Abstract
Bone mineral density (BMD) assessed by DXA is used to evaluate bone health. In children, total body (TB) measurements are commonly used; in older individuals, BMD at the lumbar spine (LS) and femoral neck (FN) is used to diagnose osteoporosis. To date, genetic variants in more than 60 loci have been identified as associated with BMD. To investigate the genetic determinants of TB-BMD variation along the life course and test for age-specific effects, we performed a meta-analysis of 30 genome-wide association studies (GWASs) of TB-BMD including 66,628 individuals overall and divided across five age strata, each spanning 15 years. We identified variants associated with TB-BMD at 80 loci, of which 36 have not been previously identified; overall, they explain approximately 10% of the TB-BMD variance when combining all age groups and influence the risk of fracture. Pathway and enrichment analysis of the association signals showed clustering within gene sets implicated in the regulation of cell growth and SMAD proteins, overexpressed in the musculoskeletal system, and enriched in enhancer and promoter regions. These findings reveal TB-BMD as a relevant trait for genetic studies of osteoporosis, enabling the identification of variants and pathways influencing different bone compartments. Only variants in ESR1 and close proximity to RANKL showed a clear effect dependency on age. This most likely indicates that the majority of genetic variants identified influence BMD early in life and that their effect can be captured throughout the life course.
Keywords: genome-wide association studies, GWASs, age-dependent effects, bone mineral density, BMD, meta-regression, RANKL, ESR1, CREB3L1, total-body DXA, fracture, genetic correlation
Introduction
Osteoporosis is a disease characterized by low bone mass and microarchitectural deterioration of bone tissue leading to increased risk of fracture.1 It is diagnosed through the measurement of bone mineral density (BMD) utilizing dual-energy X-ray absorptiometry (DXA), which is the single best predictor of fracture.1
Bone is a dynamic tissue constantly undergoing resorption and formation. Bone mass increases steadily during childhood and markedly during adolescent growth.2 Peak bone mass is attained at approximately the third decade of life. Thereafter, until about 50 years of age, BMD remains fairly stable, by virtue of the coupling between bone formation and resorption (e.g., bone remodeling). Subsequently, bone resorption exceeds the rate of bone formation, resulting in a decrease in BMD, particularly in women after the onset of menopause.3
The International Society for Clinical Densitometry (ISCD) recommends performing DXA measurements at the lumbar spine, femoral neck, and total hip to diagnose osteoporosis in postmenopausal women and men who are 50 years or older (see Web Resources). Consequently, studies of BMD determinants are frequently based on measurements at these skeletal sites. By contrast, for the assessment of bone health in children and adolescents, total body (excluding head) and lumbar spine are the preferred sites to minimize measurement artifacts resulting from changing areas in growing bones (see Web Resources). Nevertheless, in elderly individuals degenerative changes in the spine can give elevated BMD readings.4 Moreover, total body DXA scans have been obtained in many adult research cohorts, primarily to assess body composition. Therefore, the total body BMD (TB-BMD) measurement is the most appropriate method for an unbiased assessment of BMD variation in the same skeletal site from childhood to old age.
To date, nearly 80 independent genetic variants have been shown to be robustly associated with variability in bone parameters.5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 Most of these markers have been identified in studies comprising tens of thousands of adult and elderly individuals with DXA-derived BMD measurements, although a few of them have been associated with BMD specifically in studies of pediatric cohorts.7 Furthermore, several of the associated variants display significant site-specific effects, possibly reflecting differences in bone composition across skeletal sites (e.g., cortical bone versus trabecular bone) or differential response to mechanical loading.7 Moreover, genetic studies on measures from peripheral quantitative computed tomography (pQCT) and bone quantitative ultrasound, which provide additional information regarding bone size, geometry, and (micro) architecture, identified genetic variants that may have specific effects on bone properties that are poorly captured by conventional DXA measurements.8, 9
Given the complex physiological processes underlying age-related changes in BMD across the life course, it is possible that genetic studies in more refined age groups will reveal variants in unreported loci as well as age-specific genetic effects. Thus, the purpose of this study was to identify gene variants associated with TB-BMD across the lifespan and to investigate possible differences of genetic effects across age periods.
Subjects and Methods
TB-BMD GWAS Meta-analyses
Subjects, BMD Measurement, and Imputation
This study comprised 30 epidemiological studies comprising ∼66,628 individuals from populations across America, Europe, and Australia, with a variety of designs (Supplemental Data; Table S1) and participant characteristics (Table S2). In summary, most participants came from population-based cohorts of European ancestry (86%), two cohorts comprised African American individuals (2%), and four other studies held a fraction of individuals from admixed background (14%). All research aims and the specific measurements have been approved by the correspondent Medical Ethical Committee of each participating study. Written informed consent was provided by all subjects or by their parents in the case of children.
Total body BMD (g/cm2) was measured by DXA following standard manufacturer protocols. As recommended by the International Society for Clinical Densitometry, total body less head (TBLH) was the measurement used in pediatric cohorts (see Web Resources) (e.g., 0–15 years). Detailed information on the assessments performed by each study can be found in Table S1.
All individuals included in this study had genome-wide array data. Quality control of genotypes is summarized in Table S1. To enable meta-analysis, each study performed genotype imputation using the cosmopolitan (all ethnicities combined) 1000 Genomes phase 1 v.3 (March 2012) reference panel, yielding ∼30,000,000 SNPs for analysis. Three studies used the combined 1000 Genomes and the UK10K reference panels as presented in Table S1.
Association Analysis, Quality Control, and Assessment
TB(LH)-BMD was corrected for age, weight, height, and genomic principal components (derived from GWAS data), as well as any additional study-specific covariates (e.g., recruiting center), in a linear regression model. For studies with non-related individuals, residuals were computed separately by sex, whereas for family-based studies sex was included as a covariate in the model. Finally, residuals were inverse normal transformed. The analyses were performed in each study for the overall population as well as in subgroups of individuals by age strata, defined by bins of 15 years (i.e., 0–15 years, 15–30 years, 30–45 years, 45–60 years, and 60 or more years). SNP association was tested for autosomal variants, in which the additive effect of each SNP on the normalized BMD residuals was estimated via linear regression.
A centralized quality-control procedure implemented in EasyQC18 was applied to all study-specific files of association results to identify cohort-specific issues. We excluded variants if they had missing information (e.g., missing association p value, beta estimate, alleles, allele frequency), nonsensical values (e.g., absolute beta estimates or standard errors > 10, association p values > 1 or < 0, imputation quality < 0, infinite beta estimates or standard errors), minor allele frequency (MAF) less than 0.5%, or imputation quality scores < 0.4 (Impute2) or < 0.3 (Minimac). Moreover, variants were flagged if they had large allele frequency deviations from reference populations (>0.6 for admixed studies and >0.3 for ancestry-homogeneous studies).
In the first instance, no exclusion criteria based on ancestry were applied for the meta-analysis (N = 66,628). In addition, meta-analyses were carried out across age strata (minimum sample size per bin N = 200 for each study) comprising 0–15 years (N = 11,807), 15–30 years (N = 4,180), 30–45 years (N = 10,062), 45–60 years (N = 18,805), and 60 or more years (N = 22,504). Further, summary data from cohorts of European ancestry only were meta-analyzed and used in subsequent analyses. We discarded variants present in fewer than three studies. Approximately 23,700,000 markers (including SNPs and INDELS) were assessed for association. We applied the conventional genome-wide significance level (GWS, p < 5 × 10−8) for SNP discovery.
We selected SNPs that were suggestively (12,567 SNPs, p < 5 × 10−6) associated with BMD in the overall meta-analysis, present in at least two studies per age bin, and with MAF differences across these meta-analyses lower than 0.5. We clumped this dataset with an r2 ≥ 0.8, using as reference the most strongly associated SNPs with BMD and pruning remaining SNPs within 0.7 Mb of each other. Age-dependent effects were assessed using a meta-regression approach for 1,464 SNPs obtained after this selection procedure. We ran a linear regression of the SNP effect estimates onto an intercept and the median age of each subgroup (e.g., each study stratified in age bins). As proposed previously,19 standard errors of the effect estimates of each subgroup were multiplied by the square root of the genomic inflation factor when it was greater than 1. We performed the meta-regression using the Metafor package,20 and any statistical evidence of linear association was corrected for multiple testing (Bonferroni correction; 0.05/1,464 = 3.4 × 10−5). The difference between beta-estimates in children versus elderly meta-analyses (Pdiff) was tested using Easy-strata.21
Conditional analyses were undertaken based on the meta-analysis of the studies of European ancestry only (N = 56,284). Only variants in the loci that reached GWS in this meta-analysis were assessed. The Rotterdam Study I (n = 6,291) was used as reference for precise calculation of the linkage disequilibrium (LD) between the analyzed markers. We used an iterative strategy as implemented in GCTA22 to determine (1) independence of association signals within loci discovered in our study, by means of stepwise model selection procedure per chromosome (–massoc-slct routine) and (2) the novelty of the association signals discovered by our meta-analysis with regard to variants reported in previous well-powered GWASs of different bone traits (Table S3). To this end, we performed the association analysis conditional on 78 variants present in our data and associated with different bone traits (–massoc-cond routine). These 78 SNPs were selected from different GWAS publications,5, 6, 7, 8, 9, 11, 12, 13 assuring their independence to avoid collinearity issues.
Shared Genetic Architecture of TB-BMD Fracture and Other Traits
LD Score Regression Analyses
We used the LD score regression package to estimate the heritability of TB-BMD and to rule out that our results were a product of bias (e.g., residual population stratification or cryptic relatedness). LD score regression uses GWAS summary statistics and assesses the SNP heritability based on the expected relationship between LD of neighboring SNPs and strength of association under a polygenic model.23 As this methodology relies on the LD structure throughout the genome, we restricted this analysis to summary statistics from the meta-analysis of cohorts comprising only individuals from European ancestry. We used the publicly available, pre-computed LD structure data files specific to European populations of the HAPMAP 3 reference panel. An extension of this method allows estimating the genetic correlation between two traits.24 This can be performed in the LDhub pipeline, a web utility that gathers data from many different GWAS meta-analysis.25 From the 199 traits, currently available in the website, we have restricted our analysis to those traits whose heritability z-scores were larger than 4 and were analyzed only in European ancestry individuals (following the recommendations in the LD score software website [Web Resources]). Additionally, we incorporated data from a recent GWAS meta-analysis of any type of fracture in individuals from European ancestry (N = 264,267; 37,778 case subjects) (K.T., unpublished data). In total, we assessed the genetic correlation between TB-BMD and 74 traits.
Mendelian Randomization Analysis
We undertook a two-sample Mendelian randomization approach26 to estimate the causal effect of TB-BMD on any type of fracture in the Europeans samples. In short, we constructed a score based on the independent genetic variants from the TB-BMD meta-analysis (European set and excluding secondary signals); whenever the selected variant was not present in the fracture meta-analysis, the second variant with the lowest p value in the locus (p < 5 × 10−8) and r2 > 0.8 was used as proxy. Thereafter, estimates derived from the TB-BMD summary statistics were pooled using methods similar to inverse-variance weighted fixed meta-analysis using the meta R-package (Web Resources).
Search for Biological and Functional Knowledge of the Identified Association Regions
For all those SNPs outside a 500 kb window from previously known bone-associated SNPs, we did a literature search in PubMed and Web of Science to evaluate whether nearby genes (within 500 kb) were known to play a role in bone metabolism. Also, we determined whether the annotated genes underlie any human Mendelian disorder with a skeletal manifestation, had knockout mouse models with a skeletal phenotype, or were annotated to pathways critical to bone metabolism. Genomic annotation for all SNPs was made based on UCSC hg19.
DEPICT Analyses
We used DEPICT,27 a recently developed tool to prioritize genes at the associated regions, define possible pathways by enrichment testing, and identify tissue and cell types in which genes from loci associated with TB-BMD. The methodology first selects all lead SNPs below a certain threshold with respect to a target p value. We tested both the complete set of GWS SNPs and the subset of those mapping only to loci not previously reported. Enriched gene sets were group based on the degree of gene overlap into “meta gene-sets” as proposed earlier,28 and their correlation visualized using Cytoscape 3.4 (Web Resources).
Functional Annotation to MicroRNA Binding Sites
We used the PolymiRTS,28 miRdSNP,29 and microSNiPer30 databases to obtain a list of variants located in predicted microRNA binding sites on the 3′ UTRs of genes, as described in detail elsewhere.31 In summary, index SNPs (most associated variants) of the GWS loci were submitted to SNAP (Web Resources) to retrieve their high LD proxy SNPs (with r2 > 0.8, limit distance 500 kb, and CEU panel) in the 1000 Genomes project. The resulting list of SNPs was annotated to the list of microRNA binding site variants obtained from the above mentioned publicly available databases.
Functional Enrichment Analysis of Trait-Associated Variants
GWAS analysis of regulatory or functional information enrichment with LD correction (GARFIELD)32 was used to characterize the putative functional contribution of TB-BMD-associated variants mapping to non-coding regions. GARFIELD employs a non-parametric analysis to calculate fold enrichment values for regulatory marks, at given significance thresholds, and then tests them for significance via permutation testing while accounting for LD, MAF, and local gene density.32 We used data regarding DNase-I hypersensitive sites, transcription factor binding sites, histone modifications, and chromatin states (ENCODE and Roadmap Epigenomics) from 424 cell types and tissues to capture and characterize possible cell-type-specific patterns of enrichment, as provided in the GARFIELD software (Web Resources). Fold enrichment statistics were tested at the four different significance thresholds (i.e., 1 × 10−8, 1 × 10−7, 1 × 10−6, and 1 × 10−5). Multiple-testing correction was performed on the effective number of annotations used, using the default p value threshold of 1 × 10−4.
Knockout Animal Models and Gene Expression in Bone Cells
Animal Models Survey
We surveyed databases from The International Mouse Phenotyping Consortium33 together with The International Knockout Mouse Consortium34 to identify knockout models of candidate genes resulting in skeletal phenotypes. Furthermore, we mined data from The Origins of Bone and Cartilage Disease (OBCD) project,35 specialized in murine skeletal phenotypes including digital X-ray microradiography on femurs and tail vertebrae, micro-CT analysis, femur three-point bend test load-displacement curves, and tail vertebrae compression testing from knockout mice and wild-type controls at 16 weeks of age. Experiments were undertaken by the Wellcome Trust Sanger Institute Mouse Genetics Project as part of the IKMC and licensed by the UK Home Office in accordance with the Animals (Scientific Procedures) Act 1986 and the recommendations of the Weatherall report.
Gene Expression in Bone Cells
Gene expression profiles of candidate genes were examined in primary mouse osteoblasts undergoing differentiation and bone marrow-derived osteoclasts. To study murine osteoblasts, pre-osteoblast-like cells were obtained from neonatal calvaria collected from C57BL/6J. Next Generation RNA sequencing using an Illumina HiSeq 2000 was used to evaluate the transcriptome every 2 days from day 2 to 18 days after osteoblast differentiation.6 Expression of genes in murine osteoclasts was determined using publicly available data obtained using Next-Gen RNA-sequencing applied to bone marrow-derived osteoclasts obtained from 6- to 8-week-old C57BL/6 mice.36 All procedures and use of mice for the neonatal osteoblast expression studies were approved by the Jackson Laboratory Animal Care and Use Committee (ACUC), in accordance with NIH guidelines for the care and use of laboratory animals.
Gene expression profiles of candidate genes were examined in human bone marrow-derived mesenchymal stem cells differentiated into osteoblast. Total RNA (n = 3) was isolated at day 0 (MSCs) and day 4 of osteoblast differentiation.37 Also, RNA was isolated during osteoclast differentiation. Peripheral blood mononuclear cells derived from buffy coats (Sanquin) were seeded in 96-well plates (5 × 105 cells per well) as previously described.38 Total RNA (n = 3) was isolated using Trizol at day 0 (PBMCs) and at day 7 of osteoclast differentiation. Illumina HumanHT-12 v3 BeadChip human whole-genome expression arrays were used for expression profiling. The quality of isolated RNA was assessed on a 2100 Bioanalyzer (Agilent Technologies). Data were analyzed as described in detail previously.37 Genes were designated as being expressed when at least one probe coding for the gene was significantly present in at least two of the three biological replicates.
Results
TB-BMD GWAS Meta-analyses
Analyses Including All Age Strata
Our meta-analysis of TB-BMD GWAS summary statistics (N = 66,628) identified variants in 76 independent loci associated with TB-BMD at a genome-wide significant (GWS, p ≤ 5 × 10−8) level (Figure 1, Table S4). Overall, there was no evidence of a strong inflation (genomic inflation factor [λ] of 1.08, Figure S1). Yet, inflation was observed in the range of common variants (0.2 > MAF < 0.5, λ = 1.19) due to polygenicity (LD score regression intercept = 1.007). In our results, one of the signals mapping to LDLRAD3 was driven entirely by individuals of African background (MAF = 0.043 in YRI panel) since the two associated variants are monomorphic in all other populations. The low allele frequency of this variant in our study (MAF = 0.025) and our limited statistical power (N = 6,748) in non-European samples warrants independent replication efforts to exclude the possibility of a false-positive association.
Figure 1.
Manhattan Plot of Association Statistics (−log10(p Values)) for TB-BMD Overall Meta-analysis
Each dot represents a SNP and the x axis indicates its chromosomal position (built 37 NCBI). Red dots represent SNPs at GWS loci that are not within ±500 kb of leading SNPs in previous GWASs with different bone traits. Dashed horizontal red and yellow lines mark the GWS threshold (p < 5 × 10−8) and suggestive threshold (p < 1 × 10−6), respectively. Novel loci in the only-CEU analysis are not shown.
In addition, a meta-analysis comprising 56,284 individuals of European ancestry (∼84% of the study population) identified variants in two additional GWS loci (Figures S1 and S2, Table S5). Association signals mapping to these loci were close to the GWS threshold in the overall meta-analysis (p = 1 × 10−7) and showed no evidence of heterogeneity (phet > 0.1). One of them, in 12q24.21 (MED13L), has not been previously associated with bone parameters (Table 1, Figure S3), while the other in 21q22.13 (CLDN14) is not fully independent from the previously reported hip BMD association signal12 (Table S5).
Table 1.
Index SNPs of Loci Not Previously Associated with BMD
| CHR | BP | rs Number | Locus | A1 | A2 | EAF | Effect | p | N | Annotation | Closest Gene | Notes | LS-beta | LS-P | FN-beta | FN-P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 8422676 | rs2252865 | 1p36.23 | T | C | 0.32 | −0.033 | 4.72 × 10−8 | 66,075 | intronic | RERE | novel biology | −0.019 | 0.043 | −0.025 | 0.002 |
| 1 | 110475971 | rs7548588 | 1p13.3 | T | C | 0.61 | −0.037 | 9.29 × 10−8 | 66,240 | intergenic | CSF1 | osteoclast differentiation39 | −0.030 | 0.001 | −0.022 | 0.005 |
| 1 | 220038825 | rs185048405 | 1q41 | T | C | 0.54 | 0.042 | 3.07 × 10−8 | 66,540 | intronic | SLC30A10 | manganese transport40 | −0.035 | 0.076 | −0.003 | 0.878 |
| 2 | 27741072 | rs780096 | 2p23.3 | C | G | 0.44 | −0.031 | 4.58 × 10−8 | 66,578 | intronic | GCKR | calcium regulation,41 hepatic traits42 | −0.014 | 0.129 | −0.017 | 0.029 |
| 2 | 40630678 | rs10490046 | 2p22.1 | A | C | 0.76 | 0.043 | 1.43 × 10−10 | 65,961 | intronic | SLC8A1 | bone mineralization43 | 0.015 | 0.162 | 0.021 | 0.025 |
| 2 | 68962137 | rs10048745 | 2p13.3 | A | G | 0.25 | −0.039 | 6.44 × 10−8 | 66,565 | 5′ UTR | ARHGAP25 | novel biology | −0.050 | 1.03 × 10−6 | −0.036 | 5.21 × 10−5 |
| 2 | 85484818 | rs11904127 | 2p11.2 | A | G | 0.55 | −0.032 | 2.65 × 10−8 | 66,561 | intronic | TCF7L1 | factors in Wnt signaling44 | −0.021 | 0.023 | −0.015 | 0.054 |
| 2 | 198874006 | rs1595824 | 2q33.1 | T | C | 0.47 | 0.034 | 2.65 × 10−8 | 60,171 | intronic | PLCL1 | negative regulation of bone formation45 | 0.022 | 0.201 | 0.052 | 2.20 × 10−4 |
| 2 | 202799604 | rs2350085 | 2q33.2 | T | C | 0.87 | −0.064 | 3.80 × 10−14 | 66,412 | intergenic | FZD7 | factors in Wnt signaling46 | −0.042 | 0.002 | −0.044 | 1.96 × 10−4 |
| 2 | 234303405 | rs838721 | 2q37.1 | A | G | 0.44 | −0.031 | 4.48 × 10−8 | 65,516 | intronic | DGKD | calcium regulation41 | −0.016 | 0.070 | −0.014 | 0.068 |
| 5 | 112221869 | rs818427 | 5q22.2 | T | C | 0.31 | 0.034 | 2.37 × 10−8 | 66,592 | intronic | APC | bone metabolism47 | 0.004 | 0.645 | 0.008 | 0.327 |
| 5 | 122847622 | rs11745493 | 5q23.2 | A | G | 0.75 | 0.044 | 7.75 × 10−12 | 66,597 | promoter | CSNK1G3 | novel biology | 0.010 | 0.326 | 0.025 | 0.005 |
| 7 | 27989403 | rs757138 | 7p15.1 | T | G | 0.69 | −0.035 | 3.33 × 10−8 | 66,043 | intronic | JAZF1 | novel biology | −0.016 | 0.126 | −0.025 | 0.004 |
| 7 | 30957702 | rs28362721 | 7p14.3 | T | C | 0.18 | −0.059 | 6.71 × 10−14 | 66,274 | intronic | AQP1 | bone metabolism48 | −0.037 | 0.002 | −0.049 | 1.39 × 10−6 |
| 7 | 50901491 | rs1548607 | 7p12.1 | A | G | 0.69 | 0.036 | 4.18 × 10−8 | 66,564 | intergenic | GRB10 | novel biology | 0.034 | 5.59 × 10−4 | 0.005 | 0.517 |
| 7 | 99130834 | rs34670419 | 7q22.1 | T | G | 0.04 | −0.088 | 1.09 × 10−8 | 66,336 | 3′ UTR | ZKSCAN5 | DHEAS and aging mechanisms49 | −0.127 | 9.28 × 10−8 | −0.080 | 8.19 × 10−5 |
| 10 | 112245400 | rs73349318 | 10q25.2 | A | T | 0.87 | −0.047 | 2.68 × 10−8 | 66,341 | intronic | DUSP5 | osteoclast differentiation50 | −0.042 | 0.001 | −0.051 | 8.76 × 10−6 |
| 10 | 124015986 | rs10788264 | 10q26.13 | A | G | 0.48 | −0.034 | 2.61 × 10−8 | 66,565 | intergenic | TACC2 | novel biology | −0.030 | 9.64 × 10−4 | −0.029 | 1.29 × 10−4 |
| 11 | 242859 | rs55781332 | 11p15.5 | A | G | 0.78 | −0.055 | 8.07 × 10−16 | 66,198 | intronic | PSMD13 | novel biology | −0.046 | 1.76 × 10−5 | −0.026 | 0.005 |
| 11 | 35083633 | rs2553773 | 11p13 | C | G | 0.41 | −0.037 | 1.49 × 10−10 | 66,619 | intergenic | CD44 | osteoclast activity51 | −0.015 | 0.101 | −0.015 | 0.054 |
| 11 | 35981346 | rs113964474a | 11p.13∗ | A | G | 0.03 | 0.485 | 1.41 × 10−8 | 6,748 | intronic | LDLRAD3 | novel biology | – | – | – | – |
| 11 | 69299537 | rs4980659 | 11q13.3 | C | G | 0.52 | 0.033 | 1.16 × 10−8 | 66,537 | intergenic | CCND1 | target of Wnt signaling52 | 0.039 | 1.58 × 10−5 | 0.023 | 0.003 |
| 11 | 121913230 | rs725670 | 11q24.1 | A | G | 0.38 | −0.032 | 3.61 × 10−8 | 66,565 | intergenic | BLID | novel biology | −0.020 | 0.028 | −0.011 | 0.172 |
| 12 | 90334829 | rs10777212 | 12q21.33 | T | G | 0.35 | 0.045 | 5.05 × 10−14 | 66,619 | intergenic | ATP2B1 | calcium absorption53 | 0.028 | 0.003 | 0.021 | 0.010 |
| 12 | 116555786 | rs73200209b | 12q24.21 | A | T | 0.80 | 0.045 | 2.51 × 10−8 | 51,240 | intronic | MED13L | novel biology | 0.030 | 0.167 | 0.036 | 0.044 |
| 13 | 37487021 | rs556429 | 13q13.3 | A | C | 0.23 | 0.039 | 1.46 × 10−8 | 66,504 | intronic | SMAD9 | osteoblast differentiation54 | 0.023 | 0.027 | 0.013 | 0.135 |
| 15 | 38340874 | rs12442242 | 15q14 | A | G | 0.85 | −0.051 | 4.94 × 10−10 | 66,403 | intergenic | TMCO5A | novel biology | −0.046 | 3.03 × 10−4 | −0.047 | 2.26 × 10−5 |
| 15 | 51537806 | rs2414098 | 15q21.2 | T | C | 0.39 | −0.033 | 1.99 × 10−8 | 66,562 | intronic | CYP19A1 | estrogen byosynthesis55 | −0.034 | 0.007 | −0.038 | 0.001 |
| 15 | 67420680 | rs1545161 | 15q22.33 | A | G | 0.56 | 0.041 | 1.06 × 10−12 | 66,004 | intronic | SMAD3 | osteoblast differentiation56 | 0.034 | 1.27 × 10−4 | 0.035 | 5.78 × 10−6 |
| 17 | 17804725 | rs8070128 | 17p11.2 | T | C | 0.58 | −0.039 | 1.98 × 10−11 | 66,625 | intronic | TOM1L2 | novel biology | −0.033 | 4.80 × 10−4 | −0.015 | 0.052 |
| 17 | 63771079 | rs9972944 | 17q24.1 | A | G | 0.41 | 0.036 | 6.87 × 10−10 | 66,595 | intronic | CEP112 | novel biology | 0.028 | 0.003 | 0.004 | 0.576 |
| 19 | 31654615 | rs6510186c | 19q12 | T | C | 0.26 | 0.068 | 3.11 × 10−8 | 18,782 | intergenic | TSHZ3 | novel biology | 0.004 | 0.713 | 0.006 | 0.492 |
| 20 | 39103882 | rs6029130 | 20q12 | T | C | 0.30 | 0.035 | 3.50 × 10−8 | 66,497 | intergenic | MAFB | osteoclast differentiation57 | 0.027 | 0.007 | 0.015 | 0.083 |
| 21 | 28773868 | rs1452102 | 21q21.3 | T | G | 0.59 | −0.035 | 1.74 × 10−8 | 66,489 | intergenic | ADAMTS5 | endochondral ossification58 | −0.029 | 0.001 | −0.015 | 0.056 |
| 21 | 36970350 | rs9976876 | 21q22.12 | T | G | 0.45 | −0.038 | 8.01 × 10−11 | 66,514 | intronic | RUNX1 | osteoclast differentiation59 | −0.019 | 0.031 | −0.016 | 0.041 |
| 21 | 40350744 | rs11910328 | 21q22.2 | A | G | 0.84 | −0.043 | 2.99 × 10−8 | 66,298 | intergenic | ETS2 | osteoblast maturation60 | −0.028 | 0.020 | −0.028 | 0.007 |
Variants associated with TB-BMD in the all-ages combined meta-analysis that map outside ±500 kb of known index SNPs of genetic associations with different bone traits. Genomic coordinates are on build 37 of the human genome. Notes refer to annotation based on the closest gene. Associations with lumbar spine (LS) and femoral neck (FN)-BMD.9 Beta coefficients and allele frequencies (EAF) are reported for the A1 allele.
aMonomorphic in European cohorts
bReported statistics from the in the meta-analysis of European populations
cReported statistics from the meta-analysis in the 30–45 age strata
Of the 78 identified loci, variants in 35 (45%) were not located within 500 kb of known association signals nor in regions of extended LD with them (Table 1, Figure S4). Index SNPs at these 35 loci were, in general, common non-coding variants. Twenty-two of these are located in close proximity to genes likely to influence bone metabolism as shown by previous functional studies (Table 1, Figure S3), including CSF1 (MIM: 120420), important for osteoclast differentiation,39 and SMAD3 (MIM: 603109), a critical component of the TGF-β signaling pathway.56 Across these 35 signals, 31 of the index SNPs were nominally associated (p < 0.05) with either lumbar spine or femoral neck BMD in the same direction as in the previously published GEFOS GWAS meta-analysis6 (Table 1). This comparison was not possible for the rs113964474 variant, because it was not available in the GEFOS study. Moreover, we found directionally concordant effect estimates (p < 0.05) for 73 of the 78 index SNPs of known bone association signals (Table S3). The markers that failed to replicate in our study were either previously associated with lumbar spine BMD but not femoral neck BMD (rs3905706 [MPP7, 10p12.1] and rs1878526 [INSIG2, 2q14.2]), associated specifically with the hip trochanter and intertrochanteric sub-regions (rs1949542 [RP11-384F7.1, 3q13.32]), or associated with BMD only in women (rs7017914 [XKR9, 8q13.3]) or only in children (rs754388 [RIN3, 14q32.12]).
Age-Dependent Effects
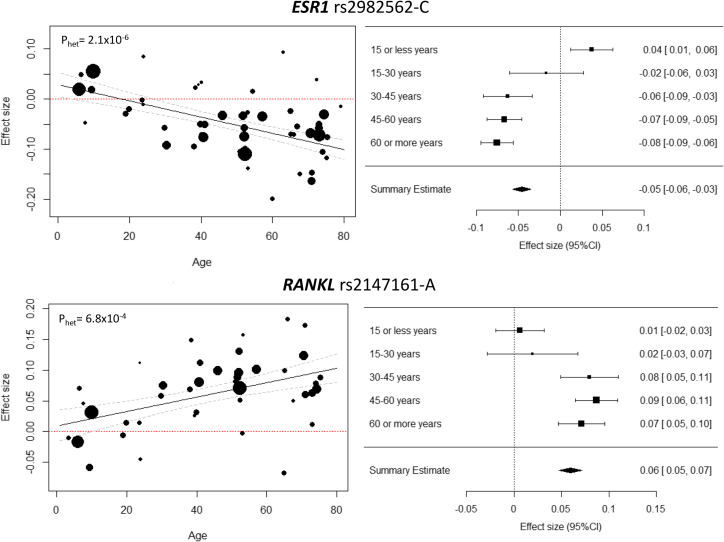
Meta-analyses across age strata resulted in the identification of variants mapping to two additional loci that were not detected in the overall meta-analysis (Figure S5; Table S6). In children (age group 0–15 years), the previously known 14q32.12 locus,7 harboring RIN3 (rs72699866, p = 1 × 10−8), and in the middle-aged (age group 45–60 years), a signal in the 19q12 locus mapping in the vicinity of TSHZ3 (rs6510186, p = 3.1 × 10−8) were identified. The rs72699866 variant leading the RIN3 signal in the youngest age stratum showed no evidence of association (p = 0.16) and high heterogeneity (phet = 6.6 × 10−5) in the overall meta-analysis. In fact, the effect of rs72699866 decreased significantly with age (ptrend = 1.69 × 10−9) (Figure S6) and showed a significant difference between the two extreme groups, i.e., children versus elderly (β0-15 = 0.099 [0.066, 0.134]; β>60 = −0.035 [−0.060, −0.010]; pdiff = 4.32 × 10−10). In contrast, the rs6510186 variant (19q12) showed nominal evidence of association and heterogeneity in the overall meta-analysis (p = 0.02; phet = 0.03). Nevertheless, no clear pattern of age dependency was identified (p = 0.2) for this SNP (Figure S6).
We also applied meta-regression analysis and found that variants mapping to 42 different loci showed nominally significant age-dependent effect (p < 0.05) (Table S7; Figure S7). In summary, 27 (64%) of the loci showed stronger effects in the older age groups. Of these, variants in the 6q25.1 (ESR1) and 13q14.11 (RANKL) loci remained significant after multiple-testing correction (p < 3.4 × 10−5) (Figure 2), while variants in 6p21.1 (RUNX2, rs148460475), 15q21.2 (CYP19A1, rs2414098), 17q21.31 (MEOX1, rs74835612), and 11p15.1 (SOX6, rs11822790) were only suggestive at p < 1 × 10−3.
Figure 2.
Age Dependence of the Genetic Variant Effect in the Meta-regression
The panels display leading SNPs from two loci exhibiting significant evidence for age influences. Heterogeneity p values (phet) are reported for the overall meta-analysis. In the left panels, each circle represents a study subgroup (i.e., study divided in age strata), with the circle size proportional to the inverse variance of the SNP main effect. In the right panels, forest plots display estimates obtained from each age-bin meta-analysis, with the symbol size proportional to the inverse variance of the SNP main effect.
Conditional Association Analyses
The stepwise conditional approach included studies comprising only individuals of European ancestry, as the method used relies on appropriate representability of the LD reference. Of the 76 GWS loci identified in the overall analysis, variants in 57 (19 previously unreported) loci were also GWS in the European-only analysis (Figure S2), likely a consequence of the lower power in this subgroup. We identified 81 SNPs independently associated with TB-BMD mapping to 58 different loci (one European-specific), 18 of which depicted multiple distinct signals attaining GWS (Table S8). These independent variants together explained 10.2% of TB-BMD variance. This proportion is slightly higher than the 7.4% TB-BMD variance explained by the 78 known variants associated with bone traits. Moreover, we identified independent signals in 13 of the 78 known bone loci after conditional analyses (Figure S2; Table S8).
Shared Genetic Architecture of TB-BMD, Fracture, and Other Traits
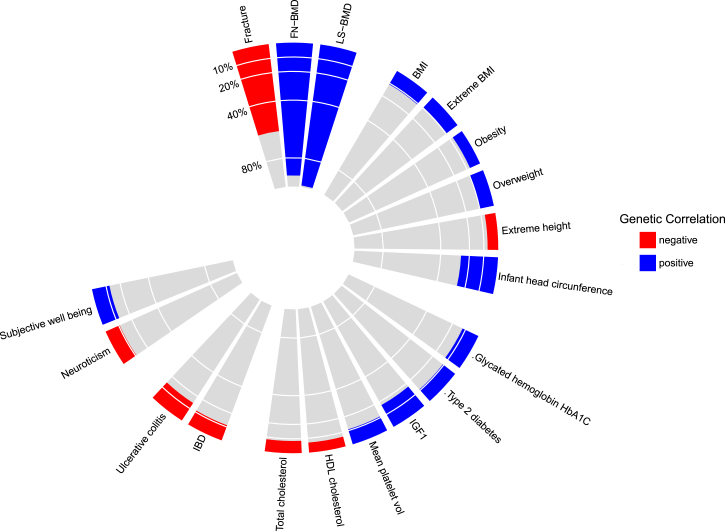
SNP heritability of TB-BMD in the European samples was estimated to be 0.259 (SE 0.017). TB-BMD was highly genetically correlated with BMD measured at other skeletal sites (ρ > 0.9). Among the non-BMD traits, all types of fracture showed the highest correlation (ρ = −0.61 [p = 1.6 × 10−27]). The MR approach indicated a strong causal relation where, per 1 standard deviation decrease in genetically determined TB-BMD, there is 56% increase in the risk of fracture (odds ratio 1.56 [1.50–1.62]). Other anthropometric, metabolic, and disease traits showed significant (yet weak) correlation with TB-BMD (Table S9, Figure 3). In contrast, other established risk factors for osteoporosis such as menopause or age of menarche showed no significant genetic correlation with TB-BMD.
Figure 3.
Genetic Correlations between TB-BMD and Other Traits and Diseases
Calculation was based on the summary statistics of the only-European meta-analysis (N = 56,284) and estimated by LD score regression implemented in LDHub. The diagram shows only traits whose correlation with TB-BMD was significant (p < 0.05).
Biological and Functional Knowledge of the Genes in BMD-Associated Loci
Loci not previously reported and their potential role in bone metabolism are summarized in Table 1. Several loci harbor genes implicated directly in bone metabolism (SLC8A1 [MIM: 182305], PLCL1 [MIM: 600597], ADAMTS5 [MIM: 605007]), affecting osteoblast or osteoclast differentiation and activity (CSF1 [MIM: 120420], DUSP5 [MIM: 603069], SMAD3 [MIM: 603109], SMAD9 [MIM: 603295], CD44 [MIM: 107269]), participating in Wnt signaling (FZD7 [MIM: 603410], TCF7L1 [MIM: 604652]), or regulating processes such as manganese or calcium absorption (GCKR [MIM: 600842], DGKD [MIM: 601826], SLC30A10 [MIM: 611146]) among others;39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60 while genes in at least 14 loci exert a potential novel role in bone biology. Rodent knockout models of several genes in the implicated loci show an altered skeletal phenotype (e.g., ostoepetrosis [Csf139], increased bone resorption [Aqp1,48 Cyp19a1,55 Cd4451], impaired skeletogenesis [Apc,47 Runx1,59 Smad356], deformities in the axial skeleton [Btg1,61 Atpaf262]). An effect on bone can be inferred for genes in other associated loci, for example, CYP19A1 (MIM: 107910) in 15q21.2 is an estrogen synthesis gene, estrogen being a key compound for bone maturation and maintenance, and ZKSCAN5 (MIM: 611272) in 7q22.1 is associated with circulating dehydroepiandrosterone sulfate (DHEAS) levels.49 DHEAS levels are positively correlated with BMD in adults and post-menopausal women.63 Across these loci, not previously reported as associated with BMD variation, we identified six exonic variants associated with TB-BMD, three of which were nonsynonymous variants all cataloged as benign both by SIFT and PolyPhen-2. We also identified 53 GWS coding variants in known loci, of which 33 are non-synonymous (Table S10). Only a low-frequency variant in LRP5 (MIM: 603506), rs4988321/A (11:68174189, MAF = 0.04), has a clinical annotation, constituting a homozygous G-to-A transition variant identified in a person with osteoporosis-pseudoglioma syndrome (OPPG [MIM: 259770]).64
DEPICT Analyses
Based on the overall meta-analysis, 53 genes were prioritized (FDR < 0.05), 15 of them mapping to loci not previously described (Table S11). Cells and tissues from the musculoskeletal system presented the largest enrichment of gene expression within the associated loci (Figure 4). These genes were overrepresented in 182 pathways clustered in 25 “meta gene-sets” (Table S12). The large majority of the clusters are involved in musculoskeletal development and bone homeostasis (Figure 4). The most significant of these implicated the regulation of cell growth and the TGFB signaling pathway and its mediating SMAD proteins.
Figure 4.
Results for Gene Set and Cell/Tissue Enrichment Analyses
Top: 25 Meta gene sets were defined from similarity clustering of significantly enriched gene sets (FDR < 5%). Each meta gene set was named after one of its member gene sets. The color of the meta gene sets represents the p value of the member set. Interconnection line width represents the Pearson correlation ρ between the gene membership scores for each meta gene set (ρ < 0.3, no line; 0.3 ≤ ρ < 0.5, narrow width; 0.5 ≤ ρ < 0.7, medium width; ρ ≥ 0.7, thick width). Bottom: Bars represent the level of evidence for genes in the associated loci to be expressed in any of the 209 medical subject heading (MeSH) tissue and cell type annotations. Highlighted in orange are these cell/tissue types significantly (FDR < 5%) enriched for the expression of the genes in the associated loci.
Restricting the DEPICT analysis to the subset of not previously reported associated regions resulted in significant enrichment of genes expressed in the musculoskeletal and immunological systems (Figure S8). Genes mapping to these loci were overrepresented in the SMAD binding pathway and TGFBR2 PPI (protein-protein interaction) subnetwork (FDR < 0.05).
Functional Annotation to MicroRNA Binding Sites
We then assessed whether the index SNPs of the 80 GWS loci detected in the main and subsequent GWAS (or their proxies in strong LD; r2 > 0.8) were located in predicted microRNA binding sites within the genes’ 3′ UTRs and thus were expected to disrupt the regulation of gene expression (Table S13). The index SNP within the 3′ UTR of ZKSCAN5 (mapping to a locus not previously identified), rs34670419 (MAF = 0.04), is predicted to create a binding site for miR-382-3p, a microRNA that is expressed in osteocytes and has been recently shown to be involved in osteogenic differentiation.65 In addition, eight proxy SNPs (mapping to PSMD13, ABCF2, GALNT3, PKDCC, REEP5, PPP6R3, AAGAB, and TOM1L2) are predicted to influence the binding of microRNAs to transcripts of their host gene.
Functional Enrichment Analysis of Trait-Associated Variants
As typically found in GWASs, the great majority of identified associations emerged from non-coding common variants and hold no direct annotation to molecular mechanisms. To assess whether there is relative enrichment of regulatory genomic marks underlying the associated variants in a cell-specific context, we used GARFIELD.32 We found relative ubiquitous enrichment for TB-BMD variants (empirical p < 2.4 × 10−4) in DNase I hypersensitive sites across the different cell types (Figure S9). Further, we found higher levels of fold-enrichment for enhancers (median 3.6, range [2.7, 4.4]) and promotors (median 3.2, range [2.9, 3.5]) than for transcribed regions (median 1.8, range [1.5, 2.2]).
Gene Expression in Bone Cells and Knockout Animal Models
From the 53 genes prioritized by DEPICT, only 49 had a mouse ortholog (Table S14). From these genes, only Mepe (osteocyte-specific) and Foxl1 were not expressed in murine osteoblast or osteoclast. Moreover, 61% of the prioritized genes were expressed in human cells in vitro during osteoblast or osteoclast differentiation (Table S14). AQP1 was the only prioritized gene mapping to a locus not previously reported showing no expression in the human bone cells differentiation experiments.
Knockout models were widely available in at least one of the different databases assessed. Nevertheless, in-depth bone phenotyping performed under the OBCD project was available for only four knockout models (Table S15). Two of these, DUSP5 and CD300LG, showed no significant bone phenotype. The TCF7L1 knockout model only showed lower cortical diameter in the femur without other clear bone phenotype. Nevertheless, TCF7L1 was shown to be expressed during osteoblastogenesis. Conversely, homozygous knockout for CREB3L1 showed a clear bone phenotype consisting of low BMC both at the vertebrae and femur together with a strong trabecular and cortical phenotype affecting bone strength (Figure S10). CREB3L1 maps to 11p11.2, a previously identified BMD locus5 harboring ARHGAP1 and LRP4 as candidates to underlie the GWAS signal in a region of extended LD.
Discussion
This meta-analysis of TB-BMD comprising up to 66,000 individuals identified variants in 36 loci not previously reported and replicated at GWS level several association signals identified by GWASs of diverse bone phenotypes. Bioinformatics analyses suggest enrichment of these 36 loci for genes expressed in the musculoskeletal system and solidly represented in the SMAD binding pathway and the TGFBR2 PPI subnetwork. We also demonstrate that for variants in few loci the size of the effect is age dependent; variants in two loci (RIN3 and TSHZ3) were identified only by the age-stratified analyses despite less power (smaller sample size); while for variants in two other loci (ESR1 and RANKL) there was significant evidence of age heterogeneity derived from a meta-regression of the genetic effects with age. Our results strengthen the evidence that genetic variants influence BMD from a young age and support the value of peak bone mass as an important determinant of bone health later in life.
Traditionally, DXA-BMD measurements performed at sites of high fracture risk (i.e., femoral neck, lumbar spine and forearm) have been used in genetic epidemiological investigations of bone health in adults. Instead, we have used BMD measurements derived from total body scans. Not only do we show a high overlap of association signals with previous GWASs of different bone traits, including DXA, pQCT, and ultrasound measurements, but we have also identified unreported loci. Five known associations failed to replicate in our studies, even though we cannot discard these associations as false positives, because these results might also indicate that variants whose effect is highly specific to skeletal sites, skeletal properties, sex, or age groups cannot be detected in our TB-BMD meta-analysis. It is plausible that more variants of this type exist and will be discovered as site-specific BMD meta-analyses are performed in increasingly powered settings. Furthermore, the genetic correlation of TB-BMD with BMD measured at other sites was close to one. Nevertheless, we found that a decrease of one standard deviation in the genetically determined TB-BMD resulted in at least 50% higher odds of suffering a fracture. Significant genetic correlations with other traits (i.e., BMI, IGF1, and ulcerative colitis) reflect the systemic context of skeletal biology and merit further study by future efforts to elucidate the underlying mechanisms.
Genes in the associated loci were highly expressed in the musculoskeletal system and overrepresented in gene sets related to bone development. The prioritized gene CREB3L1 (MIM: 616215) in 11p11.2 observed a clear bone phenotype in our mouse knockout model, which corroborates the findings of previous work showing substantial rescue of CREB3L1 deficiency with bisphosphonates and its critical role for bone formation.66 This locus, characterized by extended LD, also harbors LRP4 (MIM: 604270) whose knockout model presents with increased trabecular and cortical bone mass.67 This is in line with our conditional analysis identifying multiple independent signals in the region, making it likely that both genes are influencing bone biology. Altogether, we demonstrated that TB-BMD offers a powerful alternative to identify genetic variants associated with bone metabolism.
Variants mapping to 14q32 harboring RIN3 (MIM: 610223) were associated only at a GWS level in children (i.e., <15 years) and were only nominally significant in the elderly group (i.e., >60 years). This age-related heterogeneity may explain why this locus has not been detected in BMD meta-analyses in adults, although being identified in relation to pediatric BMD7 and Paget disease (PDB [MIM: 602080]) GWASs.68 In addition, another signal mapping to 19q12 harboring TSHZ3 (MIM: 614119) was significant in adults aged 45–60 years but not in other age groups analyzed or in previous studies, alluding to a false-positive association, so replication of this finding is necessary.
Our analyses revealed variants in the 6q25.1 (ESR1) and 13q14.11 (RANKL) loci demonstrating the most compelling evidence for age-modulation effects. The 6q25.1 locus harboring ESR1 (MIM: 133430), an important genetic factor in normal BMD variability, was not associated with BMD in children below 15 years of age, where the largest cohorts (i.e., Avon Longitudinal Study of Parents and Children [ALSPAC] and the Generation R Study) comprise predominantly pre-pubertal children. As levels of estradiol before puberty are low,69 a negligible effect of ESR1 variants on BMD is expected. Likewise, in mouse models the expression of RANKL (MIM: 602642) in bone is markedly increased with advancing age from young to adult and related to bone loss.70 Accordingly, variants influencing RANKL expression show a larger effect later in life. In general, a substantial heterogeneity of the genetic effects in the overall meta-analysis was explained by age, but the inclusion of larger sample sizes (avoiding age exclusion criteria and incrementing statistical power) leveled off the loss of power due to the heterogeneity of the genetic effects.
In brief, variants with evidence of age-specific effects were exceptional in our study. These results might reflect a lack of statistical power as only SNPs showing suggestive evidence (p < 5 × 10−6) of association with TB-BMD in the overall meta-analysis were tested for age-specific effects. This selection criteria aimed to include SNPs whose heterogeneity might have hampered their statistical significance in the overall meta-analysis, and at the same time maximize the power to discover variants with real age-dependent effects. Alternatively, these results indicate that most of the genetic variants identified so far, by us and others, influence BMD from early ages onward, and their effect persist throughout the life course. However, variants in 27 of the 42 loci (64%) showing nominal evidence for age-dependent effects had larger effects in the older groups. Nonetheless, this requires careful interpretation given the uneven sample sizes between the age groups and the criteria to select markers for the meta-regression based on significance in the overall meta-analysis. Collectively, this argues in favor of enlarging studies focused on younger populations—where the statistical power is still restricted—to discover additional genetic variants influencing BMD.
Our study has some limitations. A key disadvantage of our design is that we group the data based on age spans rather than life stages. Crucial information for this assessment, such as puberty onset in children and adolescents or menopausal status in the adults, was not available across the majority of the cohorts. Other strategies like using smaller age spans will result in even less statistical power of the discovery setting. Similarly, despite the large sample size of our study, we identified very few variants in the low-frequency spectrum (MAF < 5%), indicating that comprehensive surveys of rare variation influencing BMD still require even larger sample sizes, on top of better resources for imputation of the rarer variants, possibly needing population-specific references. Such strategies will be key to explain a larger fraction of the genetic variability of BMD phenotypes, as illustrated for other traits such as height or BMI.71 Moreover, the identified SNPs are, in the vast majority, non-coding variants, raising the possibility that the causal genes are different from the candidate genes we have prioritized based on the current biological knowledge and bioinformatic prediction tools. Additional functional studies are required to determine the potential role of the genes in the identified loci.
In conclusion, we performed a genome-wide survey for association with DXA-derived TB-BMD, combining data from five age groups including children and older individuals. In contrast to previous large-scale meta-analyses,5, 6 we used DXA-derived TB-BMD rather than measurements on specific skeletal sites prone to fracture to identify genetic factors influencing BMD variation. We demonstrate that TB-BMD is a valid phenotype for this purpose, as we replicated more than 90% of the previously reported signals. Most importantly, we identify variants in 36 loci associated with TB-BMD not previously reported by previous GWASs of bone phenotypes. Our results show steadiness in the magnitude of the genetic effects on BMD for most of the BMD-associated variants. While the contrasting skeletal physiology across different age periods is well established (i.e., endochondral ossification, linear growth, modeling, remodeling, etc.), peak bone mass acquisition remains the major determinant of variability at any age. These findings strongly support the importance of the bone accrual process in the definition of BMD status and fracture susceptibility throughout the life course.
Conflicts of Interests
B.M.P. serves on the DSMB of a clinical trial for the manufacturer (Zoll LifeCor) and on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. U.S., G.P., and K.S. are employed be deCODE genetics/Amgen Inc. M.A.N. is supported by Data Tecnica and also consults for Illumina, Inc. and the Michael J. Fox Foundation.
Acknowledgments
The authors would like to thank the many colleagues who contributed to collection and phenotypic characterization of the clinical samples, as well as genotyping and analysis of the GWAS data. Part of this work was conducted using the UK Biobank resource. Full list of acknowledgments, funding organizations, and grants are listed per cohort in the Supplemental Data.
Published: January 4, 2018
Footnotes
Supplemental Data include Supplemental Acknowledgments, Supplemental Note (Cohort Descriptions), 10 figures, and 15 tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2017.12.005.
Accession Numbers
GWAS summary data for the main and age-strata meta-analyses together with the corresponding regional plots of GWS signals have been deposited in the GEFOS website (Web Resources). Gene expression data presented in this paper can be retrieved from the Gene Expression Omnibus (GEO) as follows: murine osteoclasts (GSM1873361) and osteoblasts (GSE54461); human osteoblast differentiation (GSE54461).
Web Resources
Cytoscape, http://www.cytoscape.org/
GEFOS, http://www.gefos.org/
Meta R-package, https://github.com/guido-s/meta
OMIM, http://www.omim.org/
Supplemental Data
Estimates were derived from the overall approach. Beta coefficients and allele frequency (EAF) are reported for the A1 allele.
Estimates were derived from the all-age combined approach. Beta coefficients and allele frequency (EAF) are reported for the A1 allele.
Only suggestively associated variants (p < 5 × 10-6) in the TB-BMD overall meta-analysis were subjected to meta-regression assessment. Genomic coordinates are on build 37 of the human genome. Allele frequencies (EAF) are reported for the A1 allele. C.L-C.U, 95% Confidence interval lower and upper limit.
Based on genome-wide significant variants in the overall TB-BMD meta-analysis. These 182 gene-sets were further clustered in 25 ‘metagene-sets’ shown in Figure 4.
Data was collected for the 55 genes prioritized by DEPICT. Detailed bone phenotyping from the Origins of Bone and Cartilage Disease (OCBD) is presented in Table S15.
Knockout lines with a skeletal parameter greater than 2 standard deviations from the reference mean are highlighted in orange. The standard deviation from the reference mean for each parameter is shown with those greater than 2 highlighted (Black above the mean: Red below the mean).
References
- 1.Johnell O., Kanis J.A., Oden A., Johansson H., De Laet C., Delmas P., Eisman J.A., Fujiwara S., Kroger H., Mellstrom D. Predictive value of BMD for hip and other fractures. J. Bone Miner. Res. 2005;20:1185–1194. doi: 10.1359/JBMR.050304. [DOI] [PubMed] [Google Scholar]
- 2.Farr J.N., Khosla S. Skeletal changes through the lifespan--from growth to senescence. Nat. Rev. Endocrinol. 2015;11:513–521. doi: 10.1038/nrendo.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hendrickx G., Boudin E., Van Hul W. A look behind the scenes: the risk and pathogenesis of primary osteoporosis. Nat. Rev. Rheumatol. 2015;11:462–474. doi: 10.1038/nrrheum.2015.48. [DOI] [PubMed] [Google Scholar]
- 4.Tenne M., McGuigan F., Besjakov J., Gerdhem P., Åkesson K. Degenerative changes at the lumbar spine--implications for bone mineral density measurement in elderly women. Osteoporos. Int. 2013;24:1419–1428. doi: 10.1007/s00198-012-2048-0. [DOI] [PubMed] [Google Scholar]
- 5.Estrada K., Styrkarsdottir U., Evangelou E., Hsu Y.H., Duncan E.L., Ntzani E.E., Oei L., Albagha O.M., Amin N., Kemp J.P. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet. 2012;44:491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng H.F., Forgetta V., Hsu Y.H., Estrada K., Rosello-Diez A., Leo P.J., Dahia C.L., Park-Min K.H., Tobias J.H., Kooperberg C., AOGC Consortium. UK10K Consortium Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature. 2015;526:112–117. doi: 10.1038/nature14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kemp J.P., Medina-Gomez C., Estrada K., St Pourcain B., Heppe D.H., Warrington N.M., Oei L., Ring S.M., Kruithof C.J., Timpson N.J. Phenotypic dissection of bone mineral density reveals skeletal site specificity and facilitates the identification of novel loci in the genetic regulation of bone mass attainment. PLoS Genet. 2014;10:e1004423. doi: 10.1371/journal.pgen.1004423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paternoster L., Lorentzon M., Lehtimäki T., Eriksson J., Kähönen M., Raitakari O., Laaksonen M., Sievänen H., Viikari J., Lyytikäinen L.P. Genetic determinants of trabecular and cortical volumetric bone mineral densities and bone microstructure. PLoS Genet. 2013;9:e1003247. doi: 10.1371/journal.pgen.1003247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moayyeri A., Hsu Y.H., Karasik D., Estrada K., Xiao S.M., Nielson C., Srikanth P., Giroux S., Wilson S.G., Zheng H.F. Genetic determinants of heel bone properties: genome-wide association meta-analysis and replication in the GEFOS/GENOMOS consortium. Hum. Mol. Genet. 2014;23:3054–3068. doi: 10.1093/hmg/ddt675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medina-Gomez C., Kemp J.P., Estrada K., Eriksson J., Liu J., Reppe S., Evans D.M., Heppe D.H., Vandenput L., Herrera L. Meta-analysis of genome-wide scans for total body BMD in children and adults reveals allelic heterogeneity and age-specific effects at the WNT16 locus. PLoS Genet. 2012;8:e1002718. doi: 10.1371/journal.pgen.1002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang T.L., Guo Y., Liu Y.J., Shen H., Liu Y.Z., Lei S.F., Li J., Tian Q., Deng H.W. Genetic variants in the SOX6 gene are associated with bone mineral density in both Caucasian and Chinese populations. Osteoporos. Int. 2012;23:781–787. doi: 10.1007/s00198-011-1626-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L., Choi H.J., Estrada K., Leo P.J., Li J., Pei Y.F., Zhang Y., Lin Y., Shen H., Liu Y.Z. Multistage genome-wide association meta-analyses identified two new loci for bone mineral density. Hum. Mol. Genet. 2014;23:1923–1933. doi: 10.1093/hmg/ddt575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pei Y.F., Xie Z.G., Wang X.Y., Hu W.Z., Li L.B., Ran S., Lin Y., Hai R., Shen H., Tian Q. Association of 3q13.32 variants with hip trochanter and intertrochanter bone mineral density identified by a genome-wide association study. Osteoporos. Int. 2016;27:3343–3354. doi: 10.1007/s00198-016-3663-y. [DOI] [PubMed] [Google Scholar]
- 14.Styrkarsdottir U., Thorleifsson G., Eiriksdottir B., Gudjonsson S.A., Ingvarsson T., Center J.R., Nguyen T.V., Eisman J.A., Christiansen C., Thorsteinsdottir U. Two Rare Mutations in the COL1A2 Gene Associate With Low Bone Mineral Density and Fractures in Iceland. J. Bone Miner. Res. 2016;31:173–179. doi: 10.1002/jbmr.2604. [DOI] [PubMed] [Google Scholar]
- 15.Koller D.L., Zheng H.F., Karasik D., Yerges-Armstrong L., Liu C.T., McGuigan F., Kemp J.P., Giroux S., Lai D., Edenberg H.J. Meta-analysis of genome-wide studies identifies WNT16 and ESR1 SNPs associated with bone mineral density in premenopausal women. J. Bone Miner. Res. 2013;28:547–558. doi: 10.1002/jbmr.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielson C.M., Liu C.T., Smith A.V., Ackert-Bicknell C.L., Reppe S., Jakobsdottir J., Wassel C., Register T.C., Oei L., Alonso N. Novel Genetic Variants Associated With Increased Vertebral Volumetric BMD, Reduced Vertebral Fracture Risk, and Increased Expression of SLC1A3 and EPHB2. J. Bone Miner. Res. 2016;31:2085–2097. doi: 10.1002/jbmr.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Styrkarsdottir U., Thorleifsson G., Gudjonsson S.A., Sigurdsson A., Center J.R., Lee S.H., Nguyen T.V., Kwok T.C., Lee J.S., Ho S.C. Sequence variants in the PTCH1 gene associate with spine bone mineral density and osteoporotic fractures. Nat. Commun. 2016;7:10129. doi: 10.1038/ncomms10129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winkler T.W., Day F.R., Croteau-Chonka D.C., Wood A.R., Locke A.E., Mägi R., Ferreira T., Fall T., Graff M., Justice A.E., Genetic Investigation of Anthropometric Traits (GIANT) Consortium Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 2014;9:1192–1212. doi: 10.1038/nprot.2014.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simino J., Shi G., Bis J.C., Chasman D.I., Ehret G.B., Gu X., Guo X., Hwang S.J., Sijbrands E., Smith A.V., LifeLines Cohort Study Gene-age interactions in blood pressure regulation: a large-scale investigation with the CHARGE, Global BPgen, and ICBP Consortia. Am. J. Hum. Genet. 2014;95:24–38. doi: 10.1016/j.ajhg.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Viechtbauer W. Conducting Meta-Analyses in R with the metafor Package. J. Stat. Softw. 2010;36:1–48. [Google Scholar]
- 21.Winkler T.W., Kutalik Z., Gorski M., Lottaz C., Kronenberg F., Heid I.M. EasyStrata: evaluation and visualization of stratified genome-wide association meta-analysis data. Bioinformatics. 2015;31:259–261. doi: 10.1093/bioinformatics/btu621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J., Ferreira T., Morris A.P., Medland S.E., Madden P.A.F., Heath A.C., Martin N.G., Montgomery G.W., Weedon M.N., Loos R.J., Genetic Investigation of ANthropometric Traits (GIANT) Consortium. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012;44:369–375, S1–S3. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Patterson N., Daly M.J., Price A.L., Neale B.M., Schizophrenia Working Group of the Psychiatric Genomics Consortium LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.R., Duncan L., Perry J.R., Patterson N., Robinson E.B., ReproGen Consortium. Psychiatric Genomics Consortium. Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3 An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng J., Erzurumluoglu A.M., Elsworth B.L., Kemp J.P., Howe L., Haycock P.C., Hemani G., Tansey K., Laurin C., Pourcain B.S., Early Genetics and Lifecourse Epidemiology (EAGLE) Eczema Consortium LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burgess S., Butterworth A., Thompson S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013;37:658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pers T.H., Karjalainen J.M., Chan Y., Westra H.J., Wood A.R., Yang J., Lui J.C., Vedantam S., Gustafsson S., Esko T., Genetic Investigation of ANthropometric Traits (GIANT) Consortium Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharya A., Ziebarth J.D., Cui Y. PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2014;42:D86–D91. doi: 10.1093/nar/gkt1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong J., Tong Y., Zhang H.M., Wang K., Hu T., Shan G., Sun J., Guo A.Y. Genome-wide identification of SNPs in microRNA genes and the SNP effects on microRNA target binding and biogenesis. Hum. Mutat. 2012;33:254–263. doi: 10.1002/humu.21641. [DOI] [PubMed] [Google Scholar]
- 30.Barenboim M., Zoltick B.J., Guo Y., Weinberger D.R. MicroSNiPer: a web tool for prediction of SNP effects on putative microRNA targets. Hum. Mutat. 2010;31:1223–1232. doi: 10.1002/humu.21349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghanbari M., Franco O.H., de Looper H.W., Hofman A., Erkeland S.J., Dehghan A. Genetic Variations in MicroRNA-Binding Sites Affect MicroRNA-Mediated Regulation of Several Genes Associated With Cardio-metabolic Phenotypes. Circ Cardiovasc Genet. 2015;8:473–486. doi: 10.1161/CIRCGENETICS.114.000968. [DOI] [PubMed] [Google Scholar]
- 32.Iotchkova V., Ritchie G.R.S., Geihs M., Morganella S., Min J.L., Walter K., Timpson N.J., Dunham I., Birney E., Soranzo N. GARFIELD - GWAS Analysis of Regulatory or Functional Information Enrichment with LD correction. bioRxiv. 2016 [Google Scholar]
- 33.Dickinson M.E., Flenniken A.M., Ji X., Teboul L., Wong M.D., White J.K., Meehan T.F., Weninger W.J., Westerberg H., Adissu H., International Mouse Phenotyping Consortium. Jackson Laboratory. Infrastructure Nationale PHENOMIN, Institut Clinique de la Souris (ICS) Charles River Laboratories. MRC Harwell. Toronto Centre for Phenogenomics. Wellcome Trust Sanger Institute. RIKEN BioResource Center High-throughput discovery of novel developmental phenotypes. Nature. 2016;537:508–514. doi: 10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Angelis M.H., Nicholson G., Selloum M., White J., Morgan H., Ramirez-Solis R., Sorg T., Wells S., Fuchs H., Fray M., EUMODIC Consortium Analysis of mammalian gene function through broad-based phenotypic screens across a consortium of mouse clinics. Nat. Genet. 2015;47:969–978. doi: 10.1038/ng.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freudenthal B., Logan J., Croucher P.I., Williams G.R., Bassett J.H., Sanger Institute Mouse Pipelines Rapid phenotyping of knockout mice to identify genetic determinants of bone strength. J. Endocrinol. 2016;231:R31–R46. doi: 10.1530/JOE-16-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim K., Punj V., Kim J.M., Lee S., Ulmer T.S., Lu W., Rice J.C., An W. MMP-9 facilitates selective proteolysis of the histone H3 tail at genes necessary for proficient osteoclastogenesis. Genes Dev. 2016;30:208–219. doi: 10.1101/gad.268714.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van de Peppel J., Strini T., Tilburg J., Westerhoff H., van Wijnen A.J., van Leeuwen J.P. Identification of Three Early Phases of Cell-Fate Determination during Osteogenic and Adipogenic Differentiation by Transcription Factor Dynamics. Stem Cell Reports. 2017;8:947–960. doi: 10.1016/j.stemcr.2017.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koek W.N.H., van der Eerden B.C.J., Alves R.D.A.M., van Driel M., Schreuders-Koedam M., Zillikens M.C., van Leeuwen J.P.T.M. Osteoclastogenic capacity of peripheral blood mononuclear cells is not different between women with and without osteoporosis. Bone. 2017;95:108–114. doi: 10.1016/j.bone.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 39.Dobbins D.E., Sood R., Hashiramoto A., Hansen C.T., Wilder R.L., Remmers E.F. Mutation of macrophage colony stimulating factor (Csf1) causes osteopetrosis in the tl rat. Biochem. Biophys. Res. Commun. 2002;294:1114–1120. doi: 10.1016/S0006-291X(02)00598-3. [DOI] [PubMed] [Google Scholar]
- 40.Claro da Silva T., Hiller C., Gai Z., Kullak-Ublick G.A. Vitamin D3 transactivates the zinc and manganese transporter SLC30A10 via the Vitamin D receptor. J. Steroid Biochem. Mol. Biol. 2016;163:77–87. doi: 10.1016/j.jsbmb.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 41.O’Seaghdha C.M., Wu H., Yang Q., Kapur K., Guessous I., Zuber A.M., Köttgen A., Stoudmann C., Teumer A., Kutalik Z., SUNLIGHT Consortium. GEFOS Consortium Meta-analysis of genome-wide association studies identifies six new Loci for serum calcium concentrations. PLoS Genet. 2013;9:e1003796. doi: 10.1371/journal.pgen.1003796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Speliotes E.K., Yerges-Armstrong L.M., Wu J., Hernaez R., Kim L.J., Palmer C.D., Gudnason V., Eiriksdottir G., Garcia M.E., Launer L.J., NASH CRN. GIANT Consortium. MAGIC Investigators. GOLD Consortium Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ousingsawat J., Wanitchakool P., Schreiber R., Wuelling M., Vortkamp A., Kunzelmann K. Anoctamin-6 controls bone mineralization by activating the calcium transporter NCX1. J. Biol. Chem. 2015;290:6270–6280. doi: 10.1074/jbc.M114.602979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shy B.R., Wu C.I., Khramtsova G.F., Zhang J.Y., Olopade O.I., Goss K.H., Merrill B.J. Regulation of Tcf7l1 DNA binding and protein stability as principal mechanisms of Wnt/β-catenin signaling. Cell Rep. 2013;4:1–9. doi: 10.1016/j.celrep.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsutsumi K., Matsuda M., Kotani M., Mizokami A., Murakami A., Takahashi I., Terada Y., Kanematsu T., Fukami K., Takenawa T. Involvement of PRIP, phospholipase C-related, but catalytically inactive protein, in bone formation. J. Biol. Chem. 2011;286:31032–31042. doi: 10.1074/jbc.M111.235903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y., Dudley A.T. Noncanonical frizzled signaling regulates cell polarity of growth plate chondrocytes. Development. 2009;136:1083–1092. doi: 10.1242/dev.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miclea R.L., Karperien M., Bosch C.A., van der Horst G., van der Valk M.A., Kobayashi T., Kronenberg H.M., Rawadi G., Akçakaya P., Löwik C.W. Adenomatous polyposis coli-mediated control of beta-catenin is essential for both chondrogenic and osteogenic differentiation of skeletal precursors. BMC Dev. Biol. 2009;9:26. doi: 10.1186/1471-213X-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu Q.T., Ma Q.J., He C.Y., Wang C.X., Gao S., Hou X., Ma T.H. Reduced bone mineral density and bone metabolism in aquaporin-1 knockout mice. Chem. Res. Chin. Univ. 2007;23:297–299. [Google Scholar]
- 49.Zhai G., Teumer A., Stolk L., Perry J.R.B., Vandenput L., Coviello A.D., Koster A., Bell J.T., Bhasin S., Eriksson J., MuTHER Consortium Eight common genetic variants associated with serum DHEAS levels suggest a key role in ageing mechanisms. PLoS Genet. 2011;7:e1002025. doi: 10.1371/journal.pgen.1002025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moon S.J., Lim M.A., Park J.S., Byun J.K., Kim S.M., Park M.K., Kim E.K., Moon Y.M., Min J.K., Ahn S.M. Dual-specificity phosphatase 5 attenuates autoimmune arthritis in mice via reciprocal regulation of the Th17/Treg cell balance and inhibition of osteoclastogenesis. Arthritis Rheumatol. 2014;66:3083–3095. doi: 10.1002/art.38787. [DOI] [PubMed] [Google Scholar]
- 51.Hayer S., Steiner G., Görtz B., Reiter E., Tohidast-Akrad M., Amling M., Hoffmann O., Redlich K., Zwerina J., Skriner K. CD44 is a determinant of inflammatory bone loss. J. Exp. Med. 2005;201:903–914. doi: 10.1084/jem.20040852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krishnan V., Bryant H.U., Macdougald O.A. Regulation of bone mass by Wnt signaling. J. Clin. Invest. 2006;116:1202–1209. doi: 10.1172/JCI28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryan Z.C., Craig T.A., Filoteo A.G., Westendorf J.J., Cartwright E.J., Neyses L., Strehler E.E., Kumar R. Deletion of the intestinal plasma membrane calcium pump, isoform 1, Atp2b1, in mice is associated with decreased bone mineral density and impaired responsiveness to 1, 25-dihydroxyvitamin D3. Biochem. Biophys. Res. Commun. 2015;467:152–156. doi: 10.1016/j.bbrc.2015.09.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsukamoto S., Mizuta T., Fujimoto M., Ohte S., Osawa K., Miyamoto A., Yoneyama K., Murata E., Machiya A., Jimi E. Smad9 is a new type of transcriptional regulator in bone morphogenetic protein signaling. Sci. Rep. 2014;4:7596. doi: 10.1038/srep07596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyaura C., Toda K., Inada M., Ohshiba T., Matsumoto C., Okada T., Ito M., Shizuta Y., Ito A. Sex- and age-related response to aromatase deficiency in bone. Biochem. Biophys. Res. Commun. 2001;280:1062–1068. doi: 10.1006/bbrc.2001.4246. [DOI] [PubMed] [Google Scholar]
- 56.Borton A.J., Frederick J.P., Datto M.B., Wang X.F., Weinstein R.S. The loss of Smad3 results in a lower rate of bone formation and osteopenia through dysregulation of osteoblast differentiation and apoptosis. J. Bone Miner. Res. 2001;16:1754–1764. doi: 10.1359/jbmr.2001.16.10.1754. [DOI] [PubMed] [Google Scholar]
- 57.Kim K., Kim J.H., Lee J., Jin H.M., Kook H., Kim K.K., Lee S.Y., Kim N. MafB negatively regulates RANKL-mediated osteoclast differentiation. Blood. 2007;109:3253–3259. doi: 10.1182/blood-2006-09-048249. [DOI] [PubMed] [Google Scholar]
- 58.Tamamura Y., Otani T., Kanatani N., Koyama E., Kitagaki J., Komori T., Yamada Y., Costantini F., Wakisaka S., Pacifici M. Developmental regulation of Wnt/beta-catenin signals is required for growth plate assembly, cartilage integrity, and endochondral ossification. J. Biol. Chem. 2005;280:19185–19195. doi: 10.1074/jbc.M414275200. [DOI] [PubMed] [Google Scholar]
- 59.Soung Y., Kalinowski J., Baniwal S.K., Jacome-Galarza C.E., Frenkel B., Lorenzo J., Drissi H. Runx1-mediated regulation of osteoclast differentiation and function. Mol. Endocrinol. 2014;28:546–553. doi: 10.1210/me.2013-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li V., Raouf A., Kitching R., Seth A. Ets2 transcription factor inhibits mineralization and affects target gene expression during osteoblast maturation. In Vivo. 2004;18:517–524. [PubMed] [Google Scholar]
- 61.Tijchon E., van Ingen Schenau D., van Opzeeland F., Tirone F., Hoogerbrugge P.M., Van Leeuwen F.N., Scheijen B. Targeted Deletion of Btg1 and Btg2 Results in Homeotic Transformation of the Axial Skeleton. PLoS ONE. 2015;10:e0131481. doi: 10.1371/journal.pone.0131481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koscielny G., Yaikhom G., Iyer V., Meehan T.F., Morgan H., Atienza-Herrero J., Blake A., Chen C.K., Easty R., Di Fenza A. The International Mouse Phenotyping Consortium Web Portal, a unified point of access for knockout mice and related phenotyping data. Nucleic Acids Res. 2014;42:D802–D809. doi: 10.1093/nar/gkt977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ghebre M.A., Hart D.J., Hakim A.J., Kato B.S., Thompson V., Arden N.K., Spector T.D., Zhai G. Association between DHEAS and bone loss in postmenopausal women: a 15-year longitudinal population-based study. Calcif. Tissue Int. 2011;89:295–302. doi: 10.1007/s00223-011-9518-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gong Y., Slee R.B., Fukai N., Rawadi G., Roman-Roman S., Reginato A.M., Wang H., Cundy T., Glorieux F.H., Lev D., Osteoporosis-Pseudoglioma Syndrome Collaborative Group LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 65.Heilmeier U., Hackl M., Skalicky S., Weilner S., Schroeder F., Vierlinger K., Patsch J.M., Baum T., Oberbauer E., Lobach I. Serum miRNA Signatures Are Indicative of Skeletal Fractures in Postmenopausal Women With and Without Type 2 Diabetes and Influence Osteogenic and Adipogenic Differentiation of Adipose Tissue-Derived Mesenchymal Stem Cells In Vitro. J. Bone Miner. Res. 2016;31:2173–2192. doi: 10.1002/jbmr.2897. [DOI] [PubMed] [Google Scholar]
- 66.Sekiya H., Murakami T., Saito A., Hino S., Tsumagari K., Ochiai K., Imaizumi K. Effects of the bisphosphonate risedronate on osteopenia in OASIS-deficient mice. J. Bone Miner. Metab. 2010;28:384–394. doi: 10.1007/s00774-009-0142-y. [DOI] [PubMed] [Google Scholar]
- 67.Boudin E., Yorgan T., Fijalkowski I., Sonntag S., Steenackers E., Hendrickx G., Peeters S., De Maré A., Vervaet B., Verhulst A. The Lrp4 R1170Q homozygous knock-in mouse recapitulates the bone phenotype of sclerosteosis in humans. J. Bone Miner. Res. 2017;32:1739–1749. doi: 10.1002/jbmr.3160. [DOI] [PubMed] [Google Scholar]
- 68.Albagha O.M., Wani S.E., Visconti M.R., Alonso N., Goodman K., Brandi M.L., Cundy T., Chung P.Y., Dargie R., Devogelaer J.P., Genetic Determinants of Paget’s Disease (GDPD) Consortium Genome-wide association identifies three new susceptibility loci for Paget’s disease of bone. Nat. Genet. 2011;43:685–689. doi: 10.1038/ng.845. [DOI] [PubMed] [Google Scholar]
- 69.Courant F., Aksglaede L., Antignac J.P., Monteau F., Sorensen K., Andersson A.M., Skakkebaek N.E., Juul A., Bizec B.L. Assessment of circulating sex steroid levels in prepubertal and pubertal boys and girls by a novel ultrasensitive gas chromatography-tandem mass spectrometry method. J. Clin. Endocrinol. Metab. 2010;95:82–92. doi: 10.1210/jc.2009-1140. [DOI] [PubMed] [Google Scholar]
- 70.Cao J., Venton L., Sakata T., Halloran B.P. Expression of RANKL and OPG correlates with age-related bone loss in male C57BL/6 mice. J. Bone Miner. Res. 2003;18:270–277. doi: 10.1359/jbmr.2003.18.2.270. [DOI] [PubMed] [Google Scholar]
- 71.Yang J., Bakshi A., Zhu Z., Hemani G., Vinkhuyzen A.A., Lee S.H., Robinson M.R., Perry J.R., Nolte I.M., van Vliet-Ostaptchouk J.V., LifeLines Cohort Study Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat. Genet. 2015;47:1114–1120. doi: 10.1038/ng.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Estimates were derived from the overall approach. Beta coefficients and allele frequency (EAF) are reported for the A1 allele.
Estimates were derived from the all-age combined approach. Beta coefficients and allele frequency (EAF) are reported for the A1 allele.
Only suggestively associated variants (p < 5 × 10-6) in the TB-BMD overall meta-analysis were subjected to meta-regression assessment. Genomic coordinates are on build 37 of the human genome. Allele frequencies (EAF) are reported for the A1 allele. C.L-C.U, 95% Confidence interval lower and upper limit.
Based on genome-wide significant variants in the overall TB-BMD meta-analysis. These 182 gene-sets were further clustered in 25 ‘metagene-sets’ shown in Figure 4.
Data was collected for the 55 genes prioritized by DEPICT. Detailed bone phenotyping from the Origins of Bone and Cartilage Disease (OCBD) is presented in Table S15.
Knockout lines with a skeletal parameter greater than 2 standard deviations from the reference mean are highlighted in orange. The standard deviation from the reference mean for each parameter is shown with those greater than 2 highlighted (Black above the mean: Red below the mean).