Abstract
H/ACA ribonucleoproteins (H/ACA RNPs) are responsible for introducing many pseudouridines into RNAs, but are also involved in other cellular functions. Utilizing a purified and reconstituted yeast H/ACA RNP system that is active in pseudouridine formation under physiological conditions, we describe here the quantitative characterization of H/ACA RNP formation and function. This analysis reveals a surprisingly tight interaction of H/ACA guide RNA with the Cbf5p–Nop10p–Gar1p trimeric protein complex whereas Nhp2p binds comparably weakly to H/ACA guide RNA. Substrate RNA is bound to H/ACA RNPs with nanomolar affinity which correlates with the GC content in the guide-substrate RNA base pairing. Both Nhp2p and the conserved Box ACA element in guide RNA are required for efficient pseudouridine formation, but not for guide RNA or substrate RNA binding. These results suggest that Nhp2p and the Box ACA motif indirectly facilitate loading of the substrate RNA in the catalytic site of Cbf5p by correctly positioning the upper and lower parts of the H/ACA guide RNA on the H/ACA proteins. In summary, this study provides detailed insight into the molecular mechanism of H/ACA RNPs.
INTRODUCTION
H/ACA ribonucleoproteins (RNPs) are critically involved in many important cellular processes in archaea and eukaryotes ranging from ribosome biogenesis to spliceosome formation, mRNA modification and telomere maintenance (1). These particles are best known for their function as complex pseudouridine synthases that modify ribosomal RNA (rRNA) and small nuclear RNAs (snRNAs) (2,3), but have recently also been shown to target mRNAs, small nucleolar RNAs (snoRNAs) and other non-coding RNAs including long-noncoding RNAs (4,5). The H/ACA RNPs modifying rRNA are found in the nucleolus and contain so-called small nucleolar RNAs (snoRNAs), whereas modification of snRNAs occurs in the Cajal bodies and is mediated by small Cajal body-specific RNAs (scaRNAs) (6). In yeast, snoRNAs pseudouridylating rRNA represent the vast majority of all H/ACA RNPs, but in humans many more H/ACA guide RNAs have been identified (6). It is not known yet whether specific H/ACA RNAs are directing the pseudouridylation of mRNAs, snoRNAs and long-noncoding RNAs. In addition to this catalytic function, H/ACA snoRNPs containing the only essential snoRNA (snR30) in Saccharomyces cerevisiae contribute to ribosomal RNA processing (7). Mammalian telomerase RNPs also contain H/ACA proteins that bind to and stabilize the 3′ end of the telomerase RNA component (TERC) (1). The cellular role of so-called ‘orphan’ H/ACA RNAs has not been identified.
H/ACA RNPs are comprised of four different proteins, the catalytic protein Cbf5p and proteins Nop10p, Gar1p and Nhp2p in addition to one of many H/ACA guide RNAs (8) (Figure 1). Eukaryotic H/ACA guide RNAs are characterized by two conserved motifs, the Box H motif (hinge—ANANNA) and the Box ACA motif, that are each located immediately downstream of the first and second hairpin within the guide RNA, respectively (9). The two hairpins both contain an internal bulge called pseudouridylation pocket that base-pairs with target RNA thereby selecting the target uridine for modification (10,11). The critical cellular role of H/ACA RNPs is evident in the fact that mutations in the human genes for H/ACA proteins Cbf5p, Nop10p and Nhp2p cause an inherited disease, Dyskeratosis congenita, which presents typically with symptoms of bone-marrow failure, premature aging, increased predisposition to cancer and nail dystrophy (12). Dyskeratosis congenita has been linked to impaired telomere maintenance, presumably through the interaction of the H/ACA proteins with TERC (13,14); however, there is evidence that ribosome function is also affected due to reduced pseudouridine formation in rRNA (15,16).
Figure 1.
Schematic representation of the snR34 H/ACA RNP. One set of H/ACA core proteins (Cbf5p, Nop10p, Gar1p and Nhp2p) is believed to bind to each hairpin of the H/ACA RNA. The sequences of the conserved Box H downstream of the 5′ hairpin and the conserved Box ACA downstream of the 3′ hairpin in snR34 are indicated. Shown in the pseudouridylation pocket is the sequence of the snR34 RNA bound to short substrate RNAs used in this study. In vivo, snR34 directs pseudouridine formation at position U2826 and U2880 in 25S rRNA. The 5′ substrate is defined as the substrate targeted by the 5′ pseudouridylation pocket (left) and the 3′ substrate is defined similarly (right). The uridine that is converted to pseudouridine by Cbf5p is represented in bold.
While the cellular function of H/ACA RNPs has been well studied, only limited insight into the molecular mechanism of these RNPs has been obtained. Most investigations have focused on archaeal H/ACA small RNPs (sRNPs) which are amenable to large-scale purification, functional assays and crystallization (17,18). Historically, the eukaryotic H/ACA RNPs have been difficult to study biochemically due to protein instability (19). But in 2011, the purification and reconstitution of an active eukaryotic H/ACA RNP complex has been reported together with the crystal structure of the Cbf5p–Nop10p–Gar1p trimer (20). Interestingly, the characterization of pseudouridine formation by this H/ACA RNP complex revealed that both hairpins of the guide RNA are required for optimal activity although archaeal H/ACA sRNPs are functional with a single-hairpin guide RNA. Furthermore, the authors reported that Nhp2p is dispensable for the activity of H/ACA RNP which is surprising given that Nhp2p is an essential protein and that Nhp2p depletion leads to an overall degradation of guide RNA (21,22).
Importantly, several critical differences are evident between archaeal and eukaryotic H/ACA RNPs. First, archaea utilize a wide variety of H/ACA guide RNAs containing one to three hairpins (23,24) whereas most eukaryotes have strictly conserved guide RNAs with two hairpins (25–27). Second, archaeal guide RNAs contain a kink-turn or kink-loop motif which is specifically recognized with low nanomolar affinity by the protein L7Ae (28). In contrast, the eukaryotic protein Nhp2p, a homolog of L7Ae, does not display any kink-turn specificity (21) and, accordingly, eukaryotic H/ACA guide RNAs do not contain this motif. Third, the overall three-dimensional fold of archaeal and eukaryotic H/ACA proteins is the same (20,29), but there is significant deviation on the sequence level including the residues that are altered in Dyskeratosis congenita. For instance, eukaryotic H/ACA proteins are characterized by N- and C-terminal extensions which are predicted to be unstructured and are often rich in Lysine and Glutamate residues (Cbf5p) or Glycines and Arginines (Gar1p). At last, eukaryotic H/ACA RNPs serve many more cellular roles than only pseudouridine formation such as rRNA processing and stabilization of TERC. Together with the implication of H/ACA RNPs in human diseases, it is therefore of high interest to conduct a detailed biochemical investigation of eukaryotic H/ACA RNPs to better understand their mechanism of action in the different cellular pathways.
Here, we reconstituted H/ACA RNPs from purified components and demonstrate that this complex is highly active in pseudouridine formation under physiological conditions. In addition to measuring RNA modification, we also quantitatively assess the association of H/ACA proteins with guide RNA and the binding of substrate RNA to fully reconstituted H/ACA RNPs. This comprehensive approach allows us to reveal important aspects of H/ACA RNP assembly and activity such as extremely tight binding of guide RNA to the proteins in this complex. The protein Nhp2p and the conserved Box H and Box ACA elements are important for the function of H/ACA RNP presumably by anchoring the upper and lower regions of the guide RNA hairpins and thereby helping to position the target uridine of the substrate RNA in the catalytic pocket of the Cbf5p protein.
MATERIALS AND METHODS
Materials
[C5–3H]-UTP for in vitro transcriptions was purchased from Moravek Biochemicals, affinity chromatography from GE Healthcare, and oligonucleotides from Integrated DNA Technologies (IDT). All other enzymes and chemicals were obtained from Fisher Scientific.
Molecular cloning and site-directed mutagenesis
CBF5 and NOP10 genes from S. cerevisiae were codon optimized for expression in Escherichia coli, synthesized (Genewiz) and cloned into pETDuet-1 expression vector using restriction sites NcoI and EcoRI for CBF5 and BglII and XhoI for NOP10. A sequence encoding a C-terminal hexahistidine tag was added directly to the CBF5 sequence prior to the stop codon (20). The GAR1 and NHP2 genes were amplified from S. cerevisiae genomic DNA. GAR1 was cloned into a pGEX-5x-3 vector using the BamHI and EcoRI restriction sites to include the GST-tag at the N-terminus. NHP2 was cloned into a pET28a vector using restriction sites NheI and BamHI encoding an N-terminal hexahistidine tag. The snR34 guide RNA was amplified from S. cerevisiae genomic DNA and cloned into a pUC19 vector using the SmaI restriction site. Site-directed Quikchange mutagenesis was used to generate CBF5 D95N and to mutate the Box H and Box ACA elements. The snR5 gene together with the T7 promoter was synthesized and inserted into pUC57 (Genewiz). The gene for Pyrococcus furiousus Pf4 guide RNA including a T7 promoter was assembled from oligonucleotides and cloned in to pUC19. All plasmids were verified by sequencing (Genewiz).
Protein expression and purification
Cbf5p and Nop10p were overexpressed in E. coli BL21 DE3 cells (NEB) at 18°C whereas Gar1p and Nhp2p were independently overexpressed in E. coli Rosetta 2 BL21 DE3 pLysS (NEB) at 37°C. Cells were grown in LB medium, and protein expression was induced at an OD600 of 0.6 with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Cells were centrifuged at 5000 g for 30 min, flash frozen and stored at −80°C until further use.
Cells expressing Cbf5p and Nop10p were mixed with Gar1p-expressing cells and resuspended in 10 ml/g Buffer A (50 mM Tris–HCl pH 8.0, 1 M NaCl, 5% (v/v) glycerol, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM β-mercaptoethanol). Cells were lysed by sonication (intensity level 6, duty cycle 60%, Branson Sonifier) and then centrifuged for 45 min at 30 000 g at 4°C. The soluble cell lysate was transferred to Glutathione Sepharose resin followed by incubation for 1 h on ice. The resin was washed with Buffer A (excluding PMSF) and then incubated with Buffer A supplemented with 10 mM reduced L-glutathione to elute the proteins. Eluates were transferred to Ni2+ Sepharose resin and incubated for 1 h followed by extensive washes with Buffer B (50 mM Tris–HCl pH 8.0, 20% (v/v) glycerol, 1 mM β-mercaptoethanol, 20 mM imidazole) and elution with Buffer B containing 300 mM imidazole. Proteins were shock frozen and stored at −80°C. P. furiosus Cbf5p–Nop10p–Gar1p was purified as previously described (30).
Cells containing Nhp2p were resuspended in 5 ml/g Buffer C (50 mM Tris–HCl pH 8.0, 400 mM KCl, 0.1 mM PMSF, 5 mM β-mercaptoethanol, 5% (v/v) glycerol). Cells were lysed with egg white lysozyme (1 mg/ml) followed by the addition of sodium deoxycholate (12.5 mg/g cells). Cell lysis was completed by sonication followed by centrifugation (vide supra). Soluble cell lysate was transferred to Ni2+ Sepharose resin and incubated on ice for 1 h followed by washes with Buffer C containing 20 mM imidazole. Nhp2p was eluted with Buffer C containing 300 mM imidazole and diluted to contain 150 mM salt with Buffer C excluding KCl. Nhp2p was further purified by cation exchange chromatography (SP Sepharose) using a linear gradient from Buffer D (40 mM HEPES-KOH pH 8.0, 150 mM NaCl, 0.2 mM ethylenediaminetetraacetic acid (EDTA), 3 mM MgCl2, 2% (v/v) glycerol, 3 mM β-mercaptoethanol) to Buffer D containing 1 M NaCl. Nhp2p-containing elutions were diluted to 300 mM NaCl with Buffer D without NaCl and concentrated by ultrafiltration. Nhp2p was diluted 1:1 with glycerol and stored in −20°C.
In vitro transcription and purification of guide and substrate RNAs
The gene encoding guide RNAs (snR34, snR5, Pf4 and variants thereof) were polymerase chain reaction (PCR)-amplified including a T7 promoter (for primers, see Supplementary Table S1). For variants of the snR34 5′ hairpin, the template DNA for in vitro transcription was assembled from two partially complementary oligos in a PCR reaction. RNAs were in vitro transcribed as described (31) and purified from urea-PAGE with the Elutrap Electroelution System (Whatman) followed by ethanol precipitation. All RNA concentrations were determined by spectroscopy (A260) using extinction coefficients calculated by OligoAnalyzer 3.1 (IDT).
Substrate RNAs complementary to the snR34 pseudouridylation pockets in the 5′ and 3′ hairpin (termed 5′ and 3′ substrate, respectively) were generated by in vitro transcription using annealed oligonucleotides as template in the presence of [C5–3H]-UTP. Substrate RNA was purified with a Nucleobond AX100 column (Macherey and Nagel). Following RNA binding, the column was washed with 100 mM Tris-acetate (pH 6.3), 10 mM MgCl2, 15% ethanol, 300 mM KCl. The RNA was eluted in the same buffer containing 1150 mM KCl followed by ethanol precipitation. Scintillation counting and A260 measurements were used to quantify the RNA concentration and specific activity.
H/ACA RNP reconstitution
Guide RNAs were refolded by slowly cooling from 65°C to room temperature in Reaction Buffer (20 mM HEPES-KOH (pH 7.4), 150 mM NaCl, 0.1 mM EDTA, 1.5 mM MgCl2, 10% (v/v) glycerol, 0.75 mM dithiothreitol (DTT)) and incubated with H/ACA proteins in Reaction Buffer in a 0.9:1 or 0.45:1 ratio of guide RNA:protein for single- or dual-hairpin guide RNAs, respectively. Proteins and RNA were incubated 10 min at 30°C to allow for complex formation. Where indicated, 400 nM reconstituted H/ACA RNP was incubated with Factor Xa (0.01 U/μl, Novagen) in Reaction Buffer containing 1 mM CaCl2 for 30 min at 30°C, and cleavage of GST-Gar1p was confirmed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis.
Nitrocellulose filtration assays
Most RNAs used for filter binding were end-labeled with [32P] using common procedures. In short, RNA was dephosphorylated with Calf Intestinal Alkaline Phosphatase (NEB, 0.1 U/μl), rephosphorylated with T4 polynucleotide kinase (0.5 U/μl) in the presence of [γ-32P]ATP (5–10 μCi) and contaminating [γ-32P]ATP was removed by using a SigmaPrep spin column (Sigma-Aldrich) containing Sephadex G-25 resin or using an EZ-10 Spin Column RNA Cleanup and Concentration Kit (Bio Basic Canada). Pf4 H/ACA sRNA was transcribed in the presence of [C5–3H]-UTP.
Refolded guide RNA (0.1 or 0.05 nM for variants (Supplementary Figure S3)) was incubated with excess concentrations of proteins in Reaction Buffer for 10 min at 30°C, and the amount of RNA–protein complex formed was determined as described before (31). Binding assays with [3H]-substrate RNAs were performed by incubating increasing concentrations of substrate in the presence of 5 nM H/ACA RNPs for 3 min at 30°C.
For guide RNA binding, the dissociation constant was obtained by fitting to a quadratic equation with [RNA] = 0.1 or 0.05 nM:
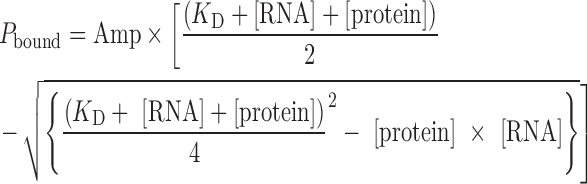 |
where, Pbound is the percentage of bound RNA and Amp is the amplitude or final level of bound RNA.
Dissociation constants (KD) for substrate RNAs were determined by fitting the binding curves to a hyperbolic function in GraphPad Prism:
 |
where, [S] is the substrate RNA concentration and Bmax is the maximum binding.
The dissociation of guide RNA from H/ACA proteins was monitored by forming H/ACA RNP complex with 0.8 nM [32P]-labeled guide RNA and 2.5 nM H/ACA proteins which is then chased by the addition of excess unlabeled guide RNA (75 nM) followed by nitrocellulose filtration at several reaction time points. Data were fit with a single-exponential decay function:
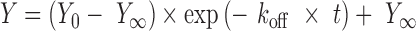 |
with Y∞ being the end level and Y0 starting level, t the time and koff the dissociation rate constant.
All experiments were repeated in duplicates or triplicates. For each individual dataset, the KD or koff was determined. These were then used to calculate average KD or koff values and the corresponding standard deviations as reported in Tables 1 and 2.
Table 1. Binding of guide RNA to H/ACA proteins forming an H/ACA RNP complex.
| Guide RNA | Protein(s) | K D, nM | k off, min−1 | t1/2, min |
|---|---|---|---|---|
| snR34 | Cbf5p–Nop10p–Gar1p–Nhp2p | 0.5 ± 0.2 | 0.13 ± 0.06 | 5.5 |
| snR5 | Cbf5p–Nop10p–Gar1p–Nhp2p | 0.7 ± 0.3 | 0.27 ± 0.08 | 3 |
| Pf4 | aCbf5p–aNop10p–aGar1p | 23 ± 6 | ||
| snR34 | Nhp2p | 600 ± 200 | ||
| snR34 | Cbf5p–Nop10p–Gar1p | 0.3 ± 0.1 | ||
| snR5 | Cbf5p–Nop10p–Gar1p | 0.6 ± 0.2 | ||
| snR34 Δ Box H | Cbf5p–Nop10p–Gar1p–Nhp2p | 0.6 ± 0.2 | ||
| snR34 Δ Box ACA | Cbf5p–Nop10p–Gar1p–Nhp2p | 0.7 ± 0.3 | ||
| snR34 5′ hairpin (HP) | Cbf5p–Nop10p–Gar1p–Nhp2p | 1.5 ± 0.8 | ||
| snR34 5′ HP Δ Box H | Cbf5p–Nop10p–Gar1p–Nhp2p | 0.5 ± 0.2 | ||
| snR34 5′ HP no 5′ extension | Cbf5p–Nop10p–Gar1p–Nhp2p | 1.0 ± 0.3 | ||
| snR34 5′ HP no pocket | Cbf5p–Nop10p–Gar1p–Nhp2p | 2.6 ± 0.8 | ||
| snR34 5′ HP small pocket | Cbf5p–Nop10p–Gar1p–Nhp2p | 1.6 ± 0.5 | ||
| snR34 5′ HP no lower stem | Cbf5p–Nop10p–Gar1p–Nhp2p | 1.6 ± 0.7 | ||
| snR34 5′ HP extended stem | Cbf5p–Nop10p–Gar1p–Nhp2p | 1.3 ± 0.5 | ||
| CrPV IRES | Cbf5p–Nop10p–Gar1p–Nhp2p | 0.1 ± 0.1 |
The dissociation constants (KD) and the dissociation rate constants (koff) were determined by nitrocellulose filtration and are stated with the corresponding standard deviation (Figures 3, 5, 6 and Supplementary Figure S3). The half-life time of the complex (t1/2) was calculated from the dissociation rate constant.
Table 2. Affinity of substrate RNAs binding to H/ACA RNPs.
| Guide RNA | Substrate RNA | Proteins | K D, nM |
|---|---|---|---|
| snR34 | 5′ substrate | Cbf5p–Nop10p–Gar1p–Nhp2p | 53 ± 22 |
| snR34 | 3′ substrate | Cbf5p–Nop10p–Gar1p–Nhp2p | 100 ± 30 |
| snR5 | 5′ substrate | Cbf5p–Nop10p–Gar1p–Nhp2p | 160 ± 50 |
| snR5 | 3′ substrate | Cbf5p–Nop10p–Gar1p–Nhp2p | 330 ± 70 |
| snR34 | 5′ substrate | Cbf5p–Nop10p–Gar1p | 50 ± 20 |
| snR34 | 3′ substrate | Cbf5p–Nop10p–Gar1p | 90 ± 20 |
| snR34 Δ H box | 3′ substrate | Cbf5p–Nop10p–Gar1p–Nhp2p | 90 ± 30 |
| snR34 Δ ACA box | 3′ substrate | Cbf5p–Nop10p–Gar1p–Nhp2p | 150 ± 30 |
The dissociation constants (KD) of substrate RNA binding to reconstituted H/ACA RNPs was determined by nitrocellulose filtration (Figure 4).
Tritium release assays
Pseudouridylation catalysis by H/ACA RNPs was monitored using a tritium release assay as previously described (31). H/ACA RNPs were reconstituted as described above. Multiple turnover assays were performed with 10-fold excess substrate over enzyme (45–150 nM) at 30°C; unless indicated differently, assays contained 50 nM H/ACA RNP and 500 nM substrate RNA. All assays have been performed in duplicates or triplicates.
RESULTS
In order to characterize the molecular mechanism of S. cerevisiae H/ACA RNPs, these complexes were reconstituted from purified proteins and in vitro transcribed snR34 guide RNA, which is a homolog of human H/ACA snoRNA U65 (32), as well as snR5, which has been used previously (20) (Figure 1 and Supplementary Figure S1). Small fragments of 25S rRNA corresponding to both target sites of snR34 and snR5 were transcribed using [C5–3H]UTP to generate radioactively labeled substrate RNA allowing observation of pseudouridine formation using a tritium release assay (Supplementary Figure S1).
Saccharomyces cerevisiae H/ACA RNPs were formed by mixing all four H/ACA proteins and snR34 guide RNA, and time courses of pseudouridine formation were recorded for both short substrate RNAs (called 5′ and 3′ substrate) being recognized by the 5′ hairpin of snR34 and the 3′ hairpin of snR34, respectively (Figure 2). For both substrates, efficient and comparable multiple-turnover pseudouridine formation was observed demonstrating the reconstituted H/ACA RNP in vitro system is highly active. Several additional control experiments verified that the presence of the GST-tag on Gar1p does not influence the pseudouridylation activity and that pseudouridine formation depends on the presence of the catalytic Asp95 residue in Cbf5p (Supplementary Figure S2A and B). Notably, the activity of the complex was significantly higher in a buffer containing 150 mM NaCl than in a previously reported buffer with 500 mM NaCl (20) (Supplementary Figure S2B). Also, only the target uridine in substrate RNA is converted to pseudouridine as the same level of tritium release was observed for both the natural substrate RNA containing four uridines and for a substrate variant containing only a single uridine at the target position (Supplementary Figure S2C). Lastly, pseudouridine formation is significantly faster when H/ACA RNPs are reconstituted with the full-length snR34 rather than with a truncated guide RNA consisting of the 5′ hairpin and the flanking H box (Supplementary Figure S2D); this observation agrees with an earlier study (20). In conclusion, the initial characterization of the purified yeast H/ACA RNP system demonstrates that the reconstituted complex is highly active under physiological, low ionic strength conditions.
Figure 2.

Pseudouridine formation by the snR34 H/ACA RNP in short substrate RNAs. An excess (1 μM) of [3H-C5]uridine-labeled 5′ substrate RNA (circles) or 3′ substrate RNA (squares) was incubated with 0.1 μM in vitro reconstituted snR34 H/ACA RNP to allow for multiple turnover pseudouridine formation. At selected time points, the percentage of pseudouridine formation was determined by the tritium release assay.
Yeast H/ACA proteins bind guide RNA with extremely high affinity
First, we focused on assessing the formation and stability of H/ACA RNPs by analyzing the affinity of the H/ACA proteins for radioactively labeled guide RNA. In brief, H/ACA proteins are incubated with a low concentration of guide RNA; protein-bound guide RNA is subsequently retained on a nitrocellulose filter and quantified by scintillation counting. Thereby, tight and efficient binding of RNA even at very low nanomolar concentrations of proteins was observed (Figure 3A). The dissociation constant (KD) for the interaction of snR34 and snR5 with the protein complex Cbf5p–Nop10p–Gar1p–Nhp2p is 0.5 nM and 0.7 nM, respectively (Table 1), indicating that the interaction between H/ACA guide RNA and proteins is extremely tight. For comparison, we also determined the affinity of the archaeal Cbf5p–Nop10p–Gar1p complex for archaeal guide RNA Pf4 which is comprised of a single hairpin (33) (Figure 3B). Filter binding experiments revealed a KD of 23 nM which is about 50-fold higher than for the interaction of yeast proteins with guide RNA (Table 1). The extremely high affinity of yeast H/ACA proteins for guide RNA allowed us to pre-form an H/ACA RNP complex using radioactively labeled guide RNA. By mixing this H/ACA RNP complex with a large excess of unlabeled guide RNA, we observed the time course of radioactive guide RNA dissociation from the protein complex in a chase experiment (Figure 3C). This experiment shows that both guide RNAs, snR34 and snR5, dissociate slowly from the Cbf5p–Nop10p–Gar1p–Nhp2p protein complex over the span of more than 10 min with a half-life time (t1/2) of 3 and 5.5 min (Table 1). Together, the quantitative characterization of the interaction of guide RNA with H/ACA proteins reveals a surprisingly tight interaction with a sub-nanomolar affinity and long half-life time in the minute range.
Figure 3.
H/ACA guide RNA binding to Cbf5p–Nop10p–Gar1p–Nhp2p proteins. To determine the affinity of H/ACA proteins for guide RNA, radioactively labeled RNA was incubated with increasing concentrations of proteins as indicated. The percentage of bound RNA was determined by nitrocellulose filtration, and fitting to a quadratic equation yielded the dissociation constant (KD). (A) Binding of 0.1 nM guide RNA snR34 (triangles) or snR5 (inverted triangles) to yeast proteins Cbf5p–Nop10p–Gar1p–Nhp2p. The dissociation constants are summarized in Table 1. (B) For comparison, the binding of 5 nM Pyrococcus furiosus guide RNA Pf9 to the archaeal protein complex aCbf5p-aNop10p-aGar1p was also characterized by nitrocellulose filtration. (C) Chase experiment to determine the dissociation rate constant for snR34 (triangles) and snR5 (inverted triangles) from the yeast H/ACA protein complex Cbf5p–Nop10p–Gar1p–Nhp2p. An RNA–protein complex was preformed using radioactively labeled RNA and mixed with a large excess of unlabeled RNA allowing the determination of the remaining bound radioactive RNA over time by nitrocellulose filtration. Single exponential decay fitting of the time course provided koff values (Table 1).
We further tested the specificity of Cbf5p–Nop10p–Gar1p–Nhp2p for H/ACA guide RNA by constructing a series of variants of the 5′ hairpin of snR34 (Supplementary Figure S3 and Table 1). Therein, a single hairpin of snR34 was used which should only bind one Cbf5p–Nop10p-Gar1p-Nhp2p complex. The 5′ hairpin of snR34 has an unstructured 5′ extension; deletion of this region did not affect binding to the H/ACA proteins. Likewise, abolishing or decreasing the single-stranded regions forming the pseudouridylation pocket as well as mutating the double-stranded region in the lower stem underneath the pseudouridylation pocket resulted in similar low nanomolar affinities to the H/ACA proteins (Table 1). We also tested these variants for their ability to direct pseudouridylation of the 5′ substrate RNA (Supplementary Figure S3). As expected, mutating the pseudouridylation pocket abolished the activity to modify substrate RNA, but all other variants of the snR34 5′ hairpin retained activity. These data suggest that Cbf5 binds H/ACA guide RNA rather unspecifically and independent of the presence of single-stranded or double-stranded regions in specific positions. We therefore tested whether Cbf5p–Nop10p-Gar1p-Nhp2p can bind to an unrelated RNA with complex structure utilizing a cricket paralysis virus internal ribosome entry site (IRES) RNA (34). Surprisingly, even this viral RNA was bound very tightly by the H/ACA protein complex with a sub-nanomolar affinity (Supplementary Figure S3G and Table 1). In conclusion, the Cbf5p–Nop10p–Gar1p–Nhp2p complex binds RNA extremely tightly, yet unspecifically.
Characterization of substrate RNA binding by H/ACA sRNPs
Since guide RNA is bound very strongly to S. cerevisiae H/ACA proteins, we were able to conduct similar filter binding experiments to determine the affinity of radioactively labeled substrate RNA to H/ACA RNPs. As the concentration of the purified Cbf5p–Nop10p–Gar1p complex did not allow titrating to high concentrations, we used a low, constant concentration of reconstituted H/ACA RNP (5 nM) which is stable under these low concentrations as evident by the low KD for guide RNA (vide supra). Substrate RNA binding was then determined upon titration with radioactively labeled substrate RNA. Control experiments performed by titrating substrate RNA alone confirmed that minimal background signal from RNA alone is recorded even at 500 nM of RNA (Figure 4A). Under these conditions, some pseudouridine formation can occur, but will be minimal as a large excess of substrate RNA over H/ACA RNP is used and the reaction is incubated for no more than 3 min. Therefore, this assay predominantly measures binding of substrate RNA to the H/ACA RNP. We measured binding of both the short 5′ and 3′ substrate RNAs to H/ACA RNPs reconstituted with snR34 and snR5, respectively, providing insight into four different types of substrate-guide RNA interactions (Figure 4). Fitting of the binding curves revealed affinities of substrate RNAs for H/ACA RNPs ranging from 50–330 nM (Table 2). Notably, of the four substrate–guide RNA interactions, three are comprised of 12 and one of 14 bp. However, these base-pairing interactions differ significantly in their GC content which ranges from 33 to 67% (Figure 4C). It is interesting to note that the affinity of the substrate binding to the H/ACA RNP correlates directly with the GC content in the base pairing region with the highest affinity (KD = 53 nM) observed for the interaction of the 5′ substrate with snR34-reconstituted H/ACA sRNPs (67% GC content).
Figure 4.
Substrate RNA binding to H/ACA RNPs. The affinity of H/ACA RNPs reconstituted with snR34 (A) or snR5 (B) for short substrate RNAs binding to either the 5′ or the 3′ pseudouridylation pocket was determined by nitrocellulose filtration. Here, a low concentration of H/ACA RNP (5 nM) was titrated with increasing concentrations of [3H]-labeled substrate RNAs. (A) Binding of 5′ (circles) and 3′ substrate RNAs (squares) to snR34 H/ACA RNPs. As control the 5′ substrate (open circles) or the 3′ substrate (open squares) was titrated into buffer. The fraction of substrate bound per H/ACA RNP was calculated, plotted against the substrate RNA concentration and fitted to a hyperbolic curve to obtain the dissociation constants (Table 2). (B) The affinity of snR5 H/ACA RNP for its 5′ (circles) and 3′ substrate RNA (squares) was also determined by nitrocellulose filtration. (C) Schematic representation of snR34 (top) and snR5 (bottom) interacting with the 5′ (left) and 3′ substrate RNAs (right) in the two pseudouridylation pockets. All four guide–substrate RNA interactions are composed of a similar number of 12–14 base-pairs, but the GC content within the base-pairing interaction differs significantly as follows: snR34-5′ substrate RNA: 67% GC; snR34-3′ substrate RNA: 43% GC; snR5-5′ substrate RNA: 42% GC; and snR5-3′ substrate RNA: 33% GC. The GC content in the base-pairing region between guide RNA and substrate RNA correlates with the affinity of H/ACA RNPs for substrate RNA (Table 2).
Nhp2p is critical for efficient pseudouridylation by H/ACA RNPs
Next, we dissected the role of the conserved protein Nhp2p for the function of H/ACA RNPs as this protein has previously been reported to be largely dispensable for H/ACA RNP activity (20). First, time courses of pseudouridine formation were recorded in the absence or presence of Nhp2p for H/ACA RNPs reconstituted with guide RNA snR34 (Figure 5A). Whereas under these conditions most pseudouridines are formed within 50 min by a complete H/ACA RNP complex, <20% of pseudouridines are observed in the absence of Nhp2p. This finding suggests that Nhp2p is critical for pseudouridylation in contrast to the earlier report (20). To assess this apparent discrepancy, we also followed pseudouridylation activity of H/ACA RNP reconstituted with the guide RNA snR5, the same guide RNA as used in the earlier study (Supplementary Figure S4). Again, pseudouridylation is significantly faster in the presence of Nhp2p. However, after 60 min of incubation (the only timepoint measured previously (20)), the 5′ substrate of snR5 was modified to almost the same level in the absence as in the presence of Nhp2p. In conclusion, the time course measurements reported here confirm that some pseudouridines can be formed in the absence of Nhp2p, albeit very slowly. The function of Nhp2p is therefore to enable fast and efficient pseudouridylation by H/ACA RNPs.
Figure 5.
Role of protein Nhp2p for H/ACA RNP function. (A) Pseudouridine formation in the 5′ (circles) and 3′ substrate (squares) (1 μM) was observed in the presence of snR34 guide RNA (100 nM) and all four proteins (filled symbols, same data as in Figure 2) or in the absence of Nhp2p (open symbols), i.e. when only the Cbf5p–Nop10p–Gar1p was added to the reaction. As before, pseudouridylation was detected by the tritium release assay. (B) Binding of snR34 (open triangles) and snR5 (open inverted triangles) guide RNA to Cbf5p–Nop10p–Gar1p in the absence of Nhp2p. Dissociation constants are summarized in Table 1. (C) The affinity of Nhp2p alone for snR34 guide RNA was determined by nitrocellulose filtration. Hyperbolic fitting yielded the dissociation constant (Table 1).
To address the question why pseudouridylation is slow in the absence of Nhp2p, we tested the hypothesis that Nhp2p binding to guide RNA stabilizes the H/ACA RNP complex. First, the affinity to bind H/ACA guide RNA was determined for the trimeric protein complex Cbf5p–Nop10p–Gar1p in the absence of Nhp2p to assess the impact of Nhp2p on complex stability. For both snR34 and snR5 guide RNA, nitrocellulose filtration revealed again sub-nanomolar affinities (Figure 5B and Table 1) suggesting that Nhp2p has no effect on guide RNA binding although both yeast and mammalian Nhp2p (as its archaeal homolog L7Ae) are RNA-binding proteins (21,28,35). For comparison, we also assessed the affinity of Nhp2p alone to snR34 guide RNA yielding a dissociation constant of 600 nM (Figure 5C and Table 1). Thus, the affinity of Nhp2p for guide RNA is 1000-fold lower than the affinity of Cbf5p–Nop10p–Gar1p for H/ACA RNA which explains why omission of Nhp2p does not destabilize the H/ACA RNP complex in vitro.
Subsequently, the hypothesis was assessed that Nhp2p plays a role in binding of the substrate RNA to the H/ACA RNP complex. As stable H/ACA RNP complexes could be formed in the absence of Nhp2p, this allowed us to conduct filter binding assays titrating with radioactively labeled substrate RNA. These experiments showed that the snR34 5′ substrate RNA binds to an H/ACA RNP complex formed without Nhp2p with the same affinities as determined in the presence of Nhp2p (Figure 4 and Table 2). We therefore conclude that neither guide RNA binding nor substrate RNA binding is affected by the omission of Nhp2p. Consequently, Nhp2p must contribute to a mechanistic step following substrate RNA binding to H/ACA RNPs as it is critical for fast pseudouridine formation.
Contribution of the conserved Box H and Box ACA elements to H/ACA RNP function
The Box H (ANANNA) and Box ACA elements are the only conserved sequence motifs in H/ACA guide RNA and bind to the PUA domain of Cbf5p as revealed in the structures of archaeal H/ACA sRNPs (29). It was previously shown that the Box H and Box ACA elements are important in vivo for the accumulation of H/ACA RNAs (9) and also for the pseudouridylation activity of H/ACA RNPs (36), but the molecular mechanism thereof remained unclear. We therefore hypothesized that these elements contribute to the function of H/ACA RNPs by enhancing binding of the guide RNA to the H/ACA proteins. To test this hypothesis, the most highly conserved adenine nucleotides in the Box H and Box ACA elements were replaced with guanines such that the conserved motifs were essentially abolished (Δ Box H = GCGAGA instead of ACAAGA; Δ Box ACA = GCG instead of ACA). First, we assessed whether these altered guide RNAs influence the overall function of H/ACA RNPs, i.e. pseudouridine formation using both 5′ and 3′ substrate RNAs (Figure 6A and B). Mutation of the Box H element reduced pseudouridine formation in the 5′ substrate RNA that binds to the 5′ hairpin adjacent to the Box H whereas modification of the 3′ substrate RNA is not affected by mutation of Box H. When the Box ACA element is altered, pseudouridylation of the 3′ substrate interacting with the adjacent hairpin is drastically decreased; surprisingly, modification of the 5′ substrate is also somewhat reduced. Thus, it seems that both the Box H and Box ACA contribute to modification of the 5′ substrate albeit moderately. In contrast, the modification of the 3′ substrate is strongly dependent on the presence of the Box ACA element, but not at all on the Box H.
Figure 6.
Role of the conserved Box H and Box ACA in guide RNA. (A) Pseudouridine formation in the 5′ substrate recognized by the pseudouridylation pocket in the first hairpin adjacent to the Box H motif using different guide RNA constructs: snR34 wild-type (circles), snR34 Δ Box H (open circles) and snR34 Δ Box ACA (open diamonds). (B) Pseudouridine formation in the 3′ substrate which binds to the second hairpin of snR34 adjacent to the Box ACA motif. snR34 wild-type (squares), snR34 Δ Box H (open triangles) and snR34 Δ Box ACA (open squares). In both (A and B), the substrate RNA was incubated in the absence of H/ACA RNP as a control (open inverted triangles). (C) The affinity of the proteins Cbf5p-Nop10-Gar1p-Nhp2p for guide RNA variants was determined by nitrocellulose filtration. Here, a representative binding curve of snR34 Δ Box ACA (open diamonds) is shown in comparison to snR34 wild-type (triangles). The dissociation constants are summarized in Table 1.
Next, we directly assessed binding of the guide RNAs lacking the Box H or Box ACA elements to the proteins Cbf5p–Nop10p–Gar1p–Nhp2p (Figure 6C). In contrast to our hypothesis, deletion of the Box H or the Box ACA did not affect the affinity for guide RNA (Table 1). In this assay, it is possible that tight binding of proteins to one of the hairpins in guide RNA leads to efficient retention of the complex on the nitrocellulose membrane thereby masking a potential effect of the Box H or Box ACA element on protein binding to the adjacent hairpin only. Therefore, we also measured guide RNA binding for the isolated 5′ hairpin of snR34 and the 5′ hairpin lacking the Box H element. However, we again observed low nanomolar affinities for both guide RNAs binding to the H/ACA proteins (Table 1). In conclusion, the Box H and Box ACA elements do not contribute significantly to the tight binding of H/ACA guide RNA to Cbf5p–Nop10p–Gar1p.
At last, we asked whether binding of substrate RNA is impaired upon mutation of the Box H or Box ACA element. We utilized the 3′ substrate since pseudouridine formation was strongly reduced upon deletion of the Box ACA element and slightly lowered for the deletion of the Box H element. However, the 3′ substrate RNAs bind with affinities of 90 and 150 nM to the snR34 Δ Box H and the snR34 Δ Box ACA guide RNAs, respectively, which are indistinguishable from the affinities of 3′ substrate RNA for snR34 wild-type (Table 2). In summary, the Box H and Box ACA elements do not affect guide RNA or substrate RNA binding, but at least the Box ACA element is required for efficient pseudouridylation in the substrate binding to the adjacent 3′ hairpin.
DISCUSSION
The characterization of yeast H/ACA RNPs represents a critical step toward gaining a thorough understanding of their functions that affect ribosome biogenesis, spliceosome formation and modification of other cellular RNAs. Using a reconstituted S. cerevisiae H/ACA RNP complex, we first show here that the Cbf5p–Nop10p–Gar1p complex interacts with H/ACA guide RNA with a remarkably high, sub-nanomolar affinity; despite the strength of this interaction, the H/ACA proteins bind RNA unspecifically. Second, the interaction of H/ACA RNPs with substrate RNA is characterized by nanomolar affinities that correlate with the GC content. Third, the efficient pseudouridylation activity of the two-hairpin H/ACA RNP is critically dependent on the presence of protein Nhp2p although Nhp2p does not contribute to guide RNA or substrate RNA binding. And fourth, the Box H and Box ACA elements have similar roles as Nhp2p since they are contributing to the modification activity of H/ACA RNPs, but not guide or substrate RNA binding.
The fact that H/ACA guide RNA is bound extremely tightly, yet unspecifically by S. cerevisiae Cbf5p–Nop10p–Gar1p provides interesting insight into both the cellular and the molecular function of H/ACA RNPs. The tight association of RNA and proteins in H/ACA RNPs indicates that these complexes are highly stable in vivo and explains why guide RNAs are not exchanged between H/ACA RNPs (35). During the in vivo assembly of H/ACA RNPs, Cbf5p is first bound by the assembly factor Shq1p in the cytoplasm which covers part of the H/ACA guide RNA binding surface on Cbf5p (37). Given the high affinity, but low specificity of Cbf5p for RNA, the binding of Shq1p is necessary to prevent Cbf5p from binding to other RNAs in the cytoplasm or nucleus before reaching the site of H/ACA guide RNA transcription. There, the assembly factor Shq1p dissociates with the help of ATPases and is replaced by the guide RNA (38). For Shq1p to effectively shield Cbf5p from other RNAs, it seems likely that Shq1p also has a high affinity for Cbf5p, and therefore Shq1p dissociation needs to be regulated and energy-dependent. The exchange of Shq1p and guide RNA is probably favored by the higher affinity of H/ACA guide RNA for Cbf5p. Thus, our findings of tight guide RNA binding in H/ACA RNPs explain crucial aspects of the cellular assembly mechanism of this complex.
The 50-fold higher affinity to guide RNA of eukaryotic Cbf5p–Nop10p–Gar1p compared to the archaeal proteins is striking since these proteins share a conserved sequence and are structurally very similar (20,29). However, the eukaryotic proteins are characterized by N- and C-terminal extensions. In the case of Cbf5p, these extensions contribute in part to the enlargement of the PUA domain (pseudouridine and archeosine transglycosylase domain) which is an RNA-binding domain (39). Hence, it is tempting to speculate that in eukaryotic H/ACA RNPs the interaction surface between guide RNA and Cbf5p might be larger and might include the N- and C-terminal extensions that are rich in lysine residues thereby strengthening the interaction between Cbf5p and guide RNA.
From an evolutionary perspective, a trade-off in guide RNA affinity seems to have occurred between the H/ACA proteins: eukaryotic Cbf5p–Nop10p–Gar1p binds guide RNA 50-fold tighter than its archaeal homologs, but Nhp2p binds RNA 100-fold weaker than the archaeal, kink-turn specific L7Ae protein (28). Or in other words, the tight binding of H/ACA guide RNA by Cbf5p–Nop10p–Gar1p is likely compensating for the low affinity of Nhp2p to guide RNA. Furthermore, in eukaryotes the weak binding of Nhp2p to H/ACA guide RNA is compensated by a stable protein-protein interaction between Nhp2p and the Cbf5p–Nop10p complex which is not observed for archaeal L7Ae (17,40).
H/ACA RNPs utilize many different guide RNAs that can each interact with at least one to three target sequences through base-pairing interactions within the two pseudouridylation pockets. The modular nature of H/ACA RNPs is their key feature in providing the flexibility to sequence-specifically target numerous cellular RNAs for modification. Obviously, H/ACA RNPs can accept many different base-pairing interactions between guide and target RNA. Indeed, our substrate RNA binding investigations revealed a nanomolar affinity of H/ACA RNPs to their target RNAs ranging from 50 to 330 nM for the four different interactions studied. It seems that this affinity is sufficiently high to select substrate RNA while also being low enough for efficient product release and multiple turnover catalysis by H/ACA RNPs. Interestingly, our data also suggest a possible correlation of substrate RNA affinity with the GC content in the guide–substrate RNA interaction. Given the recent finding of H/ACA RNP-dependent (and -independent) widespread pseudouridylation of mRNAs and other non-coding RNA (4,5), the relatively high nanomolar affinity of H/ACA RNPs for substrate RNA supports the hypothesis that an H/ACA guide RNA might have multiple target RNAs including some that bind with a lower affinity due to mismatches to the guide RNA.
Our findings suggest similar roles for protein Nhp2p and the Box H and Box ACA motifs. In contrast to our expectations, neither of these structural elements of H/ACA RNPs contributes to binding of H/ACA guide RNA, and they are also not involved in substrate binding. It is noteworthy however, that whereas Nhp2p does not contribute to the affinity of the protein complex to H/ACA guide RNA as measured in vitro (Figure 5), the Nhp2p protein is required in vivo for H/ACA RNA stability (21) and for guide RNA binding in the absence of Gar1p (35). Possibly, in absence of Nhp2p, the H/ACA RNA might bind to Cbf5p in vivo, but might be prone to degradation for example due to the exposure of RNA elements that would usually be protected by Nhp2p. Clearly, Nhp2p and the Box ACA are required for efficient pseudouridine formation although they are located far away from the active site in Cbf5p. The most plausible explanation for this observation is that both in the absence of Nhp2p and in absence of the Box ACA motif, the substrate RNA can bind to guide RNA, but the target uridine cannot be productively positioned in the catalytic site. Indeed, there is precedence for such a mechanism as observed in the crystal structure of an archaeal H/ACA sRNP complex with bound substrate RNA, but lacking the Nhp2p homolog L7Ae (41). In the absence of L7Ae, the substrate RNA forms the expected base-pairs to guide RNA; however, the upper stem of the guide RNA above the pseudouridylation pocket is slightly bent away from Cbf5p–Nop10p such that the target uridine cannot occupy the catalytic center. Further fluorescent studies supported the function of L7Ae for the long-distance placement of the target nucleotide in the active site of Cbf5p (42). Despite the striking differences in RNA specificity and affinity between archaeal L7Ae and eukaryotic Nhp2p, our data therefore suggest that these proteins have the same function within H/ACA RNPs. We propose a model where Nhp2p binds to the upper stem of H/ACA guide RNA and through its interaction with Nop10p anchors the guide RNA firmly on the H/ACA proteins. Therefore, only in the presence of Nhp2p, the target uridine can reach the catalytic center to be efficiently converted to pseudouridine (Figure 7).
Figure 7.
Model for the anchoring of H/ACA guide RNA by Nhp2p and the Box H and Box ACA elements to facilitate positioning of the target uridine into the active site of Cbf5p. Left: Mispositioning of the upper stem of H/ACA guide RNA in the absence of Nhp2p which allows substrate RNA to bind to the guide RNA, but the target uridine may reside in front of Cbf5p without reaching the active site. Right: Mispositioning upon mutating Box ACA that usually interacts with the conserved PUA domain of Cbf5p. In the absence of these motifs, the tight, yet sequence-unspecific binding of guide RNA to Cbf5p may lead to a shift of guide RNA relative to Cbf5p such that the target uridine in substrate RNA cannot reach the active site. The Box ACA element likely functions as ruler (left) to define the distance between this motif and the target uridine inserted into the active site of Cbf5p (10,11).
Similar to the function of Nhp2p, it seems that the Box H and Box ACA elements are responsible for anchoring the bottom stem of H/ACA RNA on the H/ACA proteins which is facilitated by the sequence-specific interaction of these motifs with the PUA domain of Cbf5p. Notably, this interaction does not contribute to the high affinity of the H/ACA proteins for guide RNA. As H/ACA RNPs may contain many different guide RNAs, the interaction of H/ACA proteins with guide RNA is therefore clearly sequence independent. Presumably, the large, positively charged RNA-binding surface on Cbf5p only recognizes the presence of RNA helices in an unspecific manner, namely the lower RNA helix underneath the pseudouridylation pocket as well as the two helices formed between the pseudouridylation pocket and the substrate RNA (29). Interestingly, pseudouridylation of substrate RNA is directly affected by mutation of the adjacent conserved element (5′ substrate by Box H and 3′ substrate by Box ACA). Therefore, we propose a model where, in the absence of either the Box H or Box ACA motif, the guide RNA can slide along the RNA-binding surface of Cbf5p thereby misaligning the top of the pseudouridylation pocket relative to Cbf5p such that the target uridine is no longer positioned in the active site (Figure 7). Thereby, we can provide a molecular explanation for the earlier finding that the Box H and Box ACA elements are critical in vivo for pseudouridylation activity (36). This model is consistent with the early finding that the Box H and Box ACA elements can act as rulers as there is always a defined distance of about 14 nt between this motif and the top of the pseudouridylation pocket (10,11). Notably, we also detect a long-distance effect of the Box ACA element on modification of the 5′ substrate, but not of the Box H on the 3′ substrate suggesting differential activity of the two conserved sequence elements. It is tempting to speculate that the long-range effect of the Box ACA suggests a crosstalk between the two halves of the H/ACA guide RNA. As known previously, in addition to correctly positioning the guide RNA on Cbf5p, the Box H and Box ACA motifs also contribute to the maturation, accumulation and thereby stability of guide RNA in vivo (9).
Taken together, we propose that Nhp2p anchors the upper stems of the H/ACA guide RNA hairpins while the Box H and Box ACA motifs position the lower stems of the H/ACA guide RNA onto the H/ACA proteins. Correct positioning of the guide RNA by these structural elements is essential for the efficient pseudouridylation activity of H/ACA RNPs as this allows the substrate RNA to not only base-pair with guide RNA, but to position the target uridine productively into the active site of Cbf5p (Figure 7).
In conclusion, this study reveals critical features of H/ACA RNP assembly and function regarding pseudouridine formation, the interaction of the H/ACA proteins with guide RNA and the binding of substrate RNA to the complex. Our mechanistic investigations uncovered a critical function of Nhp2p and the Box H and Box ACA elements in correctly anchoring the guide RNA within the H/ACA RNP complex and thereby indirectly helping to productively insert the target uridine in the active site. The correct positioning of a guide RNA relative to its protein interaction partners is likely a common feature of several RNP complexes. For example, in C/D RNPs that direct 2′-O-methylation of RNA, the conserved C and D as well as C’ and D’ elements form a K turn and K loop structure, respectively, which both bind L7Ae in archaea and Snu13p in yeast, and are both located at a defined distance from the methylation sites (43). Thus, both the Box H and Box ACA elements as well as the Box C and Box D elements serve as anchor points and rulers within their respective RNPs. Similarly, in CRISPR systems, the guide RNA must be properly positioned relatively to the endonucleolytic Cas enzyme to direct cleavage of the target DNA (or RNA). Here, the correct positioning is achieved by different RNA elements such as tracer RNA in type II CRISPR systems, the 5′ handle sequence in type I and type III CRISPR systems and the 3′ hairpin in type I CRISPR systems (44). In this context, our biochemical characterization of pseudouridylation, guide RNA binding and substrate RNA binding by H/ACA RNPs highlights fundamental properties of RNP systems that harness the power of proteins to catalyze chemical reactions in combination with the unique ability of RNA to sequence-specifically recognize any target nucleic acid. As we show here for H/ACA RNPs, in all cases the correct interaction of RNA and proteins is crucial for the collaborative function of these RNPs.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Phil Donkiewicz and Jessica Durand for help in establishing protein and RNA purification procedures. We are grateful to Anne Rintala-Dempsey for help with preparing figures and for critically reading the manuscript. Luc Roberts from the lab of Dr Hans-Joachim Wieden kindly provided the cricket paralysis virus IRES RNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Alberta Innovates [Strategic Research Chair 2015, Sustainability Fund 2012]; University of Lethbridge [ULRF 2014, HRAF 2015]; Banting Research Foundation (New Investigator Fund); Alberta Innovates Graduate Student Scholarship (to E.A.C.); NSERC CGS-M Scholarship (to E.K.K.); Queen Elizabeth II Graduate Scholarship (to E.K.K.). Funding for open access charge: Alberta Innovates.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kiss T., Fayet-Lebaron E., Jady B.E.. Box H/ACA small ribonucleoproteins. Mol. Cell. 2010; 37:597–606. [DOI] [PubMed] [Google Scholar]
- 2. Hamma T., Ferre-D’Amare A.R.. Pseudouridine synthases. Chem. Biol. 2006; 13:1125–1135. [DOI] [PubMed] [Google Scholar]
- 3. Ge J., Yu Y.T.. RNA pseudouridylation: new insights into an old modification. Trends Biochem. Sci. 2013; 38:210–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schwartz S., Bernstein D.A., Mumbach M.R., Jovanovic M., Herbst R.H., Leon-Ricardo B.X., Engreitz J.M., Guttman M., Satija R., Lander E.S. et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014; 159:148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carlile T.M., Rojas-Duran M.F., Zinshteyn B., Shin H., Bartoli K.M., Gilbert W.V.. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014; 515:143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Meier U.T. The many facets of H/ACA ribonucleoproteins. Chromosoma. 2005; 114:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fayet-Lebaron E., Atzorn V., Henry Y., Kiss T.. 18S rRNA processing requires base pairings of snR30 H/ACA snoRNA to eukaryote-specific 18S sequences. EMBO J. 2009; 28:1260–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meier U.T. How a single protein complex accommodates many different H/ACA RNAs. Trends Biochem. Sci. 2006; 31:311–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ganot P., Caizergues-Ferrer M., Kiss T.. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997; 11:941–956. [DOI] [PubMed] [Google Scholar]
- 10. Ni J., Tien A.L., Fournier M.J.. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997; 89:565–573. [DOI] [PubMed] [Google Scholar]
- 11. Ganot P., Bortolin M.L., Kiss T.. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997; 89:799–809. [DOI] [PubMed] [Google Scholar]
- 12. Kirwan M., Dokal I.. Dyskeratosis congenita: a genetic disorder of many faces. Clin. Genet. 2008; 73:103–112. [DOI] [PubMed] [Google Scholar]
- 13. Mitchell J.R., Wood E., Collins K.. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999; 402:551–555. [DOI] [PubMed] [Google Scholar]
- 14. Vulliamy T., Beswick R., Kirwan M., Marrone A., Digweed M., Walne A., Dokal I.. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:8073–8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yoon A., Peng G., Brandenburger Y., Zollo O., Xu W., Rego E., Ruggero D.. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science (New York, N.Y. 2006; 312:902–906. [DOI] [PubMed] [Google Scholar]
- 16. Bellodi C., Krasnykh O., Haynes N., Theodoropoulou M., Peng G., Montanaro L., Ruggero D.. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res. 2010; 70:6026–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baker D.L., Youssef O.A., Chastkofsky M.I., Dy D.A., Terns R.M., Terns M.P.. RNA-guided RNA modification: functional organization of the archaeal H/ACA RNP. Genes Dev. 2005; 19:1238–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Charpentier B., Muller S., Branlant C.. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res. 2005; 33:3133–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Normand C., Capeyrou R., Quevillon-Cheruel S., Mougin A., Henry Y., Caizergues-Ferrer M.. Analysis of the binding of the N-terminal conserved domain of yeast Cbf5p to a box H/ACA snoRNA. RNA. 2006; 12:1868–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li S., Duan J., Li D., Yang B., Dong M., Ye K.. Reconstitution and structural analysis of the yeast box H/ACA RNA-guided pseudouridine synthase. Genes Dev. 2011; 25:2409–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Henras A., Dez C., Noaillac-Depeyre J., Henry Y., Caizergues-Ferrer M.. Accumulation of H/ACA snoRNPs depends on the integrity of the conserved central domain of the RNA-binding protein Nhp2p. Nucleic Acids Res. 2001; 29:2733–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kolodrubetz D., Burgum A.. Sequence and genetic analysis of NHP2: a moderately abundant high mobility group-like nuclear protein with an essential function in Saccharomyces cerevisiae. Yeast. 1991; 7:79–90. [DOI] [PubMed] [Google Scholar]
- 23. Tang T.H., Bachellerie J.P., Rozhdestvensky T., Bortolin M.L., Huber H., Drungowski M., Elge T., Brosius J., Huttenhofer A.. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:7536–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Muller S., Leclerc F., Behm-Ansmant I., Fourmann J.B., Charpentier B., Branlant C.. Combined in silico and experimental identification of the Pyrococcus abyssi H/ACA sRNAs and their target sites in ribosomal RNAs. Nucleic Acids Res. 2008; 36:2459–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Piekna-Przybylska D., Decatur W.A., Fournier M.J.. New bioinformatic tools for analysis of nucleotide modifications in eukaryotic rRNA. RNA. 2007; 13:305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lestrade L., Weber M.J.. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006; 34:D158–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schattner P., Barberan-Soler S., Lowe T.M.. A computational screen for mammalian pseudouridylation guide H/ACA RNAs. RNA. 2006; 12:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kuhn J.F., Tran E.J., Maxwell E.S.. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5kD/Snu13p snoRNP core protein. Nucleic Acids Res. 2002; 30:931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li L., Ye K.. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006; 443:302–307. [DOI] [PubMed] [Google Scholar]
- 30. Kamalampeta R., Kothe U.. Archaeal proteins Nop10 and Gar1 increase the catalytic activity of Cbf5 in pseudouridylating tRNA. Sci. Rep. 2012; 2:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wright J.R., Keffer-Wilkes L.C., Dobing S.R., Kothe U.. Pre-steady-state kinetic analysis of the three Escherichia coli pseudouridine synthases TruB, TruA, and RluA reveals uniformly slow catalysis. RNA. 2011; 17:2074–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. King T.H., Liu B., McCully R.R., Fournier M.J.. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell. 2003; 11:425–435. [DOI] [PubMed] [Google Scholar]
- 33. Klein R.J., Misulovin Z., Eddy S.R.. Noncoding RNA genes identified in AT-rich hyperthermophiles. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:7542–7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jan E., Sarnow P.. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 2002; 324:889–902. [DOI] [PubMed] [Google Scholar]
- 35. Wang C., Meier U.T.. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 2004; 23:1857–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bortolin M.L., Ganot P., Kiss T.. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J. 1999; 18:457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Godin K.S., Walbott H., Leulliot N., van Tilbeurgh H., Varani G.. The box H/ACA snoRNP assembly factor Shq1p is a chaperone protein homologous to Hsp90 cochaperones that binds to the Cbf5p enzyme. J. Mol. Biol. 2009; 390:231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Machado-Pinilla R., Liger D., Leulliot N., Meier U.T.. Mechanism of the AAA+ ATPases pontin and reptin in the biogenesis of H/ACA RNPs. RNA. 2012; 18:1833–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cerrudo C.S., Ghiringhelli P.D., Gomez D.E.. Protein universe containing a PUA RNA-binding domain. FEBS J. 2014; 281:74–87. [DOI] [PubMed] [Google Scholar]
- 40. Trahan C., Martel C., Dragon F.. Effects of dyskeratosis congenita mutations in dyskerin, NHP2 and NOP10 on assembly of H/ACA pre-RNPs. Hum. Mol. Genet. 2010; 19:825–836. [DOI] [PubMed] [Google Scholar]
- 41. Liang B., Xue S., Terns R.M., Terns M.P., Li H.. Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex. Nat. Struct. Mol. Biol. 2007; 14:1189–1195. [DOI] [PubMed] [Google Scholar]
- 42. Liang B., Kahen E.J., Calvin K., Zhou J., Blanco M., Li H.. Long-distance placement of substrate RNA by H/ACA proteins. RNA. 2008; 14:2086–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Watkins N.J., Bohnsack M.T.. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA. 2012; 3:397–414. [DOI] [PubMed] [Google Scholar]
- 44. Jackson R.N., Wiedenheft B.. A conserved structural chassis for mounting versatile CRISPR RNA-guided immune responses. Mol. Cell. 2015; 58:722–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.








