7R, 1123IC8R, and 1123IC9R; Fig. 5C).35 Importantly, EVs released from all cell lines contained transcripts related to the development of TMZ resistance and in amounts corresponding to those found in the respective donor cells, as quantified by qPCR (Fig. 5D, E). For example, 1123IC9R cells and their derived EVs were uniquely positive for MGMT mRNA but also contained elevated amounts of Nestin and a reduced level of TGM2 transcripts, relative to their drug-responsive 1123ICS GSC counterparts. At the same time, 1123IC8R SGSCs produced EVs with high levels of Nestin and no detectable MGMT mRNA, as expected from the profile of EV donor cells (Fig. 5D, E). These observations are not an extension of global GBM signatures (Supplementary Figure S11) and are of interest as a measure of the diversity among TMZ resistance scenarios that may accompany the evolution of GSCs and can be remotely captured using EVs.
Fig. 5.
Reflection of molecular changes associated with TMZ resistance in GSC-derived EVs. (A) TMZ effects on cell viability (MTS assay). (B, C) Nanoparticle tracking analysis of EVs produced by SGSC lines in the absence (B) or presence of 100 μM TMZ (C). (D, E) Expression of transcripts implicated in TMZ resistance in GSCs and in their EVs. (D) Quantitative PCR analysis reveals changes in EV-associated TGM2, MGMT, and/or Nestin (1 = average mRNA expression in TMZ-sensitive cell lines). (E) The comparison of heatmaps between cell and EV profiles shows a great similarity. *P < 0.05 (N = 2).
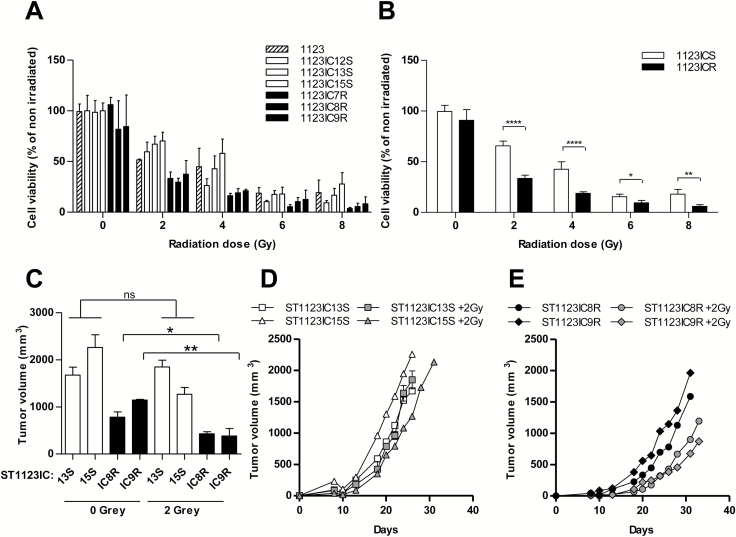
Progression of GSC Toward Temozolomide Resistance Increases Their Radiosensitivity
MES-like GSCs are thought to exhibit increased resistance to radiation therapy relative to their PN-like counterparts.24 Therefore, we chose to investigate the corresponding consequences of diminished mesenchymal characteristics associated with the development of GSC resistance to TMZ in vivo (Fig. 6). To this end, parental (GSC 1123), TMZ-naïve (GSC 1123IC12S, 1123IC13S, and 1123IC15S), and TMZ-resistant (GSC 1123IC7R, 1123IC8R, and 1123IC9R) cultures were exposed to 2, 4, 6, or 8 Gray (Gy) of ionizing radiation (Faxitron) and assayed for cell proliferation. Intriguingly, all TMZ-resistant cell lines were growth suppressed by ionizing radiation to an extent markedly greater than that exhibited by both their parental and TMZ-naïve isogenic counterparts (Fig. 6A, B). We also tested the ability of the respective cell lines to retain tumorigenic potential in vivo following mild irradiation (2 Gy) in culture (Fig. 6C–E). While this treatment exerted little effect on TMZ-naïve MES GSC cell lines, it significantly suppressed tumor formation by their TMZ-resistant counterparts (IC8R and IC9R; full names 1123IC8R and 1123IC9R). The analysis of genes involved in DNA repair (Supplementary Table S3) revealed a marked heterogeneity without a common explanatory trend (Supplementary Figures S9–S10). While the mechanisms involved remain to be elucidated, these results suggest that radiation may be effective post GBM chemotherapy, as suggested by recent clinical observations.40
Fig. 6 .
Elevated sensitivity of TMZ-resistant GSC lines to radiation. Individual (A) and averaged (B) relative (%) changes in surviving TMZ-sensitive (1123ICS, white bars) and TMZ-resistant (1123ICR, black bars) cells after exposure to 0, 2, 4, 6, and 8 Gy of radiation therapy in vitro. TMZ-resistant cell lines exhibit greater cytotoxic impact. (C–E) Impact of radiation (2 Gy) on tumor forming potential of TMZ-sensitive and -resistant GSCs. Pre-irradiated cells were injected into NSG mice. (C) Tumor volume after 26 days of growth. (D, E) Tumor growth curves of TMZ-sensitive (D) or TMZ-resistant (E) GSC lines treated (grey symbols) or not (white or black symbols) with radiation. ****P < 0.0001; **P < 0.01; *P < 0.05 (N = 3).
Discussion
Our study contains several novel observations. First, we developed a unique experimental model of TMZ resistance in GBM, which is predicated on the molecular evolution of tumor-initiating cells (GSCs) and their role in clinical tumor relapse in vivo. Second, we documented that the intrinsic drug resistance of secondary (tumor derived) GSCs is sufficient to render tumor growth refractory to the effective doses of TMZ, irrespective of the influence that other implicated factors may have, such as cellular heterogeneity, growth rates, tissue barriers, or microenvironment. Third, we observed a remarkable heterogeneity among pathways leading to the development of TMZ-resistant GSCs, both between their molecular subtypes (MES versus PN) and within them. Only a fraction of these resistant cell lines evolved to strongly express MGMT, currently the focal point in studies on TMZ resistance. Notably, MGMT-expressing cells contained lower mutational loads than their MGMT-negative TMZ-resistant isogenic counterparts. This may suggest a protective effect of this enzyme on the cellular genome, as well as existence of mechanisms that enable survival of cells that sustained drug-induced DNA damage.
In several instances the refractory GSCs exhibited multiple changes in the expression of genes previously linked to TMZ resistance, including DNA repair enzymes, drug efflux molecules, and markers of stemness. This could present challenges with regard to reversal strategies based on single markers. These respective transcripts were emitted by cells as cargo of exosome-like EVs that could serve as biomarkers. Notably, within the isogenic family of MES-like GSCs the onset of TMZ resistance correlated with loss of mesenchymal markers and led to the increased sensitivity to ionizing radiation. Based on these observations, we suggest that strategies aiming at counteracting TMZ resistance and relapse in GBM could benefit from active monitoring of multiple (rather than single) drug resistance mechanisms—for example, through the use of EV-based liquid biopsy platforms. Moreover, our study may suggest a benefit of post-TMZ radiation in a subset of GBMs. Remarkably, this prediction is borne out in results of a very recent clinical study involving neoadjuvant TMZ followed by hypofractionated accelerated radiation therapy with favorable effect on survival of GBM patients.40
There are several points of convergence between our results and the published works analyzing the hallmarks of GBM intractability and relapse. While targeted, anti-angiogenic, and immunity-based therapies have thus far made only a modest impact in this setting,23 the trimodal standard of care2 continues to offer meaningful survival advantage to GBM patients, the transiency of which remains baffling. In this regard, it is increasingly clear that GBM represents a spectrum of molecularly distinct disease subtypes, the responses of which to therapy may a priori exhibit some variability,16 as well as subtype-related distribution of markers associated with therapeutic resistance. In addition, variabilities of the blood–brain barrier, hypoxia, inflammatory responses, and infiltrative growth patterns may further diminish the impact of therapy and patient survival.20 There is also a growing appreciation for the impact that standard therapy may have on the changing biology of GBM, including therapy-related toxicities,41 mutational alterations,36,37 and a global shift toward more mesenchymal phenotype in the recurrent disease.17
Our analysis of TMZ refractory SGSCs revealed 3 classes of treatment-related changes. First, we observed a differential between TMZ-resistant phenotypes of PN-like and MES-like GSCs with respect to genes regarded as putative markers and effectors of resistance to this agent (Supplementary Table S1). For example, the most notable mRNA signals among drug-resistant MES-like GSC lines included transcripts for MGMT, APEX1, ABCC3, ABCB1, IGFBP2, and KLF8, while for PN-like GSC cells the respective changes included ABCG2, ABCC3, N-methylpurine DNA glycosylase, MLH1, GJA1, and KLF8. While some of the respective genes were also enriched in the general transcriptome of GBM samples (rather than in GSCs specifically; Supplementary Figure S11), it is unclear whether the a priori defined molecular tumor subtypes predict the profiles of GSCs or the mechanism of TMZ resistance that may be encountered at tumor relapse in the clinic.
Second, the isogenic TMZ-unresponsive SGSCs derived from individual tumors exhibited a stark diversity and multiplicity in their expression of molecular markers/effectors of TMZ resistance. In only one case was this phenotype linked to overexpression and activity of MGMT, the key marker of de novo resistance to TMZ chemotherapy. The biological basis of divergent evolutionary trajectories among SGSCs under uniform experimental treatment conditions remains unclear. Nonetheless, this observation may suggest that even within the same cellular (GSC) lineage, the repertoire of genes that can protect tumor-initiating cells from chemotherapy may carry stochastic and thereby unpredictable features and include multiple mechanisms expressed simultaneously. This complexity would likely amplify the clinical spectrum of drug resistance mechanisms that may be operative in individual GBM patients, necessitating multipronged and highly individualized diagnostic approaches. Indeed, we show that EVs isolated from TMZ-resistant SGSC lines contain mRNA signals corresponding to those of GBM cells themselves. It is of interest whether such EV-based approaches coupled with proper timing could detect the onset of TMZ resistance in GSCs in vivo and suggest countermeasures.8
The third class of events induced in GSCs during the development of TMZ resistance in vivo consisted of more uniform changes comprising a phenotypic reprogramming of these cells toward less mesenchymal characteristics. While these changes included the expression of some markers of PN-like GSCs in their MES-like counterparts (eg, Nestin), this was not a complete subtype transition. Importantly, TMZ-resistant SGSCs that have lost their MES-like characteristics also exhibited an increase in responsiveness to ionizing radiation.
This is an intriguing observation in light of the fact that some of the same DNA repair mechanisms that exacerbate the cytotoxicity of TMZ, such as MMR,4 may also lead to radiation resistance.42 Hence, selection against these mechanisms in TMZ-resistant GSCs could predict a degree of radiosensitization, in line with our observations and recent clinical experience.40
In conclusion, by defining the responses of GSCs to TMZ at the point of tumor relapse in vivo, our study reveals GSC-intrinsic contributions to the complex change that may underlie treatment failure in GBM. We propose that monitoring the multimolecular signatures of tumor derived EVs (exosomes) may represent a valuable addition to liquid biopsy in GBM and may enable predictions as to an impending TMZ resistance. While biologically based targeted therapies are under development, explorations of alternative dosing and sequencing of standard agents (radiation and chemotherapy) may be worthy of additional consideration.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
This project was supported by the Foundation Grant from the Canadian Institutes of Health Research, as well as grants MOP 11119 and 133424 and Canadian Cancer Society Research Institute, Innovation to Impact Award (all to J.R.). J.R. is the Jack Cole Chair in Paediatric Haematology/Oncology. P.D. was founded by Varian Medical Systems. Infrastructure support was provided by FRSQ (Fonds de la recherche en santé du Québec).
Conflict of interest statement. Authors declare no conflict of interest.
Supplementary Material
Acknowledgments
The authors are indebted to colleagues including Laura Montermini and Shilpa Chennakrishnaiah for helpful discussions and experimental assistance and to other members of our research team for ongoing support, and to our families for their patience and understanding.
References
- 1. Weller M, Wick W, Aldape K et al. Glioma. Nat Rev Dis Primers. 2015;1:15017. [DOI] [PubMed] [Google Scholar]
- 2. Stupp R, Mason WP, van den Bent MJ et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 3. Capdevila L, Cros S, Ramirez JL et al. Neoadjuvant cisplatin plus temozolomide versus standard treatment in patients with unresectable glioblastoma or anaplastic astrocytoma: a differential effect of MGMT methylation. J Neurooncol. 2014;117(1):77–84. [DOI] [PubMed] [Google Scholar]
- 4. Messaoudi K, Clavreul A, Lagarce F. Toward an effective strategy in glioblastoma treatment. Part I: resistance mechanisms and strategies to overcome resistance of glioblastoma to temozolomide. Drug Discov Today. 2015;20(7):899–905. [DOI] [PubMed] [Google Scholar]
- 5. Hegi ME, Diserens AC, Gorlia T et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 6. Chahal M, Abdulkarim B, Xu Y et al. O6-Methylguanine-DNA methyltransferase is a novel negative effector of invasion in glioblastoma multiforme. Mol Cancer Ther. 2012;11(11):2440–2450. [DOI] [PubMed] [Google Scholar]
- 7. Wick W, Weller M, van den Bent M et al. MGMT testing—the challenges for biomarker-based glioma treatment. Nat Rev Neurol. 2014;10(7):372–385. [DOI] [PubMed] [Google Scholar]
- 8. Shao H, Chung J, Lee K et al. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat Commun. 2015;6:6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Al-Nedawi K, Meehan B, Micallef J et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10(5):619–624. [DOI] [PubMed] [Google Scholar]
- 10. Zappulli V, Friis KP, Fitzpatrick Z, Maguire CA, Breakefield XO. Extracellular vesicles and intercellular communication within the nervous system. J Clin Invest. 2016;126(4):1198–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ricklefs F, Mineo M, Rooj AK et al. Extracellular vesicles from high-grade glioma exchange diverse pro-oncogenic signals that maintain intratumoral heterogeneity. Cancer Res. 2016;76(10):2876–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sottoriva A, Spiteri I, Piccirillo SG et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110(10):4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Snuderl M, Fazlollahi L, Le LP et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20(6):810–817. [DOI] [PubMed] [Google Scholar]
- 14. Suvà ML, Rheinbay E, Gillespie SM et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157(3):580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ceccarelli M, Barthel FP, Malta TM et al. ; TCGA Research Network. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Verhaak RG, Hoadley KA, Purdom E et al. ; Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Phillips HS, Kharbanda S, Chen R et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. [DOI] [PubMed] [Google Scholar]
- 18. Singh SK, Hawkins C, Clarke ID et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. [DOI] [PubMed] [Google Scholar]
- 19. Stiles CD, Rowitch DH. Glioma stem cells: a midterm exam. Neuron. 2008;58(6):832–846. [DOI] [PubMed] [Google Scholar]
- 20. Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29(12):1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Segerman A, Niklasson M, Haglund C et al. Clonal variation in drug and radiation response among glioma-initiating cells is linked to proneural-mesenchymal transition. Cell Rep. 2016;17(11):2994–3009. [DOI] [PubMed] [Google Scholar]
- 22. Hata AN, Niederst MJ, Archibald HL et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med. 2016;22(3):262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Campos B, Olsen LR, Urup T, Poulsen HS. A comprehensive profile of recurrent glioblastoma. Oncogene. 2016;35(45):5819–5825. [DOI] [PubMed] [Google Scholar]
- 24. Mao P, Joshi K, Li J et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci U S A. 2013;110(21):8644–8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakano I. Stem cell signature in glioblastoma: therapeutic development for a moving target. J Neurosurg. 2015;122(2):324–330. [DOI] [PubMed] [Google Scholar]
- 26. Cheng P, Wang J, Waghmare I et al. FOXD1-ALDH1A3 signaling is a determinant for the self-renewal and tumorigenicity of mesenchymal glioma stem cells. Cancer Res. 2016;76(24):7219–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Minata M, Gu C, Joshi K et al. Multi-kinase inhibitor C1 triggers mitotic catastrophe of glioma stem cells mainly through MELK kinase inhibition. PLoS One. 2014;9(4):e92546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim JK, Jin X, Sohn YW et al. Tumoral RANKL activates astrocytes that promote glioma cell invasion through cytokine signaling. Cancer Lett. 2014;353(2):194–200. [DOI] [PubMed] [Google Scholar]
- 29. Cheng P, Phillips E, Kim SH et al. Kinome-wide shRNA screen identifies the receptor tyrosine kinase AXL as a key regulator for mesenchymal glioblastoma stem-like cells. Stem Cell Reports. 2015;4(5):899–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Magnus N, Garnier D, Meehan B et al. Tissue factor expression provokes escape from tumor dormancy and leads to genomic alterations. Proc Natl Acad Sci U S A. 2014;111(9):3544–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garnier D, Magnus N, Lee TH et al. Cancer cells induced to express mesenchymal phenotype release exosome-like extracellular vesicles carrying tissue factor. J Biol Chem. 2012;287(52):43565–43572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sinha A, Ignatchenko V, Ignatchenko A, Mejia-Guerrero S, Kislinger T. In-depth proteomic analyses of ovarian cancer cell line exosomes reveals differential enrichment of functional categories compared to the NCI 60 proteome. Biochem Biophys Res Commun. 2014;445(4):694–701. [DOI] [PubMed] [Google Scholar]
- 33. Deshusses JM, Burgess JA, Scherl A et al. Exploitation of specific properties of trifluoroethanol for extraction and separation of membrane proteins. Proteomics. 2003;3(8):1418–1424. [DOI] [PubMed] [Google Scholar]
- 34. Thery C, Clayton A, Amigorena S, Raposo G. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;3:3.22. [DOI] [PubMed] [Google Scholar]
- 35. Montermini L, Meehan B, Garnier D et al. Inhibition of oncogenic epidermal growth factor receptor kinase triggers release of exosome-like extracellular vesicles and impacts their phosphoprotein and DNA content. J Biol Chem. 2015;290(40):24534–24546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang J, Cazzato E, Ladewig E et al. Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48(7):768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johnson BE, Mazor T, Hong C et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bao S, Wu Q, McLendon RE et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. [DOI] [PubMed] [Google Scholar]
- 39. Sullivan KE, Rojas K, Cerione RA, Nakano I, Wilson KF. The stem cell/cancer stem cell marker ALDH1A3 regulates the expression of the survival factor tissue transglutaminase, in mesenchymal glioma stem cells. Oncotarget. 2017;8(14):22325–22343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shenouda G, Souhami L, Petrecca K et al. A phase 2 trial of neoadjuvant temozolomide followed by hypofractionated accelerated radiation therapy with concurrent and adjuvant temozolomide for patients with glioblastoma. Int J Radiat Oncol Biol Phys. 2017;97(3):487–494. [DOI] [PubMed] [Google Scholar]
- 41. Nieder C, Andratschke NH, Grosu AL. Re-irradiation for recurrent primary brain tumors. Anticancer Res. 2016;36(10):4985–4995. [DOI] [PubMed] [Google Scholar]
- 42. Martin LM, Marples B, Coffey M et al. DNA mismatch repair and the DNA damage response to ionizing radiation: making sense of apparently conflicting data. Cancer Treat Rev. 2010;36(7):518–527. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.