Abstract
Because protein-protein interactions underpin most biological processes, developing tools that target them to understand their function or to inform the development of therapeutics, is an important task. SUMOylation is the posttranslational covalent attachment of proteins in the SUMO family (SUMO1, SUMO2, or SUMO3) and regulates numerous cellular pathways. SUMOylated proteins are recognized by proteins with SUMO-interaction motifs (SIMs) that facilitate non-covalent interactions with SUMO. Here, we describe the use of the Affimer system of peptide display for rapid isolation of synthetic binding proteins that inhibit SUMO-dependent protein-protein interactions mediated by SIMs both in vitro and in cells. Crucially, these synthetic proteins did not prevent SUMO conjugation either in vitro or in cell-based systems, enabling the specific analysis of SUMO-mediated protein-protein interactions. Furthermore, through structural analysis and molecular modelling, we explored the molecular mechanisms that may underlie their specificity in interfering with either SUMO1-mediated interactions or interactions mediated by either SUMO2 or SUMO3. Not only will these reagents enable investigation of the biological roles of SUMOylation, the Affimer technology used to generate these synthetic binding proteins could be exploited to design or validate reagents or therapeutics that target other protein-protein interactions.
Introduction
Protein-protein interactions (PPIs) underpin the majority of cellular processes (1), yet methods to target intracellular PPIs are not well established. Most common methods to identify PPIs rely on mutations or chemical inhibitors to interfere with PPIs. Mutational screens rely on changing key amino acids and determining the ability of the protein to maintain function (2), but this can result in truncated or misfolded proteins. Chemical inhibitors are small molecules that are isolated from large libraries or designed in silico to bind to particular pockets in proteins (2). However, most bioactive molecules inhibit enzymes rather than prevent PPIs. Both methods can be complicated and expensive, restricting their use and functionality. In addition, although antibodies can block PPIs, their use as PPI inhibitors is restricted to extracellular events. Therefore, rapid and robust methods for inhibiting PPIs represent a challenge of immense importance.
The development of novel molecular recognition reagents has shown promise in inhibiting PPIs (3–7). A major advantage of these systems is the ability to quickly isolate high-affinity reagents (3, 8) that tend to interact with protein-binding hotspots and block function without affecting protein abundance. This provides the potential to study the function of a discrete domain or domains of a protein without perturbing additional functions. Because proteins can act as hubs and interact with many other proteins (9), the ability to block specific individual interactions may shed light on previously unknown biological processes. Furthermore, the capacity to generate reagents to specifically block related, highly similar protein isoforms is also necessary to reveal isoform-specific functions.
Many cellular signaling processes facilitate PPIs. Examples include the recognition of a phosphorylated residue within a specific sequence motif on a protein or the recognition of protein-mediated posttranslational modifications (PTMs), such as ubiquitylation or SUMOylation. For ubiquitin-mediated interactions or interactions mediated by the related protein SUMO (small ubiquitin-related modifier), developing inhibitors that target PPIs mediated by these proteins is particularly challenging, because the PPI inhibitors must not prevent the enzymatic transfer of these modifiers onto their targets, a process that also requires the recognition of ubiquitin or SUMO (10). SUMOylation is a PTM crucial for numerous cellular processes (11–15). The covalent attachment of SUMO to its substrates is analogous to ubiquitin conjugation, involving a proteolytic processing step to reveal a C-terminal di-glycine motif on SUMO, an E1 activating enzyme, an E2 conjugating enzyme, and the formation of an isopeptide bond between the C-terminal glycine of SUMO and a lysine residue of the target substrate. The SUMO-loaded E2 may also interact with an E3 enzyme [for example, the protein inhibitor of activated STAT, (PIAS) family], although the role of SUMO E3 proteins might not be essential (10). In mammals, there are four protein isoforms of SUMO: SUMO-1, SUMO-2, SUMO-3, and SUMO-4 (although the physiological role of SUMO-4 is poorly characterized). After processing, SUMO-2 and SUMO-3 differ from one another by only three amino acids, and both are ˜47% identical to SUMO-1. Because of the similarity between SUMO-2 and -3, we refer to them as SUMO-2/3 when specificity is not clear and refer to the specific isoform when it is known. SUMO-2/3 can polySUMOylate their substrates, whereas SUMO-1 can monoSUMOylate or function as a terminator of polySUMO-2/3 chains. Despite these differences and the divergent physiological roles these modifications play, the enzyme cascade that attaches each isoform to substrates is identical; hence targeting this enzyme cascade will not discriminate between the different functions of SUMO isoforms.
Proteins that interact noncovalently with SUMO contain SUMO-interacting motifs (SIMs) (16). Examples of SIMs include those found in the promyelocytic leukaemia protein (PML), Daxx, and Sp100, all of which are proteins found in subnuclear structures known as PML nuclear bodies (PML-NBs). PML-NBs are hubs for SUMOylated proteins, and the importance of both SUMOylation and SIMs for PML-NB formation is well documented, giving rise to a model of intermolecular SUMO:SIM-dependent interactions (17–19).
To ask if we could generate PPI inhibitors able to target specific protein isoforms or discriminate between closely-related members of a protein family, we used the Affimer system (a protein aptamer technology previously known as Adhiron) to develop novel artificial binding proteins that bind to SUMO (8). The Affimer library consists of a scaffold with two variable loops that each present a short stretch of randomized amino acids from a phage display library (8, 20). Affimers are artificial protein aptamers that are evolved using phage display to generate binding reagents with high affinity and specificity for their binding partners, similar to those obtained by Monobodies, Affibodies and DARPins (reviewed in (21)). The SUMO-specific Affimers (S-Affs) we developed enabled the investigation of SUMO-dependent PPIs in vitro and in cultured cells. Through structure-guided studies, we developed a molecular understanding of the isoform specificity of S-Affs that recognize SUMO1 and those that recognize SUMO2/3 or those that recognized all tested members of the SUMO family nonselectively. Not only will these reagents enable us to dissect biological roles of the SUMO pathway, they pave the way for the study of signal transduction pathways involving the conjugation of proteins, such as ubiquitin and other ubiquitin-like proteins, and potentially inform the design of novel PPI-targeted therapeutics.
Results
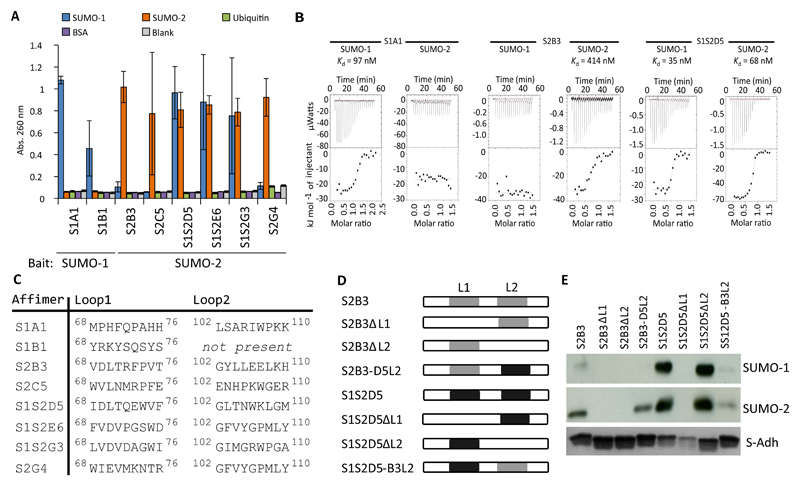
Isoform-specific SUMO Affimers contain consensus SIM domains
Affimers are thermostable small protein aptamers displaying nine randomized amino acids in each of two peptide presentation loops. The Affimer protein (previously known as Adhirons, Fig. S1A; PDB: 4N6T) is presented as a large (> 1010) phage display library (8). Using this technology in separate screens, we identified SUMO-specific Affimers (S-Affs) using either human SUMO-1 or SUMO-2 as bait (Fig. S1B-C). In the screen using SUMO-1 as bait, we isolated 30 S-Affs that demonstrated isoform-specificity by phage enzyme-linked immunosorbant assay (ELISA) (Fig. S2A). Of twelve sequenced, we identified 2 unique clones. The parallel SUMO-2 screen identified several S-Affs; however, none demonstrated isoform specificity. Therefore, we adapted the screening protocol to incorporate a negative selection step that would reject SUMO-1–specific S-Affs (see Materials and Methods). From the 48 clones identified with this updated protocol, we found 44 that bound SUMO-2 (92%) and, of these, only 8 (18%) bound SUMO-2 but not SUMO-1 (Fig. S2B). We sequenced all 8 of the SUMO-2–specific clones and identified 7 unique sequences. We selected two SUMO-1–specific S-Affs (S1A1 and S1B1 from the SUMO-1 screen), three SUMO-2_specific S-Affs (S2B3, S2G4, and S2C5 from the SUMO-2 screen), and three S-Affs that bound both SUMO isoforms (S2S1D5, S1S2E6, and S2S1G3 from the SUMO-2 screen) for purification and testing by ELISA (Fig. 1A), which confirmed the results obtained by phage display. The nomenclature of these S-Affs reflects their observed specificity. For example, S1S2D5 refers to clone D5 that bound to both SUMO-1 and SUMO-2). In addition we confirmed that the S-Affs were specific for SUMO in that they did not bind to ubiquitin or to bovine serum albumin (Fig. 1A). We also measured the binding affinities of one S-Aff from each group (SUMO-1–specific, SUMO-2–specific, and SUMO-1/2 nonspecific) with both SUMO-1 and SUMO-2 to quantify specificity by isothermal titration calorimetry (ITC) (Fig. 1B). These biochemical assays confirmed that S1A1 bound SUMO-1 (Kd 97 nM), S2B3 bound SUMO-2 (Kd 414 nM), and S1S2D5 bound both SUMO-1 and SUMO-2 (Kd 35 nM and 68 nM, respectively). We estimated the affinities of S1A1 for SUMO-2 and S2B3 for SUMO-1 to be >100-fold lower than for their cognate targets (with Kd >10 – 40 µM), because the ITC titrations showed no evidence of even low-affinity binding. Given that SUMO-2 and SUMO-3 are so similar, we anticipate that the S-Affs that bind SUMO-2 will also bind SUMO-3, although this was not explicitly tested.
Fig. 1. Identification of isoform-specific S-Affs.
(A) ELISAs in which the indicated His-tagged S-Affs were purified and incubated in ELISA plates that had been coated with SUMO-1, SUMO-2, ubiquitin, or bovine serum albumin (BSA). Control samples (blank) omitted S-Affs and coating protein. Data from 3 independent experiments was combined, and error bars indicate the standard error. Abs., absorbance. (B) Isothermal titration calorimetry analysis of the indicated S-Affs with SUMO-1 and SUMO-2. Data are representative of at least 3 experiments. (C) The amino acid sequences of the variable loop regions of the indicated S-Aff clones. The numbers represent the position of residues in the S-Aff proteins. DNA sequencing revealed that loop 2 of S1B1 was absent. (D) Schematic of S2B3 and S1S2D5 chimeras and deletion mutants. Sequences containing either loop 1 (L1) or loop 2 (L2) were deleted or swapped between S-Affs. (E) In vitro binding assay of S-Aff chimeras or deletion mutants with the indicated SUMO isoforms. The S-Affs were bound to Ni-NTA agarose and incubated with GST-SUMO-1 or GST-SUMO-2. Interacting complexes were purified and probed with antibodies against GST. Data are representative of 2 experiments.
SUMO interacts with SIMs through a binding surface located on its second 13-strand (132; Fig. S1B-C)) (16). SIM domains consist of hydrophobic residues (Ile, Val, Leu), often encompassing and surrounded by acidic residues (22). SIMs can interact with the 132 of SUMO in either a parallel or antiparallel fashion, with parallel-binding SIMs consisting of an (I/V)DLT motif that exhibits preferential binding to SUMO-2/3 (23). S2B3 and S1S2D5, which we identified with the negative selection screen, contained the (I/V)DLT motif (Fig. 1C). The other clones that we analyzed from this screen also contained partial consensus SIMs that appeared shifted C-terminally within loop 1 (for example, [I/V][D/E]V in S1S2E6, S1S2G3, and S2G4). In contrast, the SUMO-1–specific Affimers S1A1 and S1B1 lacked an obvious SIM consensus domain. Loop 2 sequences were more variable, with the exception of position +1, where Gly was overrepresented in 5/7 clones (Fig. 1C), suggesting that loop 2 residues may participate in binding and that bulkier amino acids at the +1 position may preclude this interaction or that Gly provides flexibility that favors binding. Interestingly, despite being independent clones and possessing different loop 1 residues, loop 2 of S1S2E6 and S2G4 are identical, providing additional support for a role of loop 2 in the interaction (Fig. 1C). Unexpectedly, sequencing revealed that loop 2 of S1B1 was not present (Fig. 1C); we are aware that single loop Affimers are present in the initial library owing to the cloning procedure during library generation.
Sequence analysis did not provide an obvious explanation for isoform specificity. Therefore, we created S-Affs with deleted loops and chimeras in which loop 2 sequences were swapped between S2B3 (SUMO-2–specific) and S1S205 (binds both isoforms) (Fig. 10). In vitro pulldown experiments showed that S2B3 preferentially interacted with SUMO-2 with very little interaction with SUMO-1, and S1S205 interacted with both isoforms (Fig. 1E). The increased apparent interaction between S1S205 and SUMO compared with that of S2B3 may reflect their respective affinities (Fig. 1B). Both loops were required for S2B3 binding to SUMO-2, as demonstrated by the inability of forms lacking loop1 (S2B3ΔL1) or loop2 (S2B3ΔL2) to interact with SUMO-2. Replacing loop 2 in S2B3 with that of S1S205 (S2B3-05L2) enabled SUMO-2 binding but not SUMO-1 binding (Fig. 1E). Thus, S2B3 required two loops for binding, but loop 2 from an S-Aff that bound nonspecifically to SUMO-1 and SUMO-2 (S1S205) could support SUMO-2–specific binding. In contrast, loop 2 of S1S205 was not required for S1S205 to bind to either SUMO-1 or SUMO-2 (Fig. 1E). However, when loop 2 of S2B3 was combined with loop 1 of S1S205 (S1S205-B3L2), binding to either SUMO was reduced, with a greater apparent reduction in the binding to SUMO-1. Hence, discreet properties in both loops of S2B3 contributed to isoform specificity.
S-Aff interactions are consistent with SUMO:SIM binding
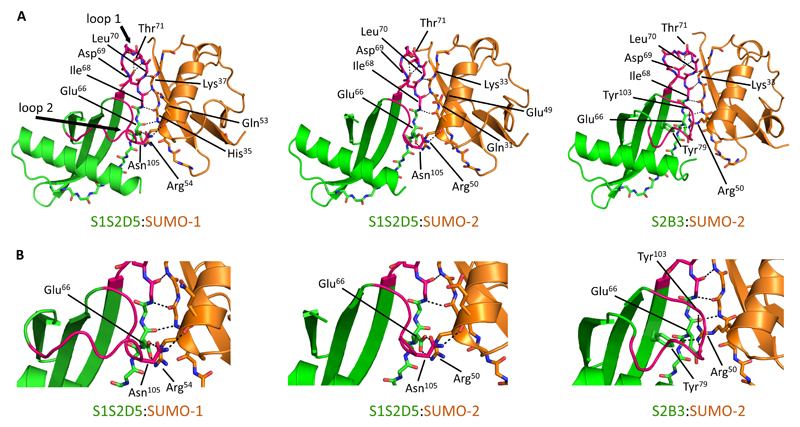
To confirm that interactions between S-Affs and SUMO are consistent with the interaction between a SIM and SUMO, we solved X-ray crystallographic structures for SUMO-1:S1S205, SUMO-2:S1S205, and SUMO-2:S2B3 (Fig. 2A and table S1). The 132 strand of the SUMO isoforms interacted with S-Aff loop 1 residues through a series of backbone hydrogen bonds, resulting in a long β-sheet structure that spanned both SUMO and the S-Aff. Importantly, these hydrogen bonds were identical to those formed between SUMO-1 and SIM peptides (PDB: 2LAS) (23) (Fig. S1D). We also observed that the carboxyl group of Asp69 in the SIM motif (66-EQIDLT-71 - italicized residues are from the Affimer scaffold) formed hydrogen bonds with both the side chain and the backbone of Thr71, presumably helping to stabilize loop 1 of each S-Aff complex. Additionally, there were hydrogen bond interactions between S1S2D5 Glu66 and an Arg at the end of the SUMO helix (Arg54 in SUMO-1 or Arg50 in SUMO-2) (Fig. 2B). Asn105 on loop 2 of S1S2D5 interacted with the C-terminal end of the SUMO helix through a hydrogen bond to Gln53 (SUMO-1) or Glu49 (SUMO-2) (Fig. 2B). We were unable to solve part of loop 2 of S1S2D5, which correlates with its minor role in interacting with SUMO isoforms (Fig. 1E). S2B3 interacted with SUMO-2 in a similar manner but with a key difference: Arg50 from SUMO-2 inserts under loop 2 of S2B3, forming a hydrogen-bonding network with the side chains of Glu66 and Tyr79 from the S2B3 13-sheet and the backbone carbonyl of Tyr103 in loop 2. We hypothesize that this hydrogen-bonding network stabilizes loop 2 and may explain the dependence on loop 2 for S2B3 binding to SUMO-2 (Fig. 1E). S2B3 has a smaller amino acid (Val68) compared to the same position of S1S2D5 (Ile68), potentially creating room for SUMO-2 Arg50 to insert under the loop 2.
Fig. 2. Binding of S-Affs to SUMO resembles SUMO:SIM interactions.
(A) X-ray crystallographic structures of SUMO-1:S1S2D5 (PDB: 5ELJ), SUMO-2:S1S2D5 (PDB: 5EQL), and SUMO-2:S2B3 (PDB: 5ELU) complexes. S-Affs are green, SUMO proteins are orange and loops are colored pink. Only loops in the S1S2D5:SUMO-1 structure are labeled for clarity. The interacting edge strands of both the S-Aff and SUMO isoforms are shown as sticks. Black dotted lines depict residues in which both the amide nitrogen and carbonyl oxygen form main chain–to–main chain hydrogen bonds between S-Aff and SUMO proteins. Carbon atoms are colored green for S-Aff and orange for SUMO. For each of the complexes, oxygen is red, nitrogen is blue, and sulfur is yellow. Loop 2 is unstructured in the S1S2D5:SUMO-2 crystal. (B) Magnified images highlighting critical interactions between S2D5 Glu66 (part of the S-Aff scaffold) with Arg residues at the end of the SUMO helix (Arg54 in SUMO-1 or Arg50 in SUMO-2); the Arg residues adopted different orientations between the two isoforms of SUMO in order to accommodate binding. This was important for establishing a hydrogen bonding network between S2B3 loop 2 Tyr103 and SUMO-2. Asn105 on loop 2 of S2D5 interacted with the C-terminal end of the SUMO helix through a hydrogen bond to Gln53 (SUMO-1) or Glu49 (SUMO-2).
Molecular dynamics simulations provide insight into the basis for isoform specificity
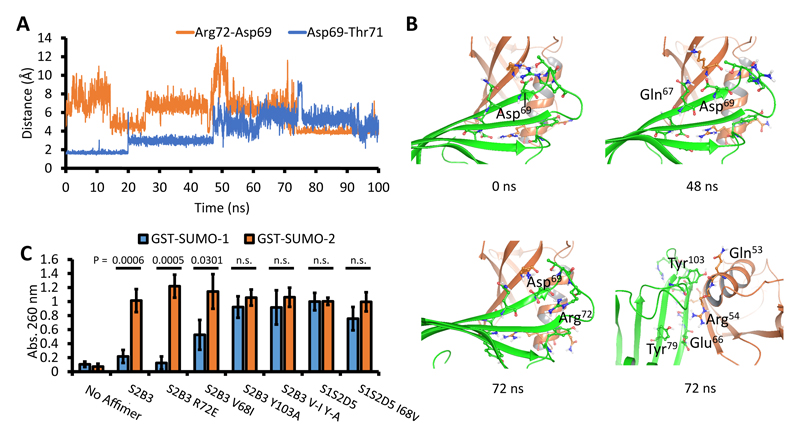
The x-ray structures did not offer any obvious explanation for the observed isoform specificity of individual S-Affs. We therefore performed long molecular dynamics (MD) simulations to understand the dynamic behavior of these proteins. Simulations of SUMO-1:S1S2D5 and SUMO-2:S1S2D5 complexes essentially reproduced the interactions observed in the crystal structures. The conserved hydrogen bonds with >90% occupancy in the 100 ns trajectories were all between the 13-strands and between Glu66 of S1S2D5 and Arg54 in SUMO-1 or Arg50 in SUMO-2 (table S2). In addition to the common conserved hydrogen bonds in the network, SUMO-2:S2B3 offered two very stable interactions: from Tyr103 on S2B3 loop 2 to Glu49 and to Arg50 on SUMO-2 (97% occupancy for both interactions). Additionally Tyr79 in the S2B3 scaffold formed a stable hydrogen bond to Arg50 in SUMO-2 (82% occupancy) within the same network. Because we were unable to generate SUMO-1:S2B3 complexes due to the specificity of this S-Aff for SUMO-2, we modelled these to try and understand why such an interaction was difficult to observe. Arg54 in the modelled SUMO-1:S2B3 complex could either adopt the conformation of Arg54 found in SUMO-1:S1S2D5 or that of Arg50 in SUMO-2:S2B3 (Fig. 2B). For MD simulations, we generated both models, one reproducing the interactions in SUMO-1:S1S2D5 called SUMO1:S2B3 and another reproducing the interactions as seen in SUMO-2:S2B3 called SUMO-1-Alt:S2B3. Despite Arg54 in SUMO-1-Alt starting with the same interactions as in SUMO-2:S2B3, it flipped its conformation in the simulation and only maintained the salt bridge to Glu66 for 100 ns. The simulation of SUMO-1:S2B3 showed that Arg54 failed to re-establish the hydrogen bond to Tyr103 on S2B3, resulting in reduced interaction between S2B3 loop-2 and SUMO-1. Events prior to and at the same time as the flip of Arg54 indicated this was caused by several amino acid substitutions. First, surface differences in the SUMO-1 and SUMO-2 β-sheets, most noticeably Lys23 and His35 on SUMO-1, are replaced by Asn19 and Gln31 on SUMO-2, which formed a fairly stable hydrogen bond network with S2B3 Gln67 (scaffold residue -1 of loop 1), whereas Gln67 did not engage SUMO-1. Secondly, the simulations showed that Arg72 in S2B3 loop 1 periodically formed a hydrogen bond with Asp69 (in the VDLT motif). Finally Gln53 on the SUMO-1 helix was substituted with Glu49 in SUMO-2, which formed a much stronger hydrogen bond with Tyr103 on S2B3 loop 2. At about 48 ns of the SUMO-1-Alt:S2B3 simulation, Asp69 on S2B3 (in the VDLT motif) briefly flipped away from its hydrogen bond interactions with Thr71 on loop 1 (Fig. 3A), and it was transiently stabilized by hydrogen bonds with Gln67 (Fig. 3B) and Lys37 on SUMO-1. At about 70 ns, Arg72 on Loop 1 approached Asp69 from the side (Fig. 3B) to form a salt bridge, and that motion bent the loop inwards and increased the distance between the stems of loop 1 from an average of 2.2 Å in the first 70 ns to an average of 3.5 Å in the final 30 ns. This movement pushed loop 2 away from the SUMO-1 helix, thus breaking the hydrogen bond between Tyr103 on loop 2 of S2B3 and Gln53 on SUMO-1 helix (Fig. 3B). At the same time, Arg54 dropped all hydrogen bond contacts. Although the hydrogen bond between Tyr103 and Gln53 were later recovered, Arg54 of SUMO-1 was only able to form the salt bridge with Glu66 and remained in that binding mode for the remaining 30 ns of simulation.
Fig. 3. Molecular dynamics simulation of SUMO-1-Alt:S2B3 identifies residues critical for isoform specificity.
(A) Distances from Asp69 to Arg72 (blue line) and to Thr71 (orange line) in the simulation are plotted against simulation time in ns. Distances of 3 Å or less indicate hydrogen bonding between the side chains of Asp69 and Thr71. Distances of around 4 Å indicate direct hydrogen bond contacts between Asp69 and Arg72. (B) Snapshots of SUMO-1-Alt:S2B3 simulation at 0 ns, which shows the starting structure around loop 1, and at 48 ns, which shows Asp69 rotated to interact with neighboring Gln67. The left snapshot at 72 ns shows Arg72 (SUMO-1-Alt) and its proximity to Asp69 (S2B3). The right snapshot at 72 ns is a rotation of the structure to illustrate that Arg54 (SUMO-1-alt) lost contact with Tyr69 and Glu66 and Tyr103 (S2B3) lost hydrogen bond with Gln53 (SUMO-1-Alt). SUMO-1-Alt is green and S2B3 is orange. (C) ELISA of S-Affs with the indicated amino acid substitutions introduced into the loop region based on predictions from the molecular dynamics simulations. S2B3 V-I Y-A is a double substitution of Val68 to Ile in loop 1 and Tyr103 to Ala in loop 2 of S2B3. Data are represented as mean absorbance at 260 nm +/standard error from 5 experiments. Student’s t-tests were used to determine statistical significance. P values below 0.05 were deemed statistically significant and ‘n.s.’ are not statistically significant.
MD simulations highlighted several residues that might be important for isoform specificity, including Arg72 in loop 1 of S2B3 and Tyr103 in loop 2 of S2B3 (Fig. 3B). In addition, we predicted that the smaller Val68 in S2B3, compared to Ile68 in S1S2D5, may facilitate an interaction between SUMO-2 Arg50 and loop 2. To test these predictions, we made selected amino acid substitutions in the loops of S2B3 and S1S2D5 and assessed binding by ELISA (Fig. 3C). Substitution of S2B3 Arg72 with Glu (S2B3 R72E) did not alter specificity of S2B3. However, substitution of Val68 with Ile (S2B3 V68I) enabled increased binding to SUMO-1, although still with reduced abundance compared to SUMO-2 binding, suggesting this residue contributed to isoform specificity. Substituting Ala for Tyr103 in S2B3 (S2B3 Y103A) completely abolished specificity, resulting in similar binding in vitro to both SUMO-1 and SUMO-2. In vitro, the double mutant (S2B3 V68I/Y103A) and S2B3 Y103A single mutant were equally effective in interacting with both SUMO isoforms. Substitution of Val for Ile68 in S1S2D5 (S1S2D5 I68V) partially reduced binding to SUMO-1 in vitro, but this difference was not large. Taken together, the isoform specificity observed in S2B3 involved both loops and the presence of Tyr103 in loop 2, confirming the predictions from the MD simulations.
S-Affs interact with SUMO in cells
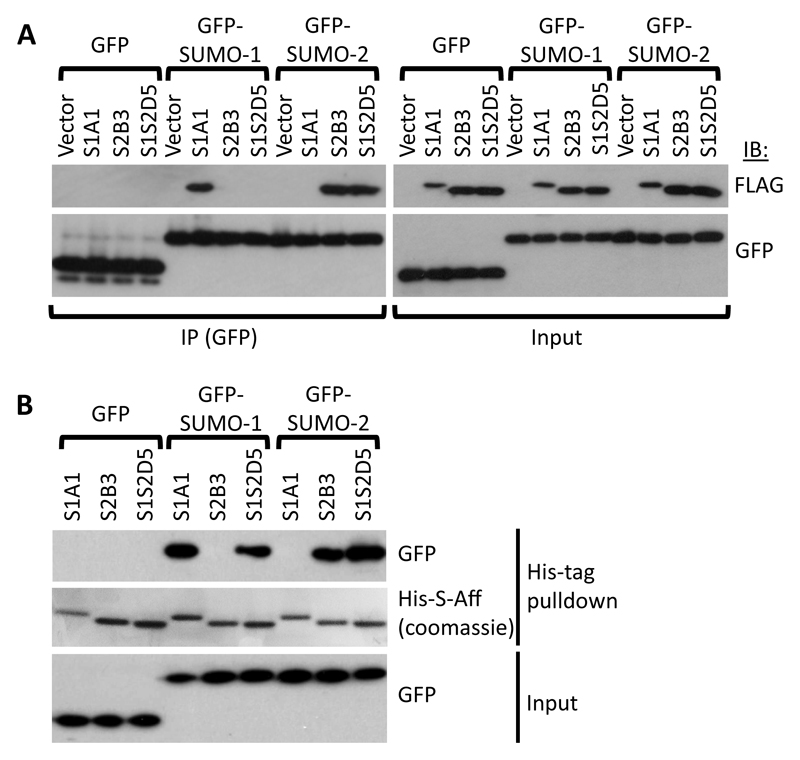
A major advantage of using protein aptamers, such as Affimers, is the ability to express them in cells. We tested whether S-Affs retained specificity when FLAG-tagged versions of the S-Affs were overexpressed in the mammalian cell line HEK293T. We expressed FLAG-S1A1 as an example of a SUMO-1–specific S-Aff, FLAG-S2B3 as a SUMO-2–specific S-Aff, and FLAG-S1S2D5 as a SUMO S-Aff that binds both SUMO-1 and SUMO-2. Expression of the S-Affs did not appear to negatively affect cellular function (Fig. S3). Immunoprecipitation (IP) experiments demonstrated that the S-Affs bound to cellular SUMO and interacted with SUMOylated proteins (Fig. S4; note that in this assay SUMO-2 cannot be distinguished from SUMO-3). Only S1A1 exhibited isoform specificity in cells, binding only to proteins SUMOylated with SUMO-1 and not showing any detectable interaction with SUMO-2/3–modified proteins. Furthermore, the detection of higher molecular weight SUMO-1–modified proteins is likely to be due to the addition of SUMO-1 to poly-SUMO-2/3 chains. Similarly, the lack of specificity with FLAG-S2B3 may reflect the recognition of proteins that are simultaneously modified with both isoforms of SUMO (24). To circumvent the dual modification with both SUMO isoforms, we expressed green fluorescent protein (GFP)-tagged SUMO-1 or GFP-tagged SUMO-2 so that sufficient unconjugated SUMO was available to assess isoform specificity. Immunoprecipitation with an antibody recognizing the GFP tag indicated that S1A1 and S2B3 exhibited the predicted specificity for SUMO-1 and SUMO-2, respectively. Unexpectedly, S1S2D5 interacted only with GFP-SUMO-2 in cells (Fig. 4A), despite various data indicating that it bound both SUMO-1 and SUMO-2 in vitro (Fig. 1, Fig. 3). It must be noted that all three S-Affs, when expressed in and purified from bacteria, S-Affretained their observed in vitro binding specificities when incubated with extracts from HEK293T cells expressing GFP-SUMO-1 or GFP-SUMO-2, (Fig. 4B); hence, the difference in S1S2D5 specificity was a result of its cellular expression.
Fig. 4. Isoform specificity of S-Affs binding to SUMO in vitro.
(A) Immunoprecipitation of GFP-tagged SUMO-1 or SUMO-2 expressed in HEK293T cells with the indicated FLAG-tagged S-Aff or in cells transfected with only the vector for the S-Aff. Proteins were immunoprecipitated (IP) with an antibody recognizing GFP and immunoblotted (IB) for either the FLAG tag or GFP. The left blot shows the coimmunoprecipitation results, and the right blot shows the proteins in the lysate prior to immunoprecipitation (Input). (B) Immunoblot showing pulldown of purified (bacterially-expressed) His-tagged S-Affs following incubation with extracts from HEK293T cells expressing GFP, GFP-tagged SUMO-1, or GFP-tagged SUMO-2S-Aff. Overexpressed proteins that interacted with His-S-Affs (top) and input samples taken prior to incubating with His-S-Affs (bottom) were detected with antibodies recognizing GFP. Pulled-down His-S-Affs were detected by Coomassie staining (middle panel). Data are representative of 2 experiments.
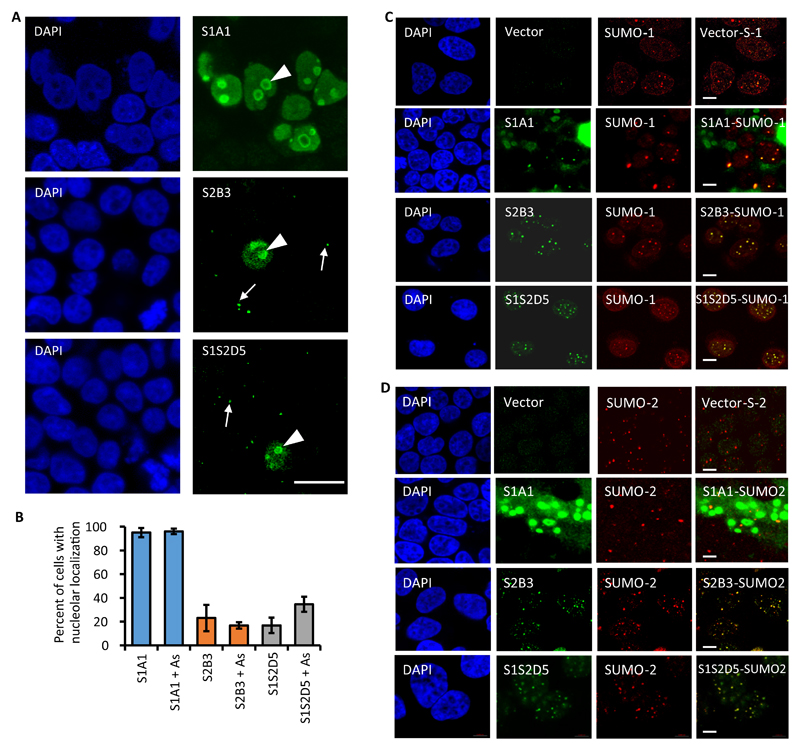
We next characterized the cellular localization of overexpressed FLAG-S-Affs in HEK293T cells. We noted that S1A1 localized to what appeared to be nucleoli in virtually all cells (Fig. 5A-B). However, although some nucleoli localization was observed for S2B3 and S1S2D5 (in 15-30% of cells, Fig. 5B) the majority was observed in small nuclear foci (Fig. 5A) that resembled PML-NBs. Arsenic treatment induces SUMOylation and an increase in the accumulation of SUMO proteins (SUMO-1 and SUMO-2/3) in PML-NBs (25). Whereas arsenic treatment did not alter the distribution of S-Affs (Fig. 5B), immunofluorescence analysis of FLAG-tagged S-Affs showed a high degree of colocalization with PML-NBs marked with either SUMO-1 or SUMO-2/3 (Fig. 5C-D). However, FLAG-S1A1 colocalized with nucleoli under both control and arsenic-treated conditions. Although we did observe S1A1 in PML-NBs upon arsenic treatment, this was often difficult to observe due to the large amount of nucleolar fluorescence (Fig. 5C-D). As noted, FLAG-S1S2D5 and S2B3 also appeared to localize with nucleoli. Other studies have demonstrated a role for both SUMO-1 and SUMO-2/3 in nucleolar function (26, 27), which may explain this observation. Furthermore, the localization of S1S2D5 resembled that of S2B3, and both differed from S1A1 (at least with regards to nucleolar localization); therefore, these data further demonstrate that S1S2D5 is more selective for SUMO-2/3 when expressed in cells than when purified from bacterial cells and used for in vitro binding assays, as suggested by immunoprecipitation experiments (Fig. 4A).
Fig. 5. S-Affs colocalize with SUMO in mammalian cells.
(A) Immunofluorescence showing FLAG-S-Affs expressed in HEK293T cells. S-Affs that colocalized with structures resembling nucleoli are denoted with a white arrowheads and those localizing with structures resembling promyelocytic leukemia nuclear bodies (PML-NBs) are denoted with white arrows. DAPI stain (blue) labels nuclei. Images shows are representative of three independent experiments. Scale bar, 20 µm. (B) Quantification of FLAG-S-Affs colocalization with nucleoli in untreated cells and in cells treated with arsenic trioxide (As). The average percent of S-Aff-positive cells displaying nucleolar staining was derived from five independent images per experiment (N = 3), with about 70-150 positive cells per image). Error bars represent the standard deviation from three independent experiments. (C,D) Immunofluorescence showing the indicated FLAG-tagged S-Affs (green) and (C) SUMO-1 (red) or (D) SUMO-2/3 (red) in S-AffHEK293T cells treated with As. Control cells were transfected with empty vector. Scale bars, 5 µm.
S-Affs do not inhibit SUMO conjugation in vitro or in cells, or SUMO deconjugation in vitro
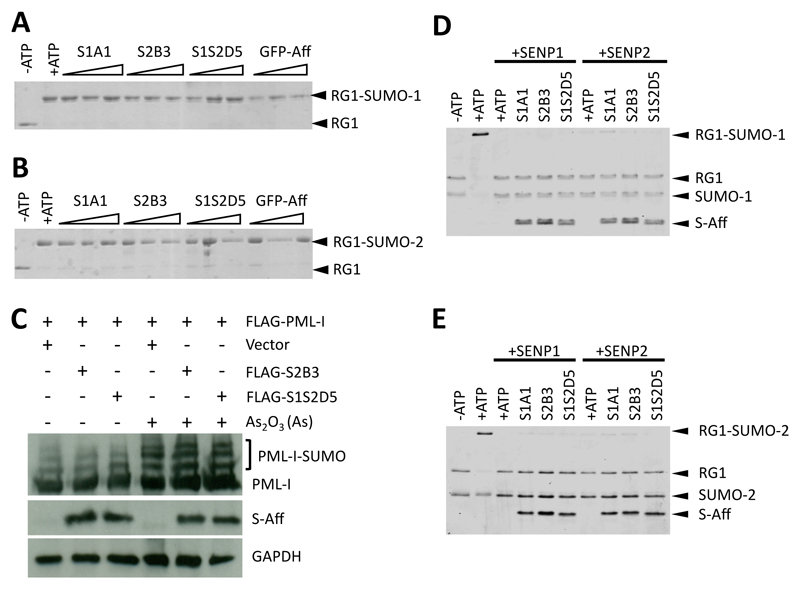
For the S-Affs to be useful for exploring SUMO:SIM interactions or as inhibitors of SUMO:SIM interactions, they must not prevent SUMO conjugation, an issue encountered with a SUMO-1–specific Monobody (28, 29). We performed in vitro SUMOylation assays in the presence of varying concentrations of S1A1, S2B3, or S1S2D5 and showed that SUMO conjugation to the substrate RanGAP1 was not blocked by any of the three S-Affs (Fig. 6A-B). Furthermore, basal SUMOylation of FLAG-tagged PML-I (isoform 1) in cells also expressing either FLAG-S2B3 or FLAG-S1S2D5 was similar to that in cells without co-expression of S-Affs. In addition, expression of FLAG-S2B3 or FLAG-S1S2D5 did not affect the amount of FLAG- PML-I SUMOylation in response to arsenic compared to control cells transfected with the vector alone (Fig. 6C), indicating that the S-Affs did not interfere with SUMOylation.
Fig. 6. S-Affs do not inhibit SUMO conjugation or deconjugation.
(A, B) In vitro SUMOylation in the presence of the indicated S-Affs for SUMO-1 (A) or SUMO-2 (B) conjugation to GST-tagged RanGAP1 fragment (RG1; amino acids 418-587 encompassing a consensus SUMOylation site). Experiments were performed in the presence of increasing concentrations of S-Aff (0.1:1, 1:1, or 10:1 ratios of S-Aff:SUMO). GFP-Adh is a control Affimer raised against green fluorescent protein (GFP) and does not bind to SUMO. Blots were probed with an antibody recognizing GFP. (C) SUMOylation of FLAG-tagged PML-I in HEK293T also expressing the indicated FLAG-tagged S-Affs. SUMOylation was induced with arsenic trioxide (As2O3). Proteins were detected with the antibody recognizing the FLAG tag or with an antibody recognizing GAPDH as a loading control. (D, E) Coomassie-stained SDS-PAGE showing in vitro assays to assess SUMO-1 (D) or SUMO-2 (E) de-SUMOylation by the SUMO deconjugases SENP1 and SENP2. All data are representative of at least 2 independent experiments.
SUMOylation is a reversible process, and several SUMO-specific proteases (SENPs) that are able to reverse SUMOylation by deconjugating SUMO from substrates have been identified. Here we assessed the effects of S-Affs on in vitro deSUMOylation by adding either recombinant SENP1 or SENP2 following SUMOylation of RanGAP-1, to reactions that contained S-Affs. These assays demonstrated that the S-Affs did not impact on deSUMOylation of either SUMO-1 (Fig. 6D) or SUMO-2 (Fig. 6E).
S-Affs specifically inhibit SIM-dependent PPIs
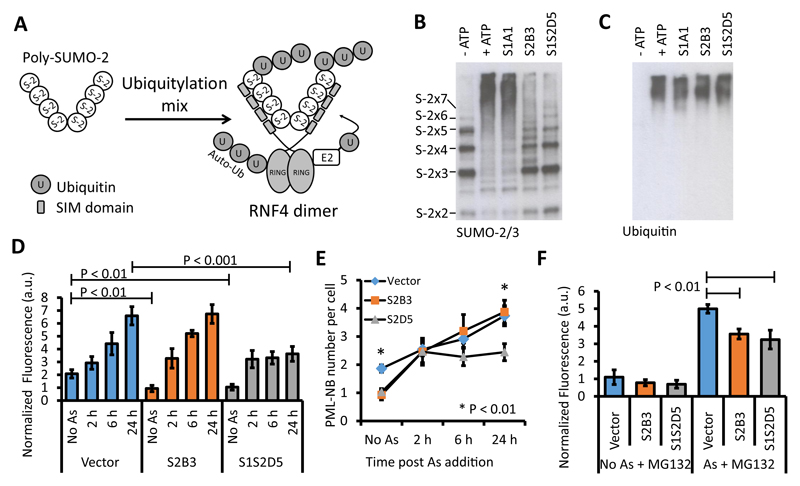
We employed an in vitro SUMO-targeted ubiquitin ligase (STUbL) assay with the E3 ubiquitin ligase RING finger protein 4 (RNF4), to test the ability of S-Affs to block SUMO-dependent PPIs. RNF4 possesses two important domains – one domain containing four SIMs that mediates binding to poly-SUMO-2 and a RING domain that mediates RNF4’s ubiquitylation activity. Enhancement of the ubiquitin E3 ligase activity of RNF4 for poly-SUMO requires binding to poly-SUMO-2 mediated by the SIMs (Fig. 7A) (30, 31). Recombinant RNF4 ubiquitylated poly-SUMO-2 in an ATP-dependent manner (Fig. 7A-B). Addition of the SUMO-1-specific S-Aff S1A1 did not inhibit poly-SUMO-2 ubiquitylation, whereas inhibition was observed upon the addition of S-Affs that recognized SUMO-2, S2B3 andS1S2D5. Furthermore, when the same samples were probed with antibodies recognizing ubiquitin, RNF4 autoubiquitylation, S-Affwhich is a RING-dependent and SIM-independent process (32) (although poly-SUMO-2 does enhance this activity (31)), was unaffected by any of the S-Affs (Fig. 7C).
Fig. 7. S-Affs block SIM-dependent protein-protein interactions.
(A) Schematic of the in vitro SUMO-targeted ubiquitin ligase (STUbL) assay. In vitro, poly-SUMO-22-8 is ubiquitylated by incubating it with recombinant RNF4, an E1 activating enzyme, a ubiquitin-conjugating enzyme, and ubiquitin. RNF4-dependent ubiquitylation of poly-SUMO-2 requires SIM-dependent binding. (B) STUbL assays in the presence of the indicated S-Affs. Reactions in the presence or absence of ATP serve as positive and negative controls, respectively. S-2X2, S-2X3, S-2X4, S-2X5, S-2X6, and S-2X7 markers indicate SUMO-2 chain lengths. The samples were immunoblotted with an antibody recognizing SUMO-2/3. (C) The same samples from panel B were analyzed by immunoblotting with an antibody recognizing ubiquitin. The detected protein represents autoubiquitylated RNF4. Data in B and C are representative of at least two experiments. (D) Quantitative immunofluorescence of PML abundance over time in HEK293T cells expressing the indicated FLAG-tagged S-Aff or transfected with the empty vector and exposed to arsenic (As). Data are representative of three independent experiments and are presented as the mean fluorescence from five fields of view +/- standard deviation. Statistical significance was determined by Student’s t-test. Statistical significance is indicated for the vector-transfected cells without arsenic treatment (Vector, No As) compared to cells transfected with each of the S-Affs without arsenic and for the vector-transfected cells with arsenic treatment compared to cells transfected with each of the S-Affs with arsenic treatment. (E) Quantification of the number of PML-NBs per cell over time in response to As in vector-transfected cells and in cells expressing the indicated FLAG-tagged S-Aff. Data are representative of three independent experiments and presented as the mean number of PML-NBs per cell +/- standard deviation and statistical significance was determined by Student’s t-test. Statistical significance is indicated for vector-expressing cells compared to S2B3- and S1S2D5-expressing cells with and without As treatment. (F) Quantitative immunofluorescence of PML abundance in cells expressing the indicated FLAG-tagged S-Aff or in cells transfected with the vector 6 h after the addition of As in the presence or absence of the proteasome inhibitor MG-132. Representative data are presented as mean fluorescence intensity +/- standard deviation, and Student’s t-tests were used to determine statistical significance. Three independent experiments were performed. For all quantitative immunofluorescence assays, values were normalized based on cell number (based on DAPI staining). a.u., arbitrary units.
S-Affs inhibit SIM-dependent PML-NB formation
PML-NB formation depends on SUMO:SIM interactions (17–19). We used PML-NB formation as a system to investigate the ability of S-Affs to inhibit SIM-dependent interactions in mammalian cells. We quantified total PML abundance (the sum total of PML fluorescence per field of view, normalized to the number of nuclei), the number of PML-NBs per cell, and the change in size (PML fluorescence intensity) of individual PML-NBs in response to cell arsenic-induced stress. We exposed HEK293T cells transfected with the empty vector, S2B3, or S1S2D5 to arsenic and monitored total PML abundance in the cells by quantitative immunofluorescence of antibody-stained PML-NBs over time. The fluorescence intensities of individual PML-NBs were calculated and where total fluorescence was reported, this represented the sum total fluorescence of all PML-NBs (normalized between samples according to DAPI). The vector-expressing cells significantly increased PML fluorescence over 24 h compared to the no-arsenic control (Fig. 7D). In S2B3-transfected cells, the basal amount of PML was reduced, but PML abundance increased in response to arsenic exposure with similar kinetics as the control cells expressing the empty vector. Cells overexpressing S1S2D5 also exhibited reduced basal PML abundance. However, S1S2D5-expressing cells exhibited an initial increase in PML abundance within 2 hours of arsenic exposure, but no further increase occurred thereafter (Fig. 7D). S1S2D5 also reduced the total number of PML-NBs per cell induced by arsenic compared with those induced in vector-expressing or S2B3-expressing cells (Fig. 7E).
To discount the possibility that the S-Affs promoted the proteasomal degradation of PML, we quantified PML fluorescence in control cells or arsenic-exposed cells transfected with the vector, S2B3, or S1S2D5 in the presence or absence of the proteasome inhibitor MG132. In the presence of MG132, both S-Affs reduced PML fluorescence (Fig. 7F). The ability of S2B3 to inhibit the arsenic-induced increase in PML fluorescence in the presence of MG132 suggested that S2B3 or its targets may be subjected to proteasomal degradation in response to arsenic. Notably, these data, together with the in vitro and cellular SUMO conjugation data shown in Fig. 6, demonstrate that S-AffS2B3 and S1S2D5 do not prevent SUMO conjugation and can be used to explore cellular processes involving SUMO:SIM interactions.
Discussion
Due to the central role of PPIs in cell biology, the ability to target them therapeutically has great potential. Tools that interfere with PPIs can also be used to probe the function of PPIs and accelerate our understanding of biochemical pathways. For this study we chose the SUMOylation system as a model. Using Affimer technology, we developed novel, isoform-specific inhibitors of SUMO-dependent PPIs. Specifically, we developed S-Aff that exhibited preferential interactions with SUMO-1 or SUMO-2 or both (we assume that those that interact with SUMO-2 will also interact with SUMO-3 due to the sequence similarity of these two SUMO isoforms). Using a combination of X-ray crystallography and MD simulations, we predicted, and then demonstrated, discrete biochemical interactions that were important for the observed isoform specificity. Additionally, Affimers can be applied to various biological systems; indeed, this technology has many advantages (speed, protein stability, high yields through simple recombinant protein expression) over alternative methods such as the production of antibodies or screening for small molecules (33). Furthermore, that Affimers (and similar aptamers) can be expressed in cells provides the ability to assess intracellular function.
Expression of the S-Affs in cells showed they colocalized with PML-NBs, the major sites of SUMOylation in the cell. Furthermore, immunoprecipitation assays showed that the S-Affs interacted with SUMOylated proteins and that they retained isoform specificity in cells. However, it appeared that overexpressed S1S2D5, which bound both SUMO-1 and -2 in vitro with similar affinities, had a reduced interaction with cellular SUMO-1, suggesting that either posttranslational modification or altered structural constraints affected its interactions with SUMO in cells. Tracts of acidic amino acids or phosphorylation sites surrounding SIMs promote SUMO-1 binding; residues close to the S1S2D5 SIM are potentially targets for phosphorylation, but there is no evidence of an acidic tract. There was however a predominance of positively charged residues (Arg and Lys) in the Affimer scaffold surrounding loop 1, which may preclude SUMO-1 binding in cells (8). Interestingly, it was recently shown that an Arg-rich region in RNF4, in combination with its SIMs, was required for binding polySUMO-2–modified KAP1 (also known as TRIM28) (34). The combination of these features in the S-Affs may therefore contribute to their specificity in cells. In addition, it appeared that the S-Affs preferentially bound to high molecular weight SUMO, suggesting that they interacted with either poly- or multi-SUMOylated substrates (Fig. S4). Although the data in Fig. 4A suggest that S-Affs do interact with monomeric SUMO, one cannot exclude the possibility that in this context the overexpressed and immobilized SUMO (with high concentrations of closely-spaced SUMO) may mimic poly-SUMOylation. Further work will be required to understand this observation.
If the S-Affs are to be suitable as PPI inhibitors, they must not block SUMO conjugation itself. This was an issue for SUMO-1–mediated interaction inhibitors developed with an analogous technology (Monobodies) (28, 29). Therefore, we established that S-Affs did not prevent SUMO conjugation in vitro or in cells. Because most of the known SUMO E3 ligases contain SIMs, which enhance protein SUMOylation, an obvious question arises: Why is total cellular SUMOylation not overtly affected in the presence of SUMO:SIM inhibition by S-Affs? It might be that, similar to in vitro conditions, SUMO E3 ligases are not essential for SUMOylation and, instead, they primarily enhance SUMO conjugation efficiency in vivo. Also, there are SUMO E3 ligases that function without SIMs (for example, PIASγ) (35). A further hypothesis is based on a recent concept that was coined ‘group modification’ in which, during responses to cellular stress, functionally-related proteins are modified locally (36, 37). Therefore, unless we stimulate a particular pathway that requires SIM-dependent SUMOylation, we would not expect to see substantial aberrations in cellular SUMOylation. Moreover, several proteins contain SIM domains that function to enhance their own SUMOylation (for example, human cytomegalovirus IE2, Daxx, and thymine DNA glycosylase); however, these are not essential for SUMOylation (17, 38, 39).
SUMO-2–specific Adhs blocked in vitro STUbL activity, confirming them as SUMO-mediated PPI inhibitors. S-Affs did not block RNF4 autoubiquitylation in vitro, demonstrating their specificity for SUMO-2 and not the similar protein ubiquitin (as also demonstrated in Fig. 1A) or any of the other ubiquitylation components. However, Rojas-Fernandez et al. (31) showed that RNF4 autoubiquitylation was enhanced by polySUMO-2 binding because this promoted dimerization of its RING domains; therefore, we were surprised that the SUMO-2–specific Adh did not abrogate this activity. However, unlike RNF4 dimerization mutants, RNF4-SIM mutants were not completely inactive, suggesting that polySUMO-2 binding enhances, but is not essential for, dimerization and autoubiquitylation (31). Furthermore, those assays were performed for a maximum of 40 min, and ours were performed for 3 h, which likely explains why we did not observe any inhibition. Therefore, we tested the ability of the S-Affs to inhibit SUMO-PPIs in cells. We showed that S1S2D5 inhibited PML-NB formation in response to arsenic. S2B3 reduced steady-state PML-NB formation in unstressed cells, but upon stimulation with arsenic, PML-NB formation appeared normal. In vitro, S2B3 had an approximately 6-fold lower affinity for SUMO-2 compared to S1S2D5, which might account for the apparent inability of S2B3 to prevent arsenic-induced PML-NB formation. We noted that even S1S2D5 failed to completely block PML-NB formation. Oxidative stress induces PML-NB nucleation, increasing their size, and SUMO:SIM interactions are important only for the expansion of PML-NBs, not their initial formation (40). Our data are consistent with SUMO:SIM interactions being required for PML-NB expansion and, furthermore, indicated that SUMO-2 is the main isoform involved in arsenic-induced PML-NB formation and expansion. Of course, alternative means of inducing PML-NB formation, such as interferon stimulation, may require additional SUMO isoforms, and these S-Affs enable the examination of such questions. Furthermore, there are six splice variants of PML (I-VI) each with a unique C-terminus. Although PML-NB formation is largely dependent on SUMO-mediated PPIs, PML-VI does not contain a SIM domain, yet it still accumulates in PML-NBs, even when expressed in the absence of endogenous PMLs (25). Therefore, this points to unique features in the C-terminus of PML for NB formation. These hypotheses merit investigation, and the S-Affs described here or others engineered with this technology provide a means of doing so.
Materials and Methods
Protein production
For purification of recombinant proteins, E. coli (BL21) were transformed and protein expression was induced in 100 ml mid-log cultures (OD600 0.6) by the addition of 0.1 mM IPTG for 5 h at 37°C. GST-SUMO-1, GST-SUMO-2 and GST-SAE1/2 expressing bacteria were lysed and sonicated in 5 ml PBS, 0.5 M (total) NaCl, 1 mM EDTA, 1% Triton X-100, Complete protease inhibitor cocktail (Roche) and purified using Glutathione Sepharose 4B (GE Healthcare) and eluted using 50 mM Tris-HCl (pH 7.6), 10 mM reduced glutathione. His-Ubc9, His-SUMO1-GG, His-SUMO2-GG and His-MBP-RNF4 (32) and His-S-Aff expressing bacteria were lysed and sonicated in 5 ml 50 mM Tris (pH 7.6), 1% Triton X-100, 300 mM NaCl, 10 mM imidazole, Complete protease inhibitor cocktail (Roche) and purified using Ni-NTA agarose (Qiagen) and eluted in 50 mM Tris-HCl (pH 7.6), 300 mM NaCl, 250 mM imidazole. PD MidiTrap G-25 columns (GE Healthcare) were used for buffer exchange of proteins used in in vitro assays and proteins were eluted in 50 mM Tris-HCl (pH 7.6), 5 mM DTT and concentrated using Vivaspin columns (with appropriate MWCO; Sartorius Stedim Biotech). Proteins used in ITC experiments and phage display were dialyzed into PBS using Slide-A-Lyzer dialysis membranes with appropriate MWCO (Thermo Scientific) following the manufacturer’s instructions.
Affimer selection
Affimer selection by phage display, phage ELISAs and confirmatory ELISAs were performed as previously reported (8). Briefly, SUMO-1 and SUMO-2 were biotinylated using EZ-link NHS-SS-biotin (Pierce), according to the manufacturer’s instructions. Biotinylation was confirmed by ELISA using streptavidin conjugated to horseradish peroxidase (HRP). We adapted the protocol to screen the Affimer phage display library in order to select isoform-specific binders for SUMO-2 (41). Here, competitive binding with 10 µg/ml non-biotinylated human SUMO-1 was applied during all phage pre-panning and phage panning steps from the second round forward. Individual colonies were confirmed as isoform-specific binders to SUMO-1 and SUMO-2 by phage ELISA. For confirmatory ELISA, the coding sequence of the S-Affs were cloned in to the NheI-NotI sites of pET11 and expressed and purified as above generating a 6xHis-tagged protein. SUMO-1, SUMO-2, ubiquitin (Enzo Life Sciences) or bovine serum albumin (BSA; Sigma) coated ELISA plates were incubated with biotinylated S-Affs followed by incubation with streptavidin-HRP.
Isothermal titration calorimetry
Typically, 0.02 mM Affimer was titrated with 0.14 mM SUMO, by 2.5 min injections at 25°C using an iTC200 system (Microcal). Data analysis was performed using Origin (OriginLab).
X-ray Crystallography
Protein complexes for crystallography were generated as follows: a shorter version of S-Aff (based on Adhiron81 (8)) was amplified and cloned into the NheI-NotI sites of pET11 which was expressed and purified as above generating a 6xHis-tagged protein. Residues 18-97 of SUMO-1 and 14-93 of SUMO-2 were amplified and cloned into the NdeI and BamHI sites of pET11 (generating a tagless SUMO expression cassettes). Truncated S-Aff was bound to Ni-NTA agarose and incubated with bacterial lysates containing expressed tagless SUMO followed by extensive washing. SUMO:S-Aff complexes were eluted and confirmed using immunoblotting.
Crystals of all S-Aff complexes were grown at 18°C using the sitting drop vapor-diffusion method. The crystallization conditions for SUMO-1:S1S2D5 and SUMO-2:S1S2D5 complexes were very similar, containing 0.1 M HEPES sodium salt pH 7.5, 10% w/v polyethylene glycol 20,000 and 0.1 M HEPES sodium salt pH 7.6, 22% w/v polyethylene glycol 3350, respectively. For the SUMO-2:S2B3 complex, the crystallization condition was 0.1 M sodium cacodylate pH 6.5, 0.2 M sodium chloride and 2.0 M ammonium sulfate.
Crystals were transferred to a cryoprotectant solution containing the mother liquor and 25% (v/v) glycerol (final concentration) before mounting in loops and flashed-cooled directly into liquid nitrogen. Data from all SUMO:S-Aff complexes were recorded to a resolution range of 2.0 to 2.5 Å from single crystals at 100 K on the macromolecular crystallography beamline stations I03 and I04 at Diamond Light Source. For the SUMO-2:S1S2D5 and SUMO-2:S2B3 data, the diffraction images were integrated using XIA2 (42), scaled and reduced using programs implemented as part of the CCP4 program suite (43). The diffraction images for the SUMO-1:S1S2D5 were integrated using XIA2 (42), scaled and truncated using the UCLA-MBI Diffraction Anisotropy Server (44). All three SUMO:S-Aff complexes have different unit cell parameters; SUMO-1:S-Aff-S1S2D5 a=36.2Å, b=70.7Å, c=83.6Å and space group P212121, SUMO-2:S-Aff-S1S2D5 a=b=74.3Å, c=59.7Å, α=β=90°, γ=120° and space group P3121, and SUMO-2/S-Aff-S2B3 a=42.0Å, b=42.3Å, c=113.57Å and space group P212121. Five percent of the reflections from each of the SUMO:S-Aff complex data were selected at random and excluded from the refinement using the program FREERFLAG (45) and constituted the Rfree set. The processing and crystallographic statistics for all the SUMO:S-Aff complexes are summarized in table S1.
Despite the different space group and unit cell parameters for each of the complexes, there was one S-Aff molecule and one SUMO molecule per asymmetric unit for each of the complexes. The crystal structure of SUMO-1:S1S2D5 was determined by molecular replacement using the program PHASER (46) with the structures of human SUMO-1 (PDB: 2UYZ (47)) and Affimer (PDB: 4N6T (8)) as the search models. One single MR solution was obtained from PHASER and after initial rounds of rigid body using REFMAC5 (48), residues around the loop regions that pertain to the different Affimer and S1S2D5 sequences were removed to prevent bias in map generation. Iterative manual model building and restrained refinement were carried out using COOT (49) and REFMAC5 allowing the two loop regions of S1S2D5 to be rebuilt. The polypeptide chains were checked against both 2Fo-Fc and Fo-Fc electron density maps during model building in COOT. Water molecules were added in COOT for peaks over 2.0ơ in the Fo-Fc map, and where appropriate hydrogen bonds could be made to surrounding residues or other water molecules. In the later stages of refinement, TLS parameters (50) based on a single-group TLS model for each monomer were calculated from the TLS Motion Determination server (http://skuld.bmsc.washington.edu/~tlsmd/) and refined in PHENIX. The final structure of SUMO-1:S1S2D5 was refined to R = 18.8% and Rfree = 23.6%. Similarly, SUMO-2:S1S2D5 was determined by MR using PHASER with the structures of human SUMO-2 (PDB: 1WM3 (47)) and the refined S1S2D5 (from SUMO-1:S1S2D5) as the search models. One single MR solution was obtained from PHASER and iterative cycles of manual model building and refinement carried out using COOT and REFMAC5. In the later stages of refinement, TLS parameters based on a single-group TLS model for each monomer were refined in PHENIX. The final structure of SUMO-2:S1S2D5 was refined to R = 21.8% and Rfree = 24.3%.
The crystal structure of the SUMO-2:S2B3 was determined by MR using PHASER with the structure of human SUMO-2 (PDB: 1WM3 (47)) and Affimer (PDB: 4N6T (8)) as the search models. One single MR solution was obtained from PHASER and iterative cycles of manual model building and refinement carried out using COOT and REFMAC5. In the later stages of refinement, TLS parameters based on a single-group TLS model for each monomer were refined in PHENIX. The final structure of SUMO-2:S2B3 was refined to R = 17.2% and Rfree = 20.7%. All structural validations were carried out with MOLPROBITY (51). The refinement statistics are summarized in Figure 2-Figure supplement table 1. The atomic coordinates and structure factor amplitudes have been deposited into the Protein Data Bank (www.pdb.org), referred to as Adhirons, and have been assigned; 5ELJ (SUMO-1:S1S2D5*), 5EQL (SUMO-2:S1S2D5*), and 5ELU (SUMO-2:S2B3). * S1S2D5 is referred to as S2D5 in the Protein Data Bank.
Molecular dynamics simulations
The complex structures of SUMO-1:S1S2D5, SUMO-2:S1S2D5 and SUMO-2:S2B3 were prepared using Protein Preparation Wizard in Maestro 10.1 (Schrödinger, LLC, New York). Briefly the bond orders were assigned and hydrogen atoms were added to the heavy atoms. Missing residue side chains and loop were filled in and their conformations were sampled with Prime (Schrödinger, LLC, New York). The N and C termini of the proteins were capped with acetyl and n-methylamine groups. The amino acid protonation states were predicted with PROPKA (52) and hydrogen bond networks were optimized by rotating Asn, Gln, and His residues and sampling hydroxyl and thiol groups. Finally the structures were minimized with restraints using the default protocol in Protein Preparation Wizard.
After the preparation, all structures were aligned using SUMO isoforms as reference. SUMO-1 and S2B3 structures were extracted from their respective complexes and merged as one complex. SUMO-2:S2B3 complex structure shows that the key Arg50 in SUMO-2 engages S2B3 through three hydrogen bonds to the side chains of Glu66 and Tyr79 and backbone carbonyl of Tyr103 which is very different compared to SUMO-2:S1S2D5 complex where Arg50 engages only the side chain of Glu66 on S1S2D5. Because it is not clear what a hypothetical interaction between SUMO-1 and S2B3 would look like, we modelled the alternative binding modes by rotating the Chi-4 angle of SUMO-1 Arg54 until the key hydrogen bonds are reproduced and we called this conformation SUMO-1-Alt.
All five S-Aff complexes were neutralized by adding sodium ions and solvated in a rectangular box of TIP4P (53) waters with a 10 Å buffer distance using System Builder in Desmond Version 4.3 (Schrödinger, LLC, New York). A default equilibration protocol consisting of a series of minimization with harmonic restraints followed by slowly heating the system from 0 to 300K using OPLS3 force field (54) and particle-mesh Ewald for long range electrostatics beyond 9 Å cutoff range. The production simulations were carried out for 100 ns at 300 K using Nose-Hoover chain thermostat (55) and at constant pressure of 1 atm using Martyna-Tobias-Klein barostat (56).
Mammalian expression vectors, transfections, cell viability, and immunoblot analysis
SUMO-Affimers were PCR amplified using the forward primer 5’-ATggatccGCCACCATGTATGAGCGTAAGAAGCGTCGTCAGCGTCGTAGGGCCGCTACCGGTGTTCGTG-3’ (incorporating a Tat nuclear localization sequence; lowercase - BamHI) and reverse primer 5’-ATgcggccgcTTACCCTAATGATGATGATGATGATGCTTGTCATCGTCATCTTTATAATCAGCGTCACCAACCGGTTTG -3’ (incorporating a FLAG-6xHis tag; lowercase - NotI), digested, ligated into and cloned using pcDNA5-FRT-TO (Life Technologies). DNA sequencing was used for verification. Transfections routinely used 1 μg plasmid DNA and Lipofectamine 2000 (Life Technologies) following the manufacturer’s instructions. Cell viability was determined using a colorimetric assay based on 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) and an electron coupling reagent (phenazine ethosulfate; PES) according to the manufacturer’s protocol (Promega). For immunoblot analysis, cells were washed in PBS and proteins extracted in lysis buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, 1% NP-40 and 1x protease inhibitor cocktail (Roche) and where stated, 40 mM NEM (to prevent deSUMOylation) for 15 min on ice and clarified by centrifugation at 12,000 xg for 10 min, 4°C. SDS-PAGE and immunoblotting of normalized protein concentrations followed standard techniques using the following antibodies: rabbit polyclonal antibodies recognizing the FLAG tag (1:1000; Sigma), mouse monoclonal antibodies recognizing GAPDH (1:5000; Abcam), rabbit polyclonal antibodies recognizing SUMO2/3 (Abcam), monoclonal antibodies recognizing SUMO1 (Abcam), mouse monoclonal antibodies recognizing PML (PG-M3, Santa Cruz Biotechnology), mouse monoclonal antibodies recognizing theGST tag (Sigma), and mouse monoclonal antibodies recognizing GFP tags (Clontech).
In vitro binding assays and immunoprecipitation
The S-Aff Loop mutants were constructed by splice overlap extension of two PCR products. The first PCR product extended from the coding sequence of the inner loop region and was generated using forward primer 1: 5’–TCTGGCGTTTTCTGCGTC–3’, and reverse primer 2: 5’–CACCGTCTTTAGCTTCCAGG–3’. The second PCR product was generated using forward primer 3: 5’-CCTGGAAGCTAAAGACGGTG–3’, and reverse primer 4: 5’–TACCCTAGTGGTGATGATGGTGATGC–3’. Using Phusion High Fidelity Polymerase (NEB), PCR reactions were as follows: 98°C for 30 s then 30 cycles of 98°C, 20 s; 56°C, 20 s; 72°C, 20 s followed by extension at 72°C for 10 min. PCR products were purified by gel extraction (Qiagen), used for 10 cycles of SOEing as above, then adding forward primer 1 and reverse primer 4 directly to the PCR sample for amplification of the final PCR product for 20 cycles as above. The PCR product was digested with NheI and NotI, gel extracted, ligated into and cloned using pET11 vector for the generation of 6xHis-tagged proteins. These were expressed as above, Ni-NTA agarose beads (10 μl per reaction) were saturated with S-Aff and in 500 μl reaction volumes, incubated with 20 μg GST-SUMO-1 or GST-SUMO-2 in PBS with an additional 350 mM NaCl (500 mM total) for 1 h at 4°C, followed by four washes in binding buffer and immunoblot analysis.
For pulldown experiments, HEK293T cells in 6-well plates were transfected with GFP, GFP-SUMO-1 or GFP-SUMO-2 for 48 h. Cells were lysed in 1 ml lysis buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, 1% NP-40 and 1x protease inhibitor cocktail (Roche) for 15 min on ice and clarified by centrifugation at 12,000 xg for 10 min, 4°C. Lysates were incubated with 10 µg of S1A1, S2B3 or S1S2D5 for 1 h at 4°C, followed by a further hour with 10 µl Ni-NTA agarose beads (Qiagen) equilibrated in lysis buffer. Complexes were washed three times in lysis buffer, eluted in 50 µl Laemmli buffer, boiled for 5 min and subject to immunoblot and Coomassie staining analysis.
For immunoprecipitations, HEK293T cells were plated into 6-well dishes and (co)-transfected with 1 μg plasmid for 48 h. Cells were washed in PBS and proteins extracted in 1 ml lysis buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, 1% NP-40 and 1x protease inhibitor cocktail (Roche) for 15 min on ice and clarified by centrifugation at 12,000 xg for 10 min, 4°C. Lysates were incubated with GFP-Trap (Chromotek) for coimmunoprecipitation of GFP-SUMO and FLAG-tagged S-Aff or agarose beads crosslinked with Goat antibodies recognizing the DDDDK epitope found in the FLAG tag (Abcam) for immunoprecipitation of endogenous SUMO with FLAG-tagged S-Aff and processed following the manufacturer’s recommendations with lysis and wash buffers containing 40 mM N-ethylmaleimide (NEM) to limit deSUMOylation. Immunoprecipitated proteins were eluted in Laemmli buffer and subject to immunoblot analysis.
SUMOylation and deSUMOylation assays
For in vitro SUMOylation assays, 20 µl reactions containing 120 ng GST-SAE1/2, 1 µg His-Ubc9, 2.5 µg His-SUMO2-GG, 2.5 µg GST-RanGAP1 (residues 418-587; Enzo Life Sciences) in 50 mM Tris-HCl (pH 7.6), 5 mM DTT, 5 mM MgCl2 and 2 mM ATP were incubated for 3 hours at 37°C. S-Affs (0.25 μg, 2.5 μg or 25 μg) were added prior to the addition of ATP. For negative controls, ATP was omitted. Reactions were stopped by the addition of 20 µl Laemmli buffer and analyzed by SDS-PAGE and Coomassie blue staining. DeSUMOylation assays were performed as above, for 1 h with the addition of 2.5 µg S-Aff (ca. 1:1 ratio). Afterwards, 0.5 µM SENP1 or SENP2 was added, followed by a further incubation of 1 h at 37°C. Reactions were stopped by adding Laemmli buffer and samples were analyzed by SDS-PAGE and Coomassie staining. For PML SUMOylation assays, HEK293T cells were co-transfected with FLAG-S-Aff and FLAG-PML-I for 16 h, followed by treatment with 1 μM As2O3 for 6 h. Cells were then processed for immunoblot analysis.
Immunofluorescence and quantitative immunofluorescence microscopy
Immunofluorescence experiments followed previously published protocols (57, 58) with the following modification: Cells were transfected with 1 μg FLAG-S-Aff for 16-20 h and treated with 1 μM arsenic trioxide (As2O3; As) in media for 2 h. For quantitative immunofluorescence, cells were treated with 5 μM As for the indicated times. Images were captured using an LSM700 laser scanning microscope (Carl Zeiss) and processed using ZEN imaging software (Carl Zeiss). For quantitative immunofluorescence, images were analyzed with a macro written in Fiji (59). Briefly cell nuclei from individual cells were isolated based on the distribution of DAPI in the images. These regions were then used to mask the original image and enable the quantification of individual PML-NBs. PML-NBs were identified using the Find Maxima function with a single pixel representing a single PML-NB. The numbers of pixels were then counted per nucleus. In analyses where the mean fluorescence intensity of the PML-NB was obtained the macro was extended to create a region from each PML-NB using the ‘Dilate’ function within Fiji. Results were imported as CSV files into Microsoft Excel for further analysis. Five fields of view per analysis were randomly selected each of which contained between 150-300 cells, as determined by DAPI staining. Cell numbers were determined by the number of DAPI-positive nuclei, and these values were used to normalize values between samples. The fluorescence intensities of individual PML-NBs were calculated and where total fluorescence were reported this represented the sum total fluorescence of all PML-NBs (normalized between samples according to DAPI). Error bars represent standard deviations. Antibodies included: mouse monoclonal antibodies specific for PML (PG-M3, Santa Cruz Biotechnology), rabbit polyclonal antibodies specific for the FLAG-tag (1:1000; Sigma), sheep polyclonal antibodies specific for SUMO1 (Enzo Life Sciences) and sheep polyclonal antibodies specific forSUMO2 (Enzo Life Sciences). Student’s t-tests were used to calculate statistical significance.
SUMO-targeted ubiquitin ligase (STUbL) assay
Assays were performed in 10 μl reaction volumes consisting of 50 mM Tris (pH 7.6), 5 mM DTT, 5 mM MgCl2, 40 ng E1 activating enzyme (Enzo Life Sciences), 20 ng UbcH5α (Enzo Life Sciences), 2.5 μg ubiquitin (Enzo Life Sciences), 20 ng poly-SUMO-22-8 (Boston Biochem), 2 mM ATP, 250 ng His-MBP-RNF4 and 5 mg S-Aff. Reactions were incubated at 37°C for 3 h and stopped with Laemmli buffer and analyzed by immunoblotting.
Supplementary Material
One sentence Summary.
Artificial binding proteins are tools for exploring cellular processes dependent on SUMOylation.
Editor’s Summary.
Artificial proteins target SUMO
SUMOylation is the covalent attachment of SUMO-1, SUMO-2, SUMO-3, or combinations thereof to target proteins to control protein function and localization. Hughes et al. screened a library of artificial proteins called Affimers to identify those that bound to SUMO-1 or SUMO-2, which is nearly identical to SUMO-3, and incorporated a negative selection step to remove SUMO-2–binding Affimers that also bound to SUMO-1. The authors identified Affimers that recognized SUMO-1, SUMO-2 and SUMO-3 (SUMO-2/3), or all three isoforms. Biochemical and cellular assays showed that these SUMO-specific Affimers (S-Affs) did not interfere with SUMO conjugation or deconjugation but did inhibit a cellular stress response that required SUMO-mediated protein interactions. In addition to generating S-Affs that will be useful tools for studying SUMO-dependent cellular processes, this study also shows the applicability of this technology for generating reagents that interfere with specific protein-protein interactions for basic research and potentially for clinical development.
Acknowledgements
The authors thank Dr Iain Manfield (University of Leeds) for technical assistance, Prof Jeremy Henley (University of Bristol), Prof Ron Hay and Ellis Jaffray (University of Dundee) for reagents and Dr Anja Berndt (University of Leeds) for advice and critical reading of the manuscript.
Funding: This work was funded in part by a Leeds Cancer Research UK (CRUK) Development Fund and The Wellcome Trust (ISSF) (DJH), The Wellcome Trust (089330), BBSRC (BB/K000306/1 and BB/M006557/1) (AW) and The University of Leeds through Biomedical Health Research Centre support for the Leeds BioScreening Technology Group (MJM, DCT).
Footnotes
This manuscript has been accepted for publication in Science Signaling. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencesignaling.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Author contributions: D.J.H., D.C.T and A.W. conceived the study. D.J.H., M.J.M, D.C.T and A.W. designed the experiments. D.J.H., C.T., A.A.S.T., N.P., C.H.T., U.M., K.Z.Z., G.H., J.D., E.F., D.C.T conducted the experiments. D.J.H., C.T., A.A.S.T., C.H.T., T.G., T.A.E., J.D., E.F., D.C.T and A.W. analyzed the data. D.J.H., D.C.T. and A.W. wrote the paper with input from all the authors.
Competing Interests: The Affimer technology is under a patent application filed by the University of Leeds and are commercialised through a license agreement by Avacta Life Sciences. The University of Leeds receives royalties on sales of reagents through Avacta Life Sciences, which are distributed according to University policy.
Data and materials availability: Structures deposited in Protein Data Bank (www.pdb.org) are referred to as Adhirons, and have been assigned the following identification numbers: 5ELJ (SUMO-1:S1S2D5*), 5EQL (SUMO-2:S1S2D5*) and 5ELU (SUMO-2:S2B3). *S1S2D5 is referred to as S2D5 in the Protein Data Bank. Material is available upon request.
References
- 1.Ruffner H, Bauer A, Bouwmeester T. Human protein-protein interaction networks and the value for drug discovery. Drug discovery today. 2007;12:709–716. doi: 10.1016/j.drudis.2007.07.011. published online EpubSep. [DOI] [PubMed] [Google Scholar]
- 2.Hopkins AL, Groom CR. The druggable genome. Nature reviews Drug discovery. 2002;1:727–730. doi: 10.1038/nrd892. published online EpubSep. [DOI] [PubMed] [Google Scholar]
- 3.Yeh JT, Binari R, Gocha T, Dasgupta R, Perrimon N. PAPTi: a peptide aptamer interference toolkit for perturbation of protein-protein interaction networks. Scientific reports. 2013;3:1156. doi: 10.1038/srep01156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wojcik J, Hantschel O, Grebien F, Kaupe I, Bennett KL, Barkinge J, Jones RB, Koide A, Superti-Furga G, Koide S. A potent and highly specific FN3 monobody inhibitor of the Abl SH2 domain. Nature structural & molecular biology. 2010;17:519–527. doi: 10.1038/nsmb.1793. published online EpubApr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kyle HF, Wickson KF, Stott J, Burslem GM, Breeze AL, Tiede C, Tomlinson DC, Warriner SL, Nelson A, Wilson AJ, Edwards TA. Exploration of the HIF-1alpha/p300 interface using peptide and Adhiron phage display technologies. Molecular bioSystems. 2015;11:2738–2749. doi: 10.1039/c5mb00284b. published online EpubSep 15. [DOI] [PubMed] [Google Scholar]
- 6.Kolonin MG, Finley RL., Jr Targeting cyclin-dependent kinases in Drosophila with peptide aptamers. Proc Natl Acad Sci U S A. 1998;95:14266–14271. doi: 10.1073/pnas.95.24.14266. published online EpubNov 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brauchle M, Hansen S, Caussinus E, Lenard A, Ochoa-Espinosa A, Scholz O, Sprecher SG, Pluckthun A, Affolter M. Protein interference applications in cellular and developmental biology using DARPins that recognize GFP and mCherry. Biology open. 2014;3:1252–1261. doi: 10.1242/bio.201410041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tiede C, Tang AA, Deacon SE, Mandal U, Nettleship JE, Owen RL, George SE, Harrison DJ, Owens RJ, Tomlinson DC, McPherson MJ. Adhiron: a stable and versatile peptide display scaffold for molecular recognition applications. Protein engineering, design & selection : PEDS. 2014;27:145–155. doi: 10.1093/protein/gzu007. published online EpubMay. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han JD, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, Dupuy D, Walhout AJ, Cusick ME, Roth FP, Vidal M. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. published online EpubJul 1. [DOI] [PubMed] [Google Scholar]
- 10.Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nature reviews Molecular cell biology. 2010;11:861–871. doi: 10.1038/nrm3011. published online EpubDec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Molecular cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. published online EpubMar 7. [DOI] [PubMed] [Google Scholar]
- 12.Feligioni M, Nistico R. SUMO: a (oxidative) stressed protein. Neuromolecular medicine. 2013;15 doi: 10.1007/s12017-013-8266-6. 707719. published online EpubDec. [DOI] [PubMed] [Google Scholar]
- 13.Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Science signaling. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- 14.Tempe D, Piechaczyk M, Bossis G. SUMO under stress. Biochemical Society transactions. 2008;36:874–878. doi: 10.1042/BST0360874. published online EpubOct. [DOI] [PubMed] [Google Scholar]
- 15.Everett RD, Boutell C, Hale BG. Interplay between viruses and host sumoylation pathways. Nature reviews Microbiology. 2013;11:400–411. doi: 10.1038/nrmicro3015. published online EpubJun. [DOI] [PubMed] [Google Scholar]
- 16.Perry JJ, Tainer JA, Boddy MN. A SIM-ultaneous role for SUMO and ubiquitin. Trends in biochemical sciences. 2008;33:201–208. doi: 10.1016/j.tibs.2008.02.001. published online EpubMay. [DOI] [PubMed] [Google Scholar]
- 17.Lin DY, Huang YS, Jeng JC, Kuo HY, Chang CC, Chao TT, Ho CC, Chen YC, Lin TP, Fang HI, Hung CC, et al. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Molecular cell. 2006;24:341–354. doi: 10.1016/j.molcel.2006.10.019. published online EpubNov 3. [DOI] [PubMed] [Google Scholar]
- 18.Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Molecular cell. 2006;24:331–339. doi: 10.1016/j.molcel.2006.09.013. published online EpubNov 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shih HM, Chang CC, Kuo HY, Lin DY. Daxx mediates SUMO-dependent transcriptional control and subnuclear compartmentalization. Biochemical Society transactions. 2007;35:1397–1400. doi: 10.1042/BST0351397. published online EpubDec. [DOI] [PubMed] [Google Scholar]
- 20.Tiede C, Bedford R, Heseltine SJ, Smith G, Wijetunga I, Ross R, AlQallaf D, Roberts AP, Balls A, Curd A, Hughes RE, et al. Affimer proteins are versatile and renewable affinity reagents. eLife. 2017;6 doi: 10.7554/eLife.24903. published online EpubJun 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helma J, Cardoso MC, Muyldermans S, Leonhardt H. Nanobodies and recombinant binders in cell biology. The Journal of cell biology. 2015;209:633–644. doi: 10.1083/jcb.201409074. published online EpubJun 08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. Specification of SUMO1- and SUMO2-interacting motifs. The Journal of biological chemistry. 2006;281:16117–16127. doi: 10.1074/jbc.M512757200. published online EpubJun 9. [DOI] [PubMed] [Google Scholar]
- 23.Namanja AT, Li YJ, Su Y, Wong S, Lu J, Colson LT, Wu C, Li SS, Chen Y. Insights into high affinity small ubiquitin-like modifier (SUMO) recognition by SUMO-interacting motifs (SIMs) revealed by a combination of NMR and peptide array analysis. The Journal of biological chemistry. 2012;287:3231–3240. doi: 10.1074/jbc.M111.293118. published online EpubJan 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH, Hay RT, Lamond AI, Mann M, Vertegaal AC. Vertegaal, In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol Cell Proteomics. 2008;7:132–144. doi: 10.1074/mcp.M700173-MCP200. published online EpubJan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hands KJ, Cuchet-Lourenco D, Everett RD, Hay RT. PML isoforms in response to arsenic: high-resolution analysis of PML body structure and degradation. Journal of cell science. 2014;127 doi: 10.1242/jcs.132290. 365375. published online EpubJan 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matafora V, D'Amato A, Mori S, Blasi F, Bachi A. Proteomics analysis of nucleolar SUMO-1 target proteins upon proteasome inhibition. Mol Cell Proteomics. 2009;8:2243–2255. doi: 10.1074/mcp.M900079-MCP200. published online EpubOct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westman BJ, Verheggen C, Hutten S, Lam YW, Bertrand E, Lamond AI. A proteomic screen for nucleolar SUMO targets shows SUMOylation modulates the function of Nop5/Nop58. Molecular cell. 2010;39:618–631. doi: 10.1016/j.molcel.2010.07.025. published online EpubAug 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berndt A, Wilkinson KA, Heimann MJ, Bishop P, Henley JM. In vivo characterization of the properties of SUMO1-specific monobodies. The Biochemical journal. 2013;456:385–395. doi: 10.1042/BJ20130241. published online EpubDec 15. [DOI] [PubMed] [Google Scholar]
- 29.Gilbreth RN, Truong K, Madu I, Koide A, Wojcik JB, Li NS, Piccirilli JA, Chen Y, Koide S. Isoform-specific monobody inhibitors of small ubiquitin-related modifiers engineered using structure-guided library design. Proc Natl Acad Sci U S A. 2011;108:7751–7756. doi: 10.1073/pnas.1102294108. published online EpubMay 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hay RT. Decoding the SUMO signal. Biochemical Society transactions. 2013;41:463–473. doi: 10.1042/BST20130015. published online EpubApr. [DOI] [PubMed] [Google Scholar]
- 31.Rojas-Fernandez A, Plechanovova A, Hattersley N, Jaffray E, Tatham MH, Hay RT. SUMO chain-induced dimerization activates RNF4. Molecular cell. 2014;53:880–892. doi: 10.1016/j.molcel.2014.02.031. published online EpubMar 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nature cell biology. 2008;10:538–546. doi: 10.1038/ncb1716. published online EpubMay. [DOI] [PubMed] [Google Scholar]
- 33.Colwill K, G. Renewable Protein Binder Working. Graslund S. A roadmap to generate renewable protein binders to the human proteome. Nature methods. 2011;8:551–558. doi: 10.1038/nmeth.1607. published online EpubJul. [DOI] [PubMed] [Google Scholar]
- 34.Kuo CY, Li X, Kong XQ, Luo C, Chang CC, Chung Y, Shih HM, Li KK, Ann DK. An Arginine-Rich Motif of Ring Finger Protein 4 (RNF4) Oversees the Recruitment and Degradation of the Phosphorylated and SUMOylated KAP1/TRIM28 during Genotoxic Stress. The Journal of biological chemistry. 2014 doi: 10.1074/jbc.M114.555672. published online EpubJun 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rytinki MM, Kaikkonen S, Pehkonen P, Jaaskelainen T, Palvimo JJ. PIAS proteins: pleiotropic interactors associated with SUMO. Cellular and molecular life sciences : CMLS. 2009;66:3029–3041. doi: 10.1007/s00018-009-0061-z. published online EpubSep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tammsalu T, Matic I, Jaffray EG, Ibrahim AF, Tatham MH, Hay RT. Proteome-wide identification of SUMO2 modification sites. Science signaling. 2014;7:rs2. doi: 10.1126/scisignal.2005146. published online EpubApr 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jentsch S, Psakhye I. Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annual review of genetics. 2013;47:167–186. doi: 10.1146/annurev-genet-111212-133453. [DOI] [PubMed] [Google Scholar]
- 38.Berndt A, Hofmann-Winkler H, Tavalai N, Hahn G, Stamminger T. Importance of covalent and noncovalent SUMO interactions with the major human cytomegalovirus transactivator IE2p86 for viral infection. J Virol. 2009;83:12881–12894. doi: 10.1128/JVI.01525-09. published online EpubDec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahashi H, Hatakeyama S, Saitoh H, Nakayama KI. Noncovalent SUMO-1 binding activity of thymine DNA glycosylase (TDG) is required for its SUMO-1 modification and colocalization with the promyelocytic leukemia protein. The Journal of biological chemistry. 2005;280:5611–5621. doi: 10.1074/jbc.M408130200. published online EpubFeb 18. [DOI] [PubMed] [Google Scholar]
- 40.Sahin U, Ferhi O, Jeanne M, Benhenda S, Berthier C, Jollivet F, Niwa-Kawakita M, Faklaris O, Setterblad F, de The H, Lallemand-Breitenbach V. Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins. The Journal of cell biology. 2014;204:931–945. doi: 10.1083/jcb.201305148. published online EpubMar 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ah-San Tang A, Tiede C, Hughes DJ, McPherson M, Tomlinson DC. Isolation of isoform-specific binding proteins (Affimers) by phage display using negative selection. Sci Signal. 2017;10 doi: 10.1126/scisignal.aan0868. eaan0868. [DOI] [PubMed] [Google Scholar]
- 42.Winter G, Dokel S, Jones AK, Scheerer P, Krauss N, Hohne W, Friedrich B. Crystallization and preliminary X-ray crystallographic analysis of the [NiFe]-hydrogenase maturation factor HypF1 from Ralstonia eutropha H16. Acta crystallographica Section F, Structural biology and crystallization communications. 2010;66:452–455. doi: 10.1107/S1744309110006196. published online EpubApr 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, et al. Overview of the CCP4 suite and current developments. Acta crystallographica Section D, Biological crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. published online EpubApr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. published online EpubMay 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brunger AT. Free R value: cross-validation in crystallography. Methods in enzymology. 1997;277 doi: 10.1016/s0076-6879(97)77021-6. 366396. [DOI] [PubMed] [Google Scholar]
- 46.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta crystallographica Section D, Biological crystallography. 2007;63:32–41. doi: 10.1107/S0907444906045975. published online EpubJan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knipscheer P, van Dijk WJ, Olsen JV, Mann M, Sixma TK. Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. The EMBO journal. 2007;26:2797–2807. doi: 10.1038/sj.emboj.7601711. published online EpubJun 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta crystallographica Section D, Biological crystallography. 2011;67:355–367. doi: 10.1107/S0907444911001314. published online EpubApr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. published online EpubApr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta crystallographica Section D, Biological crystallography. 2006;62:439–450. doi: 10.1107/S0907444906005270. published online EpubApr. [DOI] [PubMed] [Google Scholar]
- 51.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica Section D, Biological crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. published online EpubJan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Powers N, Jensen JH. Chemically accurate protein structures: validation of protein NMR structures by comparison of measured and predicted pKa values. Journal of biomolecular NMR. 2006;35:39–51. doi: 10.1007/s10858-006-9003-3. published online EpubMay. [DOI] [PubMed] [Google Scholar]
- 53.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 54.Harder E, Damm W, Maple J, Wu C, Reboul M, Xiang JY, Wang L, Lupyan D, Dahlgren MK, Knight JL, Kaus JW, et al. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. Journal of chemical theory and computation. 2015 doi: 10.1021/acs.jctc.5b00864. published online EpubNov 17. [DOI] [PubMed] [Google Scholar]
- 55.Hoover WG. Canonical dynamics: Equilibrium phase-space distributions. Physical review A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. published online EpubMar. [DOI] [PubMed] [Google Scholar]
- 56.Martyna GJ, Tobias DJ, Klein ML. Constant pressure molecular dynamics algorithms. The Journal of Chemical Physics. 1994;101:4177–4189. doi: 10.1063/1.467468. [DOI] [Google Scholar]
- 57.Baquero-Perez B, Whitehouse A. Hsp70 Isoforms Are Essential for the Formation of Kaposi's Sarcoma-Associated Herpesvirus Replication and Transcription Compartments. PLoS pathogens. 2015;11:e1005274. doi: 10.1371/journal.ppat.1005274. published online EpubNov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hughes DJ, Wood JJ, Jackson BR, Baquero-Perez B, Whitehouse A. NEDDylation Is Essential for Kaposi's Sarcoma-Associated Herpesvirus Latency and Lytic Reactivation and Represents a Novel Anti-KSHV Target. PLoS pathogens. 2015;11:e1004771. doi: 10.1371/journal.ppat.1004771. published online EpubMar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, et al. Fiji: an open-source platform for biological-image analysis. Nature methods. 2012;9 doi: 10.1038/nmeth.2019. 676682. published online EpubJul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Engh RA, Huber R. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallographica Section A. 1991;47(4):392–400. [Google Scholar]
- 61.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica Section D, Biological crystallography. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.