Abstract
The purpose of this study was to explore the relationship between SF3B1 mutations and the prognoses of patients with breast cancer. Clinical and SF3B1 mutation data from The Cancer Genome Atlas were analyzed. SF3B1 mutations were evaluated as prognostic factors in all breast cancer patients and specific subgroups through Cox regression and Kaplan-Meier analyses. We also investigated the relationship between traditional parameters and SF3B1 mutations. Receiver operating characteristics curves were used to analyze common risk factors for their sensitivity and specificity in predicting SF3B1 mutations. SF3B1 mutations were a poor prognostic factor in luminal B and progesterone receptor (PR)-negative breast cancer (P < 0.01). Age at diagnosis and estrogen receptor (ER) status were associated with SF3B1 mutations in all breast cancers (P < 0.01) and in luminal B and PR-negative subgroups (P < 0.01). The age at diagnosis and ER status combined had a higher sensitivity and specificity for predicting SF3B1 mutations than each factor alone. SF3B1 mutations are a poor prognostic factor in luminal B and PR-negative breast cancer patients. These mutations are significantly associated with age at diagnosis and ER status. SF3B1 mutations may therefore be a novel therapeutic target for breast cancer patients.
Keywords: SF3B1 mutation, luminal B, progesterone receptor-negative (PR-negative), breast cancer, prognostic parameters
INTRODUCTION
The precise excision of introns from precursor mRNAs in eukaryotes is performed by the spliceosome [1], a macromolecule composed of small nuclear RNAs associated with proteins [2]. RNA splicing, which includes constitutive and alternative splicing, is a post-transcriptional process necessary to produce mature RNA [3]. Constitutive splicing is the process of removing introns from pre-mRNA, whereas alternative splicing is the process of including or excluding exons in different combinations to create a diverse array of mRNA transcripts from a single pre-mRNA fragment. SF3b is a heptameric protein complex of the U2 small nuclear ribonucleoprotein that is essential for pre-mRNA splicing. Mutations in the largest SF3b subunit, SF3B1/SF3B155, are linked to cancer and lead to alternative branch site selection [4, 5]. The SF3B1 gene encodes subunit 1 of the splicing factor 3b, which is important for anchoring the spliceosome to the precursor mRNA [2], and is the most commonly mutated gene found in myelodysplastic syndrome [6]. The frequency of SF3B1 mutations is particularly high among the unique subtypes of myelodysplastic syndrome that are characterized by increased ring sideroblasts, in which mutation frequencies of 66.7–79% have been reported [2, 7, 8]. SF3B1 knockout mice are embryonic lethal at very early developmental stages, whereas SF3B1 heterozygous knockout mice (SF3B1+/−) exhibit mild skeletal alterations [9]. SF3B1 was found to be the second most frequently mutated gene in chronic lymphocytic leukemia (CLL) at 5–15%; SF3B1 mutations are less common in the early stages of CLL and become more prevalent in advanced disease where they tend to be associated with poor prognosis. The K700E mutation accounts for more than 50% of the variants observed, and additional codons 666, 662, and 625 were found to be hot spots for mutation [2, 10]. In addition to hematological malignancies, lower frequencies of SF3B1 mutation are also found in solid tumors such as breast cancers (1.8%), pancreatic carcinoma (4%), uveal melanoma (9.7%), and endometrial cancers (percentages not reported) [9]. Patients with uveal melanoma who harbor SF3B1 mutations are reported to have better prognoses [11, 12].
Human breast cancers are heterogeneous, and patients have varying clinical outcomes based on their diagnostic and prognostic parameters. These include morphological assessment, basal-like phenotype, and the expression statuses of estrogen receptor (ER), human epidermal growth factor receptor 2 (HER2), and progesterone receptor (PR). [13–15]. The genomic landscape of breast cancer is complex, and somatic mutations related to this disease have been extensively characterized.
RNA splicing dysfunction may be associated with the pathogenesis of breast cancer, as Maguire et al. revealed that mutations in spliceosomal component genes occur in 5.6% of breast cancers. Mutation of SF3B1 in spliceosomal component genes was the most common in breast cancers, and was detected in approximately 1.8% of cases. SF3B1 hotspot mutation K700E was detected in 16% and 6% of papillary and mucinous breast cancers, respectively [11]. These SF3B1 K700E mutations could lead to differential splicing. Alternative splicing of genes has also been shown to be associated with SF3B1 mutations in breast cancer, such as TMEM14C, RPL31, CRNDE, DYNLL1, MZB1, ICA1, RPL24, MTERFD3, OBSL1, ABCC5, UQCC, GUSBP11, ANKHD1, ADAM12, F8, and GAS8 [11, 12]. Pereira et al. also showed that recurrent K700E mutations in SF3B1 are associated with differential splicing activity in breast cancer; they found that patients presenting with mutations in SF3B1 tended to be older [14]. We performed this study to comprehensively investigate the association between SF3B1 mutations and prognoses in breast cancer patients.
RESULTS
Frequency of SF3B1 mutation in carcinoma
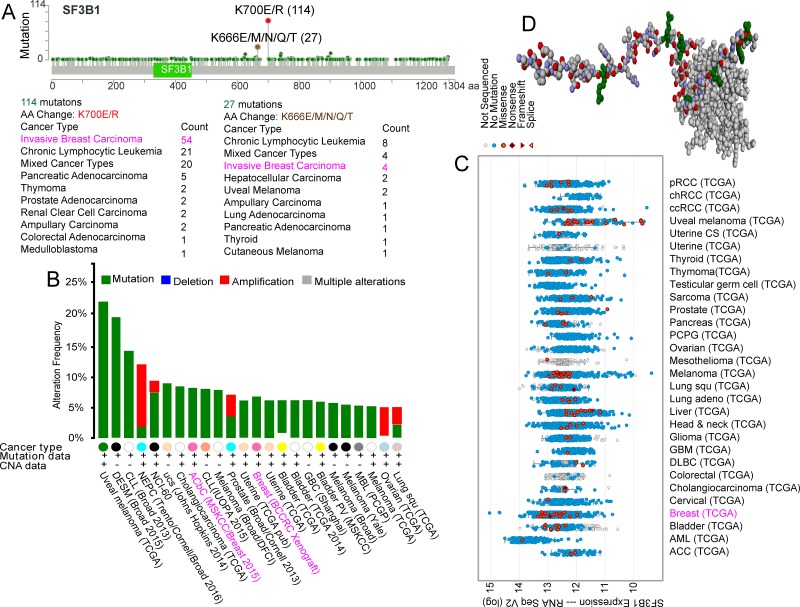
Analysis of the cBioPortal for Cancer Genomics (www.cbioportal.org/) revealed 114 SF3B1 K700E/R hotspot mutations across different types of carcinoma, 54 of which were in invasive breast carcinoma. Four patients with invasive breast carcinoma among 27 with various carcinoma types carried the SF3B1 K666E/M/N/Q/T hotspot mutation (Figure 1A). In terms of mutation frequency among different carcinomas, we found that the SF3B1 mutation ranged between 5% and 10% in breast cancer (Figure 1B). SF3B1 expression was demonstrated in different TCGA carcinoma study groups (Figure 1C). The spliceosomal protein SF3b155 structure is shown in Figure 1D [1, 4].
Figure 1. Frequency of SF3B1 mutations in carcinoma.
SF3B1 K700E/R and K666E/M/N/Q/T hotspot mutations (A), SF3B1 mutations frequency (B), and SF3B1 expression with mutations (C) in different types of carcinoma. Shown is the structure of the SF3b155 peptide complex derived from the cBioPortal for Cancer Genomics (D). [1, 4].
Clinical characteristic and prognostic factors
The median age of breast cancer patients at diagnosis was 60.09 years (range, 22–96 years). There were 2061 and 1221 patients < 65 years and ≥ 65 years, respectively. The Nottingham prognostic index (N-Index) was only found in the METABRIC data sets. The N-Index was < 4.05 in 1075 patients and ≥ 4.05 in 910 patients. Eighty-one of 3817 patients (2.12%) carried SF3B1 mutations. The clinical characteristics of the breast cancer patients are shown in Table 1. Cox univariate analysis showed that the prognostic factors significantly associated with overall survival (OS) were age, N-Index, ER status, PR status, HER2 status, menopausal status, PAM-50 and claudin-low subtype, neoplasm histologic grade, breast cancer type, and tumor stage. However, SF3B1 mutations and breast cancer laterality were not associated with OS.
Table 1. Clinical characteristics of breast cancer patients and their correlation with overall survival.
| patients (n) | percentage (n/N%) | HR 95% CI | P | ||
|---|---|---|---|---|---|
| Age (years) | |||||
| < 65 | 2061 | 54% | |||
| ≥ 65 | 1221 | 32% | 2.113 (1.893, 2.360) | 0.000 | |
| Lost | 535 | ||||
| N-index | |||||
| < 4.05 | 1070 | 28% | |||
| ≥ 4.05 | 910 | 23% | 1.957 (1.741, 2.200) | 0.000 | |
| Lost | 1837 | ||||
| SF3B1 mutation | |||||
| Yes | 81 | 2.10% | |||
| No | 3736 | 97.90% | 1.154 (0.835, 1.593) | 0.385 | |
| ER status | |||||
| Positive | 2438 | 63.90% | |||
| Negative | 762 | 20% | 0.848 (0.746, 0.965) | 0.012 | |
| Lost | 617 | ||||
| PR status | |||||
| Positive | 1844 | 48.30% | |||
| Negative | 1357 | 35.60% | 0.784 (0.703, 0.875) | 0.000 | |
| Lost | 616 | ||||
| HER2 status | |||||
| Positive | 464 | 12.20% | |||
| Negative | 2375 | 62.20% | 1.420 (1.216, 1.659) | 0.000 | |
| Lost | 978 | ||||
| Menopausal state | |||||
| Pre | 681 | 17.80% | |||
| Post | 2280 | 59.60% | 1.734 (1.487, 2.022) | 0.000 | |
| Lost | 856 | ||||
| PAM50 and claudin-low subtype | |||||
| Normal | 157 | 4.10% | |||
| luminal A | 736 | 19.30% | |||
| Luminal B | 497 | 13% | |||
| Claudin-low | 218 | 5.70% | |||
| HER2 | 242 | 6.30% | |||
| Basal-like | 222 | 5.80% | 1.075 (1.164, 1.409) | 0.000 | |
| Lost | 1745 | ||||
| Neoplasm Histologic Grade | |||||
| 1 | 181 | 4.70% | |||
| 2 | 840 | 22% | |||
| 3 | 1029 | 27% | 1.281 (1.164, 1.409) | 0.000 | |
| Lost | 1767 | ||||
| Breast cancer type | |||||
| 1 | 979 | 25.60% | |||
| 2 | 2316 | 60.70% | |||
| 3 | 347 | 9.10% | |||
| 4 | 122 | 3.20% | 1.046 (0.986, 1.109) | 0.134 | |
| Tumor stage | |||||
| 0 | 12 | 0.30% | |||
| 1 | 695 | 18.20% | |||
| 2 | 1524 | 39.90% | |||
| 3 | 387 | 10.10% | |||
| 4 | 30 | 0.80% | 1.816 (1.652, 1.997) | 0.000 | |
| Lost | 1169 | ||||
| Primary Tumor Laterality | |||||
| left | 1551 | 40.60% | |||
| right | 1423 | 37.30% | 0.930 (0.831, 1.041) | 0.210 | |
| Lost | 843 |
ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; N-index, Nottingham prognostic index; HR, hazard ratio; CI, confidence interval.
SF3B1 mutation as a prognostic factor in luminal B and PR-negative breast cancer patients
SF3B1 mutation was not associated with OS in breast cancer patients; hence, subgroup analysis was performed to further investigate the clinical value of SF3B1 mutations in these patients (Table 2). In the luminal B patient group, SF3B1 mutation was significantly associated with the OS (hazard ratio [HR]: 2.188, 95% confidence interval [CI]: 1.225–3.907, P = 0.008). In the PR-negative group, the SF3B1 mutation was also significantly associated with OS (HR: 1.845, 95% CI: 1.123–3.034, P = 0.016). Kaplan-Meier survival analysis showed that the SF3B1 mutation was not an independent predictor for OS in breast cancer patients overall (log-rank test: P = 0.385). In the luminal B and PR-negative groups, however, the SF3B1 mutation was a significantly independent prognostic factor for OS (log-rank test: P = 0.007 and P = 0.014, respectively) (Figure 2)
Table 2. SF3B1 mutation as a prognostic factor for overall survival in all patient subgroups.
| all subgroup | HR (95% CI) | P |
|---|---|---|
| < 65 years | 0.603 (0.250, 1.454) | 0.260 |
| ≥ 65 years | 1.005 (0.708, 1.426) | 0.980 |
| < 4.05 N-Index | 1.112 (0.665, 1.859) | 0.685 |
| ≥ 4.05 N-Index | 1.271 (0.830, 1.946) | 0.270 |
| ER-positive | 1.215 (0.875, 1.688) | 0.245 |
| ER-Negative | 1.420 (0.199, 10.123) | 0.726 |
| PR-Positive | 0.975 (0.637, 1.471) | 0.905 |
| PR-Negative | 1.845 (1.123, 3.034) | 0.016 |
| HER2-Positive | 1.103 (0.352, 3.454) | 0.867 |
| HER2-Negative | 1.268 (0.905, 1.776) | 0.168 |
| Menopausal state pre- | 0.049 (0.000, 15.553) | 0.304 |
| Menopausal state post- | 1.231 (0.890, 1.072) | 0.208 |
| PAM50 and claudin-low subtype | ||
| normal | 1.915 (0.465, 7.885) | 0.368 |
| luminal A | 0.876 (0.522, 1.469) | 0.615 |
| luminal B | 2.188 (1.225, 3.907) | 0.008 |
| claudin-low | 1.616 (0.397, 6.586) | 0.503 |
| HER2 | 1.443 (0.591, 3.525) | 0.421 |
| Basal-like | 1.269 (0.177, 9.101) | 0.813 |
| Neoplasm Histologic Grade | ||
| 1 | 0.049 (0.000, 362.594) | 0.506 |
| 2 | 1.049 (0.645, 1.704) | 0.848 |
| 3 | 1.386 (0.866, 2.218) | 0.173 |
| Breast cancer type | ||
| 1 | 0.615 (0.152, 2.490) | 0.495 |
| 2 | 1.209 (0.836, 1.749) | 0.314 |
| 3 | 1.590 (0.584, 4.326) | 0.364 |
| 4 | 1.166 (0.360, 3.770) | 0.798 |
| Tumor stage | ||
| 0 | / | / |
| 1 | 0.586 (0.188, 1.832) | 0.358 |
| 2 | 1.297 (0.861, 1.954) | 0.213 |
| 3 | 1.429 (0.353, 5.788) | 0.617 |
| 4 | / | / |
| Primary Tumor Laterality | ||
| left | 0.962 (0.800, 1.157) | 0.680 |
| right | 1.012 (0.595, 1.720) | 0.965 |
N-index, Nottingham prognostic index; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; HR, hazard ratio; CI, confidence interval.
Figure 2. SF3B1 mutation as prognostic factor in breast cancer patients.
Kaplan Meier curves showing overall survival in all breast cancer patients (A), as well as overall survival according to the presence of SF3B1 mutations in all breast cancer patients (B), luminal B breast cancer patients (C), and progesterone receptor (PR)-negative breast cancer patients (D).
Relationship between common prognostic factors and SF3B1 mutation
Among 11 common prognostic factors investigated, univariate analysis showed that age, ER status, PR status, menopausal state, PAM50 and claudin-low subtype, and breast cancer type were significantly associated with SF3B1 mutation in all breast cancer patients (P < 0.01) (Table 3). Age and ER status were significantly associated with SF3B1 mutation on multivariate analysis (odds ratio: 1.037, 95% CI: 1.007–1.067, P = 0.015; and odds ratio: 6.055, 95% CI: 1.253–29.253, P = 0.025; respectively). Age and ER status were significantly associated with SF3B1 mutation specifically in the luminal B and PR-negative subgroups as well (P < 0.02).
Table 3. Univariate Cox regression analysis of the association between SF3B1 mutation and common prognostic factors.
| All patients | Luminal B patients | PR-Negative patients | ||||
|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |
| Age | 3.351 (2.093, 5.364) | 0.000 | 6.414 (1.442, 28.528) | 0.015 | 5.584 (2.187, 14.262) | 0.000 |
| N-Index | 0.986 (0.581, 1.673) | 0.958 | 1.455 (0.510, 4.154) | 0.483 | 0.984 (0.385, 2.515) | 0.972 |
| ER status | 7.920 (2.490, 25.192) | 0.000 | / | / | 7.679 (2.271, 25.965) | 0.001 |
| PR | 1.750 (1.069, 2.865) | 0.026 | / | / | / | / |
| HER2 | 0.458 (0.198, 1.063) | 0.069 | / | / | 0.522 (0.153, 1.776) | 0.298 |
| Menopausal state | 2.430 (1.159, 5.092) | 0.019 | 1.880 (0.243, 14.522) | 0.545 | 3.118 (0.725, 13.421) | 0.127 |
| PAM50 and claudin-low | 0.807 (0.662, 0.982) | 0.033 | / | / | 0.818 (0.615, 1.088) | 0.168 |
| Neoplasm Histologic Grade | 0.859 (0.584, 1.263) | 0.440 | 0.852 (0.333, 2.179) | 0.738 | 0.507 (0.258, 0.994) | 0.048 |
| Breast cancer type | 1.200 (1.017, 1.417) | 0.031 | 1.643 (0.797, 3.384) | 0.178 | 1.398 (0.709, 2.754) | 0.333 |
| Tumor stage | 0.840 (0.575, 1.227) | 0.367 | 0.985 (0.413, 2.350) | 0.974 | 0.701 (0.335, 1.463) | 0.344 |
| Primary Tumor Laterality | 1.030 (0.641, 1.655) | 0.902 | 1.409 (0.466, 4.263) | 0.543 | 1.006 (0.406, 2.493) | 0.990 |
N-index, Nottingham prognostic index; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; OR, odds ratio; CI, confidence interval.
Predicting SF3B1 mutation status using age at diagnosis and ER status
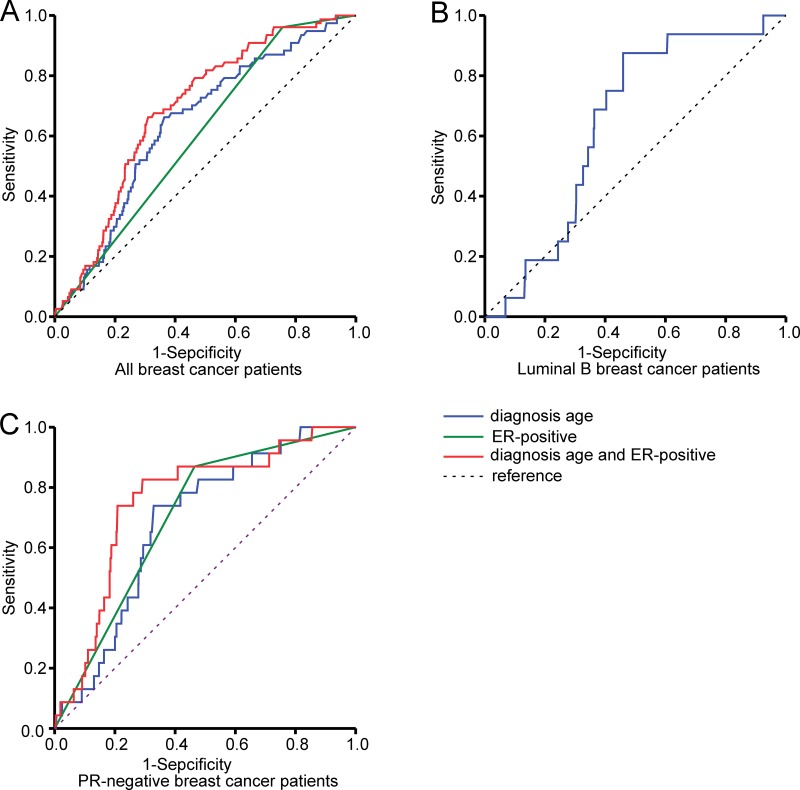
Receiver operating characteristic (ROC) curves were generated for all patients with breast cancer to identify the sensitivity and specificity of age at diagnosis and ER status in predicting SF3B1 mutation. For age at diagnosis, the ROC curves showed a sensitivity of 65.8% and specificity 64.0%, with an area under the curve (AUC) of 0.639 (95% CI: 0.582–0.696, P < 0.000). For ER status, the ROC curves showed an AUC of 0.602 (95% CI: 0.548–0.656, P = 0.002). When age at diagnosis and ER status were assessed for their combined ability to predict SF3B1 mutation, the ROC curve showed a higher sensitivity and specificity of 66.2% and 69.0%, respectively, with an AUC of 0.690 (95% CI: 0.639–0.740, P < 0.000).
For age at diagnosis in the luminal B group, ROC curves showed a sensitivity of 85.5% and specificity 54.1%, with an AUC of 0.643 (95% CI: 0.542–0.745, P = 0.051). In the PR-negative group, the ROC curve for age at diagnosis showed a sensitivity of 73.9% and specificity of 67.1%, with an AUC of 0.685 (95% CI: 0.597–0.773, P = 0.002). As for ER status, the ROC curve showed an AUC of 0.702 (95% CI: 0.612–0.793) (P = 0.001). When age at diagnosis and ER status were used in combination to predict SF3B1 mutation, the ROC curve showed a higher sensitivity and specificity of 82.6% and 70.8%, respectively, with an AUC of 0.756 (95% CI: 0.664–0.848, P < 0.000) (Figure 3).
Figure 3. Receiver operating characteristic curves for predicting SF3B1 mutation.
Shown are curves in all breast cancer patients (A), in luminal B breast cancer patients (B), and in progesterone receptor (PR)-negative breast cancer patients (C) according to age at diagnosis and estrogen receptor (ER) status.
DISCUSSION
SF3b is essential for pre-mRNA splicing, and mutations in its largest subunit (SF3B1/SF3b155) are linked to cancer [4, 16, 17]. The impact of SF3B1 mutations on patient outcomes varies according to tumor type; for instance, it is associated with poor outcomes in CLL patients but with good prognoses in uveal melanoma patients [8, 11, 18–20]. Human breast cancers are heterogeneous; Blows et al. showed that ER-positive breast cancer patients had varied outcomes and responses to therapy [21]. A study by Nik-Zainal et al. in which whole genomes from 560 breast cancers and non-neoplastic tissue were sequenced revealed 3479652 somatic base substitutions, 371993 small indels, and 77695 rearrangements [22]. The high number of gene mutations and diversity of genomic drivers may explain this disease’s clinical heterogeneity. An SF3B1 mutation can cause abnormal pre-RNA splicing that can lead to tumorigenesis, tumor drug resistance, or others detrimental features [20, 23, 24]. Therefore, SF3B1 mutation may be useful as a prognostic indicator in different tumors.
A study by Pereira et al. showed that mutations in driver genes were associated with the prognosis of breast cancer patients. For instance, MAP3K1 and GATA3 mutations were associated with longer survival, and TP53 mutations with shorter survival, in ER-positive patients but not in ER-negative patients. Conversely, PIK3CA mutations were associated with shorter survival in ER-negative patients, but not in ER-positive patients. Their study also showed associations between mutations in driver genes and clinicopathological parameters; for example, mutations in PIK3CA, GATA3, KMT2C, and CBFB were associated with lower grade tumors in ER-positive patients, while TP53 mutations were associated with higher grade tumors. GATA3, CBFB, CDH1, KMT2C, and SF3B1 mutations were also associated with age at diagnosis [14].
In our study, we further analyzed the clinic value of SF3B1 mutations in breast cancer patents. SF3B1 mutations were not significantly associated with survival outcome in breast cancer patients overall. However, these mutations were associated with worse outcomes in PR-negative and luminal B patients. In the PR-negative patient subgroup, SF3B1 mutations were associated with age at diagnosis, ER status, and histologic grade; in the luminal B subgroup, SF3B1 mutations were associated only with the age at diagnosis. The multivariate logistic regression model revealed that SF3B1 mutations were associated with age at diagnosis and ER status in all patients as well as in the PR-negative subgroup.
Because SF3B1 mutations were associated with worse outcomes in the PR-negative and luminal B subgroups, we analyzed whether these mutations were significantly associated with the age at diagnosis and ER status with the hypothesis that the age and ER status can predicting the existence of an SF3B1 mutation. We found that, when age at diagnosis and ER status were assessed in combination, the prediction of SF3B1 mutations had a slightly higher sensitivity, specificity, and AUC than the age at diagnosis in all patients. In the PR-negative subgroup, age and ER combined had a higher sensitivity (82.6%), specificity (70.8%), and AUC (0.756) in terms of predicting SF3B1 mutations than age alone.
Maguire et al. showed that SF3B1 K700E mutations are associated with differential gene splicing in breast cancer, including of TMEM14C, RPL31, CRNDE, DYNLL1, ICA1, RPL24, and MTERFD3. Cell lines carrying the SF3B1 mutation were sensitive to the SF3b complex inhibitor spliceostatin A, which suppressed tumor growth. Hence, the spliceosome SF3b complex may be a potential therapeutic target [11, 25].
In conclusion, our analysis of TCGA revealed that SF3B1 mutations are frequently found in breast cancer patients, and that they are poor prognostic indicators in PR-negative and luminal B breast cancer patients. SF3B1 mutations were found to be significantly associated with the age at diagnosis and/or ER status in PR-negative and luminal B breast cancer patients. Moreover, combining the age at diagnosis and ER status could better predict the existence of SF3B1 mutations. As demonstrated by spliceostatin A, the SF3b complex may be a novel therapeutic target for breast cancer patients with SF3B1 mutations.
MATERIALS AND METHODS
Analysis using the cancer genome atlas (tcga) database
We analyzed SF3B1 mutation data and clinic data of breast cancer patients from TCGA database (www.cbioportal.org/). We enrolled 2059, 1105, 103, and 100 invasive breast carcinoma patients from the METABRIC, TCGA (provisional), broad, and Sanger datasets, respectively, in our study. Our institutional review board approved this study, which was performed according to the principles of the Declaration of Helsinki. There were 81 patients with SF3B1 mutations among 3817 patients with invasive breast carcinoma. Breast cancers were divided into four types: invasive breast carcinoma (979), breast invasive ductal carcinoma (2316), breast invasive lobular carcinoma (347), and breast mixed ductal and lobular carcinoma (122). Fifty-three patients were not classified because of the rarity of their tumor types, including adenoid cystic breast cancer, phyllodes tumor of the breast, and unclassified breast cancer.
Risk factor analysis
The following parameters were investigated for their role in the prognosis of patients with breast cancer: age at diagnosis, N-index, ER status, PR status, HER2 status, menopausal state, PAM50 and claudin-low subtype, histological grade, breast cancer type, tumor stage, and primary tumor laterality. Additionally, SF3B1 was also analyzed as a prognostic marker. We divided the age and N-Index into dichotomous variables (< 65 vs. ≥ 65 years and < 4.05 vs. ≥ 4.05, respectively); these cutoff values were based on OS.
Statistical analysis
All dichotomous variables were analyzed by using a Cox regression models in all patients. Next, we divided all dichotomous prognostic factors into two subgroups in which the SF3B1 mutation as independent prognostic factor was evaluated. The Kaplan-Meier method was used to estimate survival function of each variable that was found to be a significant factor. Univariate analysis was performed to identify significant independent prognostic factors for OS. The Cox proportional hazards model was used to estimate the HRs and CIs of potential prognostic factors in all patients, including that of SF3B1 mutation in the aforementioned subgroups. Patients who were positive for significant prognostic factors were subjected to Kaplan-Meier analysis to clarify the role of the SF3B1 mutation on their survival. A logistic regression model was used to analyze associations between the investigated prognostic factors and SF3B1 mutation. Variables identified via univariate analysis (α = 0.25) were subjected to multivariable logistic regression analysis to determine their association with SF3B1 mutations. ROC curves were constructed to predict the sensitivity and specificity of SF3B1 mutations according to the AUC and 95% CI. We also assessed the combined effects of age at diagnosis and ER status on the occurrence of SF3B1 mutations. All analyses were performed using SPSS for Windows, version 20.0 (SPSS, Chicago, IL, USA).
Acknowledgments
We thank all patients involved in this study. We also thank cBioPortal for providing a free website that allows the retrieval of reliable public data.
Abbreviations
- AUC
area under the curve
- CI
confidence interval
- CLL
chronic lymphocytic leukemia
- ER
estrogen receptor
- HR
hazard ratio
- HER2
human epidermal growth factor receptor 2
- N-index
Nottingham prognostic index
- OR
odds ratio
- OS
overall survival
- PR
progesterone receptor
- ROC
receiver operating characteristic
- TCGA
The Cancer Genome Atlas
Footnotes
Author contributions
Xing Fu, Xing Xin, and Ding Ma conceived and designed the experiments. Ming Tian, Jia Gu, and Teng Cheng performed the experiments. Ming Tian, Jia Gu, and Xing Xin analyzed the data. Ling Feng and Xing Xin wrote the manuscript.
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.
FUNDING
The work was supported by a grant from the National Key Research & Development Program of China (2016YFC1000405, 2016YFC0902901).
REFERENCES
- 1.Schellenberg MJ, Edwards RA, Ritchie DB, Kent OA, Golas MM, Stark H, Lührmann R, Glover JN, MacMillan AM. Crystal structure of a core spliceosomal protein interface. Proc Natl Acad Sci USA. 2006;103:1266–71. doi: 10.1073/pnas.0508048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cazzola M, Rossi M, Malcovati L. Associazione Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie Mieloproliferative. Biologic and clinical significance of somatic mutations of SF3B1 in myeloid and lymphoid neoplasms. Blood. 2013;121:260–9. doi: 10.1182/blood-2012-09-399725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van den Hoogenhof MM, Pinto YM, Creemers EE. RNA Splicing: Regulation and Dysregulation in the Heart. Circ Res. 2016;118:454–68. doi: 10.1161/CIRCRESAHA.115.307872. [DOI] [PubMed] [Google Scholar]
- 4.Cretu C, Schmitzová J, Ponce-Salvatierra A, Dybkov O, De Laurentiis EI, Sharma K, Will CL, Urlaub H, Lührmann R, Pena V. Molecular Architecture of SF3b and Structural Consequences of Its Cancer-Related Mutations. Mol Cell. 2016;64:307–319. doi: 10.1016/j.molcel.2016.08.036. [DOI] [PubMed] [Google Scholar]
- 5.Golas MM, Sander B, Will CL, Lührmann R, Stark H. Molecular architecture of the multiprotein splicing factor SF3b. Science. 2003;300:980–4. doi: 10.1126/science.1084155. [DOI] [PubMed] [Google Scholar]
- 6.Dolatshad H, Pellagatti A, Fernandez-Mercado M, Yip BH, Malcovati L, Attwood M, Przychodzen B, Sahgal N, Kanapin AA, Lockstone H, Scifo L, Vandenberghe P, Papaemmanuil E, et al. Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia. 2015;29:1092–103. doi: 10.1038/leu.2014.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, Pellagatti A, Wainscoat JS, Hellstrom-Lindberg E, Gambacorti-Passerini C, Godfrey AL, Rapado I, Cvejic A, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–95. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malcovati L, Papaemmanuil E, Bowen DT, Boultwood J, Della Porta MG, Pascutto C, Travaglino E, Groves MJ, Godfrey AL, Ambaglio I, Gallì A, Da Vià MC, Conte S, et al. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood. 2011;118:6239–46. doi: 10.1182/blood-2011-09-377275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsunawa M, Yamamoto R, Sanada M, Sato-Otsubo A, Shiozawa Y, Yoshida K, Otsu M, Shiraishi Y, Miyano S, Isono K, Koseki H, Nakauchi H, Ogawa S. Haploinsufficiency of Sf3b1 leads to compromised stem cell function but not to myelodysplasia. Leukemia. 2014;28:1844–50. doi: 10.1038/leu.2014.73. [DOI] [PubMed] [Google Scholar]
- 10.Chesnais V, Kosmider O, Damm F, Itzykson R, Bernard OA, Solary E, Fontenay M. Spliceosome mutations in myelodysplastic syndromes and chronic myelomonocytic leukemia. Oncotarget. 2012;3:1284–93. doi: 10.18632/oncotarget.749. https://doi.org/10.18632/oncotarget.749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maguire SL, Leonidou A, Wai P, Marchiò C, Ng CK, Sapino A, Salomon AV, Reis-Filho JS, Weigelt B, Natrajan RC. SF3B1 mutations constitute a novel therapeutic target in breast cancer. J Pathol. 2015;235:571–80. doi: 10.1002/path.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furney SJ, Pedersen M, Gentien D, Dumont AG, Rapinat A, Desjardins L, Turajlic S, Piperno-Neumann S, de la Grange P, Roman-Roman S, Stern MH, Marais R. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013;3:1122–1129. doi: 10.1158/2159-8290.CD-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martelotto LG, De Filippo MR, Ng CK, Natrajan R, Fuhrmann L, Cyrta J, Piscuoglio S, Wen HC, Lim RS, Shen R, Schultheis AM, Wen YH, Edelweiss M, et al. Genomic landscape of adenoid cystic carcinoma of the breast. J Pathol. 2015;237:179–89. doi: 10.1002/path.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ, Tsui DW, Liu B, Dawson SJ, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JY, Lee E, Park K, Park WY, Jung HH, Ahn JS, Im YH, Park YH. Immune signature of metastatic breast cancer: Identifying predictive markers of immunotherapy response. Oncotarget. 2017;8:47400–47411. doi: 10.18632/oncotarget.17653. https://doi.org/10.18632/oncotarget.17653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alsafadi S, Houy A, Battistella A, Popova T, Wassef M, Henry E, Tirode F, Constantinou A, Piperno-Neumann S, Roman-Roman S, Dutertre M, Stern MH. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat Commun. 2016;7:10615. doi: 10.1038/ncomms10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darman RB, Seiler M, Agrawal AA, Lim KH, Peng S, Aird D, Bailey SL, Bhavsar EB, Chan B, Colla S, Corson L, Feala J, Fekkes P, et al. Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3′ Splice Site Selection through Use of a Different Branch Point. Cell Rep. 2015;13:1033–45. doi: 10.1016/j.celrep.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 18.Visconte V, Rogers HJ, Singh J, Barnard J, Bupathi M, Traina F, McMahon J, Makishima H, Szpurka H, Jankowska A, Jerez A, Sekeres MA, Saunthararajah Y, et al. SF3B1 haploinsufficiency leads to formation of ring sideroblasts in myelodysplastic syndromes. Blood. 2012;120:3173–86. doi: 10.1182/blood-2012-05-430876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gentien D, Kosmider O, Nguyen-Khac F, Albaud B, Rapinat A, Dumont AG, Damm F, Popova T, Marais R, Fontenay M, Roman-Roman S, Bernard OA, Stern MH. A common alternative splicing signature is associated with SF3B1 mutations in malignancies from different cell lineages. Leukemia. 2014;28:1355–7. doi: 10.1038/leu.2014.28. [DOI] [PubMed] [Google Scholar]
- 20.Harbour JW, Roberson ED, Anbunathan H, Onken MD, Worley LA, Bowcock AM. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet. 2013;45:133–5. doi: 10.1038/ng.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blows FM, Driver KE, Schmidt MK, Broeks A, van Leeuwen FE, Wesseling J, Cheang MC, Gelmon K, Nielsen TO, Blomqvist C, Heikkilä P, Heikkinen T, Nevanlinna H, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010;7:e1000279. doi: 10.1371/journal.pmed.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, Martincorena I, Alexandrov LB, Martin S, Wedge DC, Van Loo P, Ju YS, Smid M, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott LM, Rebel VI. Acquired mutations that affect pre-mRNA splicing in hematologic malignancies and solid tumors. J Natl Cancer Inst. 2013;105:1540–9. doi: 10.1093/jnci/djt257. [DOI] [PubMed] [Google Scholar]
- 24.Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, Council ML, Matatall KA, Helms C, Bowcock AM. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–3. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, Tani T, Horinouchi S, Yoshida M. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3:576–83. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]