Abstract
Glioblastoma (GBM) is an incurable cancer, with mean post-diagnosis survival time of 14-16 months. Metagenomic analysis by The Cancer Genome Atlas (TCGA) program has identified microRNA-182-5p (miR-182-5p or miR-182) as the only miRNA associated with favorable disease prognosis and temozolomide (TMZ) susceptibility. Previous reports have indicated that miR-182 down regulates expression of BCL2L12, c-MET, and HIF2A. However, other messenger RNA (mRNA) targets of miR-182 have not been validated which would explain its association with a favorable disease prognosis. In situ analysis revealed that protein phosphatase 1 regulatory inhibitor subunit 1C (PPP1R1C) is a putative target of miR-182. PPP1R1C protein and RNA expression as assessed by tissue microarray and quantitative real time PCR, respectively, was inversely correlated to miR-182 expression in glioblastoma patients and in the metastatic glioblastoma cell line U87-MG. Reporter assays using PPP1R1C 3′ untranslated region (UTR) showed that miR-182 can interact with the wild-type but not a miR-182-5-seed mutant. Ectopic expression of miR-182 mimic in the U87-MG cell line significantly decreased proliferation as well as suppressed in vitro migration and invasion. Opposite observations were made when the non-malignant neuronal cell line HCN-2 was transfected with anti-miR-182 antagomir. The miR-182 mimic or siRNA targeting PPP1R1C induced TMZ susceptibility indicating that decreased susceptibility to TMZ in GBM patients might be attributed to high expression of PPP1R1C. Inverse correlation of PPP1R1C mRNA and miR-182 levels in 20 GBM patients confirmed the same. Cumulatively, our results indicate that loss of miR-182 leads to increased expression of PPP1R1C which in part explain disease progression and resistance to TMZ therapy.
Keywords: MicroRNA-182, glioblastoma, PPP1R1C, protein phosphatase 1 regulatory inhibitor subunit 1C
INTRODUCTION
MicroRNAs (miRNAs) are a group of endogenous, small noncoding RNA. MiRNAs have been well characterized and are known to functionally regulate diverse cellular and pathogenic processes. MiRNAs can either bind or degrade their target mRNA or inhibit their translation; the decision to degrade mRNA or inhibit translation being determined by the degree of complementarity to the target transcript’s sequence [1]. More importantly, miRNAs have been shown to function both as tumor suppressors or oncogenes [2], even though the precise context dependent ques that make them function like this are currently unknown. Given their important role in tumorigenesis per se, a lot of research has focused on the role of miRNAs in glioblastoma (GBM).
GBM is one of the most debilitating cancers with mean survival time around 14-16 months post-diagnosis [3]. Of 470 miRNAs profiled by The Cancer Genome Atlas (TCGA) program, only miR-182 expression level has been shown to be associated with better prognosis, suggesting miR-182 might function as a tumor suppressor in the case of GBM [4]. It has been shown that miR-182 targets BCL2L12, c-MET and HIF2A in GBM and thus integrates growth, apoptosis, and differentiation programs in GBM [5]. However, our understanding of the expression and function of miRNA-182 in GBM pathogenesis is still limited. It is thus imperative to develop a more complete picture about how suppression of miR-182 observed in GBM potentiates the disease.
Our in situ analysis and subsequent experiments revealed that protein phosphatase 1 regulatory inhibitor subunit 1C (PPP1R1C) is a target of miR-182 in GBM and up regulation of PPP1R1C in GBM patients is partially responsible for observed resistance to temozolomide (TMZ) susceptibility.
RESULTS
Differential expression of miR-182 and PPP1R1C in glioblastoma cell lines
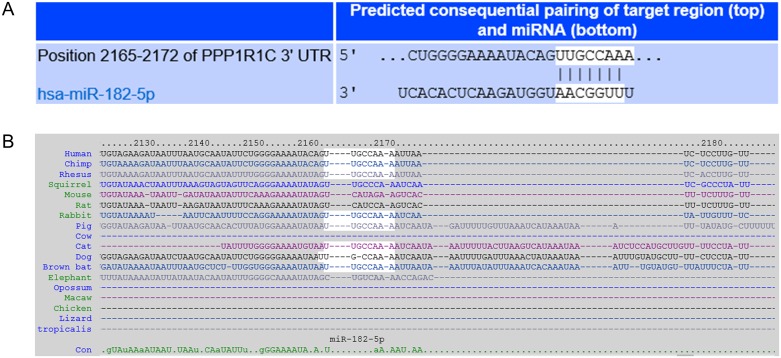
To explore the mechanism of miR-182-mediated regulation in GBM, we used two independent algorithms to in situ predict the targets for miR-182, TargetScan and microCosm. Among the predicted genes, were the previously validated Bcl2L12, c-Met and HIF2A [5]. In addition, one of the putative targets identified by both software was PPP1R1C (encoding the protein phosphatase 1 regulatory inhibitor subunit 1C) or protein phosphatase-1 (PP1) (Figure 1A and data not shown). The miR-182 seed region in the 3′UTR of PPP1R1C was conserved across different species (Figure 1B). PP-1 has been previously indicated to promote tumor growth in cervical cancer [6, 7]. However, no previous knowledge is available of PP-1 being targeted by miRNAs, hence we decided to pursue it further.
Figure 1. Prediction of PPP1R1C as a target of miR-182.
(A) Complementary 7mer-m8 seed match between miR-182 and the 3′ UTR of PPP1R1C as predicted by TargetScan software. (B) Conservation of the 7mer-m8 seed of miR-182 in the 3′UTR of PPP1R1C in indicated organisms.
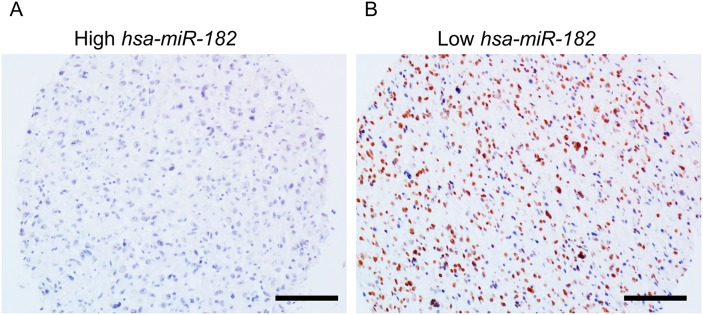
We next determined miR-182 levels in tumor tissue specimens obtained from 20 GBM patients by qRT-PCR. Representative sections with low (17/20) and high (3/20) relative miR-182 expression, compared to matched non-malignant tissue, were evaluated for PPP1R1C expression by immunohistochemistry. As shown in Figure 2, tissue specimens with high miR-182 expression were negative for PPP1R1C staining (Figure 2A), whereas those with low miR-182 expression had robust PPP1R1C staining (Figure 2B). Of note, since the number of patients with high miR-182 expression were too few for inclusion in analysis to calculate statistical significance of the observed inverse correlation between miR-182 and PPP1R1C expression.
Figure 2. PPP1R1C and miR-182 are inversely correlated in patients with GBM.
Representative immunohistochemistry images showing PPP1R1C expression in glioblastoma tissue with differential expression levels of miR-182 expression as determined by qRT-PCR. Brown staining represents positive PPP1R1C expression. Scale bar – 40 μm.
PPP1R1C is a bona-fide target of miR-182
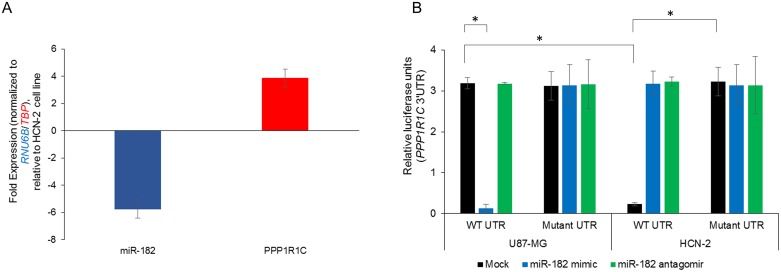
We next determined if PPP1R1C is a bona fide target of miR-182. We initially evaluated relative miR-182 and PPP1R1C expressions in the glioblastoma cell line U87-MG and the non-malignant HCN-2 cell line. U87-MG cell line had a significantly suppressed miR-182 expression (−5.76 ± 0.42 folds) and significantly increased PPP1R1C expression (+3.86 ± 0.67 folds) compared to the HCN-2 cell line (p<0.05 in each case) (Figure 3A). To test putative interaction between the 3′UTR of PPP1R1C and miR-182, luciferase reporter constructs containing the wild-type PPP1R1C 3′UTR were transfected in HCN-2 and U87-MG cells alone or in combination with miR-182 mimic or miR-182 antagomir (Figure 3B). Expression of the wild-type PPP1R1C 3′UTR containing reporter were inhibited 2.89 ± 0.03 folds (P = 0.0003) in HCN-2 cells compared with the U87-MG cells. Expression of the wild-type PPP1R1C 3′UTR containing reporter were inhibited 3.2 ± 0.05 folds (P = 0.004) in U87-MG cells transfected with the miR-182 mimic in compared to the no-mimic control. Expression of the wild-type PPP1R1C 3′UTR containing reporter was restored in HCN-2 cells transfected with miR-182 antagomir (3.43 ± 0.35 folds (P = 0.007) compared to the no-antagomir control. To confirm that the effects observed was due to miR-182 targeting the PPP1R1C 3′UTR, we generated and tested a miR-182 binding mutant of the PPP1R1C 3′UTR reporter (nucleotides 2165-2172 corresponding to miR-182 binding site). Whereas, the miR-182 binding mutant PPP1R1C reporter did not show any difference in relative luciferase activity between mock and miR-182 mimic transfected in U87-MG cells (p>0.05), the repression observed with wild-type reporter was completely ablated following mutation in the HCN-2 cell line (Figure 3B), cumulatively confirming that PPP1R1C mRNA is a bona-fide target of miR-182 in these cells.
Figure 3. PPP1R1C is a bona-fide target of miR-182 in the glioblastoma cell line U87-MG.
(A) Steady state expression of miR-182 and PPP1R1C in U87-MG and HCN-2 cell lines were determined. Data was normalized to RNU6B and TBP expression, respectively. Fold expression in U87-MG cells was determined relative to expression in HCN-2 cell line. (B) Relative luciferase activity of transiently transfected luciferase reporter constructs containing either full-length or mutated (miR-182 binding site deleted) PPP1R1C 3′ UTR in U87-MG and HCN-2 cells, either mock transfected or transfected with miR-182 mimic or miR-182 antagomir. *p<0.05. Data in ‘A’ and ‘B’ represent at least three independent experiments, each done in triplicate.
Effect of differential PPP1R1C expression on malignant behavior of glioblastoma cells
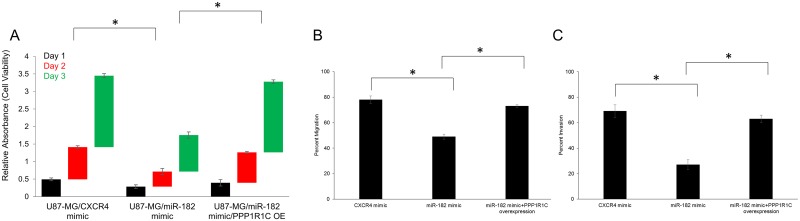
To determine whether PPP1R1C expression affects cell proliferation in GBM, cell viability assays were carried out with U87-MG cells, transfected with either CXCR4 mimic or miR-182 mimic. Transfection with miR-182 mimic suppressed cell proliferation after day 1 (CXCR vs miR-182 mimic – 0.49 ± 0.04 vs 0.29 ± 0.05, p<0.05), day 2 (CXCR vs miR-182 mimic – 0.92 ± 0.04 vs 0.43 ± 0.09, p<0.05), and day 3 (CXCR vs miR-182 mimic – 2.04 ± 0.06 vs 1.04 ± 0.09, p<0.05) (Figure 4A). MiR-182 have other targets like BCL2L12 and HIF2A, modifications of which can result in the same observation. Hence, to confirm that the observed effects on cell proliferation were due to PPP1R1C, we rescued PPP1R1C expression by overexpressing the coding sequence of PPP1R1C in U87-MG cells transfected with the miR-182 mimic. Overexpression of PPP1R1C significantly rescued the suppression of cell proliferation (day 1: 0.39 ± 0.04; day 2: 0.87 ± 0.04; day 3: 2.01 ± 0.06) (p<0.05) (Figure 4A). Our results thus confirmed that PPP1R1C potentiates cell proliferation in GBM.
Figure 4. PPP1R1C expression levels dictate cell viability, migration and invasion abilities in GBM cells.
(A) Cell viability was measured in U87-MG cells transfected with CXCR4 mimic, miR-182 mimic, or miR-182 mimic along with PPP1R1C overexpression (OE) at 24, 48, and 72 hours after transfection by the MTT assay. (B, C) Modulation of miR-182 changes cell migration and invasion abilities in the U87-MG cells. The migrated and invasive cells were photographed using a microscope, and the number of the migrated and invasive cells in every field was counted and represented as percent of total cells at the beginning of the assay. Error bars, S.D. *p<0.05. In each panel, data represent at least three independent experiments, each done in triplicate.
We scored each of the individual transfectants U87-MG cells (CXCR4 mimic, miR-182 mimic, and miR-182 mimic with PPP1R1C overexpression (OE)), for migration (Figure 4B) and invasion (Figure 4C) in standard transwell assays. Using these criteria, phase contrast imaging and quantification showed that overexpression of miR-182 inhibited in vitro migration (CXCR vs miR-182 mimic – 78% ± 3% vs 49% ± 2%, p<0.05), and invasion (CXCR vs miR-182 mimic – 69% ± 5% vs 27% ± 4%, p<0.05). Overexpression of PPP1R1C significantly rescued the migration (73% ± 4%) and invasive (63% ± 6%) potential of U87-MG cells transfected with the miR-182 mimic (p<0.05). Our results suggested that PPP1R1C potentiates in vitro migration and invasion in GBM, which occurs by repression of miR-182 expression. Cumulatively, the profound repression in relative expression of miR-182 and increased expression of PPP1R1C in GBM tissue or cell line along with its capacity to impinge in vitro migration and invasion suggested that it may drive tumorigenesis and metastatic progression in GBM [8].
MiR-182 replenishment or knock-down of PPP1R1C increases chemosensitivity of glioblastoma cells
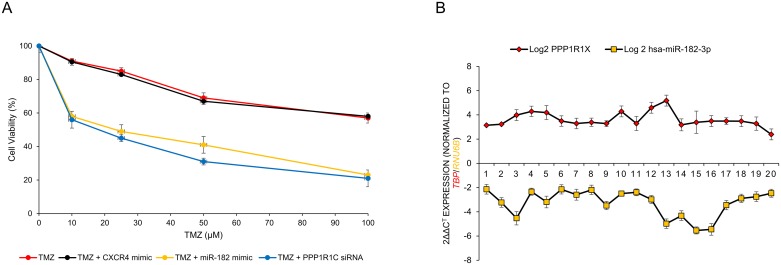
To determine the therapeutic potential of miR-182 and PPP1R1C as a chemosensitizer, we evaluated the effect of miR-182 mimic-mediated replenishment and PPP1R1C siRNA-mediated depletion on the cytotoxicity of TMZ on U87-MG cells. Transfection of miR-182 mimic significantly chemosensitized U87-MG cells (IC50 from 69 ± 3 μM in TMZ alone to 41 ± 1 μM in TMZ+miR-182 mimic, p<0.05). The PPP1R1C siRNA chemosensitized the cells even more (IC50 from 69 ± 3 μM in TMZ alone to 31 ± 3 μM in TMZ+PPP1R1C siRNA, p<0.05) (Figure 5A). Cumulatively, these results indicate that miR-182 replenishment or PPP1R1C depletion sensitizes GBM cells to the cytotoxicity of TMZ.
Figure 5. MiR-182 and PPP1R1C levels are inversely correlated in glioblastoma patients and their modulation can be used to increase sensitivity to temozolomide (TMZ).
(A) U87-MG cells were either untransfected or transiently transfected with either CXCR4 mimic, miR-182 mimic, or siRNA against PPP1R1C for 12 hours. The cells were then treated with indicated doses of TMZ for 72 hours. Cell viability was assessed by the MTT assay. Each experiment was carried out at least 3 times. Data represent mean ± SEM. Data represent at least three independent experiments, each done in triplicate. (B) MiR-182 and PPP1R1C mRNA are inversely correlated in patients with glioblastoma. Pearson correlation demonstrating the inverse relation between miR-182 and PPP1R1C in paired samples (P < 0.05, Pearson correlation r = −0.8761). Data represent at least three independent experiments, each done in triplicate.
Given that our experiments indicated that PPP1R1C is a bona-fide target of miR-182, we hypothesized that suppression of miR-182 expression might be an underlying feature of GBM pathogenesis. We determined miR-182 and PPP1R1C expression in 20 GBM patients. Our results indicated a dynamic and inverse correlation between down-regulation in the levels of miR-182 and the observed increase in the PPP1R1C in GBM tissue specimens (Figure 5B) (P <.005, Pearson correlation r = −0.8761).
DISCUSSION
In the current study, our experimental results show that miRNA-182 and PPP1R1C is downregulated and upregulated, respectively, in human GBM tissues. MiRNAs are evolutionarily conserved 21-23 nucleotides RNAs that regulate post-transcriptional gene expression either by blocking translation or degrading target messenger RNAs (mRNAs) and have been increasingly shown to function as tumor suppressors or oncogenes [9, 10]. MiRNAs can function in both normal and transformed cells and have even been shown to play a role in metastasis [11–14].
It has been recently shown that the gene repressors, HDAC1 and HDAC2, became recruited to the promoter of miR-182 and represses it in acute myeloid leukemia [15]. In addition, it was shown that HDAC inhibitors could de-repress miR-182 expression which increased sensitivity of AML cells to sapacitabine treatment [15–16]. Given that miR-182 has now been shown to target Bcl2L12, c-Met and HIF2A in GBM [5] and PPP1R1C, also in GBM, in this study, it would be interesting to evaluate HDAC inhibitors in GBM treatment. An alternative will be a combination treatment with TMZ and HDAC inhibitors to potentiate optimal treatment outcome.
Repression of PPP1R1C by using miRNA-182 mimic inhibited cell proliferation, as well as migration and invasion. Finally, modulating miR-182 or PPP1R1C levels increased sensitivity of GBM to TMZ treatment. Cumulatively, this highlights miR-182 and PPP1R1C as potential biomarkers in GBM. It will be clinically relevant and interesting to determine if exosome-mediated delivery of miR-182 will be able to restore miR-182 anti-oncogenic function in GBM cells, and whether such a design can potentially substitute or better currently utilized TMZ therapy in GBM patients.
It will be interesting to investigate in future research endeavors how miR-182 and PPP1R1C expression varies and correlates to disease progression in GBM patients that had either surgical resection, radiation therapy, or adjuvant chemotherapy. Not a lot is known about function of PPP1R1C in normal brain tissue or in cancer. There has been one finding of a CREB-PPP1R1C gene fusion associated with enrichment of insulin signaling pathway genes in breast cancer [17]. RNA-seq analysis in GBM cells overexpressing PPP1R1C will provide important indicators about function of PPP1R1C in GBM pathogenesis.
It will also be of potential interest to study if miR-182 mediated control of PPP1R1C is also functionally important in cervical cancer pathogenesis [6, 7]. Conversely, it will be important to verify if other known targets of miR-182, like FOXO1 in prostate cancer, are also deregulated in GBM. A complete understanding of miR-182 mediated regulation might emerge from experiments looking at gene expression following overexpression and knockdown of miR-182 in GBM cells.
MATERIALS AND METHODS
Tissue samples, processing, and ethical considerations
Fresh-frozen and paraffin-embedded GBM tissue specimens and corresponding adjacent non-malignant brain tissue samples were obtained from 20 Chinese patients at the Second Hospital of Hebei Medical University between 2014 and 2015. All cases were included post review by pathologist and only where complete clinical pathology and follow-up data was available. None of the 20 included patients underwent pre-operative local or systemic treatment. The study protocol was approved by the Institutional Review Board of the Second Hospital of Hebei Medical University. Freshly harvested samples were immersed in RNAlater (Life Technologies, Shanghai, China) before snap freezing within 30 minutes post-surgery. All tissue samples were stored in liquid nitrogen until further use.
Cell culture
The non-malignant encephalitis cell line HCN-2 and the glioblastoma cell line U87-MG line were obtained from ATCC (Beijing, China) and cultured in DMEM medium, containing 10% FBS (Lonza, Germany), and 100 U/mL penicillin and 0.1 mg/mL streptomycin. Cells were maintained at 37°C under a humidified atmosphere of 5% carbon dioxide.
RNA and miRNA extraction and quantitative real time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from cultured cells and tumor tissues using Trizol reagent. First strand cDNA was synthesized using the RevertAid™ First Strand cDNA synthesis Kit (Life Technologies, Shanghai, China), which was then used for real-time polymerase chain reaction (PCR) using TaqMan Gene Expression probes (Life Technologies, Shanghai, China). TBP (TaqMan Assay ID: Hs00427620_m1) was used as an internal control for assessing PPP1R1C (TaqMan Assay ID: Hs00976529_m1) transcript level. Data was normalized to TBP expression and analyzed by the -ΔΔCt method. According to the manufacturer’s instructions, miRNA from tissues and cells was extracted using the mirVana miRNA isolation kit (Life Technologies, Shanghai, China), and the expression levels of hsa-miR-182 and RNU6B were detected by TaqMan miRNA assays (Life technologies, Shanghai, China, TaqMan Assay IDs: 002334 and 001093, respectively). Data was normalized to RNU6B expression and analyzed by the -ΔΔCt method.
Gene construction
The PPP1R1C 3′ UTR clone in pMirTarget was obtained from Origene. The PPP1R1C 3′ UTR reporter was constructed by amplifying the endogenous PPP1R1C 3′ UTR from the Origene. XhoI and ApaI sites were added to the 5′ and 3′ ends of the fragment during the preceding PCR reaction and cloned into the XhoI and ApaI site on the Rr-luc-6XCXCR4 Renilla luciferase vector (Addgene). To make the PPP1R1C 3′UTR mutant construct, site-directed mutagenesis was used to delete 2165-2172 region, corresponding to the hsa-miR-182 binding site. A firefly luciferase vector was used as transfection and normalization control in all luciferase assays. The PPP1R1C plasmid encoding the coding sequence was obtained from BioClone Inc. (USA). Constructs were sequence verified before being used in experiments.
Transfection and luciferase assays
HCN-2 and U87-MG cells (4 × 104) were transiently transfected using Lipofectamine LTX (Life Technologies, Shanghai, China) as per the manufacturer’s instructions. Where indicated, cells were transfected with CXCR4 or miR-182 mimic or antagomir (Life Technologies, Shanghai, China) along with the PPP1R1C 3′UTR constructs. In indicated cases, the plasmid encoding the coding sequence of PPP1R1C was co-transfected with the miR-182 mimic in the U87-MG cells. Ninety-six hours after transfection, the renilla and firefly luciferase activities were measured consecutively using Dual-luciferase reporter assay system (Promega, Madison, Wisconsin, USA) as per manufacturer’s protocol. Each reporter plasmid was transfected at least twice in triplicate. Post-normalization the data was represented as relative fluorescence units (RFU) ± standard deviation (SD).
Cell proliferation assay
Cell proliferation in indicated cells was quantitated using a mitochondrial colorimetric assay (MTT assay, Sigma-Aldrich, St. Louis, MO, USA) as per the manufacturer’s recommendations. Results were expressed in terms of relative optical density (OD), as mean ± standard deviation.
In vitro transwell migration and invasion assays
Migration and invasion analysis in indicated cells was done using Culturex 96 well cell migration and Culturex 96 well BME cell invasion assay kits following the manufacturer’s recommendations (R&D Systems, Shanghai, China), respectively. Data obtained from both sets of experiments were used to analyze percent migration and invasion and were expressed as mean ± standard deviation.
Immunohistochemistry
This part of the study was approved by the Institutional Review Board of the Second Hospital of Hebei Medical University. Brain tissue specimens obtained from 20 patients with glioblastoma were first evaluated for miR-182 expression by qRT-PCR as described above. Tissue specimens representative of low and high miR-182 expression were stained for PPP1R1C expression (Abcam, Waltham, MA, USA; Catalogue ab111224). The stained slides were scored by a pathologist as positive and negative staining blinded to the identity of the tissue cores.
Drug treatment
U87-MG cells were either not transfected or transfected with either of CXCR4 mimic, miR-182 mimic, or siRNA targeting PPP1R1C (Silencer Select, Life Technologies, Shanghai, China) as described above. Twelve hours after transfection, the cells were subjected to treatment with indicated concentrations of temozolomide (TMZ) (Sigma Aldrich, Shanghai, China) for 72 hours. Following treatment cell viability was measured by the MTT assay as described above.
Statistical analyses
Statistical analyses were performed using SPSS version 20.0 (IBM Corporation, NY, USA). Two-sided P-values < 0.05 were considered statistically significant.
Abbreviations
- GBM
Glioblastoma
- TCGA
The Cancer Genome Atlas
- TMZ
temozolomide
- PPP1R1C
protein phosphatase 1 regulatory inhibitor subunit 1C
- UTR
untranslated region
- PP1
protein phosphatase-1
- PCR
polymerase chain reaction
- RFU
relative fluorescence units
- SD
standard deviation
- OD
optical density.
Footnotes
Author contributions
Liqiang Liu and Xiaowei Zhang conceived and designed the experiments; Chengrui Nan and Zongmao Zhao performed the experiments; Shucheng Ma analyzed the data; Wenhua Li contributed samples; Hongchao Hu and Zaohui Liang wrote the paper; all authors read and approved the final manuscript.
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.
FUNDING
This study was supported by the Key Project of Medical Science Research in Hebei Province (ZL20140147), and The Second Hospital of Hebei Medical University Research Fund Project (2h2201425).
REFERENCES
- 1.Hu S, Wilson KD, Ghosh Z, Han L, Wang Y, Lan F, Ransohoff KJ, Burridge P, Wu JC. MicroRNA-302 increases reprogramming efficiency via repression of NR2F2. Stem Cells (Dayton, Ohio) 2013;31:259–268. doi: 10.1002/stem.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 4.Kouri FM, Ritner C, Stegh AH. miRNA-182 and the regulation of the glioblastoma phenotype-toward miRNA-based precision therapeutics. Cell Cycle. 2015;14:3794–3800. doi: 10.1080/15384101.2015.1093711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kouri FM, Hurley LA, Daniel WL, Day ES, Hua Y, Hao L, Peng CY, Merkel TJ, Queisser MA, Ritner C. miR-182 integrates apoptosis, growth, and differentiation programs in glioblastoma. Genes Dev. 2015;29:732–745. doi: 10.1101/gad.257394.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng Q, Huang Y, Zeng L, Huang Y, Cai D, Zhang H. IPP5, a novel inhibitor of protein phosphatase 1, suppresses tumor growth and progression of cervical carcinoma cells by inducing G2/M arrest. Cancer Genet. 2012;205:442–452. doi: 10.1016/j.cancergen.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Zeng QY, Huang Y, Zeng LJ, Huang M, Huang YQ, Zhu QF. Sensitization of cervical carcinoma cells to paclitaxel by an IPP5 active mutant. Asian Pac J Cancer Prev. 2014;15:8337–8343. doi: 10.7314/apjcp.2014.15.19.8337. [DOI] [PubMed] [Google Scholar]
- 8.Huang Z, Cheng L, Guryanova OA, Wu Q, Bao S. Cancer stem cells in glioblastoma-molecular signaling and therapeutic targeting. Protein Cell. 2010;1:638–655. doi: 10.1007/s13238-010-0078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 11.Aleckovic M, Kang Y. Regulation of cancer metastasis by cell-free miRNAs. Biochim Biophys Acta. 2015;1855:24–42. doi: 10.1016/j.bbcan.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR, Israel MA. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 13.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 14.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 15.Lai TH, Ewald B, Zecevic A, Liu C, Sulda M, Papaioannou D, Garzon R, Blachly JS, Plunkett W, Sampath D. HDAC inhibition induces microRNA-182 which targets Rad51 and impairs HR repair to sensitize cells to sapacitabine in acute myelogenous leukemia. Clin Cancer Res. 2016;22:3537–3549. doi: 10.1158/1078-0432.CCR-15-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallis CJ, Gordanpour A, Bendavid JS, Sugar L, Nam RK, Seth A. MiR-182 is associated with growth, migration and invasion in prostate cancer via suppression of FOXO1. J Cancer. 2015;6:1295–1305. doi: 10.7150/jca.13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varadan V, Agrawal V, Kamalakaran S, Banerjee N, Miskimen K, Vadodkar A, Abu-Khalaf M, Sikov W, Harris LN, Dimitrova N. Heterogeneous gene fusions detected by RNASeq show enrichment of insulin signaling pathway genes in breast cancer. Cancer Res. 2013;73:04–07. [Google Scholar]