Abstract
Fucosyltransferase 8 (FUT8) encodes a Golgi-localized α1,6 fucosyltransferase that is essential for transferring the monosaccharide fucose into N-linked glycoproteins, a process known as “core fucosylation.” Here we describe three unrelated individuals, who presented with intrauterine growth retardation, severe developmental and growth delays with shortened limbs, neurological impairments, and respiratory complications. Each underwent whole-exome sequencing and was found to carry pathogenic variants in FUT8. The first individual (consanguineous family) was homozygous for c.715C>T (p.Arg239∗), while the second (non-consanguineous family) was compound heterozygous for c.1009C>G (p.Arg337Gly) and a splice site variant c.1259+5G>T. The third individual (consanguineous family) was homozygous for a c.943C>T (p.Arg315∗). Splicing analysis confirmed the c.1259+5G>T resulted in expression of an abnormal FUT8 transcript lacking exon 9. Functional studies using primary fibroblasts from two affected individuals revealed a complete lack of FUT8 protein expression that ultimately resulted in substantial deficiencies in total core fucosylated N-glycans. Furthermore, serum samples from all three individuals showed a complete loss of core fucosylation. Here, we show that loss of function mutations in FUT8 cause a congenital disorder of glycosylation (FUT8-CDG) characterized by defective core fucosylation that phenotypically parallels some aspects of the Fut8−/− knockout mouse. Importantly, identification of additional affected individuals can be easily achieved through analysis of core fucosylation of N-glycans.
Main Text
Congenital disorders of glycosylation (CDG) are a rapidly expanding group of metabolic disorders resulting from abnormal glycosylation of proteins or lipids.1 To date, more than 125 distinct disorders have been identified, with the majority affecting N-linked glycosylation.2
Fucosylation is the enzymatic incorporation of L-fucose into N-glycans, O-glycans and glycolipids, these processes require a family of fucosyltransferases that use guanosine diphosphate L-fucose (GDP-fucose) as a donor substrate.3 In mammals, GDP-fucose biosynthesis occurs by both de novo and salvage pathways.3 In the de novo pathway GDP-mannose is converted to GDP-fucose in a multistep reaction involving two enzymes, GDP-mannose 4,6-dehydratase (GMDS), and GDP-keto-6-deoxymannose 3,5 epimerase (TSTA3) (aka FX protein).4, 5 The majority of GDP-fucose (∼90%) in cells uses this de novo pathway.6 Chemical inhibitors or genetically modified animals targeting the de novo pathway display dramatically reduced levels of GDP-fucose and thus overall cellular fucosylation.5 Fx −/− (MIM 137020) knockout mice exhibit near complete lethality, however mice that survive to birth exhibit postnatal failure to thrive that is rescued with oral L-fucose supplementation.7 This rescue is possible because the fucose salvage pathway can use either exogenous fucose or that liberated during degradation of glycans in the lysosome to directly generate GDP-fucose, thus by-passing the de novo pathway.5 GDP-fucose is then transported into the Golgi via the multi-pass transmembrane domain transporter, SLC35C1.8 Finally, in the case of N-glycans, L-fucose is attached to the N-acetylglucosamine (GlcNAc) directly linked to an asparagine (Asn) using the α1,6 fucosyltransferase, FUT8, to form the most abundant type of fucose modification, “core fucosylation”3, 9 (Figure 1A).
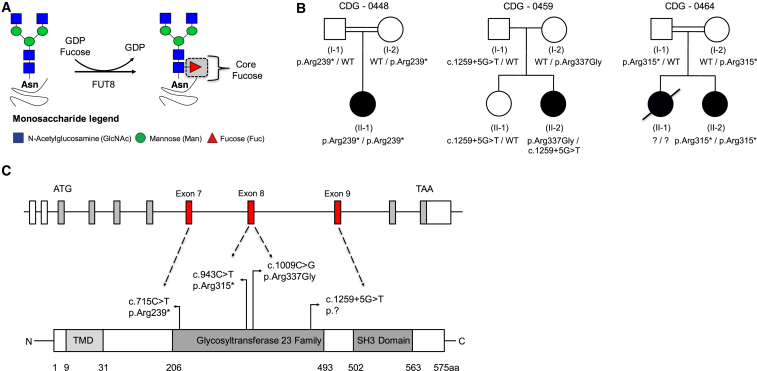
Figure 1.
Identification of FUT8 Pathogenic Variants in Three Unrelated Families
(A) Schematic of the FUT8 dependent α1,6 fucosyltransferase enzymatic reaction creating a core fucose motif.
(B) Pedigrees showing FUT8 pathogenic variants and segregation in families 0448, 0459, and 0464.
(C) Schematic of Human FUT8 with the relative positions of each pathogenic variant on mRNA (GenBank: NM_178155.2) and protein level (Uniprot: Q9BYC5).
Core fucosylation is involved in cancer metastasis,10, 11 inflammation,12, 13 and the immune system.14, 15 However, only a few genetic disorders affecting fucosylation are known. Pathogenic mutations in SLC35C1 (MIM 605881) cause leukocyte adhesion deficiency type II (LADII) or SLC35C1-CDG (MIM 266265).16, 17 In this disorder, most, but not all, affected individuals present with recurrent bacterial infections plus persistently elevated neutrophils: a few respond to oral supplementation with L-fucose.18 The neutrophil defect is thought to be due to an inability to generate the O-linked fucosylation-specific sialyl Lewis X (sLex), which is required for proper neutrophil rolling prior to extravasation.19 Mutations in POFUT1 (MIM 615327) and LFNG (MIM 609813) cause defects specifically related to abnormal NOTCH O-fucosylation.20, 21 Finally, mutations in FUCA1 (MIM 612280) cause the lysosomal storage disorder, fucosidosis (MIM 230000), which is due to an inability to degrade fucosylated glycoproteins and glycolipids.22
In this work, we detail the molecular, biochemical, and clinical characterization of three unrelated individuals, who had variants of uncertain significance (VUS) in FUT8 (MIM 602589), which we show are pathogenic mutations. Prior to diagnostic and research studies, informed consent was obtained in accordance with an approved Sanford Burnham Prebys Medical Discovery institute IRB.
Individual 1 (CDG-0448) is a female of Irish, Italian, and Native American origin born to consanguineous second-degree cousins. The pregnancy was complicated by polyhydramnios, shortened upper and lower extremities, suspicion of possible esophageal atresia or tracheesophageal fistula, intrauterine growth retardation (IUGR) (< 2nd percentile), and two-vessel cord (Table 1). She was delivered via Caesarean section due to failed induction and non-reassuring fetal heart tracing. Immediately after delivery she was transferred to the neonatal intensive care unit for possible sepsis and follow up for prenatal abnormalities. During this period, she was hypotonic and unable to tolerate breast feeding. She was noted to have severely shortened limbs, with a normal skeletal survey. A large secundum atrial septal defect (ASD) with left to right shunt was identified which required surgical repair. She had respiratory distress of unknown origins that required a tracheostomy. Newborn screening determined she had hypothyroidism. On at least two occasions she had seizure activity that did not require medication. However, more recently (∼30 months of age) she has had increased seizure frequency requiring medication. Other notable findings included dysmorphic craniofacial features, congenital neutropenia, and microcephaly (< 3rd percentile) (Table 1). All biochemical and molecular testing was normal, except for a chromosome microarray which showed large regions of homozygosity consistent with the family history of consanguinity. Whole-exome sequencing was performed by a clinical laboratory on the trio and identified that the proband was homozygous for the c.715C>T (p.Arg239∗) variant in exon 7 of FUT8 (Table 1, Figures 1B and 1C).
Table 1.
Molecular and Clinical Characterization for Three Individuals with Pathogenic Variants in FUT8
| Gender |
CDG – 0448 |
CDG – 0459 |
CDG – 0464 |
|---|---|---|---|
| Female | Female | Male | |
| Year of birth (or Age) | 2014 | 2014 | 2010 |
| Family History | No | No | Yes, deceased older brother |
| Consanguinity | Yes, 2nd cousin | No | Yes, 1st cousin |
| Ancestry | Irish, Italian, Native American | Irish-Norwegian / Irish-Ukrainian-Belgian | Arab |
| FUT8 pathogenic variants | c.715C>T (p.Arg239∗) | c.1009C>G (p.Arg337Gly) c.1259+5 G>T (N/A) |
c.943C>T (p.Arg315∗) |
| Pregnancy complications | polyhydramnios | polyhydramnios | Normal |
| IUGR | Yes (< 2nd Percentile) | Yes (3rd Percentile) | Yes (3rd percentile) |
| Feeding problems | Yes | Yes | Yes |
| Failure to thrive | Yes - Severe | Yes - Severe | Yes - Severe |
| Developmental delay | Yes - Severe | Yes - Severe | Yes - Severe |
| Dysmorphic features | High Palate, edematous eyelids and nasal bridge, wide nasal bridge, retrognathia | Buphthalmos, high broad forehead, mild lymphedema of hands and feet | Bitemporal narrowing, hirsutism, exotropia, exophthalmos, short nose, enlarged left eye with macrocornea |
| Microcephaly | Yes, 33 months < 1st percentile | Yes, 31 months < 10th percentile | Yes, 36 months < 3rd percentile |
| Intellectual disability | Yes | Yes | Yes |
| Seizures/epilepsy | Yes | Yes | Yes |
| Hypotonia | Yes | Yes | Central with peripheral spasticity |
| Skeletal abnormalities | Short stature | Short stature | Short stature, multiple contracture, severe osteopenia, dislocated hips, kyphoscoliosis |
| Cardiac abnormalities | Atrial septal defect, repaired Patent Ductus Arteriosus ligation | Not reported, normal echocardiogram | None |
| Liver abnormalities | No | No | No |
| Renal abnormalities | Nephrocalcinosis | No | No |
| Respiratory abnormalities | Tracheostomy | Reactive airway disease | Recurrent bronchopneumonia |
| Gastrointestinal abnormalities | G-tube placement | G-tube placement | Feeding difficulties with GE reflux |
| Endocrine abnormalities | Hypothyroidism | Not reported | None |
| Ocular abnormalities | No | Congenital glaucoma | No |
Individual 2 (CDG-0459) is a female of Irish-Norwegian/Irish-Ukrainian-Belgian ancestry, who was the product of a complicated pregnancy notable for polyhydramnios, severely shortened long bones, and suspected IUGR (< 3rd percentile). Complications in the perinatal period included: amniotic fluid that was dark and bloody. She began to have seizure activity within 3 hr after birth, which have continued. She was also noted to be microcephalic (< 10th percentile) and hypotonic (Table 1). Hypoglycemia was noted in the neonatal period. Like individual 1, she had difficulty feeding, severely shortened limbs, and respiratory distress characterized as reactive airway disease (Table 1). On several occasions, respiratory failure occurred during infections. Additionally, she had congenital glaucoma and recurrent apneic episodes (Table 1). Whole-exome sequencing was performed by a clinical laboratory on the trio and identified two potential VUS in FUT8, c.1009C>G (p.Arg337Gly) within exon 8 and a c.1259+5G>T splice site variant within intron 9 (Table 1, Figures 1B and 1C).
Individual 3 (CDG-0464) is a male of Arab ancestry born to consanguineous first-degree cousins, who had a family history positive for an affected male that passed away without a diagnosis. The pregnancy for CDG-0464 was not complicated by polyhydramnios, but the affected did show severe IUGR (< 3rd percentile) (Table 1). As seen in individuals 1 and 2, individual 3 had several overlapping clinical features including feeding difficulties with profound failure to thrive, seizures, hypotonia, and dysmorphic features with microcephaly (< 3rd percentile) (Table 1). Importantly, CDG-0464 also has skeletal abnormalities consisting of severe short stature, multiple contracture, severe osteopenia, dislocated hips, and kyphoscoliosis (Table 1). Respiratory complications were mainly due to recurrent episodes of bronchopneumonia (Table 1). Individual 3 passed away at seven years of age. Whole-exome sequencing was performed on the proband and identified a homozygous VUS in FUT8, c.943C>T (p.Arg315∗) within exon 8 (Table 1, Figures 1B and 1C).
Using the Genome Aggregation Database (gnomAD) of 123,136 exomes and 15,496 genomes (Ver2: accessed 11.14.2017) we determined the c.715C>T (p.Arg239∗) was seen in two carriers from 246,090 alleles (0 homozygotes) and the c.943C>T (p.Arg315∗) in three out of 246,270 alleles (0 homozygotes), while neither the c.1009C>G (p.Arg337Gly) nor the c.1259+5G>T were present in the database. In silico prediction modeling using the Combined Annotation Dependent Depletion (CADD) scoring for predicting the deleteriousness of a given variant showed that the c.715C>T (p.Arg239∗) and the c.943C>T (p.Arg315∗) had CADD scores of 36 and 38 respectively, while the c.1009C>G (p.Arg337Gly) scored a 33. These three variants are predicted to be in the top 0.1% of deleterious variants. However, the c.1259+5G>T scored a 15.7 (∼5th percentile) predicting it to be less likely to be damaging. All four variants occur within the highly-conserved glycosyltransferase 23 family domain spanning amino acids 206–493 (Figure 1C).
We compared three commercially available primary control fibroblast cultures (GM-00038, GM-03348, and GM-05381) to CDG-0448 and CDG-0459 (fibroblasts were unavailable for CDG-0464) to determine whether the variants had any effect on FUT8 protein expression. While we anticipated CDG-0448 to lack FUT8 protein, western blot analysis using two separate monoclonal antibodies (Proteintech, 66118-1-Ig and Santa Cruz Biotechnology, sc-271244) against different N-terminal and C-terminal epitopes within human FUT8 revealed no detectable protein in either affected individual’s fibroblasts (Figure 2A).
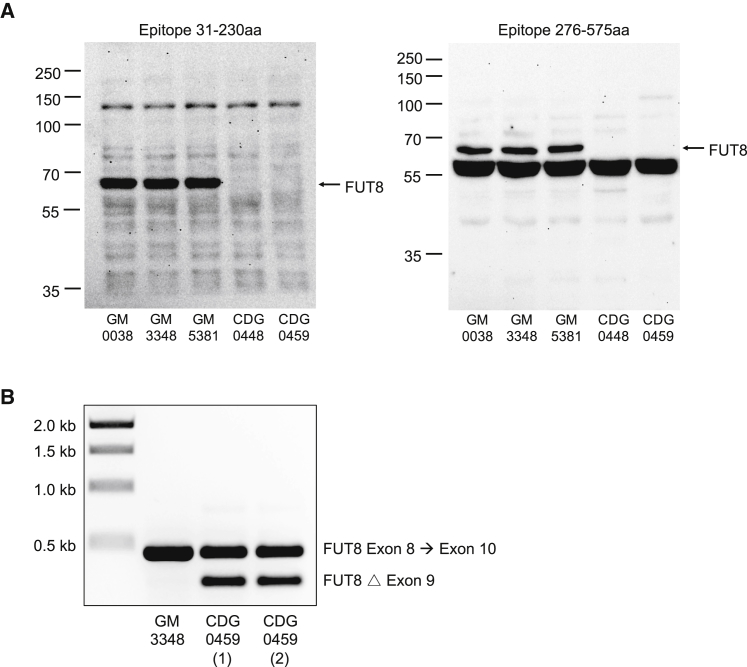
Figure 2.
Effect of FUT8 Pathogenic Variants on Protein Expression and mRNA Splicing
(A) Western blot analysis of FUT8 protein in fibroblast from three controls, CDG-0448 and CDG-0459 showing loss of FUT8 protein expression. Two different monoclonal antibodies with separate epitopes were used to detect human FUT8 protein. The first monoclonal anti-FUT8 recognizes an epitope within amino acids 31–230 and was used at a 1:500 dilution and (Santa Cruz Biotechnology, sc-271244), while the second has an epitope within amino acids 276–575 and was used at a 1:2,000 dilution (Proteintech 66118-1-Ig).
(B) Splicing analysis was performed using primers spanning exon 8 to exon 10 of FUT8 mRNA and determined that the c.1259+5G>T, found in CDG-0459, produced an abnormal transcript lacking exon 9. Two independent RNA preparations are shown for CDG-0459.
The data suggested that the c.1259+5G>T splice variant found in CDG-0459 would likely affect FUT8 splicing. To assess the potential pathogenicity of the c.1259+5G>T variant, we isolated RNA from both control and CDG-0459 fibroblasts for synthesis of cDNA using primers spanning exon 8 to exon 10 of FUT8. Whereas the control produced a single transcript with the expected PCR size (474 bp), CDG-0459 produced two products, the wild-type (474 bp) and a shorter transcript (297 bp) (Figure 2B). Sanger sequencing confirmed that the shorter transcript contained an in-frame deletion of exon 9 and is predicted to create a protein product p.Val362_Lys420del (Figure S1). Based on the known crystal structure of the FUT8 protein, deletion of these fifty-nine amino acids encoded within exon 9 results in the loss of several critical residues.23, 24 Specifically, His363, which recognizes the guanine of GDP-fucose, and Arg365, which recognizes both the β-phosphate of GDP-fucose and the fucose moiety itself, would be deleted.23, 24 This argues that the abnormal transcript produced by c.1259+5G>T would result in a non-functional protein due to loss of these critical amino acids.
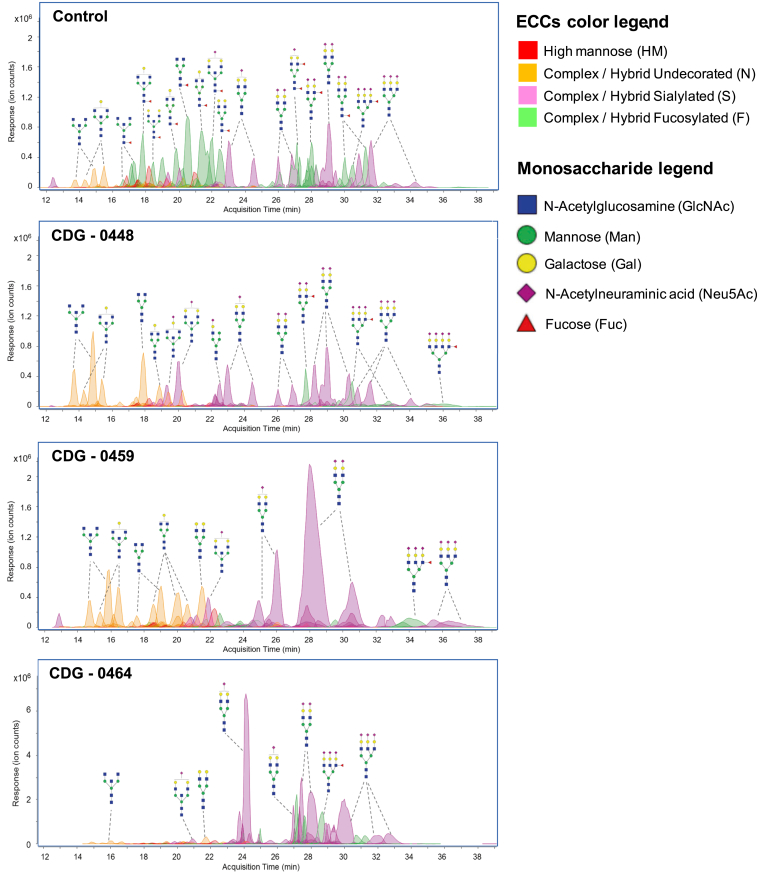
It is well established that core fucosylation of N-glycans is FUT8-dependent based on Chinese hamster ovary (CHO) knockout cell lines as well as a knockout mouse model.9, 25 Using liquid chromatography–mass spectrometry (LC-MS), we analyzed the total composition of serum N- and O-glycans from controls and the three affected individuals.26, 27 As expected, the three affected individuals had complete loss of total core fucosylated N-glycans compared to controls (N = 2) consistent with a deficiency in FUT8 (Figure 3). In addition to loss of core fucosylated glycans in the third individual (CDG-0464), other N-glycans specifically non-fucosylated neutral glycans are clearly abnormal (Figure 3). This is unlikely to result from the FUT8 deficiency, but is more likely due to other variants that affect N-glycosylation. However, a thorough review of variants identified in the exome analysis did not yield other glycosylation candidates. Importantly, the residual fucosylation of N-glycans seen in each affected individual was determined to occur on the antenna of N-glycans and not in the core, based on tandem mass spectrometry (MS/MS) fragmentation analysis (Figure S2). O-glycans were unaffected consistent with the specificity of the FUT8 protein for N-glycans (data not shown).3, 9
Figure 3.
Characterization of N-Glycans from Whole Serum using LC-MS
LC-MS chromatograms for control, CDG-0448, CDG-0459, and CDG-0464 showing the loss of multiple core fucosylated N-glycans (green shade). Unlike the other two affected individuals, CDG-0464 also had an absence of non-fucosylated neutral glycans (orange shade), which is not due to the loss of FUT8.
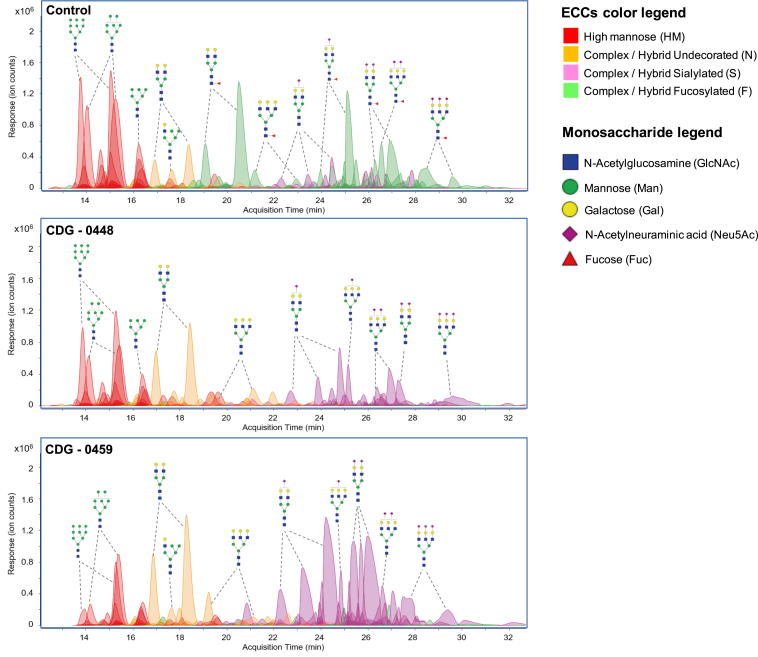
Next, we used two independent methods to further characterize the fucosylation defect in fibroblast cultures. The first was a similar LC-MS method previously mentioned to analyze serum N-glycans, with the exception that N-glycans were purified from membrane fractions isolated from fibroblasts.27 As seen in the serum N-glycans, fibroblasts from both CDG-0448 and 0459 had complete loss of core fucosylated N-glycans as compared to healthy unaffected controls (N = 3) (Figure 4).
Figure 4.
Characterization of N-Glycans from Primary Fibroblasts using LC-MS and Flow Cytometry
LC-MS chromatograms for Control, CDG-0448, and CDG-0459 showing the loss of multiple fucosylated N-glycans (green shade). The control used, GM-05381, is representative of three different control lines (GM-00038, GM-03348, GM-05381).
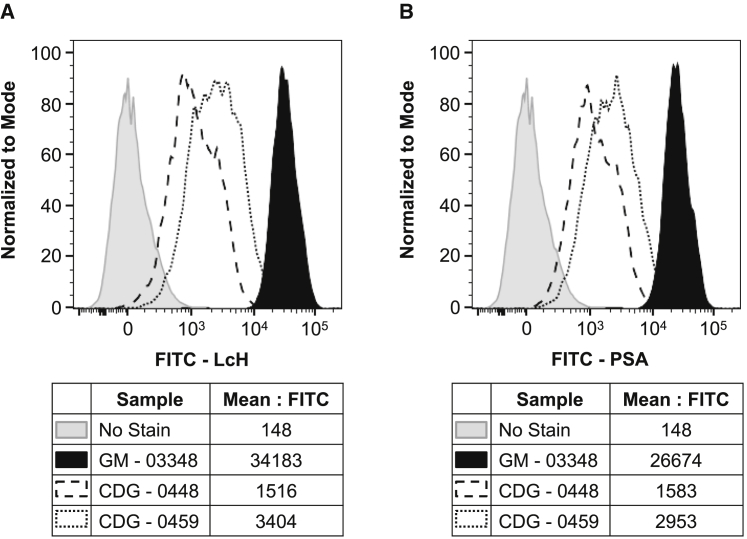
Second, we used fluorescence-activated cell sorting (FACS) with two different fluorescein isothiocyanate (FITC) labeled lectins, Lens culinaris agglutinin (LcH) and Pisum sativum agglutinin (PSA), to determine the level of core fucosylated cell surface proteins on fibroblast. Both LcH and PSA have a high preference for N-glycan structures that are core fucosylated and removal of the fucose moiety dramatically dampens lectin reactivity.28 As expected, and consistent with the LC-MS data, fibroblast samples from both CDG-0448 and 0459 displayed profound deficiencies in both LcH and PSA lectin reactivity (Figures 5A and 5B). Supplementing the fibroblast culture medium with L-fucose provided no significant benefit or improvement in core fucosylation (data not shown). However, it is possible that tissue types other than fibroblast might be able to make some FUT8 depending on how the c.1259+5G>T is utilized. While L-fucose did not improve overall core fucosylation in fibroblast from our cases, affected individuals with less severe variants might potentially be more responsive.
Figure 5.
Lectin Analysis of Cell Surface Core Fucosylated N-Glycans
(A) Flow cytometry was used to evaluate the cell surface binding of FITC-LcH (Lens culinaris agglutinin) in the two affected individuals and in a representative control culture and showed reduced lectin staining in both affected individuals.
(B) Flow cytometry analysis of cell surface FITC-PSA (Pisum sativum agglutinin) in the two affected individuals and in a representative control culture and showed reduced lectin staining in both affected individuals. GM-03348 is representative of three different control lines (GM-00038, GM-03348, GM-05381).
FUT8 encodes the only known alpha 1,6 fucosyltransferase specifically required for carrying out core fucosylation of N-glycans. Knockout mouse models have shown that, while Fut8−/− mice are born apparently normal, 70% die within the first 3 days of birth due to severe growth retardation and respiratory defects.9 Those mice that do survive continued to have growth deficits as well as progressive emphysema-like changes.9, 29 Similar to the Fut8−/− mice, our three affected individuals had severe growth retardation with shortened limbs and respiratory deficiencies. It has been shown that major growth factor receptor signaling molecules TGF-Beta1, EGF receptor and various integrins require core fucosylation for proper function and this could provide a major mechanism for both the severe growth defect and respiratory defects seen in both the Fut8−/− mice and our two subjects.9, 29 Unfortunately, biological samples from the lung of either affected individual were not available.
Given the high mortality rate seen in the Fut8−/− mouse model, it is possible that many individuals with FUT8-CDG could be missed due to prenatal or early postnatal lethality. This provides a diagnostic challenge, unless individuals have access to rapid sequencing services. This is further complicated by the fact that, unlike most CDG disorders, FUT8-CDG would be expected to and does give a normal carbohydrate-deficient transferrin (CDT) pattern (data not shown). This could potentially lead to the assumption that since the CDT results are normal, there is no glycosylation defect.
Because affected individuals described here had IUGR, severe short stature with shortened limbs and respiratory difficulties, FUT8 should be considered as a possible candidate. Analysis of total N-glycan core fucosylation could serve as an initial test prior to WES (or target panels) or be used for final diagnostic confirmation, much like serum transferrin is used for other forms of CDG. In fact, several clinical services offer LC-MS analysis of total N-glycan for this exact purpose. Importantly, we utilized this strategy and show that FUT8-CDG is easily detectable in serum samples, thus providing a diagnostic test for early detection. Furthermore, the complementary nature of both the LC-MS and FACS data, in both serum and fibroblast samples, can provide definitive confirmation of the diagnosis of FUT8-CDG. Partnering research labs with established diagnostic labs to identify additional individuals who have VUS is a useful strategy and mutually beneficial to furthering the understanding of not only FUT8-CDG, but other rare genetic disorders as well.
Acknowledgments
We would like to thank the families for their participation in this study. The SBP Medical Discovery Institute FACS core. 12G10 anti-alpha-tubulin was deposited to the DSHB by J. Frankel and E.M. Nelsen (DSHB Hybridoma Product 12G10 anti-alpha-tubulin). Deepali N. Shinde, Kelly Radtke, and Zöe Powis are employed by Ambry Genetics. Sharon F. Suchy is employed by GeneDx, Inc., a wholly-owned subsidiary of OPKO Health, Inc. The Rocket Fund and National Institutes of Health (NIH) grant R01DK099551 supported this work.
Published: January 4, 2018
Footnotes
Supplemental Data include two figures and Supplemental Methods and can be found with this article online at https://doi.org/10.1016/j.ajhg.2017.12.009.
Accession Numbers
The accession number used for the FUT8 sequence reported in this paper is GenBank: NM_178155.2
Web Resources
Combined Annotation Dependent Depletion (v1.0), http://cadd.gs.washington.edu/home
gnomAD Browser, http://gnomad.broadinstitute.org/
OMIM, http://www.omim.org/
Supplemental Data
References
- 1.Freeze H.H., Eklund E.A., Ng B.G., Patterson M.C. Neurological aspects of human glycosylation disorders. Annu. Rev. Neurosci. 2015;38:105–125. doi: 10.1146/annurev-neuro-071714-034019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaeken J., Péanne R. What is new in CDG? J. Inherit. Metab. Dis. 2017;40:569–586. doi: 10.1007/s10545-017-0050-6. [DOI] [PubMed] [Google Scholar]
- 3.Schneider M., Al-Shareffi E., Haltiwanger R.S. Biological functions of fucose in mammals. Glycobiology. 2017;27:601–618. doi: 10.1093/glycob/cwx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Overton K., Serif G.S. Synthesis of L-fucose in thyroid tissue. Biochim. Biophys. Acta. 1981;675:281–284. doi: 10.1016/0304-4165(81)90238-5. [DOI] [PubMed] [Google Scholar]
- 5.Becker D.J., Lowe J.B. Fucose: biosynthesis and biological function in mammals. Glycobiology. 2003;13:41R–53R. doi: 10.1093/glycob/cwg054. [DOI] [PubMed] [Google Scholar]
- 6.Yurchenco P.D., Atkinson P.H. Fucosyl-glycoprotein and precursor polls in HeLa cells. Biochemistry. 1975;14:3107–3114. doi: 10.1021/bi00685a011. [DOI] [PubMed] [Google Scholar]
- 7.Smith P.L., Myers J.T., Rogers C.E., Zhou L., Petryniak B., Becker D.J., Homeister J.W., Lowe J.B. Conditional control of selectin ligand expression and global fucosylation events in mice with a targeted mutation at the FX locus. J. Cell Biol. 2002;158:801–815. doi: 10.1083/jcb.200203125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puglielli L., Hirschberg C.B. Reconstitution, identification, and purification of the rat liver golgi membrane GDP-fucose transporter. J. Biol. Chem. 1999;274:35596–35600. doi: 10.1074/jbc.274.50.35596. [DOI] [PubMed] [Google Scholar]
- 9.Wang X., Inoue S., Gu J., Miyoshi E., Noda K., Li W., Mizuno-Horikawa Y., Nakano M., Asahi M., Takahashi M. Dysregulation of TGF-beta1 receptor activation leads to abnormal lung development and emphysema-like phenotype in core fucose-deficient mice. Proc. Natl. Acad. Sci. USA. 2005;102:15791–15796. doi: 10.1073/pnas.0507375102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito Y., Miyauchi A., Yoshida H., Uruno T., Nakano K., Takamura Y., Miya A., Kobayashi K., Yokozawa T., Matsuzuka F. Expression of alpha1,6-fucosyltransferase (FUT8) in papillary carcinoma of the thyroid: its linkage to biological aggressiveness and anaplastic transformation. Cancer Lett. 2003;200:167–172. doi: 10.1016/s0304-3835(03)00383-5. [DOI] [PubMed] [Google Scholar]
- 11.Agrawal P., Fontanals-Cirera B., Sokolova E., Jacob S., Vaiana C.A., Argibay D., Davalos V., McDermott M., Nayak S., Darvishian F. A Systems Biology Approach Identifies FUT8 as a Driver of Melanoma Metastasis. Cancer Cell. 2017;31:804–819. doi: 10.1016/j.ccell.2017.05.007. e807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rombouts Y., Jónasdóttir H.S., Hipgrave Ederveen A.L., Reiding K.R., Jansen B.C., Freysdottir J., Hardardottir I., Ioan-Facsinay A., Giera M., Wuhrer M. Acute phase inflammation is characterized by rapid changes in plasma/peritoneal fluid N-glycosylation in mice. Glycoconj. J. 2016;33:457–470. doi: 10.1007/s10719-015-9648-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ueda M., Kamada Y., Takamatsu S., Shimomura M., Maekawa T., Sobajima T., Fujii H., Nakayama K., Nishino K., Yamada M. Specific increase in serum core-fucosylated haptoglobin in patients with chronic pancreatitis. Pancreatology. 2016;16:238–243. doi: 10.1016/j.pan.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Li W., Yu R., Ma B., Yang Y., Jiao X., Liu Y., Cao H., Dong W., Liu L., Ma K. Core fucosylation of IgG B cell receptor is required for antigen recognition and antibody production. J. Immunol. 2015;194:2596–2606. doi: 10.4049/jimmunol.1402678. [DOI] [PubMed] [Google Scholar]
- 15.Fujii H., Shinzaki S., Iijima H., Wakamatsu K., Iwamoto C., Sobajima T., Kuwahara R., Hiyama S., Hayashi Y., Takamatsu S. Core Fucosylation on T Cells, Required for Activation of T-Cell Receptor Signaling and Induction of Colitis in Mice, Is Increased in Patients With Inflammatory Bowel Disease. Gastroenterology. 2016;150:1620–1632. doi: 10.1053/j.gastro.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Lühn K., Wild M.K., Eckhardt M., Gerardy-Schahn R., Vestweber D. The gene defective in leukocyte adhesion deficiency II encodes a putative GDP-fucose transporter. Nat. Genet. 2001;28:69–72. doi: 10.1038/ng0501-69. [DOI] [PubMed] [Google Scholar]
- 17.Lübke T., Marquardt T., Etzioni A., Hartmann E., von Figura K., Körner C. Complementation cloning identifies CDG-IIc, a new type of congenital disorders of glycosylation, as a GDP-fucose transporter deficiency. Nat. Genet. 2001;28:73–76. doi: 10.1038/ng0501-73. [DOI] [PubMed] [Google Scholar]
- 18.Marquardt T., Lühn K., Srikrishna G., Freeze H.H., Harms E., Vestweber D. Correction of leukocyte adhesion deficiency type II with oral fucose. Blood. 1999;94:3976–3985. [PubMed] [Google Scholar]
- 19.Yakubenia S., Frommhold D., Schölch D., Hellbusch C.C., Körner C., Petri B., Jones C., Ipe U., Bixel M.G., Krempien R. Leukocyte trafficking in a mouse model for leukocyte adhesion deficiency II/congenital disorder of glycosylation IIc. Blood. 2008;112:1472–1481. doi: 10.1182/blood-2008-01-132035. [DOI] [PubMed] [Google Scholar]
- 20.Li M., Cheng R., Liang J., Yan H., Zhang H., Yang L., Li C., Jiao Q., Lu Z., He J. Mutations in POFUT1, encoding protein O-fucosyltransferase 1, cause generalized Dowling-Degos disease. Am. J. Hum. Genet. 2013;92:895–903. doi: 10.1016/j.ajhg.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sparrow D.B., Chapman G., Wouters M.A., Whittock N.V., Ellard S., Fatkin D., Turnpenny P.D., Kusumi K., Sillence D., Dunwoodie S.L. Mutation of the LUNATIC FRINGE gene in humans causes spondylocostal dysostosis with a severe vertebral phenotype. Am. J. Hum. Genet. 2006;78:28–37. doi: 10.1086/498879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zielke K., Veath M.L., O’Brien J.S. Fucosidosis: deficiency of alpha-L-fucosidase in cultured skin fibroblasts. J. Exp. Med. 1972;136:197–199. doi: 10.1084/jem.136.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ihara H., Ikeda Y., Toma S., Wang X., Suzuki T., Gu J., Miyoshi E., Tsukihara T., Honke K., Matsumoto A. Crystal structure of mammalian alpha1,6-fucosyltransferase, FUT8. Glycobiology. 2007;17:455–466. doi: 10.1093/glycob/cwl079. [DOI] [PubMed] [Google Scholar]
- 24.Kötzler M.P., Blank S., Bantleon F.I., Spillner E., Meyer B. Donor substrate binding and enzymatic mechanism of human core α1,6-fucosyltransferase (FUT8) Biochim. Biophys. Acta. 2012;1820:1915–1925. doi: 10.1016/j.bbagen.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 25.Yamane-Ohnuki N., Kinoshita S., Inoue-Urakubo M., Kusunoki M., Iida S., Nakano R., Wakitani M., Niwa R., Sakurada M., Uchida K. Establishment of FUT8 knockout Chinese hamster ovary cells: an ideal host cell line for producing completely defucosylated antibodies with enhanced antibody-dependent cellular cytotoxicity. Biotechnol. Bioeng. 2004;87:614–622. doi: 10.1002/bit.20151. [DOI] [PubMed] [Google Scholar]
- 26.Song T., Aldredge D., Lebrilla C.B. A Method for In-Depth Structural Annotation of Human Serum Glycans That Yields Biological Variations. Anal. Chem. 2015;87:7754–7762. doi: 10.1021/acs.analchem.5b01340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park D., Xu G., Barboza M., Shah I.M., Wong M., Raybould H., Mills D.A., Lebrilla C.B. Enterocyte glycosylation is responsive to changes in extracellular conditions: implications for membrane functions. Glycobiology. 2017;27:847–860. doi: 10.1093/glycob/cwx041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tateno H., Nakamura-Tsuruta S., Hirabayashi J. Comparative analysis of core-fucose-binding lectins from Lens culinaris and Pisum sativum using frontal affinity chromatography. Glycobiology. 2009;19:527–536. doi: 10.1093/glycob/cwp016. [DOI] [PubMed] [Google Scholar]
- 29.Wang X., Fukuda T., Li W., Gao C.X., Kondo A., Matsumoto A., Miyoshi E., Taniguchi N., Gu J. Requirement of Fut8 for the expression of vascular endothelial growth factor receptor-2: a new mechanism for the emphysema-like changes observed in Fut8-deficient mice. J. Biochem. 2009;145:643–651. doi: 10.1093/jb/mvp022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.