Abstract
Intracranial aneurysms (IAs) are acquired cerebrovascular abnormalities characterized by localized dilation and wall thinning in intracranial arteries, possibly leading to subarachnoid hemorrhage and severe outcome in case of rupture. Here, we identified one rare nonsense variant (c.1378A>T) in the last exon of ANGPTL6 (Angiopoietin-Like 6)—which encodes a circulating pro-angiogenic factor mainly secreted from the liver—shared by the four tested affected members of a large pedigree with multiple IA-affected case subjects. We showed a 50% reduction of ANGPTL6 serum concentration in individuals heterozygous for the c.1378A>T allele (p.Lys460Ter) compared to relatives homozygous for the normal allele, probably due to the non-secretion of the truncated protein produced by the c.1378A>T transcripts. Sequencing ANGPTL6 in a series of 94 additional index case subjects with familial IA identified three other rare coding variants in five case subjects. Overall, we detected a significant enrichment (p = 0.023) in rare coding variants within this gene among the 95 index case subjects with familial IA, compared to a reference population of 404 individuals with French ancestry. Among the 6 recruited families, 12 out of 13 (92%) individuals carrying IA also carry such variants in ANGPTL6, versus 15 out of 41 (37%) unaffected ones. We observed a higher rate of individuals with a history of high blood pressure among affected versus healthy individuals carrying ANGPTL6 variants, suggesting that ANGPTL6 could trigger cerebrovascular lesions when combined with other risk factors such as hypertension. Altogether, our results indicate that rare coding variants in ANGPTL6 are causally related to familial forms of IA.
Keywords: genetics, intracranial aneurysm, ANGPTL6, whole-exome sequencing, burden test
Introduction
Intracranial aneurysms (IAs) are acquired cerebrovascular abnormalities affecting 3% of the general population, at a mean age of 50 years.1 They are characterized by a localized dilation and wall thinning in typical locations in intracranial arteries.2 The most notorious and deleterious complication of an IA is the rupture, resulting in subarachnoid hemorrhage that can lead to severe disability and death.3 Neither reliable biomarkers nor diagnostic tools are currently available to predict IA formation or evolution. Current treatments are mostly invasive and rely on microsurgical or endovascular treatment with a risk of procedural morbidity or mortality.4
Although the pathogenesis of IAs has been the subject of several studies for many years, the mechanisms underlying their formation, growth, and eventual rupture remain largely unknown.5 IAs are mostly acquired lesions resulting from a defective vascular wall response to local hemodynamic stress.6 The structural deterioration of the arterial wall involves inflammation and tissue degeneration with degradation of the extracellular matrix and smooth muscle cell apoptosis.7 Risk factors such as hypertension, female sex, increasing age, smoking, excessive alcohol consumption, and familial history of aneurysm predispose to IA formation and rupture.8 Furthermore, increasing evidence suggests a genetic component of IA formation.9 Genome-wide association studies and subsequent case-control replication analyses have identified common risk alleles for IA formation, in particular on chromosome 9 within the cyclin-dependent kinase inhibitor 2B antisense inhibitor gene, on chromosome 8 near the transcription regulator gene SOX17, and on chromosome 4 near the endothelin receptor A gene.10 However, these loci altogether may only explain around 5% of familial risk in Europe and Japan.11
Whole-exome sequencing approaches have recently been applied to families with multiple IA-affected case subjects, leading to the identification of genes associated with IA pathogenesis susceptibility, such as RNF213 (MIM: 613768)12 or THSD1 (MIM: 616821).13 While the Ring Finger Protein 213 had been previously involved in vascular-wall construction,14, 15 inactivation of the Thrombospondin Type 1 Domain Containing Protein 1 has been reported to impair the adhesion of endothelial cells to the extracellular matrix and to cause cerebral bleeding and increased mortality in zebrafish and mice.13 These recent advances provide new insights into the pathophysiology of IA and demonstrate the usefulness of familial approaches based on whole-exome sequencing to improve knowledge on the molecular mechanisms underlying IA formation and rupture.
In the present study, by combining whole-exome sequencing, identity-by-descent (IBD) analysis, gene burden testing, and functional investigations, we identified rare coding variants in the angiopoietin-like 6 gene (ANGPTL6 [MIM: 609336]) as causally related to familial forms of IA.
Material and Methods
Clinical Recruitment
Familial cases of IA are defined as at least two first-degree relatives both diagnosed with typical IA (defined as a saccular arterial dilatation of any size occurring at a bifurcation of the intracranial vasculature), without any age limitation. Index case subjects and their relatives were recruited following the French ethical guidelines for genetic research and under approval from the French Ministry of Research (no. DC-2011-1399) and the local ethical committee. Informed written consent was obtained from each individual agreeing to participate in the genetic study, to whom MRI screening and blood sampling were proposed.
The full recruiting process has been described previously.16 In brief, neuroradiological phenotyping was performed in each recruiting center by interventional neuroradiologists, neurologists, and neurosurgeons in order to recruit only case subjects with typical saccular bifurcation IA. Mycotic, fusiform-shaped, or dissecting IAs were systematically excluded, as well as IA in relation with an arteriovenous malformation and IA resulting from syndromic disorders such as Marfan disease or vascular forms of Elhers Danlos. Eye fundus, transthoracic echocardiography, non-invasive analysis of endothelial dysfunction, and Doppler echography analysis of peripheral arteries (sub clavians, radials, femorals, renals, and digestives) were carried out to check for any other vascular malformation or variation potentially linked to the presence of IA, thus constituting a syndrome yet unknown.
Whole-Exome Sequencing (WES)
Genomic DNA was extracted from peripheral blood lymphocytes using the NucleoSpin Blood kit XL (Macherey Nagel). In brief, coding exons from 3 μg of genomic DNA were captured using the SureSelect Human All Exon V4 Kit (Agilent Technologies), following the manufacturer’s protocol. DNA was sheared by acoustic fragmentation (Bioruptor Diagenode) and purified with the magnetic beads Agencourt AMPure XP (Beckmann Coulter genomics), and fragment quality was assessed (TapeStation 2200 Agilent). Exome-enriched genomes were paired-end sequenced (100-bp reads) on Illumina HiSeq 1500 (Illumina) to a mean depth above 30×. Sequence reads were mapped to the human reference genome (Broad Institute human_g1k_v37) using the Burrows-Wheeler Aligner.17 Duplicates were flagged using Picard software. Reads were realigned and recalibrated using the Genome Analysis Toolkit (GATK).18 Variant detection was performed with GATK HaplotypeCaller. Functional annotation of high-quality variants was performed using Ensembl VEPv7.4. The sequencing quality was determined with the Depth Of Coverage Walker provided in GATK. Knime4Bio19 was used for all merging and filtering steps. Variants with a sequencing depth of less than 10 or a genotype quality below 90 were excluded, as well as synonymous variants with no predicted effect on splicing sites. At last, from the resulting set of “functional” variants (as reported in Figure 1), we filtered out any variant with a minor allele frequency (MAF) higher than 0.1% in the non-Finnish European (NFE) population from the ExAC database, as well as few remaining variants reported with a minor allele frequency (MAF) higher than 10% in our in-house database of 260 whole-exome sequences from individuals with various cardiac phenotypes.
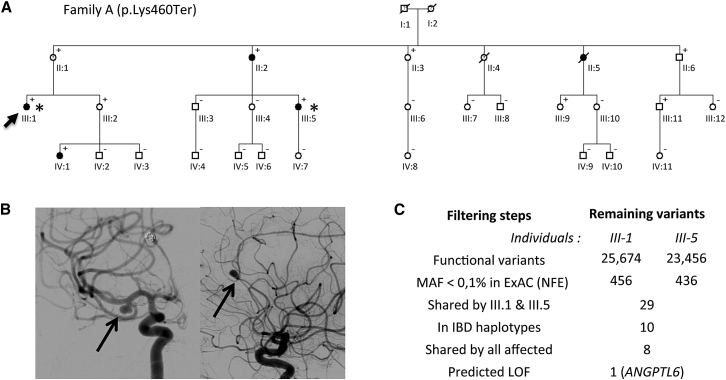
Figure 1.
Genetic Investigations in a Large Family with Multiple IA-Affected Case Subjects
(A) Pedigree of family A showing the segregation pattern of the variant ANGPTL6 c.1378A>T. Filled boxes, empty boxes, and boxes with question marks indicate IA-affected case subjects, non-affected relatives, and individuals with unknown status; plus sign (+) indicates the presence of the ANGPTL6 variant; minus sign (−) indicates its absence; arrow indicates the index case subject; and asterisks indicate the individuals included in WES analysis.
(B) Digital substracted angiographies showing two intracranial aneurysms carried by the index case III-1, one on the middle cerebral artery (left) and one on the anterior cerebral artery (right).
(C) Summary of the filtering steps applied to genetic variants detected by WES in individuals III-1 and III-5. Abbreviations: MAF, minor allele frequency; IBD, identity by descent; LOF, loss of function.
Identity-by-Descent Analysis
SNP genotyping was performed on population-optimized Affymetrix Axiom Genome-Wide CEU 1 array plates following the standard manufacturer’s protocol. Fluorescence intensities were quantified using the Affymetrix GeneTitan Multi-Channel Instrument, and primary analysis was conducted with Affymetrix Power Tools following the manufacturer’s recommendations. After genotype calling, all individuals had a genotype call rate above 97%. SNPs with an MAF < 10%, a call rate < 95%, or p < 1 × 10−5 when testing for Hardy-Weinberg equilibrium were excluded. IBD estimation was performed with IBDLD v.3.34, NoLD method.20 Shared regions were obtained by analyzing a set of independent SNPs (R2 < 0.2) using genotypes from French individuals21 as a reference panel. The IBD status at every SNP locus was obtained for each pair of individuals, based on a hidden Markov model implemented in the IBDLD program. Still using IBDLD, we estimated the kinship coefficients between pairs of IA-affected case subjects from distinct pedigrees. We invariably found values around 0.025, thus excluding non-documented close relatedness between individuals from distinct pedigrees carrying the same rare variant in ANGPTL6.
Capillary Sequencing and Burden Testing
Validation experiments for each selected variant, familial segregation analyses, and further screening for ANGPTL6 coding mutations were performed by capillary sequencing on an Applied Biosystems 3730 DNA Analyzer, using standard procedures. Sequence analyses were performed with SeqScape v.2.5. ANGPTL6 variants were numbered according to the canonical transcript (GenBank: NM_031917.2). Burden test was performed using SKAT22 and CAST,23 by comparing the proportion of individuals carrying at least one rare coding variant within ANGPTL6 among the 95 index case subjects with IA versus 404 healthy individuals with French ancestry. ANGPTL6 status in control individuals was determined by whole-genome sequencing with a mean depth of coverage above 30×. Rare variants were defined as variants with an MAF below 1% among the 7,509 whole-genome-sequenced individuals with NFE ancestry from the gnomAD database. The count of alleles with rare coding variants in ANGPTL6 among case subjects was also compared with the same allele count among the 7,509 whole-genome sequenced individuals with NFE ancestry from gnomAD,24 through the use of Fisher’s exact test.
Expression Analyses of ANGPTL6
HEK293 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Stable HEK293 cell lines were obtained by transfection of pcDNA3.1 vector encoding WT ANGPTL6 and p.Lys460Ter ANGPTL6 (G418 selection). Recombinant proteins are expressed as Nter-FLAG fusion proteins. In HEK293, recombinant human proteins were detected by both anti-FLAG (Sigma Aldrich, F-3165), anti-ANGPTL6 antibodies raised against AA 114-130 of ANGPTL6 (Adipogen, AG-25A-0030), and ELISA (kit supplied by Adipogen). In human subjects, serum ANGPTL6 levels were measured by ELISA. For transcript analysis, total RNA from stably transfected HEK293 cells was purified using Trizol (Life technology) according to the manufacturer’s instructions and then reverse-transcribed. Real-time quantitative PCR was performed using the TaqMan 7900 Sequence Detection System (Applied Biosystems). Primers used to assess ANGPTL6 mRNA expression were designed using the Primer Express 3.1 software (sequences available on request).
Results
A Nonsense Variant in ANGPTL6 Shared by Family Members with IA
The index case subject of family A (individual III-1; Figure 1A) was diagnosed after a subarachnoid hemorrhage (SAH) at the age of 51. This event revealed a ruptured anterior cerebral artery aneurysm and a second middle cerebral artery aneurysm (Figure 1B). She completely recovered from the subarachnoid hemorrhage and because of known familial history of ruptured IAs (II-2, II-5), a systematic screening was performed among relatives. Her cousin (III-5) and her niece (IV-1) were both diagnosed with two and one IAs, respectively. Her uncle (II-4) had an episode suggestive of aneurysmal SAH at the age of 36 and died before a CT scan or angiography could be performed. Her mother (II-1), who carries an ectasia measuring less than 2 mm and diagnosed as uncertain,16 was classified as phenotype unknown.
Clinical information was collected for 28 individuals from family A (Table S1). IA was diagnosed on CT angiography or conventional angiography. DNA was available for 27 of them (DNA was unavailable for II-5 who died in 1974 after a rupture of IA). Individuals with IAs (II-2, II-5, III-1, III-5, and IV-1) were all female. Noteworthy, all IA-affected case subjects except IV-1 suffered from high blood pressure.
We combined WES and IBD analysis to identify any rare genetic variant likely explaining this familial form of IA. Whole-exome sequencing applied to the first cousins III-1 and III-5 led to the detection of 25,674 and 23,456 functional sequence variants, respectively, in comparison to the human reference genome assembly (Figure 1C). After filtering out genetic variants reported with an MAF above 0.1% in the non-Finnish European (NFE) population from the ExAC database,24 we ended up with 29 rare variants shared between the first cousins, which were all manually reviewed by visual inspection of sequence reads using the Integrative Genomics Viewer.25
In parallel, IBD analysis of the complete pedigree identified 12 haplotypes shared by the 4 affected relatives. Within these chromosomal intervals, individuals III.1 and III.5 shared 10 rare, non-synonymous variants (Figure 1C). By capillary sequencing, we determined that the 4 affected relatives share 8 of these variants (Table 1), among which one nonsense variant, c.1378A>T (p.Lys460Ter), was in ANGPTL6.
Table 1.
Rare Non-synonymous Variants Shared by All Affected Members in Family A, and Rare Non-synonymous Variants in ANGPTL6 Detected in Other Families
| Family | Genomic Position (GRCh37/hg19) | Gene | Nucleotide Changea | Protein Consequencea | MAF in ExAC (NFE) | GERP Score |
Predicted Functional Impact |
Carriers / Total |
||
|---|---|---|---|---|---|---|---|---|---|---|
| SIFT | PolyPhen-2 | Affected | Unaffected | |||||||
| A | chr6:52701135C>A | GSTA5 | c.171G>T | p.Met57Ile | 0.00002 | 2.63 | deleterious | benign | 4/4 | 5/22 |
| A | chr10:73046467G>A | UNC5B | c.574G>A | p.Asp192Asn | 0.00001 | 5.43 | deleterious | possibly_damaging | 4/4 | 10/22 |
| A | chr11:64756981G>A | BATF2 | c.373C>T | p.Leu125Phe | 0.00002 | 3.54 | tolerated | possibly_damaging | 4/4 | 6/22 |
| A | chr11:65113459G>T | DPF2 | c.122G>T | p.Gly41Val | 0.00013 | −7.09 | tolerated | unknown | 4/4 | 6/22 |
| A | chr11:65746304C>T | SART1 | c.2303C>T | p.Thr768Ile | 0.00000 | 4.76 | deleterious | probably_damaging | 4/4 | 6/22 |
| A | chr19:10610439C>T | KEAP1 | c.271G>A | p.Ala91Thr | 0.00013 | 4.68 | tolerated | benign | 4/4 | 5/22 |
| A | chr19:14001265G>A | C19orf57 | c.404C>T | p.Ala135Val | 0.00025 | −3.41 | deleterious | benign | 4/4 | 5/22 |
| A | chr19:10203300T>A | ANGPTL6 | c.1378A>T | p.Lys460Ter | 0.00001 | 4.39 | LOF | 4/4 | 5/22 | |
| B + C | chr19:10206848T>A | ANGPTL6 | c.392A>T | p.Glu131Val | not found | 4.03 | deleterious | benign | 4/4 | 4/10 |
| D | chr19:10204205G>A | ANGPTL6 | c.1042C>T | p.Leu348Phe | not found | 3.67 | tolerated | probably_damaging | 1/1 | 0/1 |
| E + F | chr19:10206782G>GC+ | ANGPTL6 | c.439_457dup | p.Ala153ValfsTer66 | 0.00060 | 4.36 | LOF | 3/4 | 6/8 | |
Abbreviations are as follows: GERP, Genomic Evolutionary Rate Profiling; MAF, minor allele frequency; LOF, loss of function; ExAC (NFE), Exome Aggregation Consortium (Non-Finnish Europeans); SIFT, Sorting Intolerant From Tolerant.
The following reference sequences were used in naming mutations. They correspond to RefSeq transcripts, unless otherwise specified: GSTA5, NM_153699.1; UNC5B, NM_170744.4; BATF2, NM_001300807.1; DPF2, ENST00000531989 (from Ensembl; the variant is synonymous based on the RefSeq transcripts); SART1, NM_005146.4; KEAP1, NM_012289.3; C19orf57, NM_024323.3; ANGPTL6, NM_031917.2.
Enrichment in Rare Coding Variants within ANGPTL6 Among IA-Affected Case Subjects
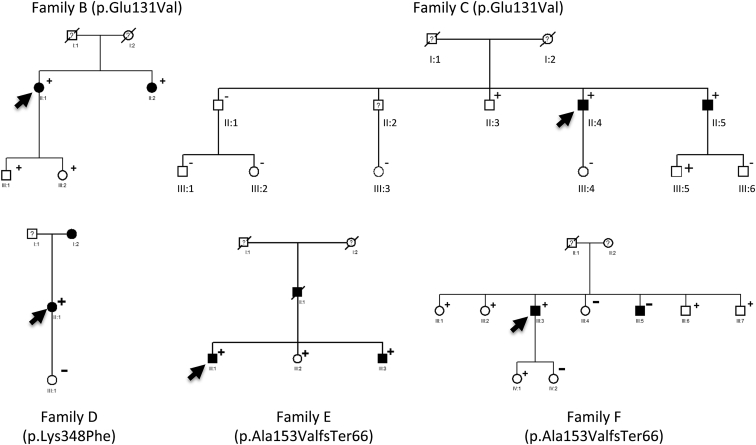
We then extended genetic screening on the coding portion of ANGPTL6 to 94 additional index case subjects with familial IA. We identified 5 additional individuals carrying rare, non-synonymous variants in ANGPTL6 predicted as damaging in silico by PolyPhen-2 and/or SIFT (Table 1): two case subjects with the same missense mutation in exon 1 leading to the c.392A>T substitution (p.Glu131Val) in ANGPTL6, one case subject with a missense mutation in exon 4 leading to the c.1042C>T substitution (p.Leu348Phe), and two case subjects with the same CGCGCTGAGCCTCGGCGGA-bp insertion leading to one premature stop codon in exon 2 (c.439_457dup [p.Ala153ValfsTer66]).
Overall, from the 6 index case subjects, family screening led to the identification of 16 relatives with diagnosed IAs (Tables 1 and S1; Figures 1A and 2). Out of the 13 family members carrying IAs and agreeing to participate in genetic research, 12 (92%) carry rare non-synonymous variants in ANGPTL6 versus 15 out of 41 (34%) unaffected ones. The only affected individual who does not carry any rare coding variant in ANGPTL6 is a 54-year-old male (III-5, family F, Figure 2) presenting with an aneurysm on the anterior communicant artery, with no reported history of smoking, high blood pressure, or any relevant associated disease.
Figure 2.
Familial Case Subjects of IA in the Presence of Rare Coding Variants in ANGPTL6
Filled boxes, empty boxes, and boxes with question marks indicate IA-affected case subjects, non-affected relatives, and individuals with unknown status; plus sign (+) indicates the presence of the ANGPTL6 variant; minus sign (−) indicates its absence; and black arrows indicate the index case subjects.
The clinical characteristics of the remaining 12 case subjects are reported in Tables 2 and S1. Seven of them (58%) carry multiple IAs (with a maximum of three). IA is located on the middle cerebral artery bifurcation in 7 case subjects (58%), on the anterior communicant artery, the anterior cerebral artery, and the internal carotid artery in 3 case subjects (25%), and on the posterior communicant artery in 2 case subjects (17%).
Table 2.
Clinical Characteristics and Exposition to Risk Factors for IA-Affected Case Subjects and Unaffected Relatives, According to ANGPTL6 Status
| Healthy Individuals; No ANGPTL6 Variant (n = 16a) | Healthy Individuals; ANGPTL6 Variant (n = 15) | IA Carriers; ANGPTL6 Variant (n = 12) | |
|---|---|---|---|
| Median age | 57 | 56 | 59 |
| Female sex | 56% | 53% | 58% |
| History of smoking | 56% (8 p.y.b) | 73% (10 p.y.b) | 67% (31 p.y.b) |
| High blood pressure | 7% | 9%c | 50%c |
| Alcohol intake > 150 g/w | 19% | 13% | 33% |
| Median BMI | 22 | 24 | 24 |
| Diabetes - dyslipidemia | 19% | 18% | 33% |
Healthy individuals aged over 35 years, without ANGPTL6 variant
p.y., median consumption in pack/years among smokers
Significant difference between affected versus healthy individuals carrying ANGPTL6 variants; p = 0.013 (chi-square test)
To further test the association of ANGPTL6 rare variants with susceptibility to familial IA, we also compared the proportions of individuals carrying at least one rare, non-synonymous variant across this gene among the 95 index case subjects enrolled in the present study (6/95; 6.32%) versus 404 healthy individuals with French ancestry (8/404; 1.98%). We found a significant enrichment in individuals carrying non-synonymous variants with an MAF below 1% in the NFE reference population, among IA-affected case subjects (SKAT, p = 0.023; see Tables S2 and S3). Similar results were found when comparing allele counts among the 95 index case subjects versus the 7,509 non-Finnish European individuals with whole-genome sequences available in the gnomAD database (Tables S2 and S3).
Reduced ANGPTL6 Secretion in the Presence of the c.1378A>T Variant
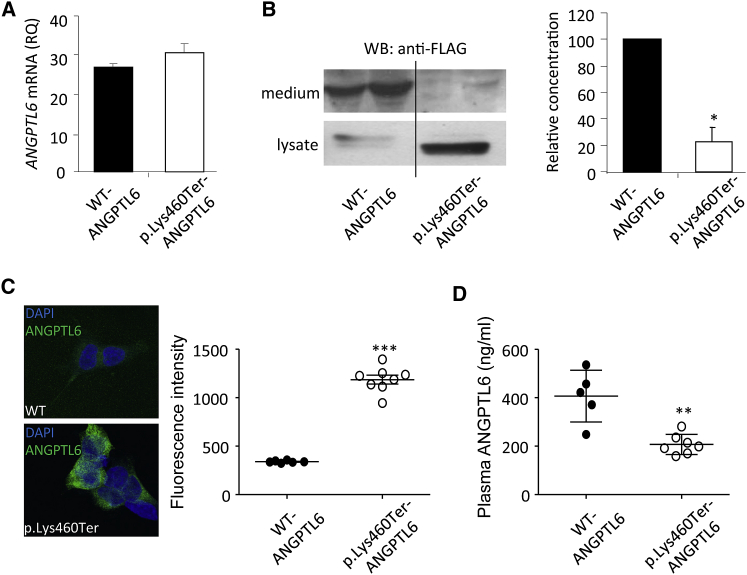
ANGPTL6 is one of the eight members of the secreted glycoprotein ANGPTL family, which share a common structure consisting of an amino-terminal coiled-coil domain, a linker region, and a carboxy-terminal fibrinogen-like domain. The c.1378A>T ANGPTL6 variant leads to the occurrence of a premature stop codon in the last exon. The corresponding transcript may thus escape the nonsense-mediated mRNA decay and is predicted to result in a protein truncated by the last 11 amino acids (p.Lys460Ter ANGPTL6). To analyze the functional properties of p.Lys460Ter ANGPTL6, we established stable cell lines expressing the wild-type (WT ANGPTL6) and mutant ANGPTL6 (p.Lys460Ter ANGPTL6). The two types of cell lines expressed similar levels of transcripts suggesting that c.1378A>T ANGPTL6 variant mRNA was not degraded (Figure 3A). Western blot using anti-FLAG antibody showed that WT ANGPTL6 was secreted in the culture medium while p.Lys460Ter ANGPTL6 was nearly undetectable in the supernatant of cells transfected with the variant (Figure 3B). Quantification of ANGPTL6 concentration by ELISA confirmed the significant reduction of the secretion of p.Lys460Ter ANGPTL6 compared to WT ANGPTL6 (Figure 3B). In agreement with this defective secretion, immunofluorescence labeling and quantification in permeabilized cells clearly showed the retention of p.Lys460Ter ANGPTL6 in the cytoplasm (Figure 3C). Although we did not directly demonstrate that c.1378A>T ANGPTL6 variant mRNA escaped nonsense-mediated mRNA decay, our data strongly suggest that the c.1378A>T ANGPTL6 variant leads to effective expression of the truncated p.Lys460Ter ANGPTL6 protein, which is not secreted. Accordingly, heterozygous subjects for the c.1378A>T ANGPTL6 variant are expected to present with decreased levels of circulating ANGPTL6. To assess this hypothesis, we performed ELISA to compare the serum concentration of ANGPTL6 in subjects from family A reported as homozygous for the WT ANGPTL6 (n = 5) versus heterozygous for the c.1378A>T ANGPTL6 (n = 7) and found a 50% reduction in the serum level of ANGPTL6 in heterozygous subjects (Figure 3D).
Figure 3.
Analysis of WT and p.Lys460Ter ANGPTL6 in Cultured Cells and Individual Sera
(A) Analysis by qPCR of ANGPTL6 transcripts in HEK293 cells encoding WT and p.Lys460Ter ANGPTL6.
(B) Analysis by western blot (anti-FLAG Ab) and ELISA of the levels of WT and p.Lys460Ter ANGPTL6 in culture media and lysates from stably transfected HEK293.
(C) Immunofluorescence labeling with anti-ANGPTL6 Ab and corresponding quantification in permeabilized HEK293 cells expressing WT and p.Lys460Ter ANGPTL6.
(D) Analysis of serum levels of ANGPTL6 in control subjects (WT ANGPTL6) and individuals harboring the p.Lys460Ter ANGPTL6 variant (heterozygous)
All results are expressed as means and error bars indicate standard deviation. Significant differences between independent groups were assessed by the Mann-Whitney test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
A similar analysis of the serum concentration of ANGPTL6 has been performed in subjects from family C. We found no reduction in the serum concentration of ANGPTL6 between heterozygous subjects for p.Glu131Val ANGPTL6 versus homozygous for the WT ANGPTL6 (Figure S1).
Discussion
In the present study, we identified one rare nonsense variant, c.1378A>T, in the last exon of angiopoietin-like 6 (ANGPTL6), which was carried by the four tested affected members of a large pedigree with multiple IA-affected case subjects. Because (1) the c.1378A>T nonsense variant is the only loss-of-function variant shared by all affected members in the family, (2) it is carried by only 5 out of the 22 unaffected relatives, and (3) ANGPTL6 is a functionally relevant candidate for playing a role in IA pathophysiology, we thereafter focused our investigations on this particular gene.
Indeed, members of the ANGPTL family have been reported as regulators of angiogenesis through their carboxyl terminus.26 In human, ANGPTL6 has been identified as a circulating pro-angiogenic factor mainly secreted from the liver, which increases endothelial permeability and stimulates endothelial cell migration.27, 28 Although members of the ANGPTL family have also been described as regulators of glucose and lipoprotein metabolisms through their amino terminus,13 a large-scale population study has shown that ANGPTL6 does not affect lipoprotein profile.29
Interestingly, it is known that genes associated with vascular malformations of the brain have demonstrated or plausible roles in angiogenesis and vascular remodelling.30 The development of the vasculature involves both vasculogenesis during embryogenesis and angiogenesis. Vasculogenesis of the cerebral vasculature occurs outside the brain, followed by capillary sprouting from the perineural plexus to penetrate the neural tube. Growth of the cerebral vasculature then entirely results from angiogenesis that requires vascular endothelial cell proliferation and migration, then vascular stabilization, corresponding to the formation of capillary tubes by endothelial cells and the recruitment of pericytes, the smooth muscle cell precursors, to their walls. All these timely organized events are tightly controlled by angiogenic factors, and aberrant expression or function of these mediators, including ANGPTL6, may lead to abnormal cerebral artery wall formation.31, 32, 33 For example, defective recruitment of pericytes leads to vessel dilation and microaneurysm.31
We then extended our genetic screen on ANGPTL6 to 94 additional index case subjects with familial IA and identified additional rare missense or frameshift variants in this gene carried by index case subjects from 5 additional families (Figure 2 and Table 1). In total, among the 6 families, 12 out of 13 (92%) family members carrying IA also carry such variants in ANGPTL6, versus 15 out of 41 (37%) unaffected ones. The only affected individual who does not carry any rare coding variant in ANGPTL6 (III-5, family F; Figure 2) presents with an IA in the anterior communicant artery, a location where IAs have been reported with probably weaker hereditability.34, 35
By burden test, we showed a significant enrichment in rare non-synonymous variants within this gene in our population of 95 index case subjects with familial IA, compared to a reference population of 404 individuals with French ancestry. This result confirms that rare coding variants in ANGPTL6 are associated with familial forms of IA.
In human, ANGPTL6 is described as a circulating protein mainly secreted from the liver and acting as an endocrine factor in the peripheral tissues.36 Here we showed a 50% reduction of ANGPTL6 serum concentration in heterozygous c.1378A>T subjects compared to their relatives who do not carry the variant. Our results in stably transfected cell lines suggest that this reduction may be caused by the inability of the truncated protein lacking the last 11 amino acids (p.Lys460Ter ANGPTL6) to be secreted. In contrast, we showed that heterozygous subjects for the c.392A>T (p.Glu131Val) variant present with similar levels of circulating ANGPTL6 as subjects who do not carry the variant. This result does not exclude that ANGPTL6 may be associated with IA through dysfunction of the secreted form of the protein. We could not assess the levels of circulating ANGPTL6 in the remaining IA-affected case subjects and relatives, due to sample unavailability (Table S1).
In this study, we have observed some individuals (n = 15) carrying possibly deleterious variations in ANGPTL6 but without IA at the moment of the study. Since all affected individuals in our study have been subjected to at least one environmental risk factor, it is possible that familial rare variants in ANGPTL6 require additional inauspicious factors to trigger IA development. A prominent and well-established environmental factor associated with the IA formation is the history of high blood pressure.8 Indeed, we found in our study a significant difference in the rate of case subjects with a history of high blood pressure in affected versus healthy individuals heterozygous for ANGPTL6 variants (Table 2). We hypothesize that ANGPTL6 variants render cerebral arterial wall vulnerable to deformation, thus promoting lesion formation and/or progression, when challenged with other deleterious genetic or environmental factors such as high blood pressure, either through direct mechanical effects on vessel walls or by triggering inflammation. In the present study, we have identified rare coding variants in ANGPTL6 causally related to IA in 6 families with multiple affected relatives. Our results point to a novel pathogenic pathway for a disease whose etiology is still poorly understood. Currently no molecular test has been established to help the diagnosis of IA. Developing biomarkers could undoubtedly facilitate early detection and risk assessment of IA. In this context, correlating the activity level of ANGPTL6 with IA risk could be of great interest. A knock-in mouse model is currently under development in order to address these prominent questions about ANGPTL6 and IA susceptibility. Discovering genetic risk factors and better understanding the molecular mechanisms responsible for IA formation will be prerequisites for the identification of new therapeutic targets.
Consortia
The ICAN Study Group includes the following investigators: Hubert Desal, Romain Bourcier, Benjamin Daumas-Duport, Bertrand Isidor, Jérôme Connault, Pierre Lebranchu, Thierry Le Tourneau, Marie Pierre Viarouge, Chrisanthi Papagiannaki, Michel Piotin, Hocine Redjem, Mikael Mazighi, Jean Philippe Desilles, Olivier Naggara, Denis Trystram, Myriam Edjlali-Goujon, Christine Rodriguez, Waghi Ben Hassen, Suzanna Saleme, Charbel Mounayer, Olivier Levrier, Pierre Aguettaz, Xavier Combaz, Anne Pasco, Emeline Berthier, Marc Bintner, Marc Molho, Pascale Gauthier, Cyril Chivot, Vincent Costalat, Cyril Darganzil, Alain Bonafé, Anne Christine Januel, Caterina Michelozzi, Christophe Cognard, Fabrice Bonneville, Philippe Tall, Jean Darcourt, Alessandra Biondi, Cristina Iosif, Elisa Pomero, Jean Christophe Ferre, Jean Yves Gauvrit, François Eugene, Hélène Raoult, Jean Christophe Gentric, Julien Ognard, René Anxionnat, Serge Bracard, Anne Laure Derelle, Romain Tonnelet, Laurent Spelle, Léon Ikka, Robert Fahed, Aymeric Rouchaud, Augustin Ozanne, Jildaz Caroff, Nidal Ben Achour, Jacques Moret, Emmanuel Chabert, Jérôme Berge, Gaultier Marnat, Xavier Barreau, Florent Gariel, Frédéric Clarencon, Mohammed Aggour, Frédéric Ricolfi, Adrien Chavent, Pierre Thouant, Pablo Lebidinsky, Brivael Lemogne, Denis Herbreteau, Richard Bibi, Laurent Pierot, Sébastien Soize, Marc Antoine Labeyrie, Christophe Vandendries, Emmanuel Houdart, Appoline Kazemi, Xavier Leclerc, Jean Pierre Pruvo, Sophie Gallas, and Stéphane Velasco.
Acknowledgments
We are grateful to the genomics and bioinformatics core facility (GenoBiRD) and to the cellular and tissue imaging core facility (MicroPICell) of Nantes for their expert services. We would like to thank the Genome Aggregation Database (gnomAD) and the groups that provided exome and genome variant data to this resource. A full list of contributing groups can be found at http://gnomad.broadinstitute.org/about. We acknowledge the Center of Biological Resources (CHU Nantes, Hôtel-Dieu, CBR, Nantes, France) as well as Martine and Marie-France Le Cunff and the Clinical Investigation Center 1413 of Nantes for their assistance in managing the ICAN and PREGO biobanks. This work was supported by the French Regional Council of Pays-de-la-Loire (VaCaRMe program to S.B., S.C., F.S., P.L., H.L.M., and R.R.), the Fondation Genavie (to G.L. and R.R.), the Agence Nationale de la Recherche (ANR-15-CE17-0008-01 to G.L.), the French Ministry of Health (clinical trial NCT02848495 to H.D.), the Société Française de Radiologie, and the Société française de Neuroradiologie (both to R.B.).
Published: January 4, 2018
Footnotes
Supplemental Data include one figure and three tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2017.12.006.
Contributor Information
Hubert Desal, Email: hubert.desal@chu-nantes.fr.
Richard Redon, Email: richard.redon@inserm.fr.
ICAN Study Group:
Hubert Desal, Romain Bourcier, Benjamin Daumas-Duport, Bertrand Isidor, Jérôme Connault, Pierre Lebranchu, Thierry Le Tourneau, Marie Pierre Viarouge, Chrisanthi Papagiannaki, Michel Piotin, Hocine Redjem, Mikael Mazighi, Jean Philippe Desilles, Olivier Naggara, Denis Trystram, Myriam Edjlali-Goujon, Christine Rodriguez, Waghi Ben Hassen, Suzanna Saleme, Charbel Mounayer, Olivier Levrier, Pierre Aguettaz, Xavier Combaz, Anne Pasco, Emeline Berthier, Marc Bintner, Marc Molho, Pascale Gauthier, Cyril Chivot, Vincent Costalat, Cyril Darganzil, Alain Bonafé, Anne Christine Januel, Caterina Michelozzi, Christophe Cognard, Fabrice Bonneville, Philippe Tall, Jean Darcourt, Alessandra Biondi, Cristina Iosif, Elisa Pomero, Jean Christophe Ferre, Jean Yves Gauvrit, François Eugene, Hélène Raoult, Jean Christophe Gentric, Julien Ognard, René Anxionnat, Serge Bracard, Anne Laure Derelle, Romain Tonnelet, Laurent Spelle, Léon Ikka, Robert Fahed, Aymeric Rouchaud, Augustin Ozanne, Jildaz Caroff, Nidal Ben Achour, Jacques Moret, Emmanuel Chabert, Jérôme Berge, Gaultier Marnat, Xavier Barreau, Florent Gariel, Frédéric Clarencon, Mohammed Aggour, Frédéric Ricolfi, Adrien Chavent, Pierre Thouant, Pablo Lebidinsky, Brivael Lemogne, Denis Herbreteau, Richard Bibi, Laurent Pierot, Sébastien Soize, Marc Antoine Labeyrie, Christophe Vandendries, Emmanuel Houdart, Appoline Kazemi, Xavier Leclerc, Jean Pierre Pruvo, Sophie Gallas, and Stéphane Velasco
Web Resources
Ensembl Genome Browser, human genome, http://useast.ensembl.org/Homo_sapiens/Info/Index
ExAC Browser, http://exac.broadinstitute.org/
gnomAD Browser, http://gnomad.broadinstitute.org/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Variant Effect Predictor, http://useast.ensembl.org/Homo_sapiens/Tools/VEP
Supplemental Data
References
- 1.Vlak M.H., Algra A., Brandenburg R., Rinkel G.J. Prevalence of unruptured intracranial aneurysms, with emphasis on sex, age, comorbidity, country, and time period: a systematic review and meta-analysis. Lancet Neurol. 2011;10:626–636. doi: 10.1016/S1474-4422(11)70109-0. [DOI] [PubMed] [Google Scholar]
- 2.Laaksamo E., Ramachandran M., Frösen J., Tulamo R., Baumann M., Friedlander R.M., Harbaugh R.E., Hernesniemi J., Niemelä M., Raghavan M.L., Laakso A. Intracellular signaling pathways and size, shape, and rupture history of human intracranial aneurysms. Neurosurgery. 2012;70:1565–1572. doi: 10.1227/NEU.0b013e31824c057e. discussion 1572–1573. [DOI] [PubMed] [Google Scholar]
- 3.Nieuwkamp D.J., Setz L.E., Algra A., Linn F.H.H., de Rooij N.K., Rinkel G.J.E. Changes in case fatality of aneurysmal subarachnoid haemorrhage over time, according to age, sex, and region: a meta-analysis. Lancet Neurol. 2009;8:635–642. doi: 10.1016/S1474-4422(09)70126-7. [DOI] [PubMed] [Google Scholar]
- 4.Pierot L., Spelle L., Vitry F., ATENA Investigators Immediate clinical outcome of patients harboring unruptured intracranial aneurysms treated by endovascular approach: results of the ATENA study. Stroke. 2008;39:2497–2504. doi: 10.1161/STROKEAHA.107.512756. [DOI] [PubMed] [Google Scholar]
- 5.Huttunen T., von und zu Fraunberg M., Frösen J., Lehecka M., Tromp G., Helin K., Koivisto T., Rinne J., Ronkainen A., Hernesniemi J., Jääskeläinen J.E. Saccular intracranial aneurysm disease: distribution of site, size, and age suggests different etiologies for aneurysm formation and rupture in 316 familial and 1454 sporadic eastern Finnish patients. Neurosurgery. 2010;66:631–638. doi: 10.1227/01.NEU.0000367634.89384.4B. discussion 638. [DOI] [PubMed] [Google Scholar]
- 6.Bacigaluppi S., Piccinelli M., Antiga L., Veneziani A., Passerini T., Rampini P., Zavanone M., Severi P., Tredici G., Zona G. Factors affecting formation and rupture of intracranial saccular aneurysms. Neurosurg. Rev. 2014;37:1–14. doi: 10.1007/s10143-013-0501-y. [DOI] [PubMed] [Google Scholar]
- 7.Frösen J. Smooth muscle cells and the formation, degeneration, and rupture of saccular intracranial aneurysm wall--a review of current pathophysiological knowledge. Transl. Stroke Res. 2014;5:347–356. doi: 10.1007/s12975-014-0340-3. [DOI] [PubMed] [Google Scholar]
- 8.Vlak M.H.M., Rinkel G.J.E., Greebe P., Algra A. Independent risk factors for intracranial aneurysms and their joint effect: a case-control study. Stroke. 2013;44:984–987. doi: 10.1161/STROKEAHA.111.000329. [DOI] [PubMed] [Google Scholar]
- 9.Bourcier R., Redon R., Desal H. Genetic investigations on intracranial aneurysm: update and perspectives. J. Neuroradiol. 2015;42:67–71. doi: 10.1016/j.neurad.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Alg V.S., Sofat R., Houlden H., Werring D.J. Genetic risk factors for intracranial aneurysms: a meta-analysis in more than 116,000 individuals. Neurology. 2013;80:2154–2165. doi: 10.1212/WNL.0b013e318295d751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yasuno K., Bakırcıoğlu M., Low S.-K., Bilgüvar K., Gaál E., Ruigrok Y.M., Niemelä M., Hata A., Bijlenga P., Kasuya H. Common variant near the endothelin receptor type A (EDNRA) gene is associated with intracranial aneurysm risk. Proc. Natl. Acad. Sci. USA. 2011;108:19707–19712. doi: 10.1073/pnas.1117137108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou S., Ambalavanan A., Rochefort D., Xie P., Bourassa C.V., Hince P., Dionne-Laporte A., Spiegelman D., Gan-Or Z., Mirarchi C. RNF213 is associated with intracranial aneurysms in the French-Canadian population. Am. J. Hum. Genet. 2016;99:1072–1085. doi: 10.1016/j.ajhg.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santiago-Sim T., Fang X., Hennessy M.L., Nalbach S.V., DePalma S.R., Lee M.S., Greenway S.C., McDonough B., Hergenroeder G.W., Patek K.J. THSD1 (Thrombospondin Type 1 Domain Containing Protein 1) mutation in the pathogenesis of intracranial aneurysm and subarachnoid hemorrhage. Stroke. 2016;47:3005–3013. doi: 10.1161/STROKEAHA.116.014161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu W., Morito D., Takashima S., Mineharu Y., Kobayashi H., Hitomi T., Hashikata H., Matsuura N., Yamazaki S., Toyoda A. Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PLoS ONE. 2011;6:e22542. doi: 10.1371/journal.pone.0022542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamada F., Aoki Y., Narisawa A., Abe Y., Komatsuzaki S., Kikuchi A., Kanno J., Niihori T., Ono M., Ishii N. A genome-wide association study identifies RNF213 as the first Moyamoya disease gene. J. Hum. Genet. 2011;56:34–40. doi: 10.1038/jhg.2010.132. [DOI] [PubMed] [Google Scholar]
- 16.Bourcier R., Chatel S., Bourcereau E., Jouan S., Marec H.L., Daumas-Duport B., Sevin-Allouet M., Guillon B., Roualdes V., Riem T., ICAN Investigators Understanding the pathophysiology of intracranial aneurysm: The ICAN Project. Neurosurgery. 2017;80:621–626. doi: 10.1093/neuros/nyw135. [DOI] [PubMed] [Google Scholar]
- 17.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindenbaum P., Le Scouarnec S., Portero V., Redon R. Knime4Bio: a set of custom nodes for the interpretation of next-generation sequencing data with KNIME. Bioinformatics. 2011;27:3200–3201. doi: 10.1093/bioinformatics/btr554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han L., Abney M. Identity by descent estimation with dense genome-wide genotype data. Genet. Epidemiol. 2011;35:557–567. doi: 10.1002/gepi.20606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karakachoff M., Duforet-Frebourg N., Simonet F., Le Scouarnec S., Pellen N., Lecointe S., Charpentier E., Gros F., Cauchi S., Froguel P., D E S I R Study Group Fine-scale human genetic structure in Western France. Eur. J. Hum. Genet. 2015;23:831–836. doi: 10.1038/ejhg.2014.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu M.C., Lee S., Cai T., Li Y., Boehnke M., Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgenthaler S., Thilly W.G. A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: a cohort allelic sums test (CAST) Mutat. Res. 2007;615:28–56. doi: 10.1016/j.mrfmmm.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hato T., Tabata M., Oike Y. The role of angiopoietin-like proteins in angiogenesis and metabolism. Trends Cardiovasc. Med. 2008;18:6–14. doi: 10.1016/j.tcm.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Santulli G. Angiopoietin-like proteins: a comprehensive look. Front. Endocrinol. (Lausanne) 2014;5:4. doi: 10.3389/fendo.2014.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oike Y., Ito Y., Maekawa H., Morisada T., Kubota Y., Akao M., Urano T., Yasunaga K., Suda T. Angiopoietin-related growth factor (AGF) promotes angiogenesis. Blood. 2004;103:3760–3765. doi: 10.1182/blood-2003-04-1272. [DOI] [PubMed] [Google Scholar]
- 29.Romeo S., Yin W., Kozlitina J., Pennacchio L.A., Boerwinkle E., Hobbs H.H., Cohen J.C. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J. Clin. Invest. 2009;119:70–79. doi: 10.1172/JCI37118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leblanc G.G., Golanov E., Awad I.A., Young W.L., Biology of Vascular Malformations of the Brain NINDS Workshop Collaborators Biology of vascular malformations of the brain. Stroke. 2009;40:e694–e702. doi: 10.1161/STROKEAHA.109.563692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hellström M., Kalén M., Lindahl P., Abramsson A., Betsholtz C. Role of PDGF-B and PDGFR-beta in recruitment of vascular smooth muscle cells and pericytes during embryonic blood vessel formation in the mouse. Development. 1999;126:3047–3055. doi: 10.1242/dev.126.14.3047. [DOI] [PubMed] [Google Scholar]
- 32.Raab S., Beck H., Gaumann A., Yüce A., Gerber H.P., Plate K., Hammes H.P., Ferrara N., Breier G. Impaired brain angiogenesis and neuronal apoptosis induced by conditional homozygous inactivation of vascular endothelial growth factor. Thromb. Haemost. 2004;91:595–605. doi: 10.1160/TH03-09-0582. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen H.L., Lee Y.J., Shin J., Lee E., Park S.O., McCarty J.H., Oh S.P. TGF-β signaling in endothelial cells, but not neuroepithelial cells, is essential for cerebral vascular development. Lab. Invest. 2011;91:1554–1563. doi: 10.1038/labinvest.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bourcier R., Lenoble C., Guyomarch-Delasalle B., Daumas-Duport B., Papagiannaki C., Redon R., Desal H. Is there an inherited anatomical conformation favoring aneurysmal formation of the anterior communicating artery? J. Neurosurg. 2017;126:1598–1605. doi: 10.3171/2016.4.JNS153032. [DOI] [PubMed] [Google Scholar]
- 35.Mackey J., Brown R.D., Jr., Moomaw C.J., Hornung R., Sauerbeck L., Woo D., Foroud T., Gandhi D., Kleindorfer D., Flaherty M.L., FIA Investigators Familial intracranial aneurysms: is anatomic vulnerability heritable? Stroke. 2013;44:38–42. doi: 10.1161/STROKEAHA.112.667261. [DOI] [PubMed] [Google Scholar]
- 36.Oike Y., Akao M., Yasunaga K., Yamauchi T., Morisada T., Ito Y., Urano T., Kimura Y., Kubota Y., Maekawa H. Angiopoietin-related growth factor antagonizes obesity and insulin resistance. Nat. Med. 2005;11:400–408. doi: 10.1038/nm1214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.