Abstract
VieA is a cyclic diguanylate phosphodiesterase that modulates biofilm development and motility in Vibrio cholerae O1 of the classical biotype. vieA is part of an operon encoding the VieSAB signal transduction pathway that is nearly silent in V. cholerae of the El Tor biotype. A DNA pull-down assay for proteins interacting with the vieSAB promoter identified the LysR-type regulator LeuO. We show that in classical biotype V. cholerae, LeuO cooperates with the nucleoid-associated protein H-NS to repress vieSAB transcription. LeuO and H-NS interacted with the vieSAB promoter of both biotypes with similar affinities and protected overlapping DNA sequences. H-NS was expressed at similar levels in both cholera biotypes. In contrast, El Tor biotype strains expressed negligible LeuO under identical conditions. In El Tor biotype vibrios, transcription of vieSAB is repressed by the quorum sensing regulator HapR, which is absent in classical biotype strains. Restoring HapR expression in classical biotype V. cholerae repressed vieSAB transcription by binding to its promoter. We propose that double locking of the vieSAB promoter by H-NS and HapR in the El Tor biotype prior to the cessation of exponential growth results in a more pronounced decline in VieA specific activity compared to the classical biotype.
Keywords: Vibrio cholerae, VieA, H-NS, LeuO, quorum sensing
Graphical Abstract
Introduction
The seventh and current cholera pandemic is characterized by the predominance of V. cholerae serogroup O1 of the El Tor biotype. A distinguishing feature of cholera caused by this biotype is a lower case-fatality rate compared to former classical biotype pandemic strains. Recently, wave 3 hypervirulent El Tor strains that produce classical cholera toxin have been identified (Kim et al., 2014, Kim et al., 2015). Genome sequencing of these strains revealed the presence of mutations in the regulatory genes hns, encoding the histone-like nucleoid structuring protein (H-NS) and vieA, encoding a cyclic diguanylic acid (c-di-GMP) phosphodiesterase (PDE) (Satchell et al., 2016). Transfer of hns and vieA single nucleotide polymorphisms (SNPs) from a wave 3 strain into a prototype wave 1 strain resulted in a hypervirulent phenotype (Carignan et al., 2016). These results suggest that natural selection of strains containing mutations in the above loci could be contributing to the increased severity and duration of recent cholera outbreaks. The vieA gene is part of an operon encoding the VieSAB signal transduction system that is differentially expressed in V. cholerae biotypes (Beyhan et al., 2006, Wang et al., 2015). This operon is expressed in the classical biotype to regulate ≈ 10% of the genome (Beyhan et al., 2006) but is nearly silent in V. cholerae of the El Tor biotype (Wang et al., 2015). Downstream targets of VieSAB include the vps and rbm genes required to make the biofilm extracellular exopolysacchide and protein matrix (Hammer & Bassler, 2009, Beyhan et al., 2006, Tischler & Camilli, 2004). The VieSAB signal transduction system consists of the hybrid sensor kinase VieS, the response regulator VieA and the auxiliary protein VieB. The VieA protein exhibits receiver, helix-turn-helix (HTH) and EAL domains, the later acting as a PDE (Tamayo et al., 2005). VieS acts through VieA to regulate gene expression by altering the c-di-GMP pool while VieB associates with VieS to inhibit VieA phosphorylation (Martinez-Wilson et al., 2008, Mitchell et al., 2015).
In El Tor biotype V. cholerae, transcription of vieSAB is silenced by H-NS and the master quorum sensing regulator HapR (Wang et al., 2015). H-NS is an abundant nucleoid-associated protein that functions as a nucleoid organizer and a transcriptional repressor with a bias for silencing the expression of laterally-acquired genes (Dorman & Kane, 2009, Winardhi et al., 2015). In V. cholerae, H-NS acts to repress the transcription of genes located within the toxin co-regulated pilus (TCP) pathogenicity island, the ctxAB operon encoding cholera toxin, as well as genes encoding hemolysin (hlyA), the multifunctional autoprocessing repeats-in-toxin (MARTX) and its transport system and genes required for the expression of the biofilm extracellular matrix (Ayala et al., 2015, Wang et al., 2015, Ayala et al., 2017). H-NS can silence transcription by direct occlusion of promoter elements, bridging DNA sequences to entrap RNA polymerase within a repression loop or binding to DNA upstream the promoter followed by oligomerization to contact and stall RNA polymerase (Fang & Rimsky, 2008, Dorman, 2013). Commonly, genes repressed by H-NS can be activated by transcription factors that antagonize H-NS binding, bridging or oligomerization (Dorman & Kane, 2009). For example, the regulator VpsT antagonizes H-NS repression at vpsA and vpsL promoters that control vibrio exopolysaccharide biosynthesis (Ayala et al., 2015); ToxT antagonizes H-NS repression at the tcpA and ctxAB promoters that control TCP and cholera toxin expression, respectively (Yu & DiRita, 2002, Stonehouse et al., 2011), and ToxR antagonizes H-NS repression at several promoters that include vpsL (Kazi et al., 2016).
The role of VieSAB in biofilm development and the potential impact of vieA SNPs in the emergence of atypical El Tor variants, prompted us to investigate the molecular basis for its differential expression in V. cholerae biotypes. An assay for proteins interacting with the vieSAB promoter identified LeuO, a stationary phase regulator belonging to the LysR family (Hernandez-Lucas & Calva, 2012). We show that in classical biotype V. cholerae, LeuO cooperates with H-NS to maintain vieSAB transcription at a basal level, which is nevertheless elevated compared to the El Tor biotype. LeuO was expressed at a very low level in El Tor biotype strains compared to the classical biotype. Instead, the quorum sensing regulator HapR, which is expressed in El Tor biotype strains, binds to the vieSAB promoter to repress its transcription.
Results
LeuO is a repressor of the vieSAB operon
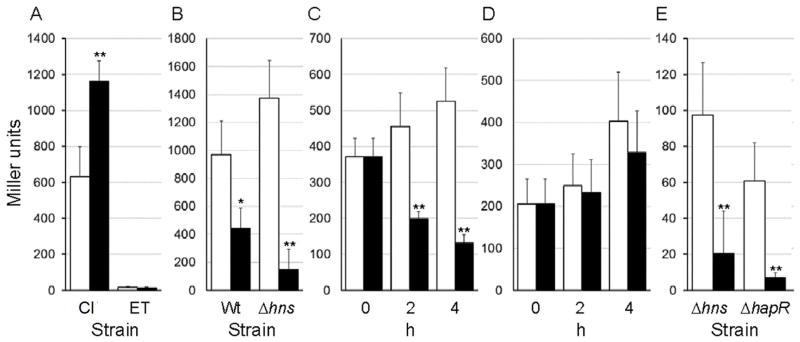
The transcription of vieSAB is repressed by H-NS in both V. cholerae biotypes. However, H-NS appeared to act as a weaker repressor in the classical biotype (Wang et al., 2015). We hypothesized that additional transcription factors present (or expressed at higher levels) in classical biotype V. cholerae could antagonize H-NS repression. Thus, we developed a DNA pull-down assay and used mass spectrometry (MS) to identify proteins capable of binding to a biotinylated vieSAB promoter probe immobilized onto streptavidin-coupled magnetic beads. We prioritized MS hits identifying DNA-binding proteins pulled-down from a lysate of strain O395 lacking H-NS but not detected (or detected to a lesser extent) in the wild type lysate. A list of MS hits is shown in Table S1. The assay identified the regulator LeuO, previously reported to antagonize H-NS repression in Escherichia coli and Salmonella enterica (De la Cruz et al., 2007, Shimada et al., 2011, Dillon et al., 2012, Espinosa & Casadesus, 2014). To investigate the role of LeuO in vieSAB transcription, we constructed leuO deletion mutants of strain O395ΔlacZ (classical biotype) and C7258ΔlacZ (El Tor biotype) and measured the expression of a vieSAB-lacZ promoter fusion in the wild type and ΔleuO mutant. In the classical biotype, the leuO mutant expressed elevated vieSAB-lacZ activity compared to wild type (Fig. 1A) suggesting that LeuO acts as a repressor. As previously reported (Wang et al., 2015), the El Tor strain expressed very low levels of vieSAB and deletion of leuO in this biotype had no effect (Fig. 1A). To confirm the effect of LeuO in the classical biotype, we constructed the plasmid pBAD-LeuO in which LeuO is expressed from the araBAD promoter. Strains O395ΔlacZ and O395ΔlacZΔhns were transformed with pBAD-LeuO or the empty vector pBAD33 and grown to stationary phase in LB medium supplemented with L-arabinose. Overexpression of LeuO significantly diminished vieSAB-lacZ activity in both strains (Fig. 1B). Next, we examined if induction of LeuO in exponentially growing cells repressed vieSAB expression. In Fig. 1C we show that induction of LeuO in mid-exponential phase results in repression of vieSAB transcription while no effect could be demonstrated in strains harboring the empty vector (Fig. 1D). We conclude that in classical biotype V. cholerae, LeuO can negatively regulate vieSAB expression regardless of the growth phase at which it is induced. We repeatedly failed to construct a ΔleuOΔhns mutant of strain O395ΔlacZ suggesting that these genes could be synthetic lethal. Consistent with previous findings (Wang et al., 2015), vieSAB expression was derepressed in El Tor biotype hns and hapR mutants (Fig. 1E). Ectopic expression of LeuO in El Tor biotype hns and hapR mutants diminished vieSAB expression (Fig. 1E). Next, we examined if LeuO repression of vieSAB transcription in classical biotype V. cholerae resulted in significantly diminished levels of VieA protein. To this end, we constructed a suicide vector encoding VieA tagged at its C-terminus with the FLAG epitope. This vector was integrated by homologous recombination into the vieA locus of strain O395ΔlacZ. The resulting co-integrate grew poorly in rich medium. In this strain, integration of vector sequences downstream of vieA may prevent or diminish expression of vieB. Thus, expression of VieA could be deleterious in the absence of the inhibitory protein VieB. The apparent toxicity was relieved by removing the C-terminal VieA HTH domain. Hence, we constructed a derivative of strain O395ΔlacZ containing a suicide vector encoding VieAΔHTH-FLAG integrated into the vieA locus (O395VieAΔHTH-FLAG). This strain was transformed with plasmids pBAD33 or pBADLeuO-6xHis. In Fig. 2 we show that overexpression of LeuO resulted in an 87 % reduction in VieA protein expression. The same blot was examined for the expression of LeuO-6xHis and TCP. No effect on TCP expression was found under these experimental conditions.
Fig. 1. LeuO represses vieSAB transcription.
A. Strains O395ΔlacZ (classical biotype, Cl) and C7258ΔlacZ (EL Tor biotype, ET) (□, open bars) and their isogenic ΔleuO mutants (■, filled bars) containing a vieSAB-lacZ promoter fusion were grown to stationary phase in LB medium B. Strain O395ΔlacZ (Wt) and its Δhns mutant containing a vieSAB-lacZ promoter fusion were transformed with pBAD33 (□, open bar) or pBAD-LeuO (■, filled bar) and grown to stationary phase in the presence of L-arabinose (0.2 %). Panels C and D. Strain O395ΔlacZ containing a vieSAB-lacZ promoter fusion was transformed with plasmid pBAD-LeuO (C) or pBAD33 (D). Strains were grown to mid-exponential phase and the cultures divided in halves. In one half, LeuO expression was induced by addition of L-arabinose (0.2 %) (■, filled bars) and the remaining half was used as a control (□, open bars). E. El Tor biotype strains AJB80 (Δhns) and AJB51ΔlacZ (ΔhapR) containing a vieSAB-lacZ promoter fusion were transformed with pBAD33 (□, open bar) or pBAD-LeuO (■, filled bar) and grown to stationary phase in the presence of L-arabinose (0.02 %). β-galactosidase activity (Miller units) was measured as an indicator of promoter activity. The error bars denote the standard deviation of at least three independent transformants. Symbols: * p < 0.05; ** p < 0.01 (unpaired T test).
Fig. 2. LeuO represses VieA protein expression.
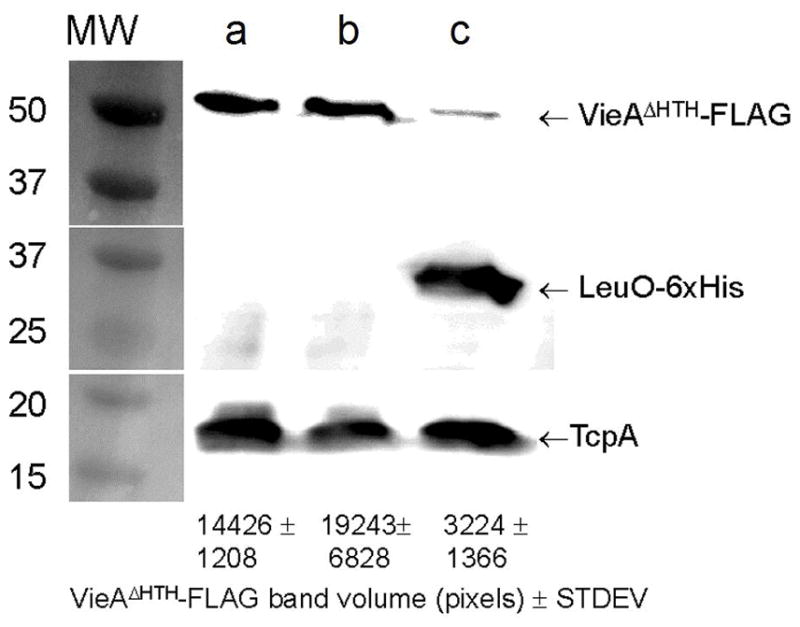
Strain O395VieAΔHTH-FLAG expressing a chromosomally-integrated allele encoding VieAΔHTH-FLAG was transformed with plasmid pBAD33 or pBADLeuO-6xHis. Triplicate cultures of each transformants were grown in LB medium to stationary phase and the expression of VieAΔHTH-FLAG, LeuO-6xHis and TcpA was determined by western blot as described in materials and methods. Lane a, O395VieAΔHTH-FLAG containing pBAD33; lane b, O395VieAΔHTH-FLAG containing pBADLeuO-6xHis grown in LB medium; lane c, O395VieAΔHTH-FLAG containing pBADLeuO-6xHis grown in LB supplemented with L-arabinose (0.02 %). Quantitation of VieAΔHTH-FLAG expression by densitometry is shown at the bottom of the figure (mean band volume ± STDEV, n = 3).
Since the PDE activity of VieA acts to diminish the c-di-GMP pool to negatively affect vibrio exopolysaccharide (vps) biosynthesis (Hammer & Bassler, 2009), we examined the effect of overexpressing LeuO in strain O395ΔlacZ and its isogenic ΔvieA mutant containing a vpsL-lacZ promoter fusion. As expected, deletion of vieA increased the expression a vpsL-lacZ fusion approximately 3-fold (Fig. 3A). Overexpression of LeuO resulted in a 12-fold increase in vpsL-lacZ expression in strain O395ΔlacZ and a 7-fold increase in the vieA mutant background (Fig. 3A). These results suggest that LeuO enhances vps expression by multiple pathways that include repression of the vieSAB operon. Both, ectopic expression of LeuO or deletion of vieA alone significantly (p < 0.01, n = 6) diminished motility in swarm agar (Fig. 3B).
Fig. 3. LeuO enhances vibrio exopolysaccharide gene expression and inhibits motility.
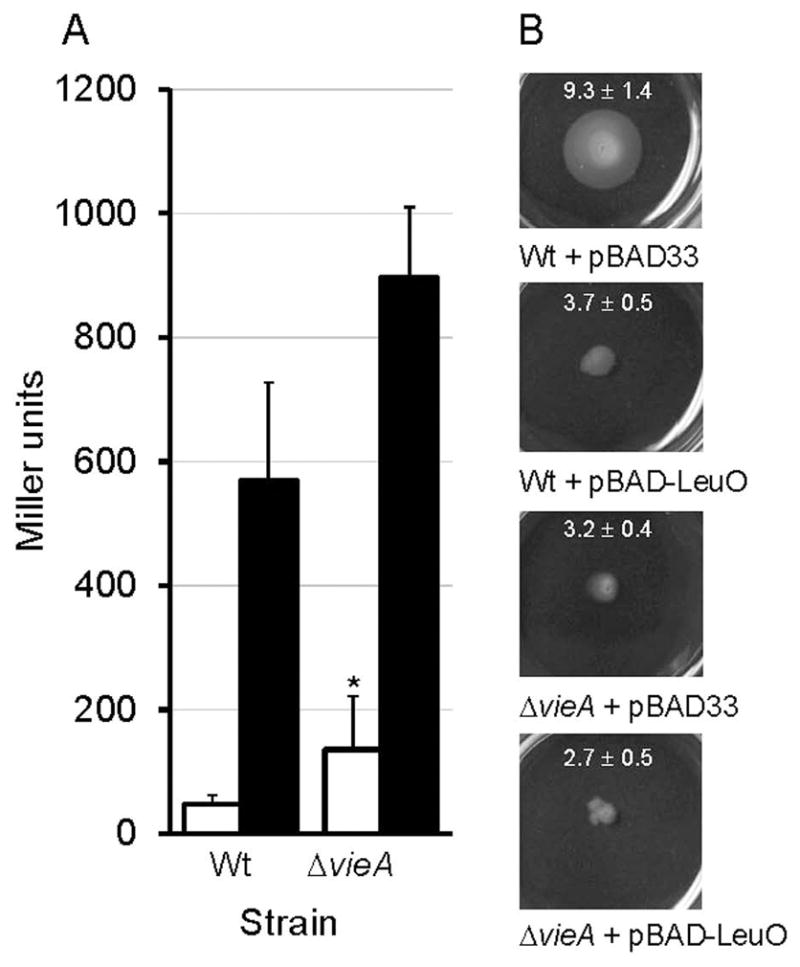
A. Strain O395ΔlacZ (Wt) and its ΔvieA isogenic mutant containing a vpsL-lacZ promoter fusion (Wang et al., 2012b) were transformed with pBAD33 (□, open bar) or pBAD-LeuO (■, filled bar) and grown to stationary phase in the presence of L-arabinose (0.2 %). β-galactosidase activity (Miller units) was measured as an indicator of promoter activity. The error bars denote the standard deviation of five independent transformants. Symbols: * p < 0.05 (ΔvieA versus Wt comparison, unpaired T- test). B. Overnight cultures of the above strains (six independent transformants per strain) were stabbed into 3 mL of swarm agar in 6-well microtiter plates. Plates were incubated 24 h at 30°C and photographed. The swarm halo (mm) ± STDEV (n = 6) are indicated within each representative picture.
Electrophoresis mobility shift assays (EMSA) confirmed that LeuO binds to the vieSAB promoter (Fig. S1). We found that LeuO and H-NS exhibited similar dissociation constant (Kd) values for this promoter (Table 1). The sequence located upstream of the vieS translation start differs in classical and El Tor biotype V. cholerae at five nucleotide positions. Thus, we estimated the Kd values of LeuO and H-NS for the same vieSAB promoter fragment amplified from strain C7258. The EMSA gels used to calculate the Kd values are shown in Fig. S1 and the Kd values for the classical and El Tor vieSAB promoters are summarized in Table 1. No differences in the affinity of LeuO and H-NS for either vieSAB promoter were found.
Table 1.
Binding and transcription inhibition of vieSAB by H-NS and LeuO
| Binding Kd (95 % CI*) | ||
|---|---|---|
|
| ||
| vieSAB promoter | ||
|
|
||
| Transcription factor | Classical biotype | El Tor biotype |
| H-NS | 44.1 (41.7–46.4) nM | 52.3 (45.8–58.8) nM |
| LeuO | 31.2 (29.9–32.6) nM | 31.2 (29.5–32.9) nM |
|
| ||
| Transcription inhibition IC50 (95% CI) | ||
|
|
||
| Transcription factor | High Eσ70 | Low Eσ70 |
|
| ||
| H-NS | 0.8 (0.6–1.2) μM | 3.6 (3.0–4.4) μM |
| LeuO | 2.8 (2.1–3.7) μM | 3.2 (2.1–4.8) μM |
Confidence interval
LeuO and H-NS cooperate to repress vieSAB transcription
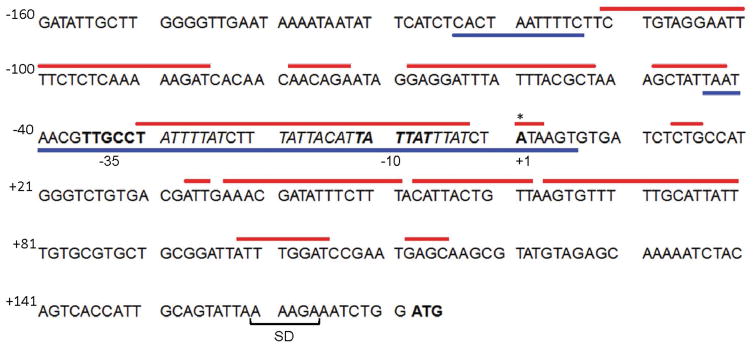
The above results prompted us to examine the interplay between LeuO and H-NS in the repression of vieSAB transcription. First, the transcription start site (TSS) of vieSAB was mapped by 5′-RACE analyses to an adenine located 171 bp upstream of the vieS start codon (Fig. 4). Then, vieSAB promoter regions protected by LeuO and H-NS were determined by DNase I footprinting. These regulators protected overlapping DNA sequences in both strands spanning the vieSAB TSS and the upstream −10 promoter element (Fig. 4). The electropherograms corresponding to the H-NS and LeuO DNase I footprints on both strands of vieSAB promoter DNA are shown in Fig. S2A, S2B and S3. A sequence matching the E. coli LeuO binding motif was identified within the vieSAB promoter DNA region protected by V. cholerae LeuO (Fig. 4). This finding is consistent with the E. coli and V. cholerae proteins sharing 94 % identity along their DNA binding domains. The above results suggested that LeuO and H-NS could have an antagonistic relationship for binding to the vieSAB promoter, which nevertheless results in transcriptional repression. We conducted a competitive EMSA assay in which vieSAB promoter DNA is pre-incubated with H-NS, treated with increasing amounts of LeuO and the final nucleoprotein complexes are sized by polyacrylamide gel electrophoresis. The above experiments suggested that LeuO could gradually displace H-NS from the promoter (Fig. 5A). The reverse experiment showed that H-NS could displace pre-bound LeuO from the promoter, though this required a higher molar excess of H-NS over LeuO (Fig. 5B). We note that protein-DNA complexes of intermediate molecular weight between the H-NS-vieSAB and LeuO-vieSAB nucleoproteins were also detected. This pattern prompted us to examine the possibility of a direct interaction between H-NS and LeuO prior to DNA binding. To this end, we tested if chitin beads coupled to an H-NS-intein-chitin-binding domain (CBD) fusion could pull-down purified LeuO from solution. In Fig. 6A, we show that the H-NS-intein-CBD bait pulled-down LeuO. However, a small amount of LeuO was also pulled-down by chitin beads coupled to the intein-CBD moiety (negative control). Thus, we further examined if the H-NS-intein-CBD bait could pull-down LeuO from a lysate of V. cholerae O395ΔlacZΔleuO transformed with a plasmid expressing LeuO tagged with the FLAG epitope (pUC-rrnB-leuO-FLAG). In Fig. 6B, we show that the H-NS-intein-CBD bait pulled-down LeuO-FLAG. No LeuO-FLAG was pulled-down in this experiment by the intein-CBD bait (Fig. 6B). Altogether, the above experiments suggest a possible interaction between H-NS and LeuO at the protein level.
Fig. 4. LeuO and H-NS bind to overlapping DNA sequences at the vieSAB promoter.
The vieSAB TSS (base +1) was determined by 5′-RACE and is indicated with an asterisk. Sequences matching −10 and −35 promoter elements are shown in bold font. The vieS start codon is shown in bold font preceded by the putative Shine-Dalgarno (SD) ribosome binding site. Red lines above the sequence represent regions protected by H-NS on both strands determined by DNase I footprint analysis. Blue lines below the sequence indicate regions protected by LeuO on both strands. A sequence matching the E. coli LeuO binding motif (Dillon et al., 2012) located within the LeuO protected region is indicated in italics.
Fig. 5. LeuO can displace H-NS from the vieSAB promoter.
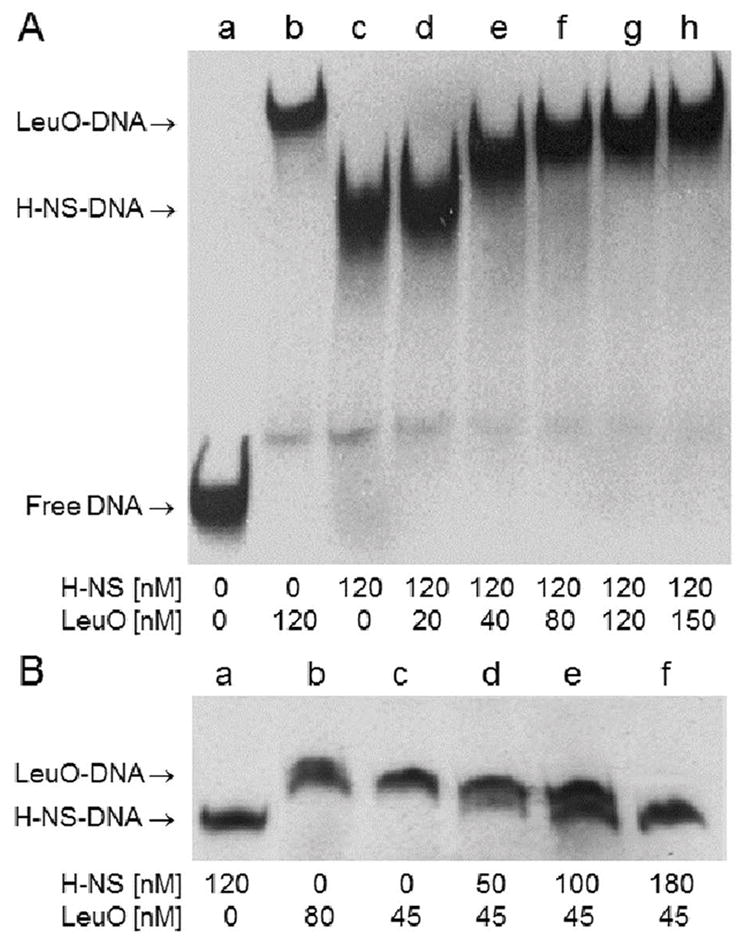
A. A competitive EMSA was conducted using a DIG-labeled DNA fragment containing the H-NS and LeuO DNase I protected regions at the vieSAB promoter and spanning nucleotides −125 to +6 relative to the TSS. The promoter DNA fragment was pre-incubated with H-NS (lane c) and subsequently treated with increasing concentrations of LeuO (lanes d through h). The mobility of free DNA (lane a) and the LeuO- and H-NS-vieSAB nucleoprotein complexes (lane b and c, respectively) are indicated at the left of the gel. B. The same DIG-labeled DNA fragment was pre-incubated with LeuO (lane c) and subsequently treated with increasing concentrations of H-NS (lanes d through f).
Fig. 6. Interaction between H-NS and LeuO.
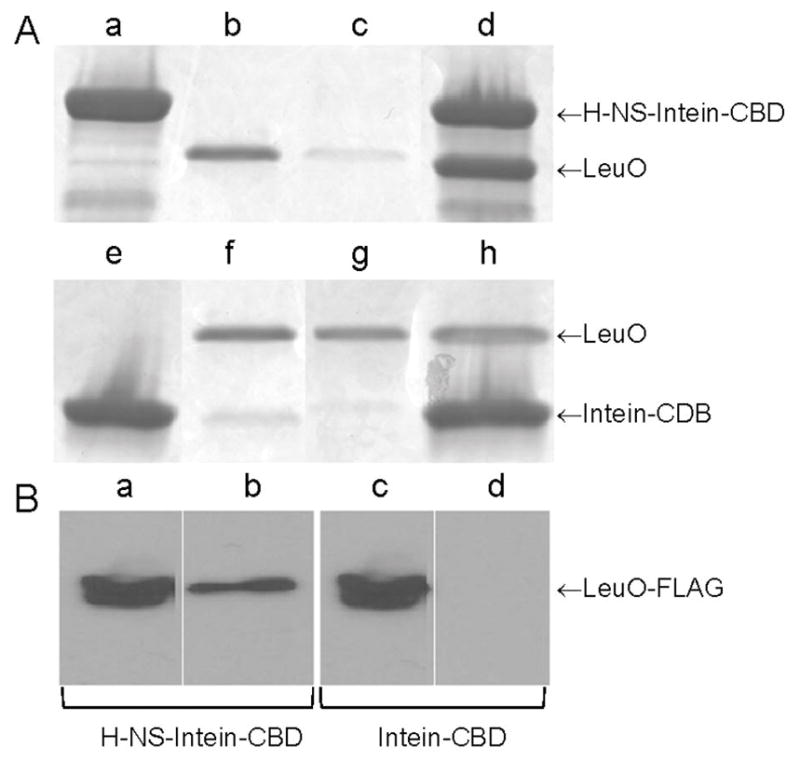
A. Fifty μL of chitin beads coupled to an H-NS-intein-CBD fusion (Wang et al., 2012a) (lane a) or to the intein-CBD moiety alone (lane e) were reacted with 20 μg of purified LeuO (lanes b,f). Pull-down was conducted using the binding buffer used for EMSA and Costar Spin-X centrifuge tube filters. Lanes c and g, flow through fractions; lanes d and h, pulled-down fractions. Proteins were separated by SDS-PAGE and stained with Coomassie brilliant blue. B. Fifty μL of chitin beads coupled to an H-NS-intein-CBD fusion (lanes a and b) or to the intein-CBD moiety alone (lanes c and d) were reacted with 0.4 mg of total proteins from a lysate of strain O395ΔlacZΔleuO containing plasmid pUC-rrnB-leuO-FLAG. Lanes a and c, cell lysate input; lanes b and d, pulled-down fractions. LeuO-FLAG was detected by western blot using the anti-FLAG M2-peroxidase monoclonal antibody.
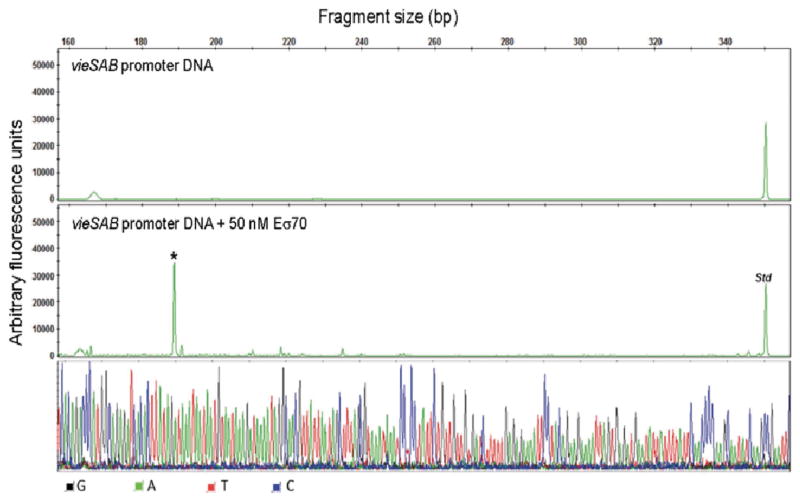
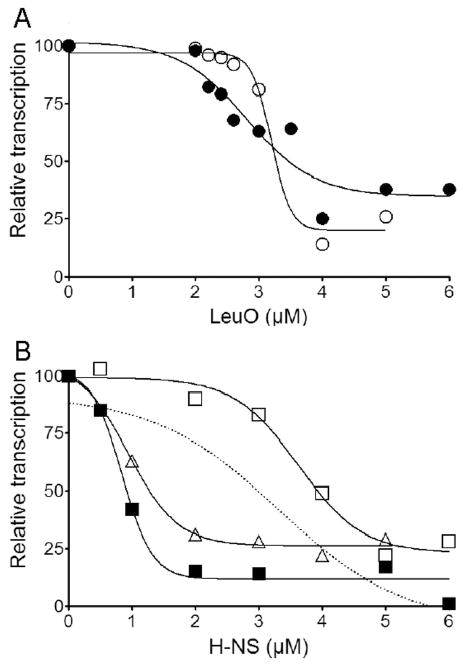
We used an in vitro transcription system to study the effect of LeuO and/or H-NS binding to the vieSAB promoter on transcription initiation. In vitro transcription was conducted at two concentrations of σ70-saturated RNA polymerase (Eσ70) and varying concentrations of LeuO and/or H-NS. In the absence of added repressors, this assay yielded 0.25–0.40 ng/μL of vieSAB cDNA at the low Eσ70 concentration and 1.63 ng/μL of vieSAB cDNA at the high Eσ70 concentration. Transcription initiation in vitro occurred at a cytosine located 173 bp upstream the vieS start codon in close agreement with the TSS determined by 5′-RACE analyses (Fig. 7). Addition of LeuO inhibited vieSAB transcription to a similar extent at both Eσ70 concentrations (Fig. 8A). In contrast, transcription inhibition by H-NS was more pronounced at the high Eσ70 concentration (Fig. 8B). We also conducted in vitro transcription at varying concentration of H-NS in the presence of a fixed concentration of LeuO (Fig. 8B, Δ). The dotted line plotted in Fig. 8B corresponds to the predicted additive inhibition of vieSAB transcription by LeuO and H-NS based on the single repressor reactions. The experimental inhibition curve (Δ), however, showed a more than additive effect suggesting that LeuO and H-NS interact with the vieSAB promoter to inhibit transcription in a synergistic manner. The 50 % inhibitory concentrations (IC50) of LeuO and H-NS are summarized in Table 1. The electropherograms supporting Fig. 8 are shown in supporting information Fig. S4, S5, and S6.
Fig. 7. In vitro transcription of vieSAB DNA.
A vieSAB DNA template spanning nucleotides −160 to +392 relative to the TSS was incubated 1 h at 37○C with or without Eσ70 (negative control). The resulting RNA products were purified and annealed with primer HEX-VieS1 complementary to the vieS open reading frame and extended as described in the methods section. The vieSAB transcription start site is indicated with an asterisk (*). A HEX-labeled DNA standard (Std) was added to each reaction before capillary electrophoresis analysis. The signal from each electropherogram peak is reported as arbitrary fluorescence units along the y axis and the transcript length in base pairs (bp) is reported at the top of the panel. The resulting nucleotide sequence corresponds to the template strand.
Fig. 8. Cooperation between H-NS and LeuO in the transcriptional silencing of vieSAB.
A. In vitro transcription of vieSAB was conducted in the presence of varying concentrations of LeuO at Eσ70 concentrations of 25 nM (○) and 50 nM (●). B. In vitro transcription of vieSAB was conducted in the presence of varying concentrations of H-NS at Eσ70 concentrations of 25 nM (□) and 50 nM (■) or at a Eσ70 concentrations of 25 nM plus a fixed concentration (3 μM) of LeuO (Δ). The dotted line indicates the predicted added effect of H-NS and LeuO on transcription inhibition based on single repressor transcription reactions.
Silencing of vieSAB expression in El Tor biotype V. cholerae is not due to elevated H-NS and/or LeuO levels
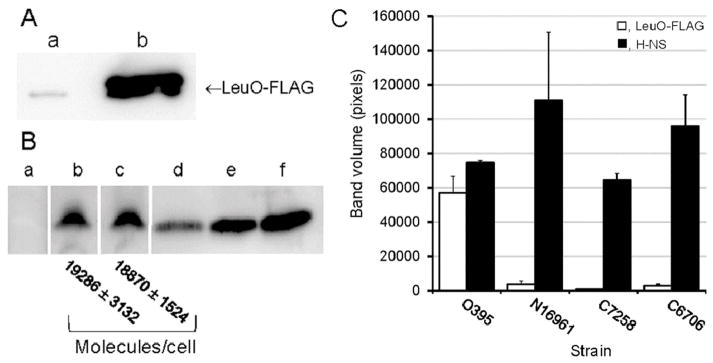
We next examined if the lower expression of vieSAB in V. cholerae of the El Tor biotype was a consequence of elevated levels of LeuO and/or H-NS. We constructed a plasmid expressing LeuO from native transcription and translation signals fused to the FLAG epitope to compare its expression in V. cholerae biotypes. Since transcription of leuO in El Tor biotype V. cholerae is repressed by H-NS (Wang et al., 2015), we confirmed that the expression of LeuO-FLAG was elevated in an hns mutant of strain C7258 (data not shown). Strain C7258 expressed negligible LeuO compared to classical biotype O395 (Fig. 9A). In contrast, both classical and El Tor biotype V. cholerae expressed similar levels of H-NS in stationary phase cultures (Fig. 9B). To exclude the possibility of strain C7258 alone harboring a mutation preventing LeuO expression, we tested the production of H-NS and LeuO-FLAG in two additional El Tor biotype strains (C6706, N16961). Both El Tor strains expressed negligible LeuO (Fig. 9C).
Fig. 9. Expression of H-NS and LeuO in V. cholerae biotypes.
A. Strains C7258ΔlacZ (El Tor, lane a) and O395ΔlacZ (classical, lane b) were transformed with vector pUC-rrnB-leuO-FLAG encoding a leuO-FLAG fusion expressed under its native promoter. The strains were grown in LB medium to stationary phase and the expression of LeuO-FLAG was determined by western blot. B. Strains C7258ΔlacZΔhns (AJB80, lane a), C7258ΔlacZ (lane b) and O395ΔlacZ (lane c) were grown in LB medium to stationary phase and the expression of H-NS was determined by western blot. Lanes d, e and f were loaded with 40, 80 and 160 ng of purified H-NS protein, respectively. The number of H-NS molecules per cell was estimated by densitometry and dilution plating. C. Quantitative densitometry analysis of LeuO and H-NS expression in classical and El Tor V. cholerae biotypes. V. cholerae strains of the classical and El Tor biotypes containing plasmid pUC-rrnB-leuO-FLAG were grown to stationary phase in LB at 37°C and the cells were collected for western blot analysis. Expression of LeuO-FLAG was detected using anti-FLAG M2-peroxidase monoclonal antibody (□, open bars). H-NS was quantified using a polyclonal antibody as described in materials and methods (■, filled bars). The bars indicate the average band volume (pixels) ± STDEV from four independent cultures.
Restoring expression of the master quorum sensing regulator HapR in classical biotype V. cholerae directly represses vieSAB transcription
A major difference between strain O395 and current pandemic El Tor biotype strains is the use of distinct signal transduction systems regulating the c-di-GMP pool. Strain O395 uses the VieA pathway to modify the c-di-GMP pool in response to unknown signal(s) (Hammer & Bassler, 2009). Instead, most El Tor biotype strains use the HapR pathway to modulate the c-di-GMP pool in response to the concentration of cholera autoinducer 1 in the extracellular milieu (Hammer & Bassler, 2009). Since a hapR mutant of El Tor biotype strain C7258 expressed elevated vieSAB-lacZ activity (Fig. 1E) (Wang et al., 2015), we tested if restoring HapR expression in the classical biotype strain O395 (known to harbor a frame shift mutation in hapR) repressed vieSAB transcription. Introduction of plasmid pC1.1 harboring the hapR gene (Jobling & Holmes, 1997) in strains O395ΔlacZ and O395ΔlacZΔhns significantly diminished vieSAB transcription (Fig. 10A). Expression of hapR was independently confirmed by measuring the production of HA/protease encoded by hapA (data not shown); a gene that has a stringent requirement for HapR (Jobling & Holmes, 1997, Silva & Benitez, 2004). To determine if HapR repressed the vieSAB operon directly, we purified HapR and conducted an EMSA. In Fig. 10B we show that HapR specifically binds to the vieSAB promoter (lanes b,c) with a Kd of 173.8 nM (95 % CI, 167.5–180.1) (Fig. S7). An excess of unlabeled vieSAB promoter DNA fully removed HapR from the DIG-labeled promoter probe (lane d) while an excess of an unlabeled DNA fragment located upstream aphA but lacking the HapR binding site (Kovacikova & Skorupski, 2002) did not (lane e). In Fig. 10C, we show the position of the predicted HapR binding site relative to the vieSAB TSS. The predicted binding site was determined using the FIMO algorithm (Grant et al., 2011) and the type-1 HapR motif described in (Tsou et al., 2009).
Fig. 10. The quorum sensing regulator HapR is a vieSAB repressor.
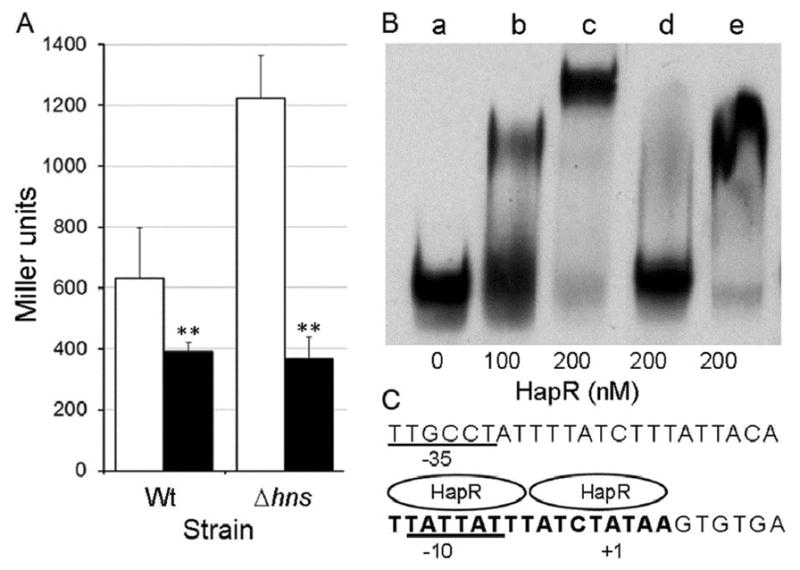
A. Strain O395ΔlacZ (Wt) and O395ΔlacZΔhns (Δhns) containing a vieSAB-lacZ fusion were transformed with parent plasmid pACY184 (□. open bars) or pC1.1 encoding HapR (■, filled bars). The strains were grown in LB and β-galactosidase activity (Miller units) was measured as an indicator of promoter activity. The error bars denote the standard deviation of three independent transformants. Symbols: ** p < 0.01 (unpaired T test). B. A DIG-labeled vieSAB promoter fragment spanning nucleotide −198 to +70 relative to the TSS was treated with increasing concentration of pure HapR (lanes b and c), 200 nM HapR in the presence of a 200-fold excess of unlabeled competitor vieSAB DNA (lane d) or 200 nM HapR in the presence of a 200-fold excess of unlabeled DNA upstream aphA lacking the HapR binding site (amplified with primers AphA51 and AphA149). C. Predicted HapR binding site (bold font) at the vieSAB promoter relative to the TSS (+1 nucleotide) and −10/−35 elements.
Discussion
Genome sequencing of atypical El Tor isolates have suggested the natural selection of strains harboring mutations in hns and vieA (Satchell et al., 2016). In the classical biotype, VieA acts to lower the c-di-GMP pool (Tischler & Camilli, 2004) to diminish vibrio exopolysaccharide gene expression (Beyhan et al., 2006) and biofilm formation, a condition that enhances vibrio survival in the environment and cholera transmission (Conner et al., 2016, Tamayo et al., 2010). In contrast, VieA is expressed at a very low level in El Tor biotype strains (Beyhan et al., 2006, Wang et al., 2015), in which the c-di-GMP pool is modulated by the LuxO/HapR quorum sensing pathway (Hammer & Bassler, 2009).
A screening for transcription factors capable of antagonizing H-NS binding to the vieSAB promoter identified the regulator LeuO. This protein belongs to the LysR family of transcription factors and consists of an N-terminal winged HTH (wHTH) motif and a C-terminal unidentified co-inducer-binding domain (Maddocks & Oyston, 2008). This protein has been shown to bind DNA as a tetramer and similar to H-NS, form oligomers capable of DNA bridging (Guadarrama et al., 2014). LeuO has been described as an H-NS antagonist (De la Cruz et al., 2007, Shimada et al., 2011, Dillon et al., 2012, Espinosa & Casadesus, 2014). In contrast, we found that it cooperates with H-NS to repress vieSAB transcription. H-NS and LeuO bound to the vieSAB promoter with similar affinities and protected overlapping DNA sequences. A competitive EMSA suggested the H-NS-vieSAB and LeuO-vieSAB nucleoproteins to predominate at low and high LeuO concentration, respectively. A pull-down experiment suggested a potential interaction between the H-NS and LeuO proteins. H-NS has been reported to form heteromeric complexes with its paralogue StpA (Johansson et al., 2001) and the Hha-family of nucleoid-associated proteins (Nieto et al., 2002) but not with non-histone-like proteins. It is noteworthy that binding of Hha to H-NS enhanced its DNA bridging function (van der Valk et al., 2017). It remains to be tested if a physical interaction between LeuO and H-NS has the above or any effect on its DNA binding pattern. The EMSA does not unequivocally discern if H-NS and LeuO bind to the vieSAB promoter in the form of heterodimers/hetero-oligomers, co-bind simultaneously as separate molecular entities or entirely displace each other from the promoter. Chromatin immunoprecipitation on chip and genomic SELEX (Systematic Evolution of Ligands with Exponential enrichment) studies conducted in S. enterica and E. coli have suggested that co-binding of H-NS and LeuO to target promoters could be a widespread partnership (Dillon et al., 2012, Ishihama et al., 2016). A compilation of V. cholerae genes repressed by H-NS and exhibiting a LeuO binding motif in the proximity of their TSS is shown in Table S2. The list includes genes encoding the regulator TcpP (tcpP), the repeats-in toxin transport system (rtxB), cholera toxin (ctxA) and hemolysin (hlyA). Therefore, LeuO may have a broader impact than anticipated in V. cholerae pathogenesis.
We developed an in vitro transcription assay to quantitatively assess the effect of H-NS and/or LeuO binding to the vieSAB promoter on transcription initiation. Transcription initiation in vitro and in vivo was in close agreement indicating that the in vitro assay provided an accurate representation of vieSAB transcription in the cell. The repressive effect of H-NS on vieSAB transcription was more pronounced at the high Eσ70 concentration. This condition is likely to prevail during exponential growth as the effective concentration of Eσ70 diminishes in the stationary phase due to sigma factor competition (Farewell et al., 1998). Furthermore, our result is consistent with the finding that Eσ70 acts as a cofactor for H-NS DNA looping-mediated repression (Shin et al., 2005). H-NS is a highly abundant protein that is expressed at similar levels during V. cholerae exponential growth and stationary phase (Wang et al., 2012b). In contrast, the expression of LeuO has been reported to occur upon bacterial entry into stationary phase (Fang et al., 2000, Dillon et al., 2012, Bina et al., 2013). Expression of LeuO in the stationary phase could compensate for H-NS acting as a weaker repressor at this physiological stage.
Inhibition of transcription initiation in the presence of H-NS and LeuO was more pronounced compared to when H-NS and LeuO were added separately at the same concentration. This brings into question the mechanism by which H-NS and LeuO may cooperate to inhibit vieSAB transcription. Based on current knowledge, several possibilities will require attention. Binding of one protein to the promoter may result in changes in DNA shape that favor binding of the second protein. For instance, the interaction of the wHTH domain of LeuO with DNAs minor groove at the vieSAB promoter could be particularly sensitive to changes in DNA shape induced upon H-NS binding (Dorman & Dorman, 2017). In addition, a physical interaction between both proteins could stabilize conformations with enhanced DNA binding, oligomerization and bridging functions.
Our results show that induction of leuO expression at mid-exponential phase results in the biosynthesis of an active LeuO protein conducive to vieSAB repression. The role of LeuO in V. cholerae virulence has been recently recognized. For instance, LeuO is part of the ToxR regulon and contributes to bile resistance (Ante et al., 2015b); it down regulates virulence gene expression in response to cyclic dipeptides (Bina et al., 2013), it represses the transcription factor CadC resulting in diminished acid tolerance (Ante et al., 2015a), and it modulates cationic antimicrobial peptide resistance (Bina et al., 2016). Since induction of LeuO can repress vieSAB transcription in both exponential and stationary phases, it could potentially function at multiple stages of the V. cholerae infective cycle provided the appropriate stimulus (i.e. bile) occurs. We found that induction of LeuO expression significantly enhanced vps gene expression and diminished motility. Comparison of the ectopic expression of LeuO in isogenic wild type and vieA strains suggested that repression of vieSAB by LeuO is partially responsible for this effect. The above findings add support to previous studies in which (i) leuO was identified in a screen for genes affecting biofilm development (Moorthy & Watnick, 2005), (ii) transcription of leuO was found to be activated by bile (Ante et al., 2015b) and (iii) bile was reported to enhance the c-di-GMP pool (Koestler & Waters, 2014).
Classical and El Tor biotype strains used in this study produced similar levels of H-NS protein. In contrast, El Tor biotype strains unexpectedly produced negligible LeuO. A possible explanation for this finding is a lower expression of ToxR under the growth conditions used. ToxR acts at the leuO promoter by antagonizing H-NS repression (Kazi et al., 2016). However, even in an El Tor hns mutant, the expression of LeuO was significantly lower compared to the classical biotype (data not shown). Thus, the low expression of vieSAB in El Tor biotype V. cholerae cannot be explained by elevated levels of H-NS, LeuO or differences in the affinity of these proteins for the vieSAB promoter. In a previous study, we observed that an El Tor biotype hapR mutant expressed elevated vieSAB (Wang et al., 2015). Here we show that restoring hapR expression in classical biotype V. cholerae results in significant repression of vieSAB transcription. Furthermore, we demonstrate that HapR binds to the vieSAB promoter at a position predicted to overlap its TSS and −10 promoter elements. ChIP-seq (Ayala et al. 2015) and DNase I footprinting suggest that H-NS also associates with this region of the vieSAB promoter to potentially obstruct the binding of other transcription factors. Consistent with this view, both HapR and LeuO exhibited a stronger repressive effect on vieSAB-lacZ expression in the absence of H-NS. Finally, we note that our screening for proteins interacting with the vieSAB promoter identified the MarR family regulator IclR and the Entner-Doudoroff repressor HexR (Table S1). However, the potential regulatory input of these factors on vieSAB transcription remains to be determined.
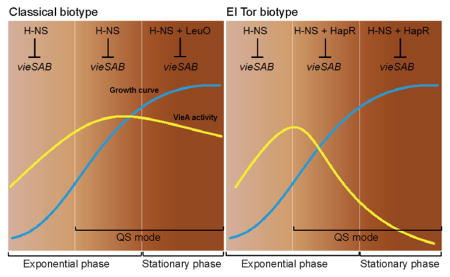
Taken together, our study shows that transcription of the vieSAB operon is subject to dual repression by H-NS and LeuO in classical biotype V. cholerae and by H-NS and HapR in the El Tor biotype. We suggest that the differential expression of vieSAB in V. cholerae biotypes results from the distinct timing at which dual repression occurs (Fig. 11). Classical biotype strains express a basal level of vieSAB controlled by H-NS during exponential growth, which can diminish in stationary phase due to the expression of LeuO. In this biotype, VieSAB expression is sufficiently high to exert its regulatory functions. In the El Tor biotype, double locking of the vieSAB promoter by HapR and H-NS prior to cessation of exponential growth, results in an exponential decline in vieSAB specific activity that does not occur in the classical biotype. This model does not exclude VieSAB playing a role in the pathogenesis of El Tor biotype strains prior to cells entering quorum sensing mode. The above regulatory connections provide a basis to study how natural selection of mutations in vieA, hns, and quorum sensing regulators could lead to the emergence of hypervirulent V. cholerae variants.
Fig. 11. Model for the differential expression of vieSAB in V. cholerae biotypes.
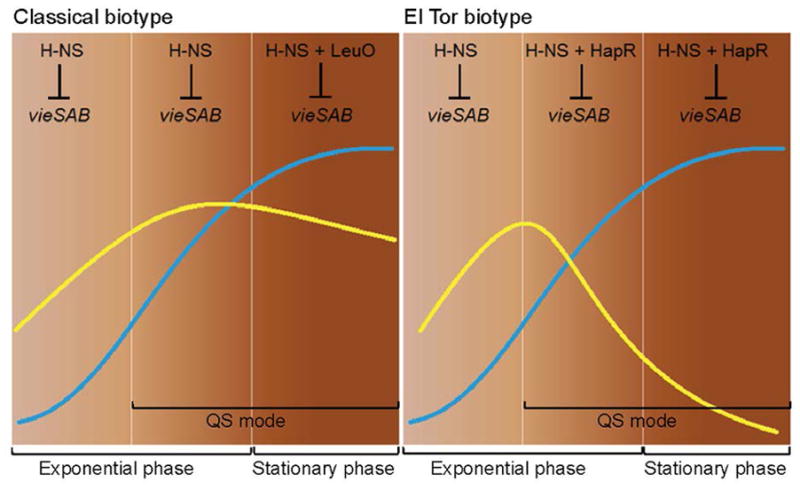
Transcription of the vieSAB operon is subject to dual repression by H-NS and LeuO in classical biotype V. cholerae and by H-NS and HapR in the El Tor biotype. In the classical biotype, H-NS determines the basal vieSAB transcription rate during exponential growth and LeuO cooperates with H-NS to further repress vieSAB transcription in stationary phase. In the El Tor biotype, the combined repression of vieSAB by H-NS and HapR prior to the cessation of exponential growth results in an exponential decline in VieA specific activity. Symbols: ⊥, transcriptional repression, QS, quorum sensing. Blue lines indicate bacterial growth curves. Yellow lines represent VieA specific activity (VieA/cell mass).
Experimental procedures
Strains and media
V. cholerae mutants used in this study were derived from the classical and El Tor biotype strains O395 and C7258, respectively. V. cholerae was grown in Luria-Bertani (LB) medium at 37°C with agitation. Escherichia coli strains TOP10 (Life Technologies) or SM10λpir (Miller & Mekalanos, 1988) were used for plasmid propagation. Culture media were supplemented with ampicillin (100 μg/ml), chloramphenicol (5 μg/ml), kanamycin (25 μg/ml), polymyxin B (100 units/ml), streptomycin (100 μg/ml), isopropyl-β-D-thiogalactopyranoside (IPTG) (0.5 mM), 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (20 μg/ml) or L-arabinose (0.02–0.2 %) as required.
Construction of mutants and plasmids
The construction of a vieSAB-lacZ and vpsL-lacZ promoter fusions was described previously (Wang et al., 2015, Wang et al., 2012b). To construct a leuO deletion mutant, the suicide vector pWM91ΔleuO (Moorthy & Watnick, 2005) was transferred by conjugation from E. coli SM10λpir to strain O395ΔlacZ or C7258ΔlacZ. The resulting leuO deletion mutants JCA9 (O395ΔlacZΔleuO) and JCA7 (C7258ΔlacZΔleuO) were isolated by allelic exchange and sucrose selection as previously described (Liang et al., 2007). Deletion of the leuO allele was confirmed by PCR with primer pair LeuUpFw/LeuDownRv and DNA sequencing. A similar approach was used to introduce a vieA deletion in strain O395ΔlacZ using the suicide vector pCVDΔVieA (Wang et al., 2015). To study the expression of LeuO in different V. cholerae strains, we constructed plasmid pUC-rrnB-leuO-FLAG encoding leuO tagged with the FLAG epitope expressed from its own promoter. To this end, the rrnBT1T2 transcription terminator was amplified with primer pair RrnBUpFw/RrnBDwRv from plasmid pACTluz-RpoS (Wang et al., 2014) and inserted in pUC19 to create pUC-rrnB. Then, the leuO promoter region and gene was amplified from V. cholerae O395 genomic DNA with primer pair LeuOFw/LeuFlagR and ligated into pUC-rrnB to obtain vector pUC-rrnB-leuO-FLAG. The plasmid pBAD-LeuO expressing LeuO from the araBAD promoter was constructed by amplifying the leuO gene with primers LeuO-F-Sac and LeuO-R-FLAG-Xba from vector pUC-rrnB-leuO-FLAG, followed by cloning in pBAD33 (Guzman et al., 1995) using the SacI and XbaI overhangs. Plasmid pBADLeuO-6xHis was constructed as described above but using primers LeuO-F-Sac and LeuO-HisR-Xba followed by cloning of the 6xhis-tagged leuO ORF in pBAD33. To construct plasmid pCVDVieAΔHTH, 1008 bp of vieA coding sequence lacking the C-terminal HTH domain was amplified using primers VieA-FLAGF-SphI and VieA-FLAGR-M-SalI, the latter oligonucleotide containing a DNA sequence encoding the FLAG epitope. The resulting PCR product was ligated into suicide vector pCVD442 (Donnenberg & Kaper, 1991) and transferred by conjugation to strain O395ΔlacZ for integration into the chromosomal vieA locus. The resulting strain (O395VieAΔHTH-FLAG), expresses a C-terminally truncated VieA protein tagged with the FLAG epitope from native transcription and translation initiation signals. Strains, plasmids and oligonucleotide primers used throughout this work are described in supporting information (Tables S3 and S4).
Screening for proteins binding to vieSAB promoter DNA
A DNA fragment encoding the vieSAB promoter was amplified with the biotinylated primer Bio-VieS and VieS-R2. The PCR product was purified using the QIAquick PCR purification kit (QIAGEN) and confirmed by DNA sequencing. A 20 μL mix containing 100 μg of streptavidin-coupled dynabeads and 2.5 μg of biotinylated DNA was incubated for 90 min at room temperature. The dynabeads-DNA complex was washed to remove unbound DNA and resuspended in 500 μL of a bacterial lysate containing 1.5 mg of total proteins. The mixture was incubated for 90 min at room temperature and the dynabeads-DNA-protein complexes were captured and washed 5 times with 500 μL of wash buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA, 0.1 M NaCl). Pulled-down proteins were eluted by resuspension and boiling 5 min in 40 μL of Laemmli sample buffer (BioRad). Samples were loaded and run 1 cm into Any kD Mini-PROTEAN TGX precast gels (BioRad), stained with Coomassie brilliant blue and processed for nanoLC-tandem mass spectrometry using an AB Sciex 5600 TripleTOF mass spectrometer. For preparing lysates, strains O395ΔlacZ and O395ΔlacZΔhns were grown in LB medium to optical density at 600 nm 0.5. Cells were harvested, resuspended in lysis buffer (20 mM HEPES pH 7.6, 1 mM EDTA, 10 mM (NH4)2SO4, 1 mM DTT, 0.1% Tween 20, 30 mM KCl, 75 mM NaCl plus Roche’s cOmplete protease inhibitor cocktail) and disrupted by sonication. The cleared lysates were adjusted to 3 mg/mL of total proteins with lysis buffer. Finally, 500 μL of the lysates were incubated with 6 μg of calf thymus DNA for 10 min prior to addition of the dynabead-DNA promoter probe.
Purification of H-NS, LeuO and HapR
H-NS and HapR were purified as described previously (Wang et al., 2011, Wang et al., 2012a). To purify LeuO, the 989 bp leuO open reading frame (ORF) was amplified with primers LeuFLAG-F and LeuOpTX-R and cloned into plasmid pTXB1 (New England BioLabs, NEB) to generate pTXB1-LeuO. The LeuO-intein-CBD fusion in pTXB1-LeuO was confirmed by DNA sequencing using T7 universal and Mxe Intein II reverse primers (NEB). LeuO was then expressed from the T7 promoter in E. coli ER2566 and purified by using the NEB IMPACT protein purification system following the kit’s accompanying protocol. The purity of recombinant H-NS, HapR and LeuO was estimated to be higher than 90 % by densitometry analysis of Coomassie-stained polyacrylamide gels using the ImageQuant TL software (GE Healthcare).
Electrophoresis mobility shift assays (EMSA)
EMSA were conducted using the second-generation digoxigenin (DIG) gel shift kit (Roche Applied Sciences) as previously described (Wang et al., 2012b, Wang et al., 2012a, Wang et al., 2014). DNA fragments encoding the vieSAB promoter were generated by PCR with the primer pair VieS-EMSA-Fw/VieS-EMSA-Rv. Gel images were acquired using the biomolecular imager ImageQuant LAS 4000 (GE Healthcare) and the binding activity was quantitated by densitometry using the ImageQuant TL software. Protein-DNA equilibrium dissociation constants (Kd) were estimated by fitting the data to the Hill equation by nonlinear regression as described previously (15). For a competitive EMSA, vieSAB promoter DNA was amplified using primer pair VieS-EMSA-F2/VieS-EMSA-R2. The DIG-labeled promoter was pre-incubated with a fixed quantity of one protein (i.e. H-NS or LeuO) to form the initial nucleoprotein complex. After 15 min at 30°C, increasing amounts of the second protein was added and incubation continued for an additional 15 min.
5′-RACE analyses
Total RNA was prepared using the RNeasy mini kit (Qiagen), RQ1 RNase-free DNase (Promega) and the RNeasy MiniElute clean-up kit (Qiagen). 5-RACE was conducted using the 5′-RACE System for Rapid Amplification of cDNA Ends kit (Invitrogen) using primer VieS-R16 for first strand cDNA synthesis. Second and nested PCR reactions were conducted using gene-specific primers VieS-R17 and VieS-R18 in combination with the abridged anchor and abridged universal amplification primers provided in the kit. The PCR products were cloned in plasmid pUC19 and the DNA sequence was determined for six independent transformants.
DNase I footprint analysis
A DNA fragment encoding the vieSAB promoter was amplified using the 6-carboxyfluorescein (FAM)- and 6-carboxy-2′,4,4′,5′,7,7′-hexachlorofluorescein (HEX)-labeled primer pair FAM-VieS-F6/HEX-VieS-R2. Protein binding, DNase I digestion and detection of fluorescently-labeled DNase I digestion products was performed as described previously (Wang et al., 2014). A template DNA fragment encoding the vieSAB promoter was amplified with the primer pair VieS-F6/VieS-R2 and ligated into vector pCR2.1 (Life Technologies) to generate pCR-VieSAB. This plasmid was used to generate a sequence ladder as described previously (Wang et al., 2014). The DNase I footprint electropherograms were aligned with the manually-generated sequence using the GeneMapper software.
In vitro transcription
A vieSAB DNA template was amplified using primer pair VieS-F6/VieS-R3 and purified with the QIAquick PCR purification kit (QIAGEN). One μg of template DNA was incubated with either H-NS or LeuO in 18 μL of RNA Polymerase Reaction Buffer (NEB) for 25 min at 30°C. Then, the mixture was supplemented with nucleotide triphosphate (NTP) mixture (0.5 mM of each NTP), 8 units of RNase inhibitor (Ambion) and either 0.5 or 1.0 units of E. coli Eσ70 (NEB) and the reaction was incubated 1 h at 37°C. The template was digested with 5 units of RQ1 RNase-Free DNase I (Promega) for 1 h at 30°C and the RNA transcript was purified and eluted in 14 μL of H2O using the QIAGEN RNeasy MinElute cleanup kit. Ten μL of RNA was annealed with 30 picomole of the HEX-labeled primer HEX-VieS1 and extended using 5 units of AMV reverse transcriptase (Promega) for 1 h at 42°C following the First-Strand Synthesis of cDNA accompanying protocol. A 352 bp HEX-labeled DNA standard generated with primer pair VieS-F6/HEX-VieS-R2 was added to each sample to a final concentration of 0.5 ng/μL for samples generated using 0.5 units of Eσ70 or 0.75 ng/μL for samples generated with 1.0 unit of Eσ70.
Quantitative analysis of in vitro transcription products
Two μL of the final samples were mixed with 7.5 μL Hi-Di formamide and 0.5 μL of GeneScan 500 LIZ size standard (Applied Biosystems) prior to analysis in a 3730 capillary sequencer. Electropherograms were generated using GeneMapper v. 4.0 software running a default microsatellite analysis. To accurately assign a nucleotide base to in vitro transcript peaks, a manually-generated sequence ladder was generated as described in the DNase I footprint analysis section but with primer HEX-VieS1 and vector pCR-VieSAB as DNA template. The GeneMapper software was used to align the transcription reaction products with the manually-generated dideoxy sequencing reactions to determine the size and start nucleotide of the vieSAB transcripts. The effect of H-NS and LeuO on vieSAB transcription was estimated from the height of vieSAB transcript peaks expressed in arbitrary fluorescent units. Transcription inhibition curves were generated with the aid of GraphPad Prism 5.0 (GraphPad, San Diego, CA) using the log (inhibitor) vs response-variable slope analysis.
Western blot analysis
Western blots were conducted using the BM bioluminescence western blotting kit (Roche Applied Science) as described previously (Wang et al., 2012a). In all experiments, bacterial samples were collected by centrifugation and adjusted to 10 OD600 units by resuspension in Laemmli sample buffer (BioRad). Twenty μL of samples were loaded onto Any kD Mini-PROTEAN TGX gels (BioRad). To measure H-NS expression, known concentrations of purified H-NS were loaded into gels alongside the bacterial samples. H-NS was quantitated by densitometry using a polyclonal antibody, immunostaining and the ImageQuant TL software as previously described (Wang et al., 2012a, Wang et al., 2012b). In parallel, colony forming units corresponding to each sample were determined by dilution plating in LB agar. To compare the expression of LeuO in V. cholerae strains, plasmid pUC-rrnB-leuO-FLAG was introduced into different V. cholerae strains and the expression of the LeuO-FLAG fusion protein was determined by western blotting using the anti-FLAG M2-peroxidase monoclonal antibody (Sigma-Aldrich). Expression of LeuO-6xHis was determined using the anti-His (C-terminal) –HRP monoclonal antibody provided by Invitrogen. Finally, expression of TCP was detected using a rabbit antiserum against the major pilus subunit TcpA as described previously (Rasmussen et al., 2011).
Supplementary Material
Acknowledgments
This study was supported by awards AI119625 and AI104993-03 (AJS), AI103693-03 (JAB) and F31AI106288 (JCA) from the National Institutes of Health. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. We are grateful to Mei Han and Caitlin Cox from the University of Alabama at Birmingham (UAB) Heflin Center for Genomic Sciences for assistance in DNA fragment analysis. We are grateful to Landon Wilson (UAB Targeted Metabolomics and Proteomics Laboratory) for assistance in the mass spectrometry analysis. The suicide vector pWM91ΔleuO was kindly provided by Paula Watnick (Harvard Medical School).
Footnotes
AJS and JAB planned research; JCA and HW conducted experiments; AJS, JAB and JCA prepared manuscript
The authors declare no conflict of interest.
References
- Ante VM, Bina XR, Bina JE. The LysR-type regulator LeuO regulates the acid tolerance response in Vibrio cholerae. Microbiology. 2015a;161:2434–2443. doi: 10.1099/mic.0.000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ante VM, Bina XR, Howard MF, Sayeed S, Taylor DL, Bina JE. Vibrio cholerae leuO transcription is positively regulated by ToxR and contributes to bile resistance. Journal of bacteriology. 2015b;197:3499–3510. doi: 10.1128/JB.00419-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala JC, Silva AJ, Benitez JA. H-NS: an overarching regulator of the Vibrio cholerae life cycle. Research in microbiology. 2017;168:16–25. doi: 10.1016/j.resmic.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala JC, Wang H, Silva AJ, Benitez JA. Repression by H-NS of genes required for the biosynthesis of the Vibrio cholerae biofilm matrix is modulated by the second messenger cyclic diguanylic acid. Molecular microbiology. 2015;97:630–645. doi: 10.1111/mmi.13058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyhan S, Tischler AD, Camilli A, Yildiz FH. Differences in gene expression between the classical and El Tor biotypes of Vibrio cholerae O1. Infection and immunity. 2006;74:3633–3642. doi: 10.1128/IAI.01750-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina XR, Howard MF, Ante VM, Bina JE. Vibrio cholerae LeuO links the ToxR regulon to expression of lipid A remodeling genes. Infection and immunity. 2016;84:3161–3171. doi: 10.1128/IAI.00445-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina XR, Taylor DL, Vikram A, Ante VM, Bina JE. Vibrio cholerae ToxR downregulates virulence factor production in response to cyclo(Phe-Pro) mBio. 2013;4:e00366–00313. doi: 10.1128/mBio.00366-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carignan BM, Brumfield KD, Son MS. Single nucleotide polymorphisms in regulator-encoding genes have an additive effect on virulence gene expression in a Vibrio cholerae clinical isolate. mSphere. 2016;1(5):e00253–16. doi: 10.1128/mSphere.00253-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner JG, Teschler JK, Jones CJ, Yildiz FH. Staying alive: Vibrio cholerae’s cycle of environmental survival, transmission, and dissemination. Microbiology spectrum. 2016;4(2) doi: 10.1128/microbiolspec.VMBF-0015-2015. VMBF-0015-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Cruz MA, Fernandez-Mora M, Guadarrama C, Flores-Valdez MA, Bustamante VH, Vazquez A, Calva E. LeuO antagonizes H-NS and StpA-dependent repression in Salmonella enterica ompS1. Molecular microbiology. 2007;66:727–743. doi: 10.1111/j.1365-2958.2007.05958.x. [DOI] [PubMed] [Google Scholar]
- Dillon SC, Espinosa E, Hokamp K, Ussery DW, Casadesus J, Dorman CJ. LeuO is a global regulator of gene expression in Salmonella enterica serovar Typhimurium. Molecular microbiology. 2012;85:1072–1089. doi: 10.1111/j.1365-2958.2012.08162.x. [DOI] [PubMed] [Google Scholar]
- Donnenberg MS, Kaper JB. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infection and immunity. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman CJ. Co-operative roles for DNA supercoiling and nucleoid-associated proteins in the regulation of bacterial transcription. Biochemical Society transactions. 2013;41:542–547. doi: 10.1042/BST20120222. [DOI] [PubMed] [Google Scholar]
- Dorman CJ, Dorman MJ. Control of virulence gene transcription by indirect readout in Vibrio cholerae and Salmonella enterica serovar Typhimurium. Environmental microbiology. 2017;19:3834–3845. doi: 10.1111/1462-2920.13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman CJ, Kane KA. DNA bridging and antibridging: a role for bacterial nucleoid-associated proteins in regulating the expression of laterally acquired genes. FEMS microbiology reviews. 2009;33:587–592. doi: 10.1111/j.1574-6976.2008.00155.x. [DOI] [PubMed] [Google Scholar]
- Espinosa E, Casadesus J. Regulation of Salmonella enterica pathogenicity island 1 (SPI-1) by the LysR-type regulator LeuO. Molecular microbiology. 2014;91:1057–1069. doi: 10.1111/mmi.12500. [DOI] [PubMed] [Google Scholar]
- Fang FC, Rimsky S. New insights into transcriptional regulation by H-NS. Current opinion in microbiology. 2008;11:113–120. doi: 10.1016/j.mib.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M, Majumder A, Tsai KJ, Wu HY. ppGpp-dependent leuO expression in bacteria under stress. Biochemical and biophysical research communications. 2000;276:64–70. doi: 10.1006/bbrc.2000.3440. [DOI] [PubMed] [Google Scholar]
- Farewell A, Kvint K, Nystrom T. Negative regulation by RpoS: a case of sigma factor competition. Molecular microbiology. 1998;29:1039–1051. doi: 10.1046/j.1365-2958.1998.00990.x. [DOI] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guadarrama C, Medrano-Lopez A, Oropeza R, Hernandez-Lucas I, Calva E. The Salmonella enterica serovar Typhi LeuO global regulator forms tetramers: residues involved in oligomerization, DNA binding, and transcriptional regulation. Journal of bacteriology. 2014;196:2143–2154. doi: 10.1128/JB.01484-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. Journal of bacteriology. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL. Distinct sensory pathways in Vibrio cholerae El Tor and classical biotypes modulate cyclic dimeric GMP levels to control biofilm formation. Journal of bacteriology. 2009;191:169–177. doi: 10.1128/JB.01307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Lucas I, Calva E. The coming of age of the LeuO regulator. Molecular microbiology. 2012;85:1026–1028. doi: 10.1111/j.1365-2958.2012.08175.x. [DOI] [PubMed] [Google Scholar]
- Ishihama A, Shimada T, Yamazaki Y. Transcription profile of Escherichia coli: genomic SELEX search for regulatory targets of transcription factors. Nucleic acids research. 2016;44:2058–2074. doi: 10.1093/nar/gkw051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling MG, Holmes RK. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Molecular microbiology. 1997;26:1023–1034. doi: 10.1046/j.1365-2958.1997.6402011.x. [DOI] [PubMed] [Google Scholar]
- Johansson J, Eriksson S, Sonden B, Wai SN, Uhlin BE. Heteromeric interactions among nucleoid-associated bacterial proteins: localization of StpA-stabilizing regions in H-NS of Escherichia coli. Journal of bacteriology. 2001;183:2343–2347. doi: 10.1128/JB.183.7.2343-2347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazi MI, Conrado AR, Mey AR, Payne SM, Davies BW. ToxR antagonizes H-NS regulation of horizontally acquired genes to drive host colonization. PLoS pathogens. 2016;12:e1005570. doi: 10.1371/journal.ppat.1005570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Lee CH, Nair GB, Kim DW. Whole-genome sequence comparisons reveal the evolution of Vibrio cholerae O1. Trends in microbiology. 2015;23:479–489. doi: 10.1016/j.tim.2015.03.010. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Lee D, Moon SH, Lee CH, Kim SJ, Lee JH, Kim JO, Song M, Das B, Clemens JD, Pape JW, Nair GB, Kim DW. Molecular insights into the evolutionary pathway of Vibrio cholerae O1 atypical El Tor variants. PLoS pathogens. 2014;10:e1004384. doi: 10.1371/journal.ppat.1004384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koestler BJ, Waters CM. Bile acids and bicarbonate inversely regulate intracellular cyclic di-GMP in Vibrio cholerae. Infection and immunity. 2014;82:3002–3014. doi: 10.1128/IAI.01664-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacikova G, Skorupski K. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Molecular microbiology. 2002;46:1135–1147. doi: 10.1046/j.1365-2958.2002.03229.x. [DOI] [PubMed] [Google Scholar]
- Liang W, Pascual-Montano A, Silva AJ, Benitez JA. The cyclic AMP receptor protein modulates quorum sensing, motility and multiple genes that affect intestinal colonization in Vibrio cholerae. Microbiology. 2007;153:2964–2975. doi: 10.1099/mic.0.2007/006668-0. [DOI] [PubMed] [Google Scholar]
- Maddocks SE, Oyston PC. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology. 2008;154:3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- Martinez-Wilson HF, Tamayo R, Tischler AD, Lazinski DW, Camilli A. The Vibrio cholerae hybrid sensor kinase VieS contributes to motility and biofilm regulation by altering the cyclic diguanylate level. Journal of bacteriology. 2008;190:6439–6447. doi: 10.1128/JB.00541-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. Journal of bacteriology. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SL, Ismail AM, Kenrick SA, Camilli A. The VieB auxiliary protein negatively regulates the VieSA signal transduction system in Vibrio cholerae. BMC microbiology. 2015;15:59. doi: 10.1186/s12866-015-0387-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorthy S, Watnick PI. Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Molecular microbiology. 2005;57:1623–1635. doi: 10.1111/j.1365-2958.2005.04797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto JM, Madrid C, Miquelay E, Parra JL, Rodriguez S, Juarez A. Evidence for direct protein-protein interaction between members of the enterobacterial Hha/YmoA and H-NS families of proteins. Journal of bacteriology. 2002;184:629–635. doi: 10.1128/JB.184.3.629-635.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen L, White EL, Pathak A, Ayala JC, Wang H, Wu JH, Benitez JA, Silva AJ. A high-throughput screening assay for inhibitors of bacterial motility identifies a novel inhibitor of the Na+-driven flagellar motor and virulence gene expression in Vibrio cholerae. Antimicrobial agents and chemotherapy. 2011;55:4134–4143. doi: 10.1128/AAC.00482-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchell KJ, Jones CJ, Wong J, Queen J, Agarwal S, Yildiz FH. Phenotypic analysis reveals that the 2010 Haiti cholera epidemic is linked to a hypervirulent strain. Infection and immunity. 2016;84:2473–2481. doi: 10.1128/IAI.00189-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Bridier A, Briandet R, Ishihama A. Novel roles of LeuO in transcription regulation of E. coli genome: antagonistic interplay with the universal silencer H-NS. Molecular microbiology. 2011;82:378–397. doi: 10.1111/j.1365-2958.2011.07818.x. [DOI] [PubMed] [Google Scholar]
- Shin M, Song M, Rhee JH, Hong Y, Kim YJ, Seok YJ, Ha KS, Jung SH, Choy HE. DNA looping-mediated repression by histone-like protein H-NS: specific requirement of Esigma70 as a cofactor for looping. Genes & development. 2005;19:2388–2398. doi: 10.1101/gad.1316305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AJ, Benitez JA. Transcriptional regulation of Vibrio cholerae hemagglutinin/protease by the cyclic AMP receptor protein and RpoS. Journal of bacteriology. 2004;186:6374–6382. doi: 10.1128/JB.186.19.6374-6382.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonehouse EA, Hulbert RR, Nye MB, Skorupski K, Taylor RK. H-NS binding and repression of the ctx promoter in Vibrio cholerae. Journal of bacteriology. 2011;193:979–988. doi: 10.1128/JB.01343-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo R, Patimalla B, Camilli A. Growth in a biofilm induces a hyperinfectious phenotype in Vibrio cholerae. Infection and immunity. 2010;78:3560–3569. doi: 10.1128/IAI.00048-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo R, Tischler AD, Camilli A. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. The Journal of biological chemistry. 2005;280:33324–33330. doi: 10.1074/jbc.M506500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler AD, Camilli A. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Molecular microbiology. 2004;53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou AM, Cai T, Liu Z, Zhu J, Kulkarni RV. Regulatory targets of quorum sensing in Vibrio cholerae: evidence for two distinct HapR-binding motifs. Nucleic acids research. 2009;37:2747–2756. doi: 10.1093/nar/gkp121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Valk RA, Vreede J, Qin L, Moolenaar GF, Hofmann A, Goosen N, Dame RT. Mechanism of environmentally driven conformational changes that modulate H-NS DNA-bridging activity. eLife. 2017:6. doi: 10.7554/eLife.27369. pii: e27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ayala JC, Benitez JA, Silva AJ. Interaction of the histone-like nucleoid structuring protein and the general stress response regulator RpoS at Vibrio cholerae promoters that regulate motility and hemagglutinin/protease expression. Journal of bacteriology. 2012a;194:1205–1215. doi: 10.1128/JB.05900-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ayala JC, Benitez JA, Silva AJ. The LuxR-type regulator VpsT negatively controls the transcription of rpoS, encoding the general stress response regulator, in Vibrio cholerae biofilms. Journal of bacteriology. 2014;196:1020–1030. doi: 10.1128/JB.00993-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ayala JC, Benitez JA, Silva AJ. RNA-seq analysis identifies new genes regulated by the histone-like nucleoid structuring protein (H-NS) affecting Vibrio cholerae virulence, stress response and chemotaxis. PloS one. 2015;10:e0118295. doi: 10.1371/journal.pone.0118295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ayala JC, Silva AJ, Benitez JA. The histone-like nucleoid structuring protein (H-NS) is a repressor of Vibrio cholerae exopolysaccharide biosynthesis (vps) genes. Applied and environmental microbiology. 2012b;78:2482–2488. doi: 10.1128/AEM.07629-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wu JH, Ayala JC, Benitez JA, Silva AJ. Interplay among cyclic diguanylate, HapR, and the general stress response regulator (RpoS) in the regulation of Vibrio cholerae hemagglutinin/protease. Journal of bacteriology. 2011;193:6529–6538. doi: 10.1128/JB.05166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winardhi RS, Yan J, Kenney LJ. H-NS Regulates gene expression and compacts the nucleoid: insights from single-molecule experiments. Biophysical journal. 2015;109:1321–1329. doi: 10.1016/j.bpj.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu RR, DiRita VJ. Regulation of gene expression in Vibrio cholerae by ToxT involves both antirepression and RNA polymerase stimulation. Molecular microbiology. 2002;43:119–134. doi: 10.1046/j.1365-2958.2002.02721.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.