Abstract
Cyanide is one of the most toxic chemicals for living organisms described so far. Its toxicity is mainly based on the high affinity that cyanide presents toward metals, provoking inhibition of essential metalloenzymes. Cyanide and its cyano-derivatives are produced in a large scale by many industrial activities related to recovering of precious metals in mining and jewelry, coke production, steel hardening, synthesis of organic chemicals, and food processing industries. As consequence, cyanide-containing wastes are accumulated in the environment becoming a risk to human health and ecosystems. Cyanide and related compounds, like nitriles and thiocyanate, are degraded aerobically by numerous bacteria, and therefore, biodegradation has been offered as a clean and cheap strategy to deal with these industrial wastes. Anaerobic biological treatments are often preferred options for wastewater biodegradation. However, at present very little is known about anaerobic degradation of these hazardous compounds. This review is focused on microbial degradation of cyanide and related compounds under anaerobiosis, exploring their potential application in bioremediation of industrial cyanide-containing wastes.
Keywords: Anaerobiosis, Biodegradation, Bioreactor, Cyanide, Cyanide-containing wastewaters, Metagenomics, Methanogenesis, Nitrilase, Nitrogenase, Thiocyanate
Introduction: Cyanide in the environment. Forms, toxicity, sources and remediation
Chemical compounds that contain the cyano group (−C≡N) are usually called “cyanides.” In the environment, these compounds may be found in different forms including volatile hydrogen cyanide (HCN), simple inorganic salts (NaCN, KCN), metal-cyanide complexes with different stability and chemical composition, cyanate (OCN−), thiocyanate (SCN−), and organic cyanides (nitriles and cyanohydrins). Free forms of cyanide (HCN and CN−) are extremely toxic compounds; cyanate, thiocyanate, and nitriles are less toxic forms; and toxicity of metal-cyanide complexes depends on their capacity to break down releasing free cyanide (Baxter and Cummings 2006; Kumar et al. 2016). Cyanide acts as a potent metabolic poison because it tightly binds to metals, provoking the inactivation of metalloenzymes. In aerobic organisms, cyanide inhibits the cytochrome c oxidase, blocking the respiratory electron transport chain, and in animals, cyanide also reacts with methemoglobin in the bloodstream (Solomonson 1981; Jaszczak et al. 2017). Anaerobic microorganisms, especially methanogens, are even more sensitive to cyanide because they contain many relevant metalloproteins that are also inhibited in the presence of this toxic compound (Smith et al. 1985; Gijzen et al. 2000). Thus, cyanide toxicity threshold may be as low as 2 ppm for some anaerobes whereas is about 200 ppm for most aerobic microorganisms (Kuyucak and Akcil 2013).
Cyanide is usually found as pollutant in wastewaters from mining, jewelry, steel and metal industrial activities, production of chemicals, and food processing, among other processes. In addition, these industrial residues often contain important cyano-derivatives like cyanate, which results from cyanide oxidation, and thiocyanate, which is formed by the reaction between cyanide and reduced sulfur species (Akcil 2003; Luque-Almagro et al. 2016). Nitriles (R−C≡N) are also produced in the manufacture of feedstock, solvents, pharmaceuticals, and organic chemicals (Banerjee et al. 2002). Accumulation of cyanide-containing wastewaters in the environment becomes a potential risk to ecosystems and human health. Therefore, these industrial residues need to be treated by physical and/or chemical methods before discharging the effluents into the environment. These physical-chemical treatments operate predominantly under aerobic conditions, and they are expensive, require complex infrastructures, need hazardous reagents or generate toxic by-products, and are usually ineffective for stable metal-cyanide complexes (Akcil, 2003; Dash et al. 2009; Novak et al. 2013; Park et al. 2017).
Despite its toxicity, cyanide is a natural compound synthesized by a variety of organisms, including bacteria, fungi, plants, and animals, in which cyanogenesis may serve as defensive or offensive mechanism (Luque-Almagro et al. 2016). The HCN synthase required for bacterial cyanogenesis is expressed during transition from exponential to stationary phase of growth under oxygen limitation in response to the FNR-like anaerobic regulator ANR (Laville et al. 1998). On the other hand, many microorganisms have evolved enzymatic pathways for cyanide degradation, transformation, or tolerance, and many of them are even able to use cyanide as a nitrogen source for growth. Therefore, cyanide biodegradation has become an efficient economically interesting alternative to the physical-chemical treatments of cyanide-containing industrial residues (Ebbs 2004; Baxter and Cummings 2006; Dash et al. 2009; Kumar et al. 2016; Luque-Almagro et al. 2016; Park et al. 2017).
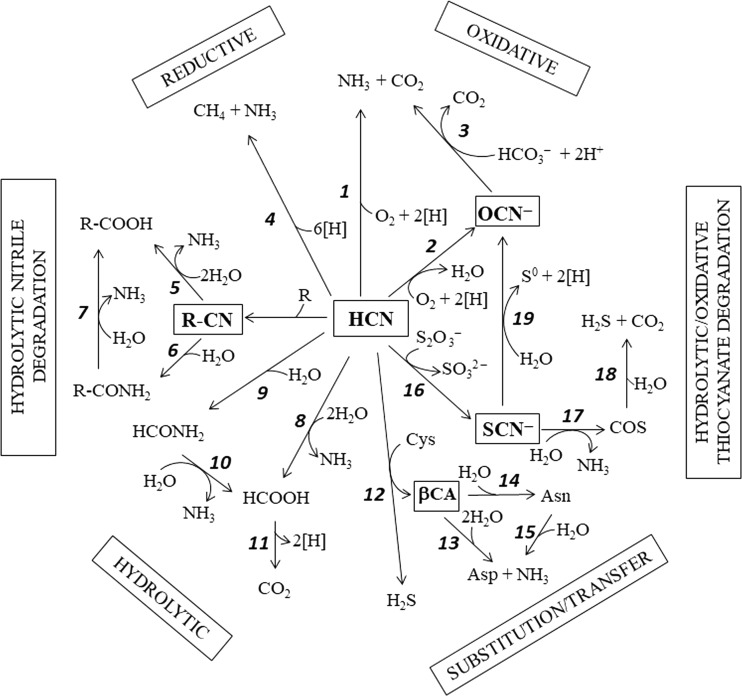
Microorganisms utilize different metabolic pathways to degrade or to assimilate cyanide. In general, these degradative routes are based in four types of enzymatic processes: hydrolytic, oxidative, reductive, and substitution/transfer reactions (Ebbs 2004; Huertas et al. 2006; Dash et al. 2009; Gupta et al. 2010; Park et al. 2017). These enzymatic degradation pathways are summarized in Fig. 1. The hydrolytic reactions are catalyzed by two different enzymes: the cyanidase (cyanide dihydratase) that transforms cyanide into formic acid and ammonia (Fig. 1; reaction 8), or the cyanide hydratase that produces formamide, which is further hydrolyzed by a formamidase (Fig. 1; reactions 9 and 10) (Martínková et al. 2015). The oxidative reactions generate carbon dioxide and ammonia either directly by the cyanide dioxygenase (Fig. 1; reaction 1) or in two-step reactions, via cyanate, catalyzed by the cyanide monooxygenase and the cyanase, respectively (Fig. 1; reactions 2 and 3) (Raybuck 1992; Ebbs 2004). The reductive pathway involves the nitrogenase required for biological nitrogen fixation, an oxygen-sensitive enzyme that also utilizes various substrates containing carbon-nitrogen triple bonds, such as hydrogen cyanide, nitriles, and isonitriles. Both molybdenum- and vanadium-nitrogenases carry out the six electrons reaction that converts HCN into methane and ammonia (Fig. 1; reaction 4) (Fisher et al. 2006; Seefeldt et al. 2013). Cyanide is also metabolized by the 3-cyanoalanine synthase (Fig. 1; reaction 12), which uses cysteine or O-acetylserine as substrate. The 3-cyanoalanine formed in this reaction can be further hydrolyzed to ammonia and aspartate in one-step reaction or with asparagine as intermediate (Fig. 1; reactions 13–15) (Howden et al. 2009). Finally, the rhodanese (thiosulfate:cyanide sulfurtransferase) catalyzes the reaction between cyanide and thiosulfate to form thiocyanate and sulfite (Fig. 1; reaction 16) (Cipollone et al. 2007). Another sulfurtransferase family enzyme, the 3-mercaptopyruvate sulfurtransferase, also transforms cyanide into thiocyanate but coupled to the conversion of mercaptopyruvate into pyruvate (Park et al. 2017).
Fig. 1.
Biochemical pathways for the biodegradation of cyanide and its derivatives. The cyano-compounds are boxed and highlighted in bold. Symbols: R-CN, nitrile (organic cyanide); R, organic compound; [H], hydrogen atom (e− + H+); βCA, β-cyanoalanine. Enzymes: 1, cyanide dioxygenase; 2, cyanide monooxygenase; 3, cyanase; 4, nitrogenase; 5, nitrilase; 6, nitrile hydratase; 7, amidase; 8, cyanidase (cyanide dihydratase); 9, cyanide hydratase; 10, formamidase; 11, formate dehydrogenase; 12, β-cyanoalanine synthase; 13, β-cyanoalanine nitrilase; 14, β-cyanoalanine hydratase; 15, asparaginase; 16, rhodanese; 17, thiocyanate hydrolase; 18, carbonyl sulfide (COS) hydrolase; 19, thiocyanate dehydrogenase
Thiocyanate, a compound much less toxic than cyanate, can be used by different bacteria as a source of energy, carbon, sulfur, or nitrogen (Sorokin et al. 2001). Thiocyanate is degraded to ammonia, carbon dioxide, and sulfide, which may be oxidized to sulfate by chemolithotrophic sulfur-oxidizing bacteria. Two different hydrolytic degradative pathways, involving either carbonyl sulfide (COS) or cyanate as intermediates, have been proposed (Kelly and Baker 1990; Sorokin et al. 2014). In the COS pathway, the initial hydrolytic cleavage of the C≡N bond by the thiocyanate hydrolase generates ammonia and carbonyl sulfide as first products (Fig. 1, reaction 17). COS is further hydrolyzed to carbon dioxide and sulfide (Fig. 1; reaction 18), which is finally oxidized to sulfate. In the cyanate pathway, it was initially proposed that an uncharacterized enzyme hydrolyzes the C−S bond converting thiocyanate into cyanate and sulfide. Then, cyanate can be hydrolyzed to ammonia and carbon dioxide by the cyanase, whereas sulfide can be oxidized to sulfate. However, very recently it has been described that the initial step of the thiocyanate degradation pathway via cyanate is an oxidation reaction catalyzed by the thiocyanate dehydrogenase (thiocyanate:cytochrome c oxidoreductase), a copper-containing enzyme that converts thiocyanate into cyanate and elemental sulfur with cytochrome c acting as electron acceptor (Fig. 1; reaction 19) (Berben et al. 2017).
Microbial degradation of nitriles usually requires hydrolytic reactions that generate ammonia and the corresponding carboxylic acid. Nitrilases catalyze this conversion in a single reaction (Fig. 1; reaction 5), while nitrile hydratases generate an amide intermediate that is further hydrolyzed by an amidase (Fig. 1; reactions 6 and 7) (Kobayashi and Shimizu 1998, 2000; Park et al. 2017).
Anaerobic degradation of cyanide
Cyanide is usually biodegraded through aerobic routes, according to the different pathways described above. However, microorganisms are also able to degrade cyanide under anaerobic conditions, although much more slowly and less successfully. The first description of an anaerobic cyanide biodegradation process was reported by Fedorak and Hrudey (1989) in methanogenic semicontinuous batch cultures. Since then, various applications based on anaerobic reactors or combining both aerobic and anaerobic processes have been developed for the treatment of different cyanide-containing wastewaters (Gijzen et al. 2000; Akcil and Mudder 2003; Chakraborti and Veeramani 2006; Novak et al. 2013; Joshi et al. 2016), although in general the anaerobic cyanide degradation process is not well understood and there is little knowledge about the microbial communities involved. In addition, abiotic anaerobic cyanide degradation may also occur when cyanide spontaneously hydrolyzes generating formic acid. In fact, a combination of simultaneous biotic and abiotic processes seems to contribute to the successful removal of cyanide in an upflow anaerobic sludge blanket (UASB) reactor (Novak et al. 2013).
Anaerobic biological treatments of wastewaters are attractive technologies that allow production of biogas with reduced biological oxygen demand (BOD) and low sludge volume and energy requirements, thus resulting in more cost-effective and energy-saving systems than aerobic procedures. In addition, anaerobic environments are not uncommon in nature, and in fact anaerobiosis prevails in most wastewater and polluted groundwater. Therefore, the identification of microorganisms able to produce methane in the presence of cyanide and the better understanding of the mechanisms involved in the anaerobic treatments of cyanide may convert this process into a feasible and efficient removal technology.
The chemical nature of the cyanide biodegradation reactions accounts for the fact that only the reductive or hydrolytic pathways may operate under anaerobic conditions (Fallon 1992). The reductive conversion of cyanide into methane and ammonia catalyzed by the nitrogenase has been described in resting cells of Klebsiella oxytoca, but not in cell-free extracts, probably due to the inactivation of the enzyme by oxygen exposure during cell disruption (Kao et al. 2003). Application of alginate and cellulose triacetate immobilized cells of K. oxytoca for the treatment of a cyanide-containing wastewater resulted in a more effective cyanide degradation, with higher tolerance to cyanide at wider ranges of pH, than when using free cells (Chen et al. 2008). This bacterium was also able to degrade the metal-cyano complex tetracyanonickelate (II) under anaerobic conditions, and nitrogenase was proposed to be the sole enzyme involved in this degradative process (Kao et al. 2004; Chen et al. 2009). However, the amount of cyanide removed by the nitrogenase in the environment is believed to be relatively small because this enzyme is rarely found in microbial populations (Gupta et al. 2010). On the other hand, it has been also demonstrated that hydrolytic reactions were responsible for anaerobic cyanide degradation in an upflow anaerobic fixed-bed reactor with activated carbon, transforming cyanide into ammonia and formic acid, which subsequently generated bicarbonate (Fallon et al. 1991; Fallon 1992). However, it was not possible to distinguish whether cyanide hydrolyzed directly or through the formation of formamide as an intermediate (Fallon 1992). Therefore, cyanide transformation analogous to hydrolytic reactions described for aerobes also occur in anaerobic systems. These hydrolytic pathways are probably the most attractive for biotechnological applications.
Nitrilases carry out the hydrolysis of the nitrile group to produce the corresponding carboxylic acid. Bacterial nitrilases show activities toward a wide range of nitriles, and are also able to degrade cyanide into ammonia and formate (Park et al. 2017). In addition, cyanide reacts chemically with different oxoacids to form cyanohydrins (hydroxynitriles), which may be hydrolyzed to ammonia and a carboxylic acid by a nitrilase enzyme, as described for the cyanide-degrading bacterium Pseudomonas pseudoalcaligenes CECT5344 (Estepa et al. 2012). Thus, nitrilases could be also good candidates for both aerobic and anaerobic cyanide remediation.
Cyanide biodegradation and methanogenesis
Methanogenesis can be maintained under a variety of feed medium conditions, which include ethanol, methanol, phenol, and toxic compounds as the primary reduced carbon sources. Many industrial wastewaters contain cyanide and related compounds, but cyanide has been usually considered highly toxic for anaerobes, especially for methanogens, resulting in minimal attention to the anaerobic treatments.
In an UASB reactor using a synthetic wastewater containing starch and fatty acids, sludge was successfully acclimatized to high cyanide concentrations (up to 125 mg/L), allowing an elevated methane production with high cyanide degradation efficiency. Cyanide inhibition on methanogenic activity was more pronounced for acetoclastic than for hydrogenotrophic methanogens, suggesting that enzymes and cofactors involved in hydrogenotrophic methane production are less sensitive to cyanide (Gijzen et al. 2000). Acclimatization of anaerobic microbes to cyanide was also used to improve the degradation rates in an anaerobic batch reactor with sludge from a wastewater treatment plant and fresh cow dung (Gupta et al. 2016). In this study, it was also observed that hydrogen-utilizing methanogens were more tolerant to cyanide than acetate-utilizing methanogens (Gupta et al. 2016). Successful biogas production and cyanide removal without methanogenesis inhibition was also reported in an UASB reactor for brewery wastewater treatment (Novak et al. 2013). Anaerobic cyanide degradation resulted from a combination of both biotic and abiotic processes, and again, the hydrogenotrophic community was less sensitive to cyanide than the acetoclastic methanogens. The phylogenetic analyses carried out by 16S rRNA sequences allowed the identification of the bacterial phylum Firmicutes and the archaeal genus Methanosarcina as relevant microbial groups involved in the anaerobic cyanide degradation associated to methane production (Novak et al. 2013).
During the production of cassava starch, large amounts of cyanide are released from cyanoglycosides by hydrolytic enzymes present in the raw cassava peel, leading to a cyanide concentration in the wastewater as high as 200 mg/L. Thus, linamarase hydrolyzes the cyanoglycoside linamarin releasing cyanide, which is detoxified to 3-cyanoalanine by the 3-cyanoalanine synthase (Cuzin and Labat 1992). When a cassava root wastewater was fermented in an anaerobic fixed-bed methanogenic reactor to produce biogas, up to 150 mg/L cyanide could be removed after biofilm establishment. All nitrogen derived from cyanide was converted into organic nitrogen by the biomass (Siller and Winter 1998a). Anaerobic degradation of this cyanide-rich agroindustrial wastewater was optimized in a two-step process with an equilibration/pre-acidification reactor followed by a methane reactor. Cell suspensions from the microbial community in the reactor generated similar amounts of ammonia and formic acid from cyanide, with little formamide accumulation. Optimal cyanide removal took place at pH 6–7.5 and temperature 25–37 °C (Siller and Winter 1998b). Sludge from an anaerobic lagoon has been also used in an UASB reactor for successful treatment of a cyanide-containing tapioca starch wastewater. High gas productivity and up to 98% cyanide removal was achieved for 25 m/L cyanide in the feed, requiring 15 days for the complete recovery of the reactor (Annachhatre and Amornkaew 2001).
Industrial wastewaters from steel manufacturing, fuel processing, coal conversion, and coking, which contain high concentrations of ammonia, phenol, thiocyanate, and cyanide, are also amenable to biodegradation by methanogenic consortia (Fedorak and Hrudey 1989). Thus, up to 98% cyanide was successfully removed during methanogenic degradation of phenol in an UASB reactor fed with 20 mg/L cyanide (Kumar et al. 2011). Sequential anaerobic-aerobic bioreactors have been also used for the treatment of complex mixtures of phenol, ammonia, thiocyanate, and cyanide (Chakraborti and Veeramani 2006). In a combined anaerobic-aerobic system treating coking wastewater with hydraulic retention time of 114 h, 81.8% chemical oxygen demand (COD), 85.6% total organic carbon (TOC), 99.9% total phenols, 98.2% thiocyanate, and 85.4% cyanide were removed (Joshi et al. 2016). Microbial diversity in both anaerobic and aerobic reactors was also analyzed resulting that phenol-degrading and hydrolytic bacteria such as Ottowia, Soehngenia, and Corynebacterium were predominant in the anaerobic sludge, whereas thiocyanate and phenol degraders belonging to Thiobacillus, Diaphorobacter, and Comamonas genera were most abundant in the aerobic sludge. Methanosarcina was the dominant archaea in the anaerobic reactor (Joshi et al. 2016). Similar removal efficiencies were obtained in a previous study using a combination of anaerobic-aerobic-anoxic bioreactors for the treatment of coke wastewater (Sharma and Philip 2015), highlighting the feasibility to apply successfully this sequential anaerobic-aerobic operation to treat complex phenol and cyanide-containing wastewaters.
The cyanidation process used for extraction of gold and other metals from ores in mining activities generates large amounts of cyanide-containing wastes that require treatment before they can be released to the environment (Luque-Almagro et al. 2016; Mekuto et al. 2016). Different approaches have been applied to remove cyanide from cyanidation and electroplating wastewater, but they operate basically under aerobic conditions (Akcil and Mudder 2003; Sirianuntapiboon et al. 2008; Kuyucak and Akcil 2013; Mekuto et al. 2016). However, laboratory and engineered wetland experiments based on aerobic and anaerobic processes have been used for the construction of a pilot field-scale passive system at a gold mine in northern Spain (Álvarez et al. 2013). In a laboratory test with two anaerobic columns operating in a continuous flow-through mode, one filled from bottom to the top with a 20-cm layer of limestone and a 100-cm layer of compost, and the other with the same content but including grained iron particles mixed with the compost substrate, the cyanidation wastewater was remediated with a 60–70% reduction of weak acid dissociable cyanides. Remediation of cyanide was slightly higher in the column with iron, probably due to the formation of complexes that could be adsorbed in the compost. In addition, the compost-based constructed wetlands detoxify the cyanidation effluents, successfully removing both weak acid dissociable cyanide and metals like copper (more than 90%). Therefore, aerobic/anaerobic wetland-based passive systems can be considered as a suitable technology for remediation of mining cyanidation effluents (Álvarez et al. 2013). However, the role of biodegradation in this system was not analyzed and probably the main mechanisms involved in cyanide removal were of physical/chemical nature, like complexation to solid phases and photodegradation.
Under anaerobic conditions, sulfate-reducing bacteria could be also used for an efficient biodegradation of cyanide and metal-containing wastewaters (Song et al. 1998; Quan et al. 2004). Thus, it has been demonstrated that removal of both free cyanide and metal-cyanide complexes (mainly with zinc, nickel, or copper) may occur under sulfate reduction conditions using a granular sludge from an UASB reactor fed with brewery wastewater and enriched with sludges from electroplating and industrial wastewater plants. Analysis of the microbial community revealed that a bacterial consortium composed of three major phylotypes including Desulfovibrio was responsible of cyanide degradation during sulfate reduction. In addition, sulfate-reducing bacteria were found to be less sensitive to cyanide than methanogenic bacteria (Quan et al. 2004). Therefore, sulfate reduction conditions may be used for a plausible treatment of industrial wastewaters containing cyanide and metal-cyanide complexes.
Biodegradation of thiocyanate and nitriles
Anaerobic biodegradation of cyanide in the presence of sulfide can produce thiocyanate. In addition, most sulfide minerals have the potential to generate thiocyanate, a process accelerated in anaerobiosis and low alkaline conditions, usually generating cyanate, nitrate, and ammonium as breakdown products (Kuyucak and Akcil 2013). Thiocyanate can be also formed by the transfer of sulfur from thiosulfate to cyanide in the reaction catalyzed by the rhodanese. It has been described that an extracellular rhodanese from Coprothermobacter is involved in anaerobic cyanide metabolism (Tandishabo et al. 2007).
Bacteria able to degrade thiocyanate have been isolated from various aerobic and anaerobic environments like soils, soda lakes, gold mine tailing, and activated sludge. These organisms can use thiocyanate as an energy, carbon, sulfur, or nitrogen source (Sorokin et al. 2001; Gould et al. 2012; Watts and Moreau 2016). Most thiocyanate-degrading chemolithotrophic bacteria oxidize aerobically the sulfide released in thiocyanate degradation, but some species like Thioalkalivibrio thiocyanodenitrificans are facultative anaerobes capable of growth anaerobically with thiocyanate as electron donor and with nitrate or nitrite as electron acceptor (Sorokin et al. 2004). Heterotrophic bacteria are also capable of thiocyanate degradation, using this compound as a source of nitrogen (Watts and Moreau 2016). There are two mechanisms for thiocyanate degradation, the carbonyl sulfide and the cyanate pathways (Fig. 1), and both are essentially aerobic. Therefore, the different systems developed for thiocyanate biodegradation are usually aerobic, like the activated sludge tailing effluent remediation (ASTER™) process (Huddy et al. 2015; Kantor et al. 2015). Most of these bioremediation systems rely on microbial co-cultures or consortia that metabolize undesirable by-products or establish potential syntrophic links, increasing the robustness of the system (Gould et al. 2012; Watts and Moreau 2016). However, the development of novel bioreactor designs, such as the utilization of several aerobic/anaerobic/anoxic reactors in series, and the better understanding of the microbial populations and processes involved in the biodegradation processes, using modern techniques of global analysis that provides a holistic view, will allow the development of more efficient and effective bioremediation approaches.
A system with two trains (A and B) of four-stage moving bed biofilm reactors, with an anoxic operating reactor in the train B, was designed for thiocyanate, cyanate, and ammonia biodegradation of gold extraction wastewater (Villemur et al. 2015). These three contaminants were completely removed and Thiobacillus strains were identified as the predominant bacteria in all reactors, although the higher content of anammox-related bacteria in train B suggests that the nitrogen dissimilation process takes place by this route (Villemur et al. 2015). A set of two continuous culture reactors, the first maintained aerobic and the second operating anaerobically, was applied at the low temperature typical for boreal climate for using the thiocyanate and thiosulfate present in a gold extraction wastewater as denitrification electron donors. Interestingly, the anaerobic reactor showed a higher diversity of microbial genera than the aerobic-operating reactor (Broman et al. 2017).
Meta-omics (metagenomics and metaproteomics) applied to study the microbial communities degrading thiocyanate and cyanide in aerobic continuous-flow bioreactors revealed the dominance of Thiobacillus strains capable of thiocyanate degradation (Kantor et al. 2015; Rahman et al. 2016; Kantor et al. 2017). A large portion of bioreactor community was autotrophic, relying on the energy generated from oxidation of sulfur compounds produced during thiocyanate degradation. Genes involved in ammonium oxidation and denitrification, as required for complete nitrogen removal, were also detected (Kantor et al. 2015).
Nitrilases, enzymes that convert organic cyanides into their respective carboxylic acid and ammonia, have acquired a relevant position in industry because they have been applied in the synthesis of numerous compounds, and are considered an economic and environmental friendly alternative to chemical methods (Gong et al. 2012; Luque-Almagro et al. 2016; Park et al. 2017). Nitrilase substrates can be aliphatics, like glutaronitrile, or aromatics, like benzonitrile (Estepa et al. 2012). Most nitrilases works aerobically under mesophilic conditions, and therefore, microbial degradation of a wide range of nitriles, using free or immobilized cells, in batch or continuous-flow bioreactors, was investigated under aerobic conditions (Kobayashi and Shimizu 2000; Kao et al. 2006; Chen et al. 2010; Maniyan et al. 2013). Nevertheless, several nitrilases can also function anaerobically at elevated temperatures. Thus, a termostable nitrilase from the hyperthermophile Pyrococcus sp. M24D13, which was isolated under strict anaerobic conditions from soil samples from Antarctica, has been purified and characterized. This nitrilase showed optimal activity at 85 °C and pH 7.5 with benzonitrile and butyronitrile, its major substrates. The enzyme also showed cyanidase activity (Dennet and Blammey 2016).
Conclusions and future perspectives
Industrial wastewaters containing cyanide and related compounds like nitriles and thiocyanate may be bioremediated under aerobic conditions, but very little information is available about anaerobic cyanide biodegradation. Reductive or hydrolytic pathways may operate under anaerobic conditions, although cyanide degradation occurs more slowly and less successfully in comparison with the aerobic biodegradation. Biogas production associated to anaerobic biological treatments of wastewaters is an attractive technology. Anaerobic environments are found in nature, and cyanide-containing spills may occur where anaerobiosis is established. Isolation of novel microorganisms able to tolerate and degrade cyanide from both aerobic and anaerobic zones, application of microbial co-cultures or consortia, and acclimatization of the microbial communities to cyanide are approaches that could improve the degradation rates of food industry, gold mining, and other industrial effluents. Development of novel bioreactor designs utilizing sequential aerobic/anaerobic/anoxic systems and application of metagenomics and metaproteomics global analysis techniques that provide a holistic view for a better understanding of the mechanisms involved in the biodegradation processes and the composition and dynamics of microbial populations will contribute to develop more efficient and effective removal technologies.
Acknowledgements
Authors thank FCC-Ámbito, SEVECO, AVENIR and MAGTEL for their fruitful collaborations.
Funding
This work was funded by Grant BIO2015-64311-R (Ministerio de Economía, Industria y Competitividad, Spain, and FEDER, UE) and Grant CVI-7560 (Junta de Andalucía, Grupo BIO-117, Spain).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Akcil A. Destruction of cyanide in gold mill effluents: biological versus chemical treatments. Biotech Adv. 2003;21(6):501–511. doi: 10.1016/S0734-9750(03)00099-5. [DOI] [PubMed] [Google Scholar]
- Akcil A, Mudder T. Microbial destruction of cyanide wastes in gold mining: process review. Biotechnol Lett. 2003;25(6):445–450. doi: 10.1023/A:1022608213814. [DOI] [PubMed] [Google Scholar]
- Álvarez R, Ordóñez A, Loredo J, Younger PL. Wetland-based passive treatment systems for gold ore processing effluents containing residual cyanide, metals and nitrogen species. Environ Sci Process Impacts. 2013;15(11):2115–2124. doi: 10.1039/c3em00410d. [DOI] [PubMed] [Google Scholar]
- Annachhatre AP, Amornkaew A. Up-flow anaerobic sludge blanket treatment of starch wastewater containing cyanide. Water Environ Res. 2001;73(5):622–632. doi: 10.2175/106143001X143358. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Sharma R, Banerjee UC. The nitrile-degrading enzymes: current status and future prospects. Appl Microbiol Biotechnol. 2002;60(1-2):33–44. doi: 10.1007/s00253-002-1062-0. [DOI] [PubMed] [Google Scholar]
- Baxter J, Cummings SP. The current and future applications of microorganism in the bioremediation of cyanide contamination. Antonie Van Leeuwenhoek. 2006;90(1):1–17. doi: 10.1007/s10482-006-9057-y. [DOI] [PubMed] [Google Scholar]
- Berben T, Overmars L, Sorokin DY, Muyzer G. Comparative genome analysis of three thiocyanate oxidizing Thioalkalivibrio species isolated from soda lakes. Front Microbiol. 2017;8:254. doi: 10.3389/fmicb.2017.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman E, Jawad A, Wu X, Chritel S, Ni G, López-Fernández M, Sundkvist JE, Dopson M. Low temperature, autotrophic microbial denitrification using thiosulfate or thiocyanate as electron donor. Biodegradation. 2017;28(4):287–301. doi: 10.1007/s10532-017-9796-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborti S, Veeramani H. Effect of HRT and recycle ratio on removal of cyanide, phenol, thiocyanate and ammonia in an anaerobic-anoxic-aerobic continuous system. Process Biochem. 2006;41(1):96–105. doi: 10.1016/j.procbio.2005.03.067. [DOI] [Google Scholar]
- Chen CY, Kao CM, Chen SC. Application of Klebsiella oxytoca immobilized cells on the treatment of cyanide wastewater. Chemosphere. 2008;71(1):133–139. doi: 10.1016/j.chemosphere.2007.10.058. [DOI] [PubMed] [Google Scholar]
- Chen CY, Kao CM, Chen SC, Chen TY. Biodegradation of tetracyanonickelate by Klebsiella oxytoca under anaerobic conditions. Desalination. 2009;249(3):1212–1216. doi: 10.1016/j.desal.2009.06.036. [DOI] [Google Scholar]
- Chen CY, Chen SC, Fingas M, Kao CM. Biodegradation of propionitrile by Klebsiella oxytoca immobilized in alginate and cellulose triacetate gel. J Hazard Mater. 2010;177(1-3):856–863. doi: 10.1016/j.jhazmat.2009.12.112. [DOI] [PubMed] [Google Scholar]
- Cipollone R, Ascenzi P, Visca P. Common themes and variations in the rhodanese superfamily. IUBMB Life. 2007;59(2):51–59. doi: 10.1080/15216540701206859. [DOI] [PubMed] [Google Scholar]
- Cuzin N, Labat M. Reduction of cyanide levels during anaerobic digestion of cassava. Int J Food Sci Technol. 1992;27(3):329–336. doi: 10.1111/j.1365-2621.1992.tb02034.x. [DOI] [Google Scholar]
- Dash RR, Gaur A, Balomajumder C. Cyanide in industrial wastewaters and its removal: a review on biotreatment. J Hazard Mater. 2009;163(1):1–11. doi: 10.1016/j.jhazmat.2008.06.051. [DOI] [PubMed] [Google Scholar]
- Dennet GV, Blammey JM. A new thermophilic nitrilase from an Antarctic hyperthermophilic microorganism. Front Bioeng Biotechnol. 2016;4:5. doi: 10.3389/fbioe.2016.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs S. Biological degradation of cyanide compounds. Curr Opin Biotechnol. 2004;15(3):231–236. doi: 10.1016/j.copbio.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Estepa J, Luque-Almagro VM, Manso I, Escribano MP, Martínez-Luque M, Castillo F, Moreno-Vivián C, Roldán MD. The nit1C gene cluster of Pseudomonas pseudoalcaligenes CECT5344 involved in assimilation of nitriles is essential for growth on cyanide. Environ Microbiol Rep. 2012;4(3):326–334. doi: 10.1111/j.1758-2229.2012.00337.x. [DOI] [PubMed] [Google Scholar]
- Fallon RD. Evidence of a hydrolytic route for anaerobic cyanide degradation. Appl Environ Microbiol. 1992;58(9):3163–3164. doi: 10.1128/aem.58.9.3163-3164.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon RD, Cooper DA, Speece R, Henson M. Anaerobic biodegradation of cyanide under methanogenic conditions. Appl Environ Microbiol. 1991;57(6):1656–1662. doi: 10.1128/aem.57.6.1656-1662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorak PM, Hrudey SE. Cyanide transformation in anaerobic phenol-degrading methanogenic cultures. Water Sci Technol. 1989;21:67–76. [Google Scholar]
- Fisher K, Dilworth MJ, Newton WE. Azotobacter vinelandii vanadium nitrogenase: formaldehyde is a product of catalyzed HCN reduction and excess ammonia arises directly from catalyzed azide reduction. Biochemistry. 2006;45(13):4190–4198. doi: 10.1021/bi0514109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijzen HJ, Bernal E, Ferrer H. Cyanide toxicity and cyanide degradation in anaerobic wastewater treatment. Water Res. 2000;34(9):2447–2454. doi: 10.1016/S0043-1354(99)00418-2. [DOI] [Google Scholar]
- Gong JS, Lu ZM, Li H, Shi JS, Zhou ZM, Xu ZH. Nitrilases in nitrile biocatalysis: recent progress and forthcoming research. Microb Cell Factories. 2012;11(1):142. doi: 10.1186/1475-2859-11-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould WD, King M, Mohapatra BR, Cameron RA, Kapoor A, Koren DW. A critical review on destruction of thiocyanate in mining effluents. Miner Eng. 2012;34:38–47. doi: 10.1016/j.mineng.2012.04.009. [DOI] [Google Scholar]
- Gupta N, Balomajumder C, Agarwal VK. Enzymatic mechanism and biochemistry for cyanide degradation: a review. J Hazard Mater. 2010;176(1-3):1–13. doi: 10.1016/j.jhazmat.2009.11.038. [DOI] [PubMed] [Google Scholar]
- Gupta P, Ahammad SZ, Sreekrishnan TR. Improving the cyanide toxicity tolerance of anaerobic reactor: microbial interactions and toxin reduction. J Hazard Mater. 2016;315:52–60. doi: 10.1016/j.jhazmat.2016.04.028. [DOI] [PubMed] [Google Scholar]
- Howden AJM, Harrison CJ, Preston GM. A conserved mechanism for nitrile metabolism in bacteria and plants. Plant J. 2009;57(2):243–253. doi: 10.1111/j.1365-313X.2008.03682.x. [DOI] [PubMed] [Google Scholar]
- Huddy RJ, Wynand A, van Zyl W, van Hille RP, Harrison STL. Characterisation of the complex microbial community associated with the ASTER™ thiocyanate biodegradation system. Miner Eng. 2015;76:65–71. doi: 10.1016/j.mineng.2014.12.011. [DOI] [Google Scholar]
- Huertas MJ, Luque-Almagro VM, Martínez-Luque M, Blasco R, Moreno-Vivián C, Castillo F, Roldán MD. Cyanide metabolism of Pseudomonas pseudoalcaligenes CECT5344: role of siderophores. Biochem Soc Trans. 2006;34(1):152–155. doi: 10.1042/BST0340152. [DOI] [PubMed] [Google Scholar]
- Jaszczak E, Polkowska Z, Narkowycz S, Namiesnik J. Cyanides in the environment-analysis-problems and challenges. Environ Sci Pollut Res. 2017;24(19):15929–15948. doi: 10.1007/s11356-017-9081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi DR, Zhang Y, Tian Z, Gao Y, Yang M. Performance and microbial community composition in a long-term sequential anaerobic-aerobic bioreactor operation treating coking wastewater. Appl Microbiol Biotechnol. 2016;100(18):8191–8202. doi: 10.1007/s00253-016-7591-8. [DOI] [PubMed] [Google Scholar]
- Kantor RS, van Zyl AW, van Hille RP, Thomas BC, Harrison STL, Bandfield JF. Bioreactor microbial ecosystems for thiocyanate and cyanide degradation unravelled with genome-resolved metagenomics. Environ Microbiol. 2015;17(12):4929–4941. doi: 10.1111/1462-2920.12936. [DOI] [PubMed] [Google Scholar]
- Kantor RS, Huddy RJ, Iyer R, Thomas BC, Brown CT, Anantharaman K, Tringe S, Hettich RL, Harrison STL, Banfield JF. Genome-resolved meta-omics ties microbial dynamics to process performance in biotechnology for thiocyanate degradation. Environ Sci Technol. 2017;51(5):2944–2953. doi: 10.1021/acs.est.6b04477. [DOI] [PubMed] [Google Scholar]
- Kao CM, Liu JK, Lou HR, Lin CS, Chen SC. Biotransformation of cyanide to methane and ammonia by Klebsiella oxytoca. Chemosphere. 2003;50(8):1055–1061. doi: 10.1016/S0045-6535(02)00624-0. [DOI] [PubMed] [Google Scholar]
- Kao CM, Lin CC, Liu JK, Chen YL, Wu LT, Chen SC. Biodegradation of the metal-cyano complex tetracyanonickelate (II) by Klebsiella oxytoca. Enzym Microb Technol. 2004;35(5):405–410. doi: 10.1016/j.enzmictec.2004.05.010. [DOI] [Google Scholar]
- Kao CM, Chen KF, Liu JK, Chou SM, Chen SC. Enzymatic degradation of nitriles by Klebsiella oxytoca. Appl Microbiol Biotechnol. 2006;71(2):228–233. doi: 10.1007/s00253-005-0129-0. [DOI] [PubMed] [Google Scholar]
- Kelly DP, Baker SC. The organosulfur cycle: aerobic and anaerobic processes leading to turnover of C1-sulfur compounds. FEMS Microbiol Rev. 1990;87(3-4):241–246. doi: 10.1111/j.1574-6968.1990.tb04919.x. [DOI] [Google Scholar]
- Kobayashi M, Shimizu S. Metalloenzyme nitrile hydratase: structure, regulation and application to biotechnology. Nat Biotechnol. 1998;16(8):733–736. doi: 10.1038/nbt0898-733. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Shimizu S. Nitrile hydrolases. Curr Opin Chem Biol. 2000;4(1):95–102. doi: 10.1016/S1367-5931(99)00058-7. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Mishra RS, Jadhav SV, Vaidya AN, Chakrabarty T. Simultaneous degradation of cyanide and phenol in upflow anaerobic sludge blanket reactor. J Environ Sci Eng. 2011;53(3):277–280. [PubMed] [Google Scholar]
- Kumar R, Saha S, Dhaka S, Kurade MB, Kang CU, Baek SH, Jeon BH. Remediation of cyanide-contaminated environments through microbes and plants: a review of current knowledge and future perspectives. Geosyst Eng. 2016;20(1):28–40. doi: 10.1080/12269328.2016.1218303. [DOI] [Google Scholar]
- Kuyucak N, Akcil A. Cyanide and removal options from effluents in gold mining and metallurgical processes. Miner Eng. 2013;50:13–29. doi: 10.1016/j.mineng.2013.05.027. [DOI] [Google Scholar]
- Laville J, Blumer C, von Schroetter C, Gaia V, Défago G, Keel C, Haas D. Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0. J Bacteriol. 1998;180(12):3187–3196. doi: 10.1128/jb.180.12.3187-3196.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque-Almagro VM, Moreno-Vivián C, Roldán MD. Biodegradation of cyanide wastes from mining and jewellery industries. Curr Opin Biotechnol. 2016;38:9–13. doi: 10.1016/j.copbio.2015.12.004. [DOI] [PubMed] [Google Scholar]
- Maniyan MN, Sjahrir F, Ibrahim AL, Cass AEG. Biodegradation of cyanide by acetonitrile-induced cells of Rhodococcus sp. UKMP-5M. J Gen Appl Microbiol. 2013;59(6):393–404. doi: 10.2323/jgam.59.393. [DOI] [PubMed] [Google Scholar]
- Martínková L, Veselá AB, Rinágelová A. Cyanide hidratases and cyanide dihydratases: emerging tools in the biodegradation and biodetection of cyanide. Appl Microbiol Biotechnol. 2015;99(21):8875–8882. doi: 10.1007/s00253-015-6899-0. [DOI] [PubMed] [Google Scholar]
- Mekuto L, Ntwampe SKO, Akcil A. An integrated biological approach for treatment of cyanidation wastewater. Sci Total Environ. 2016;571:711–720. doi: 10.1016/j.scitotenv.2016.07.040. [DOI] [PubMed] [Google Scholar]
- Novak D, Franke-Whittle IH, Pirc ET, Jerman V, Insam H, Logar RM, Stres B. Biotic and abiotic processes contribute to successful anaerobic degradation of cyanide by UASB reactor biomass treating brewery waste water. Water Res. 2013;47(11):3644–3653. doi: 10.1016/j.watres.2013.04.027. [DOI] [PubMed] [Google Scholar]
- Park JK, Sewell BT, Benedik MJ. Cyanide bioremediation: the potential of engineered nitrilases. Appl Microbiol Biotechnol. 2017;101(8):3029–3042. doi: 10.1007/s00253-017-8204-x. [DOI] [PubMed] [Google Scholar]
- Quan ZX, Bae JW, Rhee SK, Cho YG, Lee ST. Toxicity and degradation of metal-complexed cyanide by a bacterial consortium under sulfate-reducing conditions. Biotechnol Lett. 2004;26(12):1007–1011. doi: 10.1023/B:BILE.0000030048.04282.da. [DOI] [PubMed] [Google Scholar]
- Rahman SF, Kantor RS, Huddy R, Thomas BC, van Zyl AW, Harrison STL, Banfield JF. Genome-resolved metagenomics of a bioremediation system for degradation of thiocyanate in mine water containing suspended solid tailing. Microbiol Open. 2016;6(3):e446. doi: 10.1002/mbo3.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raybuck SA. Microbes and microbial enzymes for cyanide degradation. Biodegradation. 1992;3(1):3–18. doi: 10.1007/BF00189632. [DOI] [PubMed] [Google Scholar]
- Seefeldt LC, Yang ZY, Duval S, Dean DR. Nitrogenase reduction of carbon-containing compounds. Biochim Biophys Acta-Bioenerg. 2013;1827(8-9):1102–1111. doi: 10.1016/j.bbabio.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma NK, Philip L. Treatment of phenolics, aromatic hydrocarbons, and cyanide-bearing wastewater in individual and combined anaerobic, aerobic, and anoxic bioreactors. Appl Biochem Biotechnol. 2015;175(1):300–322. doi: 10.1007/s12010-014-1262-y. [DOI] [PubMed] [Google Scholar]
- Siller H, Winter J. Treatment of cyanide-containing wastewater from the food industry in a laboratory-scale fixed-bed methanogenic reactor. Appl Microbiol Biotechnol. 1998;49(2):215–220. doi: 10.1007/s002530051161. [DOI] [PubMed] [Google Scholar]
- Siller H, Winter J. Degradation of cyanide in agroindustrial or industrial wastewater in an acidification reactor or in a single-step methane reactor by bacteria enriched from soil and peels of cassava. Appl Microbiol Biotechnol. 1998;50(3):384–389. doi: 10.1007/s002530051309. [DOI] [PubMed] [Google Scholar]
- Sirianuntapiboon S, Chairattanawan K, Rarunroeng M. Biological removal of cyanide compounds from electroplating wastewater (EPWW) by sequencing batch reactor (SBR) system. J Hazard Mater. 2008;154(1-3):526–534. doi: 10.1016/j.jhazmat.2007.10.056. [DOI] [PubMed] [Google Scholar]
- Smith MR, Lequerica JL, Hart MR. Inhibition of methanogenesis and carbon metabolism in Methanosarcina sp. by cyanide. J Bacteriol. 1985;162(1):67–71. doi: 10.1128/jb.162.1.67-71.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomonson LP. Cyanide as a metabolic inhibitor. In: Vennesland B, Conn EE, Knowles CJ, Westley J, Wissing F, editors. Cyanide in biology. New York: Academic Press; 1981. pp. 11–28. [Google Scholar]
- Song YC, Piak BC, Shim HS, La SJ. Influence of electron donor and toxic materials on the activity of sulfate reducing bacteria for the treatment of electroplating wastewater. Water Sci Technol. 1998;38(4-5):187–194. [Google Scholar]
- Sorokin DY, Tourova TP, Lysenko AM, Kuenen JG. Microbial thiocyanate utilization under highly alkaline conditions. Appl Environ Microbiol. 2001;67(2):528–538. doi: 10.1128/AEM.67.2.528-538.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin DY, Tourova TP, Antipov AN, Muyzer G, Kuenen JG. Anaerobic growth of the haloalkaliphilic denitrifying sulfur-oxidizing bacterium Thialkalivibrio thiocyanodenitrificans sp. nov. with thiocyanate. Microbiology. 2004;150(7):2435–2442. doi: 10.1099/mic.0.27015-0. [DOI] [PubMed] [Google Scholar]
- Sorokin DY, Abbas B, van Zessen E, Muyzer G. Isolation and characterization of an obligately chemolithoautotrophic Halothiobacillus strain capable of growth on thiocyanate as an energy source. FEMS Microbiol Lett. 2014;354(1):69–74. doi: 10.1111/1574-6968.12432. [DOI] [PubMed] [Google Scholar]
- Tandishabo K, Takahashi A, Nakamura K, Takamizawa K. Characterization of the rhodanese enzyme in Coprothermobacter strains. Proc Int Sym EcoTopia Sci ISETS. 2007;07:1202–1203. [Google Scholar]
- Villemur R, Juteau P, Bougie V, Ménard J, Déziel E. Development of four-stage moving bed biofilm reactor train with a pre-denitrification configuration for the removal of thiocyanate and cyanate. Bioresour Technol. 2015;181:254–262. doi: 10.1016/j.biortech.2015.01.051. [DOI] [PubMed] [Google Scholar]
- Watts MP, Moreau JW. New insight into the genetic and metabolic diversity of thiocyanate-degrading microbial consortia. Appl Microbiol Biotechnol. 2016;100(3):1101–1108. doi: 10.1007/s00253-015-7161-5. [DOI] [PubMed] [Google Scholar]