Abstract
Atrial fibrillation (AF) is a common cardiac arrhythmia and a major risk factor for stroke, heart failure, and premature death. The pathogenesis of AF remains poorly understood, which contributes to the current lack of highly effective treatments. To understand the genetic variation and biology underlying AF, we undertook a genome-wide association study (GWAS) of 6,337 AF individuals and 61,607 AF-free individuals from Norway, including replication in an additional 30,679 AF individuals and 278,895 AF-free individuals. Through genotyping and dense imputation mapping from whole-genome sequencing, we tested almost nine million genetic variants across the genome and identified seven risk loci, including two novel loci. One novel locus (lead single-nucleotide variant [SNV] rs12614435; p = 6.76 × 10−18) comprised intronic and several highly correlated missense variants situated in the I-, A-, and M-bands of titin, which is the largest protein in humans and responsible for the passive elasticity of heart and skeletal muscle. The other novel locus (lead SNV rs56202902; p = 1.54 × 10−11) covered a large, gene-dense chromosome 1 region that has previously been linked to cardiac conduction. Pathway and functional enrichment analyses suggested that many AF-associated genetic variants act through a mechanism of impaired muscle cell differentiation and tissue formation during fetal heart development.
Keywords: heart, fetal, atrial fibrillation, GWAS, pathway, genetic risk score, cardiomyopathy, TTN, DMRTA2, CDKN2C
Introduction
Atrial fibrillation (AF) is characterized by high-frequency disordered electrical activity of the atria and is the most common cardiac arrhythmia encountered in clinical practice. At present, more than 30 million people are affected worldwide, including approximately six million in the United States alone, a number that is projected to double over the next two decades.1, 2 AF is a major risk factor for stroke, heart failure, and premature death.3 Existing treatment regimens are limited in their effectiveness and are very rarely curative, which makes AF a major public healthcare burden with considerable associated costs.4
In addition to having conventional risk factors, such as advanced age, obesity, and hypertension,5 AF has a substantial heritable component. In a large twin study of people of Scandinavian origin, the total heritability of AF was estimated to be 62%,6 and in the Framingham Heart Study, the risk of developing AF increased with decreasing age of onset of affected family members.7
Because genotypes are fixed during an individual’s lifetime, genetic discoveries have the potential to define causal risk factors, pathways, and therapeutic targets and hence help improve our understanding of the pathophysiology underlying AF.
Although rare mutations in genes encoding cardiac ion channels, gap junctions, and signaling molecules have been shown to contribute to AF risk in individual families,8, 9, 10 AF is generally considered a complex and polygenic disease. Despite this fact, and given the high degree of heritability, only 16 loci were associated with AF in the population at the time of analysis, and for many of these loci, the biological and therapeutic implications remain elusive.11, 12, 13, 14 Two studies have recently identified additional loci.15, 16
The Nord-Trøndelag Health (HUNT) Study in Norway has been ongoing since 1984 and, with more than 120,000 participants included over three decades,17 is one of the most comprehensive population-based health surveys ever performed.
After genotyping of more than 70,000 HUNT Study participants and dense imputation from whole-genome sequencing, we performed a genome-wide association study (GWAS) of AF and identified two risk loci. We further conducted several additional analyses, including fine-mapping of associated loci, and performed gene-set and functional enrichment analyses to highlight pathways and tissues that reinforce and expand our understanding of the biological processes underlying AF.
Subjects and Methods
Discovery Cohort
Study Participants and Definition of Phenotypes
We included individuals from the HUNT Study, which is an ongoing population-based health survey of more than 120,000 individuals in the county of Nord-Trøndelag in Norway.17 The database contains results on clinical examinations, including personal and family medical histories, and is supplemented by cross-referencing with administrative healthcare registries at the regional level. On the basis of hospital, outpatient, and emergency-room discharge diagnoses classified according to the International Classification of Disease, Ninth Revision (ICD-9; 1987–1999) or Tenth Revision (ICD-10; 1999–2016), we identified individuals with AF (ICD-9 code 427.3; ICD-10 code I48), any type of ischemic or non-ischemic cardiomyopathy (ICD-9 codes 425 and 414.8; ICD-10 codes I42 and I25.5), and dilated cardiomyopathy (DCM; ICD-10: I42). The last of these was based on the ICD-10 system only because there is no specific ICD-9 code for DCM. Participation in the HUNT Study is based on informed consent, and the study has been approved by the Data Inspectorate and the Regional Ethics Committee for Medical Research in Norway.
Quality Control of Genotype Data
A total of 69,037 samples from the HUNT Biobank in Norway were genotyped at 449,453 autosomal variants with a combined exome and GWAS chip array (HumanCoreExome-12 v.1.0, Illumina). Genotype calling was performed with GenTrain v.2.0 in GenomeStudio v.2011.1 (Illumina). Samples with <98% genotype calls, evidence of gender discrepancy, duplicates, an excess of ±6 SD of the heterozygosity rate, and individuals with non-Norwegian ancestry identified by plotting the first ten genotype-driven principal components (PCs) were excluded from further analysis (n = 2,458 [3.48%]). Variant-level quality control consisted of excluding 5,007 variants that met any of the following criteria: a cluster separation score < 0.3, <98% genotyping threshold, duplicate markers, or deviation from Hardy-Weinberg equilibrium (p < 1 × 10−7).
Genotype Imputation
The study samples were phased with SHAPEIT2.18 Genotype imputation with the Haplotype Reference Consortium19 reference panel, which included 1,023 low-pass whole-genome-sequenced HUNT samples, was conducted on the Michigan Imputation Server20 with minimac3.21
Single-Marker Association Testing
We used a linear mixed-effects model to account for cryptic population structure and relatedness to model the association between genotyped variants (when available for a particular position) or imputed variants (dosages) and AF, as implemented in BOLT-LMM.22 We assumed an additive allelic effect, and we looked at only variants with a minor allele frequency (MAF) > 0.5%. We used the Wald test in logistic regression, as implemented in EPACTS, to obtain odds ratios (ORs) and confidence intervals (CIs) for markers that reached genome-wide significance (p < 5 × 10−8). All models were adjusted for birth year, sex, genotyping batch, and PCs 1–4.
Gene-Based Burden Test
After estimating a kinship matrix, we performed a gene-based burden test by using SKAT (sequence kernel association test) and EMMAX (efficient mixed-model association expedited),23 as implemented in EPACTS, to account for relatedness and population structure. All protein-altering variants, as annotated by SnpEff,24 with a MAF < 5% were included. As a result of high computational load, the sample size was reduced to all AF individuals and a subset of the AF-free individuals. For each AF individual, we selected three AF-free individuals by using propensity-score matching based on birth year, sex, genotyping batch, and PCs 1–4. The model was adjusted for birth year, sex, genotyping batch, and PCs 1–4.
Replication Cohorts
Michigan Genomics Initiative
DNA from blood samples of surgical patients at the University of Michigan Health System was genotyped on the Illumina HumanCore Exome array. Genotypes of the Haplotype Reference Consortium were imputed into the phased Michigan Genomics Initiative genotypes, resulting in dense mapping of over 7.7 million common variants (MAF > 1%). Individuals were defined as having AF if they had at least two electronic health record (EHR) encounters with ICD-9 code 427.31. AF-free individuals were defined as persons without ICD-9 billing code 427.31 or any other billing codes related to cardiac arrhythmias or conduction disorders. We performed a genome-wide association analysis of AF in 924 AF individuals and 11,037 AF-free individuals, all of European ancestry, by using the Firth-bias-corrected logistic likelihood-ratio test,25 adjusted for age, sex, and PCs 1–4.
DiscovEHR Collaboration Cohort
The DiscovEHR study comprised a total of 50,726 adult individuals enrolled in the MyCode Community Health Initiative of the Geisinger Health System. Participants were recruited from outpatient primary care and specialty clinics from 2007 to 2016, and available EHR data covered a median of 14 years of clinical care. Samples were processed at Illumina and genotyped on the OmniExpressExome array. GWAS variants with a MAF > 1% were phased with SHAPEIT2, and IMPUTE2 was used for imputing to the 1000 Genomes Project cosmopolitan dataset (June 2014 version). Variants with an info score < 0.7 or a genotyping call rate < 99% were excluded from downstream analysis. AF and flutter case individuals (n = 5,451) were defined as European-ancestry DiscovEHR participants with at least one EHR problem-list entry or at least two diagnosis-code entries for two separate clinical encounters on separate calendar days for ICD-9 code 427.3 (AF and flutter). Control individuals (n = 30,235) were defined as European-ancestry individuals with no EHR diagnosis-code entries (problem list or encounter codes) for ICD-9 code 427 (cardiac dysrhythmias). For each variant, we tested best-guess genotypes for association with AF case-control status by using logistic regression; we included age, age2, sex, and ancestry PCs 1–4 as covariates.
UK Biobank
From 2006 to 2010, the UK Biobank recruited 40- to 69-year old individuals who were registered with a general medical practitioner within the UK National Health Service.26 We defined individuals as having AF if they had any hospital ICD-9 or ICD-10 code specific to AF or atrial flutter (427.3 or I48, respectively). All other persons were used as control individuals. We restricted analyses to those genotyped persons of European ancestry who passed the UK Biobank’s quality control. Individuals who had withdrawn consent were excluded. After exclusion, the total numbers of case and control individuals were 4,407 and 115,878, respectively. We modeled the association between genotypes of interest and AF by using a logistic regression (SNPTEST v.2.5.2) adjusted for genotype batch and PCs 1–10 under the assumption of an additive genetic model.
The Tromsø Study
We selected 1,158 AF individuals and 5,393 AF-free individuals from the ongoing Tromsø Study (Tromsø 4) for follow-up. Tromsø 4 is a population-based study of more than 27,000 people enrolled from the municipality of Tromsø in Norway between 1994 and 1995.27 Occurrences of AF were identified by the registry of hospital discharge diagnosis at the University Hospital of North Norway (diagnoses from hospitalizations and outpatient clinics) and by the National Causes of Death registry with ICD-9 codes 427.0–427.99 and ICD-10 codes I47–I48. All diagnoses of AF were confirmed by electrocardiography (ECG). DNA samples of the Tromsø Study were extracted from venous blood and genotyped with the Illumina HumanCoreExome 12v.1.1 array, and genotypes of the 1000 Genomes Project (phase 3 release 5; Minimac3) were imputed into their phased haplotypes (SHAPEIT2). Imputed variants with R2 ≤ 0.3 were excluded. Association tests of candidate single-nucleotide variants (SNVs) were performed with a logistic Wald test, which added sex and birth year as covariates.
The Mount Sinai BioMe Biobank
The Mount Sinai BioMe Biobank (BioMe) is an ongoing, prospective hospital- and outpatient-based population research program operated by the Charles Bronfman Institute for Personalized Medicine at Mount Sinai and has enrolled over 33,000 participants since September 2007. BioMe is an EHR-linked biobank that integrates research data and clinical-care information for consenting patients at the Mount Sinai Medical Center, New York, which serves diverse local upper-Manhattan communities with broad health disparities. Information on AF, age, and sex was derived from participants’ EHRs. Age was derived from the day of enrollment. BioMe participants were defined as having AF if they had an ICD-9 code specific to AF (427.31) or atrial flutter (427.32), and AF-free individuals were defined as participants who have had ECG but did not have AF or flutter ICD-9 codes. Study participants were genotyped with the Illumina HumanOmniExpressExome-8 v.1.0 BeadChip array and imputed to the 1000 Genomes Project phase 1 reference panel with IMPUTE2. Association testing was carried out under an additive genetic model with SNPTEST 2.4.1 in 291 AF individuals and 860 AF-free individuals of European ancestry with age, sex, and PCs 1–4 as covariates.
DANFIB
Individuals with AF were recruited from eight major hospitals in Copenhagen, Denmark. Diagnoses of AF were verified by ECG. Control individuals were recruited among healthy blood donors. The final study sample comprised 517 unrelated AF individuals and 350 AF-free individuals, all of European ancestry. SNVs of interest were directly genotyped by Kompetitive Allele-Specific PCR. The association between SNVs and AF was modeled with a logistic regression under the assumption of an additive genetic model.
Meta-analyses
The most statistically significant SNV at each novel locus identified in the HUNT discovery cohort was meta-analyzed with results from the replication cohorts by the inverse-variance method implemented in the software package METAL.28
Regional Plots
Regional plots were prepared with LocusZoom.29 The linkage disequilibrium (LD; r2) was calculated on the basis of the available markers in the HUNT discovery cohort.
Estimation of Heritability
To estimate the heritability of AF explained by genetic variants (SNV-based h2) without having to remove related individuals, we employed the method described by Zaitlen et al.30 and implemented it in the GCTA-REML software package.31 All directly genotyped variants that passed quality control (n = 456,297) were extracted and used for (1) constructing a genetic relationship matrix (GRM), (2) constructing an additional GRM on the basis of the first GRM,31 and finally (3) running the REML analysis on the basis of the two GRMs. The heritability explained by AF-associated loci (listed in Table S2) was calculated on the basis of ORs and risk allele frequencies obtained in the HUNT Study discovery sample as described previously.32 For both methods of estimating heritability, we assumed an AF disease prevalence of 2%.
AF Polygenic Risk Score and Power Calculations
To construct a polygenic risk score for AF, we summarized the product of genetic dosages and log(OR) for lead SNVs at all AF-associated loci available in our dataset. For previously published loci, we used the published risk estimates rather than our own to account for inflation in test statistics. To provide an interpretable per-allele risk score, we normalized values to the maximum number of risk alleles observed in our population. Power calculations were performed on the basis of observed allele frequencies and the numbers of case and control individuals as described previously.33
Identification of eQTLs with GTEx Data
We used the Genotype-Tissue Expression (GTEx) Portal v.6p,34 which holds cis-expression quantitative trait loci (cis-eQTL) data of up to 190 million SNVs across 44 tissues, to analyze eQTLs by searching for all AF-associated SNVs identified in our HUNT discovery cohort by using an eQTL significance threshold of p < 1.14 × 10−9 (5 × 10−8 per 44 tissues).
Interpretation of Genome-wide Association Loci with Predicted Gene Functions
We employed Data-Driven Expression Prioritized Integration for Complex Traits (DEPICT)35 to identify (1) the most likely causal gene at associated loci, (2) reconstituted gene sets enriched with AF loci, and (3) tissues and cell types in which genes that form associated loci are highly expressed. DEPICT uses gene expression data obtained from a panel of 77,840 mRNA expression arrays36 together with 14,461 existing gene sets defined on the basis of molecular pathways derived from experimentally verified protein-protein interactions,37 genotype-phenotype relationships from the Mouse Genetics Initiative,38 reactome pathways,39 KEGG pathways,40 and Gene Ontology terms.41 On the basis of similarities across the microarray expression data, DEPICT reconstitutes the 14,461 existing gene sets by assigning each gene in the genome a likelihood of membership in each gene set. Using these precomputed gene sets and a set of trait-associated loci, DEPICT quantifies whether any of the 14,461 reconstituted gene sets are significantly enriched with genes in the associated loci and prioritizes genes that share predicted functions with genes from the other associated loci more often than expected by chance. Additionally, DEPICT uses a set of 37,427 human mRNA microarrays to identify tissues and cell types in which genes from associated loci are highly expressed (all genes residing within a LD of r2 > 0.5 from the lead SNV).35
We ran DEPICT by using the variant with the smallest GWAS p value at each of the two novel loci that were present in DEPICT’s repository of ∼3.9 million SNVs (rs7545860 [p = 3.0 × 10−9] and rs16866373 [p = 2.2 × 10−10], respectively). Additionally, we included the lead SNVs from previously published independent AF loci (Table S2; published copy-number variants and small indels were not included because they were not available in DEPICT).11, 12, 13, 14 For the gene sets most significantly enriched with AF-associated loci (p < 5.0 × 10−5), we computed a weighted pairwise similarity on the basis of the number of overlapping genes for genes with a Z score < 4.75 (corresponding to p < 10−6) for being part of the gene set. For gene sets with no genes with a Z score < 4.75, we included the three most significant genes as suggested previously (Table S10).42
Quantification of Enrichment of Functional and Regulatory Elements at Genome-wide Association Loci
We used GWAS Analysis of Regulatory or Functional Information Enrichment with LD Correction (GARFIELD)43, 44 to systematically characterize the functional, cellular, and regulatory contribution of genetic variation implicated in AF on the basis of GWAS summary statistics from our HUNT discovery cohort and publically available regulatory maps from ENCODE,45 GENCODE,46 and the Roadmap Epigenomics Project.47 In brief, GARFIELD is a non-parametric enrichment analysis framework that takes GWAS summary statistics at various p value thresholds and calculates the fold enrichment of various regulatory elements and then tests them for statistical significance by using permutation testing while accounting for MAF, the distance to the nearest transcription start site, and the number of LD proxies.43
After enriching our GWAS summary statistics with 11 previously published AF loci that did not rise to genome-wide significance in our study (Tables 1 and S2), we computed fold-enrichment statistics at eight GWAS p value thresholds and calculated their significance at the four most significant ones (<1 × 10−8 to <1 × 10−5) by using a greedy permutation step, as suggested previously.44 Multiple-testing corrections were further performed on the basis of the number of annotations effectively used (Table S11), resulting in an experiment-wide p value threshold of 1 × 10−4.
Table 1.
AF-Associated Loci Identified in the HUNT Discovery Study and Replication Studies
| Position (hg19) | rsID | Ref/Alt | Locus |
Discovery (6,337 AF versus 61,607 AF-free Individuals) |
Replication (30,679 AF versus 278,895 AF-free Individuals) |
Combined (37,016 AF versus 340,502 AF-free Individuals) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Freq | OR (95% CI) | p Value | Freq | OR (95% CI) | p Value | OR (95% CI) | p Value | ||||
| Identified in This Study | |||||||||||
| chr1: 50,991,452 | rs56202902 | T/C | DMRTA2-FAF1-CDKN2C-RNF11 | 0.041 | 1.32 (1.20–1.44) | 2.5 × 10−9 | 0.021 | 1.16 (1.07–1.25) | 1.5 × 10−4 | 1.22 (1.14–1.31) | 1.5 × 10−11 |
| chr2: 179,489,726 | rs12614435 | A/G | TTN | 0.156 | 1.20 (1.14–1.27) | 1.1 × 10−10 | 0.155 | 1.09 (1.06–1.11) | 4.6 × 10−11 | 1.10 (1.08–1.13) | 6.8 × 10−18 |
| Known | |||||||||||
| chr1: 154,831,143 | rs34245846 | A/G | KCNN3 | 0.302 | 1.14 (1.09–1.19) | 7.1 × 10−9 | – | – | – | – | – |
| chr1: 170,193,825 | rs72700114 | G/C | PRRX1 | 0.069 | 1.26 (1.17–1.36) | 3.5 × 10−10 | – | – | – | – | – |
| chr4: 111,703,510 | rs77831929 | G/T | PITX2 | 0.180 | 1.41 (1.34–1.48) | 2.2 × 10−39 | – | – | – | – | – |
| chr10: 105,324,774 | rs60572254 | C/T | NEURL | 0.137 | 1.25 (1.19–1.33) | 1.8 × 10−14 | – | – | – | – | – |
| chr16: 73,032,032 | rs4404097 | G/A | ZHFX3 | 0.160 | 1.18 (1.12–1.24) | 6.3 × 10−10 | – | – | – | – | – |
Replication of known loci was not sought. Odds ratios (OR) are provided with respect to the alternative allele. Other abbreviations are as follows: CI, confidence interval; Freq, frequency of the alternative allele; Ref/Alt, reference and alternative alleles; rsID, reference SNP ID number.
We additionally tested for enrichment of AF risk variants in functional domains by using the software Genomic Regulatory Elements and GWAS Overlap Algorithm (GREGOR). GREGOR tests whether index variants (or their high-LD proxies) overlap genome-wide functional domains more often than expected by chance by comparing the index variants with control variants selected to match on allele frequency, the number of high-LD proxies, and distance to the nearest gene.48
Phenome-wide Association Analyses
For all AF risk loci that reached nominal significance (p < 0.05) in the HUNT discovery cohort, we tested the association between the lead SNV for each locus and 709 clinical variables, mainly comprising self-reported health issues and clinical tests, that were available to us at the time of analysis. The association tests were performed with BOLT-LMM,22 including birth year, sex, and PCs 1–4 as covariates.
Results
Discovery of AF-Associated Loci
In the discovery sample, we examined 8,952,551 genotyped or imputed SNVs with a MAF ≥ 0.5% in 67,944 individuals. We tested for an association with AF in 6,337 (9.3%) AF individuals and 61,607 AF-free individuals by using birth year, sex, genotyping batch, and ancestry PCs 1–4 as covariates while accounting for relatedness among the samples.22 The clinical characteristics of the participants in the HUNT Study discovery cohort are summarized in Table S1. Lead SNVs at previously unreported AF risk loci were further evaluated in independent replication cohorts with a combined total of 30,679 additional AF individuals and 278,895 AF-free individuals.
By examining the discovery cohort, we identified two unreported AF risk loci at chromosomal regions 1p32 and 2q31 and replicated 5 of 16 previously reported loci at genome-wide significance (p < 5 × 10−8) (Tables 1 and S2 and Figures S1 and S2). Seven of the 11 remaining known risk loci were replicated at nominal significance (p < 0.05), and all 12 replicated loci had the same direction of effect as the initial reports (p = 0.512 = 2 × 10−4).
The most statistically significant novel association was observed at the 2q31 locus (lead SNV rs12614435; OR = 1.20; 95% CI = 1.14–1.27; p = 1.10 × 10−10). This locus comprised multiple SNVs (Figure 1), including seven highly correlated missense variants (r2 ≥ 0.5; 5.0 × 10−8 > p ≥ 4.8 × 10−10) (Table S3). The associated missense variants fell within the I-, A-, and M-bands of titin (TTN), which is the largest protein in humans and responsible for the passive elasticity of heart and skeletal muscle.49 The lead SNV (rs12614435) at this locus also reached genome-wide statistical significance of association in both the replication cohort (p = 4.59 × 10−11) and the combined discovery and replication cohort (37,016 AF individuals and 340,502 AF-free individuals; p = 6.76 × 10−18) (Table 1 and Figure S3). This locus has additionally been identified as associated with AF in one study that was published while this manuscript was under review.16
Figure 1.
Regional Plots at the Two AF-Associated Loci Identified in Chromosomal Regions 1p32 and 2q31 in This Study
The dots represent SNVs, whose positions are based on genomic build GRCh37 and whose −log10(p values) are based on association tests within the HUNT discovery cohort. The SNVs reaching the highest level of statistical significance at the two loci are marked with reference numbers. The strength of the pairwise LD (r2) between the lead SNV and all other SNVs was computed on the basis of genotyped and imputed markers and is indicated by a gradient from red to green to blue. The blue line indicates estimated recombination rates.
Protein-truncating variants in TTN have been previously associated with DCM,50 and individuals with DCM have been found to have a high prevalence of AF,51 so we performed additional analyses to evaluate whether the locus around rs12614435 acts through DCM or whether it is directly associated with AF. First, excluding individuals from the discovery cohort with any type of ischemic or non-ischemic cardiomyopathy (n = 787) revealed risk estimates (OR = 1.20; 95% CI = 1.14–1.27) that were almost identical to those of the main analysis (Table 1). Second, the frequency of the G risk allele was not statistically significantly different between AF individuals with and without DCM (MAFAF+DCM = 15.5% versus MAFAF−DCM = 17.3%, pChiSq = 0.45). Third, rs12614435 was not associated with DCM (OR = 0.96; 95% CI = 0.75–1.24; p = 0.75) in the HUNT discovery cohort in a comparison of 276 DCM individuals and 67,668 DCM-free individuals. Altogether, these sensitivity analyses indicate a mechanism that does not act through overt cardiomyopathy.
The second locus we identified was in chromosomal region 1p32 (lead SNV rs56202902; OR = 1.32; 95% CI = 1.20–1.44; p = 2.50 × 10−9). This locus spans a large gene-dense region of approximately 2 Mb and includes 98 highly correlated genome-wide-significant non-coding variants (Figure 1 and Table S3). The locus reached nominal significance in the replication cohort (p = 1.45 × 10−4) and genome-wide significance in the combined discovery and replication cohort (p = 1.54 × 10−11) (Table 1 and Figure S3).
For each of these loci, the frequency of the risk allele was higher in individuals with early-onset AF than in individuals with late-onset AF. This was particularly true for the 1p32 locus, where the frequency of the risk allele was 7.0% among individuals with AF onset < 50 years of age, 5.1% in individuals with AF onset > 50 years of age, and 4.0% among AF-free individuals (Table S4). This clear increase in frequency of the risk allele by decreasing age of onset is consistent with what one would expect for a true locus and hence supports the conclusion that the associations that we report are true.
Fine-Mapping of Known Loci
We attempted to refine the association signal at known AF loci by searching for additional independent signals, coding variants, splice sites, and transcription factor binding sites.
Within 500 kb of the previously reported PRRX1 (MIM: 167420) locus on chromosome 1,11 we found an independent signal (lead SNV rs72700114; p = 3.50 × 10−10) that, in our data, showed a stronger association with AF than the previously reported lead signal at this locus (rs3903239; p = 7.6 × 10−8) (Figure S4). Interestingly, this independent lead SNV, rs72700114, falls in a fetal-heart-specific enhancer in data from the Roadmap Epigenomics Project.47
At the previously known HCN4 (MIM: 605206) locus on chromosome 15 (rs6495063; p = 1.6 × 10−5), we found a variant that was predicted as likely to affect the binding (RegulomeDB score of 2a)52 of one or more transcription factors (e.g., HNF4 and MZF1) according to evidence of transcription factor binding in combination with a matched transcription factor motif, a matched DNase footprint, and the DNase peak.52 Furthermore, the variant falls in an enhancer state in multiple tissues, including the right atrium, left and right ventricles, and fetal heart.47
At 3 of 16 previously published loci (Table S2), we found associated protein-altering variants (p < 5 × 10−5) in genes within 500 kb of reported lead SNVs (Table S5). At the two AF loci previously annotated as SYNPO2L and NEURL (MIM: 603804), both located on chromosome 10, we identified missense variants situated in the genes previously highlighted as putative functional genes for AF (SYNPO2L and NEURL).11, 12 We identified three AF-associated missense SYNPO2L variants (rs34163229, rs3812629, and rs60632610) that were in high LD (r2 > 0.78) with the previously published lead SNV (rs10824026),11 supporting SYNPO2L as the causal gene at this locus. At the NEURL locus, we found that conditioning on the NEURL missense variant that we identified (rs11191737) did not significantly change the association of the lead SNV intronic to NEURL (rs60572254; ORconditioning = 1.23; 95% CI = 1.16–1.30), indicating that the association of the lead SNV is not explained by this coding variant. This was also the case for the KCNN3 (MIM: 602983) locus, where we identified a missense variant (rs139248801; p = 2.30 × 10−5) in the gene PMVK (MIM: 607622), encoding phosphomevalonate kinase, which also did not alter the association of the lead SNV intronic to KCNN3 (rs34245846; ORconditioning = 1.12; 95% CI = 1.08–1.17).
Gene-Based Burden Test
To test for enrichment of lower-frequency variants in case or control individuals, we performed a gene-based burden test including all protein-altering variants with a MAF < 5% and tested a total of 19,099 genes in 6,337 AF individuals versus 19,011 AF-free individuals matched by birth year, sex, genotyping batch, and PCs 1–4. No genes reached our experiment-wide significance threshold (0.05/19,099 = 2.6 × 10−6) (Figure S5). We did observe, however, an enrichment of genes previously associated with rare familial forms of AF, as reported in a recent comprehensive review (prank = 0.011),53 and the most significant results were for SCN1B (MIM: 600235), SCN5A (MIM: 600163), and NKX2-5 (MIM: 600584) (Table S6). The lack of findings for individual genes might well be due to the fact that the gene-based burden test was based on genotyped and imputed markers and hence lacked sufficient coverage of rare coding variants.
Heritability
We determined the AF heritability explained by all robustly genotyped SNVs (n = 456,297 SNVs) to be 9.6% (standard error = 1.2%). The heritability explained by the two loci that we report and the 14 previously published AF risk loci that were directly genotyped or imputed in our dataset (Table S2) was 1.4%.
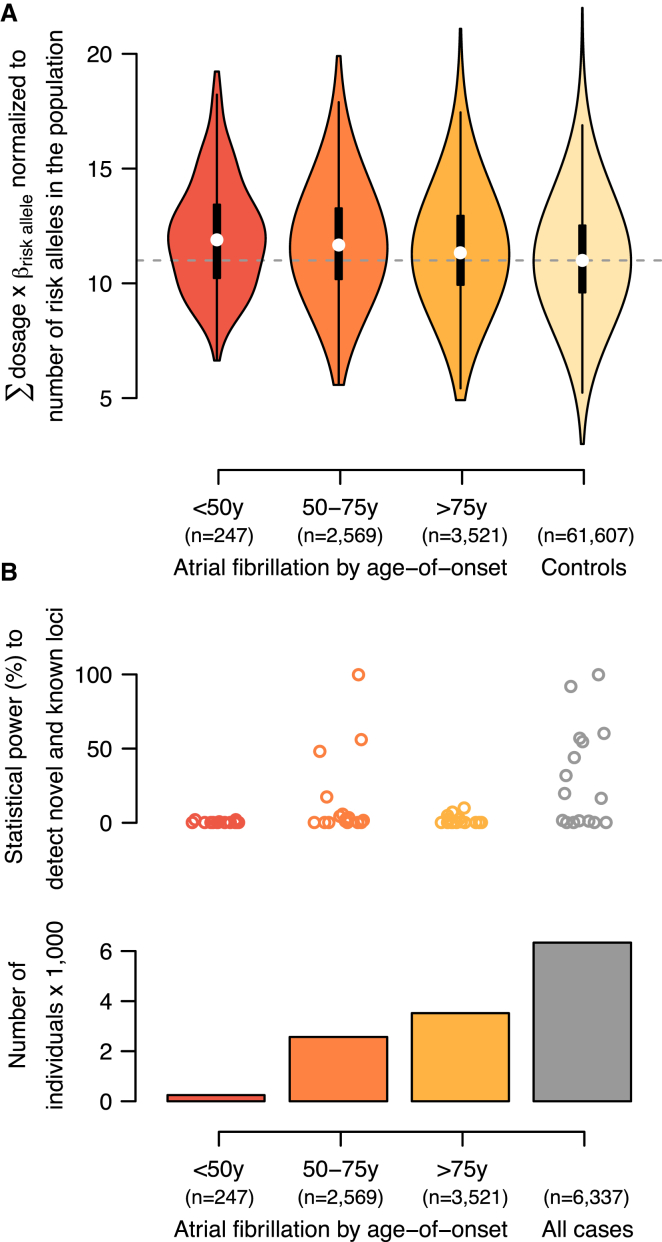
Polygenic Risk Score and Power Calculations
We calculated a polygenic risk score for AF as the sum of products of imputed dosages and log(OR) for the 16 AF risk loci that were available in our dataset (0–32 possible risk alleles in total). Figure 2A summarizes the risk-score distribution normalized to the observed number of risk alleles in the population (range 3–22). We observed that individuals with an AF onset at age <50, 50–75, or >75 years had an average of 0.93 (8.3%), 0.69 (6.5%), or 0.36 (5.0%) additional risk alleles, respectively, than control individuals (pt test < 1 × 10−8 for all three groups in comparison with control individuals). Combining all case and control individuals resulted in the largest statistical power to detect the 16 AF-associated loci; however, within the age-specific subgroups, individuals with a disease onset at 50–75 years of age contributed the most statistical power to detect association (Figure 2B).
Figure 2.
Polygenic Risk Scores and Estimates of Statistical Power for AF-Associated Loci
(A) Polygenic risk scores stratified by age of disease onset. White dots represent medians, black boxes represent interquartile ranges, black whiskers represent 1.5× the interquartile range, and colored areas show the probability density of the data. The vertical gray dotted line represents the median score for control individuals.
(B) Statistical-power estimates stratified by age of disease onset. The upper panel shows the necessary statistical power to detect risk loci (each represented by a dot) on the basis of the observed allele frequencies within subgroups, the number of case (bottom panel) and control (n = 61,607) individuals, and α = 5 × 10−8.
Expression of eQTLs at AF-Associated Loci
To identify potentially functional genes at the AF GWAS loci, we searched for AF-associated variants that were also associated with expression levels of nearby genes. From the GTEx Portal,34 we downloaded human gene expression data (cis-eQTLs) on up to ∼190 million chromosome positions across 44 tissues and searched for all 483 AF-associated SNVs (p < 5 × 10−8; Table S3). A total of 46 AF-associated SNVs reached the eQTL experiment-wide significance threshold of p < 1.14 × 10−9 (5 × 10−8 per 44 tissues) in at least one type of tissue (Table S7): 12 SNVs at the locus in chromosomal region 2q31 were associated with increased expression of FKBP7 (MIM: 607062), encoding FK506 binding protein 7, in the aortic artery or fibroblasts, and 33 SNVs at the previously published PRRX1 locus on chromosome 1 were associated with expression of the lincRNA-encoding gene RP1-79C4.4 across multiple tissues (Table S7).
AF Tissues, Biological Pathways, and Gene Sets
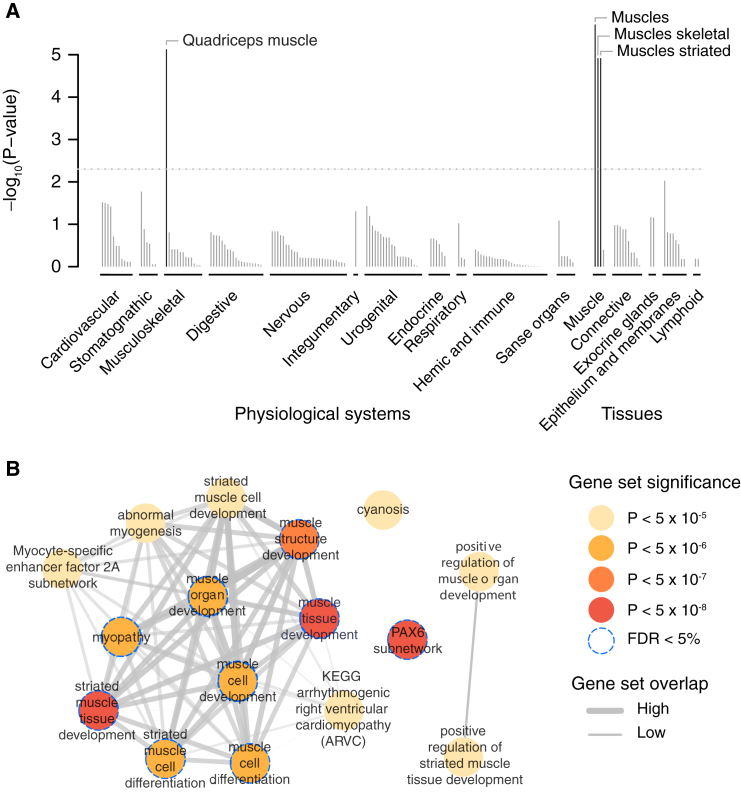
To identify tissues and cell types in which genes at AF-associated variants are more likely to be expressed than genes at randomly selected loci with the same gene density, we employed DEPICT.35 On the basis of 37,427 human microarray expression samples from a total of 209 tissues and cell types, we observed a statistically significant enrichment of various striated muscle tissues, which are the major tissue types of the myocardium (Figure 3A). Additionally, we saw a marginally significant enrichment of “heart atria” and “atrial appendage” (Table S8).
Figure 3.
Tissues and Reconstituted Gene Sets Significantly Enriched with AF-Associated Loci
(A) On the basis of expression patterns across 37,427 human mRNA microarrays, DEPICT predicted genes within AF-associated loci to be highly expressed in various muscle tissues. Tissues are grouped by type and significance. The horizontal dotted line represents the experiment-wide significance threshold (p = 5 × 10−3).
(B) Reconstituted gene sets found to be significantly (p < 5 × 10−6; FDR < 5%) and marginally (p < 5 × 10−5) significantly enriched by DEPICT are represented by nodes colored according to the permutation p value; the pairwise overlap of genes is denoted by the width of connecting lines.
We further applied DEPICT to prioritize genes at AF-associated loci and to perform a gene-set enrichment analysis through the integration of association results with predefined gene sets reconstituted according to co-expression data.35 DEPICT prioritized at least one gene (false-discovery rate [FDR] < 5%) at 13 loci (Table S9). TTN was prioritized as the most likely functional gene at the locus in chromosomal region 2q31, and DMRTA2 (MIM: 614804) was prioritized for the locus at 1p32. DMRTA2 encodes doublesex-and mab-3-related transcription factor A2. Out of 14,461 tested gene sets, DEPICT identified nine gene sets that were significantly enriched (FDR < 5%) with genes at AF-associated loci (Figure 3B and Table S10). These gene sets pointed mainly to a process related to muscle cell and tissue differentiation and development but also highlighted the PAX6 subnetwork, which has been proposed to be involved in the development of several organs and tissues.54, 55, 56
Enrichment of Regulatory Elements at Associated Loci
The vast majority of AF-associated variants identified by GWASs are located in non-coding regions of the genome, where the underlying functional mechanisms are poorly understood. To evaluate novel and previously identified AF-associated loci, we used GARFIELD43, 44 to test whether such variants were non-randomly distributed across various coding and non-coding regulatory and cell-type-specific elements throughout the genome.
Of 1,005 tissue- and cell-type-specific annotations tested from ENCODE, GENCODE, and the Roadmap Epigenomics Project (Table S11), we observed significantly increased enrichment of 31 regulatory elements at one or more GWAS p value thresholds. The most significant enrichment pattern for AF-associated index variants was observed in fetal heart tissue for DNaseI hypersensitivity peaks (Figure 4), DNaseI hypersensitivity hotspots (Figure S6), and footprints (Figure S7), comprising a total of 21 overall significant enrichment statistics and up to 31-fold enrichment (Table S11).
Figure 4.
Enrichment of AF-Associated Loci within DNaseI Hypersensitivity Sites across 424 Cell Types Available from ENCODE and the Roadmap Epigenomics Project
Enrichment tests were performed independently across markers that reached one of eight different GWAS p value thresholds (p < 1 × 10−1 to < 1 × 10−8). The radial axis shows fold enrichment calculated at each of the eight thresholds, each indicated by a different color, for each of the 424 cell types. Cell types are sorted by tissue, represented along the outside edge of the plot; font size is proportional to the number of cell types from that tissue. The outer line and dots next to the tissue labels are colored with respect to tissue type. Dots along the inside edge of the plot denote significant enrichment (if any) for a given cell type at a GWAS p value threshold < 1 × 10−5 (outermost dot) to a GWAS p value threshold < 1 × 10−8 (innermost dot). Of all tissues examined, the strongest enrichment was seen for DNaseI hypersensitivity sites in the fetal heart.
We replicated this finding by using GREGOR48 to test for enrichment of a newly published set of independent AF-associated variants15, 16 with a subset of regulatory features from the Roadmap Epigenomics Project. Using a completely independent list of 15 AF-associated loci, we replicated the significant enrichment of fetal heart tissue DNaseI hypersensitivity peaks and found an enrichment of fetal heart histone markers, indicating primed enhancers (Table S12).
All together, these results point to a congenital mechanism that occurs during fetal heart development and eventually causes AF in older hearts.
Phenome-wide Associations of AF Risk Loci
To test whether AF risk loci that reached nominal significance in the HUNT discovery cohort (Tables S1 and S2) were also associated with other phenotypes, we performed a phenome-wide association scan of 709 clinical variables, mainly comprising self-reported health issues and clinical tests, and found statistically significant associations for two loci (p < 7.1 × 10−5) (Figure S8). The AF risk allele at the locus in chromosomal region 1p32 (rs56202902) was associated with decreased alcohol intake and decreased diastolic and mean arterial blood pressure. For the HCN4 locus (rs7164883), we found that the AF risk allele was associated with decreased pulse.
Replication of Recently Discovered AF-Associated Loci
During the revision process of the present paper, two additional AF GWASs were published.15, 16 Together, the two studies report a total of 16 loci, including genome-wide-significant SNVs in the region of TTN,16 which complements our finding of an association signal with variants in high LD. In our HUNT discovery cohort, we could replicate 9 of 12 loci (p < 0.05) reported by the AFGen Consortium, which mainly included case and control subjects of European ancestry,16 whereas we could replicate only two of six loci reported for people of Japanese ancestry (Table S13).15
Discussion
In this study, we performed a genome-wide association scan for AF in a genetically homogeneous population-based sample that included 6,337 AF individuals and 61,407 AF-free individuals and follow-up in an additional 30,679 AF individuals and 278,895 AF-free individuals. We identified two risk loci for AF and confirmed association with 12 of 16 loci that were known at the time of the analyses.
TTN is a strong biological candidate gene at the 2q31 locus because of its role in maintaining the structural integrity of the sarcomere and its involvement in muscle elasticity and force transmission.49 The variants that we identified mainly span the TTN transcript, and several of the variants were predicted to alter TTN. The pathway framework DEPICT also prioritized TTN as the most likely causal gene. A less likely candidate gene at the locus is FKBP7, which was highlighted by eQTL analysis as upregulated in aortic tissue and fibroblasts. Members of this gene family are thought to exhibit peptidyl-prolyl cis-trans isomerase activity and function as molecular chaperones.
The second risk locus we identified is located in chromosomal region 1p32. The allele frequency of the risk variant in our Norwegian discovery cohort is 4.1%, providing substantially greater power to detect this non-coding risk allele than frequencies in other European populations (closer to 1%). Hence, identification of this AF risk locus emphasizes the advantage of studying genetically homogeneous or historically isolated populations in which the frequency of selected rare or low-frequency variants might have accumulated. Interestingly, the 1p32 locus has previously been associated, on a genome-wide level, with QRS amplitude and QRS duration,57, 58 both ECG-derived measurements that reflect cardiac structure and function most likely related to the development of AF.59, 60, 61 Potential functional genes at the locus include DMRTA2, which was highlighted by DEPICT, and CDKN2C (MIM: 603369), which is a member of the INK4 family of cyclin-dependent kinase inhibitors and potentially plays a role in hypoplastic left-heart syndrome via its interaction with CDK4 (MIM: 123829) and CDK6 (MIM: 603368).62, 63
On the basis of predicted gene function from multiple sources of evidence—including protein-protein interactions, phenotypic data from gene-knockout experiments in mice, and co-regulation of gene expression across thousands of physiological conditions35—DEPICT identified genes at AF-associated loci to be highly enriched in gene sets and pathways that are important for muscle cell differentiation and tissue formation. These findings were substantiated by enrichment analyses of functional and regulatory elements, which indicated that many SNVs at AF-associated loci fall within regions of open chromatin state during fetal heart development. All together, these results point to a mechanism of impaired muscle cell differentiation and tissue formation in the developing heart as important risk factors for AF in adult life. This developmental hypothesis will require additional studies to be confirmed, but it is supported by the fact that individuals with various forms of congenital cardiomyopathy, and myopathies in general, have a high prevalence of atrial tachyarrhythmias.51, 64, 65, 66 Additionally, the locus that has most consistently been associated with AF is located in chromosomal region 4q25, which is close to PITX2 (paired-like homeodomain transcription factor 2 [MIM: 601542]).11, 67 This gene is known to play a critical role in left-right asymmetry and cardiogenesis and has been shown to be involved in structural and electrical remodeling of the atrial myocardium that facilitates arrhythmia.68, 69, 70, 71
We speculate that impaired fetal heart development can make the atrium prone to structural remodeling later in life. Impaired muscle development in the fetal heart can alter the elastic properties of the atrium so that atrial compliance in response to stretch is compromised. This most likely leads to elevated left atrial pressure, which has been demonstrated to facilitate premature depolarizations from the pulmonary veins that can eventually trigger AF.72, 73
By examining a polygenic risk score constructed from these variants, we found that individuals with a lower age of AF onset carried a higher genetic burden of AF risk alleles, which is consistent with previous epidemiological reports of higher risk of AF for family members of individuals with early-onset AF.7 However, including all AF individuals irrespectively of age of onset leads to improved power by increasing the sample size. Appropriate modeling of genetic burden as indicated by the age of onset could result in further improved power.
In summary, this study identified two AF loci that both point to a mechanism of cardiac structural remodeling as a substrate of AF. Pathway and epigenetic enrichment analyses highlight a potential mechanism whereby genetic risk loci act to increase the risk of AF during fetal heart development. This hypothesis needs confirmation but might provide a foundation for directing future functional experiments to identify the functional genes and genetic mechanisms at AF risk loci.
Conflicts of Interest
E.I. is an advisor and consultant for Precision Wellness Inc. and advisor for Cellink for work unrelated to the present study.
Acknowledgments
The Nord-Trøndelag Health (HUNT) Study is a collaboration among the HUNT Research Centre (Faculty of Medicine, Norwegian University of Science and Technology), the Nord-Trøndelag County Council, the Central Norway Health Authority, and the Norwegian Institute of Public Health. The HUNT-MI study, which comprises the genetic investigations of the HUNT Study, is a collaboration between investigators from the HUNT Study and the University of Michigan School of Public Health and the University of Michigan Medical School. The authors wish to thank all study participants who contributed to the scientific research. J.B.N. was supported by grants from the Danish Heart Foundation, the Lundbeck Foundation, the A.P Møller Foundation for the Advancement of Medical Science, and Fondsbørsvekselerer Henry Hansen og Hustru Karla Hansen Født Vestergaards Legat. C.J.W. was supported by grants HL109946, HL127564, and HL130705 from the National Institutes of Health (NIH). J.H.S. and M.S.O. were supported by grants from the Danish National Research Foundation, the John and Birthe Meyer Foundation, the Arvid Nilsson Foundation, and the Research Council of Rigshospitalet, Denmark. E.I. was supported by the NIH (1R01DK106236-01A1 and 1R01HL135313-01), the Knut och Alice Wallenberg Foundation (2013.0126), the Swedish Heart-Lung Foundation (20140422), and the Göran Gustafsson Foundation. Finally, this research was conducted with the UK Biobank Resource under application number 13721.
Published: December 28, 2017
Footnotes
Supplemental Data include 8 figures and 13 tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2017.12.003.
Contributor Information
Kristian Hveem, Email: kristian.hveem@ntnu.no.
Cristen J. Willer, Email: cristen@umich.edu.
Web Resources
GTEx Portal, http://gtexportal.org
Michigan Genomics Initiative, https://michigangenomics.org/
SnpEff, http://snpeff.sourceforge.net/
OMIM, https://www.omim.org
Supplemental Data
References
- 1.Colilla S., Crow A., Petkun W., Singer D.E., Simon T., Liu X. Estimates of current and future incidence and prevalence of atrial fibrillation in the U.S. adult population. Am. J. Cardiol. 2013;112:1142–1147. doi: 10.1016/j.amjcard.2013.05.063. [DOI] [PubMed] [Google Scholar]
- 2.Chugh S.S., Havmoeller R., Narayanan K., Singh D., Rienstra M., Benjamin E.J., Gillum R.F., Kim Y.-H., McAnulty J.H., Jr., Zheng Z.-J. Worldwide epidemiology of atrial fibrillation: a Global Burden of Disease 2010 Study. Circulation. 2014;129:837–847. doi: 10.1161/CIRCULATIONAHA.113.005119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kannel W.B., Benjamin E.J. Current perceptions of the epidemiology of atrial fibrillation. Cardiol. Clin. 2009;27:13–24. doi: 10.1016/j.ccl.2008.09.015. vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim M.H., Johnston S.S., Chu B.-C., Dalal M.R., Schulman K.L. Estimation of total incremental health care costs in patients with atrial fibrillation in the United States. Circ Cardiovasc Qual Outcomes. 2011;4:313–320. doi: 10.1161/CIRCOUTCOMES.110.958165. [DOI] [PubMed] [Google Scholar]
- 5.Schnabel R.B., Sullivan L.M., Levy D., Pencina M.J., Massaro J.M., D’Agostino R.B., Sr., Newton-Cheh C., Yamamoto J.F., Magnani J.W., Tadros T.M. Development of a risk score for atrial fibrillation (Framingham Heart Study): a community-based cohort study. Lancet. 2009;373:739–745. doi: 10.1016/S0140-6736(09)60443-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christophersen I.E., Ravn L.S., Budtz-Joergensen E., Skytthe A., Haunsoe S., Svendsen J.H., Christensen K. Familial aggregation of atrial fibrillation: a study in Danish twins. Circ Arrhythm Electrophysiol. 2009;2:378–383. doi: 10.1161/CIRCEP.108.786665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lubitz S.A., Yin X., Fontes J.D., Magnani J.W., Rienstra M., Pai M., Villalon M.L., Vasan R.S., Pencina M.J., Levy D. Association between familial atrial fibrillation and risk of new-onset atrial fibrillation. JAMA. 2010;304:2263–2269. doi: 10.1001/jama.2010.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y.-H., Xu S.-J., Bendahhou S., Wang X.-L., Wang Y., Xu W.-Y., Jin H.-W., Sun H., Su X.-Y., Zhuang Q.-N. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251–254. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- 9.Gollob M.H., Jones D.L., Krahn A.D., Danis L., Gong X.-Q., Shao Q., Liu X., Veinot J.P., Tang A.S.L., Stewart A.F.R. Somatic mutations in the connexin 40 gene (GJA5) in atrial fibrillation. N. Engl. J. Med. 2006;354:2677–2688. doi: 10.1056/NEJMoa052800. [DOI] [PubMed] [Google Scholar]
- 10.Hodgson-Zingman D.M., Karst M.L., Zingman L.V., Heublein D.M., Darbar D., Herron K.J., Ballew J.D., de Andrade M., Burnett J.C., Jr., Olson T.M. Atrial natriuretic peptide frameshift mutation in familial atrial fibrillation. N. Engl. J. Med. 2008;359:158–165. doi: 10.1056/NEJMoa0706300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellinor P.T., Lunetta K.L., Albert C.M., Glazer N.L., Ritchie M.D., Smith A.V., Arking D.E., Müller-Nurasyid M., Krijthe B.P., Lubitz S.A. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat. Genet. 2012;44:670–675. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sinner M.F., Tucker N.R., Lunetta K.L., Ozaki K., Smith J.G., Trompet S., Bis J.C., Lin H., Chung M.K., Nielsen J.B., METASTROKE Consortium. AFGen Consortium Integrating genetic, transcriptional, and functional analyses to identify 5 novel genes for atrial fibrillation. Circulation. 2014;130:1225–1235. doi: 10.1161/CIRCULATIONAHA.114.009892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gudbjartsson D.F., Helgason H., Gudjonsson S.A., Zink F., Oddson A., Gylfason A., Besenbacher S., Magnusson G., Halldorsson B.V., Hjartarson E. Large-scale whole-genome sequencing of the Icelandic population. Nat. Genet. 2015;47:435–444. doi: 10.1038/ng.3247. [DOI] [PubMed] [Google Scholar]
- 14.Tsai C.-T., Hsieh C.-S., Chang S.-N., Chuang E.Y., Ueng K.-C., Tsai C.-F., Lin T.-H., Wu C.-K., Lee J.-K., Lin L.-Y. Genome-wide screening identifies a KCNIP1 copy number variant as a genetic predictor for atrial fibrillation. Nat. Commun. 2016;7:10190. doi: 10.1038/ncomms10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Low S.-K., Takahashi A., Ebana Y., Ozaki K., Christophersen I.E., Ellinor P.T., Ogishima S., Yamamoto M., Satoh M., Sasaki M., AFGen Consortium Identification of six new genetic loci associated with atrial fibrillation in the Japanese population. Nat. Genet. 2017;49:953–958. doi: 10.1038/ng.3842. [DOI] [PubMed] [Google Scholar]
- 16.Christophersen I.E., Rienstra M., Roselli C., Yin X., Geelhoed B., Barnard J., Lin H., Arking D.E., Smith A.V., Albert C.M., METASTROKE Consortium of the ISGC. Neurology Working Group of the CHARGE Consortium. AFGen Consortium Large-scale analyses of common and rare variants identify 12 new loci associated with atrial fibrillation. Nat. Genet. 2017;49:946–952. doi: 10.1038/ng.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krokstad S., Langhammer A., Hveem K., Holmen T.L., Midthjell K., Stene T.R., Bratberg G., Heggland J., Holmen J. Cohort Profile: the HUNT Study, Norway. Int. J. Epidemiol. 2013;42:968–977. doi: 10.1093/ije/dys095. [DOI] [PubMed] [Google Scholar]
- 18.Delaneau O., Zagury J.-F., Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 19.McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K., Haplotype Reference Consortium A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuchsberger C., Abecasis G.R., Hinds D.A. minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–784. doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loh P.-R., Tucker G., Bulik-Sullivan B.K., Vilhjálmsson B.J., Finucane H.K., Salem R.M., Chasman D.I., Ridker P.M., Neale B.M., Berger B. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang H.M., Sul J.H., Service S.K., Zaitlen N.A., Kong S.-Y., Freimer N.B., Sabatti C., Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Firth D. Bias reduction of maximum likelihood estimates. Biometrika. 1993;80:27–38. [Google Scholar]
- 26.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobsen B.K., Eggen A.E., Mathiesen E.B., Wilsgaard T., Njølstad I. Cohort profile: the Tromso Study. Int. J. Epidemiol. 2012;41:961–967. doi: 10.1093/ije/dyr049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaitlen N., Kraft P., Patterson N., Pasaniuc B., Bhatia G., Pollack S., Price A.L. Using extended genealogy to estimate components of heritability for 23 quantitative and dichotomous traits. PLoS Genet. 2013;9:e1003520. doi: 10.1371/journal.pgen.1003520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.So H.-C., Gui A.H.S., Cherny S.S., Sham P.C. Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet. Epidemiol. 2011;35:310–317. doi: 10.1002/gepi.20579. [DOI] [PubMed] [Google Scholar]
- 33.Skol A.D., Scott L.J., Abecasis G.R., Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 34.GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pers T.H., Karjalainen J.M., Chan Y., Westra H.-J., Wood A.R., Yang J., Lui J.C., Vedantam S., Gustafsson S., Esko T., Genetic Investigation of ANthropometric Traits (GIANT) Consortium Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fehrmann R.S.N., Karjalainen J.M., Krajewska M., Westra H.-J., Maloney D., Simeonov A., Pers T.H., Hirschhorn J.N., Jansen R.C., Schultes E.A. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat. Genet. 2015;47:115–125. doi: 10.1038/ng.3173. [DOI] [PubMed] [Google Scholar]
- 37.Lage K., Karlberg E.O., Størling Z.M., Olason P.I., Pedersen A.G., Rigina O., Hinsby A.M., Tümer Z., Pociot F., Tommerup N. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 38.Bult, C.J., Richardson, J.E., Blake, J.A., Kadin, J.A., Ringwald, M., Eppig, J.T., Baldarelli, R.M., Baya, M., Beal, J.S., Begley, D.A., et al. (2000). Mouse genome informatics in a new age of biological inquiry. In Proceedings of the IEEE International Symposium on Bio-Informatics and Biomedical Engineering, pp. 29–32.
- 39.Croft D., O’Kelly G., Wu G., Haw R., Gillespie M., Matthews L., Caudy M., Garapati P., Gopinath G., Jassal B. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raychaudhuri S., Plenge R.M., Rossin E.J., Ng A.C.Y., Purcell S.M., Sklar P., Scolnick E.M., Xavier R.J., Altshuler D., Daly M.J., International Schizophrenia Consortium Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J., LifeLines Cohort Study. ADIPOGen Consortium. AGEN-BMI Working Group. CARDIOGRAMplusC4D Consortium. CKDGen Consortium. GLGC. ICBP. MAGIC Investigators. MuTHER Consortium. MIGen Consortium. PAGE Consortium. ReproGen Consortium. GENIE Consortium. International Endogene Consortium Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iotchkova, V., Ritchie, G.R.S., Geihs, M., Morganella, S., Min, J.L., Walter, K., Timpson, N.J., Consortium, U., Dunham, I., Birney, E., et al. (2016). GARFIELD - GWAS Analysis of Regulatory or Functional Information Enrichment with LD correction. bioRxiv, https://doi.org/10.1101/085738.
- 44.Iotchkova V., Huang J., Morris J.A., Jain D., Barbieri C., Walter K., Min J.L., Chen L., Astle W., Cocca M., UK10K Consortium Discovery and refinement of genetic loci associated with cardiometabolic risk using dense imputation maps. Nat. Genet. 2016;48:1303–1312. doi: 10.1038/ng.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt E.M., Zhang J., Zhou W., Chen J., Mohlke K.L., Chen Y.E., Willer C.J. GREGOR: evaluating global enrichment of trait-associated variants in epigenomic features using a systematic, data-driven approach. Bioinformatics. 2015;31:2601–2606. doi: 10.1093/bioinformatics/btv201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Granzier H.L., Labeit S. The giant protein titin: a major player in myocardial mechanics, signaling, and disease. Circ. Res. 2004;94:284–295. doi: 10.1161/01.RES.0000117769.88862.F8. [DOI] [PubMed] [Google Scholar]
- 50.Herman D.S., Lam L., Taylor M.R.G., Wang L., Teekakirikul P., Christodoulou D., Conner L., DePalma S.R., McDonough B., Sparks E. Truncations of titin causing dilated cardiomyopathy. N. Engl. J. Med. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Spaendonck-Zwarts K.Y., van Rijsingen I.A.W., van den Berg M.P., Lekanne Deprez R.H., Post J.G., van Mil A.M., Asselbergs F.W., Christiaans I., van Langen I.M., Wilde A.A.M. Genetic analysis in 418 index patients with idiopathic dilated cardiomyopathy: overview of 10 years’ experience. Eur. J. Heart Fail. 2013;15:628–636. doi: 10.1093/eurjhf/hft013. [DOI] [PubMed] [Google Scholar]
- 52.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olesen M.S., Nielsen M.W., Haunsø S., Svendsen J.H. Atrial fibrillation: the role of common and rare genetic variants. Eur. J. Hum. Genet. 2014;22:297–306. doi: 10.1038/ejhg.2013.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.St-Onge L., Sosa-Pineda B., Chowdhury K., Mansouri A., Gruss P. Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature. 1997;387:406–409. doi: 10.1038/387406a0. [DOI] [PubMed] [Google Scholar]
- 55.Götz M., Stoykova A., Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron. 1998;21:1031–1044. doi: 10.1016/s0896-6273(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 56.Marquardt T., Ashery-Padan R., Andrejewski N., Scardigli R., Guillemot F., Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 57.Sotoodehnia N., Isaacs A., de Bakker P.I.W., Dörr M., Newton-Cheh C., Nolte I.M., van der Harst P., Müller M., Eijgelsheim M., Alonso A. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat. Genet. 2010;42:1068–1076. doi: 10.1038/ng.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van der Harst P., van Setten J., Verweij N., Vogler G., Franke L., Maurano M.T., Wang X., Mateo Leach I., Eijgelsheim M., Sotoodehnia N. 52 Genetic Loci Influencing Myocardial Mass. J. Am. Coll. Cardiol. 2016;68:1435–1448. doi: 10.1016/j.jacc.2016.07.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benjamin E.J., Wolf P.A., D’Agostino R.B., Silbershatz H., Kannel W.B., Levy D. Impact of atrial fibrillation on the risk of death: the Framingham Heart Study. Circulation. 1998;98:946–952. doi: 10.1161/01.cir.98.10.946. [DOI] [PubMed] [Google Scholar]
- 60.Okin P.M., Wachtell K., Devereux R.B., Harris K.E., Jern S., Kjeldsen S.E., Julius S., Lindholm L.H., Nieminen M.S., Edelman J.M. Regression of electrocardiographic left ventricular hypertrophy and decreased incidence of new-onset atrial fibrillation in patients with hypertension. JAMA. 2006;296:1242–1248. doi: 10.1001/jama.296.10.1242. [DOI] [PubMed] [Google Scholar]
- 61.El-Chami M.F., Brancato C., Langberg J., Delurgio D.B., Bush H., Brosius L., Leon A.R. QRS duration is associated with atrial fibrillation in patients with left ventricular dysfunction. Clin. Cardiol. 2010;33:132–138. doi: 10.1002/clc.20714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guan K.L., Jenkins C.W., Li Y., Nichols M.A., Wu X., O’Keefe C.L., Matera A.G., Xiong Y. Growth suppression by p18, a p16INK4/MTS1- and p14INK4B/MTS2-related CDK6 inhibitor, correlates with wild-type pRb function. Genes Dev. 1994;8:2939–2952. doi: 10.1101/gad.8.24.2939. [DOI] [PubMed] [Google Scholar]
- 63.Gambetta K., Al-Ahdab M.K., Ilbawi M.N., Hassaniya N., Gupta M. Transcription repression and blocks in cell cycle progression in hypoplastic left heart syndrome. Am. J. Physiol. Heart Circ. Physiol. 2008;294:H2268–H2275. doi: 10.1152/ajpheart.91494.2007. [DOI] [PubMed] [Google Scholar]
- 64.Maron B.J., Olivotto I., Bellone P., Conte M.R., Cecchi F., Flygenring B.P., Casey S.A., Gohman T.E., Bongioanni S., Spirito P. Clinical profile of stroke in 900 patients with hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2002;39:301–307. doi: 10.1016/s0735-1097(01)01727-2. [DOI] [PubMed] [Google Scholar]
- 65.Wu L., Guo J., Zheng L., Chen G., Ding L., Qiao Y., Sun W., Yao Y., Zhang S. Atrial Remodeling and Atrial Tachyarrhythmias in Arrhythmogenic Right Ventricular Cardiomyopathy. Am. J. Cardiol. 2016;118:750–753. doi: 10.1016/j.amjcard.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 66.Finsterer J., Stöllberger C. Atrial fibrillation/flutter in myopathies. Int. J. Cardiol. 2008;128:304–310. doi: 10.1016/j.ijcard.2007.12.041. [DOI] [PubMed] [Google Scholar]
- 67.Gudbjartsson D.F., Arnar D.O., Helgadottir A., Gretarsdottir S., Holm H., Sigurdsson A., Jonasdottir A., Baker A., Thorleifsson G., Kristjansson K. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 68.Faucourt M., Houliston E., Besnardeau L., Kimelman D., Lepage T. The pitx2 homeobox protein is required early for endoderm formation and nodal signaling. Dev. Biol. 2001;229:287–306. doi: 10.1006/dbio.2000.9950. [DOI] [PubMed] [Google Scholar]
- 69.Liu C., Liu W., Palie J., Lu M.F., Brown N.A., Martin J.F. Pitx2c patterns anterior myocardium and aortic arch vessels and is required for local cell movement into atrioventricular cushions. Development. 2002;129:5081–5091. doi: 10.1242/dev.129.21.5081. [DOI] [PubMed] [Google Scholar]
- 70.Ammirabile G., Tessari A., Pignataro V., Szumska D., Sutera Sardo F., Benes J., Jr., Balistreri M., Bhattacharya S., Sedmera D., Campione M. Pitx2 confers left morphological, molecular, and functional identity to the sinus venosus myocardium. Cardiovasc. Res. 2012;93:291–301. doi: 10.1093/cvr/cvr314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chinchilla A., Daimi H., Lozano-Velasco E., Dominguez J.N., Caballero R., Delpón E., Tamargo J., Cinca J., Hove-Madsen L., Aranega A.E., Franco D. PITX2 insufficiency leads to atrial electrical and structural remodeling linked to arrhythmogenesis. Circ Cardiovasc Genet. 2011;4:269–279. doi: 10.1161/CIRCGENETICS.110.958116. [DOI] [PubMed] [Google Scholar]
- 72.Nattel S. New ideas about atrial fibrillation 50 years on. Nature. 2002;415:219–226. doi: 10.1038/415219a. [DOI] [PubMed] [Google Scholar]
- 73.Bode F., Sachs F., Franz M.R. Tarantula peptide inhibits atrial fibrillation. Nature. 2001;409:35–36. doi: 10.1038/35051165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.