ABSTRACT
Bacillus cereus remains an important cause of infections, particularly in immunocompromised hosts. While typically associated with enteric infections, disease manifestations can be quite diverse and include skin infections, bacteremia, pneumonia, and meningitis. Whether there are any genetic correlates of bacterial strains with particular clinical manifestations remains unknown. To address this gap in understanding, we undertook whole-genome analysis of B. cereus strains isolated from patients with a range of disease manifestations, including noninvasive colonizing disease, superficial skin infections, and invasive bacteremia. Interestingly, strains involved in skin infection tended to form a distinct genetic cluster compared to isolates associated with invasive disease. Other disease manifestations, despite not being exclusively clustered, nonetheless had unique genetic features. The unique features associated with the specific types of infections ranged from traditional virulence determinants to metabolic pathways and gene regulators. These data represent the largest genetic analysis to date of pathogenic B. cereus isolates with associated clinical parameters.
KEYWORDS: Bacillus cereus, whole genome, pathogenesis, immunocompromised
INTRODUCTION
Bacillus cereus is a rod-shaped, Gram-positive, aerobic or facultative anaerobic bacterium that is frequently present in soil, sediments, dust, plants, and food production environments. B. cereus can form spores that are resistant to extreme environmental conditions, including both pasteurization and gamma radiation, making the elimination of this potential pathogen challenging. While widely known for disease potential in the context of food poisoning, there have been numerous reported cases of pneumonia, bacteremia, and meningitis caused by this pathogen, particularly in immunocompromised individuals (1–3). The pathogenicity of B. cereus is predominantly derived from the production of tissue-destructive exoenzymes, including hemolysins, phospholipases, and proteases. B. cereus causes not only food poisoning but also several fatal non-gastrointestinal-tract clinical infections, such as eye infections, progressive pneumonia, fulminant sepsis, and central nervous system infections, including meningitis and brain abscesses (4).
While it is widely appreciated that B. cereus has a variety of disease manifestations, to our knowledge, there has been little evidence examining whether specific genotypes are associated with different sequelae. Distinct genetic factors might be required for B. cereus to infect particular host niches. As such, genetic characterization of isolates with well-defined clinical manifestations may prove powerful in defining such associations. To understand the relationship between the genotype and clinical phenotype of B. cereus, we collected samples from patients with distinct disease manifestations, including bacteremia, sepsis, bloodstream hemolysis, meningoencephalitis, skin infection, diarrhea, and fecal colonization. Sequences of these isolates were compared, and differences in relative gene abundances and other relevant strain characteristics were analyzed, representing the largest sequenced cohort of B. cereus isolates coupled with clinical manifestations to date.
RESULTS
Clinical and phenotypic features of patients with B. cereus infections.
The clinical phenotypes of B. cereus infections are described in Table S2 in the supplemental material. Meningoencephalitis, bacteremia, sepsis, and clinical bloodstream hemolysis were categorized as invasive infections, while rectal colonization, skin infection, and diarrhea were categorized as noninvasive infections. Host characteristics of patients with invasive B. cereus infections were not significantly different from those of patients with noninvasive B. cereus infections (Table S3).
Genetic features of pathogenic B. cereus strains.
The genomic features of the assembled B. cereus strains are summarized in Table 1. The average coverage was ∼45-fold for the assemblies, and strains shared comparable genome sizes (∼5.3 Mb) and GC contents (35%), similar to the values reported previously (5). An increase of the genome size was observed for sample s18 (9.6 Mb), suggesting the presence of multiple plasmids, unique repeat elements, and/or various sizes of plasmids (6, 7). The level of continuity of the genome assembly was high for 12 samples, which had an N50 value of >10,000 and <1,000 contigs (Table 1). The assemblies of the remaining 12 samples are more fragmented. Since the sequencing depth was sufficient to cover the breadth of the genomes, the fragmentation of the assemblies may be attributed to the low GC content (8) and highly abundant repeat elements (9). In spite of the fragmentation of the assemblies, the predicted numbers of proteins were similar among the sequenced samples and comparable to those of the other sequenced B. cereus strains (5).
TABLE 1.
Genomic features of the 24 sequenced Bacillus cereus strains
| Sample | Assembly size (no. of bases) (>200 bp) | N50 | No. of contigs (>200 bp) | No. of proteins | No. of RNAs (no. of rRNAs/no. of tRNAs) | GC content (%) |
|---|---|---|---|---|---|---|
| s01 | 4,982,614 | 2,184 | 3,465 | 4,884 | 28 (8/20) | 0.35 |
| s02 | 4,554,479 | 1,776 | 4,171 | 4,442 | 25 (6/19) | 0.36 |
| s03 | 5,022,101 | 1,354 | 4,804 | 5,082 | 7 (3/4) | 0.36 |
| s04 | 4,891,265 | 2,192 | 2,962 | 5,346 | 24 (5/19) | 0.36 |
| s05 | 5,078,431 | 1,465 | 5,544 | 4,678 | 34 (5/29) | 0.35 |
| s06 | 5,001,224 | 2,070 | 3,725 | 4,776 | 21 (4/17) | 0.35 |
| s07 | 4,831,022 | 1,895 | 3,899 | 4,740 | 16 (3/13) | 0.36 |
| s08 | 4,666,508 | 1,390 | 4,644 | 4,468 | 25 (5/20) | 0.36 |
| s09 | 4,775,962 | 1,715 | 4,202 | 4,493 | 19 (3/16) | 0.35 |
| s10 | 4,815,983 | 1,344 | 5,090 | 4,793 | 32 (6/26) | 0.36 |
| s17 | 5,728,264 | 248,673 | 127 | 5,729 | 104 (16/88) | 0.40 |
| s18 | 9,643,523 | 20,399 | 639 | 10,012 | 56 (13/43) | 0.35 |
| s19 | 6,213,733 | 316,244 | 1,795 | 5,951 | 85 (10/75) | 0.36 |
| s20 | 5,107,447 | 137,978 | 63 | 5,264 | 57 (10/47) | 0.35 |
| s21 | 5,290,016 | 236,524 | 66 | 5,468 | 77 (11/66) | 0.36 |
| s22 | 5,775,796 | 467,974 | 109 | 5,795 | 77 (8/69) | 0.36 |
| s23 | 5,656,499 | 329,969 | 689 | 5,581 | 102 (12/90) | 0.35 |
| s24 | 5,708,383 | 179,036 | 895 | 5,742 | 88 (15/73) | 0.34 |
| s25 | 5,701,576 | 166,645 | 43 | 5,775 | 42 (4/38) | 0.35 |
| s26 | 6,999,176 | 173,263 | 2,832 | 6,724 | 92 (10/82) | 0.34 |
| s27 | 5,713,339 | 316,336 | 255 | 5,802 | 83 (10/73) | 0.35 |
| s28 | 5,809,653 | 178,813 | 119 | 5,925 | 90 (11/79) | 0.37 |
| s29 | 5,459,198 | 574,687 | 30 | 5,588 | 50 (3/47) | 0.34 |
| s30 | 5,625,682 | 144,042 | 194 | 5,813 | 95 (11/84) | 0.35 |
To further investigate the impact of the fragmentation of genome assemblies, we assessed the completeness of the genome assembly by the completenesses of 40 highly conserved prokaryote orthologous genes (COG). These 40 genes are single-copy phylogenetic marker genes and have been used for resolving evolutionary history across domains of life (10–12). Specifically, for one genome, predicted genes were classified as complete gene models when the gene length was within 2 standard deviations of the COG gene group length. Genes were classified as fragmented models when the genes were only partially present in the assembly. Genes absent from the genome were classified as missing genes (see Fig. S1 in the supplemental material). The majority of the genome assemblies had a high ratio of genes with complete status, ranging from 92% to 98%, except for samples s26 (80%), s1 (35%), and s22 (32%) (Fig. S1).
Phylogenetic reconstruction reveals two major clades.
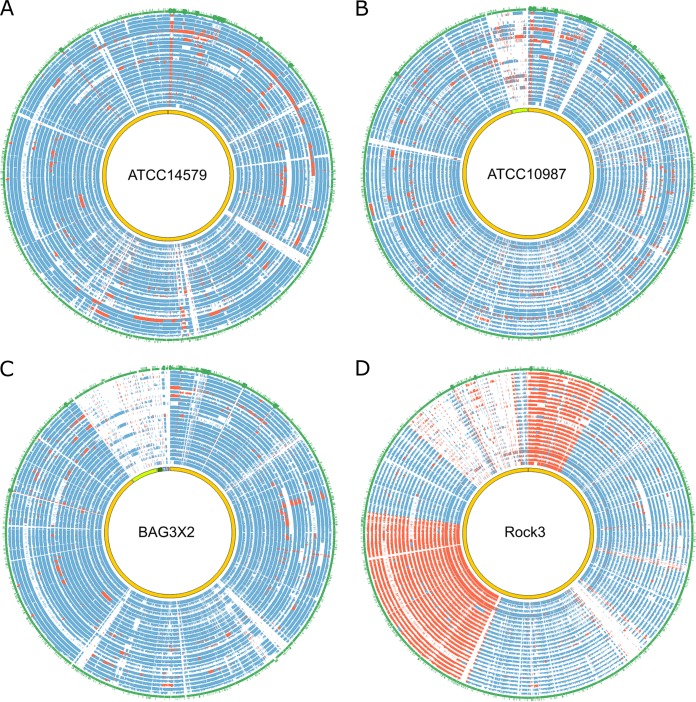
We further examined the syntenic regions present in the assemblies. The closest neighbors of the 24 strains estimated based on representative genes (13) were B. cereus BAG3X2-2 (average conservation score, 522.9), ATCC 14579 (score, 517.4), ATCC 10987 (score, 534.5) and Rock3-29 (score, 518.0). A further analysis of syntenic relationships between the samples and these close neighbors revealed a high degree of conservation in most genomic regions, particularly for ATCC 14579, ATCC 10987, and BAG3X2-2 (Fig. 1A to C). Nearly all the aligned regions displayed sequence similarities of >75%. Nonetheless, several strains contained short segments of genomic rearrangements. For example, sample s26 tended to have extended segments of genomic duplications and inversions compared to ATCC 14579 (Fig. 1A). In addition, sample s26 also contained regions sharing low similarity to the four reference strains (Fig. 1), indicating that sample s26 was evolutionarily distinct. The alignment between the clinical strains and the Rock3 strain revealed two large segments with frequent genomic inversions not present in all other comparisons (Fig. 1D).
FIG 1.
Genomic comparisons among the 24 B. cereus strains and 4 phylogenetically closely related strains. The genomes of the 24 strains are aligned against strains ATCC 14579 (A), ATCC 10987 (B), BAG3X2-2 (C), and Rock3 (D). In each plot, the innermost circles represent the genomes of the reference strains, followed by 24 circles for samples s1 to s30, displaying the sequence similarity between each sample and the reference. The height of each similarity circle reflects the degree of similarity from 0 to 100%. Colinear syntenic regions are shown in blue, while genomic regions undergoing genomic rearrangements are shown in red. Overall, the 24 samples shared a high degree of conservation with ATCC 14579, ATCC 10987, and BAG3X2-2. In contrast, frequent genomic inversions are present in comparisons to the Rock3 strain. The outermost circles depict the gene annotation for each reference strain.
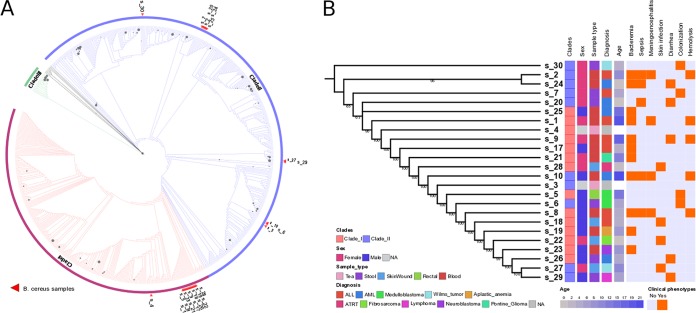
The B. cereus group has been classified into three main phylogenetic clades by multilocus sequence typing (14). The first clade (clade I) contains B. cereus and related B. anthracis and B. thuringiensis strains. The second clade (clade II) contains more distantly related B. cereus strains and B. pseudomycoides isolates. The final clade (clade III) contains the thermophilic B. cereus subspecies. In order to obtain a deeper insight into strain history, we first reconstructed phylogenetic relationships based on a collection of 673 16S RNA sequences from the 24 samples and another 649 B. cereus strains deposited in the SILVA database (15). Cytotoxic B. cereus subspecies reference genomes were used as outgroups to root the tree. In agreement with data from previous studies, three major clades were identified in the reconstructed phylogenetic tree (Fig. 2A) (14). Of the 24 samples, 13 were embedded in clade I, 12 of which were clustered tightly in a single cluster (Fig. 2A). The remaining 11 samples fell within clade II and formed three minor clusters (Fig. 2A). The supporting values for this branching pattern were generally low due to the high similarity of the 16S rRNA sequences. We further inferred their relationship by concatenated sequences of 130 single-copy genes that were conserved across all samples (Fig. 2B; see also Table S4 in the supplemental material). In the reconstructed tree, the 13 clade I strains clustered together, which was consistent with the 16S rRNA tree. However, samples s3, s6, and s10 of clade II fell in the center of clade I. Another discrepancy is that clade II samples s27 and s29 mapped to the termini of the concatenated gene tree. Notably, all five of the clade II strains that resulted in discrepancies were located in a minor cluster close to clade I in the 16S rRNA tree (Fig. 2A), suggesting that the functional proteome of these strains may have been shaped to develop a host specificity and a host range similar to those of the clade I strains. To further elucidate whether there is a causal relationship between clinical phenotypes and evolution history, the identified phenotypes were mapped to the phylogenetic tree, but no significant correlation was observed (Fig. 2B). This indicates that genetically diverse strains of B. cereus retain the capacity to cause infection in immunocompromised patients.
FIG 2.
Phylogenetic placement of the 24 B. cereus strains. (A) Maximum likelihood tree (1,000 bootstraps) constructed based on the 16S rRNA sequences from the 24 strains and 649 additional strains. The nodes with bootstrap values of >50 are depicted with gray circles. The size of each circle corresponds to the bootstrap value. The branches were colored based on the three major clades. The red triangles represent the placement of the 24 strains in the phylogenetic tree. (B) Maximum likelihood tree constructed based on the concatenated sequences of 130 single-copy genes conserved in the 24 strains. The classified clade based on the 16S rRNA tree for each strain is shown in the first column (red, clade I; blue, clade II). The right panel represents the corresponding clinical phenotypes for each strain. NA, not applicable. ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; ATRT, atypical teratoid/rhabdoid tumor.
Orthology-based analysis suggests the presence of species- and lineage-specific adaptations.
A comparative analysis of protein-coding genes was conducted to identify core or strain-specific genes. A reciprocal all-versus-all BLAST sequence analysis was used to retrieve the set of orthologous protein-coding gene groups to determine the core and strain-specific gene families. The core was defined as the gene families shared by at least 23 of the 24 strains to tolerate potential annotation inaccuracy, while the strain-specific genes were defined as the genes identified in only one strain.
A total of 1,640 protein-coding gene families shared by at least 23 strains were identified, representing the core genome (see Fig. S2A in the supplemental material). Among the total, 626 gene families were shared by all strains. A functional enrichment analysis (Table S5) based on molecular function GO (Gene Ontology) terms (16) for the core genome indicated that proteins with function related to transporter activity, such as inorganic cation transmembrane transporter activity (GO:0022890), substrate-specific transporter activity (GO:0022892), and monovalent inorganic cation transmembrane transporter activity (GO:0015077), were particularly conserved. Similarly, for the biological process category of GO terms, single-organism transport (GO:0044765), cation transport (GO:0006812), and ion transport (GO:0006811) were the most conserved functional groups. For the cellular component, the only significantly enriched gene was in the membrane-associated category. The enrichment of cation and ion transporter genes is of interest given that the pathogens involved in enteric infections must acquire these necessary cofactors from the host environment (17–19). The numbers of strain-specific genes differed among the strains, ranging from 28 to 674 (Fig. S2B). The two strains without clinical phenotypes, environmental samples s4 and s3, contained the highest number of strain-specific genes, while the other pathogenic strains contained more shared gene repertoires. Although the correlation between these variations and clinical phenotypes was not strong, the higher number of genes shared by the pathogenic strains suggests that a core subset of genes may be required to establish infection.
Identification of the genes associated with clinical phenotypes.
Gene duplications present in microbial pathogens have been associated with the host adaption process, which can be fixed in the genome through positive selection during the course of evolution (20–23). To examine the relationship between copy number variation (CNV) and clinical phenotypes, we first performed unsupervised hierarchical clustering based on the copy numbers of all the orthologous gene families (see Fig. S3A in the supplemental material). The clustering topology indicated that the strains showing clinical phenotypes of bacteremia (samples s17 and s21), skin infection (samples s22, s27, and s28), and colonization (samples s5, s6, and s7) tended to group together, with a subset of coclustering strains being associated with other clinical phenotypes. Another clustering based on the gene presence/absence profiles of the gene families revealed a similar pattern, in which the samples with bacteremia or colonization phenotypes were clustered more closely (Fig. S3B). Although the clusters were not well separated, the results implied that gains or losses of the genes were associated with disease tropism, which may underlie molecular mechanisms of the observed phenotypes.
The mixture of strains in the phylogenetic and unsupervised clustering trees indicated that the etiology of observed phenotypes could be driven by a subset of virulence genes. In order to identify these driver genes, we applied the random forest algorithm to predict the gene families that contribute most to the resultant phenotypes based on the similarity between the CNV and presence/absence profiles of the orthologous gene families. After supervised clustering using the profiles of selected gene families, the samples with a clinical hemolysis phenotype formed two well-separated groups (Fig. 3 and Table S6). One distinct allele of cardiolipin synthetase (CLS) (family 05728) was present mainly in the hemolysis samples. In addition to the presence of exclusive CLS and several hypothetical proteins, a few gene families in hemolysis and nonhemolysis samples displayed CNV. For instance, a penicillin-binding protein family (family 00005) was present in 2 to 3 copies in the hemolysis samples, while its copy number was increased to 4 to 5 copies in the other samples. Another example was an ABC transporter family (OppA) (family 00002), showing less expansion in the hemolysis samples (∼5 to 6 copies) than in the other nonhemolysis samples (9 to 10 copies). The OppA family proteins have been characterized as multifunctional proteins involved in host interaction processes, including binding oligopeptide permease (24), immunogenic cytoadhesin (25), and Mg2+-dependent ecto-ATPase (26). OppA as well as other OppA-like proteins can facilitate the import of the PapR signaling peptide (27). The skin infection strains also displayed a clear separation corresponding to their clinical phenotype (Fig. 4 and Table S6) and were characterized by enrichment for a subset of gene families. Annotation of the enriched gene families indicated that the majority were hypothetical proteins, with vancomycin B-type resistance protein (family 04769) (28) being present in all skin infection strains. A few phage-related protein families were also present, primarily in the skin infection strains. Clustering for the meningoencephalitis strains also revealed well-split clusters (Fig. 5 and Table S6). The serine phosphatase RsbUv regulator (family 00149) and lipoteichoic acid synthase LtaS (type Ia) (family 00011) protein families displayed different degrees of copy number variation in meningoencephalitis. RsbUv and LtaS are likely involved in the host interaction process (26).
FIG 3.
Supervised clustering of genes associated with hemolysis. (A) Supervised hierarchical clustering based on the copy number variation profiles of selected gene families. SAM, S-adenosyl-l-methionine; ECF, extracytoplasmic function. (B) Supervised hierarchical clustering based on the gene presence/absence profiles of selected gene families. The gene families are ordered based on the mean decrease of the Gini index value (Importance). The corresponding phenotype of each sample is shown on the top of the plot. Clustering was performed based on the Euclidean distance matrix. Numbers and phenotypes represent the grouping results following family classification with OrthoMCL.
FIG 4.
Supervised clustering of the genes associated with skin infection. (A) Supervised hierarchical clustering based on the copy number variation profiles of selected gene families. (B) Supervised hierarchical clustering based on the gene presence/absence profiles of selected gene families. The gene families are ordered based on the mean decrease of the Gini index value (Importance). The corresponding phenotype of each sample is shown on the top of the plot. Clustering was performed based on the Euclidean distance matrix. Numbers and phenotypes represent the grouping results following family classification with OrthoMCL. SMS, spermine synthase.
FIG 5.
Supervised clustering of the genes associated with meningoencephalitis. (A) Supervised hierarchical clustering based on the copy number variation profiles of selected gene families. (B) Supervised hierarchical clustering based on the gene presence/absence profiles of selected gene families. The gene families are ordered based on the mean decrease of the Gini index value (Importance). The corresponding phenotype of each sample is shown on the top of the plot. Clustering was performed based on the Euclidean distance matrix. Numbers and phenotype represent the grouping results following family classification with OrthoMCL.
Supervised clustering of the samples for the other phenotypes revealed a pattern that followed the phenotypes in general, even though ambiguities arose from a subset of samples that did not fit the overall classification patterns. For the bacteremia phenotype (Fig. S4 and Table S6), a few samples clustered with the incorrect phenotypic classification, such as sample s25, which was nested within the samples from cases without bacteremia. This indicates that while supervised clustering was successful in general, additional bacterial or host factors likely complicate more accurate clustering. Despite such ambiguities, the comparison revealed that several genes tended to be absent from the samples isolated from patients with bacteremia. For example, putative resolvase (family 04751), dehydrogenase (family 05311), and phosphonate ABC transporter phosphate-binding periplasmic component (family 05023) protein families were absent from nearly all the bacteremia samples. The resolvase gene appears to be associated with genome duplication, such as the resolvases in pathogenic Borrelia burgdorferi (29). In isolates associated with colonization, an amino acid permease family (family 07457) was found exclusively in these samples (Fig. S5A and Table S6), which was previously considered a virulence determinant in reducing host defense (30). Several genes associated with metabolic processes were absent from the colonization samples, including multiple hydrolases. d-Alanyl–d-alanine carboxypeptidase (family 04310), which removes the terminal d-alanine from UDP-MurNAc pentapeptides and serves as a target of β-lactam antibiotics (31), was also absent from the colonization samples. For the diarrhea and sepsis samples (Fig. S6 and S7 and Table S6), clustering of the CNV and presence/absence profiles did not correspond well to clinical phenotypes.
B. cereus is known to encode a number of hemolysins and toxins that play important roles in disease pathology. We next sought to determine the pattern of toxins being encoded by the respective strains with associated disease manifestations. The presence and absence of the characterized toxins and hemolysins are summarized in Table S6. The presence or absence of the respective virulence factors was determined by BLAST, with a cutoff of <1E−50 (Table S7). All strains examined harbored the cereulide synthase gene, whose dodecadepsipeptide product is an emetic toxin (32). All strains encoded at least one of the three predicted nonhemolytic enteric cytotoxins, with most strains (20/24) encoding at least two. The majority of strains (20/24) also carried both the cytotoxin K and hemolysin II genes, which were always found in the same isolates when present. While no discernible pattern between toxin profiles and disease manifestations was observed, these data indicate that the majority of the isolates encoded the traditional virulence factors required for disease.
DISCUSSION
In many bacterial pathogens, certain clonal lineages have a greater propensity to manifest as invasive infection. Due to the relative rarity of B. cereus infection and the paucity of genetic information, it is unclear as to whether particular genetic elements are associated with specific clinical manifestations. In our series of B. cereus isolates, patients had a wide spectrum of presentations, ranging from stool colonization to severe invasive infections leading to death. Previous reports (1–3) have attempted to identify patients at risk for severe B. cereus infections, such as those with neutropenia, hematologic malignancy, and chemotherapy-induced remission. However, the numbers of cases described in each of those previous reports have been small, with limited generalizability. In the present series, comparison of clinical characteristics did not reveal any significant risk factor for invasive B. cereus infections (see Table S3 in the supplemental material), possibly also related to the small sample size, but more importantly, it emphasizes the need to better understand genetic characteristics of B. cereus associated with infections.
The pathogenicity of B. cereus has been attributed primarily to toxin production. In gastrointestinal syndromes, B. cereus secretes a diarrhea-inducing protein enterotoxin or a plasmid-encoded cyclic peptide emetic toxin (4). Extragastrointestinal syndromes have been described to be related to the production of an array of other toxins that result in tissue destruction, including hemolysins, pore-forming enterotoxins, cytotoxin K, phospholipases, and others. Our data were in agreement with those findings, with bloodstream isolates being associated with a hemolytic phenotype (Fig. 2).
The hemolytic samples (isolates from patients with clinical bloodstream hemolysis) were shown to predominantly have enhanced copy numbers of penicillin-binding proteins in a high proportion of the samples analyzed (Fig. 2A). A subsequent analysis of these loci in these isolates revealed sequence differences in the individual strains, indicating that this copy variation is not an assembly artifact but indeed represents multiple alleles of the penicillin-binding proteins. Also of considerable interest is the expanded copy number of moeA, responsible for molybdenum cofactor biosynthesis (33). Molybdenum cofactor enzymes are widely distributed and have multiple cellular functions in metabolism (34). The hemolytic isolates tended to lack a component of the autoinducer 2 (AI-2) quorum sensing system compared to their nonhemolytic counterparts (Fig. 2B and 3). The difference in quorum sensing was due to the presence or absence of LsrB, the ligand-binding protein that is required for the internalization of AI-2 (35). In addition, nonhemolytic samples lacked similarity at additional genetic loci, including the small RNA (sRNA) chaperone Hfq as well as a number of transcriptional regulators of AsrR, DeoR, and the phosphate regulatory protein PhoB. These data indicate that the genetic contents of the hemolytic and nonhemolytic isolates differ considerably, but based on the differences in a number of key regulatory genes, transcriptional control may also be distinct in these isolates.
When isolates recovered from skin infection were compared to other isolates, a number of alleles were highly enriched. This included increased copy numbers of two extracellular degradation enzymes, including bacillolysin and a predicted collagenase. Multiple phage-associated genes were also highly enriched in the skin infection samples compared to the remaining data set. Interestingly, strains associated with meningoencephalitis demonstrated a distinct pattern of allelic variation compared to other isolates. This included variation with a predicted secreted ESAT-6 protein, a toxin secretion system found in multiple bacterial species, including Bacillus (36). In addition, there was considerable allelic variation in a number of transcriptional regulators, including members of the TetR, DeoR, and GntR families. In addition, there was allelic variation in the YvcQ sensor kinase genes of the YvcPQ two-component regulatory system, which is involved in mediating resistance to bacitracin in Bacillus species (37, 38). These data indicate that variation in alleles associated with transcriptional regulation, and expected alterations in transcriptional control, may be an important mediator of the tissue tropisms of B. cereus. These data represent the largest cohort of sequenced B. cereus isolates with associated patient metadata collected to date. While there is considerable genetic variation in isolates demonstrating distinct disease manifestations, specific loci were found to be enriched in or depleted from isolates with specific tissue tropisms. These data indicate that a complex combination of factors, including gene content, host status, and, likely, transcriptional regulation, are factors in the ability of B. cereus to manifest diverse diseases in immunocompromised hosts.
MATERIALS AND METHODS
Patients and samples.
B. cereus isolates were identified in clinical and environmental specimens collected during the course of routine care at St. Jude Children's Research Hospital (SJCRH) in Memphis, TN, between 1994 and 2005. Following detection by culture and identification by routine phenotypic methods, isolates were stored at −80°C. Samples were thawed, subcultured, and characterized by routine microbiological biochemical methods (see Table S1 in the supplemental material).
Three of the samples (samples s1, s2, and s17) were reported previously by our institution in a description of the clinical course of B. cereus infections in immunocompromised patients (2). Two B. cereus isolates (samples s3 and s4) were isolated from tea bag cultures tested in a study associating dietary tea ingestion and B. cereus bacteremia in pediatric oncology patients (39). Medical records were reviewed for demographic data, clinical syndrome, course, and infection outcome. Neutropenia was defined as an absolute neutrophil count (ANC) of ≤500 cells/mm3. Bacteremia was defined as growth of B. cereus in patient blood cultures. Meningoencephalitis was defined as central nervous system infection or inflammation manifesting with neurological dysfunction, such as altered mental status, seizures, and/or focal neurologic signs. This study was approved by the Institutional Review Board at SJCRH.
Sequencing and assembly of the B. cereus strains.
DNA was extracted by using Qiagen Genomic-tip 20/G (Qiagen, Hilden, Germany), prepared for sequencing by using a Paired-End Sample Preparation kit (Illumina Inc., San Diego, CA), and sequenced on the Illumina GA II system. The read quality of each sample was assessed by using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and low-quality reads were trimmed by using Trimmomatic (40). Filtered high-quality reads were assembled by using SPAdes (41) and CLC genome workbench (CLC Bio-Qiagen, Aarhus, Denmark), using different k-mer sizes. The assembly with the highest assembly qualities determined by the N50 value and assembly size was selected and merged by using CISA (42) to improve the contiguity and accuracy of the assemblies. The quality of the genome assembly was assessed by comparing the completenesses of a set of 40 highly conserved prokaryotic genes using the BUSCO pipeline (43).
Genome annotation and alignments.
Annotation of the genomes was accomplished by using the RAST annotation server (13). Based on the reciprocal BLAST best hit, the proteomes of all the strains were classified into orthologous gene families by using the OrthoMCL pipeline (44). Orthologous families were classified based on normalized scores calculated from the E values of all-versus-all BLASTp analysis (cutoff of 1E−5) for pairs of compared genomes. The normalized scores were fed into the Markov cluster algorithm to classify the genes into hypothesized orthologous and paralogous gene families using a default inflation parameter of 1.5. For the identification of syntenic regions, the genomes of B. cereus BAG3X2-2, ATCC 14579, ATCC 10987, and Rock3-29 were used as references for the genome alignment and comparisons. The pairwise similarity of the compared genomes was calculated by using MUMmer (45) to identify the anchor hits. Synteny was identified by searching colinear sequences of anchors.
Phylogeny reconstruction.
A total of 130 single-copy orthologous genes were identified through the OrthoMCL pipeline and used for subsequent analysis. These genes comprised central metabolic genes, cell division machinery, and sporulation pathways. The amino acid sequences of these 130 genes were aligned by using MUSCLE (46). The resultant alignment was concatenated, with gaps being removed by TrimAL (47), using an automated option to optimize gap trimming prior to phylogenetic tree construction. A maximum likelihood tree was constructed based on the concatenated alignment using the PROTGAMMAWAG model with 1,000 bootstraps in RAxML (48). The topology of the constructed tree was visualized by using the iTOL server (49) with data sets generated by custom scripts.
Clustering and feature selection.
Clinical phenotypes are potentially driven by the interplay between modifications of the genome content, such as a gain or loss of genes and gene family expansion or contraction, and differing mechanisms of cellular regulation. In order to identify associations between genomic changes and clinical phenotypes, we performed a random forest analysis based on the profiles of copy number variation and the presence/absence of gene families in each strain to identify a subset of genes that are most likely associated with the phenotypes. The random forest algorithm has been used extensively for associating genomic data and phenotypes (50, 51). In the present analysis, a collection of simple clustering trees (n = 5,000) was constructed based on CNVs and gene presence/absence profiles of orthologous gene families across all strains by using R. Afterwards, the candidate gene families were selected according to the vote of the ensemble of trees. The importance of each selected gene family was assessed by the mean decrease of the Gini coefficient. The Gini coefficient is a measure of the contribution of each gene family to the homogeneity in the resulting random forest. The gene families with high Gini values are important candidate gene families associated with the observed phenotypes. After this random forest selection procedure was applied, supervised clustering was performed for the selected gene families against the predefined phenotypes.
Accession number(s).
All genome assemblies were submitted to the NCBI database under the following accession numbers: NMYY00000000 for sample s1, NMYX00000000 for sample s2, NMYW00000000 for sample s3, NNAZ00000000 for sample s4, NMYV00000000 for sample s5, NMYU00000000 for sample s6, NMYT00000000 for sample s7, NMYS00000000 for sample s8, NMYR00000000 for sample s9, NMYQ00000000 for sample s10, NMYP00000000 for sample s17, NMZP00000000 for sample s18, NMYO00000000 for sample s19, NMYN00000000 for sample s20, NMYM00000000 for sample s21, NMYL00000000 for sample s22, NMYK00000000 for sample s23, NMYJ00000000 for sample s24, NMYI00000000 for sample s25, NMYH00000000 for sample s26, NMYG00000000 for sample s27, NMYF00000000 for sample s28, NMYE00000000 for sample s29, and NMYD00000000 for sample s30.
Supplementary Material
ACKNOWLEDGMENTS
We have no conflicts of interest to report.
This work was supported in part by NCI Cancer Center grant P30 CA021765 and by ALSAC.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00574-17.
REFERENCES
- 1.Dabscheck G, Silverman L, Ullrich NJ. 2015. Bacillus cereus cerebral abscess during induction chemotherapy for childhood acute leukemia. J Pediatr Hematol Oncol 37:568–569. doi: 10.1097/MPH.0000000000000413. [DOI] [PubMed] [Google Scholar]
- 2.Gaur AH, Patrick CC, McCullers JA, Flynn PM, Pearson TA, Razzouk BI, Thompson SJ, Shenep JL. 2001. Bacillus cereus bacteremia and meningitis in immunocompromised children. Clin Infect Dis 32:1456–1462. doi: 10.1086/320154. [DOI] [PubMed] [Google Scholar]
- 3.Hansford JR, Phillips M, Cole C, Francis J, Blyth CC, Gottardo NG. 2014. Bacillus cereus bacteremia and multiple brain abscesses during acute lymphoblastic leukemia induction therapy. J Pediatr Hematol Oncol 36:e197–e201. doi: 10.1097/MPH.0b013e31828e5455. [DOI] [PubMed] [Google Scholar]
- 4.Bottone EJ. 2010. Bacillus cereus, a volatile human pathogen. Clin Microbiol Rev 23:382–398. doi: 10.1128/CMR.00073-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivanova N, Sorokin A, Anderson I, Galleron N, Candelon B, Kapatral V, Bhattacharyya A, Reznik G, Mikhailova N, Lapidus A, Chu L, Mazur M, Goltsman E, Larsen N, D'Souza M, Walunas T, Grechkin Y, Pusch G, Haselkorn R, Fonstein M, Ehrlich SD, Overbeek R, Kyrpides N. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87–91. doi: 10.1038/nature01582. [DOI] [PubMed] [Google Scholar]
- 6.Okstad OA, Tourasse NJ, Stabell FB, Sundfaer CK, Egge-Jacobsen W, Risoen PA, Read TD, Kolsto AB. 2004. The bcr1 DNA repeat element is specific to the Bacillus cereus group and exhibits mobile element characteristics. J Bacteriol 186:7714–7725. doi: 10.1128/JB.186.22.7714-7725.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tourasse NJ, Helgason E, Okstad OA, Hegna IK, Kolsto AB. 2006. The Bacillus cereus group: novel aspects of population structure and genome dynamics. J Appl Microbiol 101:579–593. doi: 10.1111/j.1365-2672.2006.03087.x. [DOI] [PubMed] [Google Scholar]
- 8.Klassen JL, Currie CR. 2012. Gene fragmentation in bacterial draft genomes: extent, consequences and mitigation. BMC Genomics 13:14. doi: 10.1186/1471-2164-13-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cahill MJ, Koser CU, Ross NE, Archer JA. 2010. Read length and repeat resolution: exploring prokaryote genomes using next-generation sequencing technologies. PLoS One 5:e11518. doi: 10.1371/journal.pone.0011518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mende DR, Sunagawa S, Zeller G, Bork P. 2013. Accurate and universal delineation of prokaryotic species. Nat Methods 10:881–884. doi: 10.1038/nmeth.2575. [DOI] [PubMed] [Google Scholar]
- 11.Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 12.Creevey CJ, Doerks T, Fitzpatrick DA, Raes J, Bork P. 2011. Universally distributed single-copy genes indicate a constant rate of horizontal transfer. PLoS One 6:e22099. doi: 10.1371/journal.pone.0022099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolsto AB, Tourasse NJ, Okstad OA. 2009. What sets Bacillus anthracis apart from other Bacillus species? Annu Rev Microbiol 63:451–476. doi: 10.1146/annurev.micro.091208.073255. [DOI] [PubMed] [Google Scholar]
- 15.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and Web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gene Ontology Consortium. 2015. Gene Ontology Consortium: going forward. Nucleic Acids Res 43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson PJ, Hill KK, Laker MT, Ticknor LO, Keim P. 1999. Genetic comparison of Bacillus anthracis and its close relatives using amplified fragment length polymorphism and polymerase chain reaction analysis. J Appl Microbiol 87:263–269. doi: 10.1046/j.1365-2672.1999.00884.x. [DOI] [PubMed] [Google Scholar]
- 18.Stahler FN, Odenbreit S, Haas R, Wilrich J, Van Vliet AH, Kusters JG, Kist M, Bereswill S. 2006. The novel Helicobacter pylori CznABC metal efflux pump is required for cadmium, zinc, and nickel resistance, urease modulation, and gastric colonization. Infect Immun 74:3845–3852. doi: 10.1128/IAI.02025-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papazisi L, Rasko DA, Ratnayake S, Bock GR, Remortel BG, Appalla L, Liu J, Dracheva T, Braisted JC, Shallom S, Jarrahi B, Snesrud E, Ahn S, Sun Q, Rilstone J, Okstad OA, Kolstø AB, Fleischmann RD, Peterson SN. 2011. Investigating the genome diversity of B. cereus and evolutionary aspects of B. anthracis emergence. Genomics 98:26–39. doi: 10.1016/j.ygeno.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romero D, Palacios R. 1997. Gene amplification and genomic plasticity in prokaryotes. Annu Rev Genet 31:91–111. doi: 10.1146/annurev.genet.31.1.91. [DOI] [PubMed] [Google Scholar]
- 21.Hastings PJ. 2007. Adaptive amplification. Crit Rev Biochem Mol Biol 42:271–283. doi: 10.1080/10409230701507757. [DOI] [PubMed] [Google Scholar]
- 22.Van de Peer Y, Maere S, Meyer A. 2009. The evolutionary significance of ancient genome duplications. Nat Rev Genet 10:725–732. doi: 10.1038/nrg2600. [DOI] [PubMed] [Google Scholar]
- 23.Kondrashov FA. 2012. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc Biol Sci 279:5048–5057. doi: 10.1098/rspb.2012.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henrich B, Hopfe M, Kitzerow A, Hadding U. 1999. The adherence-associated lipoprotein P100, encoded by an opp operon structure, functions as the oligopeptide-binding domain OppA of a putative oligopeptide transport system in Mycoplasma hominis. J Bacteriol 181:4873–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henrich B, Feldmann RC, Hadding U. 1993. Cytoadhesins of Mycoplasma hominis. Infect Immun 61:2945–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hopfe M, Henrich B. 2004. OppA, the substrate-binding subunit of the oligopeptide permease, is the major ecto-ATPase of Mycoplasma hominis. J Bacteriol 186:1021–1028. doi: 10.1128/JB.186.4.1021-1028.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slamti L, Lemy C, Henry C, Guillot A, Huillet E, Lereclus D. 2016. CodY regulates the activity of the virulence quorum sensor PlcR by controlling the import of the signaling peptide PapR in Bacillus thuringiensis. Front Microbiol 6:1501. doi: 10.3389/fmicb.2015.01501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pereira SF, Goss L, Dworkin J. 2011. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol Mol Biol Rev 75:192–212. doi: 10.1128/MMBR.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moriarty TJ, Chaconas G. 2009. Identification of the determinant conferring permissive substrate usage in the telomere resolvase, ResT. J Biol Chem 284:23293–23301. doi: 10.1074/jbc.M109.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das P, Lahiri A, Lahiri A, Chakravortty D. 2010. Modulation of the arginase pathway in the context of microbial pathogenesis: a metabolic enzyme moonlighting as an immune modulator. PLoS Pathog 6:e1000899. doi: 10.1371/journal.ppat.1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Linden MP, de Haan L, Dideberg O, Keck W. 1994. Site-directed mutagenesis of proposed active-site residues of penicillin-binding protein 5 from Escherichia coli. Biochem J 303(Part 2):357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agata N, Ohta M, Mori M, Isobe M. 1995. A novel dodecadepsipeptide, cereulide, is an emetic toxin of Bacillus cereus. FEMS Microbiol Lett 129:17–20. doi: 10.1111/j.1574-6968.1995.tb07550.x. [DOI] [PubMed] [Google Scholar]
- 33.Nichols J, Rajagopalan KV. 2002. Escherichia coli MoeA and MogA. Function in metal incorporation step of molybdenum cofactor biosynthesis. J Biol Chem 277:24995–25000. doi: 10.1074/jbc.M203238200. [DOI] [PubMed] [Google Scholar]
- 34.Iobbi-Nivol C, Leimkuhler S. 2013. Molybdenum enzymes, their maturation and molybdenum cofactor biosynthesis in Escherichia coli. Biochim Biophys Acta 1827:1086–1101. doi: 10.1016/j.bbabio.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Pereira CS, de Regt AK, Brito PH, Miller ST, Xavier KB. 2009. Identification of functional LsrB-like autoinducer-2 receptors. J Bacteriol 191:6975–6987. doi: 10.1128/JB.00976-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garufi G, Butler E, Missiakas D. 2008. ESAT-6-like protein secretion in Bacillus anthracis. J Bacteriol 190:7004–7011. doi: 10.1128/JB.00458-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang S, Li X, Wang X, Li Z, He J. 2016. The two-component signal transduction system YvcPQ regulates the bacterial resistance to bacitracin in Bacillus thuringiensis. Arch Microbiol 198:773–784. doi: 10.1007/s00203-016-1239-z. [DOI] [PubMed] [Google Scholar]
- 38.Rietkotter E, Hoyer D, Mascher T. 2008. Bacitracin sensing in Bacillus subtilis. Mol Microbiol 68:768–785. doi: 10.1111/j.1365-2958.2008.06194.x. [DOI] [PubMed] [Google Scholar]
- 39.El Saleeby CM, Howard SC, Hayden RT, McCullers JA. 2004. Association between tea ingestion and invasive Bacillus cereus infection among children with cancer. Clin Infect Dis 39:1536–1539. doi: 10.1086/425358. [DOI] [PubMed] [Google Scholar]
- 40.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin SH, Liao YC. 2013. CISA: contig integrator for sequence assembly of bacterial genomes. PLoS One 8:e60843. doi: 10.1371/journal.pone.0060843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 44.Li L, Stoeckert CJ Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 49.Letunic I, Bork P. 2011. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen X, Ishwaran H. 2012. Random forests for genomic data analysis. Genomics 99:323–329. doi: 10.1016/j.ygeno.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Touw WG, Bayjanov JR, Overmars L, Backus L, Boekhorst J, Wels M, van Hijum SA. 2013. Data mining in the life sciences with Random Forest: a walk in the park or lost in the jungle? Brief Bioinform 14:315–326. doi: 10.1093/bib/bbs034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.