Abstract
The emergence of new methods for reprogramming of adult somatic cells into induced pluripotent stem cells (iPSC) led to the development of new approaches in drug discovery and regenerative medicine. Investigation of the molecular mechanisms underlying the self-renewal, expansion and differentiation of human iPSC (hiPSC) should lead to improvements in the manufacture of safe and reliable cell therapy products. The goal of our study was qualitative and quantitative proteomic characterizations of hiPSC by means of electrospray ionization (ESI)-MSe and MALDI-TOF/TOF mass spectrometry (MS). Proteomes of hiPSCs of different somatic origins: fibroblasts and peripheral blood CD34+ cells, reprogrammed by the same technique, were compared with the original somatic cells and hESC. Quantitative proteomic comparison revealed approximately 220 proteins commonly up-regulated in all three pluripotent stem cell lines compared to the primary cells. Expression of 21 proteins previously reported as pluripotency markers was up-regulated in both hiPSCs (8 were confirmed by Western blot). A number of novel candidate marker proteins with the highest fold-change difference between hiPSCs/hESC and somatic cells discovered by MS were confirmed by Western blot. A panel of 22 candidate marker proteins of hiPSC was developed and expression of these proteins was confirmed in 8 additional hiPSC lines.
Introduction
Induced pluripotent stem cells (iPSC) are an important research tool and have a potential to become a significant source of autologous cells differentiated from iPSC for therapeutic treatments. However, prior to therapeutic application appropriate characterization of human iPSC (hiPSC) is needed. To date, iPSC have been generated from numerous somatic cell types including dermal fibroblasts (Lowry et al., 2008; Takahashi et al., 2007; Yu et al., 2007), lymphocytes (Staerk et al., 2010; Loh et al., 2010), mesenchymal stem cells (Zou et al., 2011), endogenous kidney tubular renal epithelial cells (Montserrat et al., 2012), and CD34+ hematopoietic stem cells (Loh et al., 2009). It is believed that iPSC of different somatic origins may be predisposed toward re-differentiation to a particular cell lineage via “epigenetic memory” (Bar-Nur et al., 2011; Kim et al., 2010). For instance, it has been reported that hiPSC derived from hematopoietic stem cells (CD34+ cells) are particularly suitable for development of research models and treatments for hematopoietic diseases (Zou et al., 2011; Merling et al., 2013). Another recent study has shown that the hepatic lineage epigenetic memory contributed to the differentiation potential of mouse iPSC (Lee et al., 2012).
On the mRNA level hiPSC have been found to be clearly distinguishable from hESC and their expression pattern becomes closer to that of hESC after extended culture (Chin et al., 2009); hiPSC have been shown to bear residual gene expression from the donor cell type (Marchetto et al., 2009; Ghosh et al., 2010). Recent analysis of 12 established hiPSC lines has revealed epigenetic and transcriptional variations among them and has shown that these variations can have a significant impact on a cell line's ability to differentiate to a particular cell type (Bock et al., 2011).
The molecular characterization of hiPSC has been performed previously on different biological levels, including: gene expression profiling, epigenetic evaluation, the role of miRNAs in pluripotency, and genomic DNA alterations (Muller et al., 2012; Benevento and Munoz, 2012). However, quantitative proteomics has not yet been used to characterize hiPSC systematically (Munoz et al., 2011; Phanstiel et al., 2011; Kim et al., 2012; Yamana et al., 2013), and the molecular differences on the proteome level between hiPSC of different somatic origins have not been addressed. Sample preparation and MS-proteomic approaches reported previously on hiPSC vary significantly (Benevento and Munoz, 2012; Munoz et al., 2011; Phanstiel et al., 2011; Kim et al., 2012; Yamana et al., 2013), which complicates direct comparison of these studies.
The focal point of this study was the analysis of proteomes of two hiPSC lines at the earlier and later cell culture passages derived in two different laboratories and of different somatic origins: CD34+ cells circulating in peripheral blood (iNC-01) and fibroblasts of healthy donors (SB5-MP1). Both hiPSC lines were generated using the same reprogramming technique: loxP-flanked excisable polycistronic (human Oct4, Klf4, Sox2, and c-Myc) STEMCCA lentiviral vector, which generates transgene-free hiPSC lines upon Cre-mediated vector excision. iNC-01 cell line was previously used to obtain functional neutrophils (Sweeney et al., 2014) and SB5-MP1 was successfully used in differentiation into motor neurons (Grunseich et al., 2014). In parallel, we performed a quantitative global proteome analysis of H9 hESC line at the earlier and later passages, as well as of the somatic cell types (fibroblasts and peripheral blood mononuclear cells (PBMC), the cell population containing CD34+ hematopoietic stem cells).
From an analytical perspective, we applied the approach that combines application of high pressure assisted protein extraction and a combination of two LC/MS/MS techniques: electrospray ionization (ESI)-MSe and MALDI-TOF/TOF (Mindaye et al., 2013a, 2013b). Label-free quantification of proteins was performed by ESI-MSe using normalization against an internal reference standard (Silva et al., 2005, 2006). Quantitative and qualitative comparisons of hiPSC/hESC proteomes with that of somatic cells allowed the development of a protein marker panel for characterization of hiPSC, which was successfully tested in 8 more hiPSC lines of different somatic origins derived in different laboratories by different reprogramming techniques.
Materials and methods
Cells
Stem cell lines used for ESI-MSe and MALDI-TOF/TOF analyses are: iNC-01, passage (P) 36 and P53-peripheral blood CD34+ hematopoietic stem/progenitor cells derived hiPSC line, previously described (Merling et al., 2013); SB5-MP1, P22 and P28-adult fibroblasts derived hiPSC line, previously described (Grunseich et al., 2014); and H9 (WA09), P34 and P48-hESC line obtained from WiCell Research Institute. For both hiPSC lines somatic cells were transduced with the excisable STEMCCA-loxP lentivirus encoding human Oct4, Sox2, Klf4, and c-Myc (Millipore) (Sommer et al., 2009). The same donors' fibroblasts or a different healthy donors' human PBMC were used in parallel for ESI–MSe analysis. hiPSC derivation, culture, and their quality control, as well as the list of hiPSC lines used in the validation of a protein marker panel are described in the Supplementary information. The purity of both hiPSC lines was checked by immunocytochemistry, and it was shown that more than 90% of cells are expressing pluripotency markers (Supplementary Fig. 1).
The study was performed under NIAID IRB approved protocols 05-I-0213, 94-I-0073, and 09-I-0133, NINDS IRB approved protocol 00-N-0043, and FDA Research Involving Human Subjects Committee (RIHSC) approved protocol #s 13-052B and 13-053B.
Sample preparation, 2D-LC separation and ESI–MSe
The preparation of samples for MS analysis is described in the Supplementary information. 20 µg of digested protein (2 µl of each sample) was loaded and analyzed by a reverse phase (RP) nanoACQUITY™ ultrapressure liquid chromatography (UPLC) and Synapt G2 or Xevo-qTOF mass spectrometers operating in a data-independent (MSe) mode (Waters). Three technical replicates (sample injections) were done for each run in total. The HDMSe (Synapt G2) or MSe (Xevo-qTOF) data were processed and protein absolute label-free quantification (Silva et al., 2005, 2006) was performed using Protein Lynx Global Server version 2.5 (Waters). A maximum false discovery rate of 2% was allowed. All protein hits that were identified with a confidence of >95% were included in the quantitative analysis.
Label-free quantification, quantitative comparison and IPA analysis
Each protein was identified by at least three peptides that are required for quantification. Proteins identified in at least two out of three technical replicates were used for comparison. For quantitative comparison, we used combination criteria that consider both data variability and a fold change cutoff. Proteins for which the quantity didn't change by more than 30% (CV < 30%) in at least two out of three technical replicas, were considered. The “differentially expressed” proteins are proteins for which the expression level changed by ≥1.5 fold between the compared cell lines, or are uniquely expressed (identified in at least two out of three technical replicas and in only one of the two compared cell lines). A list of UniProt ID numbers of differentially expressed proteins for a given cell line was submitted to Ingenuity Pathway Analysis (IPA) (Ingenuity Systems) to determine the biological processes and canonical pathways affected by these proteins.
MALDI-MS analysis
MALDI-MS analysis was performed as described previously (Mindaye et al., 2013a, 2013b) (see brief description in the Supplementary information). Three technical replicates were performed for each sample.
Western blot analysis
Protein samples were prepared using the same protocol as for MS analysis. Protein content was measured using BCA Protein Assay kit (Pierce) and equal amount of total protein (15–20 µg/well) from each sample was resolved by SDS-PAGE, transferred to the nitrocellulose membrane using iBlot system (Life Technologies) and blotted with different primary antibodies (ABs) and IRDye secondary ABs (Li-Cor) using a standard protocol. See the Supplementary information for the list of antibodies. Odyssey Imaging System (Li-Cor) was used for signal detection.
Results
Qualitative global proteome analysis of one hESC line (H9), two hiPSC lines (SB5-MP1 and iNC-01), and their parental primary cells (fibroblasts and PBMC) was performed by both MS techniques, ESI–MSe and MALDI TOF/TOF (latter data are described in a separate section). Quantitative global proteome analysis of these hESC and hiPSC lines was performed by ESI–MSe. Two biological replicates for each cell line (one at the earlier and one at the later cell culture passage for hESC and hiPSCs) and three technical replicates were analyzed by both MS techniques. The number of proteins identified by ESI–MSe and selected for quantitative comparison in each cell line is shown in Table 1. In the table the earlier cell culture passages of pluripotent cell lines (hESC-H9-P.34, SB5-MP1-P.22, iNC-01-P.36) and the 1st biological replicate of fibroblasts and PBMC were analyzed by Xevo-qTOF mass spectrometer (in MSe mode). The later cell culture passages of pluripotent cell lines (hESC-H9-P.48, SB5-MP1-P.28, iNC-01-P.53) and the 2nd biological replicate of fibroblasts and PBMC were analyzed by SYNAPT G2 High Definition Mass Spectrometer (HDMS) (in HDMSe mode, which allows an additional, fourth dimension of peptide separation). This resulted in a higher number of proteins identified and quantified in the 2nd biological replicate for most of the cell lines (Table 1). We performed a pairwise comparison of a protein expression level among the different cell lines for each biological replicate separately, and then obtained a list of commonly differentially expressed proteins in both analyzed passages. The cut off for the expression level change between the compared cell lines was set up as ≥1.5 fold to avoid missing potentially valuable proteins.
Table 1.
Number of proteins (protein groups) identified and quantified in five different cell lines (hESC (H9), hiPSCs (SB5-MP1 and iNC-01), fibroblasts, and PBMC) by ESI-MSe.
| Cell line | hESC–H9 | hiPSC (fibro)– SB5–MP1 |
Fibroblasts | hiPSC (CD34+cells) –iNC–01 |
PBMC | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Passage | P34 | P48 | P22 | P28 | N/A | P36 | P53 | N/A | ||
|
| ||||||||||
| # of proteins identifieda | 1817 | 1624 | 954 | 1981 | 642 | 1660 | 1189 | 1522 | 939 | 1021 |
|
| ||||||||||
| # of proteins quantifiedb | 1690 | 1439 | 848 | 1785 | 547 | 1464 | 1074 | 1345 | 775 | 869 |
|
| ||||||||||
| # of proteins selected for quantitative comparisonc | 1510 | 1185 | 736 | 1448 | 320 | 1266 | 895 | 1104 | 587 | 691 |
|
| ||||||||||
| # of proteins up–regulatedd | 62 in H9 vs. SB5–MP1; | 429 in SB5–MP1 vs. fibroblasts | 540 in iNC–01 vs. PBMC | |||||||
| 149 in H9 vs. iNC–01 | ||||||||||
|
| ||||||||||
| # of proteins down–regulatedd | 50 in H9 vs. SB5–MP1; | 89 in SB5–MP1 vs. fibroblasts | 242 in iNC–01 vs. PBMC | |||||||
| 134 in H9 vs. iNC–01 | ||||||||||
Number of proteins confidently identified in at least two out of three technical replicates (sample injections).
Number of proteins depicted in the first row and identified by at least three peptides that are required for quantification.
Number of proteins depicted in the second row with CV ≤ 30% between at least two out of three technical replicates.
Proteins differentially expressed in both biological replicates (overlapped between both passages); N/A – not applicable; number of proteins used for quantitative comparison is shown in red.
Comparative proteomics of hiPSCs and their parental somatic cells
The list of proteins differentially expressed between hiPSC derived from fibroblasts (SB5-MP1) and fibroblasts is shown in Supplementary Table 1 (a–f). Eighty nine (89) proteins were commonly down-regulated and 424 proteins were commonly up-regulated in hiPSC (SB5-MP1) compared to fibroblasts in both earlier and later passages (P22 and P28) (Table 1; Supplementary Table 1 (e–f)). The list of proteins differentially expressed between hiPSC derived from CD34+ hematopoietic cells (iNC-01) and PBMC is shown in Supplementary Table 2 (a–f). Two hundred forty two (242) proteins were commonly down-regulated and 540 proteins were up-regulated in iNC-01 compared to PBMC respectively in both biological replicates (P36 and P53) (Table 1; Supplementary Table 2).
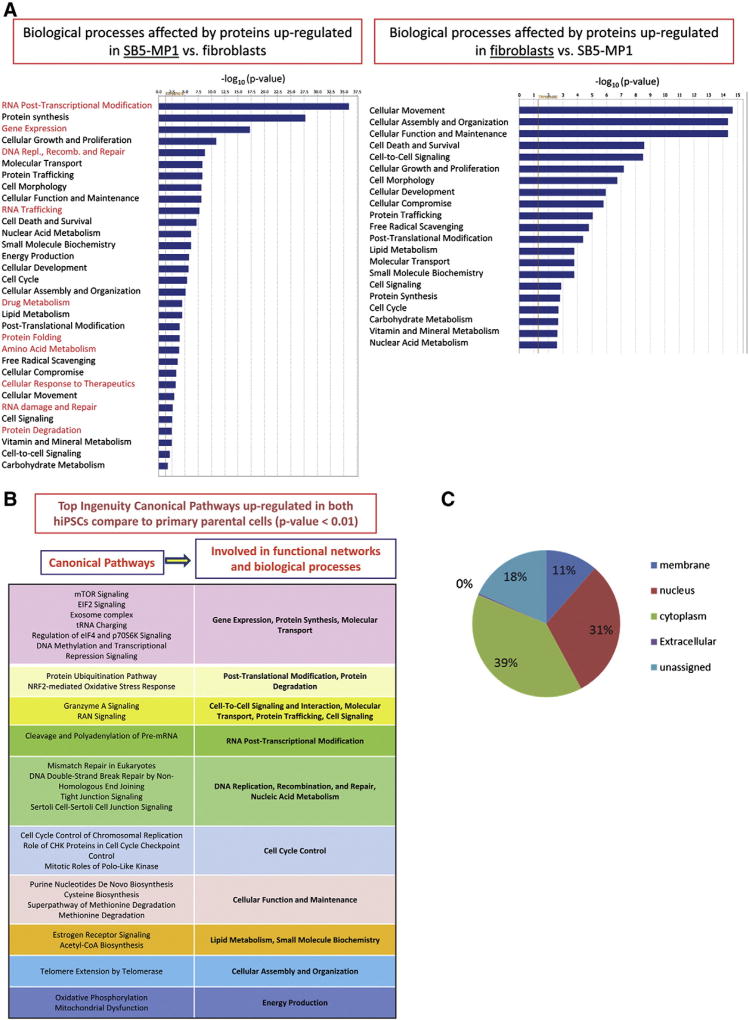
The lists of differentially expressed proteins for both hiPSC lines were analyzed using IPA. The results are summarized in Fig. 1A (SB5-MP1) and Supplementary Fig. 2 (iNC-01). Biological processes up-regulated exclusively in hiPSC or in somatic cells are shown in red. The top biological functions affected by up-regulated proteins in both hiPSCs included: RNA post-transcriptional modification, protein synthesis, gene expression, cellular growth and proliferation, DNA replication, recombination, and repair, which are characteristics of highly proliferating pluripotent stem cells. In contrast, cellular movement, cell morphology, cellular assembly and organization, cellular function and maintenance, cell death and survival, cell-to-cell signaling, etc. were top biological functions up-regulated in fibroblasts and PBMC. Overall, significant difference in the proteomic profiles was observed between somatic cells and hiPSCs.
Figure 1.
Characteristics of proteins up- or down-regulated in hiPSCs compared to somatic cells. (A): Example of Ingenuity Pathway Analysis results: biological processes affected by the proteins up- or down-regulated in SB5-MP1 vs. fibroblasts. The p-value cut-off is <0.05 (>1.3 in −log10). Biological processes affected by up-regulated proteins exclusively in hiPSC or in somatic cells are shown in red. (B): Top Ingenuity canonical pathways up-regulated in both hiPSCs compared to primary parental cells (p-value < 0.01). (C): Subcellular localization of the proteins up-regulated in both hiPSCs vs. primary cells.
Comparative proteomics of hESC (H9) and hiPSCs
The list of proteins differentially expressed between H9 and SB5-MP1 is shown in Supplementary Table 3 (a–f) and between H9 and iNC-01 — in Supplementary Table 4 (a–f). The number of proteins up- and down-regulated in SB5-MP1 compared to H9 in both analyzed passages was 50 and 62, respectively (Table 1). The number of proteins up- and down-regulated in iNC-01 compared to H9 were 134 and 149, respectively (Table 1).
IPA analysis revealed that there are only subtle differences between the functional groups affected by the differentially expressed proteins between hiPSCs and hESC (Supplementary Figs. 3 and 4). Such biological functions as cellular assembly and organization, cellular functions and maintenance, cellular growth and proliferation, protein synthesis and trafficking, cell-to-cell signaling, amino acid and lipid metabolism, cell cycle, etc. were similarly affected in hiPSC (SB5-MP1 or iNC-01) and hESC. At the same time energy production was up-regulated in both hiPSCs; protein folding and cellular response to therapeutics were up-regulated only in iNC-01 vs. H9; RNA trafficking, protein folding, protein degradation were up-regulated in H9 vs. SB5-MP1; cell signaling, RNA damage and repair, and free radical scavenging were up-regulated only in H9 vs. iNC-01.
When we analyzed the list of the main Ingenuity Canonical Pathways differentially regulated between hiPSCs and hESC (p-value < 0.0001) we found that protein kinases involved in signaling by Rho family GTPases, integrin signaling, and actin cytoskeleton signaling, were up-regulated in H9 cells; and EIF2 signaling, regulation of eIF4 and p70S6K signaling and mTOR (p-value < 0.00001) were up-regulated in both hiPSCs compared to H9 cells.
Proteomes of hiPSC lines of different somatic origins are more similar to each other than to proteome of hESC
The list of proteins differentially expressed between iNC-01 and SB5-MP1 is shown in Supplementary Table 5 (a–f). Combining proteomic analyses done at the earlier and later passages, 36 and 49 proteins were found to be up- and down-regulated in SB5-MP1 vs. iNC-01, respectively. This list was analyzed using IPA: virtually the same biological functions, mainly related to stem cell maintenance, were affected in both hiPSCs (Supplementary Fig. 5). However, post-translation modification and RNA damage and repair were up-regulated solely in SB5-MP1, and molecular transport, RNA trafficking, energy production, vitamin and mineral metabolism and carbohydrate metabolism were up-regulated only in iNC-01. We found that no particular canonical pathway was differentially regulated in any of hiPSC lines.
Quantitative comparison of proteomes of hiPSCs, hESC and somatic cells confirmed previously described and revealed novel candidates for pluripotency markers
A comparison of proteins commonly up-regulated in both hiPSCs vs. their parental somatic cells is summarized in Supplementary Table 6 (a & b). The table lists 982 proteins up-regulated in both hiPSCs in at least one biological replicate (b) and 257 proteins (26%) up-regulated in both hiPSCs in both biological replicates (a). Among these 257 proteins 221 were also up-regulated in hESC (H9) compared to primary somatic cells.
To observe global changes that occurred in hiPSC upon reprogramming we analyzed the combined list of proteins up-regulated in both hiPSCs (982 in total) using IPA and identified 25 functional networks (score 26–50) with 24–34 focus molecules contained in each network (data not shown). The top 30 Ingenuity canonical pathways involving proteins up-regulated in both hiPSCs (p-value < 0.01) are presented in Fig. 1B. Among the most statistically significant up-regulated canonical pathways with more than 15molecules involved and p-value < 0.000001 were: EIF2 signaling, regulation of eIF4 and p70S6K signaling, mTOR, the protein ubiquitination pathway, RAN signaling, and tRNA charging. In general, 6 to 90 proteins were identified to be involved in the top 30 up-regulated canonical pathways (Supplementary Table 7). The subcellular localization of the 982 proteins is displayed in Fig. 1C. 39% of these proteins are located in cytoplasm, 31% in nucleus, and only 11% are membrane proteins, of which 44% are cellular membrane proteins. Among them were EPCAM (CD326), a well-known ESC marker (Chen et al., 2011), and several other proteins, which have not been used previously for hiPSC selection: 4 F2 (CD98), TFR1 (CD71), AT1B3 (CD298), and Basigin (CD147). They can be considered as potential candidates for hiPSC surface markers.
To determine how well the results of this study compare to the published literature, we compiled a list of previously described pluripotency markers and stemness maintenance regulators and checked whether they were identified in this study and by which MS technique (ESI MS, MALDI MS or both). It has been previously shown that ESI and MALDI are complementary ionization techniques due to being biased to different types of tryptic peptides, ESI to Lys-ending, and MALDI to Arg-ending (Stapels and Barofsky, 2004; Heller et al., 2003; Bodnar et al., 2003). Supplementary Table 8 summarizes the gene expression data, the proteomics data, and the functional studies data previously obtained for hiPSC and hESC. We found that 23 previously described markers were identified exclusively by ESI-MSe (Table 2). In addition, 28 markers were identified by both MS techniques: MALDI-TOF/TOF and ESI–MSe (Table 2). In our opinion, the best case scenario is to use both ionization techniques since they complement each other. In Table 2, for example, such hiPSC marker as PODXL (TRA-1-60) was identified by ESI–MSe exclusively; SALL4 and DNMT3B were identified by both ionization techniques. In silico digestion of these proteins with peptide range of 950 to 4000 m/z (data not shown) yields peptides ending mostly by lysine (K ending peptides/R ending peptides ratios are 1.8 (PODXL), 1.4 (DNMT3B) and 1.6 (SALL4)).
Table 2.
Previously described pluripotency markers (gene names) identified in our study in three stem cell lines: hESC (H9) and hiPSCs (SB5-MP1 and iNC-01) by ESI-MSe, MALDI-TOF/TOF or both. Markers shown in red were up-regulated in both hiPSCs vs. parental somatic cells by ESI-MSe quantification.
| Identified exclusively by ESI– MSe |
Identified exclusively by MALDI– TOF/TOF |
Identified by both MS techniques |
||||
|---|---|---|---|---|---|---|
| AIBP | SMAD4 | ADCY2 | FGFR2 | SOX15 | ACTC1 | RIF1 |
| APOE | STAT3 | APC | FINC | TBX3 | ACTG2 | SALL4 |
| APOO | VSNL1 | BBS9 | GAB1 | TERT | ALPL | SERPIN B9 |
| BRIX | BCL9 | GPC6 | TGFBR1 | AXA2L | SFN | |
| CDK1 | BCL9L | GSK3A | TP53 | CALR | SMAD3 | |
| CTNNB1 | BMP2 | JAK3 | WWP2 | CXADR | TFR1 | |
| GNAS2 | BUB1 | LAD1 | DNMT3B | TPM1 | ||
| HP1–beta | CHD1 | MEF2A | DPPA4 | TUBB3 | ||
| GRB2 | CHD7 | NR2F6 | FGFR4 | |||
| KRAS | CLU | NRK | FUBP3 | |||
| MANF | CNOT3 | PDFGA | GMFB | |||
| MAPK2K2 | CNTNAP2 | PIK3C2A | GNAS | |||
| MFGM | COIA1 | PIK3C2B | GPC4 | |||
| MIF | DAG1 | PIK3R2 | HDAC2 | |||
| NEFM | DCX | PXDN | HDGF | |||
| NEST | DMD | RARA | IGF2BP1 (IMP1) | |||
| P3H1 | EHMT1 | RB1 | LAMC1 | |||
| PODXL (Tra–1–60) | EHMT2 | S1PR1 | LIN28 | |||
| RRAS2 | EP300 | SMAD2 | MAPK1 | |||
| SHC1 | FAT3 | SOX2 | PRDX4 | |||
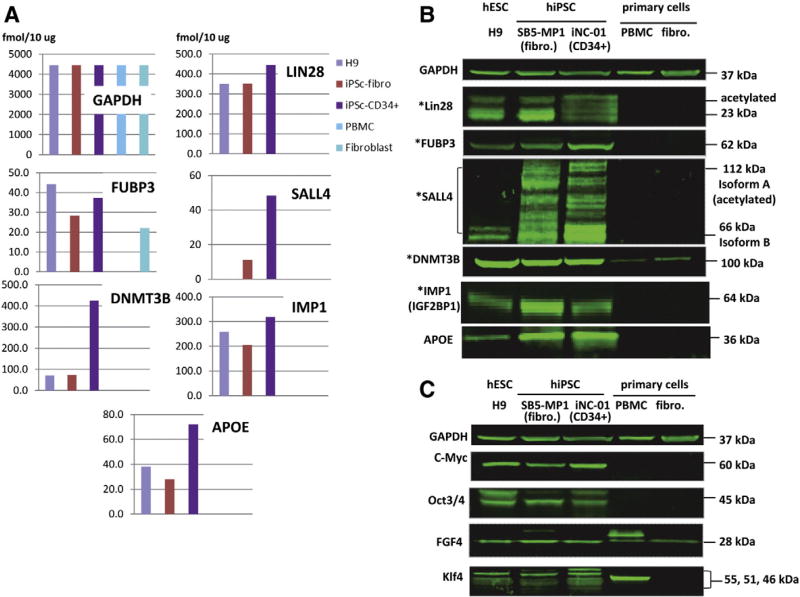
From the list of markers identified by ESI–MSe in our study, 21 proteins were found to be up-regulated in both hiPSCs compared to somatic cells (shown in red in Table 2 and in Supplementary Table 6 (a & b)). We confirmed ESI-MSe quantification (Fig. 2A) for seven of these proteins by Western blot analysis (Fig. 2B).
Figure 2.
Validation of quantification by ESI-MSe of the previously described pluripotency markers by Western blot. (A): Selected previously described markers of pluripotency quantified by ESI-MSe (TOF). Absolute quantity (fmol/10 µg) in each cell line is shown in the chart. The absolute quantity of GAPDH in each cell line estimated by ESI-MSe was found to be unchanged in all 5 cell lines. (B): Western blot analysis of three pluripotent (hESC-H9, SB5-MP1 and iNC-01) and two parental primary (PBMC and fibroblasts) cell lines. Protein quantity was normalized against GAPDH. * denotes markers also detected by MALDI-TOF/TOF. (C): Known markers of pluripotency not identified by any of MS techniques in this study but detected by Western blot.
In spite of using two complimentary MS techniques (ESI and MALDI) we could not reliably identify expression of c-Myc, FGF4, Klf4 and Oct3/4 (POU5F1) in our cells. Previous mass spectrometry-based studies also failed to identify c-Myc, FGF4, and Klf4, but identified Oct3/4 (POU5F1) (Munoz et al., 2011). Western blot analysis of 5 cell lines targeting these proteins (Fig. 2C) revealed that they all are expressed in hESC and both hiPSCs. Expression of FGF4 (also expressed in fibroblasts) and Klf4 was also detected in PBMC. The expression of Klf4 in human PBMC has been shown previously by Western blot detection (Liu et al., 2012), but the expression of FGF4 in PBMC has not been reported. In an effort to understand why we and others could not reliably identify these four well known markers of pluripotency we analyzed their amino acid sequences from the point of view of compatibility for MS-based proteomics. Supplementary Table 9 summarizes this information for the four problematical pluripotency markers and some selected proteins reliably identified by our approach. As an example, c-Myc is a midsize protein (48.8 kDa) and contains 25 Lys and 26 Arg. However, these amino acids are not evenly distributed along the c-Myc sequence. Taking into account a peptide mass range of 750–3000 Da (optimum mass range for MS identification) and assuming complete tryptic digestion of c-Myc, we could generate a maximum of 13 peptides covering 37% of the protein sequence. Further, a number of Lys residues (7) are acetylated which renders them unsuitable for digestion. Thus, a trypsin digest will not generate a sufficient number of tryptic peptides with sizes optimal for MS detection, unless one chooses to use 1 tryptic peptide for identification purposes. Correspondingly, some of these four pluripotency markers were identified in our study but did not pass the filtering criteria, such as identification scores, overall probability or FDR.
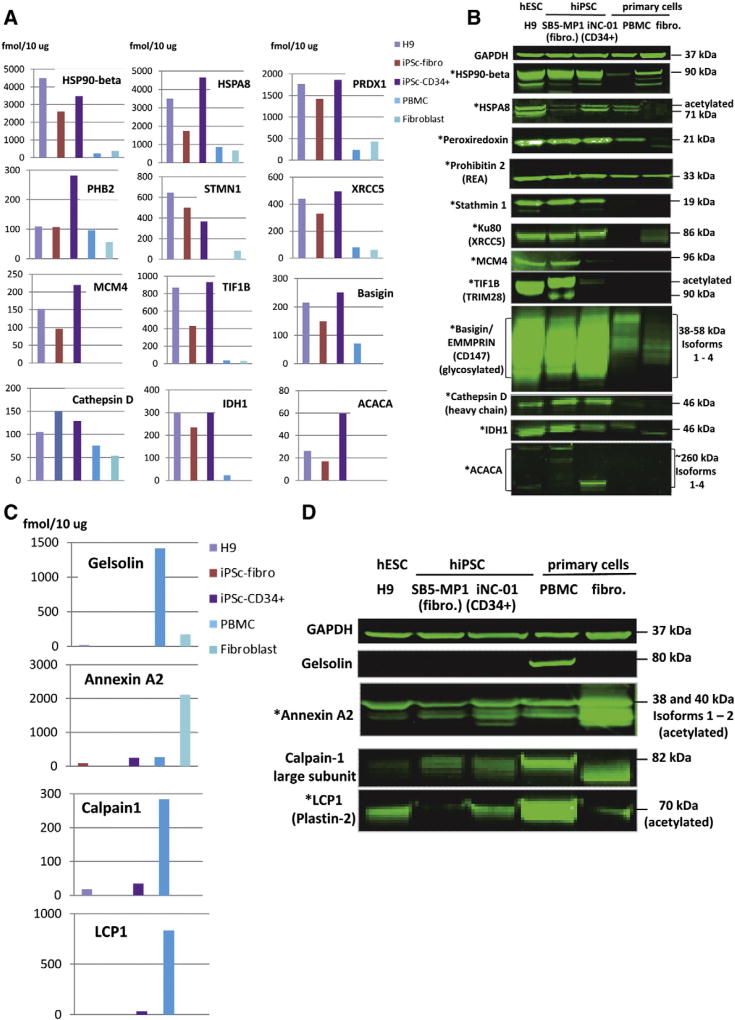
Based on the data obtained, we selected 12 proteins, including one cell surface marker (CD147), not used previously as pluripotency markers but consistently up-regulated in our experiments in early and late passage cells of both hiPSC lines, for confirmation by Western blot analysis. During the validation of ESI-MSe quantification we initially performed Western blot analysis for the identified proteins on the early and late passage cells separately and compared the results to the quantification done for each biological replicate separately (data not shown). Having similar results between cell passages, here we present the validation of quantification of these 12 selected hiPSC markers by ESI-MSe (TOF) (done on the 1st biological replicate) with Western blot (Figs. 3A and B). The list included: HSP90-beta, HSPA8, Peroxiredoxin1, Prohibitin2, Stathmin1, XRCC5 (Ku80), MCM4, TIF1-beta (TRIM28), Basigin (CD147), Cathepsin D, IDH1 and ACACA (blue bold font in Supplementary Table 6). By looking at the Supplementary Table 6 and comparing the quantity of these proteins, measured by ESI-MSe, between earlier and later passages (biological replicates 1 and 2), it becomes obvious that there is no evidence for coordinately higher expression of these proposed hiPSC markers (blue bold) or previously known pluripotency markers (red bold) in one or the other passage. In other words, we did not observe any major differences between different passages of the same hiPSC line. All selected proteins were confirmed to be up-regulated in hiPSC and hESC compared to somatic cells.
Figure 3.
Candidates to hESC/hiPSC markers found in this study by comparative quantification of five cell lines using ESI-MSe. (A): Quantification of 12 candidates to markers by ESI-MSe (TOF). Absolute quantity (fmol/10 µg) in each cell line is shown in the chart. Protein quantity was normalized against GAPDH. Absolute quantity for GAPDH measured by ESI-MSe was the same as in Fig. 2. (B): Western blot detection of the 12 candidates to markers in three pluripotent (hESC-H9, SB5-MP1 and iNC-01) and two parental primary (PBMC and fibroblasts) cell lines. Protein quantity was normalized against GAPDH. (C): Quantification of 4 candidates to “contrasting” markers by ESI-MSe (TOF). (D): Western blot detection of the candidates to “contrasting” markers. * denotes markers also detected by MALDI-TOF/TOF.
We also looked at the proteins that were dramatically down-regulated upon reprogramming and could serve as “contrasting hiPSC markers” (Supplementary Table 10 (a–d)). We selected four of them for confirmation by Western blot analysis (Figs. 3C and D, highlighted in blue bold font in Supplementary Table 10). They are Gelsolin, Calpain 1 (large subunit), LCP1 (Plastin-2) (all highly expressed in PBMC), and Annexin A2 (highly expressed in fibroblasts). For all four proteins quantification data by ESI-MSe (TOF) (Fig. 3C) correlated well with detection by Western blot (Fig. 3D).
In summary, comparative quantification between hiPSCs and somatic cells by ESI-MSe allowed confirmation of 21 previously known pluripotency markers and selection of 12 novel hESC/hiPSC protein markers and 4 somatic cell (“contrasting”) markers.
Global qualitative proteome characterization of hESC and hiPSCs of different somatic origins by MALDI MS
Since ESI and MALDI have different biases toward Arg- and Lys-ending tryptic peptides we performed global qualitative proteome characterization of these cell lines with MALDI-TOF/TOF to expand the proteome coverage of hESC and hiPSCs. All three stem cell lines were analyzed at earlier and later passages (H9: P34 and P48, SB5-MP1: P22 and P28, iNC-01: P36 and P53). The goal of this study was to identify common hiPSC/hESC markers that would be largely independent on number of passages; therefore we did not compare earlier and later passages between each other, but rather analyzed the data combined from both passages. For proteins confidently identified by MALDI MS, the Mascot score, % of the protein sequence coverage, number of total peptides and number of unique peptides used for identification are reported in Supplementary Table 11. All proteins (protein groups) identified in two different passages of each analyzed stem cell line are shown in Supplementary Table 11-a, and proteins identified in all three stem cell lines are shown in Supplementary Table 11-b; proteins that were confirmed by Western blot analysis are highlighted in red.
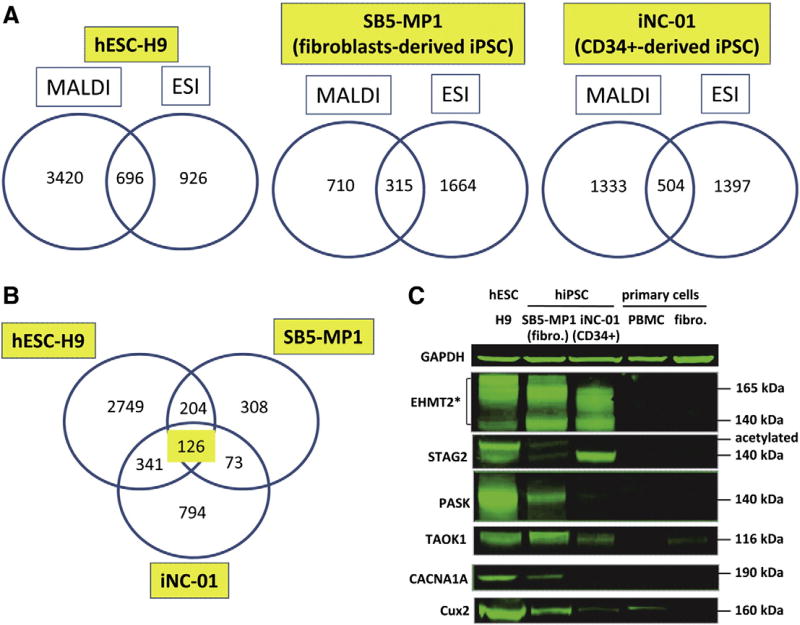
Then we compared the combined lists of proteins identified by MALDI MS and by ESI-MSe between each other for each of the cell line (Fig. 4A). We found that the number of proteins identified by each of the MS techniques exclusively (ranging from 710 to 3420 — by MALDI MS and from 926 to 1664 — by ESI-MSe) is much higher than the number of proteins identified by both MS techniques (ranging from 315 to 696) (Fig. 4A). Overall from 18 to 67% of total number of proteins in each cell line was identified uniquely either by MALDI MS or ESI-MSe, and only 12–16% of proteins were identified by both MS techniques. Similar data have been previously reported by our laboratory for human mesenchymal stemcells (Mindaye et al., 2013a). Therefore, the use of both ionization techniques allowed us to increase the proteome coverage significantly.
Figure 4.
Results of MALDI-TOF/TOF analysis of three pluripotent stem cell lines: hESC-H9, SB5-MP1 and iNC-01. (A): Number of proteins detected by MALDI-TOF/TOF exclusively, by ESI-MSe exclusively or by both techniques in each cell line in at least one out of two biological experiments. (B): Number of common proteins between three cell lines identified in at least one out of two biological experiments exclusively by MALDI-TOF/TOF (not detected by ESI-MSe). (C): One previously described (*) and 5 novel markers of hESC/hiPSC detected by MALDI-TOF/TOF exclusively were validated by Western blot analysis in three pluripotent and two parental primary (PBMC and fibroblasts) cell lines. Protein quantity was normalized against GAPDH.
In fact, 46 previously described pluripotency markers were identified exclusively by MALDI MS (Table 2, Supplementary Table 8). These include widely used hiPSC/hESC markers: BMP2, BUB1, FGFR2, Sox2, TERT, and TGFBR1. 126 proteins in total were commonly identified in all three pluripotent stem cell lines exclusively by MALDI MS (Fig. 4B; Supplementary Table 10-b). Several proteins, with functions arguably related to the establishment of the pluripotency or stemness maintenance, were confirmed by Western blot. Six proteins were confirmed to be up-regulated in hESC and hiPSCs compared to primary cells (Fig. 4C). These include: previously described EHMT2 (Sridharan et al., 2013) and 5 novel markers: TAOK1, CACNA1A, CUX2, STAG2, and PASK.
Protein markers panel developed using two MS techniques was further confirmed in eight (8) additional hiPSC lines and one additional hiPSC passage
Based on the results of the global quantitative proteome comparison of the five cell lines by mass spectrometry followed by the confirmation using Western blot we selected 20 candidates for the protein markers of hESC/hiPSC, as well as 2 “contrasting” (somatic cell)markers and combined them into a panel. Prior to further validation of the panel we looked at the expression of these 22 proteins in normal human tissues and cancer tissues. The data were obtained from “The Human Protein Atlas” (www.proteinatlas.org) and summarized in Supplementary Table 12: tissue specificity in 76 to 83 different human cell types, cancer tissue staining summary in 20 types of cancer, the main subcellular localization of each protein and summary of expression are presented. Most of the selected proteins are expressed in the majority of the normal tissues, however, some of the proteins; specifically, IDH1, HSPA8, GSN, EHMT2, CACNA1A and APOE are expressed only in certain types of tissue (Supplementary Table 12). As a matter of comparison, a well-known transcription factor Oct3/4 (POU5F1), which is essential for embryonic stem cell pluripotency, shows low to moderate expressions in 81 out of 81 analyzed cell types, with a strong immunoreactivity in Glial cells, heart and skeletal muscles, and low to moderate staining in 94% of the cancers (www.proteinatlas.org).
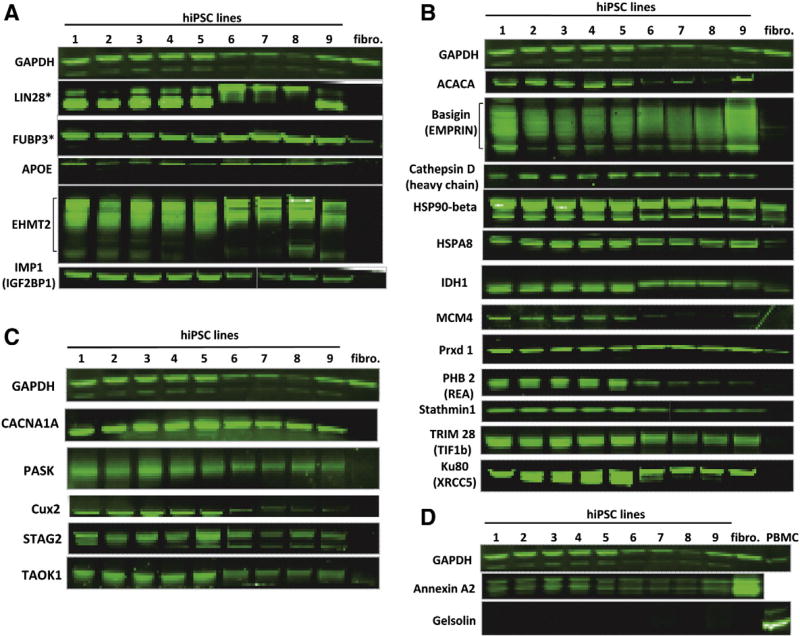
Finally, we tested the proposed protein marker panel in 9 hiPSC lines of different somatic origins, derived by different reprogramming methods in different laboratories. They included three CD34+ cell-derived hiPSC lines: iNC-06s-2E-P16 (passage 16) reprogrammed using non-integrating Sendai virus, iNC-01 at an even earlier passage — P31 and iM6-1-5-P18 both reprogrammed using STEMCCA-loxP lentivirus, which was subsequently excised; four adult fibroblasts-derived hiPSC lines iM6-3-2-P19, NC1-P38, NC8-P8, and 80-4-P20, all reprogrammed using STEMCCA-loxP lentivirus; one human umbilical vein endothelial cell (HUVEC)-derived hiPSC line NC3-P16 also reprogrammed using STEMCCA-loxP lentivirus; and one fetal fibroblast-derived hiPSC line iPS(IMR90)-1-P30, reprogrammed using four non-excisable lentiviruses (Yu et al., 2007). First we checked the expression of 5 previously described hESC/hiPSC markers in these hiPSC lines (Fig. 5A), including widely used Lin28 and FUBP3, as well as the markers that have been previously described in a literature and confirmed by MS analysis and Western blot in our study (APOE, EHMT2, and IMP1). All markers were confirmed to be up-regulated in all 9 hiPSC lines compared to fibroblasts. Then we evaluated our newly developed panel of the markers selected based on the proteomic analysis by ESI-MSe (Fig. 5B) and MALDI-TOF/TOF (Fig. 5C). All 17 selected hiPSC markers were confirmed to be highly expressed in all 9 hiPSC lines but not in fibroblasts. Finally, we confirmed that two previously selected “contrasting” markers of hiPSC (Annexin A2 and Gelsolin) were down-regulated upon reprogramming in all 9 hiPSC lines but were expressed in fibroblasts or PBMC.
Figure 5.
Qualification of the panel of 22 previously described and novel hESC/hiPSC protein markers and two opposed (somatic cell) markers in 9 hiPSC lines, primary fibroblasts and PBMC by Western blot. (A): Well-known markers (*) and proteins that were previously described in a literature as possible hESC/hiPSC markers. (B): Novel hiPSC/hESC markers found in this study by ESI-MSe quantification. (C): Novel markers detected in hESC/hiPSC in this study exclusively by MALDI-TOF/TOF. (D): “Contrasting” markers found in this study by ESI-MSe quantification. Nine hiPSC lines used in analysis (origin): 1 — iNC-06s-2E — P16 (CD34+), 2 — iNC-01 — P31 (CD34+), 3 — iM6-3-2 — P19 (adult fibroblasts), 4 — iM6-1-5 — P18 (CD34+), 5 — iPS(IMR90)-1 — P30 (fetal fibroblasts), 6 — NC1-P38 (adult fibroblasts), 7 — NC3-P16 (human umbilical vein endothelial cells (HUVECs)), 8 — NC8-P8 (adult fibroblasts), 9 — 80-4-P20 (adult fibroblasts).
In summary, all the proteins selected as candidates for the protein marker panel designed for hiPSC characterization (listed in Supplementary Table 12) were confirmed to be differentially expressed in hiPSCs (10 unique hiPSC lines were tested in total) compared to two somatic cell types (fibroblasts and PBMC) and therefore are promising candidates for the panel.
Discussion
The establishment of an appropriate quality control assays for hiPSC would be an important step in advancing their clinical translation. Our goal in this study was two-fold; on one hand we wanted to use proteomics to characterize the cellular state and biological functions activated in induced pluripotent stem cells, on the other hand to build a database of protein markers for characterization of hiPSC as a cell therapy product precursor, which could be further differentiated into any cell lineage.
When the proteomes of both hiPSCs were quantitatively compared with the proteomes of their parental primary cells ~420 to 540 up- and ~90 to 240 down-regulated proteins were found in both hiPSCs in two biological replicates. Biological functions affected by the differentially expressed proteins in both hiPSCs overlapped significantly. Coordinated up-regulation of expression of the whole set of large and small subunit ribosomal proteins, chaperons (HSP90 and HSP70) and chaperonins (CCT2, CCT7, TEBP), hnRNPs (heterogeneous nuclear ribonucleoproteins), t-RNA synthetases, and such transcriptional and splicing factors as TARDBP (TAR DNA binding protein), YBX1 (Y box binding protein), FUS (RNA-binding protein FUS) was observed in both hiPSCs compared to somatic cells.
Among the top 30 Ingenuity Canonical Pathways upregulated in both hiPSCs (p-value < 0.01) the following were found to be consistent with the data in two recent proteomics studies performed in hiPSC: EIF2 signaling, regulation of eIF4 and p70S6K signaling, protein ubiquitination, purine nucleotides de novo biosynthesis, NRF2-mediated oxidative stress response (Folmes et al., 2013); mismatch repair in eukaryotes, DNA double-strand break repair by non-homologous end joining, and cell cycle control of chromosomal replication (Sudhir et al., 2013). In line with the above studies, we have revealed up-regulation of the protein complexes involved in: RNA splicing and spliceosome formation (snRPs, hnRNP70, CDC5L); cell cycle control of chromosomal replication (MCM complex); mismatch repair and DNA double-stranded break repair by homologous recombination (MRE11A, BASC); estrogen and glucocorticoid receptor signaling (POLRs); mRNA surveillance (EXOSC).
In contrast to previously published data (Folmes et al., 2013; Menendez et al., 2011) we found that protein complexes involved in oxidative phosphorylation (OXPHOS), such as ATP synthase, NADH dehydrogenase (Complex I), Cytochrome b-c1, Cytochrome C, Cytochrome b5, H+-transporting two-sector ATPase, as well as mTOR signaling (54 molecules involved in total) were up-regulated in both hiPSCs (Supplementary Table 7). At the same time, among the markers of the transition to glycolytic metabolism we found only four up-regulated proteins: ENO2, TKT, ALDOB and PGAM1. Therefore, observations made for our hiPSC lines did not agree with previously described reprogramming-associated induction of glycolysis and down-regulation of mitochondrial reserve capacity and ATP turnover (Folmes et al., 2013; Menendez et al., 2011). This could be due to several factors, including differences between data obtained on the early (right after reprogramming) (Folmes et al., 2013; Menendez et al., 2011) and late passages of hiPSC, as well as between hiPSCs of different somatic origins. Importantly, it has been recently demonstrated that although the energy production of hiPSC favors glycolysis over OXPHOS, mitochondria in hiPSC still possess functional respiratory complexes (Zhang et al., 2011). The decoupling of glycolysis from OXPHOS was suggested to be regulated by several factors, including mitochondrial uncoupling protein 2 (UCP2) (Zhang et al., 2011). In addition, there are studies confirming that mitochondrial dynamics and maintenance of proper mitochondrial network integrity are crucial for the maintenance of pluripotency (Xu et al., 2013). Nevertheless, pivotal lipogenic enzymes acetyl-CoA carboxylase (ACACA) and fatty acid synthase (FASN) (involved in lipogenic switch) were up-regulated in our study in both hiPSCs, in line with a recent publication (Vazquez-Martin et al., 2013).
Variations on the level of genomic DNA between different pluripotent stem cells, as well as transcriptional and epigenetic profile differences can contribute to their pluripotency, stability and differentiation potential (Bock et al., 2011; Sugawara et al., 2012; Ma et al., 2014). Recent genome-wide analysis of genetically matched sets of hiPSC, hESC, and somatic cell nuclear transfer (NT) ESC has revealed that both NT-ESC and hiPSC contained a number of de novo copy variations; exome sequencing has demonstrated that hiPSC carry, on average, six non-synonymous point mutations per line (Ma et al., 2014). In addition, hiPSC retained residual DNA methylation patterns typical of parental cells (Ma et al., 2014). On the other hand, proteomic studies have shown that hiPSC proteome is almost indistinguishable from that of hESC (Munoz et al., 2011; Phanstiel et al., 2011; Kim et al., 2012). From about 2500 proteins confidently quantified in two different studies only 58 and 293 proteins were differentially expressed between hiPSC and hESC in 2 fold or less (Munoz et al., 2011; Phanstiel et al., 2011). Moreover, when the data sets from these two studies were compared, only three proteins were found to be consistently up-regulated in hESC vs. hiPSC: CRABP1, AK3 and SLC2A1 (Benevento and Munoz, 2012). Therefore, minor genomic variations or epigenetic profiles difference between different hiPSC/hESC lines may not be observed in the global proteome comparison.
In our study, quantitative comparison of each hiPSC with hESC revealed only ~50–150 proteins up- or down-regulated in both hiPSCs compared to H9 cells. CRABP1 and AK3 were also down-regulated in hiPSCs vs hESC (Supplementary Tables 3 and 4). Different proteins within the same functional network were either up- or down-regulated in hiPSCs compared to hESC, and no coordinated changes were found within any network. However, it is of interest that energy production was up-regulated in both hiPSCs vs. hESC. There were a number of canonical pathways (p-value < 0.00001) up-regulated in both hiPSCs in comparison to H9 cells, such as EIF2 signaling, regulation of eIF4 and p70S6K signaling and mTOR.
A smaller number of differentially expressed proteins between hiPSCs of different somatic origins, and less significant p-value associated with differentially regulated biological processes between them, than between hiPSCs and hESC, were observed. Besides, no statistically significant differentially regulated canonical pathways between hiPSCs were found. Based on that, we conclude that analyzed hiPSC lines are more similar to each other than to hESC. Importantly, analogous conclusion was made previously based on genome-wide expression data in mouse and human iPSC (Chin et al., 2009).
48 previously known pluripotency markers were identified by ESI-MSe in hiPSCs, from which 21 proteins were found to be up-regulated (≥1.5-fold) in both hiPSCs compared to somatic cells, and 7 of these proteins were confirmed by Western blot analysis in this study. Quantification of 12 selected novel potential pluripotency markers candidates (including one cell surface marker (Basigin, CD147)) was confirmed by Western blot analysis. Some of these proteins have been previously studied in a context of the stemness maintenance and regulation of the pluripotency. Over-expression in hESC or hiPSC of APOE, IGF2BP1 (Ghosh et al., 2010; Sarkar et al., 2012), HSPA8 (Son et al., 2005), isocitrate dehydrogenase 1 (IDH1) and Peroxiredoxin-1 (Prxd 1) (Roche et al., 2013) was demonstrated previously by gene expression or proteomic analysis. Heat shock protein 90 kDa beta (HSP90B1) has been shown to regulate pluripotency in mouse ESC and potentially regulate the folding of Oct4 and Nanog as the client proteins (Bradley et al., 2012). ACACA, has been found to be up-regulated in mouse iPSC (Vazquez-Martin et al., 2013); and tripartite motif containing 28, TRIM28 (TIF1B), has been shown to regulate the transcriptional dynamics and retroviral silencing in hESC (Wolf and Goff, 2007; Seki et al., 2010; Rowe et al., 2013).
It is well established that two different ionization techniques, MALDI and ESI, have different biases toward Arg- and Lys-ending tryptic peptides and thus complement each other (Stapels and Barofsky, 2004; Heller et al., 2003; Bodnar et al., 2003). Our MALDI MS/MS analysis revealed 46 additional previously known pluripotency markers in hiPSCs, hESC or both. From ~120 proteins commonly expressed in all three pluripotent cell lines identified exclusively by MALDI MS/MS we selected six proteins that were confirmed to be up-regulated in hESC and hiPSCs vs. primary cells by Western blot. They are: serine/threonine-protein kinase TAOK1, voltage-dependent P/Q-type calcium channel subunit alpha-1A (CACNA1A), homeobox protein cut-like 2 (CUX2), Cohesin subunit SA-2 (STAG2), PAS domain-containing serine/threonine-protein kinase (PASK) and EHMT2 (G9a methyltransferase). EHMT2 was previously described to be involved in regulation of the global histone methylation levels in hiPSC (Folmes et al., 2013). TAOK1 has been shown to be involved in p38/MAPK14 stress-activated MAPK cascade, DNA damage response and regulation of cytoskeleton stability (Raman et al., 2007; Westhorpe et al., 2010). It has also been reported to be highly expressed in the testis, and to a lower extent in the brain, placenta, colon and skeletal muscle (Yustein et al., 2003). PASK is another serine/threonine-protein kinase involved in energy homeostasis and protein translation; it is ubiquitously expressed, with slightly higher expression in the brain, prostate and testis (Schlafli et al., 2009). Interestingly, CACNA1A has been previously shown to be brain specific; mainly found in the cerebellum, cerebral cortex, thalamus and hypothalamus (Oguro-Okano et al., 1992). The functions of CUX2 have just recently been described as a likely transcription factor, which binds to DNA in a sequence-specific manner and is involved in neural specification during embryogenesis (Cubelos et al., 2010; Bachy et al., 2011); it may also be involved in regulation of the sex-specific gene expression in female liver (Conforto et al., 2012). STAG2 is a component of cohesin complex, which is required for the cohesion of sister chromatids after DNA replication (Prieto et al., 2002).
Based on our results, we propose a panel of potential hiPSC protein markers from the above-mentioned 22 proteins (Supplementary Table 12). We intentionally used three previously characterized stem cell lines (hiPSC: iNC-01, SB5-MP1 and hESC-H9) in our proteomic study. The genomic methylation profile of the two cell lines (hiPSC-iNC-01 and hESC-H9) has been previously studied and found to be indistinguishable (Merling et al., 2013). iNC-01, used to obtain functional neutrophils (Sweeney et al., 2014) and SB5-MP1, was successfully used in differentiation into motor neurons (Grunseich et al., 2014). Therefore, the selection of hiPSC markers was done using the cell lines, which underwent considerable functional assays.
This protein marker panel was evaluated in nine additional hiPSC lines or passages derived from different somatic cells and by different reprogramming techniques. All the proteins selected as the candidates for the protein marker panel for hiPSC characterization were confirmed to be differentially expressed in all analyzed hiPSCs compared to somatic cells. In addition to that, our recent proteomic analysis of hiPSC and embroid bodies (EBs) derived from them during the course of spontaneous differentiation at 24 h and 7 days revealed that the majority of the selected candidates to hiPSC markers are dramatically down-regulated upon differentiation (in EB-24 h and EB-7 days compare to hiPSC). These data (manuscript in preparation) will serve as an additional confirmation of these candidate marker proteins to be selected for hiPSC characterization panel.
Conclusions
Our study highlights the signaling pathways up-regulated in both analyzed hiPSC lines after reprogramming, some of which are consistent with previously published data, but some are contrasting to the data previously described for iPSC. In spite of their different somatic origins (adult skin fibroblasts and CD34+ hematopoietic stem cells), the biological functions affected by the up-regulated proteins in both hiPSCs overlapped significantly.
Based on the proteomic data obtained from two hiPSC lines of different somatic origins at different passage levels we selected a panel of previously described and novel hESC/ hiPSC protein markers. The possible extension, development and further evaluation of this panel should facilitate the improvement in hiPSC quality control assays and potential clinical application of hiPSC.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.scr.2015.01.009.
Supplementary Material
Acknowledgments
We thank Dr. Manfred Boehm (NIH, NHLBI) for providing several hiPSC lines for Western blot analysis. We also thank Dr. Deborah Hursh and Dr. Steve Bauer (FDA, CBER, OCTGT) for their critical reading of the manuscript.
Footnotes
Author contributions: NSP — conception and design of the study, acquisition, analysis and interpretation of data, drafting the article and final approval of the version to be published; MGK — design of the study, acquisition, analysis and interpretation of data, drafting the article and final approval of the version to be published; CG — contribution of materials and cells, revising the article and final approval of the version to be published; CS — contribution of materials and cells, revising the article and final approval of the version to be published; HM — conception and design of the study, revising the article and final approval of the version to be published; MAA — conception and design of the study, analysis and interpretation of data, drafting and revising the article and final approval of the version to be published.
There is no conflict of interest for any author to report.
References
- Bachy I, Franck MC, Li L, Abdo H, Pattyn A, Ernfors P. The transcription factor Cux2 marks development of an A-delta sublineage of TrkA sensory neurons. Dev. Biol. 2011;360:77–86. doi: 10.1016/j.ydbio.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Bar-Nur O, Russ HA, Efrat S, Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell. 2011;9:17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Benevento M, Munoz J. Role of mass spectrometry-based proteomics in the study of cellular reprogramming and induced pluripotent stem cells. Expert Rev. Proteomics. 2012;9:379–399. doi: 10.1586/epr.12.30. [DOI] [PubMed] [Google Scholar]
- Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, Gnirke A, Eggan K, Meissner A. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar WM, Blackburn RK, Krise JM, Moseley MA. Exploiting the complementary nature of LC/MALDI/MS/MS and LC/ESI/MS/MS for increased proteome coverage. J. Am. Soc. Mass Spectrom. 2003;14(9):971–979. doi: 10.1016/S1044-0305(03)00209-5. [DOI] [PubMed] [Google Scholar]
- Bradley E, Bieberich E, Mivechi NF, Tangpisuthipongsa D, Wang G. Regulation of embryonic stem cell pluripotency by heat shock protein 90. Stem Cells. 2012;30:1624–1633. doi: 10.1002/stem.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chuang CY, Lee WC, Huang HP, Wu HC, Ho HN, Chen YJ, Kuo HC. Surface marker epithelial cell adhesion molecule and E-cadherin facilitate the identification and selection of induced pluripotent stem cells. Stem Cell Rev. 2011;7:722–735. doi: 10.1007/s12015-011-9233-y. [DOI] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, Khvorostov I, Ott V, Grunstein M, Lavon N, Benvenisty N, Croce CM, Clark AT, Baxter T, Pyle AD, Teitell MA, Pelegrini M, Plath K, Lowry WE. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforto TL, Zhang Y, Sherman J, Waxman DJ. Impact of CUX2 on the female mouse liver transcriptome: activation of female-biased genes and repression of male-biased genes. Mol. Cell. Biol. 2012;32:4611–4627. doi: 10.1128/MCB.00886-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubelos B, Sebastian-Serrano A, Beccari L, Calcagnotto ME, Cisneros E, Kim S, Dopazo A, Alvarez-Dolado M, Redondo JM, Bovolenta P, Walsh CA, Nieto M. Cux1 and Cux2 regulate dendritic branching, spine morphology, and synapses of the upper layer neurons of the cortex. Neuron. 2010;66:523–535. doi: 10.1016/j.neuron.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes CD, Arrell DK, Zlatkovic-Lindor J, Martinez-Fernandez A, Perez-Terzic C, Nelson TJ, Terzic A. Metabolome and metaboproteome remodeling in nuclear reprogramming. Cell Cycle. 2013;12:2355–2365. doi: 10.4161/cc.25509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh Z, Wilson KD, Wu Y, Hu S, Quertermous T, Wu JC. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS One. 2010;5:e8975. doi: 10.1371/journal.pone.0008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunseich C, Zukosky K, Kats IR, Ghosh L, Harmison GG, Bott LC, Rinaldi C, Chen KL, Chen G, Boehm M, Fischbeck KH. Stem cell-derived motor neurons from spinal and bulbar muscular atrophy patients. Neurobiol. Dis. 2014;70:12–20. doi: 10.1016/j.nbd.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller M, Mattou H, Menzel C, Yao X. Trypsin catalyzed16O-to-18O exchange for comparative proteomics: tandem mass spectrometry comparison using MALDI-TOF, ESI-QTOF, and ESI-ion trap mass spectrometers. J. Am. Soc. Mass Spectrom. 2003;14(7):704–718. doi: 10.1016/S1044-0305(03)00207-1. [DOI] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Kim MJ, Jung H, Kim WK, Kwon SO, Son MJ, Jang IS, Choi JS, Park SG, Park BC, Han YM, Lee SC, Cho YS, Bae KH. Comparative proteomic analysis of human somatic cells, induced pluripotent stem cells, and embryonic stem cells. Stem Cells Dev. 2012;21:1272–1286. doi: 10.1089/scd.2011.0243. [DOI] [PubMed] [Google Scholar]
- Lee SB, Seo D, Choi D, Park KY, Holczbauer A, Marquardt JU, Conner EA, Factor VM, Thorgeirsson SS. Contribution of hepatic lineage stage-specific donor memory to the differential potential of induced mouse pluripotent stem cells. Stem Cells. 2012;30:997–1007. doi: 10.1002/stem.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yang T, Liu Y, Zhang H, Wang K, Liu M, Chen G, Xiao X. Kruppel-like factor 4 inhibits the expression of interleukin-1 beta in lipopolysaccharide-induced RAW264.7 macrophages. FEBS Lett. 2012;586:834–840. doi: 10.1016/j.febslet.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Loh YH, Agarwal S, Park IH, Urbach A, Huo H, Heffner GC, Kim K, Miller JD, Ng K, Daley GQ. Generation of induced pluripotent stem cells from human blood. Blood. 2009;113:5476–5479. doi: 10.1182/blood-2009-02-204800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Hartung O, Li H, Guo C, Sahalie JM, Manos PD, Urbach A, Heffner GC, Grskovic M, Vigneault F, Lensch MW, Park IH, Agarwal S, Church GM, Collins JJ, Irion S, Daley GQ. Reprogramming of T cells from human peripheral blood. Cell Stem Cell. 2010;7:15–19. doi: 10.1016/j.stem.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Morey R, O'Neil RC, He Y, Daughtry B, Schultz MD, Hariharan M, Nery JR, Castanon R, Sabatini K, Thiagarajan RD, Tachibana M, Kang E, Tippner-Hedges R, Ahmed R, Gutierrez NM, Van Dyken C, Polat A, Sugawara A, Sparman M, Gokhale S, Amato P, Wolf DP, Ecker JR, Laurent LC, Mitalipov S. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature. 2014;511:177–183. doi: 10.1038/nature13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Yeo GW, Kainohana O, Marsala M, Gage FH, Muotri AR. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS One. 2009;4:e7076. doi: 10.1371/journal.pone.0007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez JA, Vellon L, Oliveras-Ferraros C, Cufi S, Vazquez-Martin A. mTOR-regulated senescence and autophagy during reprogramming of somatic cells to pluripotency: a roadmap from energy metabolism to stem cell renewal and aging. Cell Cycle. 2011;10:3658–3677. doi: 10.4161/cc.10.21.18128. [DOI] [PubMed] [Google Scholar]
- Merling RK, Sweeney CL, Choi U, De Ravin SS, Myers TG, Otaizo-Carrasquero F, Pan J, Linton G, Chen L, Koontz S, Theobald NL, Malech HL. Transgene-free iPSCs generated from small volume peripheral blood non-mobilized CD34+ cells. Blood. 2013;121:e98–e107. doi: 10.1182/blood-2012-03-420273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindaye ST, Ra M, Lo Surdo J, Bauer SR, Alterman MA. Improved proteomic profiling of the cell surface of culture-expanded human bone marrow multipotent stromal cells. J. Proteomics. 2013a;78:1–14. doi: 10.1016/j.jprot.2012.10.028. [DOI] [PubMed] [Google Scholar]
- Mindaye ST, Ra M, Lo Surdo JL, Bauer SR, Alterman MA. Global proteomic signature of undifferentiated human bone marrow stromal cells: evidence for donor-to-donor proteome heterogeneity. Stem Cell Res. 2013b;11:793–805. doi: 10.1016/j.scr.2013.05.006. [DOI] [PubMed] [Google Scholar]
- Montserrat N, Ramirez-Bajo MJ, Xia Y, Sancho-Martinez I, Moya-Rull D, Miquel-Serra L, Yang S, Nivet E, Cortina C, Gonzalez F, Izpisua Belmonte JC, Campistol JM. Generation of induced pluripotent stem cells from human renal proximal tubular cells with only two transcription factors, OCT4 and SOX2. J. Biol. Chem. 2012;287:24131–24138. doi: 10.1074/jbc.M112.350413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller G, Tarasov KV, Gundry RL, Boheler KR. Human ESC/iPSC-based “omics” and bioinformatics for translational research. Drug Discov. Today Dis. Model. 2012;9:e161–e170. doi: 10.1016/j.ddmod.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz J, Low TY, Kok YJ, Chin A, Frese CK, Ding V, Choo A, Heck AJ. The quantitative proteomes of human-induced pluripotent stem cells and embryonic stem cells. Mol. Syst. Biol. 2011;7:550. doi: 10.1038/msb.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguro-Okano M, Griesmann GE, Wieben ED, Slaymaker SJ, Snutch TP, Lennon VA. Molecular diversity of neuronaltype calcium channels identified in small cell lung carcinoma. Mayo Clin. Proc. 1992;67:1150–1159. doi: 10.1016/s0025-6196(12)61144-6. [DOI] [PubMed] [Google Scholar]
- Phanstiel DH, Brumbaugh J, Wenger CD, Tian S, Probasco MD, Bailey DJ, Swaney DL, Tervo MA, Bolin JM, Ruotti V, Stewart R, Thomson JA, Coon JJ. Proteomic and phosphoproteomic comparison of human ES and iPS cells. Nat. Methods. 2011;8:821–827. doi: 10.1038/nmeth.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto I, Pezzi N, Buesa JM, Kremer L, Barthelemy I, Carreiro C, Roncal F, Martinez A, Gomez L, Fernandez R, Martinez AC, Barbero JL. STAG2 and Rad21 mammalian mitotic cohesins are implicated in meiosis. EMBO Rep. 2002;3:543–550. doi: 10.1093/embo-reports/kvf108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman M, Earnest S, Zhang K, Zhao Y, Cobb MH. TAO kinases mediate activation of p38 in response to DNA damage. EMBO J. 2007;26:2005–2014. doi: 10.1038/sj.emboj.7601668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche S, D'Ippolito G, Gomez LA, Bouckenooghe T, Lehmann S, Montero-Menei CN, Schiller PC. Comparative analysis of protein expression of three stem cell populations: models of cytokine delivery system in vivo. Int. J. Pharm. 2013;440:72–82. doi: 10.1016/j.ijpharm.2011.12.041. [DOI] [PubMed] [Google Scholar]
- Rowe HM, Kapopoulou A, Corsinotti A, Fasching L, Macfarlan TS, Tarabay Y, Viville S, Jakobsson J, Pfaff SL, Trono D. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res. 2013;23:452–461. doi: 10.1101/gr.147678.112. (1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar P, Collier TS, Randall SM, Muddiman DC, Rao BM. The subcellular proteome of undifferentiated human embryonic stem cells. Proteomics. 2012;12:421–430. doi: 10.1002/pmic.201100507. [DOI] [PubMed] [Google Scholar]
- Schlafli P, Borter E, Spielmann P, Wenger RH. The PASdomain kinase PASKIN: a new sensor in energy homeostasis. Cell. Mol. Life Sci. 2009;66:876–883. doi: 10.1007/s00018-009-8699-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Kurisaki A, Watanabe-Susaki K, Nakajima Y, Nakanishi M, Arai Y, Shiota K, Sugino H, Asashima M. TIF1beta regulates the pluripotency of embryonic stem cells in a phosphorylation-dependent manner. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10926–10931. doi: 10.1073/pnas.0907601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JC, Denny R, Dorschel CA, Gorenstein M, Kass IJ, Li GZ, McKenna T, Nold MJ, Richardson K, Young P, Geromanos S. Quantitative proteomic analysis by accurate mass retention time pairs. Anal. Chem. 2005;77:2187–2200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son YS, Park JH, Kang YK, Park JS, Choi HS, Lim JY, Lee JE, Lee JB, Ko MS, Kim YS, Ko JH, Yoon HS, Lee KW, Seong RH, Moon SY, Ryu CJ, Hong HJ. Heat shock 70-kDa protein 8 isoform 1 is expressed on the surface of human embryonic stem cells and downregulated upon differentiation. Stem Cells. 2005;23:1502–1513. doi: 10.1634/stemcells.2004-0307. [DOI] [PubMed] [Google Scholar]
- Sridharan R, Gonzales-Cope M, Chronis C, Bonora G, McKee R, Huang C, Patel S, Lopez D, Mishra N, Pellegrini M, Carey M, Garcia BA, Plath K. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1gamma in reprogramming to pluripotency. Nat. Cell Biol. 2013;15:872–882. doi: 10.1038/ncb2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staerk J, Dawlaty MM, Gao Q, Maetzel D, Hanna J, Sommer CA, Mostoslavsky G, Jaenisch R. Reprogramming of human peripheral blood cells to induced pluripotent stem cells. Cell Stem Cell. 2010;7:20–24. doi: 10.1016/j.stem.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapels MD, Barofsky DF. Complementary use of MALDI and ESI for the HPLC–MS/MS analysis of DNA-binding proteins. Anal. Chem. 2004;76(18):5423–5430. doi: 10.1021/ac030427z. [DOI] [PubMed] [Google Scholar]
- Sudhir PR, Kumari MP, Hsu WT, Massiot J, Chen CH, Kuo HC. Quantitative proteomics of protein complexes and their implications for cell reprograming and pluripotency. J. Proteome Res. 2013;12:5878–5890. doi: 10.1021/pr4008877. [DOI] [PubMed] [Google Scholar]
- Sugawara T, Nishino K, Umezawa A, Akutsu H. Investigating cellular identity and manipulating cell fate using induced pluripotent stem cells. Stem Cell Res. Ther. 2012;3:8. doi: 10.1186/scrt99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney CL, Merling RK, Choi U, Priel DL, Kuhns DB, Wang H, Malech HL. Generation of functionally mature neutrophils from induced pluripotent stem cells. Methods Mol. Biol. 2014;1124:189–206. doi: 10.1007/978-1-62703-845-4_12. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Vazquez-Martin A, Corominas-Faja B, Cufi S, Vellon L, Oliveras-Ferraros C, Menendez OJ, Joven J, Lupu R, Menendez JA. The mitochondrial H(+)-ATP synthase and the lipogenic switch: new core components of metabolic reprogramming in induced pluripotent stem (iPS) cells. Cell Cycle. 2013;12:207–218. doi: 10.4161/cc.23352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhorpe FG, Diez MA, Gurden MD, Tighe A, Taylor SS. Re-evaluating the role of Tao1 in the spindle checkpoint. Chromosoma. 2010;119:371–379. doi: 10.1007/s00412-010-0261-1. [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP. TRIM28 mediates primer binding sitetargeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- Xu X, Duan S, Yi F, Ocampo A, Liu GH, Izpisua Belmonte JC. Mitochondrial regulation in pluripotent stem cells. Cell Metab. 2013;18(3):325–332. doi: 10.1016/j.cmet.2013.06.005. [DOI] [PubMed] [Google Scholar]
- Yamana R, Iwasaki M, Wakabayashi M, Nakagawa M, Yamanaka S, Ishihama Y. Rapid and deep profiling of human induced pluripotent stem cell proteome by one-shot NanoLC–MS/MS analysis with meter-scale monolithic silica columns. J. Proteome Res. 2013;12:214–221. doi: 10.1021/pr300837u. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yustein JT, Xia L, Kahlenburg JM, Robinson D, Templeton D, Kung HJ. Comparative studies of a new subfamily of human Ste20-like kinases: homodimerization, subcellular localization, and selective activation of MKK3 and p38. Oncogene. 2003;22:6129–6141. doi: 10.1038/sj.onc.1206605. [DOI] [PubMed] [Google Scholar]
- Zhang J, Khvorostov I, Hong JS, Oktay Y, Vergnes L, Nuebel E, Wahjudi PN, Setoguchi K, Wang G, Do A, Jung HJ, McCaffery JM, Kurland IJ, Reue K, Lee WN, Koehler CM, Teitell MA. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J. 2011;30:4860–4873. doi: 10.1038/emboj.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Sweeney CL, Chou BK, Choi U, Pan J, Wang H, Dowey SN, Cheng L, Malech HL. Oxidase-deficient neutrophils from X-linked chronic granulomatous disease iPS cells: functional correction by zinc finger nuclease-mediated safe harbor targeting. Blood. 2011;117:5561–5572. doi: 10.1182/blood-2010-12-328161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.