Abstract
Neurofibromatosis type 1 (NF1), a common genetic disorder with a birth incidence of 1:2,000–3,000, is characterized by a highly variable clinical presentation. To date, only two clinically relevant intragenic genotype-phenotype correlations have been reported for NF1 missense mutations affecting p.Arg1809 and a single amino acid deletion p.Met922del. Both variants predispose to a distinct mild NF1 phenotype with neither externally visible cutaneous/plexiform neurofibromas nor other tumors. Here, we report 162 individuals (129 unrelated probands and 33 affected relatives) heterozygous for a constitutional missense mutation affecting one of five neighboring NF1 codons—Leu844, Cys845, Ala846, Leu847, and Gly848—located in the cysteine-serine-rich domain (CSRD). Collectively, these recurrent missense mutations affect ∼0.8% of unrelated NF1 mutation-positive probands in the University of Alabama at Birmingham (UAB) cohort. Major superficial plexiform neurofibromas and symptomatic spinal neurofibromas were more prevalent in these individuals compared with classic NF1-affected cohorts (both p < 0.0001). Nearly half of the individuals had symptomatic or asymptomatic optic pathway gliomas and/or skeletal abnormalities. Additionally, variants in this region seem to confer a high predisposition to develop malignancies compared with the general NF1-affected population (p = 0.0061). Our results demonstrate that these NF1 missense mutations, although located outside the GAP-related domain, may be an important risk factor for a severe presentation. A genotype-phenotype correlation at the NF1 region 844–848 exists and will be valuable in the management and genetic counseling of a significant number of individuals.
Keywords: neurofibromatosis type 1, NF1, genotype-phenotype correlation, missense mutation, codons 844–848, spinal NF, MPNST, plexiform neurofibroma, CSRD
Introduction
Neurofibromatosis type 1 (NF1 [MIM: 162200]), one of the most common genetic disorders with a birth incidence of 1 in 2,000–3,000,1, 2, 3 is characterized by a highly variable inter- and intrafamilial expressivity (see GeneReviews in Web Resources). It is caused by loss-of-function genetic variants in NF1 (MIM: 613113), located on chromosome 17q11.2. NF1 encodes neurofibromin, a GTPase activating protein (GAP) that downregulates the RAS signal transduction pathway through its GAP-related domain (GRD).4, 5 The most common first signs of NF1 are multiple café-au-lait macules (CALMs) in >95% of infants and skinfold freckling in >80% of children by the age of 7 years.6 Other clinical features observed in >90% of adults with NF1 are iris Lisch nodules and cutaneous neurofibromas.7 Individuals with a more severe phenotype present with plexiform and/or spinal neurofibromas, symptomatic optic pathway gliomas (OPGs), as well as specific osseous lesions, such as sphenoid wing or tibial dysplasia. Approximately 50% of NF1-affected case subjects have de novo mutations, while the remaining individuals inherit the disorder from an affected parent (see GeneReviews in Web Resources). According to the National Institutes of Health (NIH) diagnostic criteria, at least two of the aforementioned features are required to classify a person as having the clinical diagnosis of NF1.8
Due to the variability in clinical presentation, age dependency of most manifestations, the timing and number of second hits in specific cells, and the wide NF1 allelic heterogeneity, identification of specific genotype-phenotype correlations is extremely challenging. To date, more than 2,800 different germline NF1 pathogenic variants have been identified in the University of Alabama at Birmingham (UAB) cohort, with only 31 unique pathogenic variants present in ≥0.5% of all unrelated individuals (L.M.M., unpublished data). Moreover, a mild NF1 phenotype, including only CALMs and skinfold freckles, overlaps with Legius syndrome (MIM: 611431), caused by mutations in SPRED1 (MIM: 609291).9, 10
So far, only three clinically significant genotype-phenotype correlations have been reported. First, individuals with a constitutional NF1 microdeletion usually show a more severe phenotype compared to the general NF1-affected population. The NF1 microdeletion syndrome (MIM: 613675) is typically characterized by a large number of neurofibromas at a young age, dysmorphic facial features (hypertelorism, downslanted palpebral fissures, broad nasal bridge, low-set ears, micrognathia, coarse face, facial asymmetry), and developmental delay and/or intellectual disability. Individuals may present with cardiac defects as well as growth and skeletal abnormalities. NF1 microdeletions have been associated with an increased lifetime risk for malignant peripheral nerve sheath tumors (MPNSTs). The constitutional co-deletion of SUZ12 (MIM: 606245) within the common NF1-microdeletion region is thought to be a risk factor for the malignant neoplasms.11 Second, individuals with a specific single amino acid NF1 deletion (c.2970_2972del [p.Met992del]) present with a milder phenotype. These individuals have multiple CALMs with or without freckles, but no externally visible cutaneous or plexiform neurofibromas.12 A third genotype-phenotype correlation involving NF1 missense mutations affecting arginine at position 1809 is also associated with a distinct presentation,13, 14 including developmental delay and/or learning disabilities, pulmonic stenosis, and Noonan-like features, but no external plexiform neurofibromas or symptomatic OPGs. Both of these affected amino acids reside outside the GRD domain.
Another distinct form of NF1 is familial spinal neurofibromatosis (FSNF [MIM: 162210]) originally described by Pulst et al.15 in six affected members from two unrelated families. It is characterized by bilateral and histologically proven neurofibromas of all spinal dorsal roots with a paucity or absolute lack of cutaneous manifestations.16, 17 So far, only ∼100 individuals (both familial and sporadic) have been reported with this form.17 It has been suggested that individuals with the severe subtype of FSNF more frequently carry an NF1 missense or splicing mutation.18, 19, 20 Of particular interest are two families: a two-generation family with three first-degree relatives reported by Pascual-Castroviejo et al.21 and a three-generation family with three first-degree relatives reported by Burkitt-Wright et al.16 Specific NF1 missense mutations c.2542G>C (p.Gly848Arg) and c.2543G>A (p.Gly848Glu), located in the cysteine-serine-rich domain (CSRD), were present in all individuals affected by multiple spinal dorsal root neurofibromas. Despite the evidence that c.2542G>C (p.Gly848Arg) is a clearly pathogenic mutation, two recent studies using mouse models did not recapitulate the phenotype identified in humans.22, 23 Genetically engineered mice with c.2542G>C (p.Gly848Arg) mutation developed neither OPGs nor plexiform neurofibromas, demonstrating phenotypic divergence between NF1-affected individuals and mice.22, 23
In this study, we report a cohort of 129 unrelated probands and 33 affected relatives heterozygous for a constitutional missense mutation affecting one of five neighboring NF1 codons—Leu844, Cys845, Ala846, Leu847, and Gly848. These individuals have a high prevalence of a severe phenotype, including plexiform and symptomatic spinal neurofibromas, symptomatic optic pathway gliomas, other malignant neoplasms, and bone abnormalities. The current findings clearly demonstrate that missense mutations outside the GRD are not solely associated with a mild phenotype.
Material and Methods
Individuals and Phenotypic Data
A total of 162 individuals heterozygous for a missense mutation affecting one of five neighboring NF1 codons (Leu844, Cys845, Ala846, Leu847, and Gly848) were included in the study. Blood samples from 78 individuals (67 probands and 11 relatives) were originally sent to the UAB Medical Genomics Laboratory for molecular NF1 genetic testing to establish or confirm the diagnosis for NF1. This initial study was expanded to include an additional 84 individuals (62 probands and 22 relatives), molecularly diagnosed in collaborating institutions (as detailed in Table S1).
All individuals included in this study were clinically assessed using the standardized phenotypic checklist form as previously reported (Figure S1).14 The clinical data were collected at the time of mutation analysis and re-verified for accuracy by referring physicians co-authoring this paper at the time of this study. Additionally, referring physicians updated the phenotypic data at the time of this genotype-phenotype study, when available, i.e., when the individual had been seen and followed at their institution after genetic testing results were reported. The phenotypic data and age provided correspond to the latest clinical evaluation. The phenotypic checklist form consists of two parts: (1) general information including gender, date of birth, ethnicity, height, head circumference (HC), weight, fulfillment of the NIH diagnostic criteria, and mode of inheritance and (2) NF1 signs and symptoms, including CALMs, skinfold freckling, Lisch nodules, cutaneous and subcutaneous, plexiform and spinal neurofibromas, OPGs and other neoplasms, skeletal and cardiac abnormalities, development and education levels, presence/absence of Noonan syndrome features, and segmental phenotype.
Fifteen major clinical features of NF1 were selected for the genotype-phenotype correlation study (Tables 1, 2, and 3). Individuals with missing data for a particular sign and/or symptom were classified as “unknown” or “not specified” and consequently excluded from that part of the genotype-phenotype analysis. Most features were identified by physical examination; ophthalmologic examination for Lisch nodules and imaging to detect asymptomatic OPGs and spinal neurofibromas was not performed in most individuals. Brain and spine/whole-body MRI was done mainly in individuals with signs and/or symptoms indicative of OPGs or internal/spinal neurofibromas; however, depending on institutional policies, some individuals were screened by MRI despite the absence of symptoms. Noonan phenotype was diagnosed if at least two of the following features were observed: short stature, hypertelorism, low-set ears, webbed neck, ptosis, midface hypoplasia, or pulmonic stenosis. To evaluate short stature and macrocephaly, the World Health Organization (WHO) and the Center for Disease Control (CDC) growth charts and the Gerhard Nellhaus’ curve24 were used as previously described.14 Short stature and macrocephaly were defined as height below or equal to the 3rd percentile (PC ≤ 3) and as head circumference equal or above the 98th percentile (PC ≥ 98), respectively. For cognitive impairment/learning disabilities, individuals with attention deficit disorder (ADD) and/or attention deficit hyperactivity disorder (ADHD) but normal development were classified as normal.
Table 1.
Demographic and Clinical Characterization of Individuals with a Missense Mutation Affecting Codons 844–848
| Mutation [Proband:Relative] |
Codon 844 |
Codon 845 |
Codon 846 |
Codon 847 |
Codon 848 |
All Codons 844–848 | Total | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| c.2530C>T (p.Leu844Phe) [10:1]; c.2531T>A (p.Leu844His) [2:0]; c.2531T>C (p.Leu844Pro) [7:0]; c.2531T>G (p.Leu844Arg) [6:0] | c.2533T>C (p.Cys845Arg) [3:1]; c.2534G>A (p.Cys845Tyr) [8:0] | c.2536G>C (p.Ala846Pro) [1:2]; c.2537C>A (p.Ala846Asp) [5:2] | c.2540T>C (p.Leu847Pro) [58:12]; c.2540T>G (p.Leu847Arg) [8:0] | c.2542G>A (p.Gly848Arg) [6:0]; c.2542G>C (p.Gly848Arg) [8:11]; c.2543G>A (p.Gly848Glu) [7:4] | |||||||||||||||
| Mutation-positive individuals [Proband:Relative] | 26 [25:1] | 12 [11:1] | 10 [6:4] | 78 [66:12] | 36 [21:15] | 162 [129:33] | |||||||||||||
| Age group, years | ≤8 | 9–18 | ≥19 | ≤8 | 9–18 | ≥19 | ≤8 | 9–18 | ≥19 | ≤8 | 9–18 | ≥19 | ≤8 | 9–18 | ≥19 | ≤8 | 9–18 | ≥19 | all ages |
| Total | 12 | 5 | 9 | 4 | 2 | 6 | 3 | 1 | 6 | 28 | 14 | 36 | 13 | 5 | 18 | 60 | 27 | 75 | 162 |
| Proband:Relative | 12:0 | 5:0 | 8:1 | 4:0 | 2:0 | 5:1 | 2:1 | 1:0 | 3:3 | 27:1 | 12:2 | 27:9 | 6:7 | 4:1 | 11:7 | 51:9 | 24:3 | 54:21 | 129:33 |
| Age range, years | 1–8 | 9–16 | 24–55 | 1–2 | 15–16 | 19–48 | 4–5 | 18 | 33–69 | 1–8 | 9–18 | 19–72 | 1–7 | 10–17 | 19–74 | 1–8 | 9–18 | 19–74 | 1–74 |
| Male: Female | 6:6 | 4:1 | 1:8 | 1:3 | 1:1 | 1:5 | 2:1 | 0:1 | 1:5 | 10:18 | 5:9 | 19:17 | 9:4 | 2:3 | 5:13 | 28:32 | 12:15 | 27:48 | 67:95 |
| Fulfilling the NIH criteria if the family history is taken into account | 10/11 | 4/5 | 9/9 | 2/4 | 1/2 | 4/5 | 3/3 | 1/1 | 6/6 | 17/28 | 14/14 | 35/36 | 4/11 | 4/5 | 17/18 | 36/57 | 24/27 | 71/74 | 131/158 |
| Fulfilling the NIH criteria if solely taking the physical signs into account | 10/11 | 4/5 | 9/9 | 2/4 | 1/2 | 4/5 | 2/3 | 1/1 | 6/6 | 17/28 | 14/14 | 33/36 | 4/11 | 4/5 | 13/18 | 35/57 | 24/27 | 65/74 | 124/158 |
| >5 CALMs | 12/12 | 5/5 | 8/8 | 4/4 | 1/2 | 4/5 | 3/3 | 1/1 | 4/6 | 27/28 | 14/14 | 32/35 | 5/11 | 3/5 | 7/18 | 51/58 | 24/27 | 55/72 | 130/157 |
| Freckling | 10/10 | 4/5 | 6/7 | 0/4 | 1/2 | 4/5 | 2/2 | 1/1 | 5/5 | 12/23 | 13/13 | 31/34 | 4/10 | 3/5 | 8/18 | 28/49 | 22/26 | 54/69 | 104/144 |
| Lisch nodules | 2/9 | 1/4 | 4/4 | 0/1 | 0/0 | 1/2 | 0/1 | 0/1 | 2/2 | 4/19 | 3/9 | 17/19 | 2/8 | 0/5 | 6/14 | 8/38 | 4/19 | 30/41 | 42/98 |
| Skeletal abnormalitiesa | 2/11 | 2/5 | 5/9 | 2/4 | 1/2 | 2/4 | 0/2 | 0/1 | 0/5 | 3/25 | 3/14 | 17/28 | 3/11 | 3/5 | 5/18 | 10/53 | 9/27 | 29/64 | 48/144 |
| Plexiform neurofibromas | 0/11 | 2/5 | 3/9 | 0/3 | 2/2 | 2/5 | 0/2 | 1/1 | 1/2 | 6/24 | 3/13 | 19/33 | 0/11 | 1/5 | 7/17 | 6/51 | 9/26 | 32/66 | 47/143 |
| Cutaneous neurofibromasb | 1/11 | 1/5 | 7/9 | 0/4 | 0/2 | 3/4 | 0/2 | 1/1 | 4/5 | 1/26 | 4/14 | 28/33 | 1/11 | 1/5 | 5/18 | 3/54 | 7/27 | 47/69 | 57/150 |
| Subcutaneous neurofibromasb | 1/9 | 0/5 | 6/8 | 1/4 | 0/2 | 1/4 | 0/2 | 0/0 | 3/5 | 1/26 | 4/13 | 17/30 | 1/11 | 0/5 | 6/18 | 4/52 | 4/25 | 33/65 | 41/142 |
| Cutaneous and subcutaneousb | 0/9 | 0/5 | 5/8 | 0/4 | 0/2 | 1/3 | 0/2 | 0/0 | 3/5 | 0/25 | 1/13 | 17/30 | 0/11 | 0/5 | 4/18 | 0/51 | 1/25 | 30/64 | 31/140 |
| Symptomatic spinal neurofibromas | 0/10 | 0/3 | 0/8 | 0/2 | 1/2 | 0/4 | 0/2 | 0/0 | 0/2 | 1/23 | 1/13 | 2/27 | 0/11 | 1/4 | 7/16 | 1/48 | 3/22 | 9/57 | 13/127 |
| Spinal neurofibromas by MRIc | 0/1 | 0/0 | 0/5 | 0/0 | 1/2 | 1/1 | 0/1 | 0/0 | 0/1 | 1/5 | 2/6 | 3/16 | 0/1 | 2/3 | 10/11 | 1/8 | 5/11 | 14/34 | 20/53 |
| Symptomatic OPGsd | 1/11 | 1/5 | 0/9 | 0/3 | 0/2 | 0/5 | 1/3 | 1/1 | 0/3 | 2/25 | 1/13 | 2/27 | 1/11 | 1/5 | 1/13 | 5/53 | 4/26 | 3/57 | 12/136 |
| Asymptomatic OPGse | 2/6 | 1/2 | 2/4 | 0/1 | 0/2 | 0/2 | 0/1 | 0/0 | 0/3 | 1/8 | 6/9 | 4/13 | 1/4 | 0/2 | 1/6 | 4/20 | 7/15 | 7/28 | 18/63 |
| Other neoplasmsf | 1/11 | 0/4 | 1/8 | 0/2 | 0/1 | 0/4 | 0/2 | 0/1 | 0/3 | 1/24 | 3/14 | 11/34 | 2/11 | 1/5 | 1/15 | 4/50 | 4/25 | 13/64 | 21/139 |
| Cognitive impairment and/or learning disabilities | 3/11 | 3/4 | 0/6 | 1/4 | 0/2 | 3/4 | 3/3 | 0/1 | 1/5 | 10/26 | 7/13 | 12/26 | 5/11 | 5/5 | 3/17 | 22/55 | 15/25 | 19/58 | 56/138 |
| Noonan syndrome features | 0/9 | 1/5 | 1/8 | 0/2 | 1/1 | 0/4 | 0/2 | 0/1 | 0/4 | 3/27 | 0/13 | 3/26 | 1/10 | 0/5 | 0/17 | 4/50 | 2/25 | 4/59 | 10/134 |
| Short statureg | 1/7 | 0/2 | 0/4 | 0/3 | 1/1 | 0/1 | 0/2 | 0/0 | 1/2 | 0/11 | 3/10 | 4/21 | 3/10 | 0/3 | 2/14 | 4/33 | 4/16 | 7/42 | 15/91 |
| Macrocephaly | 2/11 | 1/4 | 1/2 | 1/3 | 0/1 | 0/0 | 2/2 | 0/0 | 0/2 | 8/21 | 2/11 | 10/17 | 3/11 | 1/4 | 5/9 | 16/48 | 4/20 | 16/30 | 36/98 |
| Pulmonic stenosis | 0/8 | 1/5 | 0/6 | 0/2 | 0/2 | 1/1 | 0/3 | 0/0 | 0/5 | 0/23 | 0/13 | 0/20 | 0/8 | 0/3 | 0/14 | 0/44 | 1/23 | 1/46 | 2/113 |
All bone abnormalities included, i.e., scoliosis (n = 27), pectus excavatum (n = 4), pectus carinatum (n = 6), long bone dysplasia (n = 4), pseudarthrosis (n = 2), bone cysts (n = 2), sphenoid wing dysplasia (n = 2), ulnar aplasia, dural ectasia, 4th lumbar vertebrae fragmentation, bowed long bones, tibial dysplasia, clinodactyly, postaxial polydactyly, and cherubism.
At least two cutaneous/subcutaneous neurofibromas were required to be considered as “positive for the criterion of neurofibromas.”
The frequency of both symptomatic and asymptomatic spinal neurofibromas in individuals who had done MRI examination.
The presence or absence of symptomatic OPGs was determined by ophthalmological examination and confirmed by MRI.
Including only individuals without signs of symptomatic OPGs who underwent MRI examination.
Including benign and malignant neoplasms, except for OPGs and neurofibromas.
As no specific growth curves are available for the Hispanic and Asian populations, Hispanic and Asian individuals were excluded as having short or normal stature.
Table 2.
Frequency of Clinical Features in Cohorts of Individuals with a Missense Mutation Affecting Leu844, Cys845, Ala846, Leu847, and Gly848
| NF1 Feature |
Number of Individuals (%) [95% Confidence Interval] |
||||
|---|---|---|---|---|---|
| Leu844 | Cys845 | Ala846 | Leu847 | Gly848 | |
| >5 CALMs | 25/25 (100) [86.7–100] | 9/11 (81.8) [52.3–94.9] | 8/10 (80) [49–94.3] | 73/77 (94.8) [87.4–98] | 15/34 (44.1) [28.9–60.6] |
| Skinfold frecklinga | 10/12 (83.3) [55.2–95.3] | 5/7 (71.4) [35.9–91.8] | 6/6 (100) [61–100] | 44/47 (93.6) [82.8–97.8] | 11/23 (47.8) [29.2–67] |
| Lisch nodules | 7/17 (41.2) [21.6–64] | 1/3 (33.3) [6.2–79.2] | 2/4 (50) [15–85] | 24/47 (51.1) [37.2–64.7] | 8/27 (29.6) [15.9–48.5] |
| Plexiform neurofibromasa | 5/14 (35.7) [16.3–61.2] | 4/7 (57.1) [25–84.2] | 2/3 (66.7) [20.8–93.9] | 22/46 (47.8) [34.1–61.9] | 8/22 (36.4) [19.7–57] |
| Cutaneous neurofibromasb | 7/9 (77.8) [45.3–93.7] | 3/4 (75) [30.1–95.4] | 4/5 (80) [37.6–96.4] | 28/33 (84.9) [69.1–93.4] | 5/18 (27.8) [12.5–50.9] |
| Subcutaneous neurofibromasb | 6/8 (75) [40.9–92.9] | 1/4 (25) [4.6–69.9] | 3/5 (60) [23.1–88.2] | 17/30 (56.7) [39.2–72.6] | 6/18 (33.3) [16.3–56.3] |
| Symptomatic spinal neurofibromasa | 0/11 (0) [0–25.9] | 1/6 (16.7) [3–56.4] | 0/2 (0) [0–65.8] | 3/40 (7.5) [2.6–19.9] | 8/20 (40) [21.9–61.3] |
| Spinal neurofibromas by MRIa,c | 0/5 (0) [0–43.5] | 2/3 (66.7) [20.8–93.9] | 0/1 (0) [0–79.4] | 5/22 (22.7) [10.1–43.4] | 12/14 (85.7) [60.1–96] |
| Symptomatic OPGs, age ≥5 yearsd | 1/21 (4.8) [0.9–22.7] | 0/7 (0) [0–35.4] | 2/5 (40) [11.8–76.9] | 5/47 (10.6) [4.6–22.6] | 3/24 (12.5) [4.3–31] |
| Asymptomatic OPGs, age ≥5 yearse | 4/10 (40) [16.8–68.7] | 0/4 (0) [0–49] | 0/3 (0) [0–56.2] | 11/25 (44) [26.7–62.9] | 1/10 (10) [1.8–40.4] |
| Other neoplasmsf | 2/23 (8.7) [2.4–26.8] | 0/7 (0) [0–35.4] | 0/6 (0) [0–39] | 15/72 (20.8) [13.1–31.6] | 4/31 (12.9) [5.1–28.9] |
| Skeletal abnormalities | 9/25 (36) [20.3–55.5] | 5/10 (50) [23.7–76.3] | 0/8 (0) [0–32.4] | 23/67 (34.3) [24.1–46.3] | 11/34 (32.4) [19.1–49.2] |
| Noonan syndrome features | 2/22 (9.1) [2.5–27.8] | 1/7 (14.3) [2.6–51.3] | 0/7 (0) [0–35.4] | 6/66 (9.1) [4.2–18.5] | 1/32 (3.1) [0.6–15.8] |
| Pulmonic stenosis | 1/19 (5.3) [0.9–24.6] | 1/5 (20) [3.6–62.5] | 0/8 (0) [0–32.4] | 0/56 (0) [0–6.4] | 0/25 (0) [0–13.3] |
| Short statureg | 1/13 (7.7) [13.7–33.3] | 1/5 (20) [3.6–62.5] | 1/4 (25) [4.6–69.9] | 7/42 (16.7) [8.3–30.6] | 5/27 (18.5) [8.2–36.7] |
| Macrocephaly | 4/17 (23.5) [9.6–47.3] | 1/4 (25) [4.6–69.9] | 2/4 (50) [15–85] | 20/49 (40.8) [28.2–54.8] | 9/24 (37.5) [21.2–57.3] |
| Cognitive impairment and/or learning disabilities | 6/21 (28.6) [13.8–50] | 4/10 (40) [16.8–68.7] | 4/9 (44.4) [18.9–73.3] | 29/65 (44.6) [33.2–56.7] | 13/33 (39.4) [24.7–56.3] |
| Severe phenotype, age ≥19 yearsh | 7/9 (77.8) [45.3–93.7] | 4/6 (66.7) [30–90.3] | 1/6 (16.7) [3–56.4]i | 32/36 (88.9) [74.7–95.6] | 12/18 (66.7) [43.8–83.7] |
In individuals ≥9 years.
In individuals ≥19 years.
The frequency of both symptomatic and asymptomatic spinal neurofibromas in individuals who had undergone MRI examination.
The presence or absence of symptomatic OPGs was determined by ophthalmological examination and confirmed by MRI.
Including only individuals without signs of symptomatic OPGs who underwent MRI examination.
Including benign and malignant neoplasms, except for OPG and neurofibromas.
As no specific growth curves are available for the Hispanic and Asian populations, Hispanic and Asian individuals were excluded as having short or normal stature.
Individual was classified as having a severe phenotype if at least one of the following features was observed: plexiform and/or symptomatic spinal neurofibroma, symptomatic OPG, malignant neoplasm, or osseous lesions.
Among individuals with a missense mutation affecting codon 846, the status of plexiform and spinal neurofibromas was known only for 2/6 individuals (UG-R0781-S and UG-R665-F), thus a severe phenotype cannot be excluded in the remaining four individuals with missing data.
Table 3.
Comparison of Clinical Features of the Studied Group with the NF1 Arg1809 Cohort, the NF1 Met992del Cohort, and Large-Scale Previously Reported Cohorts of Individuals with “Classic” NF1
| NF1 Feature |
Number of Individuals (%) |
p Value (2-Tailed Fisher’s Exact Test) |
|||||
|---|---|---|---|---|---|---|---|
| aa 844–848 | Arg1809a | Met992delb | Previously Reported NF1 Cohorts | aa 844–848 versus Arg1809 | aa 844–848 versus Met992del | aa 844–848 versus “Classic” NF1 | |
| >5 CALMs | 130/157 (82.8) | 157/169 (92.9) | 46/47 (97.9) | 1,537/1,728 (89)c | 0.0060∗ ➘ | 0.0067∗ ➘ | 0.0263 ➘ |
| Skinfold freckling | 104/144 (72.2) | 95/161 (59) | 32/47 (68.1) | 1,403/1,667 (84.2)c | 0.0164 ➚ | 0.0007∗∗ ➘ | |
| Lisch nodules | 42/98 (42.9) | 12/120 (10) | 3/38 (7.9) | 729/1,237 (58.9)c | <0.0001∗∗ ➚ | <0.0001∗∗ ➚ | 0.0028∗ ➘ |
| Major external plexiform neurofibromasd | 36/92 (39.1) | 0/105 (0) | 0/41 (0) | 120/648 (18.5)e,f | <0.0001∗∗ ➚ | <0.0001∗∗ ➚ | <0.0001∗∗ ➚ |
| Cutaneous neurofibromasg | 47/69 (68.1) | 0/57 (0) | 0/18 (0) | 656/723 (90.7)f,h,i,j | <0.0001∗∗ ➚ | <0.0001∗∗ ➚ | <0.0001∗∗ ➘ |
| Subcutaneous neurofibromasg | 33/65 (50.8) | 0-5/57 (0-8.8)k | ND | 297/515 (57.7)f,i,j | <0.0001∗∗ ➚ | ||
| Symptomatic spinal neurofibromasd,l | 12/79 (15.2) 13/127 (10.2) |
0/40 (0) 0/76 (0) |
1/41 (2.4) 1/47 (2.1) |
2/119 (1.7)e 36/2,058 (1.8)e,f,m |
0.0080∗ ➚ 0.0022∗ ➚ |
0.0341 ➚ | 0.0004∗∗ ➚ <0.0001∗∗ ➚ |
| Symptomatic OPGs, age ≥ 5 yearsl,n | 11/104 (10.6) 12/136 (8.8) |
0/114 (0) 0/139 (0) |
0/46 (0) 0/47 (0) |
7/180 (3.9)e,o 64/1,650 (3.9)c |
0.0002∗∗ ➚ 0.0002∗∗ ➚ |
0.0186 ➚ 0.0384 ➚ |
0.0404 ➚ 0.0125∗ ➚ |
| Asymptomatic OPGs, age ≥ 5 yearsl,p | 16/52 (30.8) 18/63 (28.6) |
0/35 (0) 0/38 (0) |
ND | 2/45 (4.4)o 70/519 (13.5)q,r,s |
0.0001∗∗ ➚ <0.0001∗∗ ➚ |
0.0012∗∗ ➚ 0.0043∗ ➚ |
|
| Other malignant neoplasmst | 13/139 (9.4) | 2/155 (1.3)u | 0/47 (0) | 18/523 (3.4)f | 0.0023∗ ➚ | 0.0409 ➚ | 0.0061∗ ➚ |
| Skeletal abnormalitiesd,l | 38/91 (41.8) 48/144 (33.3) |
14/72 (19.4) 21/126 (16.7) |
8/41 (19.5) 9/47 (19.2) |
14/96 (14.6)e 144/948 (15.2)e,f,j,v |
0.0025∗ ➚ 0.0020∗ ➚ |
0.0174 ➚ | <0.0001∗∗ ➚ <0.0001∗∗ ➚ |
| Scoliosisg | 20/64 (31.3) | 6/48 (12.5) | 2/18 (11.1) | 51/236 (21.6)h,j | 0.0241 ➚ | ||
| Noonan syndrome features | 10/134 (7.5) | 46/148 (31.1) | 4 (all from 1 family) | 57/1,683 (3.4)c | <0.0001∗∗ ➘ | 0.0276 ➚ | |
| Pulmonic stenosis | 2/113 (1.8) | 14/132 (10.6) | 4/47 (8.5) | 25/2,322 (1.1)w | 0.0076∗ ➘ | ||
| Short stature | 15/91 (16.5) | 32/111 (28.8) | 5/47 (10.6) | 109/684 (15.9)e,i | 0.0451 ➘ | ||
| Macrocephaly | 36/98 (36.7) | 31/107 (29) | 4/45 (8.9) | 239/704 (33.9)e,i | 0.0005∗∗ ➚ | ||
| Cognitive impairment and/or learning disabilities | 56/138 (40.6) | 80/159 (50.3) | 8/47 (17) | 190/424 (44.8)e,f | 0.0042∗ ➚ | ||
Statistically significant p values with false discovery rates of 0.05 (indicated by ∗) and 0.01 (indicated by ∗∗) after correction for multiple testing using Benjamini-Hochberg procedure (see details in Table S10). After applying the Benjamini-Hochberg correction, p ≤ 0.0125 remained statistically significant at FDR of 0.05, while p values ≤ 0.0012 were still be considered as significantly different at FDR of 0.01. The black arrows indicate the statistically significant differences of the NF1 clinical features prevalence between the studied group and the cohort(s) used for the comparison, with the up and down arrows representing an increase and a decrease of the prevalence in the studied group, respectively. Abbreviation: ND, no data
Based on data from Pinna et al.,13 Rojnueangnit et al.,14 Nyström et al.,25 Ekvall et al.,26 and Santoro et al.27
Based on data from Upadhyaya et al.12
Previous NF1 cohort used for comparison: Friedman and Birch.32
In individuals ≥9 years in this study and Arg1809, ≥10 years in Met992del and other studies.
Previous NF1 cohort used for comparison: Huson et al.7
Previous NF1 cohort used for comparison: McGaughran et al.34
In individuals ≥19 years in this study and Arg1809, ≥20 years in Met992del and other studies.
Previous NF1 cohort used for comparison: Khosrotehrani et al.38
Previous NF1 cohort used for comparison: Plotkin et al.39
Five individuals with few (1–6) small, subcutaneous “possible” neurofibromas, none were biopsied and therefore none have been histologically confirmed.14
Second value is the frequency of a particular feature regardless of the individuals’ age.
Previous NF1 cohort used for comparison: Thakkar et al.35
The presence or absence of symptomatic OPGs was determined by ophthalmological examination and confirmed by MRI.
Previous NF1 cohort used for comparison: Van Es et al.31
Including only individuals without signs of symptomatic OPGs who underwent MRI examination.
Previous NF1 cohort used for comparison: Listernick et al.30
Previous NF1 cohort used for comparison: Blazo et al.37
Previous NF1 cohort used for comparison: Blanchard et al.40
Only malignant neoplasms, hence excluding neurofibromas and OPGs, have been taken into account.
Breast cancer (n = 1) and Ewing sarcoma (n = 1) were found in the NF1 Arg1809 cohort, no follow-up information on these individuals was available.14
Previous NF1 cohort used for comparison: Cnossen et al.33
Previous NF1 cohort used for comparison: Lin et al.36
To establish a genotype-phenotype association, we used the same approach as previously described.14 We compared the phenotypes of individuals with missense mutations affecting codons 844–848 with the cohort of 169 individuals with missense mutations affecting p.Arg1809,13, 14, 25, 26, 27 47 individuals heterozygous for c.2970_2972del (p.Met992del) mutations,12 and previously described large-scale NF1-affected individual cohorts with “classic” NF1.7, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40
This study was approved by the Institutional Review Boards of all participating institutions offering clinical genetic testing.
Molecular Analysis
In the Medical Genomics Laboratory at UAB, comprehensive NF1 mutation screening using an RNA-based approach complemented by DNA-dosage analysis was performed as previously described.41, 42 The status of the specific familial mutation in relatives was ascertained by bidirectional Sanger sequencing (ABI PRISM 3730, Life Technologies).
The nomenclature of the mutations is based on NF1 mRNA sequence GenBank: NM_000267.3 according to the recommendations of the Human Genome Variation Society (HGVS). For exon numbering we used the NCBI numbering, followed by the historical numbering in square brackets originally developed by the NF1 community.42
In Silico Prediction of Effect of Missense Mutations
Eight software programs were used to predict the effects of missense variants: two online in silico prediction tools (CADD v.1.3 and PolyPhen-2) and six complementary tools (Grantham Difference, SIFT v.4.0.3, SpliceSiteFinder-like, MaxEntScan, NNSplice v.0.9, and Human Splicing Finder v.2.4.1) embedded in Alamut visual software v.2.9.0 (Interactive Biosoftware). The presence or absence of the variants was checked in population databases, including the Genome Aggregation Database (gnomAD), 1000 Genomes, and the Exome Variant Server (EVS) as well as in disease databases: the Leiden Open Variation Database (LOVD), ClinVar, and the Human Gene Mutation Database (HGMD) (last accessed May 2017). Evolutionary conservation for human neurofibromin GenBank: NP_000258.1 residues 804–950 was evaluated using Clustal software v.2.0.12. The palindromic sequences and quadruplex forming G-Rich sequences (QGRS) were identified by Palindrome search and QGRS Mapper, respectively.
Interpretation of variant pathogenicity was performed based on the American College of Medical Genetics and Genomics (ACMG) recommendations.43
Statistical Analysis
For univariate analysis, two-tailed Fisher’s exact test was used to compare categorical variables with a p value < 0.05 considered as statistically significant. The resulting p values were adjusted for multiple comparisons using Benjamini-Hochberg (B-H) procedure with false discovery rates (FDRs) of 0.05 and 0.01. The 95% confidence interval (CI) was also calculated when appropriate. All statistical analyses were performed with GraphPad and VassarStats softwares.
Results
Description of Missense Mutations Affecting Codons 844–848
Exon 21 [16] is the largest NF1 exon (441 nucleotides), and in it we identified, besides the missense variants affecting the codons 844–848, a total of 19 different missense variants in 35 unrelated individuals from the UAB cohort. Fourteen of these alterations were classified as variants of uncertain significance (8/19) or likely benign (6/19) and reported 1–3 times in the UAB cohort (Figure S2). Only five variants were classified as pathogenic (4/19) or likely pathogenic (1/19) according to the current recommendations.43 Region 844–848 in exon 21 [16] stood out due to its high frequency of variants compared with the neighboring codons, indicating functional importance (Figures S2 and S3). A similar distribution and spectrum of missense alterations in the NF1 exon 21 [16] was observed in the publicly available databases (ClinVar, LOVD, and HGMD). Besides a clear cluster of recurrent variants in codons 844–848, other alterations spread over the entire exon 21 [16] were mostly classified as variants of uncertain significance and reported 1–2 times in these databases (Figure S2). The frequency of this cluster of variants in aa 844–848 is ∼0.8% (67/8,400) in unrelated NF1 mutation-positive individuals from the UAB cohort, second only to the p.Arg1809 (∼1.2%), and therefore represents a significant hotspot for missense mutations within NF1.
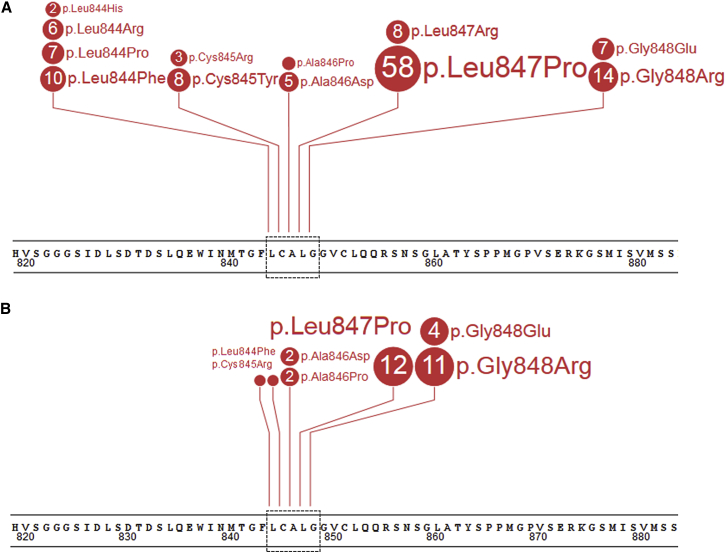
In the 129 unrelated individuals reported here, we identified 12 different NF1 missense alterations affecting one of five neighboring codons in exon 21 [16] (Table 1 and Figure 1). Within the group of individuals with p.Gly848Arg, two different substitutions were observed: c.2542G>A (6/14) and c.2542G>C (8/14). Detailed characteristics of the identified missense mutations are shown in Tables S2–S4 and Figure 1. All variants identified in this study with confirmed origin of the variant were submitted to the LOVD and ClinVar databases. Based on the data accumulated in this report (Tables S1 and S2), these variants can all be classified as pathogenic (Table S4) according to current recommendations.43
Figure 1.
Spectrum of Missense Mutations Affecting NF1 Codons 844–848 in the Cohort of 129 Probands and 33 Relatives
Shown are 129 probands (A) and 33 relatives (B). Each number in circle corresponds with the total number of individuals heterozygous for a specific mutation. The black dotted lines on the panels present the region 844–848. The figure was prepared using the ProteinPaint application.44
Among the aforementioned variants, 8/12 were present in the LOVD database with 5/8 classified as pathogenic (c.2533T>C [p.Cys845Arg], c.2536G>C [p.Ala846Pro], c.2537C>A [p.Ala846Asp], c.2540T>C [p.Leu847Pro], and c.2543G>A [p.Gly848Glu]) and 3/8 as variants of uncertain significance (c.2534G>A [p.Cys845Tyr], c.2540T>G [p.Leu847Arg], and c.2542G>C [p.Gly848Arg]). Eight of the 12 were present in ClinVar, including 3/8 classified as pathogenic (c.2531T>G [p.Leu844Arg], c.2540T>C [p.Leu847Pro], and c.2542G>C [p.Gly848Arg]), 1/8 as likely pathogenic (c.2534G>A [p.Cys845Tyr]), 1/8 as a variant of uncertain significance (c.2533T>C [p.Cys845Arg]), and 3/5 with no significance provided (c.2530C>T [p.Leu844Phe], c.2531T>C [p.Leu844Pro], and c.2543G>A [p.Gly848Glu]) (Tables S2 and S3). One individual (UAB-R4444) with c.2531T>A (p.Leu844His) carried another novel alteration (c.2524G>A); assuming both variants reside in cis, this alteration should be described as c.2524_2531delinsAGCTTCCA (p.Gly842_Leu844delinsSerPheHis). None of these variants, except for c.2531T>G (p.Leu844Arg), has been reported in 129,639 unrelated controls of the gnomAD and EVS databases or in the 1000 Genomes Project; c.2531T>G (p.Leu844Arg) was reported once in Latino (the variant’s frequency in all populations is 0.00041%). Based on in silico analysis, all alterations are predicted to be deleterious (SIFT) and probably or possibly damaging (PolyPhen-2). Additionally, CADD classified all variants as more likely to have deleterious effects (range: 22.6 to 31). In contrast to results of in silico analysis, suggesting a possible effect of two identified alterations (c.2542G>A and c.2543G>A) on splicing through creation of a novel exonic splice acceptor sequence, transcript analysis and sequencing showed a minor effect on splicing only for c.2542G>A in three individuals (UAB-R9493, UAB-R1474, and UAB-R0008), i.e., low levels of r.2410_2543del. The other individuals with c.2542G>A screened with an RNA-based approach (UAB-R3513 and UAB-R4476) in whom no missplicing was observed also carried the nearby benign variant c.2544G>A (p.Gly848=) (rs17883704) with both variants proven to reside in cis through next-generation sequencing. As missplicing was observed only in individuals carrying c.2542G>A in the absence of rs17883704 (Figure S4), rs17883704 is hypothesized to have a modifying effect. All missense mutations, except for c.2536G>C (p.Ala846Pro), were proven to be de novo in at least one proband; a total of 26 probands with unaffected parents were proven to have a de novo mutation, but formal confirmation of paternity/maternity by identity testing was pursued only for individuals tested in the Netherlands (ROT-R02233, ROT-R22853, and ROT-R17435). Additionally, 7/12 missense mutations (c.2530C>T [p.Leu844Phe], c.2533T>C [p.Cys845Arg], c.2536G>C [p.Ala846Pro], c.2537C>A [p.Ala846Asp], c.2540T>C [p.Leu847Pro], c.2542G>C [p.Gly848Arg], and c.2543G>A [p.Gly848Glu]) segregated with the phenotype (at least one individual per family) in 23 affected first-degree relatives from 15 families (Tables S1 and S2 and Figure S5). Finally, all missense mutations affecting amino acids 844–848 are located in a highly conserved region of the CSRD (amino acids 543–909; Figure S6). Besides cysteine at position 845 that is conserved up to zebrafish, all remaining amino acids are evolutionarily conserved up to Drosophila melanogaster (Ala846 and Gly848) and even to yeast IRA1 and/or IRA2 (Leu844 and Leu847). In chimpanzee, rat, and mouse all amino acids from 775 to 856 are fully evolutionarily conserved. None of these variants has been functionally characterized.
Demographic and Clinical Characterization of the Studied Cohort
A total of 162 individuals from 129 unrelated families were enrolled in the study, including 37/129 (28.7%) familial and 89/129 (69%) sporadic case subjects; 3/129 (2.3%) individuals had an unknown family history (ROT-R13734, ROT-R89874, and CAR-R8012M6). Detailed demographic and clinical descriptions of the individuals included in the study are shown in Tables 1 and S1 and Figure S5.
The complete phenotypic checklist forms were collected from 151/162 individuals (93.2%). Of these, 125/151 (82.8%) fulfilled the NIH diagnostic criteria and 118/151 (78.2%) fulfilled the NIH diagnostic criteria if family history was excluded as a criterion. Among 26/151 individuals who did not fulfill the NIH diagnostic criteria (with 20/26 being ≤8 years), multiple CALMs-only (>5) were present in 16/26, <6 CALMs-only were present in 8/26, and 2/26 did not have any pigmentary manifestations but had externally visible plexiform neurofibromas (UAB-R9135 and UG-R5831) (Table S5). CALMs-only (<6) were observed mostly in individuals with a missense mutation at codon 848 (5/8 with c.2542G>C [p.Gly848Arg], 1/8 with c.2542G>A [p.Gly848Arg], 1/8 with c.2543G>A [p.Gly848Glu], and 1/8 with c.2534G>A [p.Cys845Tyr]).
Among 102 individuals ≥9 years, more than 5 CALMs and skinfold freckling were present in 79.8% (79/99) and 80% (76/95), respectively (Table 1). Both clinical features were found in 71.6% (68/95) of case subjects. Out of 20 individuals ≥9 years with only few or absolute lack of CALMs (Table S1), 11 case subjects fulfilled the NIH diagnostic criteria based on presence of other clinical signs, such as skinfold freckles, Lisch nodules, neurofibromas, and/or osseous lesions (UG-R0781, UAB-R3618-M, MIL-R192/982-F, UAB-R4476, MIL-R999/399, MIL-R999/399-M, ROT-R95424, UG-R923-S, UAB-R3237, MAN-R95417G, and MAN-R95417G-C). Among these individuals, 8/11 (72.7%) carried a missense mutation at codon 848. Lisch nodules were reported less frequently (42/98 all ages, but in 34/60 ≥9 years).
Cutaneous and subcutaneous neurofibromas were found in 68.1% (47/69 ≥19 years) and 50.8% (33/65 ≥19 years) of the case subjects, respectively. Thirty adults had both types of tumors (30/64 ≥19 years, 46.9%). Ten individuals ≥17 years had >100 cutaneous and/or subcutaneous nodules, including a 47-year-old man previously reported45 with >1,400 neurofibromas (individual counts of externally visible neurofibromas; BRA-R38) and a 17-year-old woman (ROT-R1CMUL) with >500 cutaneous neurofibromas, >100 subcutaneous neurofibromas, and >100 intradermal neurofibromas. Nine out of ten individuals with a very high number of neurofibromas carried a missense mutation at codon 847: c.2540T>G (p.Leu847Arg) (2/9) or c.2540T>C (p.Leu847Pro) (7/9, including two individuals with metastasized MPNSTs). In 16 case subjects with “several” neurofibromas, a more precise estimated number was not reported. Eight individuals (UAB-R5776, UAB-R3618, UAB-R4624, UAB-R7447, UAB-R1002, UAB-R1037-M, UAB-R3237, PAD-R500-C1) were reported to have a single cutaneous or subcutaneous nodule (none histopathologically confirmed); these individuals were considered as “negative for the criterion of neurofibromas” as ≥2 cutaneous/subcutaneous neurofibromas are required according to the NIH clinical criteria.
45% of the individuals ≥9 years had known plexiform neurofibromas (41/92 ≥9 years; 47/143 all ages), including externally visible (n = 36) and internal (n = 5) tumors. For six case subjects, the information was not provided whether plexiform neurofibromas were identified clinically or by MRI. Among all individuals with plexiform neurofibromas, 31/47 presented with one plexiform tumor and 16/47 with ≥2 plexiform neurofibromas. Plexiform tumors were found in the head, face, and neck area (35.7%, 25/70), limbs (34.3%, 24/70), trunk (17.1%, 12/70), back (n = 3), abdomen (n = 3), pelvis (n = 2), and chest (n = 1).
Symptomatic spinal neurofibromas visible by MRI were found in 15.2% of individuals (12/79 ≥9 years; 13/127 all ages). Forty asymptomatic individuals received MRI screening, leading to the identification of another seven case subjects with spinal tumors (Table S6). Approximately one-third of the individuals with spinal tumors (6/20) had fewer than 6 CALMs and no skinfold freckling, whereas in 60% (12/20) plexiform neurofibromas were observed (with 11/12 being externally visible).
Symptomatic OPGs, confirmed by MRI imaging, were found in 11/104 of individuals older than 5 years (10.6%), whereas asymptomatic OPGs were present in 16/52 additional individuals who underwent MRI examination (30.8% ≥5 years). In 19 of 27 symptomatic and asymptomatic OPGs, the detailed information about the tumor’s location was collected, involving optic nerves (2 symptomatic OPGs and 7 asymptomatic OPGs), chiasm (1 symptomatic OPG and 1 asymptomatic OPG), or both locations (6 symptomatic OPGs and 2 asymptomatic OPGs). Three children were diagnosed with a symptomatic OPG (PAD-R300) or asymptomatic OPGs (UAB-R3714 and UAB-R3513) before age 4 years (Table S7).
Skeletal abnormalities were frequently reported (48/144 all ages) and included scoliosis (27/144 all ages, but 20/64 ≥19 years) and pectus anomalies (10/144 all ages: pectus carinatum 6/10 and excavatum 4/10). In addition, long bone dysplasia (n = 4), pseudarthrosis (n = 2), tibial dysplasia (n = 1), bone cysts (n = 2), sphenoid wing dysplasia (n = 2), ulnar aplasia, likely representing the severe end of ulnar pseudarthrosis with bone resorption and absence of ulnar bone (n = 1), dural ectasia (n = 1), 4th lumbar vertebrae fragmentation (n = 1), bowed long bones (n = 1), clinodactyly (n = 1), postaxial polydactyly (n = 1), and cherubism (n = 1) were observed in the studied group.
Noonan syndrome features were observed in 10/134 (7.5%) individuals. One previously reported individual (UAB-R624) with a family history of PTPN11-positive (MIM: 176876) Noonan syndrome (MIM: 163950) had a severe phenotype of pulmonic stenosis and aortic coarctation, dysmorphic features (high forehead, hypertelorism, downslanting palpebral fissures, short neck with a low posterior hair line), short stature, pectus carinatum, >5 CALMs, axillary and inguinal freckling, plexiform and cutaneous neurofibromas, and symptomatic OPG with signs of hydrocephalus.46 Besides the familial PTPN11 c.1529A>G (p.Gln510Arg) inherited from the individual’s father, the NF1 missense mutation c.2531T>G (p.Leu844Arg) was found de novo in the proband (Figure S5). In other individuals with Noonan syndrome features (UAB-R2696, UAB-R5001, UAB-R3725, and UAB-R4676) no pathogenic or likely pathogenic variants in Noonan-related disorders genes (PTPN11 [MIM:176876], SPRED1 [MIM:609291], BRAF [MIM: 164757], CBL [MIM: 165360], HRAS [MIM: 190020], KRAS [MIM: 190070], MAP2K1 [MIM: 176872], MAP2K2 [MIM: 601263], NRAS [MIM: 164790], RAF1 [MIM: 164760], SHOC2 [MIM: 602775], SOS1 [MIM: 182530], RIT1 [MIM: 609591], RASA2 [MIM: 601589], and SOS2 [MIM: 601247]) were identified. Cardiovascular abnormalities observed in the studied group included hypertension (n = 7, one related to renal artery stenosis), pulmonic stenosis (n = 2), mitral valve stenosis, atrial septal defect, ventricular septal defect, Moyamoya disease, pericarditis carcinomatosa, mitral valve insufficiency, mild pulmonic insufficiency, and hypertrophic cardiomyopathy (each observed in a single individual). Short stature (PC ≤ 3) and macrocephaly (PC ≥ 98) were found in 15/91 (16.5%) and 36/98 (36.7%), respectively. Of the 138 case subjects with provided developmental data, 56 individuals had abnormal development presenting with at least one of the following forms of cognitive impairment and/or learning difficulties: learning disabilities (n = 43), developmental delay (n = 30), speech delay (n = 8), ADD (n = 8), ADHD (n = 10), motor delay (n = 5), autism spectrum (n = 2), or Asperger syndrome (n = 1). Seven individuals had significant global developmental delay with/without speech delay, learning difficulties, and/or AD(H)D, including one with a full scale intelligence quotient (FSIQ) score 59. Additionally, three individuals were reported to have frequent migraine headaches and two had epilepsy and/or psychiatric problems.
For 139/162 individuals, data on the presence or absence of tumors other than neurofibromas and OPGs was available. 13 of 139 (9.4%) individuals were diagnosed with malignant neoplasms (Table S8), including embryonal rhabdomyosarcoma (3/13), MPNST (7/13, including one woman with MPNST and BRCA1/2-negative breast cancer), colon cancer (1/13), medullary thyroid carcinoma (1/13), and juvenile myelomonocytic leukemia (JMML) (1/13). Individuals ≥14 years old with c.2540T>C (p.Leu847Pro) had a higher number of malignant neoplasms compared to individuals carrying other missense mutations in the studied region (p = 0.0448; Table S9). Moreover, this mutation was present in most case subjects with MPNST (5/7), except for one each carrying c.2543G>A (p.Gly848Glu) or c.2530C>T (p.Leu844Phe). Four of seven individuals with MPNST died before age 30 years (Table S8). Hypothalamic glioma (n = 1), lipoma (n = 1), cerebral tumors (n = 3), non-ossifying fibroma (n = 2), and odontogenic fibroma (n = 1) were also reported.
The frequency of clinical features in individuals heterozygous for missense mutations affecting one of five neighboring codons 844–848 is presented in Table 2. A lower number of CALMs, freckling, and cutaneous neurofibromas was observed in case subjects with missense mutations at codon 848 (all p < 0.0001; Table S9); however, these individuals had a higher prevalence of symptomatic spinal neurofibromas (p = 0.0012; Table S9).
Taken together, a severe phenotype, including at least one of the following features (plexiform and/or symptomatic spinal neurofibromas, symptomatic OPGs, malignant neoplasm, or osseous lesions) was observed in 75% of adult NF1-affected individuals (56/75 ≥19 years; Table 2).
Comparison of Clinical Features Observed in the Studied Cohort with Individuals Heterozygous for p.Arg1809 and p.Met992del Mutations and Cohort of Individuals with “Classic” NF1 Phenotype
Comparison of clinical features of the studied group with the NF1 p.Arg1809 and p.Met992del cohorts as well as previously described large-scale cohorts of individuals with “classic” NF1 is shown in Table 3. The complete list of adjusted p values with FDRs at 0.05 and 0.01 after B-H correction for multiple testing is presented in Table S10. All p values ≤ 0.0125 and p values ≤ 0.0012 remained statistically significant after applying the B-H correction at FDRs of 0.05 and 0.01, respectively.
In the current study, we observed a significantly higher number of major external plexiform neurofibromas compared with the NF1 p.Arg1809 and the NF1 p.Met992del cohorts, as well as classic NF1-affected population (all p < 0.0001; statistically significant after B-H correction at FDR of 0.01). Importantly, while none of the individuals carrying the p.Arg1809 and p.Met992del had external plexiform, cutaneous, and/or subcutaneous neurofibromas, ∼71% of the individuals ≥19 years with a missense mutation affecting codons 844–848 had cutaneous and/or subcutaneous neurofibromas (p < 0.0001; statistically significant after B-H correction at FDR of 0.01) and ∼39% of the individuals ≥9 years had externally visible plexiform neurofibromas (p < 0.0001; statistically significant after B-H correction at FDR of 0.01). Compared with p.Arg1809, p.Met992del, and classic NF1-affected cohorts, at least 5-fold greater prevalence of symptomatic spinal neurofibromas was reported in the studied group (0%–2.1% versus 10.2%) which was statistically significant at FDR of 0.01 for the general NF1-affected population (p < 0.0001) and at FDR of 0.05 for the p.Arg1809 cohort (p = 0.0022).
Symptomatic and asymptomatic OPGs were more frequent compared to individuals with p.Arg1809, p.Met992del, and classic NF1, with symptomatic and asymptomatic OPGs statistically increased after B-H correction at FDR of 0.05 in the 844–848 cohort compared to the classic NF1-affected cohorts (p = 0.0125 and p = 0.0043, respectively) and at FDR of 0.01 compared with the p.Arg1809 cohort (p = 0.0002 and p < 0.0001, respectively). The overall prevalence of malignant neoplasms, other than neurofibromas and OPGs, was also higher in the studied group compared to a large cohort of classic NF1-affected individuals (9.4% versus 3.4%; p = 0.0061, statistically significant at FDR of 0.05 after B-H correction).
Additionally, the aa 844–848 cohort had a significantly increased frequency of skeletal abnormalities compared to individuals with p.Arg1809 and classic NF1 phenotypes (both statistically significant after B-H correction at FDR of 0.05), regardless of the age. Scoliosis was reported more frequently compared with p.Arg1809 individuals (31.3% versus 12.5% in ≥ 19 years), but this difference was not statistically significant after B-H correction.
The prevalence of CALMs was lower than in p.Arg1809 and p.Met992del cohorts (both significant at FDR of 0.05 after B-H correction), while skinfold freckles occurred more commonly in classic NF1-affected cohorts than in the studied group (significant at FDR of 0.01 after B-H correction). Noonan syndrome features were significantly less frequent in the studied group compared to individuals with p.Arg1809 (significant at FDR of 0.01 after B-H correction). In line with this finding, pulmonic stenosis was very rarely observed in the cohort (1.8% versus 10.6% in the p.Arg1809 cohort; significant at FDR of 0.05 after B-H correction). All cohorts, except for the p.Met992del, shared a similar frequency of cognitive impairment and/or learning difficulties (∼45%).
Discussion
We present 162 individuals heterozygous for a constitutional NF1 missense mutation in one of five neighboring codons 844–848 who have a high prevalence of a severe NF1 phenotype, including plexiform and/or symptomatic spinal neurofibromas, symptomatic OPGs, and other malignant neoplasms, as well as bone abnormalities. The frequency of the cluster of these mutations is ∼0.8% (67/8,400) in unrelated NF1 mutation-positive individuals from the UAB cohort, second only to the p.Arg1809 (∼1.2%) among the missense variants.
One of the most severe complications in NF1-affected individuals are clinically apparent plexiform neurofibromas affecting 15%–30% of the NF1-affected general population.7, 34, 47, 48, 49, 50 In this study, externally visible plexiform neurofibromas were found in ∼39% of individuals ≥9 years, therefore significantly higher compared with p.Arg1809 and p.Met992del and classic NF1-affected cohorts (significant at FDR of 0.01 after B-H correction; Tables 3 and S10). Individuals in this study did not undergo whole-body MRI; therefore, the frequency provided here is a likely underestimate, as internal asymptomatic plexiform neurofibromas were not accounted for.
As plexiform neurofibromas have been suggested to be associated with a higher lifetime risk for the development of MPNSTs,50, 51, 52, 53 the finding of MPNSTs in 5% (7/139) of the affected in our cohort, which is twice as high as reported by Huson et al. in the South-East Wales cohort,28, 29 is in line with expectations.
Approximately 24%–40% of NF1-affected individuals develop spinal neurofibromas,35, 39, 52 but they are most often asymptomatic and not detectable by physical examination. The estimated prevalence of symptomatic spinal neurofibromas in the general NF1-affected population is less than 2%.7, 34, 35 In the current study, a high number of individuals with symptomatic spinal neurofibromas was reported, compared to the classic NF1-affected cohorts (statistically significant at FDR of 0.01 after B-H correction): 13/127 (10.2%) for all ages and 12/79 (15.2%) for ≥9 years. Kluwe et al. suggested that spinal neurofibromas cause symptoms mainly in older case subjects (mean age 32.8 years),18 but 4 of 13 symptomatic individuals in our cohort were below age 18 (range: 7–17 years). In 40 individuals who underwent MRI examination, an additional seven case subjects with asymptomatic spinal neurofibromas were found. Among all affected individuals, five belonged to two previously reported multi-generation families (UG-R923 and MAN-R95417G) where the spinal tumors segregated within the family.16, 21 For two relatives of these probands, the spinal neurofibromas were recognized only after MRI, although the tumor burden was extensive. None of the individuals had >5 CALMs, including 2/5 who had <6 CALMs and 3/5 had none. This rare form of NF1 is called familial spinal neurofibromatosis (FSNF).
Plexiform and spinal tumors as well as subcutaneous neurofibromas are associated with a severe NF1 phenotype and may result in significant morbidity in children and adults.54, 55 OPGs, the most common brain tumors in children, are another complication in the general NF1-affected population.56 The overall prevalence of OPGs in the NF1-affected population is ∼11%–20%,39, 50, 57 but only ∼30% of these individuals have clinically symptomatic OPGs and present with impaired visual acuity, visual field loss, abnormal color vision, squint, proptosis, and/or hypothalamic dysfunction.49 Most symptomatic OPGs are diagnosed before age 7 years57 with the mean age of 5 years.58 In the studied group, symptomatic OPGs were found in 11/104 (10.6%) of individuals ≥5 years, which is more frequent compared with p.Arg1809 and p.Met992del cohorts (none of the individuals had OPGs) and with classic NF1-affected population (3.9%); however, after applying the B-H correction, only the result of comparison with p.Arg1809 cohort and the general NF1-affected population remained statistically significant at FDR of 0.05 (Tables 3 and S10). Furthermore, there was a higher prevalence of asymptomatic OPGs in 16/52 (30.8%) individuals ≥5 years who underwent MRI examination (statistically significant at FDR of 0.01).
Individuals with NF1 are at higher risk to develop specific malignancies compared with the general population, significantly increasing mortality.59, 60 Besides the high-grade gliomas, the most common malignancies in NF1-affected children are rhabdomyosarcomas, JMML, and neuroblastomas, but accurate estimates on prevalence are not available due to the rarity of these tumors.61, 62 Based on the data provided by Sung et al. and Crucis et al.,63, 64 the prevalence of rhabdomyosarcomas in children with NF1 is estimated at 0.4%–0.5%, while Chang and Shannon reported that the individual risk of JMML in NF1 is ∼0.04%.65 In the studied group, three NF1-affected children younger than 5 years developed embryonal rhabdomyosarcomas, including one individual, now >26 years, who survived both a rhabdomyosarcoma and astrocytoma grade II, diagnosed at the age 2 and 15 years, respectively. Furthermore, one 5-year-old girl (out of 50 children ≤8 years) presented with <6 CALMs and JMML. This girl was heterozygous for two pathogenic NF1 mutations in the blood, c.2542G>A (p.Gly848Arg) as well as c.1246C>T (p.Arg416∗), with p.Gly848Arg being the first hit given the absence of p.Arg416∗ in buccal swabs, indicating somatic mosaicism for p.Arg416∗. A UK population-based hospital admission and death certificate study found that individuals with NF1 have, after excluding the well-established risks of nervous systems tumors, a 2.7-fold increased risk of developing cancers of the esophagus, stomach, colon, liver, lung, bone, thyroid, malignant melanoma, non-Hodgkin lymphoma, chronic myeloid leukemia, breast, and ovary.66 In the current study, we noted recurrent malignant tumors, such as MPNSTs (7/139; 5%) (Tables S1 and S8). Among these individuals, one 44-year-old woman previously described with the missense mutation c.2540T>C (p.Leu847Pro) had MPNST, BRCA1/2-negative (MIM: 113705 and 600185) breast cancer as well as a high number of cutaneous neurofibromas (>100).67 In addition, one individual developed a medullary thyroid carcinoma and three first-degree relatives of a Belgian proband with c.2540T>C (p.Leu847Pro) died from malignancies (a metastasized colon adenocarcinoma and two MPNSTs, both deceased before age 26). Taken together, the overall prevalence of malignant neoplasms in the studied group was substantially higher than in the published datasets of the general NF1-affected population (significant at FDR of 0.05 after B-H correction; Tables 3 and S10). Furthermore, specifically mutation p.Leu847Pro seems to confer a high predisposition to develop malignant tumors compared to other missense variants reported in this study (p < 0.0448; Table S9), although the CADD score of this variant is not the highest among the studied region (only 26.1; Table S2). Given the predominance of the p.Leu847Pro mutations in the studied cohort (70/162 individuals), larger datasets are required to further refine the increased tumor risk associated with the other mutations within the studied region.
Skeletal abnormalities, including long bone dysplasia with or without pseudarthrosis, scoliosis, sphenoid wing dysplasia, bone cysts, including cherubism, non-ossifying fibromas and osseous giant cell lesions, hand anomalies, anterior chest wall anomalies, and short stature, can lead to serious clinical consequences and significant morbidity.68 We observed a clear overall increase in the number of skeletal anomalies compared with p.Arg1809 (FDR of 0.05 after B-H correction) and the general NF1-affected population (FDR of 0.01 after B-H correction). As many as 33.3% of the NF1-affected individuals (48/144) presented with one or more osseous lesion, scoliosis (n = 27), and pectus anomalies (n = 10) being most frequent (18.8% and 6.9%, respectively). The overall frequency would be higher if individuals with short stature (40.3%; 58/144) are included. Rarely reported complications possibly associated with NF1 status included cherubism, chronic arthritis of multiple joints with elbow contractures, clinodactyly of the 3th–5th toes, postaxial polydactyly, and ulnar aplasia, likely representing the severe end of ulnar pseudarthrosis with bone resorption and absence of the ulnar bone. Interestingly, the latter has been reported only in two NF1-affected case subjects.69 Mild to moderate scoliosis was reported in only 18% of NF1-positive individuals with bilateral neurofibromas of all spinal roots;17 however, in our study we observed co-occurrence of scoliosis and spinal tumors in 45% (9/20) of individuals with confirmed symptomatic or asymptomatic spinal neurofibromas (not necessarily affecting all dorsal roots) (Table S6). An additional 11 individuals had scoliosis without evidence of spinal neurofibromas by MRI (Table S1).
Cohorts of individuals with NF1 missense mutations affecting codons 844–848 and classic NF1-affected population shared a similar frequency for short stature and macrocephaly. Noonan syndrome features were rarely observed in the studied group compared with the p.Arg1809 cohort (significant at FDR of 0.01 after B-H correction). In line with previous studies,7, 34, 39, 70 intellectual disability, developmental delay, and/or learning difficulties were frequently observed in the current study (40.6%).
Among the 129 unrelated probands with a missense mutation affecting codons 844–848, p.Leu847Pro and p.Gly848Arg are the most recurrent variants, found in 58 and 14 unrelated individuals, respectively (Table S2 and Figure 1). Both alterations are associated with a severe NF1 phenotype, including a high prevalence of plexiform neurofibromas and skeletal abnormalities, compared to the general NF1-affected population. However, missense mutations at p.Gly848 predispose with a greater frequency to symptomatic or asymptomatic spinal tumors, which were found in ∼70% of probands carrying the p.Gly848Arg or p.Gly848Glu mutations (9/13 ≥9 years, but in 9/10 ≥9 years who received MRI screening), which is slightly higher than in individuals presenting with a severe phenotype caused by a total NF1 deletion (8/13 ≥9 years).71 Several of the severely affected individuals with a missense mutation at p.Gly848 had only few or no pigmentary skin findings. So far, ∼100 case subjects have been reported with the true “spinal NF” phenotype17 and these individuals more frequently carry a splice site or missense mutation spread over the entire NF1 coding region.18, 19, 20 So far, no single mutation has been correlated with this severe clinical presentation. We provide the specific genotype-phenotype association between a particular NF1 mutation and the spinal phenotype. Individuals with missense mutations at p.Gly848 appear to constitute a distinct group of NF1-affected individuals with a high prevalence of symptomatic spinal neurofibromas and a clear decrease of pigmentary manifestations (CALMs and skinfold freckles) as well as cutaneous neurofibromas (Tables 2 and S9). Because of the limited number of individuals ≥9 years old with the missense mutations at codons 844–846, it is still difficult to establish a genotype-phenotype correlation among these cohorts; however, so far these variants also seem to be associated with a severe phenotype, including a high prevalence of plexiform neurofibromas in the p.Cys845 and p.Ala846 cohorts (57.1% and 66.7%, respectively) and OPGs in p.Leu844 cohort (∼24% for both symptomatic and asymptomatic OPGs in ≥5 years). At this moment, it cannot be excluded that two specific genotype-phenotype correlations exist within this small region of NF1 with the NF1 codon 847 associated with an increased risk for malignant neoplasia and the NF1 codon 848 associated with a high prevalence of symptomatic spinal neurofibromas. The current study, however, intended to show that the whole region of 844–848 codons stood out due to its high frequency of variants compared with the neighboring codons, indicating functional importance. In addition, the cluster of missense mutations here described, although located outside the GRD important for RAS regulation, is clearly associated with a severe phenotype, not reported so far in literature. As the current study necessarily still underestimates the internal tumor burden, as systematic whole-body imaging was not performed, close clinical management seems warranted for individuals presenting with a missense variant affecting aa 844–848.
As NF1 is known for its variable expressivity and age dependency, it is challenging to establish genotype-phenotype correlations. Although we performed a comparative analysis on a large well-described cohort using a standardized phenotypic data collection form, one limitation of the study is that clinical information was collected by physicians from different referral centers, although all were NF1 specialists. Data in this and the previously reported p.Arg1809 cohort were “double-checked” through verification of the originally submitted phenotypic checklist forms and subsequent update of the clinical notes, so data should be highly accurate.
Clinical variability, both inter- and intrafamilial, has been widely reported in the past two decades.72, 73, 74 Although significant progress has been made over the last 20 years, the mechanisms underlying this phenotypic heterogeneity only gradually start to be unraveled. The factors contributing to the phenotypic variability include (1) age dependency of some of the NF1 features,29, 75, 76 (2) timing, cell of origin, and number of second hits in specific cells, resulting in presence and number of CALMs, freckling, tibial dysplasia, neurofibromas, and other tumors,77 (3) post-zygotic mosaicism for the first NF1 hit in mosaic individuals,77 (4) the enormous NF1 allelic heterogeneity,78 (5) occasional presence of two different NF1 pathogenic variants segregating within a family (see MAD-R9.232; Table S1 and Figure S5) or the occurrence of two independent mutations, one in NF1 and the other in a different gene, within an individual (see UAB-R624 with the NF1/PTPN11 mutations and UF-R1 with the NF1/KIT mutations; Table S1), (6) modifying genes,79 and (7) environmental factors (e.g., number of pregnancies).80 To date, two studies have identified potential modifying genes, unlinked to the NF1 locus, associated with the severity of NF1 presentation.81, 82 Pasmant et al. demonstrated that a high number of plexiform neurofibromas has been significantly associated with allele T of SNP rs2151280 of ANRIL (MIM: 613149).81 Pemov et al. reported a correlation of two common SNPs (rs4660761 and rs7161), located between DPH2 (MIM: 603456) and ATP6V0B (MIM: 603717), as well as of SNP rs1800934 in MSH6 (MIM: 600678) with the number of CALMs.82 Further studies are needed to confirm these findings.
Missense mutations affecting NF1 codons 844–848 described in this study are clearly pathogenic and individuals with these missense mutations have a statistically higher risk of developing spinal neurofibromas, plexiform neurofibromas, and OPGs. Functional studies in mutant mice harboring the missense mutation c.2542G>C (p.Gly848Arg) did not recapitulate this human phenotype, as neither optic pathway gliomas nor plexiform neurofibromas developed.22, 23 Western blot analysis showed that c.2542G>C (p.Gly848Arg) resulted in 38%–50% reduction of neurofibromin levels.22, 23 These mutations reside outside the GRD (amino acids 1,217–1,511), known to have tumor-suppressor activity through downregulation of members of the Ras family of small GTP-binding proteins. Although NF1 was cloned in 1990, the cellular functions performed by this huge 2,818-amino acid multi-domain protein are still incompletely understood. The cluster of recurrent missense mutations involving aa 844–848 described in the current study are located within the CSRD (amino acids 543–909), located N-terminal to the GRD. The CSRD domain, originally described by Fahsold et al.,83 is likely functionally important, which is further implied by the presence of multiple missense variants in this segment of the gene in NF1-affected individuals. The 3D structure of this region has not been resolved and its precise functions and interactors have not been described. Ras GAP activity is enhanced through phosphorylation by Protein Kinase Cα of serine and threonine residues within this domain.84 Based on the 2D modeling of the CSRD using PredictProtein server,85 the region 831–847 might form the C-part of a helix and be buried in the protein. Missense mutations affecting codons 844–848, especially those substituting smaller hydrophobic amino acids to large ones, may result in breaking of the helix and exposure of the buried protein domain, consequently affecting the function of the protein. No functional studies confirming the aforementioned bioinformatics analysis have been performed, however. In any case, missense mutations in this region seem to act through a loss-of-function mechanism and not gain-of-function or dominant-negative, at least in melanocytes and JMML. Indeed, the c.2540T>C (p.Leu847Pro) was observed as a “second hit” in one CALM, biopsied from a 13.5-year-old girl with >5 CALMs and skinfold freckling carrying the NF1 constitutional mutation c.5547−1G>A (Table S11), confirming that two hits are required to cause a phenotypic effect. Additionally, we reported a 5-year-old girl with JMML (UAB-R9493; Table S1) who carried two pathogenic NF1 mutations in the blood: c.2542G>A (p.Gly848Arg) as a “first hit” mutation and c.1246C>T (p.Arg416∗) as a “second hit.” There is a need to improve our understanding of the physiological functions of neurofibromin and to determine how each domain regulates the function of this protein.
Six amino acids in the region aa 804–950 are evolutionarily conserved down to yeast (IRA1 and IRA2), Leu844, Gly849, Leu852, Glu924, Leu933, and Phe934 (Figure S6) and would therefore be expected to be of particular functional importance.86 Only Leu844 and Leu933 have, however, been observed in NF1-affected individuals to predispose to recurrent missense mutations (HGMD, LOVD, ClinVar, and our cohort). The tumorigenic potential of aa 844 is further highlighted by identification of somatic mutations in the COSMIC database: one glioma with c.2531T>C (p.Leu844Pro), one glioma and four malignant melanomas with c.2530C>T (p.Leu844Phe).
Palindromic structures belong to the non-B DNA structures and are often the site of replication errors resulting in substitutions.87 The NF1 missense mutation hotspot (aa 844–848) is located in the highly conserved amino acid region, suggesting that it is functionally important. The genomic sequence encoding the human NF1 aa 845–853 is a part of two palindromic structures (Figure S7); therefore the high rate of recurrent missense mutations affecting Leu847 and Gly848 may partially be due these being both located in the loop of the palindrome. In NF1 exon 21 [16], other palindromic nucleotide sequences, specifying the amino acid residues aa 828–832, aa 865–868, aa 908–911, and aa 933–937 are observed, resulting in four additional stem-loop structures. However, these structures do not predispose to recurrent missense mutations as none were found either in the UAB, HGMD, or LOVD cohort, except for c.2798T>C (p.Leu933Pro), whose location does not include the loop of the palindrome. The complex interplay between functional significance and genomic architecture needs to be considered when analyzing the recurrence of mutations.
Although only a few clear genotype-phenotype correlations have been so far reported,11, 12, 13, 14 the data presented here show that additional clinically relevant NF1 genotype-phenotype correlations exist. A renewed interest in such studies is needed to come to a timely unfolding of additional correlations, as so far only the surface has been scratched. This will require close collaboration between NF1 clinicians and molecular geneticists. The lack of discovery of more specific genotype-phenotype correlations may be partly due to the methodological approach, including lumping mutations in large categories (truncating versus microdeletion, splice, missense mutations).88, 89 Identification of mutation-specific genotype-phenotype correlations depends on the dataset size with a large number of individuals, preferentially postpubertal, carrying the same non-truncating constitutional mutation, with the associated phenotype recorded in a standardized way. As there are only a limited number of truly recurrent non-truncating mutations, prioritization on individuals carrying such recurrent mutations is indicated. Although each of the recurrent mutation affects only a small percentage of NF1-affected individuals (3%–8% with the microdeletion type I, ∼0.8% with p.Met992del, ∼1.2% with the p.Arg1809 missense mutation, and ∼0.8% for the cluster of missense mutations affecting codons 844–848), together they may affect counseling and surveillance in a significant fraction of the NF1-affected population.
In conclusion, the present findings indicate that missense mutations affecting one of five neighboring codons 844–848 located outside the GAP-related domain are an important risk factor for a severe phenotype in NF1-affected individuals. We report that these individuals have a high prevalence of plexiform and/or spinal neurofibromas, symptomatic and asymptomatic OPGs, malignant neoplasms, and skeletal abnormalities. A severe phenotype was observed in 75% of adult NF1-affected individuals with these mutations, clearly demonstrating that missense mutations outside the GRD can be associated with a severe clinical presentation. The current study identified a genotype-phenotype correlation in this region that may be valuable in the management and genetic counseling of a significant number of NF1-affected individuals. These data suggest that there is a potential need for increased disease surveillance in individuals with these mutations enabling genotype-driven personalized medicine.
Acknowledgments
We thank the individuals and their families for participating in this study. This work was supported by the Children’s Tumor Foundation by the Isaac and Sadie Fuchs Genotype-Phenotype Study (to L.M.M.) and by internal funds from the Medical Genomics Laboratory at UAB. Parts of this work were presented during the 17th European Neurofibromatosis Meeting (September 8–11, 2016, Padova-Abano Terme, Italy) and the Children’s Tumor Foundation NF Conference (June 10–13, 2017, Washington, DC, USA).
M.K. is also affiliated with the Department of Biology and Medical Genetics at the Medical University of Gdansk in Poland. D.G.R.E. is supported by the all Manchester NIHR Biomedical Research Centre as an NIHR Senior investigator. M.U. acknowledges Sheila Palmer-Smith and Ian Frayling for their support.
Published: December 28, 2017
Footnotes
Supplemental Data include 7 figures and 11 tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2017.12.001.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
Clustal, http://www.clustal.org/clustal2/
GeneReviews, Friedman, J.M. (2014). Neurofibromatosis 1. http://www.ncbi.nlm.nih.gov/books/NBK1109
gnomAD Browser, http://gnomad.broadinstitute.org/
GraphPad, https://www.graphpad.com/
HGVS, http://varnomen.hgvs.org
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
Palindrome search, http://bioinfo.cs.technion.ac.il/projects/Engel-Freund/new.html
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
QGRS Mapper, http://bioinformatics.ramapo.edu/QGRS/index.php
VassarStats, http://vassarstats.net
Supplemental Data
References
- 1.Lammert M., Friedman J.M., Kluwe L., Mautner V.F. Prevalence of neurofibromatosis 1 in German children at elementary school enrollment. Arch. Dermatol. 2005;141:71–74. doi: 10.1001/archderm.141.1.71. [DOI] [PubMed] [Google Scholar]
- 2.Evans D.G., Howard E., Giblin C., Clancy T., Spencer H., Huson S.M., Lalloo F. Birth incidence and prevalence of tumor-prone syndromes: estimates from a UK family genetic register service. Am. J. Med. Genet. A. 2010;152A:327–332. doi: 10.1002/ajmg.a.33139. [DOI] [PubMed] [Google Scholar]
- 3.Uusitalo E., Leppävirta J., Koffert A., Suominen S., Vahtera J., Vahlberg T., Pöyhönen M., Peltonen J., Peltonen S. Incidence and mortality of neurofibromatosis: a total population study in Finland. J. Invest. Dermatol. 2015;135:904–906. doi: 10.1038/jid.2014.465. [DOI] [PubMed] [Google Scholar]
- 4.Ballester R., Marchuk D., Boguski M., Saulino A., Letcher R., Wigler M., Collins F. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell. 1990;63:851–859. doi: 10.1016/0092-8674(90)90151-4. [DOI] [PubMed] [Google Scholar]
- 5.DeClue J.E., Cohen B.D., Lowy D.R. Identification and characterization of the neurofibromatosis type 1 protein product. Proc. Natl. Acad. Sci. USA. 1991;88:9914–9918. doi: 10.1073/pnas.88.22.9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peltonen S., Pöyhönen M. Clinical diagnosis and atypical forms of NF1. In: Upadhyaya M., Cooper D.N., editors. Neurofibromatosis Type 1. Molecular and Cellular Biology. Springer-Verlag Berlin Heidelberg; 2012. pp. 17–30. [Google Scholar]
- 7.Huson S.M., Harper P.S., Compston D.A. Von Recklinghausen neurofibromatosis. A clinical and population study in south-east Wales. Brain. 1988;111:1355–1381. doi: 10.1093/brain/111.6.1355. [DOI] [PubMed] [Google Scholar]
- 8.National Institutes of Health Consensus Development Conference Neurofibromatosis. Conference statement. Arch. Neurol. 1988;45:575–578. [PubMed] [Google Scholar]
- 9.Brems H., Chmara M., Sahbatou M., Denayer E., Taniguchi K., Kato R., Somers R., Messiaen L., De Schepper S., Fryns J.P. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat. Genet. 2007;39:1120–1126. doi: 10.1038/ng2113. [DOI] [PubMed] [Google Scholar]
- 10.Messiaen L., Yao S., Brems H., Callens T., Sathienkijkanchai A., Denayer E., Spencer E., Arn P., Babovic-Vuksanovic D., Bay C. Clinical and mutational spectrum of neurofibromatosis type 1-like syndrome. JAMA. 2009;302:2111–2118. doi: 10.1001/jama.2009.1663. [DOI] [PubMed] [Google Scholar]
- 11.Kehrer-Sawatzki H., Mautner V.F., Cooper D.N. Emerging genotype-phenotype relationships in patients with large NF1 deletions. Hum. Genet. 2017;136:349–376. doi: 10.1007/s00439-017-1766-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Upadhyaya M., Huson S.M., Davies M., Thomas N., Chuzhanova N., Giovannini S., Evans D.G., Howard E., Kerr B., Griffiths S. An absence of cutaneous neurofibromas associated with a 3-bp inframe deletion in exon 17 of the NF1 gene (c.2970-2972 delAAT): evidence of a clinically significant NF1 genotype-phenotype correlation. Am. J. Hum. Genet. 2007;80:140–151. doi: 10.1086/510781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinna V., Lanari V., Daniele P., Consoli F., Agolini E., Margiotti K., Bottillo I., Torrente I., Bruselles A., Fusilli C. p.Arg1809Cys substitution in neurofibromin is associated with a distinctive NF1 phenotype without neurofibromas. Eur. J. Hum. Genet. 2015;23:1068–1071. doi: 10.1038/ejhg.2014.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rojnueangnit K., Xie J., Gomes A., Sharp A., Callens T., Chen Y., Liu Y., Cochran M., Abbott M.A., Atkin J. High incidence of Noonan syndrome features including short stature and pulmonic stenosis in patients carrying NF1 missense mutations affecting p.Arg1809: genotype-phenotype correlation. Hum. Mutat. 2015;36:1052–1063. doi: 10.1002/humu.22832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pulst S.M., Riccardi V.M., Fain P., Korenberg J.R. Familial spinal neurofibromatosis: clinical and DNA linkage analysis. Neurology. 1991;41:1923–1927. doi: 10.1212/wnl.41.12.1923. [DOI] [PubMed] [Google Scholar]
- 16.Burkitt Wright E.M., Sach E., Sharif S., Quarrell O., Carroll T., Whitehouse R.W., Upadhyaya M., Huson S.M., Evans D.G.R. Can the diagnosis of NF1 be excluded clinically? A lack of pigmentary findings in families with spinal neurofibromatosis demonstrates a limitation of clinical diagnosis. J. Med. Genet. 2013;50:606–613. doi: 10.1136/jmedgenet-2013-101648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruggieri M., Polizzi A., Spalice A., Salpietro V., Caltabiano R., D’Orazi V., Pavone P., Pirrone C., Magro G., Platania N. The natural history of spinal neurofibromatosis: a critical review of clinical and genetic features. Clin. Genet. 2015;87:401–410. doi: 10.1111/cge.12498. [DOI] [PubMed] [Google Scholar]
- 18.Kluwe L., Tatagiba M., Fünsterer C., Mautner V.F. NF1 mutations and clinical spectrum in patients with spinal neurofibromas. J. Med. Genet. 2003;40:368–371. doi: 10.1136/jmg.40.5.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Messiaen L., Riccardi V., Peltonen J., Maertens O., Callens T., Karvonen S.L., Leisti E.L., Koivunen J., Vandenbroucke I., Stephens K., Pöyhönen M. Independent NF1 mutations in two large families with spinal neurofibromatosis. J. Med. Genet. 2003;40:122–126. doi: 10.1136/jmg.40.2.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Upadhyaya M., Spurlock G., Kluwe L., Chuzhanova N., Bennett E., Thomas N., Guha A., Mautner V. The spectrum of somatic and germline NF1 mutations in NF1 patients with spinal neurofibromas. Neurogenetics. 2009;10:251–263. doi: 10.1007/s10048-009-0178-0. [DOI] [PubMed] [Google Scholar]
- 21.Pascual-Castroviejo I., Pascual-Pascual S.I., Velazquez-Fragua R., Botella P., Viaño J. Familial spinal neurofibromatosis. Neuropediatrics. 2007;38:105–108. doi: 10.1055/s-2007-985136. [DOI] [PubMed] [Google Scholar]
- 22.Li K., Turner A.N., Chen M., Brosius S.N., Schoeb T.R., Messiaen L.M., Bedwell D.M., Zinn K.R., Anastasaki C., Gutmann D.H. Mice with missense and nonsense NF1 mutations display divergent phenotypes compared with human neurofibromatosis type I. Dis. Model. Mech. 2016;9:759–767. doi: 10.1242/dmm.025783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toonen J.A., Anastasaki C., Smithson L.J., Gianino S.M., Li K., Kesterson R.A., Gutmann D.H. NF1 germline mutation differentially dictates optic glioma formation and growth in neurofibromatosis-1. Hum. Mol. Genet. 2016;25:1703–1713. doi: 10.1093/hmg/ddw039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nellhaus G. Head circumference from birth to eighteen years. Practical composite international and interracial graphs. Pediatrics. 1968;41:106–114. [PubMed] [Google Scholar]
- 25.Nyström A.M., Ekvall S., Strömberg B., Holmström G., Thuresson A.C., Annerén G., Bondeson M.L. A severe form of Noonan syndrome and autosomal dominant café-au-lait spots - evidence for different genetic origins. Acta Paediatr. 2009;98:693–698. doi: 10.1111/j.1651-2227.2008.01170.x. [DOI] [PubMed] [Google Scholar]
- 26.Ekvall S., Sjörs K., Jonzon A., Vihinen M., Annerén G., Bondeson M.L. Novel association of neurofibromatosis type 1-causing mutations in families with neurofibromatosis-Noonan syndrome. Am. J. Med. Genet. A. 2014;164A:579–587. doi: 10.1002/ajmg.a.36313. [DOI] [PubMed] [Google Scholar]
- 27.Santoro C., Maietta A., Giugliano T., Melis D., Perrotta S., Nigro V., Piluso G. Arg(1809) substitution in neurofibromin: further evidence of a genotype-phenotype correlation in neurofibromatosis type 1. Eur. J. Hum. Genet. 2015;23:1460–1461. doi: 10.1038/ejhg.2015.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huson S.M., Compston D.A., Clark P., Harper P.S. A genetic study of von Recklinghausen neurofibromatosis in south east Wales. I. Prevalence, fitness, mutation rate, and effect of parental transmission on severity. J. Med. Genet. 1989;26:704–711. doi: 10.1136/jmg.26.11.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huson S.M., Compston D.A., Harper P.S. A genetic study of von Recklinghausen neurofibromatosis in south east Wales. II. Guidelines for genetic counselling. J. Med. Genet. 1989;26:712–721. doi: 10.1136/jmg.26.11.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Listernick R., Charrow J., Greenwald M., Mets M. Natural history of optic pathway tumors in children with neurofibromatosis type 1: a longitudinal study. J. Pediatr. 1994;125:63–66. doi: 10.1016/s0022-3476(94)70122-9. [DOI] [PubMed] [Google Scholar]
- 31.Van Es S., North K.N., McHugh K., De Silva M. MRI findings in children with neurofibromatosis type 1: a prospective study. Pediatr. Radiol. 1996;26:478–487. doi: 10.1007/BF01377205. [DOI] [PubMed] [Google Scholar]
- 32.Friedman J.M., Birch P.H. Type 1 neurofibromatosis: a descriptive analysis of the disorder in 1,728 patients. Am. J. Med. Genet. 1997;70:138–143. doi: 10.1002/(sici)1096-8628(19970516)70:2<138::aid-ajmg7>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 33.Cnossen M.H., de Goede-Bolder A., van den Broek K.M., Waasdorp C.M., Oranje A.P., Stroink H., Simonsz H.J., van den Ouweland A.M., Halley D.J., Niermeijer M.F. A prospective 10 year follow up study of patients with neurofibromatosis type 1. Arch. Dis. Child. 1998;78:408–412. doi: 10.1136/adc.78.5.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGaughran J.M., Harris D.I., Donnai D., Teare D., MacLeod R., Westerbeek R., Kingston H., Super M., Harris R., Evans D.G. A clinical study of type 1 neurofibromatosis in north west England. J. Med. Genet. 1999;36:197–203. [PMC free article] [PubMed] [Google Scholar]
- 35.Thakkar S.D., Feigen U., Mautner V.F. Spinal tumours in neurofibromatosis type 1: an MRI study of frequency, multiplicity and variety. Neuroradiology. 1999;41:625–629. doi: 10.1007/s002340050814. [DOI] [PubMed] [Google Scholar]
- 36.Lin A.E., Birch P.H., Korf B.R., Tenconi R., Niimura M., Poyhonen M., Armfield Uhas K., Sigorini M., Virdis R., Romano C. Cardiovascular malformations and other cardiovascular abnormalities in neurofibromatosis 1. Am. J. Med. Genet. 2000;95:108–117. doi: 10.1002/1096-8628(20001113)95:2<108::aid-ajmg4>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 37.Blazo M.A., Lewis R.A., Chintagumpala M.M., Frazier M., McCluggage C., Plon S.E. Outcomes of systematic screening for optic pathway tumors in children with Neurofibromatosis Type 1. Am. J. Med. Genet. A. 2004;127A:224–229. doi: 10.1002/ajmg.a.20650. [DOI] [PubMed] [Google Scholar]
- 38.Khosrotehrani K., Bastuji-Garin S., Riccardi V.M., Birch P., Friedman J.M., Wolkenstein P. Subcutaneous neurofibromas are associated with mortality in neurofibromatosis 1: a cohort study of 703 patients. Am. J. Med. Genet. A. 2005;132A:49–53. doi: 10.1002/ajmg.a.30394. [DOI] [PubMed] [Google Scholar]
- 39.Plotkin S.R., Bredella M.A., Cai W., Kassarjian A., Harris G.J., Esparza S., Merker V.L., Munn L.L., Muzikansky A., Askenazi M. Quantitative assessment of whole-body tumor burden in adult patients with neurofibromatosis. PLoS ONE. 2012;7:e35711. doi: 10.1371/journal.pone.0035711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blanchard G., Lafforgue M.P., Lion-François L., Kemlin I., Rodriguez D., Castelnau P., Carneiro M., Meyer P., Rivier F., Barbarot S., Chaix Y., NF France network Systematic MRI in NF1 children under six years of age for the diagnosis of optic pathway gliomas. Study and outcome of a French cohort. Eur. J. Paediatr. Neurol. 2016;20:275–281. doi: 10.1016/j.ejpn.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 41.Messiaen L.M., Callens T., Mortier G., Beysen D., Vandenbroucke I., Van Roy N., Speleman F., Paepe A.D. Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum. Mutat. 2000;15:541–555. doi: 10.1002/1098-1004(200006)15:6<541::AID-HUMU6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 42.Messiaen L.M., Wimmer K. Mutation analysis of the NF1 gene by cDNA-based sequencing of the coding region. In: Cunha K.S.G., Geller M., editors. Advances in Neurofibromatosis Research. Nova Science Publishers, Inc.; 2012. pp. 89–108. [Google Scholar]
- 43.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou X., Edmonson M.N., Wilkinson M.R., Patel A., Wu G., Liu Y., Li Y., Zhang Z., Rusch M.C., Parker M. Exploring genomic alteration in pediatric cancer using ProteinPaint. Nat. Genet. 2016;48:4–6. doi: 10.1038/ng.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cunha K.S., Oliveira N.S., Fausto A.K., de Souza C.C., Gros A., Bandres T., Idrissi Y., Merlio J.-P., de Moura Neto R.S., Silva R. Hybridization capture-based next-generation sequencing to evaluate coding sequence and deep intronic mutations in the NF1 gene. Genes (Basel) 2016;7:133. doi: 10.3390/genes7120133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bertola D.R., Pereira A.C., Passetti F., de Oliveira P.S., Messiaen L., Gelb B.D., Kim C.A., Krieger J.E. Neurofibromatosis-Noonan syndrome: molecular evidence of the concurrence of both disorders in a patient. Am. J. Med. Genet. A. 2005;136:242–245. doi: 10.1002/ajmg.a.30813. [DOI] [PubMed] [Google Scholar]
- 47.Tonsgard J.H., Kwak S.M., Short M.P., Dachman A.H. CT imaging in adults with neurofibromatosis-1: frequent asymptomatic plexiform lesions. Neurology. 1998;50:1755–1760. doi: 10.1212/wnl.50.6.1755. [DOI] [PubMed] [Google Scholar]
- 48.Waggoner D.J., Towbin J., Gottesman G., Gutmann D.H. Clinic-based study of plexiform neurofibromas in neurofibromatosis 1. Am. J. Med. Genet. 2000;92:132–135. [PubMed] [Google Scholar]
- 49.Ferner R.E., Huson S.M., Thomas N., Moss C., Willshaw H., Evans D.G., Upadhyaya M., Towers R., Gleeson M., Steiger C., Kirby A. Guidelines for the diagnosis and management of individuals with neurofibromatosis 1. J. Med. Genet. 2007;44:81–88. doi: 10.1136/jmg.2006.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duong T.A., Sbidian E., Valeyrie-Allanore L., Vialette C., Ferkal S., Hadj-Rabia S., Glorion C., Lyonnet S., Zerah M., Kemlin I. Mortality associated with neurofibromatosis 1: a cohort study of 1895 patients in 1980-2006 in France. Orphanet J. Rare Dis. 2011;6:18. doi: 10.1186/1750-1172-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Evans D.G., Baser M.E., McGaughran J., Sharif S., Howard E., Moran A. Malignant peripheral nerve sheath tumours in neurofibromatosis 1. J. Med. Genet. 2002;39:311–314. doi: 10.1136/jmg.39.5.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tucker T., Wolkenstein P., Revuz J., Zeller J., Friedman J.M. Association between benign and malignant peripheral nerve sheath tumors in NF1. Neurology. 2005;65:205–211. doi: 10.1212/01.wnl.0000168830.79997.13. [DOI] [PubMed] [Google Scholar]
- 53.McCaughan J.A., Holloway S.M., Davidson R., Lam W.W. Further evidence of the increased risk for malignant peripheral nerve sheath tumour from a Scottish cohort of patients with neurofibromatosis type 1. J. Med. Genet. 2007;44:463–466. doi: 10.1136/jmg.2006.048140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Korf B.R. Plexiform neurofibromas. Am. J. Med. Genet. 1999;89:31–37. doi: 10.1002/(sici)1096-8628(19990326)89:1<31::aid-ajmg7>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 55.Prada C.E., Rangwala F.A., Martin L.J., Lovell A.M., Saal H.M., Schorry E.K., Hopkin R.J. Pediatric plexiform neurofibromas: impact on morbidity and mortality in neurofibromatosis type 1. J. Pediatr. 2012;160:461–467. doi: 10.1016/j.jpeds.2011.08.051. [DOI] [PubMed] [Google Scholar]
- 56.Jahraus C.D., Tarbell N.J. Optic pathway gliomas. Pediatr. Blood Cancer. 2006;46:586–596. doi: 10.1002/pbc.20655. [DOI] [PubMed] [Google Scholar]
- 57.Listernick R., Ferner R.E., Liu G.T., Gutmann D.H. Optic pathway gliomas in neurofibromatosis-1: controversies and recommendations. Ann. Neurol. 2007;61:189–198. doi: 10.1002/ana.21107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nicolin G., Parkin P., Mabbott D., Hargrave D., Bartels U., Tabori U., Rutka J., Buncic J.R., Bouffet E. Natural history and outcome of optic pathway gliomas in children. Pediatr. Blood Cancer. 2009;53:1231–1237. doi: 10.1002/pbc.22198. [DOI] [PubMed] [Google Scholar]
- 59.Rasmussen S.A., Friedman J.M. NF1 gene and neurofibromatosis 1. Am. J. Epidemiol. 2000;151:33–40. doi: 10.1093/oxfordjournals.aje.a010118. [DOI] [PubMed] [Google Scholar]
- 60.Uusitalo E., Rantanen M., Kallionpää R.A., Pöyhönen M., Leppävirta J., Ylä-Outinen H., Riccardi V.M., Pukkala E., Pitkäniemi J., Peltonen S., Peltonen J. Distinctive cancer associations in patients with neurofibromatosis type 1. J. Clin. Oncol. 2016;34:1978–1986. doi: 10.1200/JCO.2015.65.3576. [DOI] [PubMed] [Google Scholar]
- 61.Brems H., Beert E., de Ravel T., Legius E. Mechanisms in the pathogenesis of malignant tumours in neurofibromatosis type 1. Lancet Oncol. 2009;10:508–515. doi: 10.1016/S1470-2045(09)70033-6. [DOI] [PubMed] [Google Scholar]
- 62.Patil S., Chamberlain R.S. Neoplasms associated with germline and somatic NF1 gene mutations. Oncologist. 2012;17:101–116. doi: 10.1634/theoncologist.2010-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sung L., Anderson J.R., Arndt C., Raney R.B., Meyer W.H., Pappo A.S. Neurofibromatosis in children with Rhabdomyosarcoma: a report from the Intergroup Rhabdomyosarcoma study IV. J. Pediatr. 2004;144:666–668. doi: 10.1016/j.jpeds.2004.02.026. [DOI] [PubMed] [Google Scholar]
- 64.Crucis A., Richer W., Brugières L., Bergeron C., Marie-Cardine A., Stephan J.L., Girard P., Corradini N., Munzer M., Lacour B. Rhabdomyosarcomas in children with neurofibromatosis type I: A national historical cohort. Pediatr. Blood Cancer. 2015;62:1733–1738. doi: 10.1002/pbc.25556. [DOI] [PubMed] [Google Scholar]
- 65.Chang T., Shannon K. NF1 mutations in hematologic cancers. In: Upadhyaya M., Cooper D.N., editors. Neurofibromatosis Type 1. Molecular and Cellular Biology. Springer-Verlag Berlin Heidelberg; 2012. pp. 469–485. [Google Scholar]
- 66.Seminog O.O., Goldacre M.J. Risk of benign tumours of nervous system, and of malignant neoplasms, in people with neurofibromatosis: population-based record-linkage study. Br. J. Cancer. 2013;108:193–198. doi: 10.1038/bjc.2012.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McPherson J.R., Ong C.K., Ng C.C., Rajasegaran V., Heng H.L., Yu W.S., Tan B.K., Madhukumar P., Teo M.C., Ngeow J. Whole-exome sequencing of breast cancer, malignant peripheral nerve sheath tumor and neurofibroma from a patient with neurofibromatosis type 1. Cancer Med. 2015;4:1871–1878. doi: 10.1002/cam4.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stevenson D.A., Yang F.C. The musculoskeletal phenotype of the RASopathies. Am. J. Med. Genet. C. Semin. Med. Genet. 2011;157C:90–103. doi: 10.1002/ajmg.c.30296. [DOI] [PubMed] [Google Scholar]
- 69.Vargiami E., Zafeiriou D.I., Bantouraki M. Ulnar hypoplasia and neurofibromatosis type I. J. Pediatr. 2004;145:859. doi: 10.1016/j.jpeds.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 70.Krab L.C., de Goede-Bolder A., Aarsen F.K., Pluijm S.M., Bouman M.J., van der Geest J.N., Lequin M., Catsman C.E., Arts W.F., Kushner S.A. Effect of simvastatin on cognitive functioning in children with neurofibromatosis type 1: a randomized controlled trial. JAMA. 2008;300:287–294. doi: 10.1001/jama.300.3.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mautner V.F., Kluwe L., Friedrich R.E., Roehl A.C., Bammert S., Högel J., Spöri H., Cooper D.N., Kehrer-Sawatzki H. Clinical characterisation of 29 neurofibromatosis type-1 patients with molecularly ascertained 1.4 Mb type-1 NF1 deletions. J. Med. Genet. 2010;47:623–630. doi: 10.1136/jmg.2009.075937. [DOI] [PubMed] [Google Scholar]
- 72.Easton D.F., Ponder M.A., Huson S.M., Ponder B.A. An analysis of variation in expression of neurofibromatosis (NF) type 1 (NF1): evidence for modifying genes. Am. J. Hum. Genet. 1993;53:305–313. [PMC free article] [PubMed] [Google Scholar]
- 73.Szudek J., Joe H., Friedman J.M. Analysis of intrafamilial phenotypic variation in neurofibromatosis 1 (NF1) Genet. Epidemiol. 2002;23:150–164. doi: 10.1002/gepi.1129. [DOI] [PubMed] [Google Scholar]
- 74.Sabbagh A., Pasmant E., Laurendeau I., Parfait B., Barbarot S., Guillot B., Combemale P., Ferkal S., Vidaud M., Aubourg P., members of the NF France Network Unravelling the genetic basis of variable clinical expression in neurofibromatosis 1. Hum. Mol. Genet. 2009;18:2768–2778. doi: 10.1093/hmg/ddp212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.DeBella K., Szudek J., Friedman J.M. Use of the national institutes of health criteria for diagnosis of neurofibromatosis 1 in children. Pediatrics. 2000;105:608–614. doi: 10.1542/peds.105.3.608. [DOI] [PubMed] [Google Scholar]
- 76.Williams V.C., Lucas J., Babcock M.A., Gutmann D.H., Korf B., Maria B.L. Neurofibromatosis type 1 revisited. Pediatrics. 2009;123:124–133. doi: 10.1542/peds.2007-3204. [DOI] [PubMed] [Google Scholar]
- 77.Messiaen L., Xie J. NF1 germline and somatic mosaicism. In: Upadhyaya M., Cooper D.N., editors. Neurofibromatosis Type 1. Molecular and Cellular Biology. Springer-Verlag Berlin Heidelberg; 2012. pp. 151–172. [Google Scholar]
- 78.Messiaen L.M., Wimmer K. NF1 mutational spectrum. In: Kaufmann D., editor. Neurofibromatoses. Karger; Monogr. Hum. Genet. Basel: 2008. pp. 63–77. [Google Scholar]
- 79.Pasmant E., Vidaud D., Wolkenstein P. Modifier genes in NF1. In: Upadhyaya M., Cooper D.N., editors. Neurofibromatosis Type 1. Molecular and Cellular Biology. Springer-Verlag Berlin Heidelberg; 2012. pp. 269–285. [Google Scholar]
- 80.Terry A.R., Barker F.G., 2nd, Leffert L., Bateman B.T., Souter I., Plotkin S.R. Neurofibromatosis type 1 and pregnancy complications: a population-based study. Am. J. Obstet. Gynecol. 2013;209:46.e1–46.e8. doi: 10.1016/j.ajog.2013.03.029. [DOI] [PubMed] [Google Scholar]
- 81.Pasmant E., Sabbagh A., Vidaud M., Bièche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25:444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 82.Pemov A., Sung H., Hyland P.L., Sloan J.L., Ruppert S.L., Baldwin A.M., Boland J.F., Bass S.E., Lee H.J., Jones K.M., NISC Comparative Sequencing Program Genetic modifiers of neurofibromatosis type 1-associated café-au-lait macule count identified using multi-platform analysis. PLoS Genet. 2014;10:e1004575. doi: 10.1371/journal.pgen.1004575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fahsold R., Hoffmeyer S., Mischung C., Gille C., Ehlers C., Kücükceylan N., Abdel-Nour M., Gewies A., Peters H., Kaufmann D. Minor lesion mutational spectrum of the entire NF1 gene does not explain its high mutability but points to a functional domain upstream of the GAP-related domain. Am. J. Hum. Genet. 2000;66:790–818. doi: 10.1086/302809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mangoura D., Sun Y., Li C., Singh D., Gutmann D.H., Flores A., Ahmed M., Vallianatos G. Phosphorylation of neurofibromin by PKC is a possible molecular switch in EGF receptor signaling in neural cells. Oncogene. 2006;25:735–745. doi: 10.1038/sj.onc.1209113. [DOI] [PubMed] [Google Scholar]
- 85.Rost B., Yachdav G., Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32 doi: 10.1093/nar/gkh377. W321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ng P.C., Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kamat M.A., Bacolla A., Cooper D.N., Chuzhanova N. A role for non-B DNA forming sequences in mediating microlesions causing human inherited disease. Hum. Mutat. 2016;37:65–73. doi: 10.1002/humu.22917. [DOI] [PubMed] [Google Scholar]
- 88.Sabbagh A., Pasmant E., Imbard A., Luscan A., Soares M., Blanché H., Laurendeau I., Ferkal S., Vidaud M., Pinson S. NF1 molecular characterization and neurofibromatosis type I genotype-phenotype correlation: the French experience. Hum. Mutat. 2013;34:1510–1518. doi: 10.1002/humu.22392. [DOI] [PubMed] [Google Scholar]
- 89.van Minkelen R., van Bever Y., Kromosoeto J.N., Withagen-Hermans C.J., Nieuwlaat A., Halley D.J., van den Ouweland A.M. A clinical and genetic overview of 18 years neurofibromatosis type 1 molecular diagnostics in the Netherlands. Clin. Genet. 2014;85:318–327. doi: 10.1111/cge.12187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.