Abstract
Iron-sulfur cluster biogenesis is a complex, but highly regulated process that involves de novo cluster formation from iron and sulfide ions on a scaffold protein and delivery to final targets via a series of Fe-S cluster binding carrier proteins. The process of cluster release from the scaffold/carrier for transfer to the target proteins is mediated by a dedicated Fe-S cluster chaperone system. In human cells, the chaperones include heat shock protein HSPA9 and the J-type chaperone Hsc20. While the role of chaperones has been somewhat clarified in yeast and bacterial systems, many questions remain over their functional roles in cluster delivery and interactions with a variety of human Fe-S cluster proteins. One such protein, Nfu, has recently been recognized as a potential interaction partner of the chaperone complex. Herein, we examined the ability of human Nfu to function as a carrier by interacting with the human chaperone complex. Human Nfu is shown to bind to both chaperone proteins with binding affinities similar to those observed for IscU binding to the homologous HSPA9 and Hsc20, while Nfu can also stimulate the ATPase activity of HSPA9. Additionally, the chaperone complex was able to promote Nfu function by enhancing the second-order rate constants for Fe-S cluster transfer to target proteins and providing directionality in cluster transfer from Nfu by eliminating promiscuous transfer reactions. Together, these data support a hypothesis in which Nfu can serve as an alternative carrier protein for chaperone-mediated cluster release and delivery in Fe-S cluster biogenesis and trafficking.
Keywords: Iron-sulfur clusters, heat shock chaperones, cluster trafficking, thermodynamics, ATPase
Graphical abstract

Biosynthesis of iron-sulfur (Fe-S) clusters, a common metallocofactor, occurs in a highly regulated process with the assistance of a dedicated chaperone system consisting of HSPA9 and Hsc20. The complex role of these chaperones is currently unclear; however, we show that they can interact with and regulate the activity of the Fe-S cluster protein Nfu in cellular cluster trafficking.
Introduction
Studies of the isc (iron-sulfur cluster) operon have revealed the presence of both DnaK and DnaJ chaperones [4, 5], suggesting that the corresponding encoded products were potentially involved in iron-sulfur (Fe-S) cluster biosynthesis [6]. The DnaK-type chaperone is designated HscA in E.coli, Ssq1 in yeast and HSPA9 in humans, and belongs to the Hsp70 family of proteins that function to assist in protein folding or refolding through ATP hydrolysis [7–9]. This family of chaperones typically consists of two domains: an N-terminal nucleotide-binding domain that exhibits ATPase activity, and a C-terminal substrate-binding domain [10, 11]. The K-type chaperones have been found to interact with encoded J-type co-chaperones to facilitate function by stimulating the ATPase activity of the chaperone and enhance substrate binding affinity [7, 9, 10, 12]. These J-type co-chaperones are classified as ‘small Jac’s’ (small J-type accessory chaperones), and the Fe-S cluster related protein is termed HscB in E. coli, Jac1 in yeast, and Hsc20 in humans [9, 13]. Unlike other J-type co-chaperones, Hsc20 lacks a cysteine-rich zinc finger, and the mass (~20 kD) is different from the classic DnaJ chaperone (~45 kD) [12, 14, 15]. Moreover, HSPA9/Hsc20 could not functionally replace the bacterial DnaK/DnaJ system [16–18], such that disruption of the hscA gene in E. coli did not produce gross phenotypic changes, in contrast to the growth defect when DnaK was deleted [17, 19, 20]. Despite these functional differences, recent work has implicated HSPA9 in relation to Myelodysplastic syndromes (MDS) [21, 22], where deficiency of HSPA9 results in ineffective hematopoiesis and delayed maturation of erythroid precursors [23, 24], suggesting that eukaryotic HSPA9 and its role in Fe-S cluster biogenesis has important implications for downstream protein function.
The potential involvement of the chaperone complex in erythroid differentiation via Fe-S cluster biogenesis has fueled interest in better understanding its specific role in cluster biosynthesis. The HSPA9/Hsc20 system is highly conserved; as such, research on the chaperone system incorporates studies in Azotobacter vinelandii, E. coli, yeast, and humans. Current understanding suggests that the co-chaperone, Hsc20, binds to the iron-sulfur cluster scaffold/carrier protein IscU via a conserved LVF motif [25] and recruits HSPA9, which can bind to both Hsc20 and IscU [7, 9, 26]; however, the binding site for IscU may be blocked depending on whether HSPA9 is bound to ATP or ADP. In this complex, IscU and Hsc20 can synergistically stimulate the ATPase activity of HSPA9 [27–29], with a proposed subsequent change in HSPA9 structure that impacts the interaction with Hsc20. The resulting Hsc20-IscU interaction shifts the conformation of IscU to the disordered (D) state, thereby releasing the Fe-S cluster and allowing it to bind to HSPA9 [26, 30, 31]. The Fe-S cluster is subsequently released in a process that delivers it to the target protein. Target specificity is believed to be conferred by Hsc20, which has been found to bind to partners such as succinate dehydrogenase (SDH) and glutaredoxin 5 (Grx5) via LYR motifs present on the partner proteins [9, 32]. However, Grx5 has also been shown to interact with yeast Ssq1, suggesting that this transfer reaction is facilitated through interaction with both proteins [33]. The LYR motif has allowed for identification of other potential targets by pull-down experiments, but these need to be confirmed biochemically [9].
In the context of the co-chaperone complex, IscU and Hsc20 can stimulate the ATPase activity of HSPA9, and in turn, the chaperone complex promotes Fe-S transfer from IscU to downstream targets. In A. vinelandii, the presence of chaperones increases the rate constant for Fe-S cluster transfer from IscU 22-fold for ferredoxin [34] and 670-fold for Grx5 [35]; whereas in E. coli, transfer from IscU to apo ferredoxin was increased on the scale of 5–10 fold [36, 37]. Although the components of the Fe-S cluster biosynthesis pathway and the corresponding chaperones are highly conserved, these differences in rate enhancement suggest organism-specific control over the process.
In addition to the Fe-S cluster targets mentioned above, the iron-sulfur cluster protein Nfu contains a version of the LYR motif, specifically an IYK motif in its N-terminal domain that may permit an interaction with the chaperone system, particularly with Hsc20, as observed for Grx5 and SDH [38, 39]. Furthermore, the Nfu protein has been found to interact with HSPA9 in both the human [21, 39] and yeast [40] systems, based on pull-down assays and genetic knockouts, respectively. The human Nfu protein plays a variety of functional cellular roles, including histone cell cycle regulation [41], thioredoxin-like activity in the apo form [42, 43], assembly and transfer of a [2Fe-2S] cluster [2], assembly of a [4Fe-4S] cluster [44, 45], and transfer of an Fe-S cluster into apo aconitase [38]. The role of Nfu in Fe-S cluster binding and transfer is essential, as mutations on Nfu are implicated in causing Multiple Mitochondrial Dysfunctions Syndrome 1 (MMDS1) [46–48]. Structurally, Nfu consists of two domains, an N- and a C-terminal domain [42–44, 49], where the latter contains a highly conserved CXXC motif [44] required for Fe-S cluster binding. The two protein domains can be separately isolated, which has provided thermodynamic and structural insights into the system. The N-terminal domain (N-Nfu) exists in a highly ordered state, while the C-terminal domain (C-Nfu) is molten globular [43, 49]. Bringing the two domains together helps to stabilize the C-terminal domain and the full-length protein maintains a well-folded structure [49].
Although Nfu and the chaperones have each been studied in depth, as separate entities, the biochemical connection between them has not yet been examined. Herein, we have sought to understand the thermodynamic and functional connections between these proteins (human Nfu, human Hsc20, and human HSPA9) in the context of Fe-S cluster biosynthesis.
Results
ATP concentration-dependent ATPase activity of human HSPA9
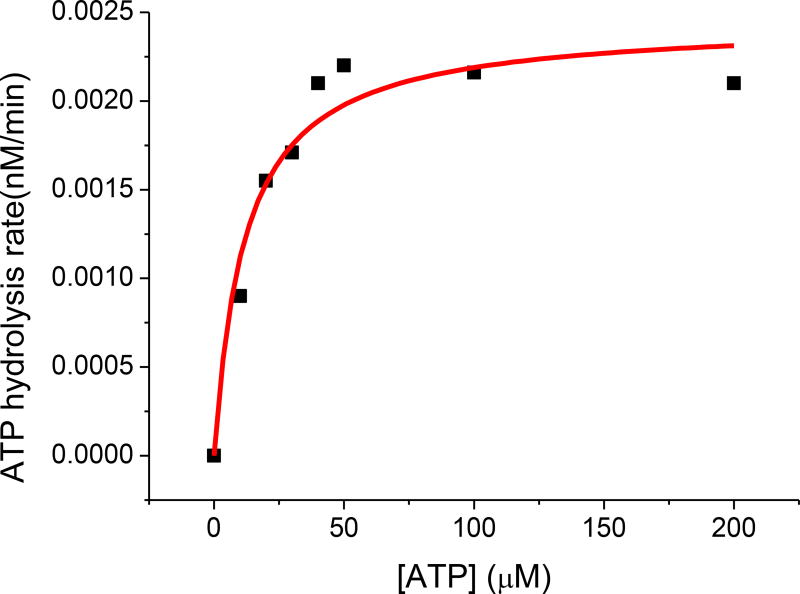
The ATPase activity of the human HSPA9 chaperone was monitored by use of the Molecular Probe EnzChek kit [50]. A standard curve was generated by varying the concentration of potassium phosphate from 2 to 100 µM (data not shown). To monitor the ATPase activity of HSPA9, a 1 µM solution of human HSPA9 in HKM buffer was degassed and mixed with the necessary components of the EnzChek reaction kit. Reaction was initiated by addition of increasing concentrations of ATP (0~200 µM), and the absorbance at 360 nm was recorded prior to conversion to the rate of ATP hydrolysis based on the standard curve. ATPase activity was plotted against the ATP concentration and fit to the Michaelis-Menten equation to obtain Km and Vmax parameters (Fig. 1). The Km value of 12 µM is similar to the previously reported Km for HscA, the HSPA9 homolog from Escherichia coli [51], while the Vmax of 0.27 nM/min demonstrates that the protein is active and at levels consistent with a previous characterization of this protein family [52]. This characterization also demonstrated the concentration of ATP (1 mM) required to saturate HSPA9 in the stimulation experiments.
Figure 1.
ATP concentration-dependent ATPase activity of human HSPA9. Fitting to the Michaelis-Menten equation (solid red curve) yielded a Km = 12 µM, and a maximum ATP hydrolysis rate of 0.272 nM/min. Error bars represent the standard deviation, where n=3.
Stimulation of HSPA9 ATPase activity by human Nfu
To date, only the E. coli IscU has been studied for its ability to stimulate the ATPase activity of an HSPA9-type chaperone [26, 53, 54]. However, growing evidence suggests that Nfu can interact with components of the chaperone system, with examples from yeast Ssq1 [40] and human HSPA9 [21]. To better examine this potential interaction, we have analyzed the functional properties of both HSPA9 and Nfu, and their mutual interdependence.
The relative stimulation of HSPA9 ATPase activity by human Nfu was plotted as a function of Nfu concentration, for both the full length and the separate domains. ATPase activity was observed to increase with increasing Nfu concentration, and the data was fit to a one site binding function (1),
| (1) |
where y describes the relative stimulation of HSPA9 ATPase activity and [Nfu] represents the concentration of Nfu (or one of the truncated Nfu forms). Vmax represents the maximum ratio of stimulation, KD is the apparent binding affinity for the interaction, and kATP represents the basal ATPase activity in the absence of Nfu.
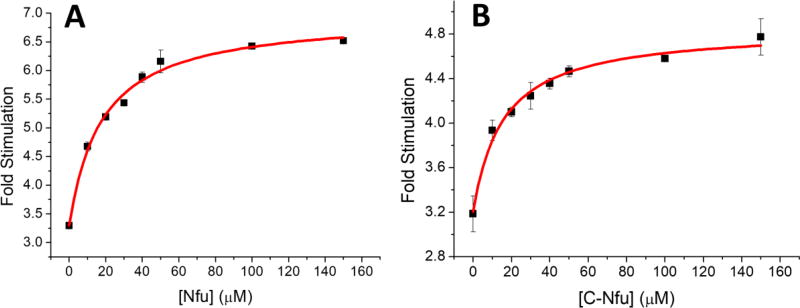
Addition of Nfu was observed to stimulate the ATPase activity of HSPA9 up to 3.7 fold (Fig. 2a); while the C-Nfu form increased activity 2.4 fold (Fig. 2b). Binding affinities were also obtained from the stimulation plots and yielded an apparent affinity of 50 µM for both full-length Nfu (Fig. 2a) and C-Nfu (Fig. 2b) for interaction with human HSPA9. Moreover, N-Nfu alone did not demonstrate any stimulation of the ATPase activity of HSPA9 (data not shown). The fitting parameters determined from the stimulation assays are summarized in Table 1.
Figure 2.
ATPase activity of HSPA9 when stimulated by different concentrations of full-length human Nfu (A) or C-Nfu (B). Data were reported based on the ratio of stimulation over basal ATPase activity. Solid red curves represent fitting to equation 1. The fitting parameters are shown in Table 1. Error bars represent the standard deviation, where n=3.
Table 1.
Summary of parameters obtained for HSPA9 ATPase activity following stimulation by Nfu in the presence or absence of Hsc20 at 25 °C. The parameters are based on equation (1).
| Vmax (fold) | KD (µM) | kATP (fold) | |
|---|---|---|---|
| Full-length Nfu/HSPA9 | 3.78 ± 0.31 | 50.1 ± 12.2 | 0.91 ± 0.14 |
| Full-length Nfu with Hsc20/HSPA9 | 3.67 ± 0.16 | 17.9 ± 2.62 | 3.30 ± 0.11 |
| C-Nfu/HSPA9 | 2.44 ± 0.21 | 50.7 ± 12.7 | 1.00 ± 0.08 |
| C-Nfu with Hsc20/HSPA9 | 1.64 ± 0.09 | 15.7 ± 2.79 | 3.20 ± 0.06 |
Stimulation of the ATPase activity of HSPA9 by Nfu in the presence of Hsc20
Because HSPA9 and Hsc20 have been shown to function together in several key roles [9, 16, 53], we also investigated the impact of Nfu on ATPase activity from the HSPA9/Hsc20 complex. Human Hsc20 was overexpressed in E. coli with an N-terminal His-tag to facilitate purification. Purification of Hsc20 by affinity chromatography was confirmed with SDS-PAGE and its migration position was consistent with the predicted mass of 21.7 kD (data not shown).
Another HSPA9 ATPase stimulation experiment was performed at a saturating concentration of 50 µM Hsc20 [55], since 20 µM has been shown to saturate the ATPase activity of 2 µM HSPA9 (data not shown). The experiments were carried out as described above, and data analyzed relative to the basal activity of the HSPA9/Hsc20 complex. Full length Nfu was able to stimulate HSPA9 ATPase activity in the complex from approximately 3-fold (Hsc20/HSPA9 complex relative to HSPA9 alone) to 7-fold (Fig. 3a), while C-Nfu increased activity from approximately 3-fold to 5-fold (Fig. 3b). Stimulation by either form of Nfu in the presence of Hsc20, does not demonstrate the synergistic effect observed by IscU stimulation of HscA in the presence of HscB [53], suggesting a different interaction mechanism for Nfu. The apparent binding affinities were also determined from the fit, with full-length Nfu displaying an apparent affinity of 17.9 µM (Fig. 3a) and C-Nfu of 15.7 µM (Fig. 3b). Similar to the case of HSPA9 alone, N-Nfu was unable to stimulate the HSPA9/Hsc20 complex (data not shown). Fitted parameters are summarized in Table 1.
Figure 3.
ATPase activity of HSPA9 when stimulated with different concentrations of (A) full-length human Nfu or (B) C-Nfu, in the presence of Hsc20. Data were reported based on the fold of stimulation over basal ATPase activity. Solid red curves represent fitting to equation 1. The fitting parameters are shown in Table 1. Error bars represent the standard deviation, where n=3.
Quantitation of human Nfu binding to human HSPA9 (ADP- or AMPPNP-bound form) by isothermal titration calorimetry (ITC)
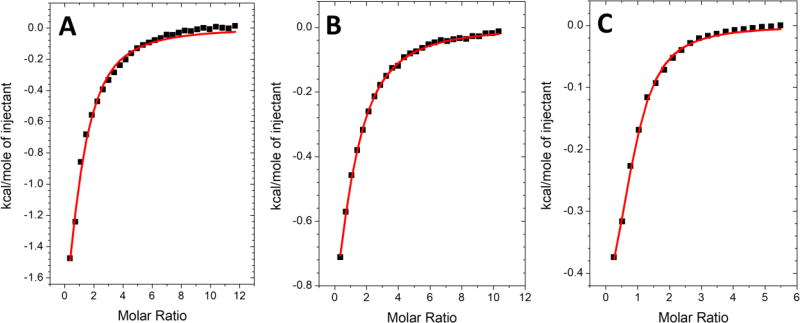
Given the stimulation of HSPA9 activity by both full-length and C-terminal truncated Nfu, we examined these two proteins for a direct protein-protein interaction. By use of isothermal titration calorimetry (ITC), human Nfu was titrated into a solution of HSPA9 in the presence of ADP to prevent any conformational changes that may result from ATPase activity. In the ADP-form, Nfu and HSPA9 interact in a 1:1 ratio with a binding affinity of 8.3 µM (Fig. 4a; Table 2). Similarly, C-Nfu also binds HSPA9 at a single binding site, but with a lower affinity of 18 µM (Fig. 4b; Table 2). Binding of N-Nfu to HSPA9 was also investigated; however, no change in heat was observed, consistent with no direct binding interaction (data not shown) and correlates with the lack of ATPase stimulation.
Figure 4.
Representative calorimetric analysis of the binding of HSPA9 (ADP-bound) with (A) full-length NFU and (B) C-Nfu. The heat of binding was integrated and fit to a one-site binding model to yield the parameters described in Table 2.
Table 2.
ITC parameters for Nfu binding to HSPA9 in the presence of 1 mM ADP or 1 mM AMPPNP at 25 °C.
| binding sites | KD (µM) | ΔH (kcal/mol) | ΔS (cal/mol K) | |
|---|---|---|---|---|
| Full-length Nfu/HSPA9 (ADP) | 0.9 ± 0.1 | 8.3 ± 1.4 | −6.3 ± 0.7 | 2.3 |
| Full-length Nfu / HSPA9 (AMPPNP) | 0.9 ± 0.2 | 36 ± 4 | −4.9 ± 0.1 | 3.7 |
| C-Nfu/HSPA9 (ADP) | 1.0 ± 0.2 | 18 ± 2 | −7.3 ± 1.3 | 2.8 |
| C-Nfu / HSPA9 (AMPPNP) | 1.0 ± 0.4 | 51 ± 6 | −3.2 ± 0.2 | 8.9 |
Chaperones with ATPase activity alternate between high substrate-binding affinity in the ADP form, and low substrate-binding affinity in the ATP form [56]. However, binding of Nfu to the ATP-loaded form of HSPA9 cannot be measured directly as a result of the intrinsic ATPase activity of HSPA9. Therefore, a non-hydrolyzable analog of ATP (AMPPNP) was used in which the oxygen atom in ATP that connects the β and γ phosphate is replaced by nitrogen. In the presence of AMPPNP, ITC experiments demonstrated the binding affinity to increase from 8.3 µM and 18 µM, to 36.5 µM and 51 µM, respectively, for human NFU and the truncated C-terminal domain (Table 2), indicating that both full length Nfu and the C-terminal domain can bind the ADP form of HSPA9 3-fold more tightly than HSPA9 in the presence of an ATP analog. These binding affinities are in line with prior observations for the interaction of HscA with IscU, with a KD of 39 µM in the absence of HscB in the ATP-bound form measured via surface plasmon resonance (SPR) [29]. N-Nfu was unable to interact directly with HSPA9 in the AMPPNP-bound form (data not shown).
Quantitation of human Nfu binding to human Hsc20 by ITC
As the co-chaperone of HSPA9, Hsc20 stimulates its activity and function [9, 16]. Because the presence of Hsc20 in the ATPase assays promoted the stimulation of HSPA9 by Nfu and increased the apparent binding affinity, we sought to characterize the binding interaction between Nfu (or C-Nfu) and Hsc20. By use of ITC, both full length and C-Nfu were found to interact with Hsc20 at 1:1 ratio with binding affinities of 10 µM and 11 µM, respectively (Table 3). Although Nfu can bind to Hsc20 with a relatively low affinity, the binding is not as tight as the interaction between IscU and HscB of 0.10 µM [29], which further supports the notion that there is a different mode of interaction between Nfu and Hsc20. As before, the N-terminal domain alone did not demonstrate binding to Hsc20.
Table 3.
ITC parameters for Nfu binding to Hsc20 at 25 °C.
| binding sites | KD (µM) | ΔH (kcal/mol) | ΔS (cal/mol K) | |
|---|---|---|---|---|
| Full-length Nfu/Hsc20 | 0.93 ± 0.06 | 10 ± 1 | −0.9 ± 0.1 | 20 |
| C-Nfu/Hsc20 | 0.94 ± 0.06 | 11 ± 2 | −1.7 ± 0.1 | 17.1 |
Quantitation of full-length human Nfu binding to the human HSPA9/Hsc20 complex by ITC
ATPase stimulation experiments described herein suggest that the binding affinity between NFU and HSPA9 is enhanced in the presence of Hsc20. ITC was again used to analyze the interaction of Nfu with the HSPA9/Hsc20 complex by titration with Nfu in the presence of ADP. The HSPA9/Hsc20 complex was pre-formed via overnight dialysis at concentrations sufficient to enable complex formation. Addition of Nfu yielded a binding affinity of 0.3 µM (Fig. 5a; Table 4), which is significantly tighter than the affinity of 8.3 µM obtained for Nfu binding to HSPA9 alone. When AMPPNP was present with the HSPA9/Hsc20 complex, Nfu was seen to bind with an affinity of 9.4 µM (Fig. 5b; Table 4), resulting in an increased binding affinity of approximately 4-fold compared to Nfu binding to HSPA9 alone (Table 2). This was consistent with the ATPase activity results and demonstrates thermodynamically that Nfu can bind more tightly to the HSPA9/Hsc20 complex than to HSPA9 alone. Again these affinities correlate well with those obtained for IscU interacting with the HscA/HscB complex [29]. While the interaction with Hsc20 is weaker, Nfu can interact with HSPA9 as strongly as IscU can bind to HscA.
Figure 5.
Calorimetric analysis of the binding of full-length Nfu to the HSPA9/Hsc20 complex in the presence of 1 mM ADP (A) or 1 mM AMPPNP (B). The binding enthalpy was integrated and fit to a one-site binding model (solid red curve) to yield the parameters described in Table 4.
Table 4.
ITC parameters obtained for full-length Nfu binding to the HSPA9/Hsc20 complex in the presence of either ADP or AMPPNP at 25 °C.
| binding sites | KD (µM) | ΔH (kcal/mol) | ΔS (cal/mol K) | |
|---|---|---|---|---|
| full-length Nfu/HSPA9/Hsc20 (ADP) | 0.95 ± 0.02 | 0.3 ± 0.1 | −3.4 ± 0.1 | 18.3 |
| full-length Nfu/HSPA9/Hsc20 (AMPPNP) | 0.985 ± 0.04 | 9.43 ± 0.07 | −1.7 ± 0.1 | 17.4 |
Interaction of a mixture of N- and C-Nfu with HSPA9 and the HSPA9/Hsc20 complex
Although N-Nfu alone did not demonstrate any stimulation of HSPA9, or binding to either of the chaperones under investigation, it may serve a specific function when present with the C-terminal domain. This notion is further supported by previous research demonstrating that a mixture of the two domains results in structural conformational changes similar to those found with the full-length protein [49], which are likely required for proper function. Thus, the ATPase stimulation assay and ITC experiments were performed with a mixture of the separately purified N- and C-Nfu domains (Fig. 6 and 7) at a concentration that would promote formation of the full-length protein as a complex of the two separate domains [49]; however, both the kinetic and thermodynamic results matched those obtained with the C-terminal domain alone (Tables 1–3, 5–6). These data suggest that either the N-terminal domain is only required in the context of the full length protein, or that the two domains can only come together functionally via the linker region that is missing from the separate constructs, thereby preventing proper formation of the full length protein. In either case, the mixture of the two separate domains is unable to functionally match the full-length protein in these experiments.
Figure 6.
ATPase activity of HSPA9 with N-Nfu when stimulated by different concentrations of C-Nfu (A) and with Hsc20 present in (B). Data were reported based on the fold of stimulation over basal ATPase activity. The fitting parameters are detailed in Table 5. Error bars represent the standard deviation, where n=3.
Figure 7.
Calorimetric analysis of the binding of a mixture of N- and C-Nfu to (A) HSPA9 with 1 mM ADP, (B) HSPA9 with 1 mM AMPPNP, or (C) the HSPA9/Hsc20 complex. The heat of binding was integrated and fit to a one binding site model yielding the parameters described in Table 6.
Table 5.
Summary of the parameters for HSPA9 ATPase activity following stimulation by C-Nfu in the presence of N-Nfu.
| Vmax (fold) | KD (µM) | kATP (fold) | |
|---|---|---|---|
| C-Nfu stimulation of HSPA9 with N-Nfu present | 2.30 ± 0.24 | 35.3 ± 11.7 | 0.96 ± 0.13 |
| C-Nfu stimulation of HSPA9 with N-Nfu and Hsc20 present | 1.77 ± 0.07 | 13.9 ± 1.85 | 3.10 ± 0.05 |
Table 6.
ITC parameters for C-Nfu binding to HSPA9, or Hsc20, in the presence of N-Nfu.
| binding sites | KD (µM) | ΔH (kcal/mol) |
ΔS (cal/mol K) |
|
|---|---|---|---|---|
| C-Nfu/N-Nfu/HSPA9 (ADP) | 0.88 ± 0.14 | 14.9 ± 1.67 | −3.82 ± 0.67 | 9.28 |
| C-Nfu/N-Nfu/HSPA9 (AMPPNP) | 0.907 ± 0.05 | 30.3 ± 2.82 | −2.00 ± 0.13 | 14.1 |
| C-Nfu/N-Nfu/Hsc20 | 0.79 ± 0.03 | 10.0 ± 1.22 | −0.57 ± 0.03 | 21.0 |
Stimulation of the transfer activity of full-length Nfu by the HSPA9/Hsc20 complex
Since Nfu stimulates the ATPase activity of HSPA9 and directly interacts with both HSPA9 and Hsc20, we investigated the impact of the chaperones on the functional properties of full-length Nfu. Our laboratory has previously reported the chemical reconstitution of a [2Fe-2S] cluster on human Nfu and shown that this iron-sulfur cluster can be quantitatively delivered to downstream target proteins, such as ferredoxins and glutaredoxins, with robust second-order rate constants obtained by monitoring the reaction by use of circular dichroism [1–3]. The HSPA9/Hsc20 chaperone system has been previously shown to stimulate the transfer of a [2Fe-2S] cluster from the scaffold/carrier protein IscU to various targets [9, 26], with the range of increase from 5–10 fold in E. coli [36, 37] to 22–670 fold in A. vinelandii [34, 35]. Given the proposed role for Nfu as an alternative scaffold/carrier [46, 57, 58], and our demonstrated functional and thermodynamic connections, we also examined the ability of the chaperones to increase the rate of Fe-S cluster transfer from Nfu.
Cluster transfer reactions were performed anaerobically using a 1:1 ratio of holo:apo protein with the chaperone complex, MgATP and DTT present. Reactions were carried out at a 1:1 ratio, due to limitations of detection sensitivity and protein solubility. Although these values were not obtained under pseudo first-order kinetic conditions, we have developed a method to provide estimates for second-order rate constants in cluster transfer reactions that are consistent with previously published literature values [34, 59, 60]. Our previous work provides test cases that demonstrate the apparent second-order rate constant obtained in this manner to agree with the actual rate constant determined from more traditional concentration dependence measurements [2, 61]. Prior studies of HscA/HscB complex formation with the scaffold/carrier protein IscU have indicated that substoichiometric chaperone complex can maximally stimulate of cluster transfer [36, 37]; as such, prior kinetic CD experiments have utilized between 2- and 4-fold excess of cluster-bound IscU to HscA/HscB complex [35]. Similar ratios were utilized in this work to examine cluster transfer from Nfu. Likewise, the concentrations of MgATP were adapted from earlier experiments that monitored the kinetics of cluster transfer with the chaperone system using CD [34–37, 62]. However, varying the concentration of chaperones outside of this ratio cannot drive a reaction to occur that did not already work at the optimal ratio [63], consistent with these being saturating concentrations.
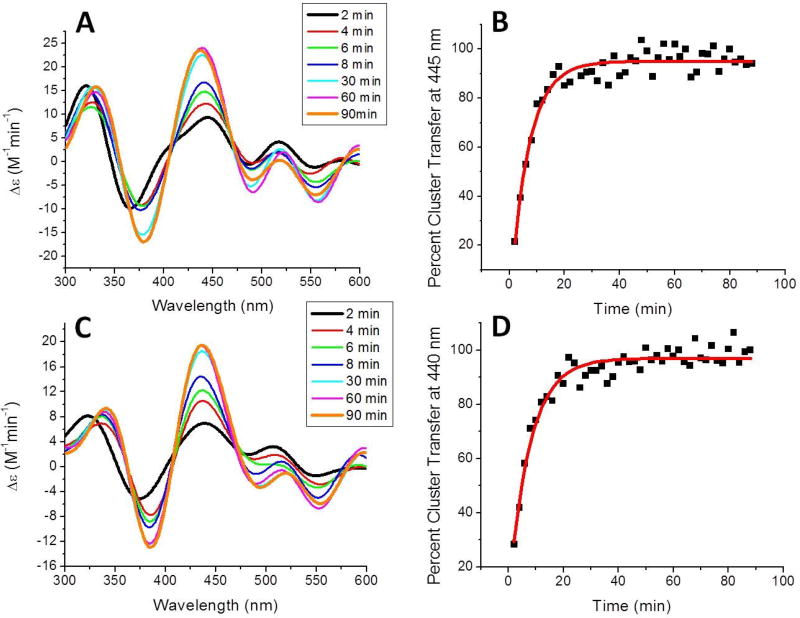
As such, transfer from holo human Nfu to apo ferredoxin 1 (Fdx1) was monitored every 2 min for a total of 90 min and the increase in CD signal at 445 nm (Fig. 8a), indicative of formation of holo Fdx1, was measured and converted to percent cluster transfer (Fig. 8b). The second-order rate constant was determined by use of DynaFit based on the concentration of [2Fe-2S] cluster initially present in holo Nfu to yield a second-order rate constant of 8800 ± 720 M−1min−1 (Table 7), which is almost 2-fold faster than transfer in the absence of chaperones [2]. Transfer to apo ferredoxin 2 (Fdx2) was monitored in the same way, but at the Fdx2 specific wavelength of 440 nm (Fig. 8c), to yield a second-order rate constant of 6050 ± 2700 M−1min−1 (Table 7), slightly increasing the second-order rate constant observed in the absence of chaperones [2]; however, the increase is negligible when considering the error values on the measurements (Table 7).
Figure 8.
Time course for cluster transfer from holo Nfu to apo ferredoxin 1 (Fdx1) (A) or apo ferredoxin 2 (Fdx2) (C) in the presence of 22 µM HspA9, 22 µM Hsc20, 40 mM MgCl2 and 2 mM ATP. Reactions were monitored by UV-vis CD spectroscopy under anaerobic conditions in a semi-micro 1 cm cuvette at room temperature. MgATP and the holo protein were added last to initiate reaction and CD spectra were recorded every 2 min for a total of 90 min. The change in extinction, based on the initial concentration of holo Nfu-bound [2Fe-2S] cluster, was monitored over time and conversion to percent cluster transfer for determination of cluster transfer kinetics. Fitting with DynaFit [88] (solid red curve) yielded second-order rated constants of 8800 ± 720 M−1min−1 for transfer to Fdx1 (B) and 6050 ± 2700 M−1min−1 for Fdx2 (D).
Table 7.
Comparison of second-order rate constants determined for transfer from holo Nfu to apo targets in the presence and absence of the HSPA9/Hsc20 complex [1–3].
| second-order rate constant without chaperones (M−1min−1) |
second-order rate constant with chaperones (M−1min−1) |
|
|---|---|---|
| human Nfu to human IscU | No Transfer | No Transfer |
| human Nfu to human Fdx1 | 4700 ± 820 | 8800 ± 720 |
| human Nfu to human Fdx2 | 3850 ± 1240 | 6050 ± 2700 |
| human Nfu to human Grx2 | 3700 ± 77 | No Transfer |
| human Nfu to S. cerevisiae Grx3 | 36200 ± 7700 | 34400 ± 4500 |
| human Nfu to human Grx5 | 35100 ± 2000 | 28500 ± 7500 |
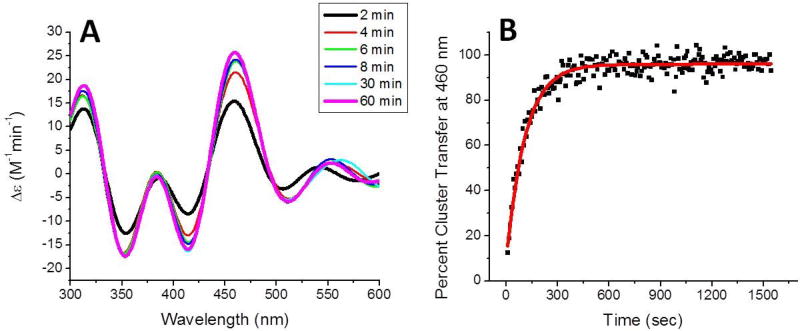
Interestingly, the clear stimulation pattern that was observed in the transfer from holo Nfu to the apo Fdx1 was not present for transfer to apo glutaredoxins. Initially, the transfer of a [2Fe-2S] cluster from holo Nfu to apo glutaredoxin 5 (Grx5) was examined in the absence of chaperones, as this reaction has not yet been characterized. Transfer was relatively fast, such that after the first 2 min, the spectra demonstrated complete formation of holo Grx5 (Fig. 9a). Therefore, the reaction was monitored at 10 s intervals at 460 nm to yield a second-order rate constant of 35100 ± 2000 M−1min−1 (Fig. 9), which is in line with the rate-constant for transfer to the yeast glutaredoxin 3 homolog (Table 7) [3].
Figure 9.
Time course for cluster transfer from holo to apo glutaredoxin 5 (Grx5) (A) in the absence of the chaperone complex. The reaction was monitored by UV-vis CD spectroscopy under anaerobic conditions in a semi-micro 1 cm cuvette at room temperature. Holo Nfu was added to initiate the reaction and CD spectra were recorded every 2 min for a total of 60 min. The CD spectra resembled Grx5 within the first 2 min, so data was collected only for 10 nm around the 460 nm peak. The change in extinction, based on the initial concentration of holo Nfu-bound [2Fe-2S] cluster, was monitored over time and conversion to percent cluster transfer (B) for determination of cluster transfer kinetics. Fitting with DynaFit [88] (solid red curve) yielded a second-order rated constant of 35100 ± 2000 M−1min−1.
Since we have previously characterized transfer to other glutaredoxin homologs [1, 3], we also examined their kinetic properties in the presence of chaperones. Holo Nfu was first added to apo glutaredoxin 2 (Grx2) with chaperones present; however, no change in CD signal was observed that would suggest a transfer reaction to occur, at least not within the time frame of 2 h (data not shown). Addition of holo Nfu to apo glutaredoxin 3 (Grx3) in the presence of the HSPA9/Hsc20 complex did result in a change in CD signal (Fig. 10a), but the second-order rate constant was the same as that observed in the absence of chaperones, within error limits, with a value of 34400 ± 4500 M−1min−1 (Fig. 10b, Table 7). Interestingly, CD spectra obtained for holo Grx3, following cluster delivery in the presence of chaperones, looks slightly different than the holo spectra obtained in the absence of chaperones (Fig. 10a) [3, 61]. The signature peak at 353 nm is enhanced, while the feature at 460 nm is lower in intensity, which may indicate either an interaction between Grx3 and Nfu that is chaperone mediated, or a minimal modification to the cluster coordination environment. Likewise, transfer to apo Grx5 was monitored in the same way to yield a second-order rate constant that agreed with the value obtained in the absence of chaperones, when considering the errors involved; however, no spectral changes were observed (Fig. 10c–d; Table 7).
Figure 10.
Time course for cluster transfer from holo Nfu to apo glutaredoxin 3 (Grx3) (A) and apo glutaredoxin 5 (Grx5) (C) in the presence of in the presence of 22 µM HspA9, 22 µM Hsc20, 40 mM MgCl2 and 2 mM ATP. Reactions were monitored by UV-vis CD spectroscopy under anaerobic conditions in a semi-micro 1 cm cuvette at room temperature. MgATP and the holo protein were added last to initiate the reaction and CD spectra were recorded every 2 min for a total of 90 min. The change in extinction, based on the initial concentration of holo Nfu-bound [2Fe-2S] cluster, was monitored over time and conversion to percent cluster transfer for determination of cluster transfer kinetics. Fitting with DynaFit [88] (solid red curve) yielded second-order rated constants of 34400 ± 4500 M−1min−1 for Grx3 (B) and 28500 ± 7500 M−1min−1 for Grx5 (D).
Discussion
In recent years the involvement of chaperones in the catalytic cycle of Fe-S cluster biogenesis has been of increasing interest, but has primarily focused on the bacterial or yeast homologs of the iron-sulfur cluster scaffold/carrier IscU and a limited number of downstream targets. Herein, we have analyzed a second Fe-S cluster binding protein, Nfu, that can not only stimulate the ATPase activity of HSPA9, but also binds to both chaperone components and displays accelerated second-order cluster transfer rate constants. These interactions can serve to delineate Fe-S cluster delivery pathways from holo Nfu.
Both full-length Nfu, and the two truncated forms representing the separate N- and C-terminal domains [43, 49], were examined for ATPase stimulation and partner binding to better understand the role of chaperones in promoting Nfu cluster transfer reactivity. Both full-length and C-terminal Nfu were able to stimulate the ATPase activity of HSPA9 (Fig. 2), and the full-length construct stimulated the activity to almost the same level as IscU (Table 1) [29]. However, unlike IscU, addition of the co-chaperone in the ATPase assay did not show a synergistic increase in stimulation (Fig. 3; Table 1) [29], where the addition of IscU and HscB increased the activity 100-fold over the stimulation of either HscB or IscU alone; this suggests that there is a difference between the two mechanisms. N-terminal Nfu was unable to either stimulate HSPA9 activity, or bind to either of the chaperones, indicating that the C-terminal domain is the functional domain for chaperone interaction and stimulation. The structural properties of human Nfu may explain the apparent lack of any contribution from the N-terminal domain. The latter is well-structured, while the C-terminal domain displays molten globule-like characteristics [43, 49], even in the context of the full-length protein, which suggests that the inherent flexibility of this C-terminal domain promotes interaction with the chaperone protein(s) [8, 10]. However, in a 1:1 mixture of the N- and C-terminal domains, the presence of the N-terminal domain enhances the stability of the C-terminal domain [43, 49]. Therefore, we postulate that the modest enhancement in ATPase activity of HSPA9 and the modest increase in binding affinity for the interaction between HSPA9 and full-length Nfu, when compared to the C-terminal domain alone (Tables 1 and 3), can be attributed to the stabilized conformation of the C-terminal domain. Structural stabilization of the C-terminal domain of Nfu in the presence of the N-terminal domain likely alters the structural conformation of HSPA9 to promote ATPase activity and enhance binding affinity, which in turn promotes cluster trafficking.
This hypothesis is further supported by the measured binding affinities. Full-length Nfu was able to bind to HSPA9 with tighter affinities under every condition examined (Table 3). Furthermore, the affinities determined by use of ITC for the full-length protein correlate well with the previously determined affinities for IscU binding to HscA [29]. As such Nfu may function in a catalytic cycle similar to what has been identified for IscU [9, 10, 26, 64].
However, postulating a mechanism of a catalytic cycle for Nfu and the chaperones is challenging, since the binding locations for the HSPA9, Hsc20 and Nfu interactions are currently unknown. For IscU, the conserved sequence for interaction with HSPA9 has been identified as LPPVK [54, 65], which lies close to the Fe-S cluster binding site [9]. However, Nfu does not contain an LPPVK motif, and while there are a total of 15 proline residues in human Nfu, there are no adjacent proline residues. Moreover, the NMR solution structure of the C-terminal domain [38] does not shown any prolines in close proximity, confirming that Nfu binding to HSPA9 must arise via a different motif. IscU binding to Hsc20 is promoted by an LVF motif [25], but again, Nfu does not possess such a motif in either the primary protein sequence or in the tertiary structure of the C-terminal domain [38]. An L(I)YR motif has been identified for Hsc20 binding to targets for cluster delivery [32], and Nfu does show a similar motif in the N-terminal domain with an IYK motif [38]. However, the N-terminal domain does not appear to promote the binding interaction with Hsc20, since both the full length protein and C-Nfu demonstrated the same binding affinity for Hsc20 (Table 3). Therefore, additional work must be completed to determine the molecular mechanism of binding between Nfu and HSPA9 and Hsc20, in order to develop a catalytic cycle for cluster assembly and transfer involving Nfu. Furthermore, the dimeric nature of Nfu [2, 38] will also need to be considered to accurately compare and contrast the Fe-S cluster chemistry of Nfu with that of IscU.
Although the molecular details remain incomplete, it is clear that Nfu can functionally complement IscU as a scaffold/carrier protein to complete Fe-S cluster transfer to target proteins. Addition of the chaperones only served to modestly stimulate transfer from holo Nfu to apo Fdx1 (Fig. 8a–b; Table 7), increasing the second-order rate constant approximately 2-fold. In transfer reactions to apo targets Fdx2, Grx3 and Grx5, the HSPA9/Hsc20 complex maintained the second-order rate constants obtained previously (Table 7), with a slight increase in the average value for Fdx2. The HSPA9/Hsc20 complex appears to be driving the specificity of cluster transfer reactions. By enhancing the rate of transfer to Fdx1 approximately 2-fold, the co-chaperone system is defining a preferred kinetic pathway for Fe-S cluster transfer from holo Nfu. Likewise, there is now a preferred pathway for Fe-S cluster transfer when examining mitochondrial glutaredoxins. Transfer from holo Nfu to apo Grx2, which is observed in the absence of chaperones [1], no longer occurs when they are present. Although transfer to apo Grx5 is not kinetically enhanced in the presence of chaperones (Fig. 9; Table 7), there is a preferred pathway from holo Nfu, since the alternative route is blocked by the chaperone interaction. A similar trend is observed for transfer to Grx3, in that there is no kinetic rate enhancement in the presence of the Hsc chaperones (Fig. 10; Table 7). While recognizing that this transfer is occurring between isolated proteins from two different organisms, and that Grx3 is primarily a cytosolic protein [66], nevertheless it speaks to the evolutionary conservation of the monothiol glutaredoxins and their interaction networks for Fe-S cluster transfer [67–70], because similar second-order rate constants and trends were observed (Table 7). Nevertheless, these interaction networks will need to be confirmed through additional in vitro and in vivo characterization
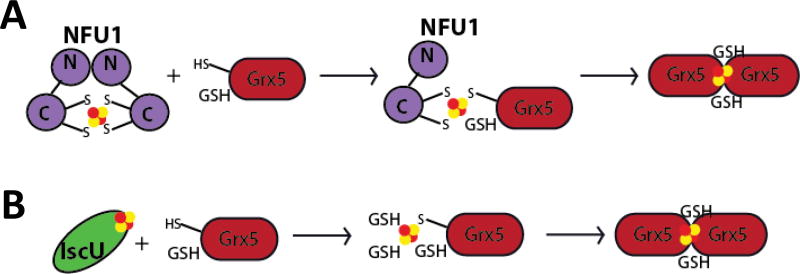
Since Nfu interaction with the chaperones primarily dictates preferred interaction networks, these results stand in contrast to the IscU-chaperone interaction. Kinetic experiments, conducted in A. vinelandii, demonstrated that addition of HscA/HscB stimulated cluster transfer from IscU to the homologous Fdx from 36 M−1min−1 to 800 M−1min−1 [34]. Similarly, in the Azotobacter system the second-order rate constant for transfer to Grx5 from IscU increased from 30 to 20,000 M−1min−1 [35]. However, it is significant that our second-order rate constants were much higher prior to addition of the chaperones, suggesting that they may not need to contribute much enhancement to transfer an Fe-S cluster from holo human Nfu to partner proteins. These differences may simply reflect the nature of the cluster scaffold/carrier and the molecular mechanism of Fe-S cluster transfer. Nfu is primarily a dimeric protein when an Fe-S cluster is bound (Fig. 11a) [2, 38, 71]; while IscU exists in a monomer state in the holo form (Fig. 11b) [72, 73]. Since these two scaffold/carrier-type proteins are in different oligomeric states, their mechanisms of transfer must differ to allow efficient cluster delivery. Likewise, the transfer process may also depend on the particular target protein. A monomeric target protein, such as a ferredoxin [74], may receive cluster via one pathway, while a dimer, such as a glutaredoxin [75–77], may prefer a transient mixed dimer intermediate. In depth structural characterization is required to delineate the precise pathway and the role of the chaperones therein.
Figure 11.
Representative models for cluster transfer from scaffold proteins. The chaperone proteins are not shown for simplicity. (A) Dimeric holo Nfu can transfer a [2Fe-2S] cluster to monomeric apo Grx5 with a bound GSH. Together, they form a transient mixed dimer intermediate that disappears when a second Grx5 molecule with a GSH displaces the Nfu monomer to form the dimeric holo Grx5. (B) Monomeric holo IscU can transfer a [2Fe-2S] cluster to apo Grx5 with a bound GSH. Additional GSH molecules can bind to the cluster to stabilize the bound cluster until a second Grx5 molecule can bind and form the dimeric holo form with two exogenous GSH ligands.
In summary, we have begun to elucidate and characterize the molecular mechanisms and involvement of various partner proteins in the complex mechanisms of Fe-S cluster biosynthesis and trafficking. The interaction between Nfu and the Hsc chaperones was identified previously [21, 40], and in this work, we have analyzed the functional implications of this interaction. In this paper, we have shown that Nfu can interact with the chaperones to stimulate the ATPase activity of HSPA9, in a manner that serves to regulate cluster transfer from Nfu [9, 26] to enhance or inhibit Fe-S cluster transfer to various targets, and provide preferential pathways for cluster delivery.
Materials and Methods
Materials
BL21 (DE3) cells and pET 28 vectors were obtained from Novagen (Madison, WI). EnzChek phosphate kit was obtained from Invitrogen (Carlsbad, CA). ATP and ADP were obtained from Sigma (St. Louis, MO). PD10 desalting columns were purchased from GE Healthcare (Wauwatosa, WI). Ferric chloride, sodium sulfide, DTT (Dithiothreitol), TCEP (Tris (2-carboxy-ethyl) phosphine) and L-cysteine were purchased from Fisher (Waltham, MA). The gene for Homo sapiens glutaredoxin 5 (Grx5) located in the pET28b(+) vector between the NdeI and HindIII restriction sites and lacking the first 31 amino acids (Δ1-31) that correspond to the mitochondrial targeting sequence [78], was ordered from GenScript (Piscataway, NJ).
Cloning of human Hsc20
Human Hsc20 was cloned into an appropriate expression vector as follows. Restriction sites NdeI and XhoI were introduced into the pET-28b(+) plasmid for ligation via the following primers, which were also utilized to amplify the coding region of hsc20 without the mitochondrial targeting sequence (residues 1-71): 5’-GGA AGG CCA TAT GGA CTA CTT CAG CCTTAT GGA CTG CAA CC-3’ and 5’-ACC GCT CGA GCC ACA ATT AAA GGG GAA TCT TCT TTA ACT T-3’. After amplifying the gene hsc20 from the human cDNA library by PCR, both PCR product and pET-28 plasmid were digested with NdeI and XhoI (Invitrogen, Waltham, MA) at 37°C. The digested products were cleaned on 0.8% agarose gel and extracted by use of a Qiagen gel extraction kit. Subsequently, both digested products were mixed and ligated by T4 ligase. The entire ligation product was used to transform the competent DH5α cells and sequencing was confirmed by the Plant Microbe Genomics Facility at the Ohio State University.
Overexpression of human Hsc20
The Hsc20 construct in pE28b(+) was transformed into a BL21 (DE3) E. coli cells. The transformed cells were grown in 10 mL of Luria Bertani (LB) broth media containing kanamycin (50 µM), and incubated at 37 °C overnight. Subsequently, the cell culture was transferred to 3 L of fresh LB media containing kanamycin, and incubated at 37 °C until the OD600 reached 0.8. Subsequently, 1 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG) was added for human Hsc20 induction. The cells were cultured for another 10 h at 30 °C before harvesting by centrifugation at 4330 g for 15 min. Cell pellets were frozen at −80 °C for future use.
Purification of human Hsc20
The cell pellet was resuspended in 50 mM HEPES, 100 mM NaCl, 1 mM PMSF, pH 7.5 and incubated with 1 mg/mL lysozyme at 4 °C for 30 min. The cells were lysed by sonication, and the lysate was centrifuged at 28,928 g for 30 min to remove all cell debris. The cleared supernatant was loaded onto a Ni-NTA column, equilibrated with 50 mM HEPES, 100 mM NaCl and washed with the equilibration buffer plus 50 mM imidazole, before elution with the same buffer plus 250 mM imidazole. Human Hsc20 was concentrated by ultrafiltration (Amicon, Billerica, MA) and analyzed for purity on a 12% SDS-PAGE gel. Concentrated protein was dialyzed at 4 °C against 50 mM HEPES and 100 mM NaCl, pH 7.5. The concentration of Hsc20 was determined from the calculated extinction ε280 = 7680 M−1cm−1 and the protein stored at −80°C for future use.
Additional protein expression and purification protocols
Human HSPA9 was expressed and purified as previously described [52]. Purification of human IscU (Hs IscU) and Thermatoga maritima (Tm Nifs) were also performed as previously reported [79–81]. The expression vector for human ferredoxin-1 (Hs Fdx1) was kindly provided by J. Markley and protein was expressed and purified according to literature procedures [74]. Purification for human ferredoxin-2 (Hs Fdx2) was performed as previously reported [82]. The ferredoxins were purified as holo proteins and were then subsequently converted to apo forms by treatment with 100 mM EDTA, 5 mM DTT and 8 M urea in a buffered solution, pH 7.5. To isolate the apo ferredoxins, the colorless solution was passed through a PD-10 column to remove EDTA, DTT and urea. Apo protein concentration was determined by use of the Bradford assay, and absorbance at 280 nm by use of calculated extinction coefficients.
A construct of human Grx2 (comprising residues 56-161) with a tobacco etch virus cleavable N-terminal His6-tag in expression vector pNic-Bsa4 was kindly provided by Drs. Kavanagh, Muller-Knapp, and Oppermann, and protein was expressed and purified as previously reported [83]. Expression and purification of full-length human Nfu, the N-terminal domain construct, and the C-terminal domain construct were performed from BL21 (DE3) competent cells as previously described [42, 43]. Yeast Grx3 (Δ1–35)[84] in pET28b(+) was expressed in Escherichia coli BL21 (DE3) and purified as described [61]. Human Grx5 (Q86SX6, residues 32-157) with an open reading frame containing the Grx domain, but lacking the first 31 amino acids (Δ1–31) [78] was cloned into pET28b(+) between NdeI and HindIII restriction sites and expressed in Escherichia coli BL21 (DE3). The transformed cells were grown in 10 mL of Luria Bertani (LB) broth media containing kanamycin (50 µM) overnight at 37 °C, transferred to 3 L of fresh LB media containing kanamycin, and incubated at 37 °C until the OD600 reached 0.8.
Protein expression was induced by addition of isopropyl β-D-1-thiogalactopyranoside (IPTG) (300 µM) and the cell culture incubated at 30 °C for 10 h. Cell pellets were collected by centrifugation at 4330 g for 15 min, suspended in 30 mL of buffer containing 50 mM Hepes, 100 mM NaCl, pH 7.5, and lysed by use of a dismembranator. The lysate was centrifuged at 28,928 g for 30 min, the supernatant subsequently applied to a Ni-NTA column, and the protein eluted with buffer containing 50 mM Hepes, 100 mM NaCl, 0.25 M imidazole, pH 7.5, prior to analysis on a 12% SDS-PAGE gel that was then visualized by Coomassie Blue staining. Dialysis was carried out at 4 °C against 50 mM Hepes, 100 mM NaCl, pH 7.5. Protein concentration was determined by use of the Bradford assay and absorbance at 280 nm, using calculated extinction coefficients.
Reconstitution of full-length human Nfu
NifS-mediated in vitro reconstitution of Nfu [85] was completed as previously described. Briefly, argon purged ferric chloride and L-cysteine were added to an anaerobic mixture of approximately 200 µM purified Nfu, 2 µM Tm NifS, and 5 mM DTT to final concentrations of 1.6 mM FeCl3 and 3.2 mM L-cysteine. The final solution was incubated for 1 h with stirring at room temperature, before separation of excess iron and sulfide through a PD-10 column that was equilibrated with an argon-purged solution of 50 mM HEPES, 100 mM NaCl, pH 7.5. Reconstituted protein was eluted with 3.5 ml of the equilibration buffer. The protein concentration was determined via the Bradford assay and reconstitution of protein was confirmed by absorbance at 330 nm and 420 nm on a Cary WinUV Spectrophotometer.
Iron quantification
A solution of holo protein at 50 µM in 200 µL was acidified by concentrated HCl (60 µL) and heated to 100 °C for 15 min. The resulting suspension was centrifuged at 14,000 rpm for 2 min and the supernatant (100 µL) was diluted with Tris-HCl (0.5 M, 1.3 mL, pH 8.5). Solutions of sodium ascorbate (0.1 mL, 5%) and bathophenanthroline-disulfonate (0.4 mL, 0.1%) were sequentially added to the neutralized reaction solution with mixing between each addition. The solution was incubated at 25 °C for 1 h and iron was quantified by measuring the absorbance at 535 nm on a UV-vis spectrophotometer and calculated from a calibration curve made with 0.01–0.2 mM FeCl3 standard solutions [86, 87].
ATP concentration-dependent ATPase activity of human HSPA9
The intrinsic ATPase activity of human HSPA9 with different concentrations of ATP was measured by use of the Molecular Probe EnzChek phosphate kit (Invitrogen) as previously reported [50]. This colorimetric assay monitors ATP hydrolysis via the formation of 2-amino-6-mercapto-7-methylpurine, which results the coupling of inorganic phosphate with the enzyme purine nucleoside phosphorylase (PNP) and its substrate 2-amino-6-mercapto-7- methylpurine riboside (MESG). The mixture for the measurement contained 1 µM HSPA9, MESG (2-amino-6-mercapto-7-methyl purine riboside) substrate, 1 mM DTT, and PNP (purine nucleoside phosphorylase) in HKM buffer (50 mM HEPES buffer, 150 mM KCl, 10 mM MgCl2 pH 7.5). ATP at different concentrations (0–200 µM) was injected to initiate the reaction. The UV-Visible signal at 360 nm was monitored at 25 °C on a Cary Win-UV spectrophotometer and correlated to phosphate concentration by use of a standard curve generated from different concentrations of potassium phosphate buffer (0~100 µM). Data was fit to the Michaelis-Menten equation by use of Origin 7.0 (Northampton, MA).
Stimulation of the ATPase activity of HSPA9 by human Nfu
Based on previous HSPA9 stimulation assays [50, 53], the ATP hydrolysis rates were measured at 25°C in HKM buffer by use the Molecular Probe EnzChek phosphate assay kit (Invitrogen). Full length Nfu, C-Nfu and N-Nfu were all monitored for their ability to stimulate the ATPase activity of HSPA9 in a reaction consisting of 1 µM HSPA9, MESG (2-amino-6-mercapto-7 -methylpurine riboside), substrate PNP (purine nucleoside phosphorylase), 1 mM DTT. The concentration of human Nfu (all forms) was added at different concentrations (0~150 µM). To initiate the reaction, 1 mM ATP was added and the absorbance at 360 nm was monitored at 25 °C over time on a Cary Win-UV spectrophotometer. The rate of ATP hydrolysis in the presence of either full length or truncated Nfu was converted to the fold of stimulation based on the basal intrinsic ATPase activity of human HSPA9. Binding affinities were obtained by plotting the amount of stimulation against the concentration of Nfu and fitting the data using Origin 7.0.
To monitor the stimulation of HSPA9 ATPase activity in the presence of Hsc20 and Nfu, the ATPase assay was carried out as described above by varying the concentration of Nfu (C-Nfu or full length), but in the presence of 50 µM Hsc20 in the cuvette and mixed with the other components before the addition of ATP. For N-Nfu, 150 µM Hsc20 was present, and the remaining conditions were the same. Stimulation of ATPase activity was based on the measurement of HSPA9 ATPase activity in the presence of Hsc20 (specified above), but in the absence of any forms of Nfu. Since the N-terminal domain has been shown to stabilize the C-terminal domain [49], the stimulation of HSPA9 ATPase activity by the C-terminal domain was performed as described earlier, following pre-incubation of 1 µM HSPA9 with 150 µM N-Nfu. The ATPase hydrolysis rate was converted with reference to the basal activity of HSPA9 in the presence of N-Nfu. The extent of stimulation was plotted against the concentration of the C-terminal domain of NFU, and the impact of the N- and C-terminal Nfu mix in the presence of Hsc20 was also examined. The reaction mixture contained the above components and included 150 µM N-Nfu and 150 µM Hsc20. The concentration of C-Nfu was varied and data analyzed as described above.
Quantitation of human Nfu binding to human HSPA9 by isothermal titration calorimetry (ITC)
All binding affinity measurements were carried out on VP-ITC (MicroCal, Northampton, MA) system at 25°C. Nfu and HSPA9 were dialyzed at 4 °C against HKM buffer (50 mM HEPES buffer, 150 mM KCl, 10 mM MgCl2 pH 7.5) in the presence of an ATP analog (see below). All reagents and solutions were thoroughly argon-purged before loading of either the syringe or the sample cell. Both Nfu and HSPA9 were treated with 1 mM TCEP (triscarboxyehthylphosphine) to reduce any disulfide bonds.
To determine the affinity in the ADP-bound state of HSPA9, either full length Nfu at 650 µM or truncated Nfu at 680 µM was titrated in 10 µL increments and a 300 sec interval into the sample cell at 25 °C, which contained 11 µM HSPA9 in the presence of 1 mM ADP in the HKM buffer from dialysis. The titration continued until the signal reached saturation. The heat of dilution was subtracted as background and the data were fit to a one site binding model using Origin 7.0.
In order to examine the affinity in the ATP bound state, the non-hydrolyzable analog AMPPNP was used at 1 mM in HKM buffer. Both Nfu and HSPA9 were incubated with 1 mM TCEP and argon-purged before loading the instrument. Full length Nfu (1 mM) or truncated Nfu (1.5 mM) was titrated into 20 µM HSPA9 using the same conditions as for the ADP-bound form. Data were again fit to a one-site binding model using Origin 7.0.
The N- and C-terminal Nfu mix was also examined by ITC. The experimental method was as described above, but 150 µM N-Nfu was pre-incubated with HSPA9 and the specific nucleotide in the ITC sample cell before C-Nfu was injected.
Quantitation of human Nfu binding to human Hsc20 by ITC
The experiments were carried out on VP-ITC (MicroCal) at 25 °C with all buffers and protein samples thoroughly argon-purged. Both proteins were dialyzed in the same buffer (50 mM HEPES buffer, 150 mM KCl, 10 mM MgCl2 pH 7.5) and incubated with 1 mM TCEP for disulfide bond reduction. Human Nfu at 0.62 mM was titrated in 10 µL increments with a 300 sec interval into 0.022 mM Hsc20 at 25 °C. More than 4 equivalents of Nfu were titrated into the cell to guarantee the complete formation of Nfu-Hsc20 complex. The heat of dilution was subtracted as background and the integrated and baseline corrected data was then fit to a one site binding model using Origin 7. Truncated Nfu containing the C-terminal domain only (1.1 mM) was titrated in to human Hsc20 (0.03 mM) while all other conditions were maintained.
The N- and C-terminal Nfu mix was also examined by ITC. The experimental method was as described above, but 150 µM N-Nfu was pre-incubated with HSPA9 and Hsc20 in the ITC sample cell before the C-Nfu was injected.
Quantitation of human Nfu binding to human HSPA9/Hsc20 complex by ITC
Binding affinities were measured by use of a VP-ITC (MicroCal) instrument at 25 °C as described above. All proteins were dialyzed in HKM buffer in the presence of either 1 mM ADP or 1 mM AMPPNP. Both HSPA9 and Hsc20, at 9 µM, were pre-incubated overnight at 4 °C at concentrations sufficient to allow for complex formation. Prior to loading the instrument, all proteins were incubated with 1 mM TCEP and argon-purged.
To monitor binding in the presence of ADP, 354 µM full length Nfu was titrated into the HSPA9/Hsc20 mixture in 10 µL injections with a 300 sec interval at 25 °C. The heat of dilution was subtracted as the background and the data fit to a one-site binding model using Origin 7.0. The same conditions were used in the presence of 1 mM AMPPNP to monitor binding of Nfu to the complex.
Likewise, the N- and C-Nfu mix was also examined. The experimental method was as described earlier, but 150 µM N-Nfu was pre-incubated with HSPA9, Hsc20, and the specific nucleotide in the ITC sample cell before C-Nfu was injected.
Cluster transfer monitored by circular dichroism (CD)
CD spectra were recorded using a Jasco J815 spectropolarimeter (Easton, MD). The time course for cluster transfer from holo iron-sulfur cluster proteins to apo proteins was monitored under anaerobic conditions at 25°C by use of UV-vis CD spectroscopy in small volume 1-cm quartz cuvettes. CD scans from 300 to 600 nm were collected to identify signature peaks from cluster-bound protein at a scan rate of 200 nm/min with a 2 min interval between acquisitions. Transfer reactions occurring over faster time scales were monitored with a 10 sec interval over a 10 nm range (see specific wavelengths below). Reactions were carried out in 50 mM HEPES, 100 mM NaCl, pH 7.5 and were prepared by degassing a mixture of apo protein in 5 mM DTT with 22 µM HSPA9 and 22 µM Hsc20. The reaction was transferred to an anaerobic cuvette via a gas tight syringe and was initiated via the addition of degassed MgCl2 and ATP, to final concentrations of 40 mM and 2 mM respectively, and degassed holo protein. The concentration of [2Fe-2S] cluster for each holo protein was determined via iron quantitation following reconstitution. Reactions were carried out in 1:1 ratio of holo to apo protein, with each at 45 µM. Data were processed by use of JASCO Spectramanager II Analysis software and analyzed by use of the DynaFit chemical kinetics and equilibria program by BioKin (Watertown, MA) [88]. The deconvolution function from Spectramanager II analysis software was used for analysis of bands in the spectra that contained overlapping Lorentzian curves having the same full width at half maximum value that accurately distinguishes the peak positions for each band.
A second-order rate constant for cluster transfer was obtained via a method similar to the iron-sulfur cluster transfer method of Johnson and colleagues [59, 60]. Proteins were mixed in a 1:1 ratio (20–100 µM) and the peaks at 330 nm for IscU, 365 nm for Grx2, 455 nm for Grx3, 460 for Grx5, 445 nm for Fdx1 and 440 nm for Fdx2 were monitored by CD. Data were subsequently converted to percent cluster transfer, which was determined from a set of control spectra of the holo proteins at a series of known concentrations recorded after reconstitution. The subsequent plot of percent cluster transfer versus time was analyzed using the DynaFit program [88] to determine second-order rate constants for the various reactions by best-fit simulation to second-order kinetics.
Acknowledgments
We thank Dr. Marina Bakhtina for her assistance with circular dichroism. This work was supported by a grant from the National Institutes of Health [AI072443]. Christine Wachnowsky was supported by an NIH Chemistry/Biology Interface training grant (T32 GM095450), as well an Ohio State University Presidential Fellowship.
Abbreviations
- GSH
Glutathione
- IscU
Iron-sulfur cluster scaffold protein
- CD
Circular Dichroism
- Fdx
Ferredoxin
- Grx
Glutaredoxin
- ITC
Isothermal titration calorimetry
Footnotes
Author Contributions
CW, YL, and TY conducted experiments. CW, TL, TY, and JAC analyzed and interpreted the data and wrote the manuscript.
References
- 1.Fidai I, Wachnowsky C, Cowan JA. Mapping cellular Fe–S cluster uptake and exchange reactions – divergent pathways for iron–sulfur cluster delivery to human ferredoxins. Metallomics. 2016;8:1283–1293. doi: 10.1039/c6mt00193a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wachnowsky C, Fidai I, Cowan JA. Iron-sulfur cluster exchange reactions mediated by the human Nfu protein. J Biol Inorg Chem. 2016;21:825–836. doi: 10.1007/s00775-016-1381-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wachnowsky C, Fidai I, Cowan JA. Cytosolic iron-sulfur cluster transfer: a proposed kinetic pathway for the reconstitution of glutaredoxin 3. FEBS Lett. 2016;590:4531–4540. doi: 10.1002/1873-3468.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frazzon J, Dean DR. Formation of iron-sulfur clusters in bacteria: an emerging field in bioinorganic chemistry. Curr Opin Chem Biol. 2003;7:166–173. doi: 10.1016/s1367-5931(03)00021-8. [DOI] [PubMed] [Google Scholar]
- 5.Zheng L, Cash VL, Flint DH, Dean DR. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J Biol Chem. 1998;273:13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- 6.Strain J, Lorenz CR, Bode J, Garland S, Smolen GA, Ta DT, Vickery LE, Culotta VC. Suppressors of superoxide dismutase (SOD1) deficiency in Saccharomyces cerevisiae. Identification of proteins predicted to mediate iron-sulfur cluster assembly. J Biol Chem. 1998;273:31138–31144. doi: 10.1074/jbc.273.47.31138. [DOI] [PubMed] [Google Scholar]
- 7.Cai K, Frederick RO, Kim JH, Reinen NM, Tonelli M, Markley JL. Human mitochondrial chaperone (mtHSP70) and cysteine desulfurase (Nfs1) bind preferentially to the disordered conformation, whereas co-chaperone (Hsc20) binds to the structured conformation of the iron-sulfur cluster scaffold protein (IscU) J Biol Chem. 2013;288:28755–28770. doi: 10.1074/jbc.M113.482042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maio N, Rouault TA. Mammalian Fe–S proteins: definition of a consensus motif recognized by the co-chaperone HSC20. Metallomics. 2016;8:1032–1046. doi: 10.1039/c6mt00167j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vickery LE, Cupp-Vickery JR. Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron-sulfur protein maturation. Crit Rev Biochem Mol Biol. 2008;42:95–111. doi: 10.1080/10409230701322298. [DOI] [PubMed] [Google Scholar]
- 11.Cupp-Vickery JR, Peterson JC, Ta DT, Vickery LE. Crystal structure of the molecular chaperone HscA substrate binding domain complexed with the IscU recognition peptide ELPPVKIHC. J Mol Biol. 2004;342:1265–1278. doi: 10.1016/j.jmb.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 12.Vickery LE, Silberg JJ, Ta DT. Hsc66 and Hsc20, a new heat shock cognate molecular chaperone system from Escherichia coli. Protein Sci. 1997;6:1047–1056. doi: 10.1002/pro.5560060511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silberg JJ, Hoff KG, Vickery LE. The Hsc66-Hsc20 chaperone system in Escherichia coli: Chaperone activity and interactions with the DnaK-DnaJ-GrpE system. J Bacteriol. 1998;180:6617–6624. doi: 10.1128/jb.180.24.6617-6624.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szabo A, Korszun R, Hartl FU, Flanagan J. A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. Embo J. 1996;15:408–417. [PMC free article] [PubMed] [Google Scholar]
- 15.Cupp-Vickery JR, Vickery LE. Crystal structure of Hsc20, a J-type co-chaperone from Escherichia coli. J Mol Biol. 2000;304:835–845. doi: 10.1006/jmbi.2000.4252. [DOI] [PubMed] [Google Scholar]
- 16.Vickery LE, Silberg JJ, Ta DT. Hsc66 and Hsc20, a new heat shock cognate molecular chaperone system from Escherichia coli. Protein Sci. 1997;6:1047–1056. doi: 10.1002/pro.5560060511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawula TH, Lelivelt MJ. Mutations in a gene encoding a new Hsp70 suppress rapid DNA inversion and bgl activation, but not proU derepression, in hns-1 mutant Escherichia coli. J Bacteriol. 1994;176:610–619. doi: 10.1128/jb.176.3.610-619.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seaton BL, Vickery LE. A gene encoding a DnaK/hsp70 homolog in Escherichia coli. Proc Natl Acad Sci. 1994;91:2066–2070. doi: 10.1073/pnas.91.6.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bukau B, Walker GC. Cellular defects caused by deletion of the Escherichia coli dnaK gene indicate roles for heat shock protein in normal metabolism. J Bacteriol. 1989;171:2337–2346. doi: 10.1128/jb.171.5.2337-2346.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paek KH, Walker GC. Escherichia coli dnaK null mutants are inviable at high temperature. J Bacteriol. 1987;169:283–290. doi: 10.1128/jb.169.1.283-290.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shan Y, Cortopassi G. Mitochondrial Hspa9/Mortalin regulates erythroid differentiation via iron-sulfur cluster assembly. Mitochondrion. 2016;26:94–103. doi: 10.1016/j.mito.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 22.Vergillo JA, Bagg A. Myelodysplastic syndromes. Contemporary biologic concepts and emerging diagnostic approaches. Am J Clin Pathol. 2003;119:S58–77. doi: 10.1309/T8DF-XEXM-YLHW-51DG. [DOI] [PubMed] [Google Scholar]
- 23.Chen TH-P, Kambal A, Krysiak K, Walshauser MA, Raju G, Tibbitts JF, Walter MJ. Knockdown of Hspa9, a del(5q31.2) gene, results in a decrease in hematopoietic progenitors in mice. Blood. 2011;117:1530–1539. doi: 10.1182/blood-2010-06-293167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Craven SE. Loss of Hspa9b in zebrafish recapitulates the ineffective hematopoiesis of the myelodysplastic syndrome. Blood. 2005;105:3528–3534. doi: 10.1182/blood-2004-03-1089. [DOI] [PubMed] [Google Scholar]
- 25.Majewska J, Ciesielski SJ, Schilke B, Kominek J, Blenska A, Delewski W, Song JY, Marszalek J, Craig EA, Dutkiewicz R. Binding of the chaperone Jac1 protein and cysteine desulfurase Nfs1 to the iron-sulfur cluster scaffold Isu protein Is mutually exclusive. J Biol Chem. 2013;288:29134–29142. doi: 10.1074/jbc.M113.503524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JH, Bothe JR, Alderson TR, Markley JL. Tangled web of interactions among proteins involved in iron–sulfur cluster assembly as unraveled by NMR, SAXS, chemical crosslinking, and functional studies. Biochim Biophys Acta. 2015;1853:1416–1428. doi: 10.1016/j.bbamcr.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoff KG, Cupp-Vickery JR, Vickery LE. Contributions of the LPPVK motif of the iron-sulfur template protein IscU to interactions with the Hsc66-Hsc20 chaperone system. J Biol Chem. 2003;278:37582–37589. doi: 10.1074/jbc.M305292200. [DOI] [PubMed] [Google Scholar]
- 28.Silberg JJ, Hoff KG, Tapley TL, Vickery LE. The Fe/S assembly protein IscU behaves as a substrate for the molecular chaperone Hsc66 from Escherichia coli. J Biol Chem. 2001;276:1696–1700. doi: 10.1074/jbc.M009542200. [DOI] [PubMed] [Google Scholar]
- 29.Hoff KG, Silberg JJ, Vickery LE. Interaction of iron sulfur cluster assembly protien IscU with the Hsc66/Hsc20 molecular chaperone system of Escherichia coli. Proc Natl Acad Sci. 2000;97:7790–7795. doi: 10.1073/pnas.130201997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim JH, Tonelli M, Markley JL. Disordered form of the scaffold protein IscU is the substrate for iron-sulfur cluster assembly on cysteine desulfurase. Proc Natl Acad Sci. 2012;109 doi: 10.1073/pnas.1114372109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markley JL, Kim JH, Dai Z, Bothe JR, Cai K, Frederick RO, Tonelli M. Metamorphic protein IscU alternates conformations in the course of its role as the scaffold protein for iron–sulfur cluster biosynthesis and delivery. FEBS Lett. 2013;587:1172–1179. doi: 10.1016/j.febslet.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maio N, Singh A, Uhrigshardt H, Saxena N, Tong W-H, Rouault Tracey A. Cochaperone binding to LYR motifs confers specificity of iron sulfur cluster delivery. Cell Metab. 2014;19:445–457. doi: 10.1016/j.cmet.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uzarska MA, Dutkiewicz R, Freibert SA, Lill R, Muhlenhoff U. The mitochondrial Hsp70 chaperone Ssq1 facilitates Fe/S cluster transfer from Isu1 to Grx5 by complex formation. Mol Biol Cell. 2013;24:1830–1841. doi: 10.1091/mbc.E12-09-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandramouli K, Johnson MK. HscA and HscB stimulate [2Fe-2S] cluster transfer from IscU to apoferredoxin in an ATP-dependent reaction. Biochemistry. 2006;45:11087–11095. doi: 10.1021/bi061237w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shakamuri P, Zhang B, Johnson MK. Monothiol glutaredoxins function in storing and transporting [2Fe-2S] clusters assembled on IscU scaffold proteins. J Am Chem Soc. 2012;134:15213–15216. doi: 10.1021/ja306061x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonomi F, Iametti S, Morleo A, Ta D, Vickery LE. Studies on the mechanism of catalysis of iron-sulfur cluster transfer from IscU[2Fe2S] by HscA/HscB chaperones. Biochemistry. 2008;47:12795–12801. doi: 10.1021/bi801565j. [DOI] [PubMed] [Google Scholar]
- 37.Bonomi F, Iametti S, Morleo A, Ta D, Vickery LE. Facilitated transfer of IscU–[2Fe2S] clusters by chaperone-mediated ligand exchange. Biochemistry. 2011;50:9641–9650. doi: 10.1021/bi201123z. [DOI] [PubMed] [Google Scholar]
- 38.Cai K, Liu G, Frederick Ronnie O, Xiao R, Montelione Gaetano T, Markley John L. Structural/functional properties of human NFU1, an intermediate [4Fe-4S] carrier in human mitochondrial iron-sulfur cluster biogenesis. Structure. 2016;24:2080–2091. doi: 10.1016/j.str.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maio N, Kim KS, Singh A, Rouault TA. A single adaptable cochaperone-scaffold complex delivers nascent iron-sulfur clusters to mammalian respiratory chain complexes I–III. Cell Metab. 2017;25:945–953.e946. doi: 10.1016/j.cmet.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schilke B, Voisine C, Beinert H, Craig E. Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1999;96:10206–10211. doi: 10.1073/pnas.96.18.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ganesh S. The Lafora disease gene product laforin interacts with HIRIP5, a phylogenetically conserved protein containing a NifU-like domain. Hum Mol Gen. 2003;12:2359–2368. doi: 10.1093/hmg/ddg253. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Cowan JA. Iron sulfur cluster biosynthesis. Human NFU mediates sulfide delivery to ISU in the final step of [2Fe-2S] cluster assembly. Chem Commun. 2007:3192–3194. doi: 10.1039/b704928e. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, Cowan JA. Iron–sulfur cluster biosynthesis: characterization of a molten globule domain in human NFU. Biochemistry. 2009;48:7512–7518. doi: 10.1021/bi9002524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tong WH, Jameson GNL, Huynh BH, Rouault TA. Subcellular compartmentalization of human Nfu, an iron-sulfur cluster scaffold protein, and its ability to assemble a [4Fe-4S] cluster. Proc Natl Acad Sci USA. 2003;100:9762–9767. doi: 10.1073/pnas.1732541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melber A, Na U, Vashisht A, Weiler BD, Lill R, Wohlschlegel JA, Winge DR. Role of Nfu1 and Bol3 in Iiron-sulfur cluster transfer to mitochondrial clients. eLife. 2016;5:e15991. doi: 10.7554/eLife.15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cameron Jessie M, Janer A, Levandovskiy V, MacKay N, Rouault Tracey A, Tong W-H, Ogilvie I, Shoubridge Eric A, Robinson Brian H. Mutations in iron-sulfur cluster scaffold genes NFU1 and BOLA3 cause a fatal deficiency of multiple respiratory chain and 2-oxoacid dehydrogenase enzymes. Am J Hum Genet. 2011;89:486–495. doi: 10.1016/j.ajhg.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Navarro-Sastre A, Tort F, Stehling O, Uzarska Marta A, Arranz José A, del Toro M, Labayru MT, Landa J, Font A, Garcia-Villoria J, et al. A fatal mitochondrial disease is associated with defective NFU1 function in the maturation of a subset of mitochondrial Fe-S proteins. Am J Hum Genet. 2011;89:656–667. doi: 10.1016/j.ajhg.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tort F, Ferrer-Cortes X, Ribes A. Differential diagnosis of lipoic acid synthesis defects. J Inherit Metab Dis. 2016;39:781–793. doi: 10.1007/s10545-016-9975-4. [DOI] [PubMed] [Google Scholar]
- 49.Li J, Ding S, Cowan JA. Thermodynamic and structural analysis of human NFU conformational chemistry. Biochemistry. 2013;52:4904–4913. doi: 10.1021/bi400320s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Webb MR. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc Natl Acad Sci U S A. 1992;89:4884–4887. doi: 10.1073/pnas.89.11.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silberg JJ, Vickery LE. Kinetic characterization of the ATPase cycle of the molecular chaperone Hsc66 from Escherichia coli. J Biol Chem. 2000;275:7779–7786. doi: 10.1074/jbc.275.11.7779. [DOI] [PubMed] [Google Scholar]
- 52.Luo W-I, Dizin E, Yoon T, Cowan JA. Kinetic and structural characterization of human mortalin. Protein Expr Purif. 2010;72:75–81. doi: 10.1016/j.pep.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoff KG, Silberg JJ, Vickery LE. Interaction of the iron-sulfur cluster assembly protein IscU with the Hsc66/Hsc20 molecular chaperone system of Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:7790–7795. doi: 10.1073/pnas.130201997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim JH, Tonelli M, Frederick RO, Chow DCF, Markley JL. Specialized Hsp70 chaperone (HscA) binds preferentially to the disordered form, whereas J-protein (HscB) binds preferentially to the structured form of the iron-sulfur cluster scaffold protein (IscU) J Biol Chem. 2012;287:31406–31413. doi: 10.1074/jbc.M112.352617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoon T. Functional and structural studies of human frataxin :an iron chaperone protein for mitochondrial iron-sulfur cluster and heme biosyntheses Ph.D. The Ohio State University, Columbus; 2005. [Google Scholar]
- 56.Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 57.Tong WH, Jameson GNL, Huynh BH, Rouault TA. Subcellular compartmentalization of human Nfu, an iron-sulfur cluster scaffold protein, and its ability to assemble a [4Fe-4S] cluster. Proc Natl Acad Sci USA. 2003;100:9762–9767. doi: 10.1073/pnas.1732541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yabe T. The Arabidopsis chloroplastic NifU-like protein CNFU, which can act as an iron-sulfur cluster scaffold protein, is required for biogenesis of ferredoxin and photosystem I. Plant Cell. 2004;16:993–1007. doi: 10.1105/tpc.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao H, Subramanian S, Couturier J, Naik SG, Kim S-K, Leustek T, Knaff DB, Wu H-C, Vignols F, Huynh BH, et al. Arabidopsis thaliana Nfu2 accommodates [2Fe-2S] or [4Fe-4S] clusters and is competent for in vitro maturation of chloroplast [2Fe-2S] and [4Fe-4S] cluster-containing proteins. Biochemistry. 2013;52:6633–6645. doi: 10.1021/bi4007622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mapolelo DT, Zhang B, Randeniya S, Albetel AN, Li H, Couturier J, Outten CE, Rouhier N, Johnson MK. Monothiol glutaredoxins and A-type proteins: partners in Fe-S cluster trafficking. Dalton Trans. 2013;42:3107–3115. doi: 10.1039/c2dt32263c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fidai I, Wachnowsky C, Cowan JA. Glutathione-complexed [2Fe-2S] clusters function in Fe-S cluster storage and trafficking. J Biol Inorg Chem. 2016;21:887–901. doi: 10.1007/s00775-016-1387-2. [DOI] [PubMed] [Google Scholar]
- 62.Wu S-P, Mansy SS, Cowan JA. Iron-sulfur cluster biosynthesis. Molecular chaperone DnaK promotes IscU-bound [2Fe-2S] cluster stability and inhibits cluster transfer activity. Biochemistry. 2005;44:4284–4293. doi: 10.1021/bi0483007. [DOI] [PubMed] [Google Scholar]
- 63.Wachnowsky C, Wesley NA, Fidai I, Cowan JA. Understanding the molecular basis for Multiple Mitochondrial Dysfunctions Syndrome 1 (MMDS1) - Impact of a disease-causing Gly208Cys substitution on structure and activity of NFU1 in the Fe/S cluster biosynthetic pathway. J Mol Biol. 2017;429:790–807. doi: 10.1016/j.jmb.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maio N, Rouault TA. Iron–sulfur cluster biogenesis in mammalian cells: New insights into the molecular mechanisms of cluster delivery. Biochim Biophys Acta. 2015;1853:1493–1512. doi: 10.1016/j.bbamcr.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoff KG, Ta DT, Tapley TL, Silberg JJ, Vickery LE. Hsc66 substrate specificity is directed toward a discrete region of the iron-sulfur cluster template protein IscU. J Biol Chem. 2002;277:27353–27359. doi: 10.1074/jbc.M202814200. [DOI] [PubMed] [Google Scholar]
- 66.Chung WH, Kim KD, Roe JH. Localization and function of three monothiol glutaredoxins in Schizosaccharomyces pombe. Biochem Biophys Res Commun. 2005;330:604–610. doi: 10.1016/j.bbrc.2005.02.183. [DOI] [PubMed] [Google Scholar]
- 67.Cheng N-H, Zhang W, Chen W-Q, Jin J, Cui X, Butte NF, Chan L, Hirschi KD. A mammalian monothiol glutaredoxin, Grx3, is critical for cell cycle progression during embryogenesis. FEBS J. 2011;278:2525–2539. doi: 10.1111/j.1742-4658.2011.08178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Molina MM, Bellí G, de la Torre MA, Rodríguez-Manzaneque MT, Herrero E. Nuclear monothiol glutaredoxins of Saccharomyces cerevisiae can function as mitochondrial glutaredoxins. J Biol Chem. 2004;279:51923–51930. doi: 10.1074/jbc.M410219200. [DOI] [PubMed] [Google Scholar]
- 69.Molina-Navarro MM, Casas C, Piedrafita L, Bellí G, Herrero E. Prokaryotic and eukaryotic monothiol glutaredoxins are able to perform the functions of Grx5 in the biogenesis of Fe/S clusters in yeast mitochondria. FEBS Lett. 2006;580:2273–2280. doi: 10.1016/j.febslet.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 70.Ojeda L, Keller G, Muhlenhoff U, Rutherford JC, Lill R, Winge DR. Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J Biol Chem. 2006;281:17661–17669. doi: 10.1074/jbc.M602165200. [DOI] [PubMed] [Google Scholar]
- 71.Yabe T, Yamashita E, Kikuchi A, Morimoto K, Nakagawa A, Tsukihara T, Nakai M. Structural analysis of Arabidopsis CNfu protein: An iron–sulfur cluster biosynthetic scaffold in chloroplasts. J Mol Biol. 2008;381:160–173. doi: 10.1016/j.jmb.2008.05.072. [DOI] [PubMed] [Google Scholar]
- 72.Shi R, Proteau A, Villarroya M, Moukadiri I, Zhang L, Trempe J-F, Matte A, Armengod ME, Cygler M. Structural basis for Fe–S cluster assembly and tRNA thiolation mediated by IscS protein–protein interactions. PLoS Biol. 2010;8:e1000354. doi: 10.1371/journal.pbio.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yan R, Kelly G, Pastore A. The scaffold protein IscU retains a structured conformation in the Fe-S cluster assembly complex. ChemBioChem. 2014;15:1682–1686. doi: 10.1002/cbic.201402211. [DOI] [PubMed] [Google Scholar]
- 74.Xia B, Cheng H, Bandarian V, Reed GH, Markley JL. Human ferredoxin: overproduction in Escherichia coli, reconstitution in vitro, and spectroscopic studies of iron-sulfur cluster ligand cysteine-to-serine mutants. Biochemistry. 1996;35:9488–9495. doi: 10.1021/bi960467f. [DOI] [PubMed] [Google Scholar]
- 75.Johansson C, Kavanagh KL, Gileadi O, Oppermann U. Reversible Sequestration of Active Site Cysteines in a 2Fe-2S-bridged Dimer Provides a Mechanism for Glutaredoxin 2 Regulation in Human Mitochondria. J Biol Chem. 2006;282:3077–3082. doi: 10.1074/jbc.M608179200. [DOI] [PubMed] [Google Scholar]
- 76.Johansson C, Roos Annette K, Montano Sergio J, Sengupta R, Filippakopoulos P, Guo K, von Delft F, Holmgren A, Oppermann U, Kavanagh Kathryn L. The crystal structure of human GLRX5: iron–sulfur cluster co-ordination, tetrameric assembly and monomer activity. Biochem J. 2011;433:303–311. doi: 10.1042/BJ20101286. [DOI] [PubMed] [Google Scholar]
- 77.Wang L, Li Y, Jacquot JP, Rouhier N, Xia B. Characterization of poplar GrxS14 in different structural forms. Protein Cell. 2014;5:329–333. doi: 10.1007/s13238-014-0042-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Banci L, Brancaccio D, Ciofi-Baffoni S, Del Conte R, Gadepalli R, Mikolajczyk M, Neri S, Piccioli M, Winkelmann J. [2Fe-2S] cluster transfer in iron-sulfur protein biogenesis. Proc Natl Acad Sci USA. 2014;111:6203–6208. doi: 10.1073/pnas.1400102111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Foster M, Mansy S, Hwang J, Penner-Hahn J, Surerus K, Cowan J. A mutant human IscU protein contains a stable [2Fe-2S]2+ center of possible functional significance. J Am Chem Soc. 2000;122:6805–6806. [Google Scholar]
- 80.Mansy SS, Xiong Y, Hemann C, Hille R, Sundaralingam M, Cowan JA. Crystal structure and stability studies of C77S HiPIP: a serine ligated [4Fe-4S] cluster. Biochemistry. 2002;41:1195–1201. doi: 10.1021/bi011811y. [DOI] [PubMed] [Google Scholar]
- 81.Nuth M, Yoon T, Cowan JA. Iron-Sulfur Cluster Biosynthesis: Characterization of Iron Nucleation Sites for Assembly of the [2Fe-2S]2+ Cluster Core in IscU Proteins. J Am Chem Soc. 2002;124:8774–8775. doi: 10.1021/ja0264596. [DOI] [PubMed] [Google Scholar]
- 82.Qi W, Li J, Cowan JA. Human ferredoxin-2 displays a unique conformational change. Dalton Trans. 2013;42:3088–3091. doi: 10.1039/c2dt32018e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Qi W, Cowan JA. Mechanism of glutaredoxin-ISU [2Fe-2S] cluster exchange. Chem Commun. 2011;47:4989–4991. doi: 10.1039/c0cc05079b. [DOI] [PubMed] [Google Scholar]
- 84.Li H, Mapolelo DT, Dingra NN, Naik SG, Lees NS, Hoffman BM, Riggs-Gelasco PJ, Huynh BH, Johnson MK, Outten CE. The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry. 2009;48:9569–9581. doi: 10.1021/bi901182w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Krebs C, Agar JN, Smith AD, Frazzon J, Dean DR, Huynh BH, Johnson MK. IscA, an alternate scaffold for Fe-S cluster biosynthesis. Biochemistry. 2001;40:14069–14080. doi: 10.1021/bi015656z. [DOI] [PubMed] [Google Scholar]
- 86.Wu S-p, Wu G, Surerus KK, Cowan JA. Iron-sulfur cluster biosynthesis. Kinetic analysis of [2Fe-2S] cluster transfer from holo ISU to apo Fd: role of redox chemistry and a conserved aspartate. Biochemistry. 2002;41:8876–8885. doi: 10.1021/bi0256781. [DOI] [PubMed] [Google Scholar]
- 87.Moulis J-M, Meyer J. Characterization of the selenium-substituted 2[4Fe-4Se] ferredoxin from Clostridium pasteurianum. Biochemistry. 1982;21:4762–4771. doi: 10.1021/bi00262a037. [DOI] [PubMed] [Google Scholar]
- 88.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: Application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]