Abstract
Anatomical connectivity and lesion studies reveal distinct functional heterogeneity along the dorsal-ventral axis of the hippocampus. The immediate early gene Arc is known to be involved in neural plasticity and memory and can be used as a marker for cell activity that occurs, for example, when hippocampal place cells fire. We report here, that Arc is expressed in a greater proportion of cells in dorsal CA1, CA3 and dentate gyrus (DG), following spatial behavioral experiences compared to ventral hippocampal subregions (dorsal CA1= 33%; ventral CA1= 13%; dorsal CA3 = 23%; ventral CA3 = 8%; and dorsal DG = 2.5%; ventral DG = 1.2%). The technique used here to obtain estimates of numbers of behavior-driven cells across the dorsal-ventral axis, however, corresponds quite well with samples from available single unit recording studies. Several explanations for the two-to-three-fold reduction in spatial behavior-driven cell activity in the ventral hippocampus can be offered. These include anatomical connectivity differences, differential gain of the self-motion signals that appear to alter the scale of place fields and the proportion of active cells, and possibly variations in the neuronal responses to non-spatial information within the hippocampus along its dorso-ventral axis.
Introduction
Early studies examining the behavioral effects of lesions to the dorsal versus ventral regions of the hippocampus suggested that these regions are functionally distinct (e.g., Kimura and Douglas, 1958; Hughes, 1965; Nadel, 1968). Studies since then have confirmed and extended these initial observations and suggest that there are both gradual and discrete transitions of functional organization across the long axis of the hippocampus (e.g., Fanselow and Dong, 2010; Small et al., 2011; Strange et al., 2014) that divide it into at least three separate domains. Lesions that are restricted to the dorsal hippocampus impair spatial memory in small environments (Moser et al., 1993; Moser et al., 1995), while ventral hippocampal lesions do not. It has been proposed that the most ventral region is more critical for emotional behavior and stress responses including contextual fear learning (Hunsaker et al., 2008; Bannerman et al., 2004).
Electrophysiological data are also consistent with the idea that the hippocampus does not function as a unitary, homogeneous structure with respect to its dorso-ventral axis (John and Killam, 1959; Hughes, 1965; Moser and Moser 1998; Strange et al., 2014). Recordings from neurons in the dorsal hippocampus have revealed that the firing of the principle cells in this structure is tuned to specific locations in the environment (O'Keefe and Dostrovsky, 1971), and this elevated activity has been called a “place field”. In the intermediate third to ventral pole region, fewer neurons are active during foraging and those that are responsive exhibit larger place fields (Jung et al., 1994; Maurer et al., 2005; Kjelstrup et al., 2008). Therefore, the ventral hippocampus may perform spatial functions similar to dorsal hippocampus but at a larger spatial scale. Consistent with the idea that the hippocampus can represent environments at multiple spatial scales, Kjelstrup et al. (2008) have shown that the relative size of place fields in area CA3 expands almost linearly from a scale of < 1 m in size near the dorsal hippocampal pole to ~ 10 m near the ventral pole.
There are also gradients in hippocampal-cortical-subcortical connectivity in rats along the dorso/ventral axis of the hippocampus (Amaral and Witter, 1989) consistent with the idea of gradated functional organization. For example, inputs from infralimbic and prelimbic cortices reach the ventral parts of the hippocampus via the ventromedial entorhinal cortex (EC), whereas, projections from the prelimbic cortex primarily influence the intermediate-dorso-ventral hippocampus via intermediate EC (Dolorfo and Amaral, 1998; Witter et al., 2000; Strien et al., 2009). The dorsal hippocampus receives visuo-spatial inputs from the dorsolateral entorhinal cortex (Swanson and Cowan, 1977), while the ventral hippocampus receives afferent projections from the ventromedial entorhinal cortex and subcortical inputs from hypothalamus and amygdala (Hargreaves et al., 2005). Therefore, cingulate areas involved in spatial processing (retrosplenial cortex) project to more dorsal regions and cingulate areas involved in emotional regulation (infralimbic and prelimbic) project primarily to ventral hippocampal regions, as do hypothalamic and amygdalar projections. Furthermore, neuromodulatory projections also show changes in density along the long axis, with stronger projections of monoamine systems to the ventral hippocampus (Strange et al., 2014).
The differences in gene transcription along the dorsal-ventral axis of the hippocampus also supports the notion of functional differentiation of this structure (Leonardo et al., 2006; Lein et al., 2007; Frisoni et al., 2008; Dong et al., 2009; Christensen et al., 2010). Using next- generation RNA sequencing it was revealed that there is a continuous gene-expression gradient resulting in prominent heterogeneity of CA1 pyramidal cells along the dorsal-ventral axis (Cembrowski et al., 2016). A handful of studies have compared gene expression differences in dorsal versus ventral hippocampal subregions, (Czerniawski et al., 2011) including in response to altered cognitive demands, which results in remapping in dorsal CA1 but not ventral CA1, (Schmidt et al., 2012) in response to fear conditioning (Czerniawski et al., 2011), in response to recent and remote spatial memory retrieval (Gusev et al., 2005) or following spatial versus non-spatial recognition memory tasks (Beer et al., 2014).
Based upon behavioral, anatomical and gene expression studies, Fanselow and Dong (2010) suggest that the hippocampus can be functionally segregated into 3 compartments along its longitudinal axis (Thompson et al., 2008; Dong et al., 2009): the dorsal region primarily performs functions associated with spatial exploration, navigation and locomotion, while the ventral compartment performs functions related to stress, emotion and affect. According to these authors, the intermediate zone has partly overlapping functions involved in translating cognitive and spatial information into motivation and survival behaviors.
The IEG Arc is rapidly induced following exploratory behaviors and has been widely used as a marker of neuronal activity (e.g., Guzowski et al., 1999; Guzowski et al., 2005; Kubik et al., 2007; Bramham et al., 2008; Miyashita et al., 2008). The proportions of cells that express Arc in the dorsal hippocampus, following spatial foraging tasks in environments of a size typically used in recording experiments (e.g., Muller and Kubie, 1987; Jeffery et al., 1997; Leutgeb et al., 2004; Rich et al., 2014; Witharana et al., 2016) is similar to the proportions of cells that express place fields. Furthermore, there are several experiments that suggest a lower proportion of active cells in the ventral hippocampus than in the dorsal region using both recording and gene expression methodology (Jung et al., 1994; Maurer et al., 2005; Kjelstrup et al., 2008).
The present study is the first to examine activity-induced gene expression in CA1, CA3 and dentate gyrus of the same rats following spatial exploratory behavior, in both the dorsal and more ventral regions of the hippocampus.
Materials and Methods
Subjects and behavioral procedures
Adult male (9–12 mo old) Fisher 344 rats (Harlan Sprague Dawley, Indianapolis, IN) were used in accordance with NIH guidelines and Animal Care and Use Committee at the University of Arizona. Animals were individually caged with free access to food and water. Rats (n=30) were assigned to one of the three groups: caged controls (n=10), spatial exploration group (n=10) and maximal electroconvulsive shock (n=10). The rats assigned to exploration group sampled the same environment twice for 5 min with either a 30, 60 or 120 min rest interval in their home cages. The rest interval time was different as these rats were also used for another experiment (assessing the time-course of Arc expression). The size of the environment was 61×61cm2 with a 30 cm high surrounding wall, the floor of the box was divided into a 3×3 grid. Each rat was moved to the center of a new square every 15 sec in a pseudorandom way to ensure adequate spatial sampling of the entire environment. Another group of rats were allowed to explore a rectangular track (6×24 inch) for 5 min, allowed a 20 min rest interval and given another 5 min exploration on the track different room (n=3). Lastly, another cohort of animals received maximal electroconvulsive shock treatment using a UGO Basile ECT unit (Via Giuseppe Di Vittorio 2–21036, Gemonio-Varee, Italy). The duration of shock was 1sec, at 85 mA current, 100Hz with a 0.5ms square wave pulse. Rats were sacrificed 30 min later after receiving MECS.
Brain extraction and dissection
Following the behavioral treatment rats were anesthetized with 5% isoflurane and decapitated with a rodent guillotine. Brains were rapidly removed, hemisected with the right hemisphere quickly frozen in isopentane cooled over an ethanol/dry ice bath and stored at –70° C until sectioning for in situ hybridization. Twenty micron thick sections were cut after blocking the hemi-brains such that all the experimental groups and negative controls (caged) and positive controls (MECS) were included on the same slide to minimize technical variability.
Fluorescence in situ hybridization
Riboprobes were generated from the full length Arc c-DNA (~ 3 K bp in length, described in Lyford et al. (1995) using a commercial RNA transcription kit (Maxiscript; Ambion, Austin, Texas) and RNA labeling nucleotide mix containing digoxigenin-tagged UTP (Roche Molecular Biochemicals, Nutley, N.J.). Fluorescence in situ hybridization was performed as described in detail by (Guzowski et al., 1999). Briefly, slides containing the sections were thawed to room temperature, fixed with freshly prepared buffered paraformaldehyde (4%) and treated with 0.5% acetic anhydride/1.5% triethanolamine. Incubated in methanol and acetone (1:1) for 5 min and equilibrated in 2× SSC. Sections were incubated with 100 µl 1× prehybridization buffer (Sigma, St. Louis, MS) for 30 min at room temperature. Approximately 100 ng of riboprobe was diluted in 1× hybridization buffer (Amersham, Piscataway, NJ), heat denatured at 90° C, chilled on ice and applied to each section. A coverslip was placed on each slide and incubated overnight at 56° C. Post hybridization washes started with 2× SSC, increased in stringency to 0.5× SSC at 56° C. RNase A (10 µg/ml) at 37° C was used to dissociate any single stranded RNA. After quenching the endogenous peroxidases with 2% H2O2, slides were blocked with NEN blocking agent (Perkin Elmer, Boston, MA) and incubated with an anti-digoxigenin Ab conjugated with HRP (Roche Molecular Biochemicals, Nutley, NJ) overnight at 4° C. Slides were washed with Tris-buffered saline containing 0.05% Tween-20 and the HRP-antibody conjugate was detected using a Cyanine-3 (CY3) tyramide signal amplification kit (Perkin Elmer, Boston, MA). Coverslips were applied to the slides after counterstaining with Sytox (Molecular Probes, Eugene, OR) with a small amount of Vectashield antifade media (Vector Labs, Burlingame, CA) and sealed with nail polish.
Confocal microscopy and cellular analysis
Stained sections were imaged using a Zeiss 510 Metaseries laser confocal microscope with a Plan Apo 40× oil immersion objective, N.A. 1.3 or a Plan Neofluar 10× objective. Laser settings, detector gain and offsets were kept constant after initial optimization for each slide. Three different subregions (CA1, CA3 and dentate gyrus) of the right hemisphere of the hippocampus from the dorsal (ranging from −2.64 mm to −3.48 relative to Bregma) and ventral (ranging from −5.64 mm to −6.12 relative to Bregma) regions were imaged (Figure 1). At these coordinates the “dorsal” region sampled corresponds to Fanselow and Dong’s (2010) “dorsal” region, and the more ventral coordinates used here to their “intermediate” region which overlaps with the ventral hippocampus as described by Bannerman (1999) and Moser et al. (1995). Areas of analysis from the CA1 or CA3 subregion were optically sectioned at ~ 0.75 µm in the z-plane. Three different CA1 and CA3 sites within each of the pyramidal cell fields were imaged in triplicate for each animal. On average, the number of dorsal CA1 and CA3 cells counted per animal was 450. The number ventral CA1 and CA3 cells counted per animal was also approximately 450 on average. Cells were counted by an experimenter blind to the conditions using Metamorph image analysis software. Image analysis was done as described earlier (Penner et al., 2011; Guzowski et al., 1999; Chawla et al., 2005) using Metamorph imaging software (Universal Imaging). Only whole neurons found in the middle 20% of the confocal stack were included in the analysis. Cells were classified as either 1) negative or 2) intranuclear Arc-positive only with one or two intense foci present in at least 3 planes.
Figure 1.
Diagrams of coronal sections taken from a rat brain atlas (Paxinos and Watson, 1998). (A) Depicts approximate regions of the dorsal (~Bregma 3.2 to 4.5) and (B) more ventral hippocampus regions (~Bregma 4.8 to 6.0) that were studied.
Because Arc expression in the dentate gyrus (DG) is so sparse, the entire region was imaged from two to three slides per animal using the 10× objective and the confocal pinhole opened wide such that all the labeled cells were visible in a single optical plane. Images were connected via a montaging procedure. Specifically, the overlapping confocal images were first collected and used for offline reconstruction using MetaMorph image analysis software to prune common areas between images as follows: Two reconstructed dentate gyrus sections/rat from the middle planes of overlapping 10× Z-stacks were used for the analysis. The area of the granule cell layer and the total number of neurons was assessed in each reconstructed flat image. Approximately, 2500 cells in the suprapyramidal blade and ~ 2000 cells in the infrapyramidal blade were analyzed in the dorsal hippocampus and ~ 2000 suprapyramidal blade cells and ~2000 infrapyramidal blade cells were analyzed in the ventral hippocampus for each rat. The area was used to estimate the total number of neurons using a correction factor that represented the total neurons per square micron. This factor was derived from 92 Z-stacks from 10 different rats collected with a 40× objective for the dorsal hippocampus. The factor for ventral hippocampus was derived from 57 stacks from 5 different animals. The total number of neurons/stack was counted and the area of the granule cell layer (in µm2) from the middle plane was calculated (Chawla et al., 2005; Ramirez-Amaya et al., 2005). Utilizing this factor, the percent of neurons with Arc mRNA in the dentate gyrus of each rat was calculated according to the following formula:
p= the number of Arc (+) neurons in a given reconstructed flat image,
Ap= the area (in µm2) of the DG, as measured from the reconstructed flat image,
N= the total number of cells from all 40× Z-stacks,
A= the total area (in µm2) of the DG from the middle planes of all 40× Z-stacks.
Statistical Analysis
The average % cells showing Arc expression in the nuclear compartment for CA1and CA3 pyramidal cells, and DG granule cells was used as the dependent measure. A two-way ANOVA was conducted on these data using behavior (exploration vs control) and region (dorsal vs ventral) as the two factors.
Results
Spatial behavior induced more Arc mRNA expression in the dorsal as compared to more ventral hippocampal CA1 and CA3 subregions
Although we analyzed Arc-positive cells using the catFISH method (Guzowski et al., 1999), the rest intervals between the first 5 min behavior and the second 5 min exposure was greater than the usual 20 min, Arc mRNA translocate to cytoplasm within 5–15 min of transcription. Therefore, Arc mRNA induced by the second exploration session was measured as transcription foci in the nucleus, rather than cytoplasmic Arc signal. First of all there was no difference in the proportion of cells that expressed Arc as a function of behavioral treatment. All rats were sacrificed 5 min after the last exploratory treatment, and only the nuclear compartment was analyzed for Arc expression in this study. Some animals, however, had an initial exposure to this treatment at 30, 60 and 120 min prior to the last exploration. There was, however, no effect of differential timing of prior exploration treatment on the numbers of cells that expressed Arc 5 min before sacrifice (dorsal CA1, 60 vs 120min [F [1, 16] = 2.10; p = 0.17]; ventral CA1 only had one earlier time point at 30 min; dorsal CA3, 60 vs 120min [F [1, 14] = 1.18; p = 0.30]; ventral CA3 only had one earlier time point at 30 min).
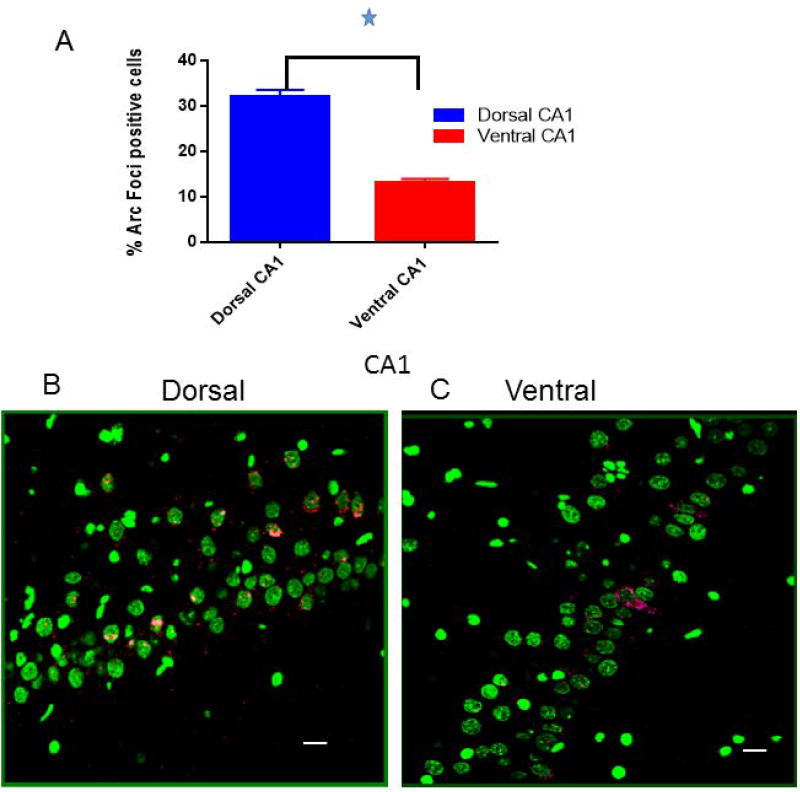
Following exploration, dorsal CA1 expressed significantly more Arc mRNA positive cells (32%+/− 1.5) compared to the ventral CA1 (13% +/− 0.9); A two-way ANOVA revealed that there was a significant main effect of behavior relative to caged controls (F [1,41] = 67.83, p < 0.0001) and a main effect of region (F [1,41] = 26.43, p < 0.0001) (Figure 2A). Similarly, dorsal CA3 had 22% +/−1.8 Arc positive cells whereas the ventral CA3 expressed 8.1% +/− 0.9 Arc mRNA positive cells. A two-way ANOVA revealed a main effect of behavior vs caged controls (F [1, 36] = 15.13; p < 0.0004) and a main effect of region, (F [1,36] = 23.44; p < 0.00041) (Figure 3A).
Figure 2.
(A) The proportion of Arc mRNA-positive cells is significantly higher in dorsal CA1 pyramidal cells (32% +/− 1.5) compared to more ventral CA1 pyramidal cells (13% +/− 0.9). (B) Representative confocal images from dorsal and (C) more ventral CA1 hippocampal subregions, respectively. Calibration bar = 20 µm. Cell nuclei are counterstained with Sytox shown here in green and Arc mRNA-positive transcription foci are in red. The insets in B and C show a magnified view of the Arc transcription foci in CA1 pyramidal cells in the dorsal and more ventral areas respectively. The white arrows correspond to the same cell in the main image and the magnified image.
Figure 3.
(A) The proportion of Arc mRNA-positive cells is significantly higher in dorsal CA3 pyramidal cells (22% +/− 1.8) compared to more ventral CA3 pyramidal cells (8.1% +/− 0.9). (B) Representative confocal images from dorsal and (C) ventral CA3 hippocampal subregions, respectively. Calibration bar = 20 µm. Cell nuclei are counterstained with Sytox shown here in green and Arc mRNA positive Foci are in red. The insets in B and C show a magnified view of the Arc transcription foci in CA3 pyramidal cells in the dorsal and more ventral areas respectively. The white arrows correspond to the same cell in the main image and the magnified image.
More granule cells express Arc mRNA in the dorsal than ventral hippocampus in response to spatial behavior
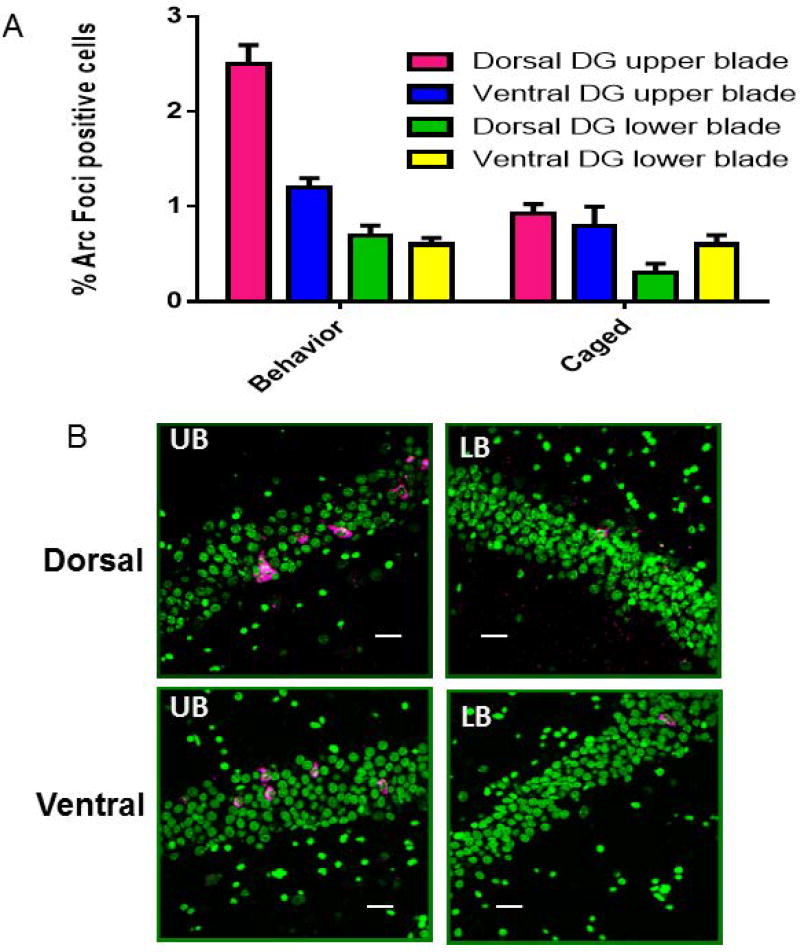
Spatial behavior resulted in a greater number of Arc positive cells in both dorsal and ventral hippocampus suprapyramidal blade (sp) compared to caged controls. There was no difference between behavior-treated animals and caged rats in the infrapyramidal blade (ip) of DG in the ventral hippocampus, Figure 4A. The percentage of granule cells that expressed Arc mRNA in the present experiment in the dorsal DG (suprapyramidal and infrapyramidal blades) is comparable to previous reports (Chawla et al., 2005; Ramírez-Amaya et al., 2005; Alme et al., 2010; Gheidi et al., 2012). A significant difference in the proportions of granule cells that express Arc mRNA in the suprapyramidal blade (2.5% +/− 0.2) was seen compared with ventral DG ub (1.2%+/− 0.13); (F [1,28] = 48.87, p < 0.0001) (Figure 4A). A significant effect of suprapyramidal vs infrapyramidal DG blade was also seen (F [1,28] = 22.04, p < 0.0001). In addition, in the dorsal region we visually observed more Arc mRNA expression in the crest of the DG, where the suprapyramidal and infrapyramidal blades meet as has been previously described (Chawla et al., 2005).
Figure 4.
(A) The proportion of Arc mRNA-positive granule cells is significantly higher in the suprapyramidal blade (SPB) of dorsal dentate gyrus following exploratory behavior (2.5% +/− 0.2) as compared to the suprapyramidal blade of the ventral dentate gyrus (1.2% +/− 0.1). (B) Confocal images taken from the dorsal and ventral suprapyramidal blades of the dentate gyrus (left panels) and infrapyramidal blades of the dentate gyrus (right panels). A significant difference was observed between behavior-treated animals and caged control rats in the suprapyramidal blade of dentate gyrus. No significant difference was observed between behavior-treated animals and caged control rats in the infrapyramidal blade (IPB) of dentate gyrus. Calibration bar = 20 µm. Arc mRNA- positive cells are shown in red and granule cell nuclei are counterstained with Sytox, shown in blue. The insets in B show a magnified view of the Arc transcription foci in granule cells in the dorsal and more ventral areas respectively. The white arrows correspond to the same cell in the main image and the magnified image.
Discussion
The main finding of the present study is that behavioral induction of Arc expression differs along the dorsal-ventral axis of the hippocampus in all 3 subregions examined - CA1, CA3 and DG. The general finding was a 2–3 fold reduction in the proportion of cells that express Arc in response to exploratory behavior in more ventral regions. This finding held within all individual animals, providing a direct comparison of behavior-driven expression. Consistent with previous reports, spatial behaviors induce robust Arc mRNA expression in dorsal hippocampal pyramidal cells compared to caged control animals (e.g., Guzowski et al., 1999; Guzowski et al., 2001; Vazdarjanova et al., 2002; Ramírez-Amaya et al., 2005; Guzowski et al., 2006; Penner et al., 2011; Hartzell et al., 2013). More ventrally located pyramidal cells did show behaviorally-induced Arc expression above that of caged control animals, but proportionally fewer cells were active compared to dorsal regions. The extent to which Arc expression differences in more ventral regions of the hippocampus arise from lower firing rates in those regions (Maurer et al., 2005), remains to be examined.
Our results are in agreement with Van et al. (2000). In this experiment performance on a 8-arm radial maze was used in combination with immunohistochemistry for cFos to examine protein levels during a spatial working memory task. In their study all hippocampal subfields showed increased cFos activation in response to task performance, with the dorsal hippocampus revealing increased cFos protein levels relative to the ventral hippocampus. Our study confirms that this difference in cFos protein also occurs at the RNA for the immediate early gene Arc along the longitudinal axis. Arc expression differences between the dorsal and most temporal subregions was also reported by Beer et al. (2014) following a spontaneous object recognition task that included both spatial and non-spatial components of recognition memory. Similar to our data, the number of cells that expressed Arc in their study was reduced in the ventral CA1 and CA3 regions compared to dorsal hippocampal subregions in the spatial version of the task. Interestingly, in the non-spatial version of the task Arc expression was similar across dorsal and ventral CA3 subregions.
Several factors may contribute to the difference observed between the dorsal and more ventral regions of the hippocampus. First, there are substantial connectivity differences along the dorsal/ventral axis. Afferents from cortical and subcortical areas differ, with the dorsal hippocampus receiving inputs from the dorso-lateral entorhinal cortex while the ventral hippocampus receives fibers from the ventro-medial entorhinal cortex along with subcortical inputs from the hypothalamus and amygdala (e.g., Hargreaves et al., 2005; McNaughton et al., 2006). Second, hippocampal efferents to the subiculum, entorhinal perirhinal, retrospenial, and cingulate cortices, may contribute to the regional dissociation along the septo-temporal axis (Swanson and Cowan, 1977; Jay and Witter, 1991). Moreover, non-spatial factors like reward and emotion may have a more powerful effect on activity in the more temporal regions of the hippocampus.
In addition, data from lesions to the dorsal or ventral hippocampus show different effects on a number of behaviors such as those in the Morris water maze task, linear track and fear conditioning (Moser et al., 1993; Hock and Bunsey, 1998; Bannerman et al., 1999; Kjelstrup et al., 2002; Jarrard et al. 2012; Nadel et al., 2013). This is consistent with the idea that detailed representation in the dorsal hippocampus is essential for precise spatial navigation while the coarse representation in the ventral hippocampus can additionally reflect non-spatial aspects of behavior. Furthermore, there is a spatial scaling gradient in the firing of CA1 pyramidal cells along the dorsal/ventral axis of the hippocampus (Jung et al., 1994; Maurer et al., 2005) similar to the gradient found in entorhinal cortex (Sargolini et al., 2006). Neurons in the more dorsal regions show smaller firing fields and greater spatial resolution, which decreases towards ventral hippocampal regions. Admittedly, neither extreme dorsal or ventral hippocampal regions have been extensively sampled in either recording studies or in behavior-driven gene expression experiments including the present study. It will be necessary to evaluate extreme septal and temporal poles in order to determine whether there is a true functionally relevant gradient along the longitudinal axis.
Additionally, analysis of the functional and structural connectivity of the hippocampus suggests a distinction between a posterior medial cortical system and an anterior temporal cortical system (Ranganath and Richey; 2012). The posterior medial system includes the parahippocampal and retrosplenial cortices and the anterior temporal system includes the perirhinal cortex. The anatomical and functional characteristics of these systems suggest that they are core components of two separate large-scale networks that support different types of memory. This organization supports functional dissociations along the longitudinal axis of the hippocampus, with unique topographical gradients across its long axis (Small et al., 2001), including differences in contributions to emotional processing (Fanselow and Dong, 2010) and scale of contextual representation (Strange et al., 2014). Furthermore, reviews of the role that the hippocampus has been shown to play hippocampal in memory (e.g., Eichenbaum et al., 2016) identify a broad range of dimensions by which the hippocampal region can map cognition. These include egocentric visual space, social context, conceptual space and non-spatial dimensions, and each of these may be differentially mapped along the long axis of the hippocampus.
The finding that there is a two to three-fold reduction in more ventral regions of the hippocampus in the proportion of pyramidal cells in CA1 and CA3 and granule cells in the dentate gyrus that express Arc in response to exploratory behavior, provides a framework to further explore possible transcriptomic and functional differences along the long axis of the hippocampus.
Acknowledgments
Grant Sponsor: Supported by the Evelyn F. McKnight Brain Institute, the National Institutes of Health AG09219 and MH060123 and NSF1631465
References
- Alme CB, Buzzetti RA, Marrone DF, Leutgeb JK, Chawla MK, Schaner MJ, Bohanick JD, Khoboko T, Leutgeb S, Moser EI, et al. Hippocampal granule cells opt for early retirement. Hippocampus. 2010;20:1109–1123. doi: 10.1002/hipo.20810. [DOI] [PubMed] [Google Scholar]
- Amaral DG, Witter MP. The three-dimensional organization of the hippocampal formation: a review of anatomical data. Neuroscience. 1989;31:571–591. doi: 10.1016/0306-4522(89)90424-7. [DOI] [PubMed] [Google Scholar]
- Bannerman DM, Rawlins JNP, McHugh SB, Deacon RMJ, Yee BK, Bast T, Zhang W-N, Pothuizen HHJ, Feldon J. Regional dissociations within the hippocampus--memory and anxiety. Neurosci Biobehav Rev. 2004;28:273–283. doi: 10.1016/j.neubiorev.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Bannerman DM, Yee BK, Good MA, Heupel MJ, Iversen SD, Rawlins JN. Double dissociation of function within the hippocampus: a comparison of dorsal, ventral, and complete hippocampal cytotoxic lesions. Behav. Neurosci. 1999;113:1170–1188. doi: 10.1037//0735-7044.113.6.1170. [DOI] [PubMed] [Google Scholar]
- Beer Z, Chwiesko C, Sauvage MM. Processing of spatial and non-spatial information reveals functional homogeneity along the dorso-ventral axis of CA3, but not CA1. Neurobiol Learn Mem. 2014;111:56–64. doi: 10.1016/j.nlm.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Cembrowski MS, Bachman JL, Wang L, Sugino K, Shields BC, Spruston N. Spatial Gene-Expression Gradients Underlie Prominent Heterogeneity of CA1 Pyramidal Neurons. Neuron. 2016;89:351–368. doi: 10.1016/j.neuron.2015.12.013. [DOI] [PubMed] [Google Scholar]
- Chawla MK, Guzowski JF, Ramirez-Amaya V, Lipa P, Hoffman KL, Marriott LK, Worley PF, McNaughton BL, Barnes CA. Sparse, environmentally selective expression of Arc RNA in the upper blade of the rodent fascia dentata by brief spatial experience. Hippocampus. 2005;15:579–586. doi: 10.1002/hipo.20091. [DOI] [PubMed] [Google Scholar]
- Christensen T, Bisgaard CF, Nielsen HB, Wiborg O. Transcriptome differentiation along the dorso-ventral axis in laser-captured microdissected rat hippocampal granular cell layer. Neuroscience. 2010;170:731–741. doi: 10.1016/j.neuroscience.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Czerniawski J, Ree F, Chia C, Ramamoorthi K, Kumata Y, Otto TA. The importance of having Arc: expression of the immediate-early gene Arc is required for hippocampus-dependent fear conditioning and blocked by NMDA receptor antagonism. J. Neurosci. 2011;31:11200–11207. doi: 10.1523/JNEUROSCI.2211-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolorfo CL, Amaral DG. Entorhinal cortex of the rat: topographic organization of the cells of origin of the perforant path projection to the dentate gyrus. J. Comp. Neurol. 1998;398:25–48. [PubMed] [Google Scholar]
- Dong HW, Swanson LW, Chen L, Fanselow MS, Toga AW. Genomic-anatomic evidence for distinct functional domains in hippocampal field CA1. Proc. Natl. Acad. Sci. U.S.A. 2009;106:11794–11799. doi: 10.1073/pnas.0812608106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas KD. Effects of selective hippocampal damage on avoidance behaviour in the rat. Can J Psychol Can Psychol. 1958;12:213–218. doi: 10.1037/h0083740. [DOI] [PubMed] [Google Scholar]
- Eichenbaum H, Amaral DG, Buffalo EA, Buzsáki G, Cohen N, Davachi L, Frank L, Heckers S, Morris RGM, Moser EI, et al. Hippocampus at 25. Hippocampus. 2016;26:1238–1249. doi: 10.1002/hipo.22616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanselow MS, Dong H-W. Are the dorsal and ventral hippocampus functionally distinct structures? Neuron. 2010;65:7–19. doi: 10.1016/j.neuron.2009.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisoni GB, Ganzola R, Canu E, Rüb U, Pizzini FB, Alessandrini F, Zoccatelli G, Beltramello A, Caltagirone C, Thompson PM. Mapping local hippocampal changes in Alzheimer’s disease and normal ageing with MRI at 3 Tesla. Brain. 2008;131:3266–3276. doi: 10.1093/brain/awn280. [DOI] [PubMed] [Google Scholar]
- Gheidi A, Satvat E, Marrone DF. Experience-dependent recruitment of Arc expression in multiple systems during rest. J. Neurosci. Res. 2012;90:1820–1829. doi: 10.1002/jnr.23057. [DOI] [PubMed] [Google Scholar]
- Gusev PA, Cui C, Alkon DL, Gubin AN. Topography of Arc/Arg3.1 mRNA expression in the dorsal and ventral hippocampus induced by recent and remote spatial memory recall: dissociation of CA3 and CA1 activation. J. Neurosci. 2005;25:9384–9397. doi: 10.1523/JNEUROSCI.0832-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat. Neurosci. 1999;2:1120–1124. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Timlin JA, Roysam B, McNaughton BL, Worley PF, Barnes CA. Mapping behaviorally relevant neural circuits with immediate-early gene expression. Curr. Opin. Neurobiol. 2005;15:599–606. doi: 10.1016/j.conb.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Miyashita T, Chawla MK, Sanderson J, Maes LI, Houston FP, Lipa P, McNaughton BL, Worley PF, Barnes CA. Recent behavioral history modifies coupling between cell activity and Arc gene transcription in hippocampal CA1 neurons. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1077–1082. doi: 10.1073/pnas.0505519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, Setlow B, Wagner EK, McGaugh JL. Experience-dependent gene expression in the rat hippocampus after spatial learning: a comparison of the immediate-early genes Arc, c-fos, and zif268. J. Neurosci. 2001;21:5089–5098. doi: 10.1523/JNEUROSCI.21-14-05089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves EL, Rao G, Lee I, Knierim JJ. Major dissociation between medial and lateral entorhinal input to dorsal hippocampus. Science. 2005;308:1792–1794. doi: 10.1126/science.1110449. [DOI] [PubMed] [Google Scholar]
- Hartzell AL, Burke SN, Hoang LT, Lister JP, Rodriguez CN, Barnes CA. Transcription of the immediate-early gene Arc in CA1 of the hippocampus reveals activity differences along the proximodistal axis that are attenuated by advanced age. J. Neurosci. 2013;33:3424–3433. doi: 10.1523/JNEUROSCI.4727-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock BJ, Bunsey MD. Differential effects of dorsal and ventral hippocampal lesions. J. Neurosci. 1998;18:7027–7032. doi: 10.1523/JNEUROSCI.18-17-07027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes KR. Dorsal and ventral hippocampus lesions and maze learning: influence of preoperative environment. Can J Psychol. 1965;19:325–332. doi: 10.1037/h0082915. [DOI] [PubMed] [Google Scholar]
- Hunsaker MR, Fieldsted PM, Rosenberg JS, Kesner RP. Dissociating the roles of dorsal and ventral CA1 for the temporal processing of spatial locations, visual objects, and odors. Behav. Neurosci. 2008;122:643–650. doi: 10.1037/0735-7044.122.3.643. [DOI] [PubMed] [Google Scholar]
- Jarrard LE, Luu LP, Davidson TL. A study of hippocampal structure-function relations along the septo-temporal axis. Hippocampus. 2012;22:680–692. doi: 10.1002/hipo.20928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay TM, Witter MP. Distribution of hippocampal CA1 and subicular efferents in the prefrontal cortex of the rat studied by means of anterograde transport of Phaseolus vulgaris-leucoagglutinin. J. Comp. Neurol. 1991;313:574–586. doi: 10.1002/cne.903130404. [DOI] [PubMed] [Google Scholar]
- Jeffer KJ, Donnet JG, Burgess N, Okeefe JM, Jeffery KJ, Donnet JG, Burges N, Okeefe JM. Directional control of hippocampal place fields. Exp Brain Res. 1997;117:131–142. doi: 10.1007/s002210050206. [DOI] [PubMed] [Google Scholar]
- John ER, Killam KF. Electrophysiological correlates of avoidance conditioning in the cat. J. Pharmacol. Exp. Ther. 1959;125:252–274. [PubMed] [Google Scholar]
- Jung MW, Wiener SI, McNaughton BL. Comparison of spatial firing characteristics of units in dorsal and ventral hippocampus of the rat. J. Neurosci. 1994;14:7347–7356. doi: 10.1523/JNEUROSCI.14-12-07347.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjelstrup KB, Solstad T, Brun VH, Hafting T, Leutgeb S, Witter MP, Moser EI, Moser M-B. Finite scale of spatial representation in the hippocampus. Science. 2008;321:140–143. doi: 10.1126/science.1157086. [DOI] [PubMed] [Google Scholar]
- Kjelstrup KG, Tuvnes FA, Steffenach H-A, Murison R, Moser EI, Moser M-B. Reduced fear expression after lesions of the ventral hippocampus. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10825–10830. doi: 10.1073/pnas.152112399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Leonardo ED, Richardson-Jones JW, Sibille E, Kottman A, Hen R. Molecular heterogeneity along the dorsal-ventral axis of the murine hippocampal CA1 field: a microarray analysis of gene expression. Neuroscience. 2006;137:177–186. doi: 10.1016/j.neuroscience.2005.08.082. [DOI] [PubMed] [Google Scholar]
- Leutgeb S, Leutgeb JK, Treves A, Moser M-B, Moser EI. Distinct ensemble codes in hippocampal areas CA3 and CA1. Science. 2004;305:1295–1298. doi: 10.1126/science.1100265. [DOI] [PubMed] [Google Scholar]
- Maurer AP, Vanrhoads SR, Sutherland GR, Lipa P, McNaughton BL. Self-motion and the origin of differential spatial scaling along the septo-temporal axis of the hippocampus. Hippocampus. 2005;15:841–852. doi: 10.1002/hipo.20114. [DOI] [PubMed] [Google Scholar]
- McNaughton BL, Battaglia FP, Jensen O, Moser EI, Moser M-B. Path integration and the neural basis of the “cognitive map”. Nat. Rev. Neurosci. 2006;7:663–678. doi: 10.1038/nrn1932. [DOI] [PubMed] [Google Scholar]
- Moser E, Moser MB, Andersen P. Spatial learning impairment parallels the magnitude of dorsal hippocampal lesions, but is hardly present following ventral lesions. J. Neurosci. 1993;13:3916–3925. doi: 10.1523/JNEUROSCI.13-09-03916.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser MB, Moser EI, Forrest E, Andersen P, Morris RG. Spatial learning with a minislab in the dorsal hippocampus. Proc. Natl. Acad. Sci. U.S.A. 1995;92:9697–9701. doi: 10.1073/pnas.92.21.9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M-B, Moser EI. Functional differentiation in the hippocampus. Hippocampus. 1998;8:608–619. doi: 10.1002/(SICI)1098-1063(1998)8:6<608::AID-HIPO3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Muller RU, Kubie JL. The effects of changes in the environment on the spatial firing of hippocampal complex-spike cells. J. Neurosci. 1987;7:1951–1968. doi: 10.1523/JNEUROSCI.07-07-01951.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadel L. Dorsal and ventral hippocampal lesions and behavior. Physiol Behav. 1968;3:891–900. [Google Scholar]
- Nadel L, Hoscheidt S, Ryan LR. Spatial cognition and the hippocampus: the anterior-posterior axis. J Cogn Neurosci. 2013;25:22–8. doi: 10.1162/jocn_a_00313. [DOI] [PubMed] [Google Scholar]
- O’Keefe J, Dostrovsky J. The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res. 1971;34:171–175. doi: 10.1016/0006-8993(71)90358-1. [DOI] [PubMed] [Google Scholar]
- Penner MR, Roth TL, Chawla MK, Hoang LT, Roth ED, Lubin FD, Sweatt JD, Worley PF, Barnes CA. Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiol. Aging. 2011;32:2198–2210. doi: 10.1016/j.neurobiolaging.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Amaya V, Vazdarjanova A, Mikhael D, Rosi S, Worley PF, Barnes CA. Spatial exploration-induced Arc mRNA and protein expression: evidence for selective, network-specific reactivation. J. Neurosci. 2005;25:1761–1768. doi: 10.1523/JNEUROSCI.4342-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganath C, Ritchey M. Two cortical systems for memory-guided behaviour. Nat. Rev. Neurosci. 2012;13:713–726. doi: 10.1038/nrn3338. [DOI] [PubMed] [Google Scholar]
- Rich PD, Liaw H-P, Lee AK. Large environments reveal the statistical structure governing hippocampal representations. Science. 2014;345:814–817. doi: 10.1126/science.1255635. [DOI] [PubMed] [Google Scholar]
- Sargolini F, Fyhn M, Hafting T, McNaughton BL, Witter MP, Moser M-B, Moser EI. Conjunctive representation of position, direction, and velocity in entorhinal cortex. Science. 2006;312:758–762. doi: 10.1126/science.1125572. [DOI] [PubMed] [Google Scholar]
- Schmidt B, Satvat E, Argraves M, Markus EJ, Marrone DF. Cognitive demands induce selective hippocampal reorganization: Arc expression in a place and response task. Hippocampus. 2012;22:2114–2126. doi: 10.1002/hipo.22031. [DOI] [PubMed] [Google Scholar]
- Small SA, Nava AS, Perera GM, DeLaPaz R, Mayeux R, Stern Y. Circuit mechanisms underlying memory encoding and retrieval in the long axis of the hippocampal formation. Nat. Neurosci. 2001;4:442–449. doi: 10.1038/86115. [DOI] [PubMed] [Google Scholar]
- Small SA, Schobel SA, Buxton RB, Witter MP, Barnes CA. A pathophysiological framework of hippocampal dysfunction in ageing and disease. Nat. Rev. Neurosci. 2011;12:585–601. doi: 10.1038/nrn3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange BA, Witter MP, Lein ES, Moser EI. Functional organization of the hippocampal longitudinal axis. Nat. Rev. Neurosci. 2014;15:655–669. doi: 10.1038/nrn3785. [DOI] [PubMed] [Google Scholar]
- Swanson LW, Cowan WM. An autoradiographic study of the organization of the efferent connections of the hippocampal formation in the rat. J. Comp. Neurol. 1977;172:49–84. doi: 10.1002/cne.901720104. [DOI] [PubMed] [Google Scholar]
- Thompson CL, Pathak SD, Jeromin A, Ng LL, MacPherson CR, Mortrud MT, Cusick A, Riley ZL, Sunkin SM, Bernard A, et al. Genomic anatomy of the hippocampus. Neuron. 2008;60:1010–1021. doi: 10.1016/j.neuron.2008.12.008. [DOI] [PubMed] [Google Scholar]
- van Strien NM, Cappaert NLM, Witter MP. The anatomy of memory: an interactive overview of the parahippocampal-hippocampal network. Nat. Rev. Neurosci. 2009;10:272–282. doi: 10.1038/nrn2614. [DOI] [PubMed] [Google Scholar]
- Vann SD, Brown MW, Erichsen JT, Aggleton JP. Fos imaging reveals differential patterns of hippocampal and parahippocampal subfield activation in rats in response to different spatial memory tests. J. Neurosci. 2000;20:2711–2718. doi: 10.1523/JNEUROSCI.20-07-02711.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazdarjanova A, McNaughton BL, Barnes CA, Worley PF, Guzowski JF. Experience-dependent coincident expression of the effector immediate-early genes arc and Homer 1a in hippocampal and neocortical neuronal networks. J. Neurosci. 2002;22:10067–10071. doi: 10.1523/JNEUROSCI.22-23-10067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witharana WKL, Cardiff J, Chawla MK, Xie JY, Alme CB, Eckert M, Lapointe V, Demchuk A, Maurer AP, Trivedi V, et al. Nonuniform allocation of hippocampal neurons to place fields across all hippocampal subfields. Hippocampus. 2016;26:1328–1344. doi: 10.1002/hipo.22609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witter MP, Wouterlood FG, Naber PA, Van Haeften T. Anatomical organization of the parahippocampal-hippocampal network. Ann. N. Y. Acad. Sci. 2000;911:1–24. doi: 10.1111/j.1749-6632.2000.tb06716.x. [DOI] [PubMed] [Google Scholar]