Abstract
We describe the cloning, expression and characterization of the first truly non-specific adenine DNA methyltransferase, M.EcoGII. It is encoded in the genome of the pathogenic strain Escherichia coli O104:H4 C227–11, where it appears to reside on a cryptic prophage, but is not expressed. However, when the gene encoding M.EcoGII is expressed in vivo - using a high copy pRRS plasmid vector and a methylation–deficient E. coli host—extensive in vivo adenine methylation activity is revealed. M.EcoGII methylates adenine residues in any DNA sequence context and this activity extends to dA and rA bases in either strand of a DNA:RNA-hybrid oligonucleotide duplex and to rA bases in RNAs prepared by in vitro transcription. Using oligonucleotide and bacteriophage M13mp18 virion DNA substrates, we find that M.EcoGII also methylates single-stranded DNA in vitro and that this activity is only slightly less robust than that observed using equivalent double-stranded DNAs. In vitro assays, using purified recombinant M.EcoGII enzyme, demonstrate that up to 99% of dA bases in duplex DNA substrates can be methylated thereby rendering them insensitive to cleavage by multiple restriction endonucleases. These properties suggest that the enzyme could also be used for high resolution mapping of protein binding sites in DNA and RNA substrates.
INTRODUCTION
DNA methyltransferases (MTases) are ubiquitous in all kingdoms of life (1). In bacteria and archaea, they are most often associated with providing protection against restriction enzymes (2). Many prokaryotic DNA MTases, however, have no counterpart restriction enzyme and the biological functions of such ‘orphan’ MTases have only been determined in a few cases. For instance, the Dam MTase of E. coli is known to be involved in mismatch repair and control of the replication cycle (3), the CcrM MTase from Caulobacter crescentus has been shown to be involved in cell cycle regulation (4) and the MamA MTase of Mycobacterium tuberculosis functions as a sequence-specific regulator of transcription with a potential role in adaptation to hypoxia (5). Some bacteriophage genomes are also known to encode DNA MTases where it is assumed that they provide protection against commonly encountered restriction systems (6). Because MTases contain distinctive amino acid sequence motifs, a large number of putative MTase genes can be found using bioinformatic analyses (7,8), but usually their larger role within the metabolism of their bacterial hosts are unknown.
Until recently, DNA MTases were quite difficult to characterize in terms of their recognition sequences due to the tedious biochemical procedures necessary to rigorously identify the sequences surrounding the methylated base. However, with the introduction of Pacific Biosciences SMRT sequencing (9,10), this situation has changed and it has now become very simple to determine recognition sequences for DNA MTases either by expressing them in a non-methylating strain of E. coli (11) or even easier, by performing computational motif analysis of raw genome sequence data (12). This has resulted in the discovery of very many new MTases, especially those associated with Type I and Type III restriction systems (13), but also some with unexpected specificity such as the one characterized in detail here (14).
A DNA MTase, M.HaeV with greatly reduced specificity BA (B = C, G or T) was described by Drozdz et al. (15), who also noted that several similar enzymes could be found in GenBank on the basis of sequence similarity. Noteworthy in this regard is a DNA adenine methyltransferase, M.CsaII, which recognizes the sequence AB (12). Although this enzyme has a related recognition sequence, its protein sequence is quite distinct from that of M.HaeV. Nevertheless, it too has many related genes in GenBank.
During our analysis of the DNA MTases present in the E. coli strain, O104:H4 C227-11, which was responsible for a severe outbreak of hemorrhagic uremia in Europe, we expressed and characterized the specificities of each of the DNA MTases present in this system (14). Two of those proved completely non-specific for all A residues, although in the initial experiments where in vivo methylation by cloned genes was reported, only 70% of the A residues were methylated. However, no sequence specificity for that modification was detected in the genome. We have subsequently characterized one of those enzymes, M.EcoGII in detail, and find that it is indeed non-specific and, under appropriate conditions, is able to methylate >85% of A-residues in a DNA substrate in vivo and close to 100% in vitro.
MATERIALS AND METHODS
Materials
Restriction endonucleases, T4-DNA ligase, Phusion-HF DNA polymerase, proteinase K, S-adenosylmethionine (SAM), Hi-Scribe in vitro transcription kit and competent E. coli cells were from New England Biolabs Inc. (Ipswich, MA, USA). Tritiated SAM (specific activity 55–85Ci/mmol) was acquired from Perkin Elmer. Plasmid DNAs and PCR products were purified using spin-column purification reagents from Qiagen and New England Biolabs Inc. All synthetic DNA and RNA oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA, USA).
Expression and purification of recombinant M.EcoGII
The M.EcoGII gene was amplified from E. coli O104:H4 C227–11 genomic DNA by PCR using Phusion-HF DNA polymerase, restricted with SbfI and BamHI endonucleases and ligated to PstI-BamHI-restricted pRRS plasmid DNA (14). This vector, however, proved to be unsuitable for recombinant expression of M.EcoGII, most likely due to unregulated expression from the vector lac promoter. We therefore subcloned the M.EcoGII gene in several inducible expression vectors to identify an optimal expression platform. M.EcoGII enzyme was expressed using a pBAD24 (16) arabinose-inducible vector in Escherichia coli ER3037 cells (E. coli B fhuA2 [lon] ompT gal sulA11 Δ(mcrC-mrr)114::IS10 R(mcr-73::miniTn10–TetS)2 R(zgb-210::Tn10 )(TetS) endA1 [dcm]). Cells were harvested 2 h after arabinose induction and the M.EcoGII enzyme was purified from E. coli cell-free extracts using an ÅKTA-FPLC system (Pharmacia/GE) via sequential chromatography using DEAE-Sepharose, Heparin Hyper-D, Source-Q, Source-S, Heparin-TSK and Superdex-75.
Biochemical characterization
To assess the extent of M.EcoGII methylation activity achievable in vivo the pRRS:M.EcoGII vector was transferred to E. coli ER2796 (17) which lacks dam, dcm and EcoKI MTase activities. Plasmid DNA samples were isolated from stationary-phase ER2796 cultures after growth at 37°C and 200 rpm for 20 h, followed by hydrolysis to nucleosides and LC–MS analysis (see below). In vitro methylation activity of M.EcoGII on various substrates was assayed by radiometric and non-radiometric methods. Radiometric assays of M.EcoGII activity on single- and double-stranded DNA, single stranded RNA and RNA/DNA-hybrid oligonucleotide substrates used 50 mM HEPES buffer, pH 7.0, containing 1 mM EDTA and 3.7 μM 3H-SAM. Larger scale non-radiometric assays with single- and double-stranded DNA, RNA/DNA-hybrid or in vitro transcribed RNA substrates used either 50 mM HEPES buffer or 1× CutSmart buffer (50 mM potassium acetate/20 mM Tris-acetate/10 mM magnesium acetate, pH 7.9) containing 1 mM EDTA and SAM concentrations varying from 80–320 μM.
RNA products of in vitro transcription reactions were treated with DNAse I to remove template DNA and RNA was recovered by ethanol precipitation, then dissolved in DEPC-treated water prior to methylation. Five μg of RNA transcript was methylated in vitro for two hours at 37°C in a 1 ml assay containing 1× CutSmart buffer containing 320 μM SAM, 1 mM EDTA and 1 μM M.EcoGII. Methylated RNA was recovered by phenol extraction, followed by two cycles of ethanol precipitation and resuspension in DEPC-treated water.
Qualitative analyses of M.EcoGII activity on double-stranded plasmid and genomic DNA substrates employed restriction-protection assays using a selection of restriction endonucleases that are known to be insensitive to, or inhibited by, adenine-methylation (13).
Quantitative analyses of M.EcoGII activity in vivo and in vitro using DNA and RNA substrates using an LC–MS assay
The relative abundances of unmethylated (dA and rA) and methylated (m6dA and m6rA) bases in M.EcoGII-methylated substrates were determined using liquid chromatography and mass spectrometry (LC–MS). DNA and/or RNA samples were converted to nucleosides using a proprietary mixture of nucleases and phosphatases (New England Biolabs Inc.) based on the method of Hashimoto et al. (18). LC–MS/MS analysis was performed in duplicate by injecting digested polynucleotide samples on an Agilent 1290 UHPLC equipped with a G4212A diode array detector and a 6490A Triple Quadrupole Mass Detector operating in the positive electrospray ionization mode. UHPLC was carried out using a Waters XSelect HSS T3 XP column (2.1 × 100 mm, 2.5 μm) with the gradient mobile phase consisting of methanol and 10 mM aqueous ammonium formate (pH 4.4). Data acquisition was performed in the dynamic multiple reaction monitoring (DMRM) mode. Each nucleoside was identified in the extracted chromatogram associated with its specific MS/MS transition: dA [M+H]+ at m/z 252 →136, m6dA [M+H]+ at m/z 266 →150, rA [M+H]+ at m/z 268 →136, and m6rA [M+H]+ at m/z 282 →150. External calibration curves with known amounts of the nucleosides were used to calculate their ratios within the samples analyzed.
Bioinformatic analyses
M.EcoGII was first identified using the SEQWARE program as described previously (11,12). Flanking sequences were examined for the presence of phage genes and also compared with the flanking sequences of M.EcoGI,which is very closely related in sequence to M.EcoGII. BLAST analyses at NCBI were used to detect homologs in other genomes. The M.EcoGII protein sequence was analyzed with the PHYRE2 prediction server (using default parameters) for comparison with structures of each of the five β-class DNA MTase structures deposited in the PDB (19).
RESULTS
Cloning, expression and purification
During initial attempts to express the gene for M.EcoGII using the multicopy plasmid pRRS, in which the gene is constitutively active, we noticed that clones grew slowly and were somewhat unstable. We attributed this to potentially high levels of methylation of the E. coli chromosome, which we assumed would affect gene expression, with deleterious effects for the cell. However, transfer of the gene to an arabinose-inducible pBAD24 vector was sufficient to achieve stable recombinant expression. The enzyme was purified to near homogeneity as described in Materials and Methods and the results of SDS-PAGE and native MS analyses of the purified M.EcoGII are presented in Supplementary Figure S1.
Analyses of DNA methylation by M.EcoGII in vivo and in vitro
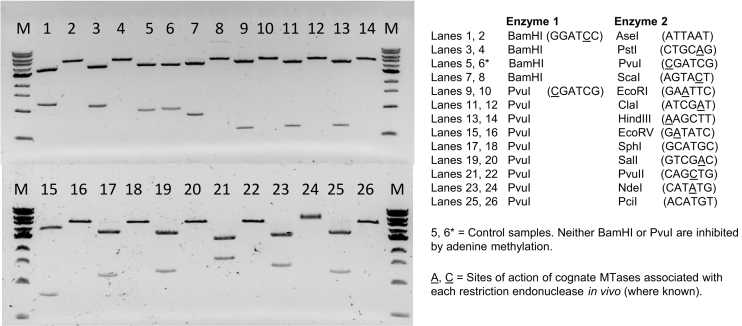
To assess the specificity of M.EcoGII-methylation and its effectiveness at inhibiting restriction endonucleases, we carried out restriction analyses using pBR322 plasmid DNA and an enzyme known to be insensitive to dA methylation (either BamHI or PvuI) in combination with one of twelve additional enzymes that cleave different six base-pair sequences, the activities of which are known to be blocked by adenine methylation (13). Duplicate assays were set-up using unmethylated pBR322 DNA (isolated from E. coli ER2796 cells) and equivalent samples that had been methylated using M.EcoGII at 37°C for 1 h in vitro. The results, shown in Figure 1, confirm that the activities of all twelve enzymes are blocked by the action of M.EcoGII while the equivalent unmethylated samples are completely restricted. The modified adenine residues present in these restriction sites are embedded in multiple different flanking sequences, consistent with the notion that M.EcoGII methylates dA residues in many sequence contexts. The results for EcoRI, NdeI, PstI and SalI—each of which is known to be protected from restriction in vivo by the action of a cognate adenine MTase acting at defined positions in each strand of their 6-base pair (bp) palindromic recognition sequences—collectively demonstrate that M.EcoGII methylates dA bases in all possible dinucleotide contexts. The results of a similar analysis using BamHI and six restriction endonucleases that cleave four base-pair sequences (AluI, MluCI, MseI, NlaIII, RsaI and TaqI) are presented in Supplementary Figure S2. As previously, each unmethylated plasmid DNA sample was completely restricted, whereas the equivalent M.EcoGII-methylated samples yielded only the full-length linear product of BamHI restriction.
Figure 1.
In vitro methylation of plasmid DNA by M.EcoGII inhibits cleavage activities of multiple restriction endonucleases. pBR322 plasmid DNA was prepared from E. coli ER2796 (a non-methylating strain lacking dam, dcm and M.EcoKI activities) and a 10 μg sample of this DNA was subsequently methylated in vitro using purified M.EcoGII enzyme (1 μM). Unmethylated and M.EcoGII-methylated DNA samples were each incubated with pairs of restriction endonucleases comprising one enzyme that is known to be insensitive to adenine-methylation (either BamHI or PvuI) and a second that is known to be inhibited by this modification. Note that BamHI was only used in the control samples (lanes 5 and 6) and in combination with AseI, PstI and ScaI restriction. The latter three sites are located very close to the PvuI site of pBR322 and would therefore yield products that are not distinguishable from linear pBR322. Lanes 1, 3, 5…..25: unmethylated pBR322 DNA isolated from E. coli ER2796 cells. Lanes 2, 4, 6….26: equivalent DNAs after in vitro methylation with recombinant M.EcoGII. M1 = 1 kb DNA Ladder 0.5–10.0 kb (NEB).
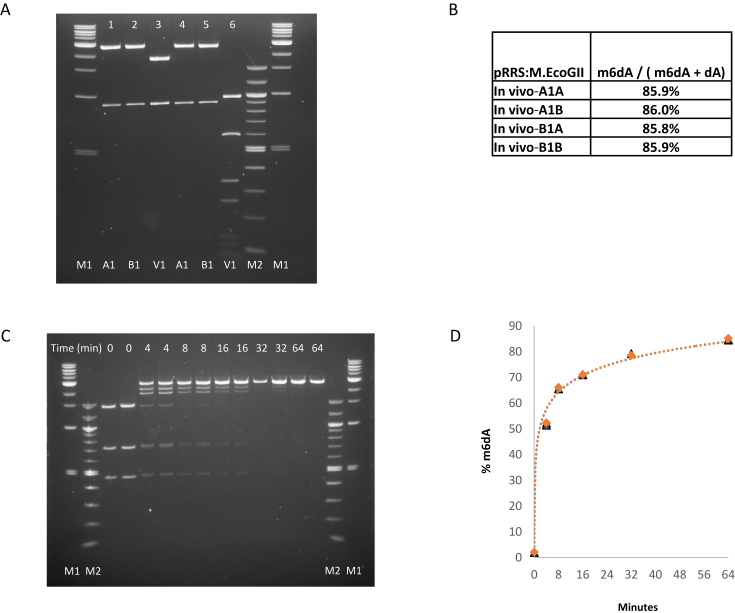
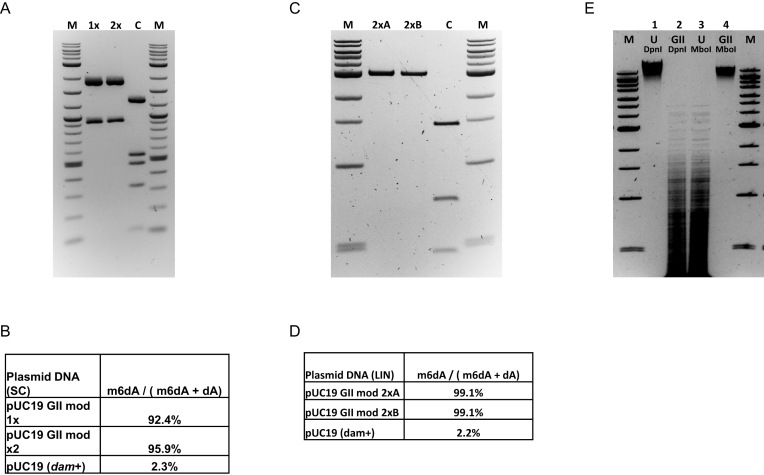
We then determined the extent of M.EcoGII-catalyzed methylation that could be achieved in vivo using an LC–MS-based assay and found that 86% of total dA in plasmid pRRS:M.EcoGII DNA isolated from stationary phase ER2796 cultures was in the form of m6dA (Figure 2A, B). We also determined the time course of M.EcoGII methylation of dam+ pUC19 plasmid DNA at 37°C using a restriction-protection assay (Figure 2C) followed by LC–MS analysis of the products. The latter shows that more than 50% of total dA was converted to m6dA after 4 minutes, increasing to 85% m6dA after 64 minutes (Figure 2D). Additional in vitro experiments using pUC19 DNA and an excess of enzyme yields DNA wherein m6dA constitutes greater than 92% of total dA, increasing to 96% m6dA if the methylated DNA is spin-column purified then subjected to a second round of methylation in vitro (Figure 3A andB). Control pUC19 DNA, without M.EcoGII treatment, contains 2.3% m6dA representing complete methylation of the 15 dam methylation sites present in the 2686 bp plasmid. In equivalent experiments, using linear pUC19 plasmid DNA prepared by SmaI restriction as a substrate for M.EcoGII methylation, the proportion of methylated dA can exceed 99%, possibly indicating that relaxed DNA is a better substrate for M.EcoGII than supercoiled plasmid DNA (Figure 3C and D).
Figure 2.
Extent of M.EcoGII methylation of plasmid DNA in vivo and in vitro. (A) In vivo assay. Plasmid pRRS:M.EcoGII, a high-copy replicon that expresses M.EcoGII, was introduced into methylation-deficient ER2796 E. coli cells by transformation. Plasmid DNA was recovered from stationary phase cultures of two independent isolates after growth at 37°C for 20 h. pRRS:M.EcoGII and unmethylated pRRS vector control DNAs were restricted with either PvuI alone (lanes 1, 2 and 3) or PvuI plus MboI (lanes 4, 5 and 6). As PvuI activity is insensitive to m6A modification all samples are fully restricted by PvuI (lanes 1–6) but only the unmethylated pRRS vector control DNA is sensitive to MboI restriction (lane 6). M1 = 1 kb DNA Ladder 0.5–10.0 kb (NEB). M2 = 100 bp DNA ladder 0.1–1.5 kb (NEB). (B) LC–MS analysis of pRRS:M.EcoGII plasmid isolates. DNA samples were converted to nucleosides and analyzed in duplicate using LC–MS. In each sample, 86% of the dA bases were methylated in vivo. (C) Analysis of M.EcoGII methylation activity in vitro. Duplicate assays containing 10 μg pUC19 plasmid DNA and 320 μM SAM were set-up on ice and 0.1 ml aliquots of each were removed and snap-frozen in a dry-ice/ethanol bath (as unmethylated control samples). The remainder of each sample was placed in a 37°C water-bath, M.EcoGII enzyme (1 μM) was added and additional 0.1 ml samples were removed and snap-frozen after 4, 8, 16, 32 and 64 minutes of incubation at 37°C. Methylated DNAs were restricted with BamHI and TaqI. M1 = 1 kb DNA Ladder 0.5–10 kb and 100bp DNA Ladder 0.1–1.5 kb (NEB). (D) Time-course of M.EcoGII methylation in vitro. After 4 minutes, 50% of dA is present as m6dA, increasing to over 84% after 64 min. LC–MS data for the individual assays and time points are presented in Supplementary Table S2.
Figure 3.
M.EcoGII methylates up to 99% of the dA residues in plasmid pUC19 DNA substrates and can be used for genome-wide methylation in vitro. (A) Supercoiled pUC19 plasmid DNA (20 μg) was methylated with M.EcoGII enzyme (1 μM) and 320 μM SAM in vitro, purified by phenol extraction and ethanol precipitation (lane 1x), then an aliquot was re-methylated using the same protocol (Lane 2x). Each experimental sample and the unmodified dam+ control DNA (lane C) were restricted using PvuI and BspHI endonucleases. BspHI cleaves DNA between the first and second nucleotide of TCATGA sequences and its activity is blocked by methylation of either of the dA bases. M = 2-log DNA Ladder 0.1–10.0 kb (NEB). (B) LC–MS data for experimental and control DNA samples. (C) pUC19 plasmid DNA was linearized by SmaI restriction and duplicate samples (2 μg) were subjected to two cycles of methylation using M.EcoGII (1 μM) and 160 μM SAM in vitro. Methylated DNAs were recovered by phenol extraction followed by ethanol precipitation. An aliquot of each sample (lanes 2xA, 2xB) and unmethylated (dam+) control DNA (lane C) were restricted using TaqI endonuclease. (D) LC–MS data for experimental and control DNA samples. (E) Genomic DNA was isolated from E. coli ER2796 cells (which lacks all E. coli DNA MTase activities) and a 20 μg sample was methylated in vitro using M.EcoGII (2 μM) and 320 μM SAM. Lanes 1, 3: unmethylated (ER2796) gDNA samples. Lanes 2, 4: M.EcoGII-methylated (ER2796) gDNA samples. Lanes 1, 2: DpnI (GATC) restricted samples (DpnI requires adenine methylation of both DNA strands for efficient cleavage activity). Lanes 3, 4: MboI (GATC) restricted samples (MboI activity is inhibited by adenine hemi or complete methylation). M = 1 kb-Extend DNA Ladder 0.5–48.5 kb (NEB).
M.EcoGII can also be used for genome-wide methylation of unmethylated bacterial DNA isolated from E. coli ER2796, yielding products that are refractory to MboI cleavage but are completely restricted by DpnI (Figure 3E). MboI activity is inhibited by hemimethylation of adenine at GATC sequences, whereas DpnI is a methyl-dependent enzyme that requires methylation of adenines in both strands of the GATC site for efficient cleavage and is only partially active at hemimethylated sites (13).
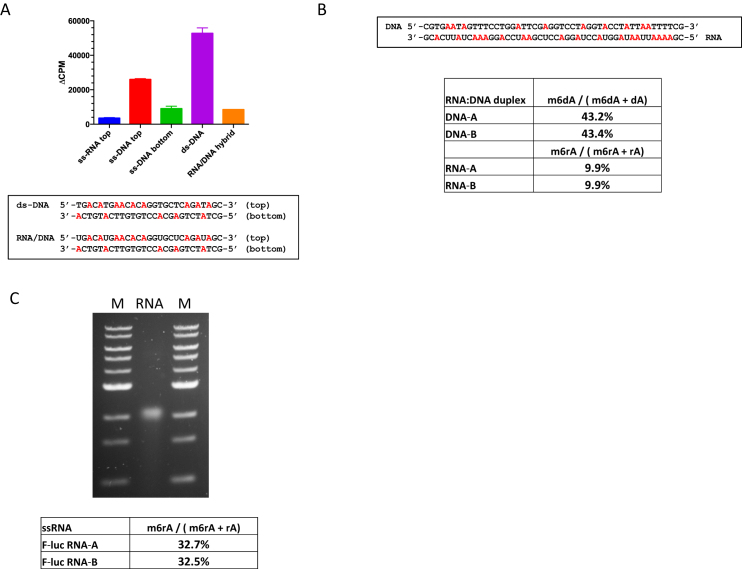
Activities of M.EcoGII on ssDNA, DNA–RNA hybrid and ssRNA substrates
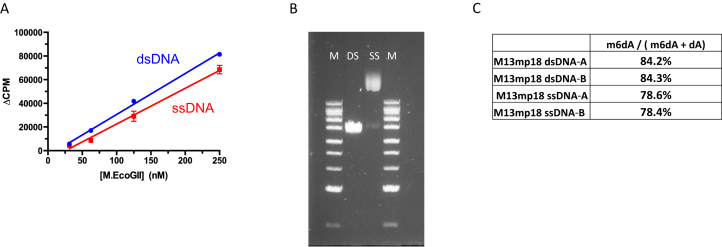
Radiometric assays, using tritiated SAM as methyl donor, were used to compare the activity of M.EcoGII (25–250 nM) using single-stranded or duplex oligonucleotide substrates each of which contained a single dA residue (Figure 4A). Robust activity was evident in both cases, with methylation of the ssDNA substrate corresponding to about 80% of that observed for the equivalent duplex. To confirm these observations we methylated 2 μg of M13mp18 single-stranded virion DNA and an equivalent amount of double-stranded M13mp18 RF duplex DNA with M.EcoGII in vitro. Methylated DNA samples were hydrolyzed to nucleosides and analyzed using LC–MS revealing that 78% and 84% of dA bases were methylated in the single-stranded and duplex substrates, respectively (Figure 4B and C).
Figure 4.
M.EcoGII methylates single-stranded and duplex DNA substrates with near equivalent efficiency in vitro. (A) Radioactive assay of M.EcoGII activity using double- and single-stranded oligonucleotide substrates (2μM) containing a single dA nucleotide. Assays used 50mM HEPES buffer, pH 7.0, 0.1 mM EDTA and 3.7 μM [3H-SAM] and were incubated for 10 min at 37°C. (B) and (C) M.EcoGII methylation of single- and double-stranded M13mp18 bacteriophage DNAs in vitro and quantitation of m6dA content using LC–MS. DNA samples (2 μg) were methylated in vitro using M.EcoGII (2 μM). M = 1 kb-Extend DNA Ladder 0.5-to 48.5 kb (NEB).
Additional experiments using the radiometric assay provided evidence that M.EcoGII can also methylate DNA–RNA hybrid and ssRNA substrates in vitro (Figure 5A). To confirm these activities we incubated M.EcoGII with a 48 base-pair oligonucleotide duplex containing 10 dA residues and 18 rA residues in the DNA and RNA strands, respectively. LC–MS analyses of the reaction products revealed that 43% of total dA bases were converted to m6dA and 10% of total rA bases were in the form of m6rA (Figure 5B). To quantify activity of M.EcoGII on ssRNA we prepared a 1.7 kb transcript that encodes the 5′-end of a luciferase mRNA using in vitro transcription. The M.EcoGII-treated RNA sample was degraded to nucleosides and duplicate samples were analyzed (using the LC–MS assay) revealing that >32% of rA bases were converted to m6rA (Figure 5C). M.EcoGII, therefore, appears to methylate RNA substrates less efficiently than duplex DNA. This likely reflects suboptimal enzyme substrate interactions arising from the conformational heterogeneity of single-stranded RNA - and potential for forming stable secondary structures - compared to duplex DNA.
Figure 5.
Activity of M.EcoGII on RNA:DNA hybrid duplex and single-stranded RNA substrates in vitro. (A) Radioactive assays of M.EcoGII activity using 27mer ssRNA, ssDNA, dsDNA and RNA:DNA hybrid substrates (2 μM) in vitro. Assays used 50mM HEPES buffer, pH 7.0, 0.1mM EDTA and 3.7 μM [3H-SAM] and were incubated for 10 min at 37°C. (B) Methylation of a synthetic 48mer DNA:RNA hybrid oligonucleotide substrate (0.5 μM) containing 10 dA bases in the DNA strand and 18 rA bases in the RNA strand, Assay used 50 mM HEPES buffer pH 7.0, 1.0 mM EDTA, 320 μM SAM and M.EcoGII (1 μM). After incubation at 37°C for 60 minutes m6dA and m6rA products were quantified using LC–MS. (C) M.EcoGII-methylation of a 1.7 kb in vitro transcribed RNA (5 μg) followed by agarose gel electrophoresis and analysis of m6rA content using LC–MS. Reaction conditions were identical to those used with the DNA:RNA hybrid substrate but incubation at 37°C was extended to 120 min. M = 1 kb-DNA Ladder 0.5-to 10.0 kb (NEB).
Bioinformatic analyses
M.EcoGII, is a member of a very large family of DNA MTases found in different strains of E. coli and located in what appears to be a well-defined prophage. It is extremely similar (333 of 349 amino acids are identical) to M.EcoGI, which also appears to be encoded on a prophage in the same strain. A very similar MTase is found in the E. coli phage 1720A-02 and also in a number of known Enterobacterial phages. A few representative examples are listed in Supplementary Table S1.
The sequences of M.EcoGII, the close homolog from the same strain (M.EcoGI), and the additional homologs listed in Supplementary Table S1 appear to be typical examples of the β-class of DNA methyltransferases which contain a canonical N-terminal catalytic (‘DPPY’) motif followed by a centrally-located target recognition domain (TRD) and a C-terminal S-adenosylmethionine-binding (‘FxGxG’) motif. We do not find evidence of additional domains or novel sequence elements in M.EcoGII that might be potentially associated with non-specific methylation of dA and rA bases and are skeptical that any need be invoked. This is because we assume that—in strictly energetic terms—base-flipping to present an extrahelical adenine to a MTase active site is no more challenging if that adenine be present at the fifth position of a PstI site (CTGCAG), the second position of a dam site (GATC), or a solitary A in any sequence context as is the case with M.EcoGII. We also note that—in contrast to M.EcoGII and M.EcoGI—the promiscuous adenine MTases described previously by Drozdz et al. (15) are of the α-class wherein the motif sequences are reversed (i.e. N-terminal ‘FxGxG’, central ‘TRD’ and C-terminal ‘DPPY’), demonstrating that two distinct enzyme architectures can catalyze promiscuous methylation of dA bases in DNA.
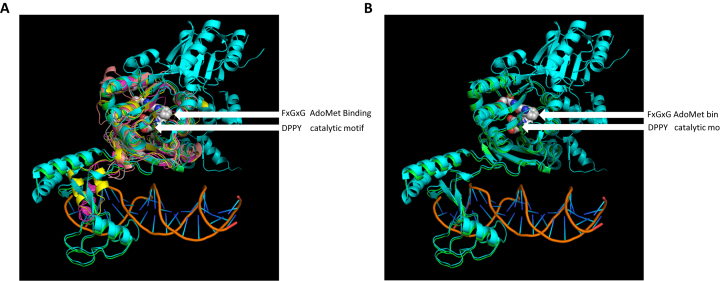
The structures of five β-class MTases, M1.MboII, M.RsrI, M.PvuII, M.HpyAVI and EcoP15I modA have been determined by X-ray crystallography but only the latter was determined in the presence of a DNA substrate. We therefore used PHYRE2 to thread the M.EcoGII sequence onto the known MTase structures with the various models being aligned to the M.EcoP15I-DNA complex. In all cases the M.EcoGII core domain, including the canonical FxGxG (AdoMet-binding) and DPPY (catalytic) motifs, model well (Figure 6, panel A, center). In the context of the putative target recognition domain (Figure 6, panel A, lower left) structural models were only predicted using the M.RsrI, M1.MboII and M.EcoP15I structural templates. Of these, only the M.EcoGII model derived from threading using the M.EcoP15I template predicts significant structural concordance in the form of conserved secondary structural elements (Figure 6, panel B). As a consequence, we believe that the molecular details of M.EcoGII function are best addressed by X-ray crystallography. However, the M.EcoGII crystals that we have prepared to date are of insufficient quality to permit structure determination.
Figure 6.
Predictions of M.EcoGII structure using PHYRE2 analyses. (A) Alignment of predicted structural models for M.EcoGII to the M.EcoP15I:DNA structure (PDB: 4zcf). EcoP15I_ModA (cyan) and the DNA are experimentally determined structures. The various predicted M.EcoGII structure models were derived from threading onto available MTase structures as follows: green - EcoP15I_ModA (pdb:4zcf, CAGCAG); magenta - M1.MboII (pdb:1g60, GAAGA); yellow—M.RsrI (pdb:1nw6, GAATTC); wheat—M.PvuII (pdb:1boo, CAGCTG); gray—M.HpyAVI (pdb:5hfj, GAGG). The AdoMet binding residues (FxGxG motif) and catalytic residues (DPPY motif) of the M.EcoGII models are shown as white, red and blue spheres. Note lack of consensus among the predicted structures for the putative DNA binding/TRD domain (lower left, contacting DNA) versus the high degree of consensus for the methyltransferase structural core (upper middle). (B) Alignment of the EcoP15I_ModA:DNA crystal structure and the predicted M.EcoGII structure derived from threading. The experimentally-determined TRD domain of EcoP15I_ModA (cyan) and the predicted M.EcoGII model (green) appear to be similar with several secondary structural elements in common.
It appears likely that these phage encoded MTases are being exploited to overcome host restriction systems during infections, which may account for their widespread distribution. However, once the phage becomes lysogenic in its host, the MTase appears to be rendered transcriptionally silent as judged by the failure to detect extensive methylation of the chromosome in the native strain from which it was originally isolated. Presumably, this is to avoid excessive methylation of the host chromosome, which might be expected to interfere with transcription and replication. Indeed, E. coli strains overexpressing M.EcoGII on a plasmid grow more slowly than the parent strain lacking the plasmid and its encoded MTase.
DISCUSSION
In this work we describe the enzyme M.EcoGII which appears to be the first truly non-specific adenine DNA methyltransferase. The enzyme is able to methylate dA residues in any sequence context as demonstrated by inhibition of multiple restriction endonucleases with known methylation sensitivities. Using supercoiled plasmid DNA, we demonstrate—using LC–MS analyses of methylated DNA products—that up to 96% of dA bases can be methylated in vitro, increasing to 99% for linearized equivalent (i.e. relaxed) plasmid substrates. We find that M.EcoGII methylates single-stranded oligonucleotide and M13 mp18 virion DNA substrates almost as efficiently as equal quantities of double-stranded oligonucleotide or M13 mp18 RF DNA. Finally, we show that M.EcoGII has robust activity on single-stranded RNA substrates prepared using in vitro transcription and on both strands of RNA/DNA hybrid oligonucleotide substrates, albeit to a lesser extent than what is achievable using single-stranded or duplex DNA.
While, in vitro, we have been able to achieve very high levels of methylation, in an in vivo context it is likely that even partial methylation is more than sufficient for a phage to overcome host restriction systems. The strategy of completely methylating one or more of the bases in a phage DNA genome, has been described previously, including the well-known case of T-even phages, which incorporate 5-hydroxymethylcytosine into their genome during replication and then further decorate it with glucosyl residues following replication (20,21). There are some phages infecting many different genera of bacteria, which use a variety of different modifications, presumably also to overcome restriction barriers (22,23). Xanthomonas phage XP12 has only C5-methylcytosine in its genome, although these are incorporated during replication (24). Other phages frequently pick up individual MTases such as the GATC MTase and others with more specific recognition sequences (25,26). The phenomenon is especially common among mycobacteriophages, which frequently have a variety of different DNA MTases in their genomes (13,27).
One interesting finding is that neither M.EcoGI nor M.EcoGII is expressed in the genome of E. coli strain, O104:H4 C227–11 during normal growth. At the time it was thought that they might just be following the usual practice of prophage genes, which tend not to be expressed while the prophage is in its lysogenic state, but get turned on when the prophage excises. In this case, our attempts to obtain constitutive expression of M.EcoGII, and the subsequent deleterious effects on the health of the cell, indicate why its expression in the genome is switched off. It could have a very adverse effect on growth of the cell and so is likely just switched on as the prophage is ready to excise and launch a new infection.
The fact that M.EcoGII is non-specific, immediately suggests that this property might be exploited in a variety of different situations. For instance, restriction barriers are very common in bacteria and archaea, but often the precise specificity of the restriction system is unknown. Since many systems employ Type I and III restriction enzymes as the principal barrier to control against infection, a non-specific dA MTase would be most useful. This is because in most Type I and III systems, protection against the action of the restriction enzyme is achieved by dA methylation (13). In principle, many ‘difficult-to-transform’ strains of bacteria reported in the literature might be rendered amenable to genetic manipulation. In practice, however, this appears not to be the case because M.EcoGII-catalyzed methylation of plasmid DNAs inhibits bacterial transformation. For example, using competent cells prepared by calcium chloride treatment, we find that M.EcoGII-modified pBR322 DNA (containing 80% m6dA) transforms non-restricting laboratory strains of E. coli 4–5 orders of magnitude less efficiently than dam-methylated pBR322 control DNA.
Another potential application for this MTase will be in protection experiments to detect the binding sites for proteins that interact with DNA and RNA substrates. In the former case, less specific DNA MTases have been used for this purpose (28), but that of course greatly limits the accuracy with which the binding site can be defined.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Brian Anton, Bill Jack and Geoff Wilson for careful reading of the manuscript; Katherine Marks, Mala Samaranayake and Ashley Luck for helpful discussions regarding purification and functional analyses of M.EcoGII, Colleen McClung for native MS analysis of the purified M.EcoGII enzyme and Dr Matt Waldor for providing the E. coli DNA that was the original source of the M.EcoGII gene.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
New England Biolabs; National Institutes of Health [R44 GM100560 to R.J.R., R01 GM049245 to X.C.]. Funding for open access charge: New England BioLabs Inc., Ipswich, MA, USA.
Conflict of interest statement. Some of the authors (I.A.M., R.D.M., Y.L., A.F., I.C., N.D., R.J.R.) are full time employees of New England Biolabs, a company that sells research reagents such as DNA methyltransferases.
REFERENCES
- 1. Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. ChemBioChem. 2002; 3:274–293. [DOI] [PubMed] [Google Scholar]
- 2. Loenen W.A., Dryden D.T., Raleigh E.A., Wilson G.G., Murray N.E.. Highlights of the DNA cutters: a short history of the restriction enzymes. Nucleic Acids Res. 2014; 42:3–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marinus M.G., Casadesus J.. Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol. Rev. 2009; 33:488–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reisenauer A., Kahng L.S., McCollum S., Shapiro L.. Bacterial DNA methylation: a cell cycle regulator. J. Bacteriol. 1999; 181:5135–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shell S.S., Prestwich E.G., Baek S-H., Shah R.R., Sassetti C. M., Dedon P.C., Fortune S.M.. DNA methylation impacts gene expression and ensures hypoxic survival of Mycobacterium tuberculosis. PLoS Pathog. 2013; 9:e1003419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hattman S., Malygin E.G.. Bacteriophage T2Dam and T4Dam DNA-[N6-adenine]-methyltransferases. Prog. Nucleic Res. Mol. Biol. 2004; 77:67–126. [DOI] [PubMed] [Google Scholar]
- 7. Posfai J., Bhagwat A.S., Posfai G., Roberts R.J.. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989; 17:2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klimasauskas S., Timinskas A., Menkevicius S., Butkiene D., Butkus V., Janulaitis A.. Sequence motifs characteristic of DNA [cytosine-N4] methylases: similarity to adenine and cytosine-C5 DNA-methylases. Nucleic Acids Res. 1989; 17:9823–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eid J., Fehr A., Gray J., Luong K., Lyle J., Otto G., Peluso P., Rank D., Baybayan P., Bettman B. et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009; 323:133–138. [DOI] [PubMed] [Google Scholar]
- 10. Flusberg B.A., Webster D.R., Lee J.H., Travers K.J., Olivares E.C., Clark T.A., Korlach J., Turner S.W.. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods. 2010; 7:461–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clark T.A., Murray I.A., Morgan R.D., Kislyuk A.O., Spittle K.E., Boitano M., Fomenkov A., Roberts R.J., Korlach J.. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 2012; 40:e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Murray I.A., Clark T.A., Morgan R.D., Boitano M., Anton B.P., Luong K., Fomenkov A., Turner S.W., Korlach J., Roberts R.J.. The methylomes of six bacteria. Nucleic Acids Res. 2012; 40:11450–11462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roberts R.J., Vincze T., Posfai J., Macelis D.. REBASE – a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2015; 43:D298–D299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fang G., Munera D., Friedman D.I., Mandlik A., Chao M.C., Banerjee O., Feng Z., Losic B., Mahajan M.C., Jabado O.J. et al. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nature Biotech. 2012; 30:1232–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Drozdz M., Piekarowicz A., Bujnicki J.M., Radlinska M.. Novel non-specific DNA adenine methyltransferases. Nucleic Acids Res. 2012; 40:2119–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guzman L-C, Belin D., Carson M.J., Beckwith J.. Tight regulation, modulation and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995; 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kong H., Lin L.F., Porter N., Stickel S., Byrd D., Posfai J., Roberts R.J.. Functional analysis of putative restriction-modification system genes in the Helicobacter pylori J99 genome. Nucleic Acids Res. 2000; 28:3216–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hashimoto H., Pais J.E., Zhang X., Saleh L., Fu Z.Q., Dai N., Correa I.R. Jr, Zheng Y., Cheng X.. Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature. 2014; 506:391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kelley L.A., Mezulis S., Yates C. M., Wass M.N., Sternberg M.J.E.. The Phyre2 web portal for protein modelling, prediction and analysis. Nat. Protoc. 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hattman S. DNA methylation of T-even bacteriophages and of their non-glucosylated mutants: its role in P1-directed restriction. Virology. 1970; 42:367–359. [DOI] [PubMed] [Google Scholar]
- 21. Kuo T.-T., Huang T.C., Teng M.H.. 5-Methylcytosine replacing cytosine in the deoxyribonucleic acid of a bacteriophage for Xanthomonas oryzae. J. Mol. Biol. 1968; 34:373–375. [DOI] [PubMed] [Google Scholar]
- 22. Warren R.A.J. Modified bases in bacteriophage DNA. Ann. Rev. Microbiol. 1980; 34:137–158. [DOI] [PubMed] [Google Scholar]
- 23. Weigele P., Raleigh E.A.. Biosynthesis and unction of modified bases in bacteria and their viruses. Chem. Rev. 2016; 116:12655–12687. [DOI] [PubMed] [Google Scholar]
- 24. Kuo T.-T., Tu J.. Enzymatic synthesis of deoxy-5-methyl-cytidylic acid replacing deoxycytidylic acid in Xanthomonas oryzae phage Xp12 DNA. Nature. 1976; 26:615. [DOI] [PubMed] [Google Scholar]
- 25. Murphy J., Mahony J., Ainsworth S., Nauta A., van Sinderen D.. Bacteriophage orphan DNA methyltransferases: insights from their bacterial origin, function and occurrence. Appl. Environ. Micro. 2013; 79:7547–7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gunthert U., Trautner T.A.. DNA methyltransferases of Bacillus subtilis and its bacteriophages. Curr. Top. Microbiol. Immunol. 1984; 108:11–22. [DOI] [PubMed] [Google Scholar]
- 27. Hatfull G.F. Molecular genetics of mycobacteriophages. Microbiol. Spectr. 2014; 2:1–36. [PMC free article] [PubMed] [Google Scholar]
- 28. Hoose S.A., Kladde M.P.. DNA methyltransferase probing of DNA-protein interactions. Methods Mol. Biol. 2006; 338:225–244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.