Abstract
Prions are infectious misfolded proteins that assemble into oligomers and large aggregates, and are associated with neurodegeneration. It is believed that the oligomers contribute to cytotoxicity, although genetic and environmental factors have also been shown to have additional roles. The study of the yeast prion [PSI+] has provided valuable insights into how prions form and why they are toxic. Our recent work suggests that SDS-resistant oligomers arise and remodel early during the prion formation process, and lysates containing these newly formed oligomers are infectious. Previous work shows that toxicity is associated with prion formation and this toxicity is exacerbated by deletion of the VPS5 gene. Here, we show that newly made oligomer formation and infectivity of vps5Δ lysates are similar to wildtype strains. However using green fluorescent protein fusions, we observe that the assembly of fluorescent cytoplasmic aggregates during prion formation is different in vps5Δ strains. Instead of large immobile aggregates, vps5Δ strains have additional population of small mobile foci. We speculate that changes in the cellular milieu in vps5Δ strains may reduce the cell's ability to efficiently recruit and sequester newly formed prion particles into central deposition sites, resulting in toxicity.
Keywords: Prion, Sup35, Yeast, oligomer, infectivity, Vps5
Introduction
Prions are misfolded aggregated proteins that are infectious. Several proteins have been shown to form prions in yeast, including Sup35p, Ure2p, Rnq1p, Swi1p, Cyc8p, and Mot3p (reviewed in Liebman and Chernoff, 2012). In each case, the monomeric protein misfolds into an alternate stable conformation that in turn is able to convert normal copies of the protein to a misfolded form that is prone to aggregation. The process by which these proteins initially misfold and aggregate to form infectious particles is still unclear. Work in yeast has provided important insights into how prions form and when they become infectious.
Spontaneous prion formation in yeast is quite rare. Analysis of the Sup35 prion, or [PSI+], found that the spontaneous rate of formation is less than 1 in 1 million cells per generation (Lund and Cox, 1981; Allen et al., 2005; Lancaster et al., 2010). Despite this low level, several approaches can increase the rate of de novo formation. An expansion of an oligopeptide repeat within the N-terminal prion forming region of Sup35 increases prion formation (Liu and Lindquist, 1999). These extra repeats have been suggested to more readily direct the assembly of Sup35 proteins into ordered complexes. Prion formation has also been shown to be increased by exposure to environmental stress (Tyedmers et al., 2008). It is thought that the accumulation of misfolded proteins, including misfolded Sup35p, under these stress conditions may initiate prion formation. However, the most dramatic increase in de novo prion formation occurs by the overexpression of the Sup35 protein in a process called prion induction (Chernoff et al., 1993; Derkatch et al., 1996). This increase requires the presence of a second prion, such as the prion form of the Rnq1 protein, [PIN+] (otherwise known as [RNQ+]), or overexpressed glutamine and asparagine-rich proteins (Derkatch et al., 1997; Derkatch et al., 2001; Osherovich and Weissman, 2001). Using this prion induction method, Derkatch et al. (1996) observed that the frequency of [PSI+] colonies after SUP35 overexpression in [psi-][PIN+] cultures was higher than 30%. The elevated Sup35 protein concentration likely increases the probability of protein misfolding (Chernoff et al., 1993; Derkatch et al., 1996), and the presence of a second prion allows for heterologous cross seeding for the de novo formation of [PSI+] (Derkatch et al., 2001; Osherovich and Weissman, 2001; Arslan et al., 2015; Keefer et al., 2017).
Newly formed prion aggregates
The laboratory of Susan Liebman was the first to visualize prion induction in vivo (Zhou et al., 2001). The prion domain of Sup35p, which consists of the N-terminal and middle domain of the protein, can be fused to Green Fluorescent Protein (Sup35PrD-GFP). In cells lacking the [PSI+] prion, these fusion proteins are evenly distributed throughout the cytoplasm resulting in diffuse fluorescence. However, overexpression of this construct can induce [PSI+] formation in [PIN+] cells (Derkatch et al., 2001; Zhou et al., 2001). Observations of overnight cultures overexpressing the fusion protein show that a small percentage of cells formed large intracellular ring, line, and dot-like aggregates (Zhou et al., 2001). Since then, several studies have used periodic “snapshots” to infer how these aggregates are made. Small fluorescent foci initially appear, with some located near the vacuole. Later snapshot observations suggest that these small foci are replaced with the ring, line, and dot-like aggregates (Arslan et al., 2015), which are retained in the mother cell during cell division (Mathur et al., 2010). Isolation of cells that contain these newly formed aggregates can give rise to a proportion of progeny that are [PSI+], whereas sibling cells that lack fluorescent aggregates always give rise to progeny that lack the prion (Ganusova et al., 2006).
Since much of what we know about Sup35PrD-GFP ring, line, and dot-like aggregate formation during prion induction is due to temporal extrapolation, we recently employed 4D live cell imaging in order to continuously capture the initial formation of the Sup35PrD-GFP aggregate (Sharma et al., 2017). We found that cells displaying diffuse cytoplasmic fluorescence developed one or several small foci (which we called “early foci”) that quickly assembled into larger aggregates. While this assembly could result in rings, lines or dot-like structures, the frequency in which early foci formed each structure was similar.
The formation of SDS-resistant oligomers and infectivity of newly formed particles
It was originally observed that during prion induction, Sup35p forms large SDS-resistant oligomers that migrate differently than Sup35p oligomers associated with the propagating [PSI+] prion (Salnikova et al., 2005). In vitro studies showed that lysates containing these newly made oligomers were able to convert monomeric Sup35p to an aggregated form, suggesting that these newly formed oligomers can seed aggregation (Salnikova et al., 2005). However, the ability of these lysates to convert [psi-] cells to [PSI+] in vivo, thereby showing that these newly made prion oligomers are infectious, was unknown.
To begin to understand oligomer formation and infectivity, we looked at how the size of SDS-resistant oligomers changes during prion formation, and how these changes are correlated with the ability to convert [psi-] cultures to [PSI+] (Sharma et al., 2017). Using semi-denaturing detergent agarose gel electrophoresis (SDD-AGE; Kryndushkin et al., 2003), we resolved SDS-resistant Sup35PrD-GFP oligomers at different time points of the induction process. At early time points of induction, when cultures have diffuse Sup35PrD-GFP fluorescence, Sup35PrD-GFP forms a single band that is larger than the monomeric protein. At later time points, when approximately 20% of the cells exhibited ring, line, or dot-like aggregates, larger molecular weight smears are detected suggesting that Sup35PrD-GFP undergoes assembly into oligomers of diverse sizes. Endogenous Sup35p also forms SDS-resistant oligomers during induction that are different sizes than established [PSI+] oligomers (Sharma et al., 2017). These data suggest that between the time of prion formation and prion propagation, Sup35p oligomers must be remodeled or changed.
Next, we asked whether lysates from induced cultures could convert [psi-] cells to [PSI+]. The “protein-only” hypothesis proposed that misfolded prion proteins are infectious. In yeast, proof for this hypothesis has been demonstrated by several studies that show recombinant prionogenic proteins, either incubated to form fibrils or seeded with lysates from yeast cells containing prions, can convert non-prion containing cells to into the prion state (King and Diaz-Avalos, 2004; Tanaka et al., 2004; Patel and Liebman, 2007; Du et al., 2010). Even with strong proof for the protein only hypothesis, little was known about when newly formed prion particles gain their infectivity.
To uncover when induced cultures become infectious, we obtained lysates from different time points of prion induction. We found that the transfection of fresh lysates from cells that overexpressed Sup35PrD-GFP for 16 to 24 hours, and thus contained ring, line, or dot-like aggregates, were able to transform [psi-] cells into [PSI+] colonies. Conversely, cultures lacking cells with aggregates, such as uninduced cells or Sup35PrD-GFP overexpressed in the [pin-] background, could not convert [psi-] cells to the prion state (Sharma et al., 2017). Our data suggest that during the induction process, lysates contain infectious material.
An unanswered question is how newly made prion particles gain their infectivity. In vitro formed Sup35 fibers seeded with [PSI+] lysates can convert [psi-] to [PSI+] cells. Yet sonication of [PSI+] lysates prior to seeding significantly enhanced infectivity (King and Diaz-Avalos, 2004). It is thought that this shearing by sonication mimics how prions are propagated in vivo. The Hsp104p chaperone has been shown to be required for [PSI+] propagation (Chernoff et al., 1995) and in conjunction with several other chaperones appears to shear and fragment larger prion aggregates into smaller seeds, or propagons, that can be inherited by progeny (reviewed in Liebman and Chernoff, 2012; Cox and Tuite, 2017). Inactivation of Hsp104p by low levels of guanidine-HCl blocks [PSI+] shearing, which limits the production of these smaller propagons (Eaglestone et al., 2000; Wegrzyn et al., 2001; Ness et al., 2002; Byrne et al., 2009). Hsp104p may also play an important role for ensuring the de novo formation of [PSI+]. During prion induction, Hsp104p has been shown to colocalize with newly formed Sup35PrD-GFP rings structures (Arslan et al., 2015). It is possible that Hsp104p immediately initiates the shearing of newly formed aggregates during de novo induction, which generates heritable, infectious propagons. Further infectivity studies will be required to understand whether Hsp104p plays a role in the infectivity of newly formed prion particles.
Prion induction-associated toxicity and newly made prion aggregates
The presence of [PSI+] alone does not have any adverse effect on cell growth, but increasing Sup35p expression in [PSI+] cells reduces viability (Derkatch et al., 1996). This [PSI+] associated toxicity has been shown to be relieved by the expression of the C-terminal translational termination domain of Sup35p or the Sup45 protein, suggesting that toxicity is due to the sequestration of these essential proteins into the prion aggregate (Vishveshwara et al., 2009). Other studies have shown that prion associated toxicity can also be relieved by the overexpression of chaperones, such as Sis1p and Ssb1p (Douglas et al., 2008; Keefer and True, 2016). Therefore, the sequestration of essential proteins into aggregates and changes in the protein quality control machinery could influence [PSI+] associated toxicity.
Toxicity has also been shown to be associated with prion induction. Cells containing newly formed aggregates are less viable than those with diffuse fluorescence (Ganusova et al., 2006). Overexpression of the C-terminal region of Sup35p was shown to suppress this prion induction-associated toxicity (Vishveshwara et al., 2009), suggesting that sequestration of the essential Sup35 protein into the newly formed aggregates results in its loss of function. It was also shown that the deletion of non-essential genes that code for proteins associated with cytoskeleton organization and biogenesis, response to stress, and cell budding decrease prion induction-associated toxicity (Tyedmers et al., 2008). Therefore similar to prion associated toxicity, prion induction-associated toxicity may be due to sequestration of essential proteins as well as several other factors.
Examining genetic mutants that enhance prion induction-associated toxicity could provide important clues to the causes of cell death. We previously characterized a genetic mutant that has increased prion induction-associated toxicity. Cells lacking the VPS5 open reading frame (YOR069w) form fewer ring, line, and dot-like structures compared to wildtype cells. Of the few vps5Δ cells containing these aggregates, we found that these cells were also significantly less viable than wildtype cells containing aggregates (Manogaran et al., 2011). It is possible that the inability to form ring and dot aggregates in vps5Δ cells may be due to toxicity.
VPS5 codes for a sorting nexin 1 homolog, a member of the retromer complex that mediates vesicle transport by ensuring the recycling of late endosome cargo to the Golgi (Nothwehr and Hindes, 1997; Seaman et al., 1998). The retromer complex has also been implicated in having dual roles in cargo recycling and indirectly affecting Ypt7-dependent vacuole tethering and fusion (Liu et al., 2012). Strains lacking the VPS5 open reading frame exhibit vacuolar protein-sorting defects (Horazdovsky et al., 1997) and decreased autophagy in response to certain stress conditions (Dengjel et al., 2012). Interestingly, deletion of VPS5 also results in the loss of a second open reading frame on the opposite DNA strand (YOR068c; Fig 1A). This other open reading frame, VAM10, appears to be required for the Sec18p independent priming stages of vacuole fusion (Kato and Wickner, 2003).
Figure 1.
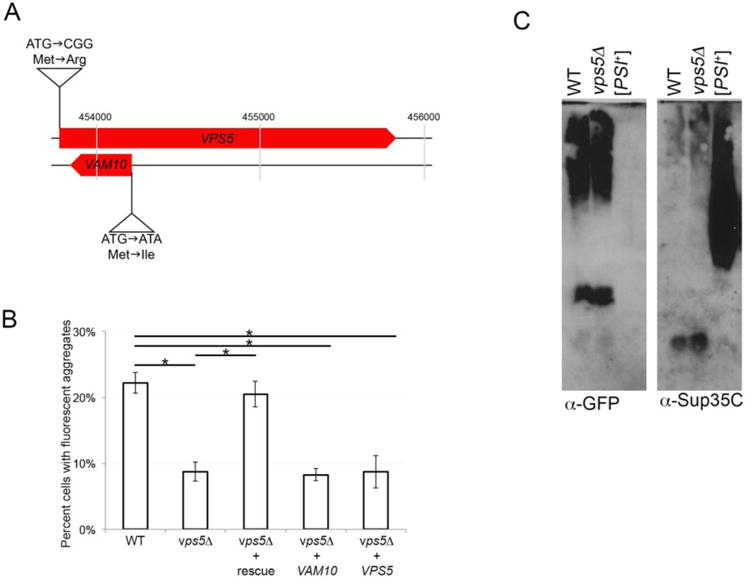
vps5Δ strains have reduced aggregate formation frequency, yet show no change in SDS-resistant oligomers. A. The VPS5 open reading frame (YOR069w) and VAM10 open reading frame (YOR068c) are located on chromosome 15 in the yeast genome. Site directed mutagenesis was performed to generate plasmids that contain a mutation in the initiator methionine of either VPS5 or VAM10. Two nucleotide substitutions replaced the initiation methionine with an arginine in the VPS5 open reading frame, while leaving the VAM10 open reading frame untouched. In a second plasmid, a single nucleotide substitution at the beginning of the VAM10 open reading frame leads to a mutation that changes methionine for isoleucine, while maintaining the same wildtype amino acid (serine) in the VPS5 sequence encoded by the opposite strand. All plasmids were sequenced in both directions to confirm the engineered mutation and the opposite open reading frame sequence. B. Plasmids containing wildtype versions of both genes (rescue), or mutated versions that maintain wildtype versions of only one gene (VAM10 or VPS5) were transformed into vps5Δ [PIN+] 74D-694 strains (Manogaran et al., 2011) along with a plasmid containing a copper inducible Sup35PrD-GFP allele. Sup35PrD-GFP was overexpressed for 24 hours in wildtype, vps5Δ, or vps5Δ strains with the indicated plasmid. The number of cells containing ring, line, or dot-like aggregates was counted from a population of at least 300 cells from three independent transformants. Standard deviation is shown. Statistically significant differences from wildtype or vps5Δ strains were determined by unpaired two-tailed t-test * p<0.005. C. Sup35PrD-GFP was overexpressed in wildtype and vps5Δ strains for 24 hours. Cultures were lysed and immediately subjected to SDD-AGE immunoblots using anti-GFP antibody (left) to detect Sup35PrD-GFP and anti-Sup35C antibody to detect full length Sup35p (BE4; right) according to Sharma et al., 2017. [PSI+] lysates are run for the detection of established [PSI+] oligomers.
To begin, we asked which gene (VPS5 or VAM10) was responsible for the reduced number of cells containing Sup35PrD-GFP ring, line, or dot-like aggregates during prion induction. Single-rescue plasmids, that maintain the wildtype polypeptide sequence for one gene while eliminating the initiation methionine codon of the other gene on the opposite strand, were introduced into mutants lacking both VPS5 and VAM10 open reading frames. We will refer to this deletion strain as vps5Δ below. We found that the introduction of wildtype versions of both genes was able to rescue the low level of Sup35PrD-GFP ring, line, and dot-like aggregates in vps5Δ strains (Fig. 1b). However, introduction of either individual wildtype gene showed the same low ring, line, and dot-like aggregate formation frequency as strains lacking both open reading frames. Our data suggest that Sup35PrD-GFP aggregation seems to require both genes and may involve a common pathway. Since both genes appear to play a role in vacuolar fusion, it is possible that impairment of vacuole fusion may underlie this change in aggregation state.
We next determined whether there were other differences between vps5Δ and wildtype strains that could explain the observed prion induction-associated toxicity. We found that overexpression of Sup35PrD-GFP in vps5Δ and wildtype strains produced Sup35PrD-GFP and endogenous Sup35 oligomers of similar sizes (Fig. 1C). Transfection of lysates from these induced strains were able to convert [psi-] recipient strains into [PSI+] (Table 1). However, conversion caused by vps5Δ lysates was approximately half of the conversion caused by parallel wildtype lysates (Table 1). Since ring, line, and dot-like aggregate formation in vps5Δ strains is half that of wildtype (Fig. 1B), the reduction in conversion is possibly correlated with less available infectious protein rather than cell death caused by a toxic particle.
Table 1.
Transfection of de novo formed Sup35PrD-GFP oligomers from vps5Δ strains convert [psi-] to [PSI+]. Freshly obtained lysates from cultures overexpressing Sup35PrD-GFP for 24 hours in either the [pin-][psi-] (which cannot induce [PSI+]) or [PIN+][psi] (which can induce [PSI+]) background were transfected into [psi-][PIN+] recipient cultures. Numbers indicate the percent of transfectants (approximately 130-250 transfectants scored) that were converted to [PSI+]. Induced vps5Δ cultures were also lysed and transfected at the same time as the WT controls. Experiments were performed as previously described (Sharma et al., 2017). Binomial comparison of conversion frequencies between WT [PIN+] and vps5Δ [PIN+] show that the two values are significantly different (p < 0.0001)
| No lysate | Donor lysates prepared after Sup35PrD-GFP overexpression for 24 hours | |||
|---|---|---|---|---|
| WT [pin-] | WT [PIN+] | vps5Δ [PIN+] | ||
| Recipient wildtype [psi-] [PIN+] | 0% | 0.5% | 28.7% | 15.9% |
Next, we explored whether there were any differences in aggregate formation. We previously found that early foci can assemble into large ring, line and dot-like aggregates by four different pathways in wildtype cells (Sharma et al., 2017). We were able to follow the progression from early foci to large ring, line, and dot-like aggregates in 33 individual vps5Δ cells. Unlike wildtype cells, it appeared the probability of vps5Δ cells to form aggregates by the four pathways was not equally likely (Fig. 2A). We also observed that the physical appearance of aggregates was quite different. In wildtype strains, diffuse fluorescence within the cytoplasm is initially observed upon Sup35PrD-GFP overexpression. The formation of early foci and the subsequent assembly into larger aggregates is correlated with a dramatic reduction in the diffuse cytoplasmic fluorescence, suggesting that the majority of soluble Sup35PrD-GFP is recruited into the large aggregates. While a similar reduction in diffuse fluorescence is observed in vps5Δ during the formation of large aggregates, many of the cells had an additional population of small anomalous aggregates not observed in wild type (Fig. 2B and C). Approximately 35% of these vps5Δ with ring and dot aggregates had these anomalous aggregates (data not shown), many of which were mobile even after several hours of video recording.
Figure 2.
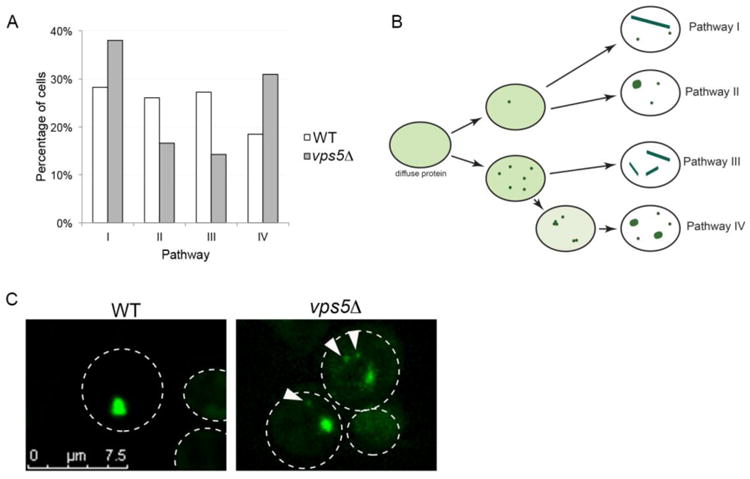
Sup35PrD-GFP forms additional small anomalous aggregates during prion induction. A. Sup35PrD-GFP was overexpressed in vps5Δ cells for 18 hours, and then imaged using 8-well glass slides for an additional 6-12 hours by 4D microscopy. Because of the reduced aggregate formation in vps5Δ, we selectively captured fields of cells in which in the initial stages of early foci formation could be captured. Of the 149 cells imaged, we were able to view aggregate formation in 33 cells (17 cells in G1, and 16 cells in G2/M phase). We followed the formation of early foci into larger aggregates and categorized them into four distinct pathways previously characterized for wildtype cells by Sharma et al., 2017. Statistical analysis using Chi-square goodness of fit tests indicate that while wildtype cells have an equal probability for each pathway, vps5Δ cells do not (p < 0.05). B. Diagrammatic representation of the four pathways in vps5Δ strains. While the pathways were similar between wildtype and vps5Δ strains, we noticed small anomalous aggregates, many of which were mobile, associated with pathways I, II, and IV in vpsΔ. C. Representative images of wildtype (WT) and vps5Δ strains are shown. Arrows indicate small anomalous aggregates. All images were subjected to 3D deconvolution using Autoquant deconvolution algorithms (Media Cybernetics) and are shown as maximum projection images.
While it is unclear whether there is a direct correlation between the presence of these small anomalous aggregates and the higher toxicity in vps5Δ strains, it is possible these aggregates could be a contributing factor. One could speculate that the changes in vacuolar fusion or autophagy caused by the loss of the VPS5 open reading frame may allow for the buildup of these anomalous aggregates, which could be detrimental to cell viability. Vacuolar defects in vps5Δ strains could also disrupt the cell's ability to direct newly made prion particles to central protein holding places in the cell, such as the vacuole associated insoluble protein deposit, IPOD. Since prion proteins have been shown to accumulate at IPOD (Kaganovich et al., 2008; Mathur et al., 2010; Saarikangas and Barral, 2016), the sequestration and retention of these anomalous aggregates to a central location like IPOD in vps5Δ strains could be compromised.
These studies have begun to uncover the cellular nuances that direct prion formation and toxicity. Emerging evidence from mammalian systems has suggested that oligomeric species contribute to toxicity in human amyloid-based neurodegenerative diseases (Huang et al., 2013; Kayed and Lasagna-Reeves, 2013; Gerson et al., 2016; Salahuddin et al., 2016). However, genetic and environmental factors have also been shown to exacerbate neurodegenerative progression of these diseases (Bertram and Tanzi, 2005; Campdelacreu, 2014; Agrawal et al., 2017). Therefore, these data suggest that the cellular context may direct the degree of toxicity caused by oligomers. Work with yeast prions allows for a quick and tractable system to uncover which cellular factors influence toxicity associated with oligomers. Studies focused on [PSI+] induction-associated toxicity will likely tease apart how both the newly made oligomers and cellular pathways contribute to cellular toxicity.
Acknowledgments
We thank Susan Liebman for the gifts of plasmids and strains used in this study. The Sup35C (BE4) antibody was a gift from Viravan Prapapanich and Susan Liebman. We would also like to thank Jane Dorweiler, Doug Lyke, and Emily Davis for critical reading of the manuscript. This work was supported by a grant from the National Institutes of Health (GM109336) to A.L.M., and B.T.W. and E. R. L. were supported by the Marquette University Honors Research Fellowship.
Literature Cited
- Agrawal S, Berggren KL, Marks E, Fox JH. Impact of high iron intake on cognition and neurodegeneration in humans and in animal models: a systematic review. Nutr Rev. 2017 doi: 10.1093/nutrit/nux015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen KD, Wegrzyn RD, Chernova TA, Muller S, Newnam GP, Winslett PA, Wittich KB, Wilkinson KD, Chernoff YO. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+] Genetics. 2005;169:1227–1242. doi: 10.1534/genetics.104.037168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arslan F, Hong JY, Kanneganti V, Park SK, Liebman SW. Heterologous aggregates promote de novo prion appearance via more than one mechanism. PLoS genetics. 2015;11:e1004814. doi: 10.1371/journal.pgen.1004814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Tanzi RE. The genetic epidemiology of neurodegenerative disease. J Clin Invest. 2005;115:1449–1457. doi: 10.1172/JCI24761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne LJ, Cole DJ, Cox BS, Ridout MS, Morgan BJ, Tuite MF. The number and transmission of [PSI] prion seeds (Propagons) in the yeast Saccharomyces cerevisiae. PLoS One. 2009;4:e4670. doi: 10.1371/journal.pone.0004670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campdelacreu J. Parkinson disease and Alzheimer disease: environmental risk factors. Neurologia. 2014;29:541–549. doi: 10.1016/j.nrl.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Derkach IL, Inge-Vechtomov SG. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet. 1993;24:268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Cox B, Tuite M. The life of [PSI] Curr Genet. 2017 doi: 10.1007/s00294-017-0714-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengjel J, Hoyer-Hansen M, Nielsen MO, Eisenberg T, Harder LM, Schandorff S, Farkas T, Kirkegaard T, Becker AC, Schroeder S, et al. Identification of autophagosome-associated proteins and regulators by quantitative proteomic analysis and genetic screens. Mol Cell Proteomics. 2012;11:M111014035. doi: 10.1074/mcp.M111.014035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas PM, Treusch S, Ren HY, Halfmann R, Duennwald ML, Lindquist S, Cyr DM. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc Natl Acad Sci U S A. 2008;105:7206–7211. doi: 10.1073/pnas.0802593105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Crow ET, Kang HS, Li L. Distinct subregions of Swi1 manifest striking differences in prion transmission and SWI/SNF function. Mol Cell Biol. 2010;30:4644–4655. doi: 10.1128/MCB.00225-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone SS, Ruddock LW, Cox BS, Tuite MF. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI(+)] of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2000;97:240–244. doi: 10.1073/pnas.97.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganusova EE, Ozolins LN, Bhagat S, Newnam GP, Wegrzyn RD, Sherman MY, Chernoff YO. Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol Cell Biol. 2006;26:617–629. doi: 10.1128/MCB.26.2.617-629.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerson JE, Mudher A, Kayed R. Potential mechanisms and implications for the formation of tau oligomeric strains. Crit Rev Biochem Mol Biol. 2016;51:482–496. doi: 10.1080/10409238.2016.1226251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horazdovsky BF, Davies BA, Seaman MN, McLaughlin SA, Yoon S, Emr SD. A sorting nexin-1 homologue, Vps5p, forms a complex with Vps17p and is required for recycling the vacuolar protein-sorting receptor. Mol Biol Cell. 1997;8:1529–1541. doi: 10.1091/mbc.8.8.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Lian F, Wen Y, Guo C, Lin D. Prion protein oligomer and its neurotoxicity. Acta Biochim Biophys Sin (Shanghai) 2013;45:442–451. doi: 10.1093/abbs/gmt037. [DOI] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Wickner W. Vam10p defines a Sec18p-independent step of priming that allows yeast vacuole tethering. Proc Natl Acad Sci U S A. 2003;100:6398–6403. doi: 10.1073/pnas.1132162100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayed R, Lasagna-Reeves CA. Molecular mechanisms of amyloid oligomers toxicity. J Alzheimers Dis. 2013;33(1):S67–78. doi: 10.3233/JAD-2012-129001. [DOI] [PubMed] [Google Scholar]
- Keefer KM, Stein KC, True HL. Heterologous prion-forming proteins interact to cross-seed aggregation in Saccharomyces cerevisiae. Sci Rep. 2017;7:5853. doi: 10.1038/s41598-017-05829-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefer KM, True HL. Prion-Associated Toxicity is Rescued by Elimination of Cotranslational Chaperones. PLoS genetics. 2016;12:e1006431. doi: 10.1371/journal.pgen.1006431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- Lancaster AK, Bardill JP, True HL, Masel J. The spontaneous appearance rate of the yeast prion [PSI+] and its implications for the evolution of the evolvability properties of the [PSI+] system. Genetics. 2010;184:393–400. doi: 10.1534/genetics.109.110213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman SW, Chernoff YO. Prions in yeast. Genetics. 2012;191:1041–1072. doi: 10.1534/genetics.111.137760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Lindquist S. Oligopeptide-repeat expansions modulate ‘protein-only’ inheritance in yeast. Nature. 1999;400:573–576. doi: 10.1038/23048. [DOI] [PubMed] [Google Scholar]
- Liu TT, Gomez TS, Sackey BK, Billadeau DD, Burd CG. Rab GTPase regulation of retromer-mediated cargo export during endosome maturation. Mol Biol Cell. 2012;23:2505–2515. doi: 10.1091/mbc.E11-11-0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund PM, Cox BS. Reversion analysis of [psi-] mutations in Saccharomyces cerevisiae. Genetical research. 1981;37:173–182. doi: 10.1017/s0016672300020140. [DOI] [PubMed] [Google Scholar]
- Manogaran AL, Hong JY, Hufana J, Tyedmers J, Lindquist S, Liebman SW. Prion formation and polyglutamine aggregation are controlled by two classes of genes. PLoS genetics. 2011;7:e1001386. doi: 10.1371/journal.pgen.1001386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur V, Taneja V, Sun Y, Liebman SW. Analyzing the birth and propagation of two distinct prions, [PSI+] and [Het-s](y), in yeast. Mol Biol Cell. 2010;21:1449–1461. doi: 10.1091/mbc.E09-11-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness F, Ferreira P, Cox BS, Tuite MF. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol Cell Biol. 2002;22:5593–5605. doi: 10.1128/MCB.22.15.5593-5605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Hindes AE. The yeast VPS5/GRD2 gene encodes a sorting nexin-1-like protein required for localizing membrane proteins to the late Golgi. J Cell Sci. 1997;110(Pt 9):1063–1072. doi: 10.1242/jcs.110.9.1063. [DOI] [PubMed] [Google Scholar]
- Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- Patel BK, Liebman SW. “Prion-proof” for [PIN+]: infection with in vitro-made amyloid aggregates of Rnq1p-(132-405) induces [PIN+] J Mol Biol. 2007;365:773–782. doi: 10.1016/j.jmb.2006.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas J, Barral Y. Protein aggregation as a mechanism of adaptive cellular responses. Curr Genet. 2016;62:711–724. doi: 10.1007/s00294-016-0596-0. [DOI] [PubMed] [Google Scholar]
- Salahuddin P, Fatima MT, Abdelhameed AS, Nusrat S, Khan RH. Structure of amyloid oligomers and their mechanisms of toxicities: Targeting amyloid oligomers using novel therapeutic approaches. Eur J Med Chem. 2016;114:41–58. doi: 10.1016/j.ejmech.2016.02.065. [DOI] [PubMed] [Google Scholar]
- Salnikova AB, Kryndushkin DS, Smirnov VN, Kushnirov VV, Ter-Avanesyan MD. Nonsense suppression in yeast cells overproducing Sup35 (eRF3) is caused by its non-heritable amyloids. J Biol Chem. 2005;280:8808–8812. doi: 10.1074/jbc.M410150200. [DOI] [PubMed] [Google Scholar]
- Seaman MN, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma J, Wisniewski BT, Paulson E, Obaoye JO, Merrill SJ, Manogaran AL. De novo [PSI +] prion formation involves multiple pathways to form infectious oligomers. Sci Rep. 2017;7:76. doi: 10.1038/s41598-017-00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Madariaga ML, Lindquist S. Prion switching in response to environmental stress. PLoS Biol. 2008;6:e294. doi: 10.1371/journal.pbio.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishveshwara N, Bradley ME, Liebman SW. Sequestration of essential proteins causes prion associated toxicity in yeast. Mol Microbiol. 2009;73:1101–1114. doi: 10.1111/j.1365-2958.2009.06836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn RD, Bapat K, Newnam GP, Zink AD, Chernoff YO. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol Cell Biol. 2001;21:4656–4669. doi: 10.1128/MCB.21.14.4656-4669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Derkatch IL, Liebman SW. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI(+)] and [PIN(+)] Mol Microbiol. 2001;39:37–46. doi: 10.1046/j.1365-2958.2001.02224.x. [DOI] [PubMed] [Google Scholar]


