Abstract
Background
Specificity protein 1 (Sp1) is involved in the transcription of several genes implicated in tumor maintenance. We investigated the effects of Mithramycin A (MTA), an inhibitor of SP1 DNA binding, on radiation response.
Methods
Clonogenic survival after irradiation was assessed in tumor cell lines (A549, UM-UC3) and one human fibroblast line (BJ) after Sp1 knockdown or MTA treatment. DNA damage repair was evaluated using γH2AX foci formation, and mitotic catastrophe was assessed using nuclear morphology. Gene expression was evaluated using PCR arrays. In vivo tumor growth delay was used to evaluate the effects of MTA on radiosensitivity.
Results
Targeting of Sp1 with siRNA or MTA sensitized A549 and UMUC-3 to irradiation, with no effect on BJ radioresponse. MTA did not alter γH2AX foci formation after irradiation in tumor cells, but did enhance mitotic catastrophe. Treatment with MTA suppressed transcription of genes involved in cell death. MTA administration to mice bearing A549 and UM-UC3 xenografts enhanced radiation-induced tumor growth delay.
Conclusions
These studies support Sp1 as a target for radiation sensitization and confirm MTA as a radiation sensitizer in human tumor models.
Keywords: radiation, mithramycin A, Sp1, mitotic catastrophe
Introduction
Specificity protein 1 (Sp1) is a transcription factor required for the expression of numerous genes important in proliferation, apoptosis, angiogenesis, DNA damage response, invasion, and metastasis [1]. In addition to a reported role in DNA damage repair [2–6], Sp1 transcriptionally regulates several genes involved in radiation response [6]. Mithramycin A (MTA) is an antibiotic with anti-neoplastic activity in diverse cancer histologies [7–9] that is attributed to its capacity to inhibit RNA synthesis [10]. MTA preferentially binds to GC-rich regions of DNA, thus preventing binding of transcription factors, such as Sp1 [11] and Egr1 [12, 13], that play critical roles in cell survival, cell growth, cell differentiation, and tumor progression [14].
To evaluate pharmacologic targeting of Sp1 as a strategy for enhancing tumor cell radiation response, we first confirmed that silencing of Sp1 sensitized cancer cell lines to irradiation; the response of a human fibroblast line was not affected. Pharmacologic inhibition of Sp1 activity with MTA increased apoptosis and mitotic catastrophe and sensitized tumor cells to irradiation in vitro and in vivo. The expression of several proteins transcriptionally regulated by Sp1 and implicated in cell death pathways were reduced after MTA exposure, suggesting MTA reduced the threshold for cell death in a multi-target fashion.
Material and Methods
Detailed methods are included in supplementary materials.
Cell culture and treatment
The A549 (human lung carcinoma), UM-UC-3 (human transitional cell carcinoma), and BJ (human foreskin fibroblast) cell lines were obtained from American Type Culture Collections (ATCC, Manassas, VA) and used within 15 passages. Cells were cultured at 37°C in a humidified atmosphere with 5% CO2, and maintained in RPMI 1640 medium with 2 mM L-glutamine and HEPES (ATCC) supplemented with 10% fetal bovine serum (Gemini Bio-products, West Sacramento, CA).
MTA, from Tocris Bioscience (Bristol, UK), was prepared in DMSO (vehicle, Sigma-Aldrich, St. Louis, MO) and used at final concentrations of 25–50 nM). Radiation was delivered using an XRAD 320 (Precision X-Ray Inc., N. Branford, CT) at a dose rate of 2.21 Gy/min (additional details provided in the supplemental methods).
Short interfering RNA treatment
The smart interfering RNA (ON-TARGETplus siRNA) SMARTpool containing four pooled siRNA targeting Sp1 (GE Dharmacon, Lafayette, CO) and a non-specific control pooled siRNA were transfected using DharmaFECT 1 (GE Dharmacon).
Clonogenic survival assay
Exponentially growing cells were plated. After attachment (5 h after plating), cells were treated with vehicle or MTA for 1 h prior to irradiation. Media was replaced 24h after irradiation. Cells were stained with crystal violet after 7–14 days, and the number of colonies (≥ 50 cells) was determined. Dose enhancement factors (DEF) were calculated by taking the ratio of the radiation dose resulting in 10% survival (control: drug-treated). Toxicity was calculated as the ratio of the plating efficiency of cells treated with MTA versus vehicle alone. In separate studies, cells transfected with Sp1 specific or control siRNA were plated and irradiated (24 h post-transfection) and clonogenic survival was determined as described above.
Gene expression analysis
The expression of 84 cell death-associated genes was determined using RT2 Profiler PCR arrays (Human Apoptosis, Qiagen, Valencia, CA) as per manufacturer’s instructions using RNA isolated from A549 cells treated for 24 h with MTA (50nM) or vehicle. RNA was purified with the RNEasy system and reverse-transcribed using the RT2 First Strand Kit (Qiagen). Gene expression was derived by the 2−ΔΔCT method using the Qiagen data analysis center (Qiagen), and normalized by the expression of β-2-microglobulin, glyceraldehyde-3-phosphate dehydrogenase, and the large ribosomal protein P0. Gene expression differences were considered significant when fold regulation was ≥ 1.5 and ≤ −1.5 and p≤0.05 by paired T-test.
Cell cycle and ploidy analysis
DNA content and mitotic fraction were determined from duplicate experiments using a FACSCaliber cytometer (BD Biosciences, San Jose, CA) and FlowJo software (Tree Star, Inc., Ashland, OR). Briefly, mitotic fractions of single cell suspensions were detected with anti-phospho-serine-10 histone H3 antibody (Millipore, Billerica, MA) and CruzFluor™ 488 labeled anti-rabbit IgG (Santa Cruz Biotechnology, Dallas, TX). DNA content was determined with propidium iodide (PI, Sigma-Aldrich). For ploidy analysis, cellular DNA was stained with FxCycle Violet (Life Technologies). DNA content was determined using a FACSCanto II cytometer and FlowJo software.
Apoptosis
Apoptosis was detected using an annexin V-FITC (fluorescein isothiocyanate) apoptosis kit (Sigma-Aldrich) per the manufacturer’s protocol. Cells were immediately transferred to ice and apoptosis was analyzed using a FACSCanto II cytometer (BD Biosciences) and FlowJo software.
γH2AX immunofluorescence
Briefly, fixed cells were incubated in anti-γH2AX antibody (Millipore) followed by a fluorescein isothiocyanate (FITC) conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA). Nuclei were stained with 4′, 6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich). Slides were imaged using a Leica DM LB2 fluorescent microscope and digital images were captured with QCapture (QImaging, BC, Canada). The number of γH2AX foci per cell was manually scored in 150 cells per condition.
Mitotic catastrophe
After methanol fixation (−20°C for 15 min) and permeabilization (0.5% Triton X-100), cells were incubated with anti-α-tubulin antibody (Sigma-Aldrich) followed by a FITC-conjugated secondary antibody (Invitrogen, Carlsbad, CA). Nuclei were counterstained with DAPI. The presence of fragmented nuclei and/or the presence of two or more distinct nuclear lobes in a single cell were scored in 150 cells per condition.
Immunoblotting
Cells and tumor xenografts were lysed in radioimmunoprecipitation assay buffer (Thermo Scientific, Grand Island, NY) supplemented with protease and phosphatase inhibitors (Roche, Basel, CH). Protein concentrations were measured by bicinchoninic acid assay (Thermo Scientific). Equal amounts of proteins were separated by SDS-PAGE (Thermo Scientific) and transferred to polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA). Immunoblotting was performed with primary antibodies against XIAP, survivin, Fas, Caspase-2, IGF1R, Bcl-w (Cell Signaling, Danvers, MA), and Actin (EMD-Millipore, Billerica, MA).
Tumor xenograft model
Animal procedures were institutionally approved and in accordance with the guidelines of the Institute of Laboratory Animal Resources, National Research Council. A549 or UM-UC-3 cells were injected subcutaneously into the hind leg of athymic nude mice (NCI Frederick, Frederick, MD). Tumor volumes were measured three times weekly.
Radiation was delivered with a Therapix DXT300 x-ray irradiator (Pantak, Inc., East Haven, CT) using 2.0 mm Al filtration (300 kV peak) at a dose rate of 1.9 Gy/min (additional details included in the supplemental methods). Mice received a single intraperitoneal injection of MTA or vehicle 1 h prior to irradiation (IR). For multi-fraction radiation, mice received MTA or vehicle 1 h prior to the first and third fraction of IR.
Tumor growth to 1000 mm3 was scored for individual animals. The mean growth delay was calculated as the mean number of days for treated tumors to grow to 1000 mm3 minus mean number of days for the vehicle group to reach the same size. The DEF was calculated as the ratio of the normalized tumor growth delay in mice treated with MTA + IR versus that obtained with IR alone.
Statistical Analysis
All in vitro experiments were conducted in triplicate unless otherwise noted. Comparisons between conditions were performed with ANOVA with Tukey’s correction for multiple comparisons. A p-value less than 0.05 was considered statistically significant.
Results
Targeting Sp1 enhances tumor cell radiosensitivity
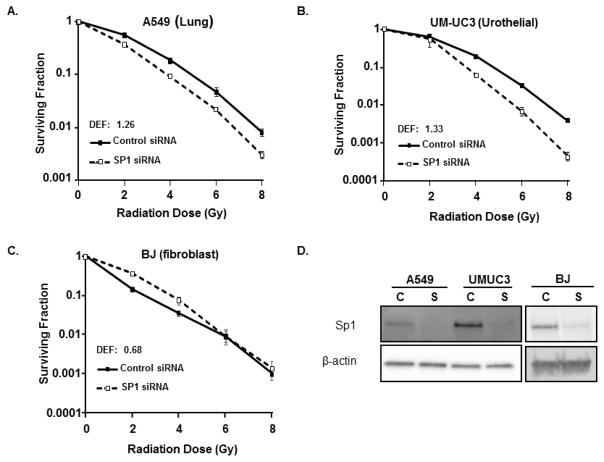
To confirm the role of Sp1 in radiation response, clonogenic assays were performed in the A549, UMUC-3, and BJ cell lines after Sp1 knockdown. Sp1 knockdown reduced clonogenic capacity after irradiation in the A549 and UMUC-3 tumor cells lines (Figure 1A), but had no impact on BJ radioresponse. SP1 knockdown was confirmed by immunoblotting (Figure 1B, Supplemental table 1).
Figure 1. SP1 knockdown enhances sensitivity of tumor cells to radiation.
The A549, UMUC-3, and BJ (fibroblast) cell lines were treated with control or SP1 siRNA. (A–C) Colony forming efficiency of transfected cells was determined. (column, mean; bars standard error). (D) SP1 knockdown was confirmed by immunoblotting (bottom panel). C: control siRNA, S: SP1 siRNA.
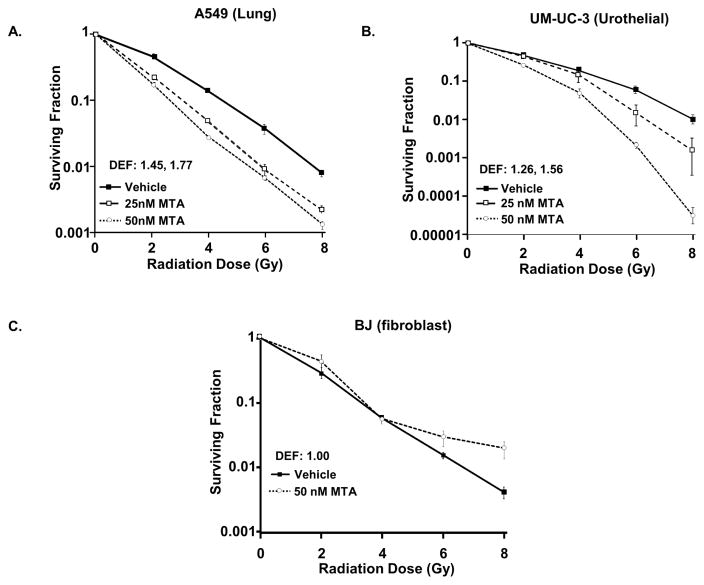
MTA was used to determine the effect of pharmacological inhibition of Sp1 in combination with radiation. MTA increased A549 and UM-UC-3 radiation sensitivity with DEF values 1.77 and 1.56, respectively (Figure 2, Supplemental table 1). The BJ fibroblast cell line was not sensitized to irradiation by MTA, as expected based on siRNA studies (DEF 1.0).
Figure 2. MTA enhances sensitivity of tumor cells to radiation.
A549 (A) and UM-UC3 (B) cells and BJ fibroblasts (C) were treated with MTA 1 h prior to and 24 h after irradiation. Colony forming efficiency was determined. (Points, mean; bars standard error). DEF: dose enhancement factor.
Effects of Mithramycin A on DNA repair and cell cycle
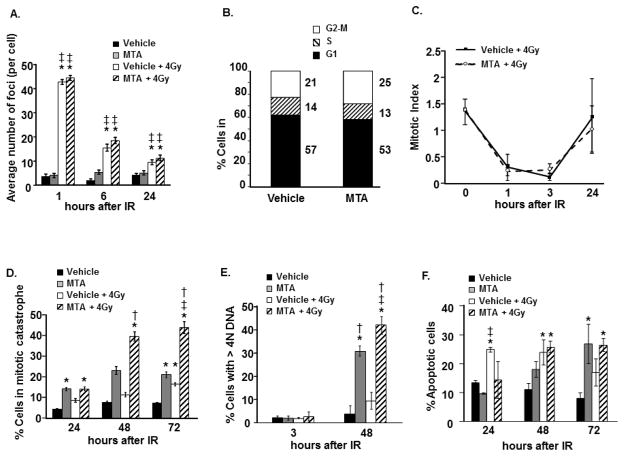
The A549 cell line was used to investigate the processes by which MTA enhances radiation response. Prior studies have documented the role of Sp1 in the repair of DSBs [5]. To determine if MTA altered DSB repair, a lethal event after ionizing radiation exposure, γH2AX foci induction and resolution was evaluated. No difference in γH2AX foci induction or resolution was observed between irradiated A549 cells treated with MTA or vehicle (Figure 3A), suggesting that MTA mediated radiosensitization does not involve inhibition of DSB repair. Because Sp1 is known to regulate the transcription of a number of cell cycle proteins [1], cell cycle analysis was performed to determine if MTA treatment altered cell cycle distribution at the time of irradiation. One hour MTA pre-treatment resulted in minimal change in the G2/M population (Figure 3B).
Figure 3. MTA enhances mitotic catastrophe and polyploidy.
A549 cells were treated with MTA or vehicle 1 h prior to irradiation (4Gy). MTA was removed after 24 h (panels A, C, D, E, F). (A) γH2AX was detected by immunocytochemistry and scored. (B) A549 cells treated with MTA or vehicle for 1 h and DNA was labeled with PI to determine the percentage of cells in the G2-M phase of the cell cycle (4N DNA content). (C) Mitotic index was assessed using PI and anti-phospho H3 antibodies. (D) After immunocytochemistry for tubulin with DAPI counterstain, the percentage of cells with nuclear fragmentation or bi-lobed nuclei was determined for each treatment condition. (E) DNA content was assessed by flow cytometry using FxCycle Violet. (F) At indicated time points cells were collected and extracellular phosphatidylserine (a marker for apoptosis) was detected by FITC conjugated annexin V binding. Cell membrane integrity was monitored by PI staining. For (A) columns, mean; bars ± standard error. *, significantly different than vehicle; ‡, significantly different than MTA. For (D), (E), and (F) columns, mean; bars ± standard deviation. *, significantly different than vehicle; ‡, significantly different than MTA; †, significantly different than vehicle + 4Gy.
Another potential source of radiosensitization is abrogation of the G2 checkpoint [15]. Flow cytometric analysis of phosphorylated histone H3 in the 4N cell population after irradiation was used to quantify G2 checkpoint activation [16]. Irradiation resulted in a rapid reduction in the mitotic index, reaching a maximum decrease at 3 h, indicating activation of the early G2 checkpoint (Figure 3C). There was no evidence that MTA treatment altered G2 checkpoint activation or recovery when compared to vehicle.
Mithramycin enhances mitotic catastrophe and apoptosis
To assess the rate of mitotic catastrophe, a common mechanism of cell death after radiation [17], the number of cells with abnormal nuclei was scored. There was a significant increase in mitotic catastrophe 72 h after cells were treated with MTA + IR compared to either treatment alone, suggesting MTA mediated radiosensitization involves enhanced mitotic catastrophe (Figure 3D). The observed increase in cells with >4N DNA content after MTA + IR relative to MTA or IR treatment alone is also consistent with induction of mitotic catastrophe (Figure 3E).
The importance of apoptosis in the radiation response of solid tumors is controversial [18, 19]. Apoptosis was measured in A549 cells exposed to MTA and/or 4 Gy of radiation. As shown in Figure 3F, MTA treatment and radiation both increase the number of apoptotic cells individually; however, the combination treatment was not significantly different than either treatment alone, indicating that MTA mediated radiation sensitization is likely not apoptosis dependent.
MTA reduces the expression of cell death pathway targets
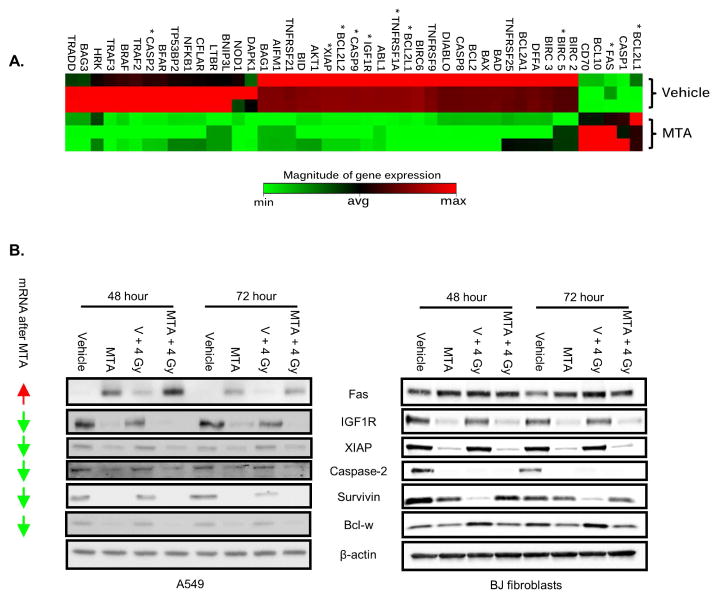
To explore the effects of MTA on cell death pathways, differential expression of 84 cell death-associated genes was assayed after 24 h of MTA exposure. A total of 45 genes were differentially expressed between vehicle and MTA treated A549 cells, with 40 of those down-regulated (Table 1). Unsupervised hierarchical clustering (Figure 4A) was used to organize evaluated genes based on common patterns of expression between sample groups.
Table 1.
Effects of MTA on Gene Expression
| Symbol | Fold Regulation (MTA vs. vehicle) | p-value |
|---|---|---|
| BIRC6 | −39.3377 | 0.0001 |
| DAPK1 | −31.5852 | 0.0481 |
| TRAF2 | −24.6101 | 0.0030 |
| TNFRSF1A | −19.533 | 0.0002 |
| IGF1R | −15.5392 | 0.0002 |
| BCL2L1 | −9.8117 | 0.0002 |
| BRAF | −9.7665 | 0.0044 |
| BCL2L2 | −7.7696 | 0.0003 |
| XIAP | −7.7517 | 0.0003 |
| TRAF3 | −7.7517 | 0.0047 |
| CASP9 | −7.7338 | 0.0003 |
| CASP2 | −6.181 | 0.0056 |
| ABL1 | −6.1667 | 0.0003 |
| BFAR | −6.1667 | 0.0057 |
| TNFRSF9 | −4.8945 | 0.0004 |
| BID | −3.9028 | 0.0012 |
| AKT1 | −3.8491 | 0.0011 |
| NFKB1 | −3.0977 | 0.0155 |
| BAG1 | −3.0905 | 0.0013 |
| NOD1 | −3.0905 | 0.0362 |
| AIFM1 | −3.0834 | 0.0013 |
| CFLAR | −3.0834 | 0.0174 |
| TP53BP2 | −3.0692 | 0.0149 |
| TNFRSF21 | −3.055 | 0.0015 |
| HRK | −2.4586 | 0.0334 |
| BAD | −2.4529 | 0.0011 |
| BCL2 | −2.4473 | 0.0013 |
| DIABLO | −2.4473 | 0.0014 |
| CASP8 | −2.4473 | 0.0015 |
| BAX | −2.4416 | 0.0013 |
| LTBR | −2.4416 | 0.0210 |
| BNIP3L | −2.436 | 0.0224 |
| TNFRSF25 | −1.9695 | 0.0295 |
| BCL2A1 | −1.9514 | 0.0296 |
| DFFA | −1.9424 | 0.0249 |
| BIRC3 | −1.9424 | 0.0257 |
| TRADD | −1.9379 | 0.0324 |
| BAG3 | −1.9379 | 0.0348 |
| BIRC2 | −1.5417 | 0.0456 |
| BIRC5 | −1.5417 | 0.0499 |
| BCL2L11 | 1.6232 | 0.0105 |
| CD70 | 2.5946 | 0.0148 |
| BCL10 | 2.5946 | 0.0154 |
| FAS | 4.1186 | 0.0087 |
| CASP1 | 8.4688 | 0.0151 |
Figure 4. Gene expression profiling with MTA.
(A) A549 cells were treated with vehicle or MTA 24 h. Gene expression was determined by quantitative PCR. Differentially expressed genes are clustered based on common patterns between sample groups. Genes implicated in mitotic catastrophe are marked with a “*”. (B) A549 and BJ cells were treated with MTA for 1 h followed by IR. After 24 h media was replaced. Lysates were collected at 48 and 72 h after IR. Arrows: mRNA expression change with MTA treatment (up vs. down relative to vehicle).
To validate these findings, we concentrated on 6 genes that have been implicated in sensitivity to both apoptosis and mitotic catastrophe. Treatment of A549 cells with MTA for 24 h reduced IGF1R, XIAP, Caspase-2, survivin, and Bcl-w protein levels, a finding that persisted with combined MTA and radiation exposure (Figure 4B). Simultaneously, the expression of Fas, a molecule involved in positive regulation of cell death, increased with MTA treatment, radiation, and the combination. Similar studies in the BJ fibroblast line revealed no obvious changes in Fas expression and reduced caspase-2 expression in the MTA and radiation treatment groups. Survivin expression in BJ fibroblasts also exhibited a different pattern than that observed in A549 cells, with less impressive reduction in expression after MTA treatment, and reduced expression after irradiation.
Mithramycin enhances radiation response in vivo
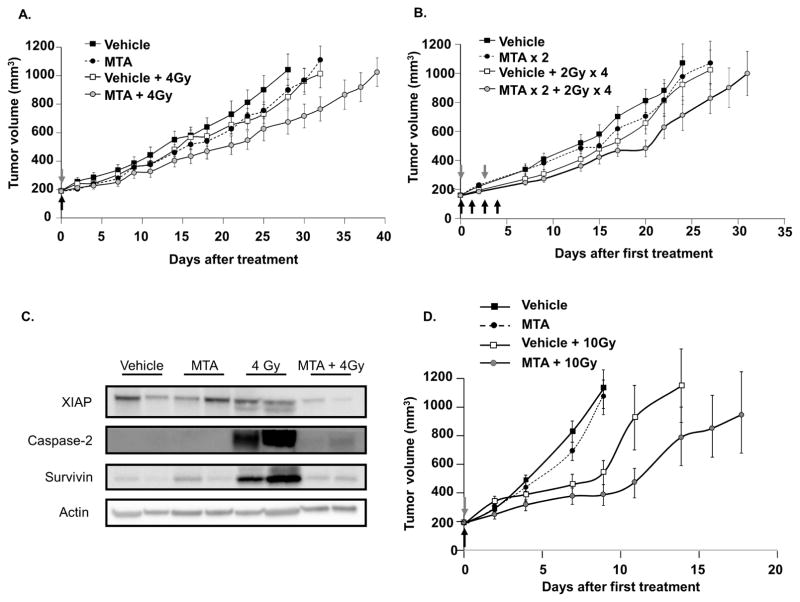
To translate our in vitro findings to an in vivo model, tumor growth delay of A549 xenografts was assessed. Mice bearing A549 xenografts were randomized into 4 groups on the first day of treatment: vehicle; MTA; vehicle + IR; and MTA + IR. In a second experiment, treatment groups included vehicle, fractionated IR, MTA x 2, and MTA x 2 + fractionated IR. The growth rates for the A549 tumors exposed to each treatment are shown in Figure 5A and C, respectively. For the single fraction study, MTA and IR delayed xenograft growth by 0.6 and 5.6 days, respectively. In mice that received the MTA + IR combination, xenograft growth was delayed by 9.2 days. Thus, the growth delay of the combined treatment was more than the sum of the growth delays of individual treatments (DEF of 1.6 for MTA).
Figure 5. MTA enhances tumor xenograft response to radiation.
Mice bearing A549 (A–B) or UMUC3 (C) xenografts were randomized to treatment with vehicle, MTA, IR, or MTA + IR. MTA (1 mg/kg) was given by IP injection 1 hour prior to IR. Points, mean; bars, SE. Grey arrows: MTA treatment, Black arrows: IR treatment. (D) A549 tumors were collected from mice 72 h after treatment as in (A) and protein extracts were subjected to immunoblotting.
A similar response was observed with fractionated exposures in the A549 xenograft model (Figure 5C). MTA and IR delayed tumor growth by 5 and 4.8 days respectively. With the MTA + IR combination, tumor growth was delayed by 9.6 days.
The UMUC-3 xenograft model was tested to confirm findings in a second histology (Figure 5D). MTA and IR treatments delayed UMUC3 xenograft growth by 0.9 and 9 days respectively, and for MTA + IR growth was delayed by 14 days (DEF 1.5).
Immunoblotting of protein isolated from A549 xenograft tumors collected at 72 h after treatment confirmed a reduction in XIAP and survivin expression with MTA treatment (Figure 5B). The in vivo expression of XIAP was unchanged by IR, whereas radiation increased survivin expression. With the combination of MTA and IR, both survivin and XIAP were reduced to or below levels observed with MTA treatment alone.
Discussion
MTA and its derivatives have shown therapeutic efficacy in a variety of pre-clinical models of cancer [7, 9, 20–22]. In this study, MTA significantly increased the sensitivity of cancer cell lines to radiation both in vitro and in vivo. In contrast, the radiosensitivity of a normal fibroblast cell line was not impacted by MTA treatment. Treatment of tumor cells with MTA reduced the expression of several genes that are implicated in resistance to apoptosis and mitotic catastrophe, a finding consistent with the observed increase in apoptosis after MTA treatment and the increase in mitotic catastrophe after MTA + IR relative to IR or MTA alone. These data support the use of MTA to selectively enhance the sensitivity of tumors to irradiation, and suggest that this efficacy may be multi-target in nature.
Sp1 has been implicated in the transcriptional control of several genes involved in DNA DSB repair, including ATM [23], CHEK2 [24], and DNA-PK/Ku/70/80 [25]. Sp1 also plays a transcription-independent role in DNA DSB repair [5, 6]. Indeed, silencing of Sp1 in some studies results in impaired DNA damage repair and sensitization to IR [5]. In contrast, we observed no difference in the rate of DNA DSB repair after IR with MTA treatment. This is consistent with the proposed mechanism of MTA, in which MTA impairs Sp1 binding to specific promotor DNA sequences, but does not directly alter Sp1 phosphorylation or non-transcriptional functions. Employing longer pre-IR incubation periods with MTA may lead to greater sensitization to IR through transcription-dependent mechanisms.
Mitotic catastrophe is a major mechanism of cell death in solid tumors following exposure to IR [17]. XIAP and survivin are two members of the inhibitor of apoptosis (IAP) protein family that suppress caspase activation [26], are transcriptionally regulated by Sp1 [20, 27], and have been reported to regulate mitotic catastrophe in different contexts [20, 28]. A decrease in the expression of XIAP and survivin was observed with MTA treatment and persisted in the setting of combined treatment with MTA and IR. Thus, MTA treatment may reduce the high basal levels of the protective XIAP and survivin, thus decreasing resistance to cell death after radiation exposure [29–31].
The observed increase in polyploid cells with MTA treatment in addition to irradiation, suggests that mitotic slippage may also contribute to mitotic catastrophe with the combination of MTA and IR. Indeed, Sp1 knockdown results in centrosome amplification, chromosome misalignment, multipolar mitotic spindle formation, micronuclei formation and an increase in aneuploidy and polyploidy [32], consistent with our findings.
Although the effects of MTA that lead to polyploidy and mitotic catastrophe are likely multi-factorial given the broad range of proteins transcriptionally regulated by Sp1, a decrease in survivin expression at the transcript and protein level in A549 cells was observed. Survivin is a key component of the chromosomal passenger complex, which regulates key mitotic events [33] and plays an integral role in chromosome segregation and ordered mitosis [34, 35]. Thus, reduction in survivin expression with MTA treatment may encourage mitotic slippage and aberrant mitosis.
The capacity to selectively enhance tumor compared to normal tissues is a critical consideration. Studies conducted with the BJ human dermal fibroblast cell confirmed that MTA and Sp1 silencing provide tumor selective sensitization to irradiation. Consistent with these findings, prior studies have demonstrated a protective capacity of MTA in normal tissues exposed to irradiation and oxidative stress [12]. The tumor selectivity of MTA radiation enhancement may reflect the relative importance of the genes transcriptionally repressed by MTA [1, 13]. We observed similar patterns of expression of cell death related proteins in A549 and BJ cells with MTA treatment, with a few notable exceptions. The expression of survivin after MTA and/or IR was notably different between fibroblasts and A549. Based on the importance of survivin in mitotic catastrophe, chromosome segregation, and apoptosis, this is a particularly interesting finding.
Mithramycin is currently in clinical trials as a single agent for the treatment of malignancy (NCT01624090, NCT02859415). Few modern data on mithramycin related toxicity is available, although older studies have suggested nausea, thrombocytopenia, and tumor necrosis are possible toxicities of treatment [36]. A number of analogues of mithramycin have been developed to reduce toxicity and enhance potency in regards to EWS-FLI1 and SP1 inhibition [37–39]. These data support the study of mithramycin analogues as radiation modifiers.
Conclusions
In summary, we demonstrate that Sp1 targeting with MTA selectively enhances radiation response in vitro and in vivo. This effect corresponds to an increase in tumor cell mitotic catastrophe after irradiation in the presence of MTA and occurs in the setting of reduced expression of multiple cell death-associated proteins.
Supplementary Material
Summary.
In this manuscript, the ability of MTA to selectively enhance tumor response to radiation is established. Treatment with MTA prior to irradiation increases mitotic catastrophe and reduces the expression of several genes related to cell death.
Acknowledgments
Acknowledgements/Research Support: This research was supported by the Intramural Research Program (Center for Cancer Research) of the National Institutes of Health, National Cancer Institute (ZIA BC 011552). This research was also made possible in part through the NIH Medical Research Scholars Program, a public-private partnership supported jointly by the NIH and generous contributions to the Foundation for the NIH from the Doris Duke Charitable Foundation, the Howard Hughes Medical Institute, the American Association for Dental Research, the Colgate-Palmolive Company, and other private donors. No funds from the Doris Duke Charitable Foundation were used to support research that used animals.
Footnotes
Conflicts of interest: The authors declare that they have no conflicts of interest to report.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Beishline K, Azizkhan-Clifford J. Sp1 and the ‘hallmarks of cancer’. FEBS J. 2015;282(2):224–58. doi: 10.1111/febs.13148. [DOI] [PubMed] [Google Scholar]
- 2.Yang CR, et al. Coordinate modulation of Sp1, NF-kappa B, and p53 in confluent human malignant melanoma cells after ionizing radiation. FASEB J. 2000;14(2):379–90. doi: 10.1096/fasebj.14.2.379. [DOI] [PubMed] [Google Scholar]
- 3.Meighan-Mantha RL, et al. Ionizing radiation stimulates octamer factor DNA binding activity in human carcinoma cells. Mol Cell Biochem. 1999;199(1–2):209–15. doi: 10.1023/a:1006958217143. [DOI] [PubMed] [Google Scholar]
- 4.Gueven N, et al. Site-directed mutagenesis of the ATM promoter: consequences for response to proliferation and ionizing radiation. Genes Chromosomes Cancer. 2003;38(2):157–67. doi: 10.1002/gcc.10261. [DOI] [PubMed] [Google Scholar]
- 5.Olofsson BA, et al. Phosphorylation of Sp1 in response to DNA damage by ataxia telangiectasia-mutated kinase. Mol Cancer Res. 2007;5(12):1319–30. doi: 10.1158/1541-7786.MCR-07-0374. [DOI] [PubMed] [Google Scholar]
- 6.Beishline K, et al. Sp1 facilitates DNA double-strand break repair through a nontranscriptional mechanism. Mol Cell Biol. 2012;32(18):3790–9. doi: 10.1128/MCB.00049-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bataller M, et al. Mithramycin SK modulates polyploidy and cell death in colon carcinoma cells. Mol Cancer Ther. 2008;7(9):2988–97. doi: 10.1158/1535-7163.MCT-08-0420. [DOI] [PubMed] [Google Scholar]
- 8.Brown JH, Kennedy BJ. Mithramycin in the Treatment of Disseminated Testicular Neoplasms. N Engl J Med. 1965;272:111–8. doi: 10.1056/NEJM196501212720301. [DOI] [PubMed] [Google Scholar]
- 9.Grohar PJ, et al. Identification of an inhibitor of the EWS-FLI1 oncogenic transcription factor by high-throughput screening. J Natl Cancer Inst. 2011;103(12):962–78. doi: 10.1093/jnci/djr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller DM, et al. Mithramycin selectively inhibits transcription of G-C containing DNA. Am J Med Sci. 1987;294(5):388–94. doi: 10.1097/00000441-198711000-00015. [DOI] [PubMed] [Google Scholar]
- 11.Blume SW, et al. Mithramycin inhibits SP1 binding and selectively inhibits transcriptional activity of the dihydrofolate reductase gene in vitro and in vivo. J Clin Invest. 1991;88(5):1613–21. doi: 10.1172/JCI115474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao DY, et al. Silencing Egr1 Attenuates Radiation-Induced Apoptosis in Normal Tissues while Killing Cancer Cells and Delaying Tumor Growth. Mol Cancer Ther. 2015 doi: 10.1158/1535-7163.MCT-14-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vizcaino C, et al. Genome-wide modulation of gene transcription in ovarian carcinoma cells by a new mithramycin analogue. PLoS One. 2014;9(8):e104687. doi: 10.1371/journal.pone.0104687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur J Cancer. 2005;41(16):2438–48. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Cliby WA, et al. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. Embo J. 1998;17(1):159–69. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu B, et al. Two molecularly distinct G(2)/M checkpoints are induced by ionizing irradiation. Mol Cell Biol. 2002;22(4):1049–59. doi: 10.1128/MCB.22.4.1049-1059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eriksson D, Stigbrand T. Radiation-induced cell death mechanisms. Tumour Biol. 2010;31(4):363–72. doi: 10.1007/s13277-010-0042-8. [DOI] [PubMed] [Google Scholar]
- 18.Crowther AJ, et al. Radiation Sensitivity in a Preclinical Mouse Model of Medulloblastoma Relies on the Function of the Intrinsic Apoptotic Pathway. Cancer Res. 2016;76(11):3211–23. doi: 10.1158/0008-5472.CAN-15-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abend M. Reasons to reconsider the significance of apoptosis for cancer therapy. Int J Radiat Biol. 2003;79(12):927–41. doi: 10.1080/09553000310001632958. [DOI] [PubMed] [Google Scholar]
- 20.Lee TJ, et al. Mithramycin A sensitizes cancer cells to TRAIL-mediated apoptosis by down-regulation of XIAP gene promoter through Sp1 sites. Mol Cancer Ther. 2006;5(11):2737–46. doi: 10.1158/1535-7163.MCT-06-0426. [DOI] [PubMed] [Google Scholar]
- 21.Rao M, et al. Mithramycin Depletes Specificity Protein 1 and Activates p53 to Mediate Senescence and Apoptosis of Malignant Pleural Mesothelioma Cells. Clin Cancer Res. 2016;22(5):1197–210. doi: 10.1158/1078-0432.CCR-14-3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sleiman SF, et al. Mithramycin is a gene-selective Sp1 inhibitor that identifies a biological intersection between cancer and neurodegeneration. J Neurosci. 2011;31(18):6858–70. doi: 10.1523/JNEUROSCI.0710-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gueven N, et al. Epidermal growth factor sensitizes cells to ionizing radiation by down-regulating protein mutated in ataxia-telangiectasia. J Biol Chem. 2001;276(12):8884–91. doi: 10.1074/jbc.M006190200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang S, et al. A variant in the CHEK2 promoter at a methylation site relieves transcriptional repression and confers reduced risk of lung cancer. Carcinogenesis. 2010;31(7):1251–8. doi: 10.1093/carcin/bgq089. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig DL, et al. Ku80 gene expression is Sp1-dependent and sensitive to CpG methylation within a novel cis element. Gene. 1997;199(1–2):181–94. doi: 10.1016/s0378-1119(97)00366-1. [DOI] [PubMed] [Google Scholar]
- 26.Dubrez L, Berthelet J, Glorian V. IAP proteins as targets for drug development in oncology. Onco Targets Ther. 2013;9:1285–304. doi: 10.2147/OTT.S33375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Y, Giacalone NJ, Lu B. Terameprocol (tetra-O-methyl nordihydroguaiaretic acid), an inhibitor of Sp1-mediated survivin transcription, induces radiosensitization in non-small cell lung carcinoma. J Thorac Oncol. 2011;6(1):8–14. doi: 10.1097/JTO.0b013e3181fa646a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arora V, et al. Degradation of survivin by the X-linked inhibitor of apoptosis (XIAP)-XAF1 complex. J Biol Chem. 2007;282(36):26202–9. doi: 10.1074/jbc.M700776200. [DOI] [PubMed] [Google Scholar]
- 29.Hehlgans S, et al. Double targeting of Survivin and XIAP radiosensitizes 3D grown human colorectal tumor cells and decreases migration. Radiother Oncol. 2013;108(1):32–9. doi: 10.1016/j.radonc.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 30.Yang CT, et al. Adenovirus-mediated transfer of siRNA against survivin enhances the radiosensitivity of human non-small cell lung cancer cells. Cancer Gene Ther. 2010;17(2):120–30. doi: 10.1038/cgt.2009.55. [DOI] [PubMed] [Google Scholar]
- 31.Yang W, et al. Survivin downregulation by siRNA/cationic liposome complex radiosensitises human hepatoma cells in vitro and in vivo. Int J Radiat Biol. 2010;86(6):445–57. doi: 10.3109/09553001003668006. [DOI] [PubMed] [Google Scholar]
- 32.Astrinidis A, et al. The transcription factor SP1 regulates centriole function and chromosomal stability through a functional interaction with the mammalian target of rapamycin/raptor complex. Genes Chromosomes Cancer. 2010;49(3):282–97. doi: 10.1002/gcc.20739. [DOI] [PubMed] [Google Scholar]
- 33.Carmena M, et al. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol. 2012;13(12):789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li F, et al. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396(6711):580–4. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 35.Lens SM, et al. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J. 2003;22(12):2934–47. doi: 10.1093/emboj/cdg307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baum M. A clinical trial of mithramycin in the treatment of advanced malignant disease. Br J Cancer. 1968;22(2):176–83. doi: 10.1038/bjc.1968.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osgood CL, et al. Identification of Mithramycin Analogues with Improved Targeting of the EWS-FLI1 Transcription Factor. Clin Cancer Res. 2016;22(16):4105–18. doi: 10.1158/1078-0432.CCR-15-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou C, et al. Structures of mithramycin analogues bound to DNA and implications for targeting transcription factor FLI1. Nucleic Acids Res. 2016;44(18):8990–9004. doi: 10.1093/nar/gkw761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nunez LE, et al. A novel mithramycin analogue with high antitumor activity and less toxicity generated by combinatorial biosynthesis. J Med Chem. 2012;55(12):5813–25. doi: 10.1021/jm300234t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.