Abstract
CRISPR technology has made genome editing widely accessible in model organisms and cells. However, conditional gene inactivation in diploid cells is still difficult to achieve. Here, we present CRISPR-FLIP, a strategy that provides an efficient, rapid, and scalable method for bi-allelic conditional gene knockouts in diploid or aneuploid cells such as pluripotent stem cells, 3D organoids and cell lines by co-delivery of CRISPR/Cas9 and a universal conditional intronic cassette.
Introduction
Analysing gene function is a crucial step in our understanding of normal physiology and disease pathogenesis. In cell-based models, loss-of-function studies require inactivation of both copies of the gene. Gene knockouts in cell lines were achieved by loss-of-heterozygosity1 or serial gene targeting approaches2. The development of site-specific nucleases has greatly facilitated functional studies in cells due to the fact that both copies of a gene can be efficiently inactivated in a single step3. Recently, the CRISPR/Cas9 gene editing technology4–7 has become the tool of choice for gene knockout studies due to its simplicity and robustness. Cas9 nuclease is an RNA-guided nuclease that is highly efficient in inducing a double-strand break (DSB) at a genomic site of interest. These DSBs can be repaired by error-prone non-homologous end joining (NHEJ) to generate gene-inactivating mutations or, in the presence of a donor template, the DSBs can be repaired by homology-directed repair (HDR) to generate more precise and complex alleles8. While simple constitutive knockouts are useful and informative, it is desirable to engineer conditional loss-of-function models, particularly for genes essential for cell viability or embryonic development. Here, we describe a simplified, one-step method for engineering conditional loss-of-function mutations in diploid cells.
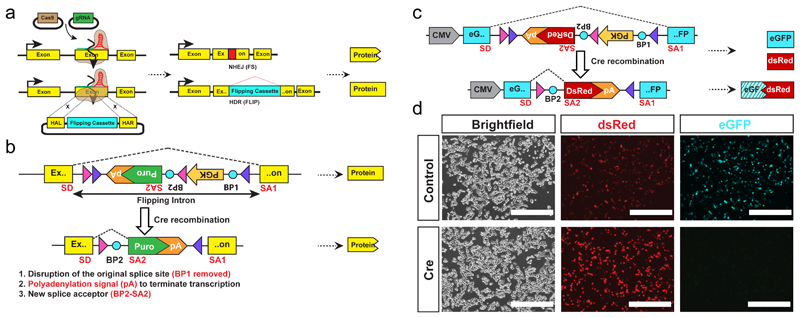
Existing methods for engineering conditional mutations in cultured cells9–12 rely on the inclusion of a drug selection cassette that must be removed in a second step to ensure proper expression of the targeted conditional allele (Supplementary Fig 1a,b). These methods were not designed for the generation of conditional loss-of-function models in a single step, particularly where the target gene is essential for cell growth or viability. To overcome these limitations, our strategy combines an invertible intronic cassette (FLIP), similar to COIN12, with high efficiency Cas9-assisted gene editing. Critically, the non-mutagenic orientation of the FLIP cassette expresses a puromycin resistance gene (puroR) allowing selection of correct nuclease-assisted targeting into the exon of one allele and simultaneous enrichment of cells that inactivate the second allele by nuclease-mediated NHEJ (Fig 1a). Upon exposure to Cre recombinase the FLIP cassette is inverted to a mutagenic configuration that activates a cryptic splice acceptor and polyadenylation signal (pA) and disrupts the initial splicing acceptor resulting in the complete loss of gene function (Fig. 1b and Supplementary Fig. 2a). In contrast to COIN which requires the removal of the drug selection cassette, our FLIP cassette permits the generation of conditional mutant cells in one step.
Figure 1. FLIP cassette strategy for bi-allelic conditional gene modification.
(a) Schematic drawing of the FLIP cassette strategy for bi-allelic conditional gene modification.
(b) The design of the FLIP cassette.
SD – splice donor, SA1, SA2 – splice acceptor, Purple triangles - LoxP1 sites Pink triangles – Lox5171 sites BP1, BP2 (blue circles) – branching point, pA - polyadenylation signal.
(c) Schematic of the FLIP cassette containing a DsRed reporter gene
(d) Images of HEK 293 cells transfected with the FLIP cassette. Both eGFP and DsRed proteins are expressed (top row). After Cre recombination the eGFP expression is disrupted, and only DsRed expression is maintained (bottom row). Scale bar 400 µm.
Initially we inserted a FLIP cassette variant containing a dsRed2 reporter in place of puroR into a CMV-eGFP (enhanced green fluorescent protein) expression plasmid (CMV-eGFP[FLIP], Fig. 1c). Following transient transfection of HEK293 cells with CMV-eGFP[FLIP], both green and red fluorescence was observed, demonstrating that insertion of the FLIP cassette in the non-mutagenic orientation is inert (Fig. 1d). This was further confirmed by flow cytometry analysis showing similar level of eGFP expression from both CMV-eGFP and CMF-eGFP[FLIP] (Supplementary Fig. 3). The Cre recombined CMV-eGFP[FLIP] showed loss of eGFP expression, suggesting the inactivation of eGFP expression in the inverted, mutagenic orientation of FLIP cassette (Fig. 1c,d).
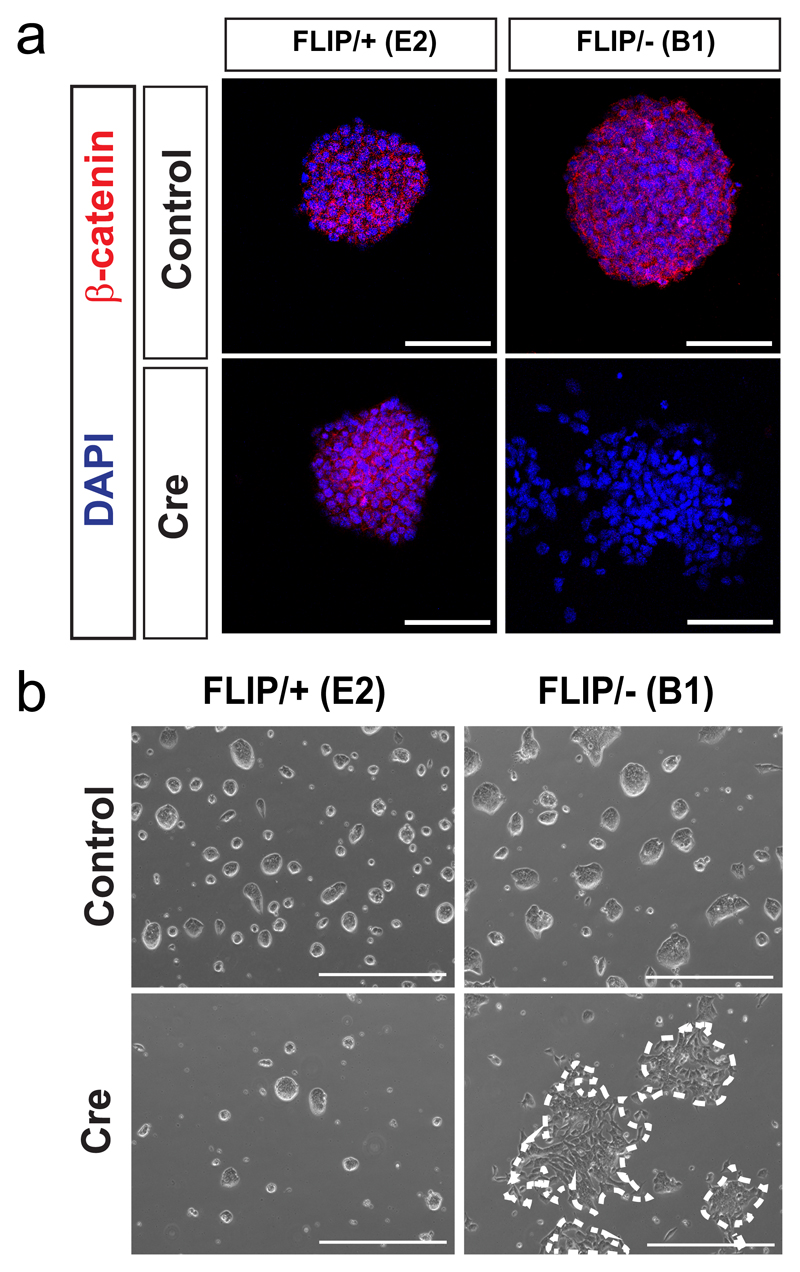
Next, we employed CRISPR/Cas9 endonuclease in mouse embryonic stem cells (mESCs) to introduce the puroR FLIP cassette into one allele of β-catenin (Ctnnb1) via HDR and to simultaneously induce a frameshift mutation by NHEJ in the second β-catenin allele (Fig. 1a, Supplementary Fig. 4a). β-catenin is an important gene for the morphology and efficient self-renewal of mESCs13,14. A donor vector containing the puroR FLIP cassette flanked by ~1 kb homology arms was inserted in exon 5 of β-catenin by co-transfection of mESCs with Cas9 and gRNA expression plasmids. Following selection in puromycin, drug-resistant colonies were genotyped by PCR to confirm correct integration of the FLIP cassette and then assayed for NHEJ events in the second allele by Sanger sequencing (Supplementary Fig. 2b, 4b, c). From 64 clones, 14 clones (21.9%) were correctly targeted, among which 4 clones carried a frame-shift mutation in the second allele (Supplementary table 1). The recovery of β-catenin compound mutant clones (FLIP targeted/NHEJ frameshift; FLIP/-) with wildtype morphology strongly suggests that the insertion of the FLIP cassette does not disrupt the function of β-catenin in the non-mutagenic orientation. Upon expression of Cre recombinase in Ctnnb1FLIP/- clones, we observed a loss of β-catenin expression in cells (Fig. 2a, Supplementary Fig. 4d). Moreover, compared to control (Ctnnb1FLIP/+) cells treated with Cre recombinase, the Ctnnb1FLIP/- cells became scattered and lost their dome-like morphology (Fig 2b). In addition, we performed quantitative RT-PCR analysis to determine the splicing efficiency of the FLIP intron in comparison to the neighbouring intron 7 of β-catenin. Our data demonstrate highly efficient splicing of the FLIP intron. Thus the FLIP cassette is inert to gene activity in the non-mutagenic orientation (Supplementary Fig. 5 and Supplementary table 2).
Figure 2. Insertion of the FLIP cassette in the endogenous Ctnnb1 gene of mouse embryonic stem cells.
(a) Immunofluorescence of β-catenin before and after Cre transfection.
(b) Representative bright field images of the ESC clones before (top) and after (bottom) Cre transfection. Scale bar 400μm.
We additionally targeted Apc, Esrrb, Nfx1, Sox2, Tcf7l2, Trim13, and Trim37 in mESCs; ARID1A and TP53 in human HEK293 cells; and TP53 in human induced pluripotent stem cells (Supplementary Fig. 6-9). The FLIP intron targeting efficiency ranged from 19.8% to 40.6% in mESCs (Supplementary table 1, please note that non-targeted clones are a result of random integration of the puro cassette). Importantly, for all genes, FLIP/- clones were obtained (Supplementary table 1, Supplementary Fig. 6-9). To induce gene knockout, a Cre expressing plasmid was transfected to ES clones with an average transfection efficiency higher than 95% (Supplementary Fig. 10) and conditional inactivation of gene expression was confirmed by Western blot and immunofluorescence for Esrrb, Sox2, Trim13, and Trim37 (Supplementary Fig. 6d,6h,6i, 7m,7q).
We further modified our FLIP intronic cassette to generate a reversible conditional allele. The region containing the cryptic splice acceptor and pA is flanked by two FRT sites (Supplementary Fig. 11a, FLIP-Flp Excision (FLIP-FlpE)). When inserted into eGFP, the intronic FLIP-FlpE cassette permits the expression of eGFP like the original FLIP cassette (Supplementary Fig. 3, 11b). Upon Cre recombination the FLIP-FlpE cassette turns into the mutagenic orientation, which blocks the eGFP expression. Next, the added FRT sites enables the mutagenic FLIP-FlpE cassette to be excised by Flp recombinase, thus allowing the revival of eGFP expression (Supplementary Fig. 11a,b). The FLIP-FlpE cassette was inserted in the 5th exon of the mouse β-catenin allele. The Ctnnb1FLIP-FlpE/ FLIP-FlpE (FLIP-FlpE homozygote) mutant clone went through a series of recombination, first by Cre and then Flp. At each step, the mutant showed wildtype, mutant (after Cre), and again wildtype (after Cre and Flp) morphology, respectively (Supplementary Fig. 11e). Accordingly, we observed loss and gain of β-catenin expression (Supplementary Fig. 11f, g), suggesting that with a simple modification the FLIP intronic cassette can also be used for ‘switchable’ gene expression.
To extend our application, we inserted the FLIP-FlpE cassette into the 16th exon of the mouse Apc allele in intestinal organoids expressing CreERT2 under the Villin promoter (Supplementary Fig. 12a). Apc is a component of the destruction complex acting in the Wnt pathway and its deletion causes hyperactive Wnt signalling and makes organoids adopt a cystic morphology15. ApcFLIP-FlpE/- clones (Supplementary Fig 12b, c) initially showed budding morphology when cultured in standard ENR (Egf, Noggin, Rspondin) media. Upon treatment with 4-hydroxytamoxifen (4-OHT), for Cre activation, the organoids adopt a cystic morphology due to the loss of Apc (Supplementary Fig. 12d). In addition to the application of CRISPR-FLIP to intestinal organoids, FLIP-targeted ES clones can be used to generate other cell types e.g. mouse embryonic fibroblast (MEF) (Supplementary Fig. 13).
Discussion
Our strategy requires the presence of a CRISPR site overlapping or nearby the insertion site of the FLIP cassette, imposing constraints on the exons than can be targeted. To maximize the potential for a null mutation, the target exon must be common to all transcripts and lie within the first 50% of the protein-coding sequence. Additionally, based on the minimum size of mammalian exons (50 bp)16, we set the size of the split exons to be at least 60 bp. Finally, for optimal splicing, we chose insertion points that match the consensus sequence for mammalian splice junctions (minimally MAGR (A/CAG/Pu))17. Using this set of rules, we used bioinformatics to estimate the number of suitable FLIP insertion sites in the protein-coding genes in the mouse and human genomes. Our bioinformatics analysis revealed 1,171,712 FLIP insertion sites and corresponding gRNA binding sites covering 16,460 genes in the mouse genome and 1,171,787 FLIP insertion sites and corresponding gRNA binding sties covering 15,177 genes in the human genome. (Supplementary table 3,4). Although haploinsufficient genes impose a limitation to our strategy, as one allele is already null in FLIP/- clones, the generation of FLIP/FLIP clones provide an option for haploinsufficient genes.
Recently developed methods used to achieve higher HDR-mediated targeting efficiency are likely to further increase the efficiency of our CRISPR-FLIP method18. The FLIP targeting vectors only require short homologous arms (less than 1 kb) which makes the assembly of targeting vectors easy and scalable. The FLIP cassette is invariable and can be generically applied to any gene, including non-coding RNA genes. The CRISPR-FLIP technology is widely applicable to many diploid and aneuploid cell types including mESCs, fibroblasts, 3D organoids, hiPSCs, and cell lines (e.g. 293 cells).
Online methods
dsRed FLIP cassette inserted in the eGFP cDNA
The FLIP cassette inserted in the middle of eGFP and containing a dsRed2 reporter gene was synthesized and ordered from GenScript. The split eGFP cDNA and the FLIP cassette were cloned into the mammalian expression vector pCDNA4TO (Invitrogen) using BamHI (R0136S, NEB) and XhoI (R0146S, NEB) for pre-recombined form. The vector was subsequently transformed into Cre expressing bacteria (A111, Gene bridges) to generate the Cre-recombined form. Correct clones were confirmed with restriction digest BamHI (R0136S, NEB) and XhoI (R0146S, NEB) and Sanger sequencing. The FLIP-FlpE cassette was also synthesized and inserted into the same site of the eGFP expression vector.
FLIP cassette containing selection marker genes
The FLIP cassette was PCR amplified and cloned into Pjet1.2 vector (ThermoFisher Scientific, K131). Replacement of dsRed was done through restriction digest excision using EcoRI (R3101S, NEB) and Acc65l (RO599S, NEB) followed by insertion of PCR amplified selection marker genes using Infusion cloning (638909, Clontech). The FLIP cassette including selection marker gene was then transferred to the vector pUC118 (3318, Clontech) using the restriction enzymes SacI (R0156S, NEB) and PstI (R0140S, NEB) and Mighty cloning (6027, Takara).
Addition of homologous arms to the FLIP cassette – FLIP targeting vector generation
Homologous arms around an intron insertion site were amplified by high fidelity Phusion DNA polymerase (M0530S, NEB). After PCR product purification, both homologous arms and FLIP cassette-containing vector were mixed with the type II restriction enzyme SapI and T4 DNA ligase (M0202T, NEB). After 25 cycles of 37°C and 16°C, the reaction mixture was directly used for E.Coli transformation. DNA was extracted (27106, Qiagen) and analysed with restriction digest to identify correctly assembled FLIP donor vectors.
Cas9 and gRNA plasmids
Human codon optimized Cas9 (41815, Addgene) and empty gRNA vector (41824, Addgene) were obtained from Addgene.
Cell culture
Human embryonic kidney (HEK) 293 cells
Human embryonic kidney 293 cells were cultured in media consisting of DMEM, high glucose (11965092, Thermofisher Scientific) supplemented with 10% foetal bovine serum (Thermofisher Scientific), 1x penicillin-streptomycin according to the manufacturer’s recommendation (P0781, Sigma). The cells were tested negative for mycoplasma.
Embryonic stem cells (ESCs)
Murine E14 Tg2a embryonic stem (mES) cells were cultured feeder-free on 0.1% gelatin-coated dishes in serum+LIF+2i (Chiron and PD03) composed of GMEM (G5154, Sigma), 10% foetal bovine serum (Gibco), 1x non-essential amino acids according to the manufacturer’s recommendation (11140, Thermofisher Scientific), 1 mM sodium pyruvate (113-24-6, Sigma), 2 mM L-glutamine (25030081, Thermofisher Scientific), 1x penicillin-streptomycin according to the manufacturer’s recommendation (P0781, Sigma) and 0.1 mM 2-mercaptoethanol (M7522, Sigma), 20 ng/ml murine LIF (Hyvonen lab, Cambridge), 3 μM CHIR99021 and 1 μM PD0325901 (Stewart lab, Dresden). BOBSC19 human induced pluripotent stem (hiPS) cells were cultured feeder-free on dishes coated with Synthemax II (3535, Corning) in TeSR-E8 media (05940, Stem Cell Technologies). ESCs were kept in a tissue culture incubator at 37°C and 5% CO2. Cells were split in a 1:10 – 1:15 ratio every 3-4 days depending on confluence. All cells were tested negative for mycoplasma.
Intestinal organoid culture
Mouse small intestinal organoids were cultured as previously described20.
Cell electroporation
For targeting of mESCs 1x106 cells were collected and resuspended in magnesium and calcium free phosphate buffered saline (D8537, Sigma). A total of 50 µg of DNA consisting of the targeting vector, Cas9 and gRNA in a 1:1:1 ratio were added to the cells and then transferred to a 4 mm electroporation cuvette (Biorad). Electroporation was performed using the Biorad Gene Pulser XCell’s (165-2660, Biorad) exponential program and the following settings: 240 V, 500 uF, unlimited resistance. For targeting of human iPS cells, 2x106 cells were dissociated with Accutase (SCR005, Millipore) and resuspended in nucleofection buffer (Solution 2, LONZA). A total of 12 µg of DNA consisting of 4 µg Cas9 plasmid, 4 µg of each gRNA plasmid and 4 µg of targeting vector was added to the cells and transferred to a 100 µl nucleofection cuvette (LONZA). Nuclefection was performed with the AMAXA Human Nucleofector Kit 2 (LONZA Cat # VPH-5022) using the B-016 program. The cells were plated and cultured for 1 day in TeSR-E8 media (05940, STEM CELL technologies) containing ROCK inhibitor (Y-27632, Stem Cell Technologies) to promote survival of transfected cells. For targeting of HEK293 cells, the cells were cultured until they reached 50-60% confluence. A total of 8 µg of DNA consisting of targeting vector, Cas9 and gRNA in a 1:1:1 ratio was transfected using Lipofectamine-2000 (11668019, Invitrogen) according to the manufacturer’s instructions.
Plasmid transfection
1 µg of pCAGGS-Cre-IRES-Puro and/or pCAGGS-Flp-IRES-Puro plasmid vector and 3 µl of Lipofectamine-2000 (11668019, Invitrogen) were mixed according to the manufacturer’s protocol, applied to 2x106 recently seeded (less than 30 min) cells/ 6-well and incubated overnight. Media was refreshed the following morning. Ctnnb1FLIP/+ and Ctnnb1FLIP/- with CreERT2 clones were generated by transfecting 0.66 μg of PiggyBac CreERT2 expressing plasmid (with hygromycin 50 μg/ml) together with 0.33 μg of transposase using Lipofectamine-2000 as described above. Cre-recombinase was activated by adding 4-OHT with a final concentration of 1 μM for 48h.
Western blot
Following transfection ESCs were cultured for 2-5 days and then lysed in buffer containing complete protease-inhibitor cocktail tablets (11697498001, Roche) and centrifuged at 13,000 rpm for 15 min at 4°C. Protein concentration was measured with Bradford assay (5000204, Biorad) and equal amounts were loaded on a 10% acrylamide gel and run at 120 V for 1.5-2hrs. The proteins were subsequently transferred to an Immobilon-FL PVDF 0.45 µm membrane (IPFL00010, Millipore) at 90 V for 1hr 15 min. The following primary antibodies and dilutions were used to detect the indicated proteins: Rabbit monoclonal antibody against β-Catenin (1:1000, 8480S, Cell Signaling), mouse monoclonal against alpha Tubulin antibody (1:5000, ab7291, Abcam), mouse monoclonal antibody against beta-actin (1:5000, ab8226, Abcam), mouse monoclonal antibody against Esrrb, (1:1000, PP-H6705-00, Bio-Techne), rat monoclonal antibody against Sox2, (1:500, 14-9811-80, eBioscience), mouse monoclonal antibody against Trim 13 (1:500, sc-398129, Santa Cruz), mouse monoclonal antibody against Trim37 (1:500, sc-514828, Santa Cruz) and rabbit monoclonal against Vinculin (1:3000, ab19002, Abcam). The membrane was washed and the indicated horseradish-peroxidase conjugated secondary antibodies were applied: horse anti-mouse IgG (1:5000, Cell Signaling) and goat anti-rabbit (1:5000, Cell Signaling) and goat anti-rat HRP conjugated (1:5000, SC2032, Santa Cruz). Detection was achieved using ECL prime Western blotting Detection system (RPN2133, GE Healthcare). All original blot images are shown (Supplementary Fig. 14).
Immunofluorescence
Cells were cultured in Ibid tissue culture dishes (IB-81156, Ibid) coated with 0.1% gelatin, washed twice with calcium and magnesium free PBS and fixed in 4% PFA for 20 min at RT. The cells were permeabilised in 0.5% Triton X-100 (T8787, Sigma) in PBS for 15 min at RT. Subsequently, blocking was performed in 5% donkey serum (D9663, Sigma) and 0.1% Triton X-100 for 1hr at RT. The following primary antibodies in blocking buffer were applied for the indicated protein: Sox2, (1:500, 14-9811-80, eBioscience) and β-Catenin (1:1000, 4627, Cell Signaling). Primary antibodies were incubated overnight at 4°C. Subsequently excess primary antibody was washed away and anti-rat Alexa Flour 594® conjugated antibody (1:1000, A21209, Abcam) was added for Sox2, and incubated for 1h at RT. Excess secondary antibody was washed away and DAPI (1:1000, D9542, Sigma) was added and incubated for 10 min at RT. Cells were washed and mounted in RapiClear (RCCS002, Sunjin lab).
Chimeric embryo generation and ESC-derived fibroblast establishment
Sox2FLIP/FLIP mESCs transfected with pPyCAG-eGFP-IRES-Zeo plasmid were aggregated. Chimerae were generated by standard aggregation using F1 embryos and transplanted into pseudopregnant recipient mice of C57BL/6J strain. E13.5 embryos were beheaded and dissected to remove all organs, including genital ridges. The remaining body was cut into small pieces, trypsinised, and plated on gelatin in serum+LIF media containing selecting reagents. GFP expression confirms that the MEFs are derived from the Sox2FLIP/FLIP mESCs. All animal work was performed in accordance with Home Office guidelines and regulations at the University of Cambridge, UK.
Quantitative RT-PCR
Total RNA was extracted using RNwasy Mini kit (74104, Qiagen) with an on-column DNase digestion (79254, Oiagen). Reverse transcription was performed using 250 ng of RNA using M-MLV Reverse Transcriptase (M1701, Promega). Quantitative PCR reactions were performed in triplicates using iQ SYBR Green Supermix according to the manufacturer’s protocol (1708882, Biorad) with the primers in Supplementary table 2 and the StepOnePlus Real Time PCR System (Applied Biosystems). Average gene expression was normalized to exon5 and error bars represent ± standard deviation.
Flow cytometry analysis
HEK 293 cells were co-transfected with eGFP, eGFP[FLIP] or eGFP[FLIPFlpE] and a BFP reporter as described previously, and harvested 24h post transfection. mESCs were transfected with eGFP as describes above and harvested 48h post transfection. Cells were analysed using BD LSRFortessa (BD Biosciences) and Flowjo software.
Supplementary Material
Editorial summary.
The combination of knocking one allele out with CRISPR-mediated NHEJ and targeting the other with a conditionally inactivating cassette allows rapid generation of conditional alleles.
Acknowledgements
pPyCAG-eGFP-IRES-Zeo plasmid was kindly provided by Austin Smith (WT-MRC Cambridge Stem Cell Institute, UCAM) and pCAGGS-Cre-IRES-Puro and pCAGGS-Flp-IRES-Puro plasmid vectors were kindly provided by B. Hendrich (WT-MRC Cambridge Stem Cell Institute, UCAM). We thank Masaki Kinoshita for advice regarding antibodies. A.A-R. and K.T. are supported by the Medical Research Council, A.M.is supported by Wntsapp, Marie Curie ITN, J.F. and J.C.R.S. are supported by the Wellcome Trust. W.C.S received core grant support from the Wellcome Trust to the Wellcome Trust Sanger Institute. B-K.K. and R.M. are supported by a Sir Henry Dale Fellowship from the Wellcome Trust and the Royal Society [101241/Z/13/Z] and receives core support grant from the Wellcome Trust and MRC to the WT - MRC Cambridge Stem Cell Institute.
Footnotes
Data availability
Mammalian expression plasmids are available at Addgene. pUC118-FLIP-Puro (#84538 for generation of conditional knockouts), pUC118-FLIP-FlpE-Puro (#84539 for generation of reversible conditional knockouts), pUC118-mCtnnb1-FLIP-Puro (#84540, FLIP vector for conditional knockout of Ctnnb1), pUC118-mCtnnb1-FLIP-FlpE-Puro (#84541, FLIP-FlpE vector for reversible conditional knockout of Ctnnb1) and gRNA-mCtnnb1 (#84542).
Author contributions
A.A-R., WC.S. and B-K.K. wrote the manuscript. A.A-R, J.F, WC.S. and B-K.K. designed the FLIP cassette targeting vector. A.A-R., WC.S. and B-K.K. designed and discussed the experiments. A.A-R., R.M., and J.K. targeted mESCs and performed WB. A.A.R. performed IF. K.A. and A.M. targeted hiPSCs. A.M. targeted HEK 293 cells. A.A.R. and A.M. performed FACS. A.A.R. performed the organoid experiments. S.P. and T.G. performed the bioinformatics analysis. K.T. derived Sox2FLIP/FLIP MEFs. J.C.R. S supervised K.T. WC.S. and B-K.K. supervised the project.
Competing financial interest
The authors declare no financial interest.
References
- 1.Mortensen RM, Conner DA, Chao S, Geisterfer-Lowrance AA, Seidman JG. Production of homozygous mutant ES cells with a single targeting construct. Mol Cell Biol. 1992;12:2391–5. doi: 10.1128/mcb.12.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.te Riele H, Maandag ER, Clarke A, Hooper M, Berns A. Consecutive inactivation of both alleles of the pim-1 proto-oncogene by homologous recombination in embryonic stem cells. Nature. 1990;348:649–51. doi: 10.1038/348649a0. [DOI] [PubMed] [Google Scholar]
- 3.Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–51. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 4.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mali P, et al. RNA-guided human genome engineering via Cas9. Science (80-. ) 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho SW, Kim S, Kim JM, Kim J-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–2. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 7.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–55. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–6. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 10.Abuin A, Bradley A. Recycling selectable markers in mouse embryonic stem cells. Mol Cell Biol. 1996;16:1851–6. doi: 10.1128/mcb.16.4.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tate PH, Skarnes WC. Bi-allelic gene targeting in mouse embryonic stem cells. Methods. 2011;53:331–8. doi: 10.1016/j.ymeth.2010.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Economides AN, et al. Conditionals by inversion provide a universal method for the generation of conditional alleles. Proc Natl Acad Sci U S A. 2013;110:E3179–88. doi: 10.1073/pnas.1217812110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anton R, Kestler HA, Kühl M. Beta-catenin signaling contributes to stemness and regulates early differentiation in murine embryonic stem cells. FEBS Lett. 2007;581:5247–54. doi: 10.1016/j.febslet.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Lyashenko N, et al. Differential requirement for the dual functions of β-catenin in embryonic stem cell self-renewal and germ layer formation. Nat Cell Biol. 2011;13:753–61. doi: 10.1038/ncb2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato T, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–72. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 16.Dominski Z, Kole R. Selection of splice sites in pre-mRNAs with short internal exons. Mol Cell Biol. 1991;11:6075–83. doi: 10.1128/mcb.11.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burset M, Seledtsov IA, Solovyev VV. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 2000;28:4364–75. doi: 10.1093/nar/28.21.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chu VT, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 19.Yusa K, et al. Targeted gene correction of α1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478:391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.