Abstract
KRAS is one of the most mutated genes in human cancer. It is controlled by a G4 motif located upstream of the transcription start site. In this paper, we demonstrate that 8-oxoguanine (8-oxoG), being more abundant in G4 than in non-G4 regions, is a new player in the regulation of this oncogene. We designed oligonucleotides mimicking the KRAS G4-motif and found that 8-oxoG impacts folding and stability of the G-quadruplex. Dimethylsulphate-footprinting showed that the G-run carrying 8-oxoG is excluded from the G-tetrads and replaced by a redundant G-run in the KRAS G4-motif. Chromatin immunoprecipitation revealed that the base-excision repair protein OGG1 is recruited to the KRAS promoter when the level of 8-oxoG in the G4 region is raised by H2O2. Polyacrylamide gel electrophoresis evidenced that OGG1 removes 8-oxoG from the G4-motif in duplex, but when folded it binds to the G-quadruplex in a non-productive way. We also found that 8-oxoG enhances the recruitment to the KRAS promoter of MAZ and hnRNP A1, two nuclear factors essential for transcription. All this suggests that 8-oxoG in the promoter G4 region could have an epigenetic potential for the control of gene expression.
INTRODUCTION
Cancer cells are characterized by high metabolic rates, normally associated with an increased level of reactive oxygen species (ROS) (1,2). Anion superoxide (O2−), hydrogen peroxide (H2O2) and hydroxyl radical (•OH) are produced by endogenous and exogenous sources (3). Among the endogenous sources, the mitochondrial electron transport chain, which reduces oxygen to water, is the major source of cellular ROS (4). In suspended mitochondria about 0.12–2% of oxygen consumed in the respiration is converted into O2− (3,4). However, anion superoxide is also produced enzymatically in essential metabolic pathways, including NADH oxidase, xanthine oxidase, lipo- and cyclo-oxygenases (5). Furthermore, O2− is reduced by superoxide dismutase to H2O2, which is then converted to •OH via a non-enzymatic Fenton reaction (6). All these chemical and enzymatic reactions push the ROS level more up in high metabolic rate cancer cells than in normal cells. An enhanced ROS level may damage DNA, RNA, lipids and proteins, and may also alter the intracellular signal transduction, for instance through NF-kB (7,8). A key protein of the antioxidant network is Nrf2, a (b-Zip)-type transcription factor that binds to antioxidant response elements in gene promoters and induces the expression of protective genes of the antioxidant response (9–11). Nrf2, being upregulated in pancreatic ductal adenocarcinoma (PDAC) cells, increases the capacity of the cells to control oxidative stress, a necessary condition for optimal cell proliferation (12–14).
The primary genetic lesions causing pancreatic cancer are somatic mutations in the KRAS gene. About 90% of PDAC carries KRAS G12D, i.e. a KRAS allele with a point mutation G→D in exon 1, codon 12 (15–17). The activity of mutant KRAS G12D is required in all stages of carcinogenesis (initiation, progression and metastasis) and the inactivation of mutant KRAS G12D reverses the transformation process (18–20). Recent studies have reported that KRAS G12D reduces the level of ROS in pancreatic cancer cells via Nrf2 (12,14). Since there is a correlation between mutant KRAS and Nrf2, we interrogated if in pancreatic cancer cells, the expression of KRAS is in some way influenced by oxidative stress. It is well known that oxidation of DNA occurs mainly at guanine, as it has the lowest oxidation energy among nucleobases (21). GG steps are preferred sites for oxidation, with 5′ G being particularly reactive (22). In the promoter of the KRAS oncogene there is a G-rich element called 32R that is critical for transcription and able to fold into a G-quadruplex structure (23). 32R is located between −148 and −116 bp from transcription start site (TSS) and is recognized by several transcription factors including MAZ and hnRNP A1 (24–27). Polymerase-stop assays, footprinting and circular dichroism showed that 32R is highly polymorphic in nature, as it can fold into three alternative G4 structures (28,29). In this study, we have found by chromatin immunoprecipitation (ChIP) combined with quantitative polymerase chain reaction (qPCR) that the 32R region is more oxidized than other G-rich regions that are unable to fold into G4. The presence of 8-oxoG in 32R may lower the stability of the G-quadruplex, depending on where the damage is located inside the sequence. When the oxidation is situated in the major 11-nt loop of the KRAS G4, the TM and folding are practically not affected. But when 8-oxoG is located in a G-tetrad, both TM and folding are strongly modified. We also investigated how guanine oxidation impacts the binding of the transcription factors to the regulatory G4 motif of KRAS. Our data show that 8-oxoG modulates the binding of the nuclear factors to the KRAS promoter and also strengthens the recruitment of MAZ and hnRNP A1 to the promoter. Finally, the results are discussed in terms of possible role of 8-oxoG as an epigenetic regulator in the transcription of oncogenic KRAS.
MATERIALS AND METHODS
Oligonucleotides and reagents
Unmodified oligonucleotides used in this study have been obtained from Microsynth (CH). 8-oxoG-substituted oligonucleotides were synthesized from 8-oxo-dG CEP from Berry & Associates in 1-μmol scale on solid support by standard procedure, except using concentrated ammonia in the presence of 2-mercaptoethanol (0.25 M) in the deprotection step as described by Bodepudi et al. (30). The oligonucleotides were purified by reverse-phase high pressure (or high performance) liquid chromatography on a Water system 600, equipped with a C18 column (XBridge OST C18, 19 × 1000 mm, 5 μm). The composition of the oligonucleotides was verified by Matrix Assisted Laser Desorption Ionisation-Time of Flight (MALDI-TOF) (Supplementary Table S1). Luteolin, was purchased from Alfa Aesar (D), 8-oxoguanine (8-oxoG) and 8-oxodeoxyguanosine from Cayman Chemicals (MI, USA), GTP from Euroclone (I), hydrogen peroxide solution 30% (w/w) from BDH (UK).
Cell cultures
Human pancreatic cancer (Panc-1, MIA PaCa-2, BxPC3) and non-cancer human embryonic kidney 293 cells were maintained in exponential growth in Dulbecco’s modified Eagle’s medium (DMEM) containing 100 U/ml penicillin, 100 mg/ml streptomycin, 20 mM L-glutamine and 10% foetal bovine serum (Euroclone, I). The cell lines have been genotyped by Microsynth (CH) and their identity confirmed.
Recombinant proteins
Recombinant MAZ and hnRNP A1 were obtained with a high degree of purity as previously described (24,26). Recombinant OGG1 with His-Tag at the N-terminus was expressed in Escherichia coli bacteria transformed with plasmid pET20 hOGG1. The bacteria were grown for 2 h at 37°C to an absorbance at 600 nm of 0.8–1 units before induction with isopropyl 1-thio-β-d-galactopyranoside (0.4 mM final concentration). The cells were allowed to grow overnight at 25°C, and then centrifuged at 5000 rpm at 4°C. The supernatant was removed and the pellet resuspended in Lysis buffer (50 mM NaH2PO4, 300 mM NaCl and 10 mM imidazole) added with 0.2 mM PMSF (phenylmethylsulphonyl fluoride). The bacteria were lysed by sonication [3 × (30 s sonication/1 min off)], added with 0.05% Tween 20 (Sigma-Aldrich, MO, USA) and the lysate centrifuged for 10 min, 4°C, 104 rpm. Ni-NTA resin (Qiagen, D) was added to the supernatant and the mixture was shaken for 1 h, 4°C. The mixture was then centrifuged for 5 min at 1700 rpm and the pellet washed two times with Wash Buffer (50 mM NaH2PO4, 300 mM NaCl and 20 mM imidazole). The OGG1 bound to the resin was eluted with a buffer composed by 50 mM NaH2PO4, 300 mM NaCl and 600 mM imidazole. OGG1 concentration was determined by Bradford method and the purity was confirmed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) (Supplementary Figure S1). Finally, the protein was concentrated and desalted by using the Ultracel YM-3 Microcon Centrifugal Filter Devices (Millipore, MA, USA).
Chromatin immunoprecipitation (ChIP) and qPCR analysis
ChIP was carried out as described in ref. 31, by using the ChIP-IT® Express Shearing Kit (Active Motif, CA, USA). In brief, Panc-1 cells (8 × 105) were seeded in 6-well plates and after 24 h some plates were treated with 10 μM luteolin for 24 h in DMEM or with 1 mM hydrogen peroxide for 15 min in serum-free DMEM. The cells were then washed with phosphate buffered saline (PBS) and fixed for 10 min in serum-free DMEM containing 1% formaldehyde. After fixing, the cells were washed with cold PBS and added with Glycine Stop-Fix Solution to arrest the fixing reaction. The cells were washed again with cold PBS, treated with Scraping Solution and centrifuged at 2500 rpm for 10 min at 4°C. The pellet was resuspended in ice-cold lysis buffer supplemented with PMSF, PIC (protease inhibitor cocktail) and incubated for 30 min on ice. The cells were transferred to an ice-cold dounce homogenizer for 20 strokes to release the nuclei. The homogenate was centrifuged for 10 min at 5000 rpm, 4°C, to pellet the nuclei. The nuclei were resuspended in Shearing Buffer and the chromatin sheared by sonication [10 × (30 s pulse on/30 s pulse off)] on Bioruptor Plus (Diagenode, BG) into DNA fragments of about 300–400 bp. The sheared chromatin was centrifuged at maximum speed for 15 min, 4°C. The chromatin concentration was determined with a spectrophotometer by ultraviolet (UV) absorption (260 nm) and 10 μg treated overnight at 4°C with 1 μg antibody specific for 8-oxoG (Bioss Antibodies, MA, USA) or MAZ or OGG1 (Santa Cruz Biotechnology, TX, USA) or hnRNP-A1 (Sigma-Aldrich, MO, USA). In addition to the antibodies, the mixtures were added with Protein G Magnetic Beads, ChIP buffer-1 and PIC, following the Active Motif Kit protocol. After incubation the mixtures were span and the chromatin bound to the antibody collected with a magnetic bar. The collected beads were washed once with ChIP buffer-1 and twice with ChIP buffer-2. The beads were then re-suspended in Elution Buffer AM2 and let to incubate on shaking for 15 min at room temperature. The beads were treated with Reverse Crosslinking Buffer and the supernatant with the chromatin was collected. The DNA fragments were amplified by qPCR using primers specific for genomic KRAS (accession number NG_007524): G4-plus 5′-GTACGCCCGTCTGAAGAAGA-3′ (nucleotides (nt) 4889–4908, 0.2 μM), G4-minus 5′-GAGCACACCGATGAGTTCGG-3′ (nt 4958–4977, 0.1 μM), Ctr-1-plus 5′-ACAAAAAGGTGCTGGGTGAGA-3′ (nt 12–32, 0.2 μM), Ctr-1-minus 5′-TCCCCTTCCCGGAGACTTAAT-3′ (nt 248–268, 0.2 μM), Ctr-2-plus 5′-CTCCGACTCTCAGGCTCAAG-3′ (nt 7536–7555, 0.15 μM), Ctr-2-minus 5′-CAGCACTTTGGGAGGCTTAG-3′ (nt 7692–7711, 0.15 μM). Ctr-1 is located in a non-coding region, G4-region is in the promoter, Ctr-2 is in an intron. qPCR reactions were carried out with a CFX-96 real-time PCR apparatus controlled by Optical System software (version 3.1) (Bio-Rad Laboratories, CA, USA) on 1 μl of immunoprecipitated chromatin or input, which were mixed to Sybr Green mix following manufacturer instructions (Kapa Sybr Fast QPCR Mix, Kapa Biosystems, MA, USA) and primers. For G4-region amplification cycles were: 3 min at 95°C, 40 cycles 30 s at 95°C and 40 s at 59°C. For controls amplification cycles were: 3 min at 95°C, 40 cycles 10 s at 95°C and 30 s at 57°C (control-1) or 61°C (control-2). All reactions have been validated before amplification for each target and couple of primers. The Ct-values (number of cycles required for the fluorescent signal to cross the threshold) given by the instrument (Bio-Rad CFX-96) were used to evaluate the difference between sample and input. The adjusted Ct input was obtained by Ct input − log2 (input dilution factor). Then ΔCt = Ct sample − Adjusted Ct input. The % Input for each sample was calculated as follows: % Input = 100 × 2∧(-ΔCt). The % Input was obtained for the G4 region and also for the non-G4 regions (Ctr-1 and Ctr-2). The enrichment in 8-oxoG or MAZ or hnRNPA1 or OGG1 of the G4 region respect to the non-G4 regions was determined by the ratio [% Input (G4 region))]/[% Input (non-G4 region)]. In average, from three to seven samples were used for each experiment. The t-test analysis was performed with Sigma Plot 10.1(UK).
Circular dichroism and UV-melting
Circular Dichroism (CD) spectra were obtained on a JASCO J-600 spectropolarimeter, equipped with a thermostated cell holder, with 5 μM oligonucleotides solutions in 50 mM Tris–HCl, pH 7.4, 100 mM KCl. The spectra were recorded in 0.5 cm quartz cuvette at room temperature and 90°C. The spectra are reported as ellipticity (mdeg) versus wavelength (nm). Each spectrum was recorded three times, smoothed and subtracted to the baseline.
UV-melting analysis was performed using the Jasco V-750 UV-visible spectrophotometer equipped with a Peltier temperature control system (ETCS-761) (Jasco, JP). The spectra were analyzed with Spectra Manager (Jasco, JP). Oligonucleotides (5 μM) were annealed in 100 mM KCl, 50 mM Na-cacodylate pH 7.4 (10 min at 95°C, overnight at room temperature). The melting curves were recorded at 295 nm in a 0.5 cm path length quartz cuvette heating (20–90°C) and cooling (90–20°C) at a rate of 0.5°C/min. The thermodynamic parameters for the folding of the wild-type and modified oligonucleotides into G4 were obtained from the UV-melting curves. The ‘DNA-Melting Analysis’ program (Jasco, JP), which analyzed the melting curves according to a standard all-or-none model, gave the ΔH ° and ΔS° values. The free energy of quadruplex formation was calculated according to: ΔG ° = −RT ln K = ΔH ° − TΔS °.
PAGE assay
8-OxoG-substituted oligonucleotides end-labeled with [γ-32P]adenosine triphosphate (ATP) (Perkin Elmer) and T4 polynucleotide kinase (Thermo Fisher Scientific, USA), were annealed in duplex or quadruplex as follows: the duplex was obtained annealing (5 min at 95°C, overnight at room temperature) the 8-oxoG-substituted oligonucleotides and 32R with complementary 32Y in 50 mM Tris–HCl, pH 7.4, 100 mM NaCl; the quadruplexes were obtained in 50 mM Tris–HCl, pH 7.4, 100 mM KCl (5 min at 95°C, overnight at room temperature). Radiolabeled duplex and quadruplex (2 nM) were incubated at 37°C with increasing amounts of OGG1 (1 and 5 μM), in 20 mM Tris–HCl pH 8, 50 mM NaCl, 1 mM ethylenediaminetetraaceticacid (EDTA), 0.1 mg/ml bovine serum albumin, 1 mM Na3VO4, 5 mM NaF and 0.01% Phosphatase Inhibitor Cocktail. After 15 min, the reactions were stopped by adding to the mixtures 8 μl stop solution (90% formamide, 50 mM EDTA). The samples were then denatured for 5 min at 95°C and run for 1 h on a denaturing 20% polyacrylamide gel, prepared in Tris-borate-EDTA (TBE) and 7 M urea, pre-equilibrated at 55°C in an electrophoretic apparatus (C.B.S Scientific Company, CA, USA). After running the gel was fixed in a solution containing 10% acetic acid and 10% methanol, dried and exposed to film for auto-radiography (Aurogene, I).
Electrophoresis mobility shift assay (EMSA)
Duplexes and quadruplexes have been prepared as described in previous section. PAGE purified 8-oxoG-substituted oligonucleotides were end-labeled with [γ-32P] ATP and T4 polynucleotide kinase (30 pmol). The corresponding duplexes were obtained by annealing (10 min at 95°C and overnight at room temperature) the modified oligonucleotides with the complementary strand in 50 mM Tris–HCl pH 7.4, 100 mM NaCl. Protein–oligonucleotide interactions were analyzed by electrophoresis mobility shift assays (EMSA). End-labeled duplexes or G-quadruplexes were incubated in 20 μl solutions containing 50 mM Tris–HCl, pH 7.4, 100 mM NaCl (for duplexes) or 100 mM KCl (for G-quadruplexes), 1 mM EDTA, 0.01% Phosphatase Inhibitor Cocktail I (Sigma-Aldrich, MO, USA), 5 mM NaF, 1 mM Na3VO4, 2.5 ng/μl poly [ dI−dC], 1 mM Dithiothreitol (DTT) and 8% glycerol with increasing amounts of recombinant OGG1 (0.3 and 0.6 μg) or MAZ or hnRNP A1 (2.5–5 μg), time and temperature are indicated in figure legends. The mixtures were analyzed in 5% polyacrylamide gels prepared in TBE at 20°C. After running, the gels were dried and exposed overnight to auto-radiography (Aurogene, I) at −80°C.
DMS-footprinting experiments
Dimethylsulphate (DMS)-footprinting experiments were performed using PAGE purified 8-oxoG-substituted oligonucleotides (24 nM), end-labeled with [γ-32P] ATP. The oligonucleotides were incubated overnight at 37°C, in 50 mM Tris–HCl, pH 7.4, 1 μg sonicated salmon sperm DNA, 1 mM EDTA, 100 mM KCl or 100 mM LiCl, as specified in the figure legend. DMS dissolved in ethanol (DMS:ethanol, 2/38, vol/vol) was added to the DNA solution (2 μl to a total volume of 50 μl) and left to react for 1 min at room temperature. The reactions were stopped by adding to the mixtures 5 μl of stop solution (1.5 M sodium acetate, pH 5.2, 1 M β-mercaptoethanol and 16 ng/μl salmon sperm DNA). DNA was precipitated with four volumes of ethanol and resuspended in piperidine 1 M. After cleavage at 90°C for 20 min, the reactions were stopped on ice and the DNA precipitated with 0.3 M sodium acetate, pH 5.2 and three volumes of ethanol. The DNA samples were resuspended in 90% formamide and 50 mM EDTA, denatured at 90°C and run for 2 h on a denaturing 20% polyacrylamide gel, prepared in TBE and 8 M urea, pre-equilibrated at 55°C in a Sequi-Gen GT Nucleic Acids Electrophoresis Apparatus (Bio-Rad, CA, USA), which was equipped with a thermocouple that allows a precise temperature control. After running, the gel was fixed in a solution containing 10% acetic acid and 10% methanol, dried at 80°C and exposed to film (CL-XPosure Thermo scientific, MA, USA) for auto-radiography. Lane scan and analysis was performed with Image Quant TL software (Image Scanner, Amersham, UK).
Pull-down assay with Panc-1 extract
A total of 0.5 mg of nuclear Panc-1 extract (1.3 mg/ml) were incubated for 1.5 h at 37°C with 80 nM biotinylated 32R or 96 in 20 mM Tris–HCl, pH 7.4, 50 mM KCl, 8% glycerol, 1 mM DTT, 0.1 mM ZnAc, 5 mM NaF, 1 mM Na3VO4 and 2.5 ng/μl poly[dI-dC]. A total of 100 μg of Streptavidin MagneSphere Paramagnetic Particles (Promega, I) were added and let to incubate for 1 h at 4°C. The beads were captured with a magnet and washed two times. The proteins were denatured and eluted with Laemmli buffer (4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.004% bromophenol blue and 0.125 M Tris–HCl). Then they were separated in 10% SDS-PAGE and blotted into nitrocellulose at 70 V for 2 h. The nitrocellulose membrane was blocked for 1 h with 5% fat dried milk in PBS and 0.05% Tween (Sigma-Aldrich, USA) at room temperature. The primary antibodies used were: anti-MAZ (clone 133.7, IgG mouse, Santa Cruz Biotechnology, USA) diluted 1:200, anti-hnRNP A1 (clone 9H10, IgG mouse, Sigma-Aldrich, USA) diluted 1:2000 and anti PARP-1 (polyclonal antibody, IgG rabbit, Cell Signalling Technology, USA) diluted 1:200. The membranes were incubated overnight at 4°C with the primary antibodies, then washed with 0.05% Tween in PBS and incubated for 1 h with the secondary antibodies conjugated to horseradish peroxidase: anti-mouse IgG (diluted 1:5000) and anti-rabbit IgG (diluted 1:5000) (Calbiochem, Merck Millipore, D). The signal was developed with Super Signal ®West PICO, and FEMTO (Thermo Fisher, USA) and detected with ChemiDOC XRS, Quantity One 4.6.5 software (BioRad Laboratories, USA).
RESULTS AND DISCUSSION
8-oxoG in KRAS is more abundant in G4 than in non-G4 regions
Cancer cells have relatively high levels of ROS that may damage DNA, RNA and proteins. Oxidative damage to DNA occurs mainly on guanine (21), in particular at the 5′ guanine of GG runs (22). G-rich quadruplex motifs, being composed by several runs of guanines, are effective hotspots for guanine oxidation (32). The formation of 7,8-dihydro-8-oxoguanine (or 8-oxoG) in these sequence motifs may be favored by the particular folded structure that they assume under physiological conditions (23,29). Indeed, four consecutive G-runs separated by few bases can form a G-quadruplex or G4 structure stabilized by tetrads of guanines. A G4-Seq conducted on the human genome found >7 × 105 potential G4 motifs, mainly located in functional regions including promoters, 5′-Untranslated region (UTRs) and splicing sites (33). A subsequent G4-ChIP-Seq analysis conducted on chromatin revealed a lower number (about 104) of G4 motifs folded into a G-quadruplex. The critical point of these studies is that not all G4 motifs are folded under cellular conditions. Interestingly, many folded G4 motifs are present in oncogenes including CMYC and KRAS (34). Recently, Burrows and co-workers developed an elegant method, ‘8-oxoG-Seq’, to sequence 8-oxoG in the mouse genome. They found ∼104 regions of 8-oxoG enrichment in WT mouse embryonic fibroblast, in particular where there are G4 motifs (gene promoters and UTRs) (35). We therefore asked if 8-oxoG has an epigenetic potential in gene regulation and focused on the KRAS oncogene, which harbors upstream of the TSS a G-rich sequence with regulatory functions.
Human KRAS displays three G4-motifs that, according to their distance from TSS, can be named G4-proximal (previously named 32R, −148/−116), G4-middle (−207/−175) and G4-far (−260/−226) (23,25,28,29,36). Sequence 32R (69% GC), which has been extensively studied in our laboratory, is sensitive to nucleases and shows a complex structural polymorphism (23,25,37). DMS-footprinting and CD experiments showed that 32R folds into a parallel G4 with a thymidine bulge in one strand and a (1/1/11) topology (23,25). Moreover, a truncated portion of 32R, comprising the first four G-runs from the 5′-end, folds into a (1/1/4) G-quadruplex (28,38). We reported that 32R is recognized by several nuclear proteins, including MAZ, PARP-1, Ku70 and hnRNP A1 (25–27). The role of MAZ and/or hnRNP A1 on KRAS transcription regulation has been investigated in our laboratory and also by Chu et al. (24,26,27,39). The data showed that both transcription factors upon binding to 32R unfold the G4 structure and favor the transcription process.
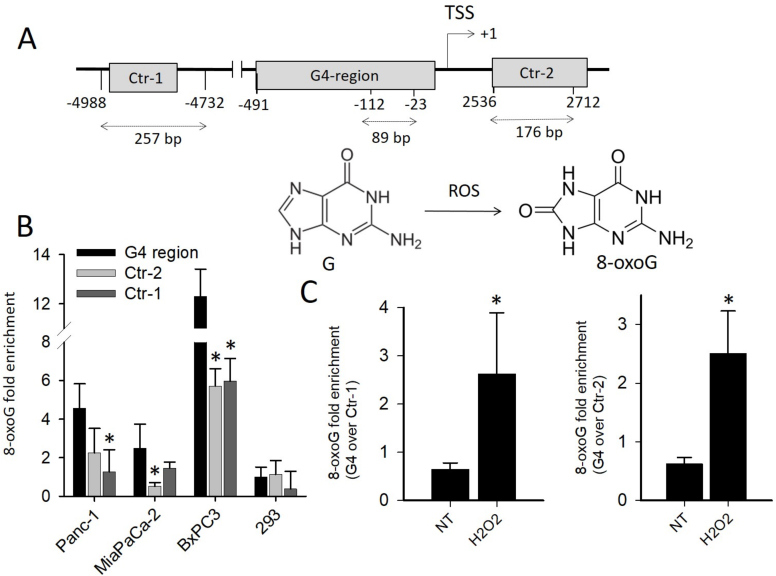
In order to understand, if the guanines of the G4 motifs upstream TSS are prone to oxidation, we carried out ChIP qPCR experiments. We measured the basal level of 8-oxoG in the KRAS promoter region including 32R and compared it to other G-rich regions which are unable to fold into G4: Ctr-1 (containing a segment with 56% CG) and Ctr-2 (with 65% CG), both located >2000 bp from 32R (Figure 1A). Preliminary semi-quantitative ChIP PCR experiments carried out with pancreatic Panc-1 cancer cells clearly showed that 8-oxoG is more abundant in G4 than in non-G4 regions (Supplementary Figure S2). However, to determine the difference in guanine oxidation between G4 and non-G4 regions, we performed quantitative ChIP qPCR in three pancreatic cancer cell lines (Panc-1 with KRAS G12D, MIA PaCa-2 with KRAS G12V and BxPC3 with wild-type KRAS) and in one non-tumor cell line (human embryonic kidney 293 cells, HEK-293) (Figure 1B). It turned out that the basal level of 8-oxoG in the cancer cells is up to 12-fold higher than in normal HEK-293 cells. This correlates with the fact that cancer cells have higher levels of ROS than normal cells (1,2). Our analysis also confirmed that in the three cancer cells analyzed, the G4-containing regions are more exposed to guanine oxidation (up to 4-fold) than the non-G4 regions Ctr-1 and Ctr-2. As Ctr-1 and Ctr-2 have a CG content >50% and contain several GG runs, the difference in 8-oxoG between G4 and non-G4 regions cannot be ascribed to a low presence of guanines in the non-G4 regions. Previous studies reported that the secondary structure adopted by DNA affects the reactivity of guanine toward oxidative stress and that the guanines in a G-quadruplex are more keen to oxidation than the guanines in a duplex (40,41). Another interesting observation is the higher level of 8-oxoG in BxPC3 cells compared to Panc-1 and MIA PaCa-2 cells. This can be rationalized with the fact that only the latter cells carry a hyperactivated mutant KRAS which is known to constitutively stimulate the expression of Nrf2, a gene that activates the detoxification program bringing down ROS and thus 8-oxoG (9–11). As previously reported (12,14), we also found that there is a direct link between KRAS and Nrf2, as the overexpression of KRAS in Panc-1 cells brought about an increase of Nrf2 (Supplementary Figure S3).
Figure 1.
(A) Relative distance from TSS of G4 and non-G4 sequences used in quantitative real-time ChIP experiments. The length of each amplified DNA fragment is indicated. The structures of guanine and 8-oxo-7,8-dihydroguanine (8-oxoG) are shown; (B) ChIP qPCR showing the basal level of 8-oxoG in G4 and in non-G4 regions Ctr-1 and Ctr-2 in pancreatic cancer cells harboring mutated KRAS (Panc-1 and MIAPaCa-2) or wild-type KRAS (BxPC3) and in non-cancer HEK-293 cells. The histogram shows the fold enrichment of 8-oxoG in pancreatic cancer cells compared to 8-oxoG in G4 region of 293 cells; (C) ChIP qPCR showing the relative level of 8-oxoG in G4 compared to non-G4 regions (Ctr-1 and Ctr-2). The ordinate reports the ratio between the level of 8-oxoG in G4 and in non-G4 region, in Panc-1 cells treated with 1 mM H2O2. The asterisk (*) means P < 0.05 (n = 4), a Student’s t-test was performed.
Finally, to make sure that the level of 8-oxoG correlates with the cellular amount of ROS, we performed ChIP qPCR assays with Panc-1 cells treated with H2O2. As expected, the treatment with H2O2 raised 8-oxoG in the G4 region ∼ 4-fold more than in the non-G4 regions Ctr-1 and Ctr-2 (Figure 1C).
8-OxoG affects the folding of the G-rich 32R promoter sequence
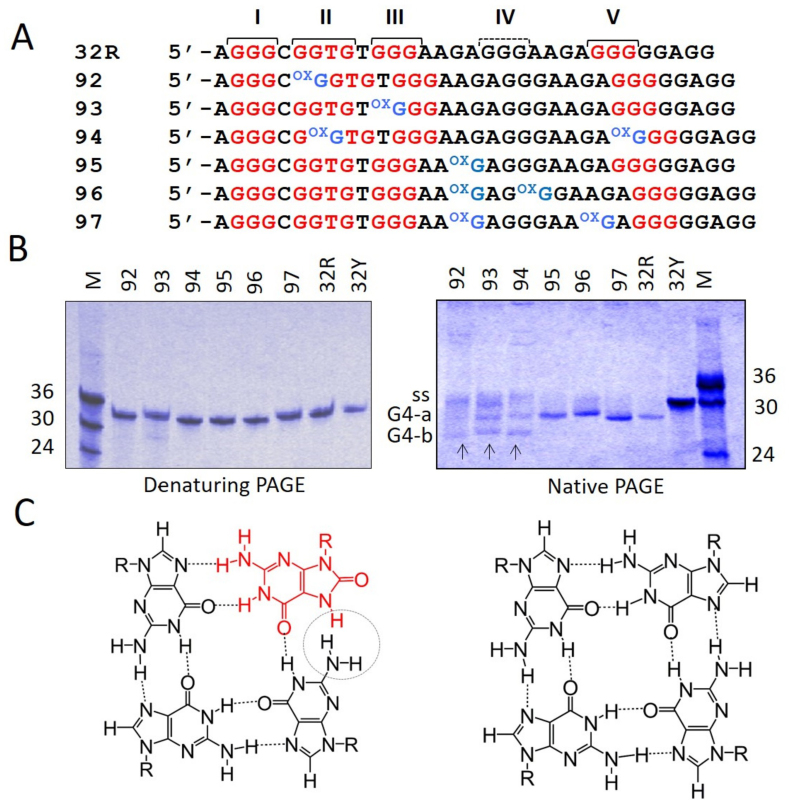
DMS-footprinting and CD studies, reported by ourselves (23,25) and others (36), showed that 32R folds into a parallel G4 formed by the G-runs I, II, III and V (Figure 2A). Distinctive features of this G-quadruplex are a large 11-nt loop and a strand with a thymidine bulge (Supplementary Figure S4). On the basis of this putative structure, we designed mimics of 32R, carrying 8-oxoG at specific positions: in the G-tetrads (oligonucleotides 92, 93 and 94) or in the 11-nt loop (95, 96 and 97). Under denaturing conditions (7 M urea), the 8-oxoG-substituted oligonucleotides migrated expectedly as wild-type 32R or as its complementary strand 32Y. By contrast, under native conditions (100 mM KCl) the oligonucleotides carrying one or two 8-oxoG in the 11-nt loop (95, 96 and 97) migrated in the same way as 32R, with a single band running faster than the unstructured oligonucleotide 32Y (Figure 2B). This suggests that when the oxidized guanine is in the 11-nt loop, the folding of the G4-motif is not affected [i.e. it is similar to that of wild-type 32R, (1/1/11) G4]. Instead, when 8-oxoG is placed in the G-runs II, III or V, which are involved in the formation of the G-tetrads (92, 93 and 94), the oligonucleotides migrated with 2-folded structures: one running as the G4 of wild-type 32R (band G4-a) and one running faster (band G4-b) (Figure 2B). It should be borne in mind that 8-oxoG is expected to destabilize the G-tetrad arrangement, as the N7 of diketo 8-oxoG becomes a hydrogen donor and sterically clashes with the amino group of a neighboring guanine (Figure 2C). This means that 8-oxoG can hardly stay in a G-tetrad, as the two H-bonding pattern is replaced by a single H-bonding pattern. It is therefore reasonable to assume that the incorporation of 8-oxoG in a G-tetrad destabilizes the G-quadruplex. Indeed, single substitutions of guanine with 8-oxoG have been reported to do so (42).
Figure 2.
(A) Sequences of 32R and of the designed oligonucleotides with one or two 8-oxoG either in the major 11-nt loop of KRAS G4 (92, 93 and 94) or in G-tetrads (95, 96 and 97). The G-runs I–V are pointed out; (B) 20% PAGE of 32R and 8-oxoG-substituted oligonucleotides in denaturing (left) and native (right) gels. As reference oligonucleotides of 24, 30 and 36 nt have been loaded. G4-a and G4-b are due to G4 structures, ss indicates unstructured oligonucleotides. Gels were repeated three times; (C) Structure of a G-tetrad with 8-oxoG (left) and of a canonical G-tetrad (right).
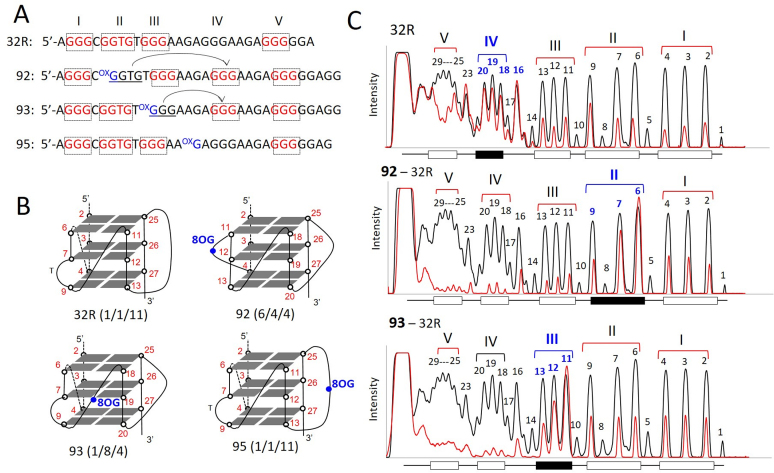
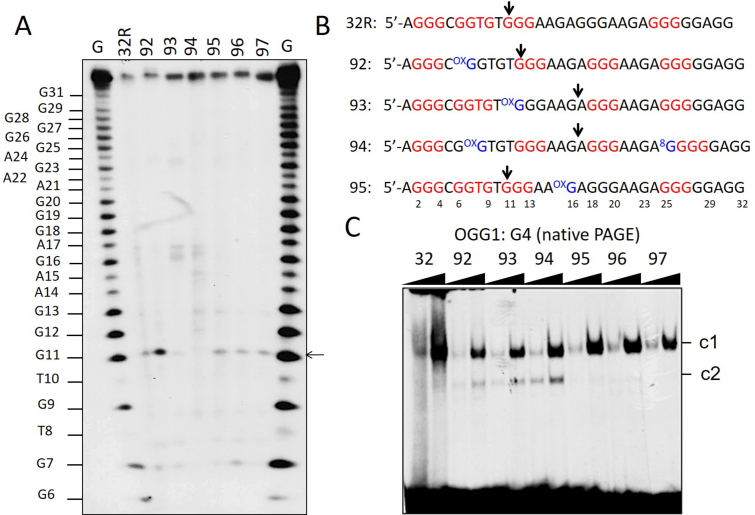
An insight into the structure of the human KRAS G-quadruplex with 8-oxoG substitutions was obtained by DMS-footprinting. As 32R contains 5 G-runs, it is likely that the G-run carrying 8-oxoG is excluded from the formation of the G-tetrads and replaced by the redundant fifth G-run present in 32R, as observed with G4 motifs found in oncogene promoters and telomeres (Figure 3A and B) (43,44). This occurs through an alternative folding of 32R. As illustrated in Figure 3C, wild-type 32R shows its typical DMS footprinting in 100 mM KCl with all guanines protected from DMS, except G16, G18–20 and G23. This cleavage pattern is consistent with the formation of a (1/1/11)-G4 by the G-runs I, II, III and V (23,25). The DMS-footprinting of oligonucleotide 92, which was designed with 8-oxoG in G-run II, clearly shows a different cleavage pattern. In keeping with the ‘fifth G-run’ hypothesis, its folding involves the G-runs I, III, IV and V, giving rise to (6/4/4) G-quadruplex. It can actually be seen that G-run II (G6-G7-G9) is reactive to DMS, while G-run IV (G18-G19-G20) is not. This DMS reactivity pattern demonstrates that the fifth G-run (G-run IV) has indeed replaced G-run II carrying 8-oxoG, through a re-modulation of the folding in order to exclude the oxidized guanine from the G4 scaffold. The resulting (6/4/4) G4, having loops <11 nt, is more compact than wild-type (1/1/11) G4 and that explains why it runs faster in a polyacrylamide gel (band G4-b, Figure 2B). Oligonucleotide 93, carrying 8-oxoG in G-run III, shows a similar behavior as 92. In this case, G-run III is more reactive to DMS, suggesting that it is replaced by G-run IV (G18-G19-G20), which appears protected. We also analyzed oligonucleotide 95 with 8-oxoG in the 11-nt loop, that should not impact on the folding. In keeping with this prediction, the footprinting of 95 shows that the bases cleaved are those located in the 11-nt loop, as observed with the wild-type 32R sequence (Supplementary Figure S5). A similar behavior has been observed with the critical G4 motif of CMYC (45). DMS footprinting and RNA polymerase stop assays showed that a simple 8-oxoG induces a change in the folding to exclude the oxidized G-run from the formation of the G-tetrads.
Figure 3.
(A) Sequences of 32R, 92, 93 and 95 showing the G-runs involved in G4 formation (indicated in red). Compared to 32R, 92 and 93 undergo a different folding in order to exclude 8-oxoG from a G-tetrad; (B) Structure of the putative G-quadruplexes with 8-oxoG; (C) Comparison of the DMS-footprinting of unstructured 32R with those of structured 32R and 92, 93 analogs. Note that the fifth G-run of 32R replaces the G-run with 8-oxoG through an alternative folding. Experiment was repeated three times.
Fleming and co-workers have observed that there are many oncogenes with G4 motifs carrying an extra G-run that could relieve a damaged guanine through a structural transition (44). The authors hypothesized that the extrusion of the damage into a loop should be necessary for the activation of the base excision repair mechanism. However, the evolutionary selection of a fifth G-run in regulatory G4 motifs recognized by transcription factors might also find its rationale in the fact that 8-oxoG could act as a transcription regulator by modulating the binding and recruitment of transcription factors to promoter sequences (see infra).
Circular dichroism and UV-melting of the G4 structures with 8-oxoG
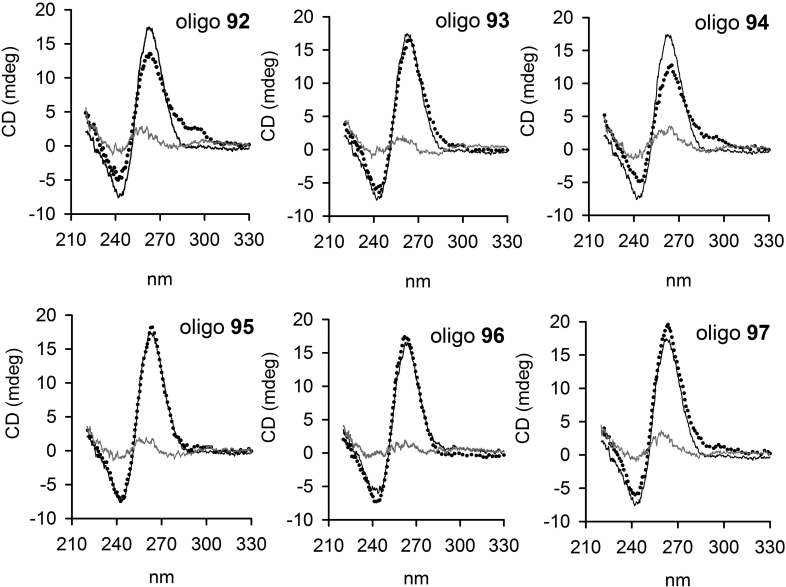
Next, we asked if the presence of 8-oxoG in 32R may affect the strand directionality of the G-quadruplex. To address this question, we performed circular dichroism experiments, as CD is a spectroscopic technique sensitive to DNA secondary structures. In Figure 4, we compared the CD spectra of each 8-oxoG-substituted sequence, obtained at 20 and 90°C, with the CD of 32R. At 20°C, all the spectra are characterized by a strong and positive ellipticity at 264 nm and a negative ellipticity at 245 nm, which are typical of a parallel or type I G-quadruplex (46). At 90°C, the intensity of the 264-nm ellipticity is dramatically reduced, suggesting that at 20°C the 8-oxoG-substituted oligonucleotides are structured. Oligonucleotides 92 (6/4/4) and 94 (6/4/5) show also a weak ellipticity at 295 nm, that may point to the formation of an alternative parallel/antiparallel G4. Instead, the CD spectra of 95, 96, 97 and 32R, forming a G4 with the same (1/1/11) topology, exhibit almost identical CD spectra.
Figure 4.
CD spectra of 32R at 20°C (black) and of 8-oxoG-substituted oligonucleotides at 20°C (dotted spectra) and 90°C (gray spectra), in 50 mM Tris–HCl, pH 7.4, 100 mM KCl. Each panel compare the CD spectrum of 32R with that of an oligonucleotide with 8-oxoG. The ordinate reports the ellipticity signal expressed in mdeg.
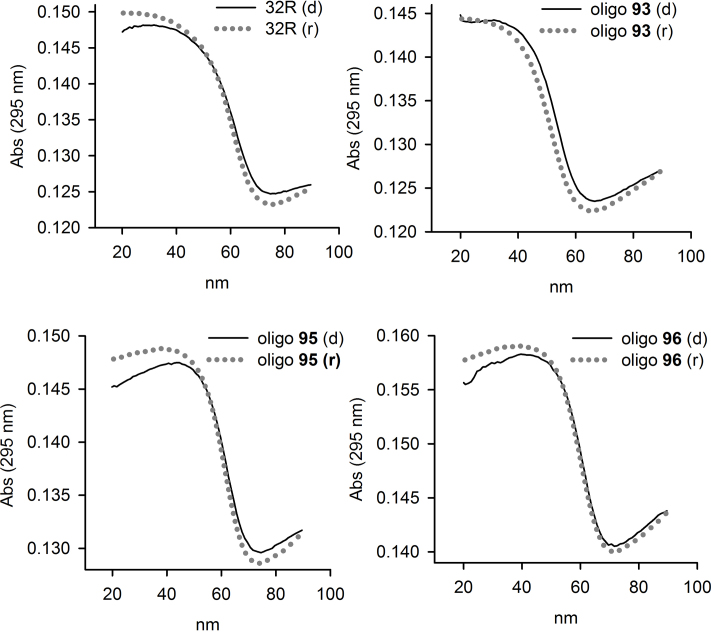
As the thermal differential spectra of 32R and 8-oxoG-substituted oligonucleotides show a negative band at 295 nm, we followed their melting by measuring the absorbance at 295 nm as a function of temperature (47). Typical melting curves are shown in Figure 5. At heating/cooling rates of 0.5°C/min, we obtained curves that are superimposable, indicating that the melting/annealing proceeded through equilibrium states. The TM’s, determined by the dAbs/dT versus T plots, are reported in Table 1. The data show that when the oxidized guanine is located in the 11-nt loop (95, 96 and 97), the damage is well tolerated, as the oxidized G4s show TM values nearly similar to that of 32R (ΔTM ∼ 1–2°C). So, in terms of stability, CD spectra, electrophoretic mobility and DMS-footprinting demonstrate that 32R and 95, 96 and 97 form the same G-quadruplex. In contrast, when the oxidized guanine is inserted in a G-tetrad, the TM’s of the resulting G4s (92, 93, 94) are significantly lower than that of the wild-type G4 (ΔTM ∼ 10–11°C), in agreement with previous data obtained with the telomeric sequence (42). However, in our case, the decrease of the TM’s is not due to altered stacking interactions between the G-tetrads, but to a change in the folding involving the fifth G-run.
Figure 5.
Denaturing and annealing UV-melting curves of 32R and 8-oxoG substituted oligonucleotides in 50 mM cacodylate pH 7.4, 100 mM KCl. The curves have been obtained by measuring the absorbance at 295 nm as a function of temperature, at a heating/cooling rates of 0.5°C/min. Denaturing, black filled curves; renaturing, dotted gray curves.
Table 1. Thermodynamic parameters for the KRAS 8-oxoG substituted oligonucleotides.
| Oligo | T M a (°C) | ΔTMb (°C) | Number 8-oxoG | ΔH°c kcal/mol | ΔS°c cal/K mol | ΔG°c,d kcal/mol |
|---|---|---|---|---|---|---|
| 92 | 50.7 | 11.4 | 1 in G-tetrad | −61.6 ± 0.8 | −165 ± 2 | −10.4 ± 0.2 |
| 93 | 52.8 | 9.3 | 1 in G-tetrad | −74.2 ± 1.3 | −202 ± 4 | −11.5 ± 0.1 |
| 94 | 52.6 | 9.5 | 2 in G-tetrad | −56.0 ± 0.9 | −147 ± 3 | −10.4 ± 0.2 |
| 95 | 61.5 | 0.6 | 1 in 11-nt loop | −78.7 ± 1.0 | −208 ± 1 | −14.2 ± 0.1 |
| 96 | 61.1 | 1.0 | 1 in 11-nt loop | −80.1 ± 1.3 | −215 ± 4 | −13.4 ± 0.1 |
| 97 | 63.7 | −1.6 | 2 in 11-nt loop | −85.1 ± 1.0 | −227 ± 3 | −14.7 ± 0.1 |
| 32R | 62.1 | −66.1 ± 1.3 | −172 ± 4 | −12.8 ± 0.2 |
adata obtained in 50 mM cacodylate pH 7.4, 100 mM KCl.
bΔTM = TM (32R) − TM (8-oxoG oligo).
cThermodynamic parameters obtained from analysis of melting cuves.
dΔG = ΔH − TΔS, T = 310 K.
The thermodynamic parameters of G-quadruplex formation were obtained from the melting profiles by using a ‘DNA Melting Analysis’ software (Jasco, JP). As the melting curves proceeded in a two-state manner, we could analyze them with a standard all-or-none model. The data reported in Table 1 show that the ΔG of the G-quadruplexes with one or two 8-oxoGs in the major 11-nt loop (95, 96 and 97) is ∼1–2 kcal/mol more favorable than the ΔG of wild-type G4. This increase of stability is enthalpic in origin, and may probably arise from more efficient stacking interactions between 8-oxoG and the surrounding bases in the loop. In contrast, when the damage is inserted in a G-tetrad (92, 93 and 94) the sequences fold into an alternative G-quadruplex with a ΔG about ∼2 kcal/mol less favorable than the ΔG of wild-type G4.
OGG1 is recruited to the KRAS G4 motif region carrying 8-oxoG
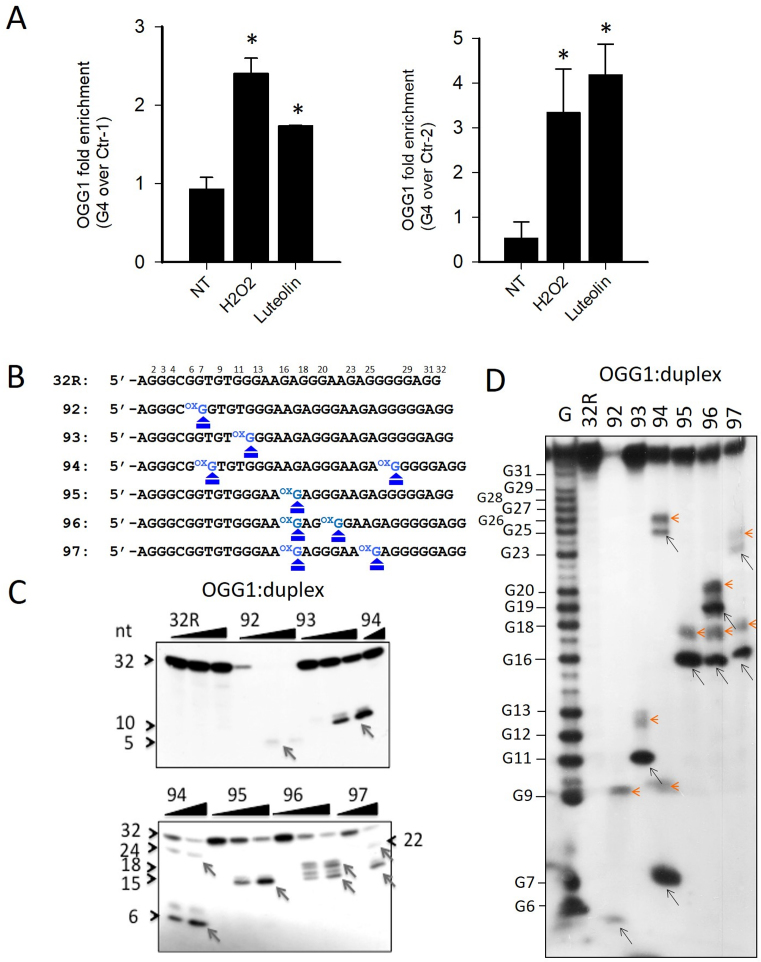
The enhanced level of 8-oxoG in the G4 region of KRAS upstream of TSS suggested to investigate whether the base-excision repair (BER) pathway is activated. BER is initiated by DNA glycosylases, which recognize and remove the oxidized guanines. The resulting apurinic site is then cleaved and the single-strand break processed by a short- or long-patch BER. The first enzyme of this pathway is OGG1, which behaves in vitro as a bifunctional DNA glycosylase excising 8-oxoG and cleaving the abasic site, while in vivo it behaves as a monofunctional glycosylase, with APE1 performing the lyase function (48–52). In order to see if OGG1 is recruited to the KRAS promoter when the level of 8-oxoG is increased by oxidative stress, we performed ChIP qPCR assays. We found that when Panc-1 cells are treated with H2O2 or luteolin, an inhibitor of Nrf2 that causes an increase of cellular ROS (53,54), the recruitment of OGG1 to the KRAS promoter increases more in G4 than in the non-G4 regions Ctr-1 and Ctr-2 (Figure 6A). We then produced and purified recombinant OGG1 to test its capacity to excise 8-oxoG and cleave the abasic site. Increasing amounts of enzyme were incubated, for 15 min at 37°C, with 32P-ATP labeled 8-oxoG-substituted oligonucleotides, transformed into duplexes with the complementary strand and the products were analyzed in a denaturing PAGE gel (Figure 6B and C). The enzyme did not show any activity on the wild-type 32R duplex, but it cleaved the duplexes carrying one or two 8-oxoGs. With duplexes 92, 93 and 95, which have only one 8-oxoG insertion, OGG1 gave only one main cleaved fragment; with duplexes 94, 96 and 97, designed with two 8-oxoG lesions, it produced two fragments, as expected. Note that the bands are doublets, because after having removed 8-oxoG, OGG1 cleaves the apurinic site by both β and δ eliminations (55). In a native gel, we detected only at 4°C a weak binding of OGG1 to the designed duplexes, because the enzyme destabilizes the complexes (Supplementary Figure S6) by cleaving the substrates. In order to confirm that the cleavage catalyzed by OGG1 occurs exactly at the duplex sites where guanine has been replaced with 8-oxoG, we analyzed the products of the enzymatic reaction in a sequencing gel (Figure 6D). In lane 1, we report a G-reaction that determines the sequence of the duplex substrate. The pattern obtained is in nice agreement with Figure 6B and C. Each duplex substrate gave the fragment of the expected length, indicating that the cleavage occurs exactly at the place where the damaged guanine is. In the sequencing gel, the doublets appear clearer showing that the two fragments differ for two/three nucleotides, as a results of successive β and δ eliminations (55).
Figure 6.
(A) ChIP qPCR showing that after treatment of Panc-1 cells with 1 mM H2O2 or 10 μM luteolin, the recruitment of OGG1 to the KRAS promoter increases more at G4 region than non-G4 regions (Ctr-1 and Ctr-2). Asterisk (*) indicate P < 0.05 (n = 4), a Student’s t-test was performed; (B) Primary sequences of 32R and designed 8-oxoG-substituted oligonucleotides where the positions of 8-oxoG are indicated; (C) The panel shows in a denaturing PAGE that OGG1 excises 8-oxoG in the duplexes formed by the designed 8-oxoG oligonucleotides and the complementary strand; (D) Sequencing 18% PAGE showing that OGG1 (1 μM) cleaves the radiolabeled duplexes (2 nM) exactly at the positions where there is 8-oxoG. The experiments in (C) and (D) were repeated three times.
Then, we analyzed the catalytic activity of OGG1 on 32R and 8-oxoG- substituted oligonucleotides in the G4 conformation. The radiolabeled oligonucleotides were let to fold in KCl buffer and incubated with OGG1. Almost no cleaved products were detected in a denaturing gel, suggesting that the enzyme does have no or only slight activity against G4, as previously reported by Zhou et al. (56) (not shown). However, when we analyzed in a sequencing gel the mixtures between OGG1 and the designed 8-oxoG G-quadruplexes, we observed that the enzyme had a weak activity against specific guanines in the G4s (Figure 7A and B). The G-quadruplexes with 1/1/11 topology showed a small cleavage at G11 (a guanine of an external G-tetrad, Supplementary Figure S4), while G4s with 1/8/4 and 6/4/5 topologies (93 and 94) showed a weak cleavage in the loop, at G16/A17. This cleavage activity does not occur at oxidized guanines, is G4 specific and was not detected in the duplexes (see Figure 6C and D). To rule out that the OGG1 cleavage could be due to specific depurinations occurring during oligomer synthesis or handling (57), yielding abasic sites recognized by OGG1, we treated the oligonucleotides with hot piperidine and re-purified them by electrophoresis. The weak activity of OGG1 was detected even after this treatment (Supplementary Figure S7). On a native gel, OGG1 binds to the KRAS G-quadruplexes in a non-productive manner, forming stable G4-OGG1 complexes (Figure 7C). It is worth noting that oligonucleotides 92, 93 and 94, that fold in two G-quadruplexes (see infra), form two OGG1:G4 complexes: one involving the (1/1/11)-G4 and the other the (6/4/4)-G4 (92), 1/8/4 G4 (93) and (6/4/5) G4 (94). Together, these experiments demonstrate that OGG1 recognizes both the duplex and folded conformations of 32R, but only with the duplex substrate the enzyme is able to excise 8-oxoG.
Figure 7.
(A) Sequencing 18% PAGE showing the effect of OGG1 (1 μM) on the G-quadruplexes. A very weak cleavage activity is detected at specific guanines: G11 in the G4s formed by 32R, 95, 96 and 97; G16/A17 in the G4s formed by 93, G4-(1/8/4) and 94, G4-(6/4/5). This cleavage activity is not correlated with 8-oxoG; (B) Sequences of the G4 motifs 32R, 92, 93, 94 and 95 showing the positions where the G4s are cleaved by OGG1 (96 and 97 behave as 95); (C) Native PAGE showing that OGG1 binds to radiolabeled 32R and 8-oxoG oligonucleotides (20 nM) in the G-quadruplex form. Note that 92, 93 and 94 form two complexes (c1 and c2) as they form in solution two quadruplexes. OGG1 (0.3 and 0.6 μg) and oligonucleotides (20 nM) have been incubated 45 min at 37°C prior to PAGE. The experiment in A was repeated three times, that in C two times.
Guanine oxidation and DNA folding modulate the binding of MAZ and hnRNP A1 to the KRAS promoter
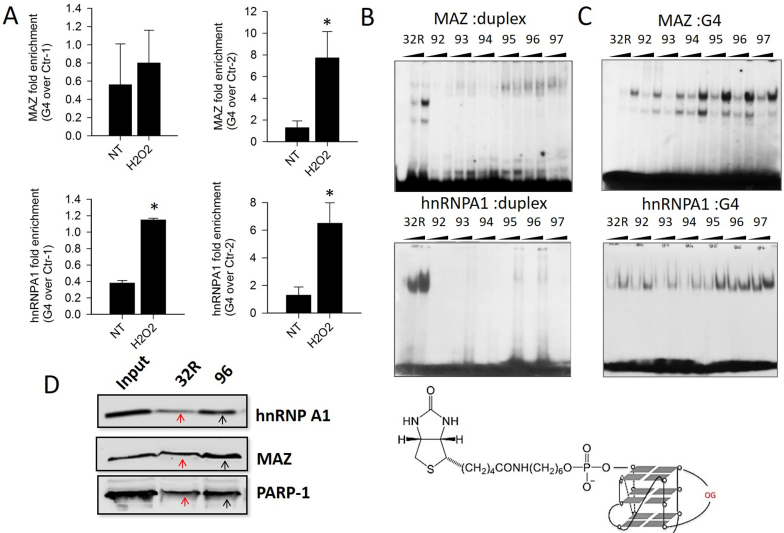
We previously have demonstrated that 32R in G4 conformation is recognized by several transcription factors including MAZ and hnRNP A1 (25). The consensus sequence of MAZ is 5′-GGG(A/C)GG (58). There are two binding sites for MAZ at the 5′ and 3′ ends of 32R. We found that MAZ activates the transcription of KRAS and HRAS (24,59). The KRAS G4 is also recognized by hnRNP A1, a protein of 34 kDa that has a wide range of functions including telomere biogenesis, RNA stability and control of transcription (60). The essential role of this nuclear factor in the transcription of KRAS has been demonstrated (26,27,39). Both MAZ and hnRNP A1, upon binding to the KRAS G4, destabilize the structure and facilitate, in the presence of the complementary strand, the transformation of G4 into duplex (26,59). As the guanines in 32R are exposed to oxidation, we asked if 8-oxoG modifies somehow the recruitment and binding of MAZ and hnRNP A1 to the KRAS promoter. To address the first point, we performed ChIP qPCR with Panc-1 cells treated with H2O2. The results show that the treatment increases the recruitment of MAZ and hnRNP A1 more to G4 than non-G4 regions, as a result of a cellular increase of 8-oxoG (Figure 8A). These data suggest that an increase of oxidation favors the recruitment to the promoter of MAZ and hnRNP A1, two proteins that activate transcription. A ChIP–reChIP assay (61), based on two independent rounds of immunoprecipitations with antibodies specific for MAZ/hnRNP A1 and 8-oxoG, was performed to further confirm the co-localization of the transcription factors and 8-oxoG in the chromatinized DNA fragment carrying the G4 motif (Supplementary Figure S8).
Figure 8.
(A) ChIP qPCR showing that the recruitment of MAZ and hnRNP A1 to the promoter G4 region is higher than to non-G4 regions (Ctr-1 and Ctr-2), following cell treatment with 1 mM H2O2. Asterisk (*) indicates P < 0.05 (n = 4), a Student’s t-test was performed; (B) The panels show the binding of MAZ and hnRNP A1 (2.5 and 5 μg) to 20 nM radiolabeled 32R and 8-oxoG oligonucleotides in duplex. The binding of MAZ and hnRNP A1 to the duplexes bearing 8-oxoG is strongly inhibited; (C) The panels show the binding of MAZ and hnRNP A1 to G-quadruplexes 32R and analogs bearing 8-oxoG. The proteins bind to the G4s, even though they harbor 8-oxoG. The G4s with 8-oxoG in the 11-nt loop (95, 96 and 97) bind MAZ much more than wild-type G4. Before EMSA, the G4s or duplexes have been incubated with MAZ for 1 h at 37°C and with hnRNP A1 for 30 min at 25°C; (D) Streptavidin-biotin pull-down assay with nuclear Panc-1 extract and biotinylated 32R and 96 used as DNA baits (the structure is shown). Cellular proteins bound to G4 were pulled down with streptavidin magnetic beads and analyzed by western blot. The experiments in (B) and (C) were repeated three times.
To know if the binding of MAZ and hnRNP A1 to the KRAS promoter is affected by 8-oxoG, we performed EMSA assays with recombinant proteins and 8-oxoG oligonucleotides in G4 or duplex conformation. Figure 8B reports native gels on the binding of MAZ and hnRNP A1 to wild-type 32R and 8-oxoG-substituted oligonucleotides in the duplex conformation. It shows that the interaction between the proteins and the duplexes carrying one or two 8-oxoG modifications is strongly inhibited. This is in keeping with the finding that the oxidation of both guanines in the consensus sequence of cAMP responsive element-binding protein (CREB) strongly decreased the protein binding (62). In contrast, when 32R and 8-oxoG oligonucleotides are in the G4 conformation, MAZ and hnRNP A1 bind to the DNA target despite it carries 8-oxoG lesions. It is worth noting that the binding of MAZ to 8-oxoG G-quadruplexes with (1/1/11) topology (95, 96, 97) increases by ∼5-fold compared to the binding of MAZ to 32R G4. The binding of hnRNP A1 also appears ∼4-fold more robust with the G-quadruplexes bearing 8-oxoG in the 11-nt loop. To confirm the finding that the transcription factors have more affinity for KRAS G4 when it is oxidized in the major loop, we covalently linked biotin to 32R and 96, carrying two 8-oxoGs in the 11-nt loop. We used the conjugates as DNA baits in streptavidin–biotin affinity precipitation experiments with a nuclear Panc-1 extract. The presence of MAZ and hnRNP A1 in the proteins binding to the DNA baits was detected and quantified by western blots (Figure 8D). The results are in keeping with those obtained with the recombinant proteins: in the presence of all nuclear proteins, oxidized G4 (96) shows ∼2-fold higher affinity than wild-type G4 for MAZ and hnRNP A1 [also for PARP-1, a protein that binds to 32R (25)]. Considering the G4 unfolding activity of MAZ and hnRNP A1, these findings are likely to have an impact on the transcription regulation mechanism, as proposed in the following paragraph.
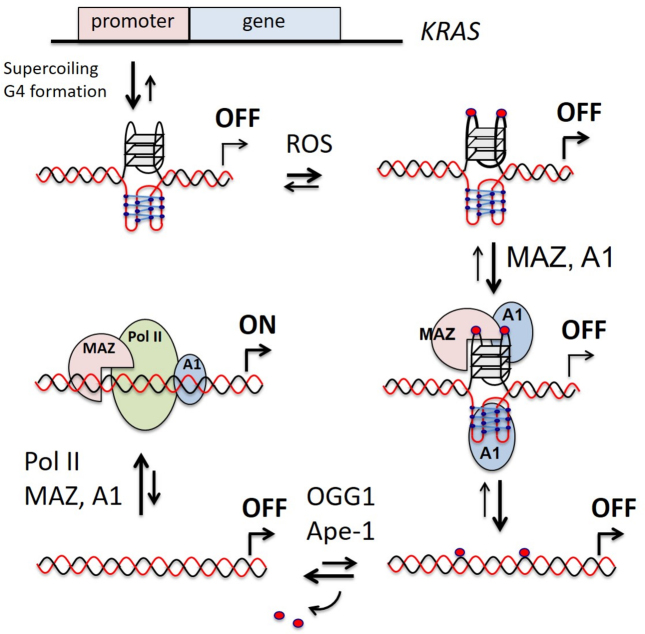
A transcription model for oncogene KRAS involving 8-oxoG, OGG1, hnRNP A1 and MAZ
The results of this study, together with previously reported data, add a new element to the molecular mechanism controlling KRAS transcription in pancreatic cancer cells: the oxidation of guanine at the critical G4 motif upstream TSS. The high metabolic rate of cancer cells causes an enhanced level of ROS that favors the oxidation of guanine. An increased level of 8-oxoG going beyond the repairing capacity by the cell, would have a negative impact on KRAS expression, as the binding of MAZ and hnRNP A1 to the promoter in the duplex conformation would be strongly inhibited. To prevent this, pancreatic cancer cells express high levels of Nrf2: a protein that stimulates the detoxification pathway to keep the oxidative damage at levels compatible with an optimal cell growth. Indeed, in agreement with previous work (63), we found that the inhibition of Nrf2 brought about an increase of ROS in Panc-1 cells (Supplementary Figure S9). The damage caused to DNA by oxidative stress can modulate gene expression in different ways. While the insertion of a single 8-oxoG in a promoter non-G4 sequence of a reporter gene was found to affect negatively transcription (64), when the oxidized guanine was inserted in the G4-motif of VEGF composed of five G-runs, the expression of Renilla luciferase increased by 3-fold. To rationalize this behavior, Fleming et al. (65) proposed that gene expression increases because 8-oxoG is excised by OGG1, yielding an abasic site that would favor the folding into a G4 looping out the oxidized G-run. This conformation should facilitate the binding of Ape1 to G4, but without cleaving efficiently the abasic site (66). Finally, the Ref-1 domain of Ape1 would recruit other nuclear factors and stimulate transcription. In addition to this interesting mechanism, Boldogh and co-workers (67) found that in the TNF-α promoter, ROS preferentially oxidizes the guanines in a G-rich region adjacent to a NF-kB-binding site. The authors sustained that the binding of OGG1 to the oxidized G-rich region would promote the recruitment of NF-kB to its binding site, with the consequence of activating transcription. The former mechanism has a more general character and could rationalize also the data of our study. However, also another mechanism can be postulated, in keeping with the original idea that G-quadruplex behaves as a transcription repressor (Figure 9). If we assume that the picture found for HRAS (59), based on a mutation analysis of the promoter, also holds for KRAS, the promoter G4 motif is normally folded into G4 and transcription is kept to a basal low level. The folding of the G4 motif is favored by DNA supercoiling, which provides sufficient energy to locally unwind the double helix (68,69). This induces the formation of a G-quadruplex on the purine-rich strand and, most likely, an i-motif on the pyrimidine strand (70). Although the i-motif shows in vitro higher stability under slightly acidic conditions, both cellular crowding and supercoiling are expected to stabilize this unusual structure at physiological conditions too (69,71,72). The relatively high oxidative stress in pancreatic cancer cells may induce the oxidation of certain guanines, particularly the DNA motifs composed by blocks of guanines (73) and folded into G4 (40). Our data show that the presence of one or two oxidized guanines in the major 11-nt loop of the KRAS G-quadruplex increases the binding of MAZ, hnRNP A1 and also PARP-1 to the G-quadruplex. As hnRNP A1 recognizes also the i-motif (71), both strands of the G4 motif could interact with nuclear proteins. The proteins upon binding should destabilize the folded structures and facilitate the reconstitution of the double helix (59,71). In this scheme, 8-oxoG would act as an epigenetic marker boosting the recruitment to the promoter of the nuclear factors essential for KRAS transcription. The oxidized guanines in the reconstituted duplex will be efficiently repaired via the BER pathway involving OGG1, that excises 8-oxoG, and Ape1, that cleaves the abasic site (48,74). The excision of 8-oxoG from the G4 motif in double helix will increase the affinity of the nuclear factors for the promoter, with the result of activating transcription. This mechanism is supported by the fact that KRAS transcription strongly depends on MAZ (24,59) and hnRNP A1 (26,27,39). It also depends on OGG1, as its downregulation in Panc-1 cells by siRNA determines a parallel decrease of KRAS transcription (Supplementary Figure S10).
Figure 9.
A model for KRAS transcription regulation involving 8-oxoG. A1 and Pol II stand for hnRNP A1 and RNA Pol II.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Magnar Bjorås (University of Oslo) for providing plasmid pET20hOGG1 expressing human OGG1.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Associazione Italiana per la Ricerca sul Cancro (AIRC) [IG 2013, project code 14301]. Funding for open access charge: AIRC [14301].
Conflict of interest statement. None declared.
REFERENCES
- 1. Liou G.-Y., Storz P.. Reactive oxygen species in cancer. Free Radic. Res. 2010; 44:1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sosa V., Moliné T., Somoza S., Paciucci R., Kondoh H.. Oxidative stress and cancer: an overview. Ageing Res. Rev. 2012; 12:376–390. [DOI] [PubMed] [Google Scholar]
- 3. Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009; 417:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Giorgio M., Trinei M., Migliaccio E., Pelicci P.G.. Hydrogen peroxide: a metabolic by-product or a common mediator of ageing signals. Nat. Rev. Mol. Cell Biol. 2007; 8:722–728. [DOI] [PubMed] [Google Scholar]
- 5. St-Pierre J., Buckingham J., Roebuck S.J., Brand M.D.. Topology of superoxide production from different sites in mitochondrial electron transport chain. J. Biol. Chem. 2002; 277:44784–44790. [DOI] [PubMed] [Google Scholar]
- 6. Lloyd R.V., Hanna P.M., Mason R.P.. The origin of the hydroxyl radical oxygen in the Fenton reaction. Free Radic. Biol. Med. 1997; 22:885–888. [DOI] [PubMed] [Google Scholar]
- 7. Trachootham D., Lu W., Ogasawara M.A., Nilsa R.D., Huang P.. Redox regulation of cell survival. Antioxid. Redox Signal. 2008; 10:1343–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klaunig J.E., Kamendulis L.M., Hocevar B.A.. Oxidative stress and oxidative damage in carcinogenesis. Toxicol. Pathol. 2010; 38:96–109. [DOI] [PubMed] [Google Scholar]
- 9. Jaramillo M.C., Zhang D.D.. The merging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013; 27:2179–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mitsuishi Y., Motohashi H., Yamamoto M.. The Keap1–Nrf2 system in cancers: stress response and anabolic metabolism. Front. Oncol. 2012; 2:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hayes J.D., McMahon M., Chowdhry S., Dinkova-Kostova A.T.. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid. Redox Signal. 2010; 13:1713–1748. [DOI] [PubMed] [Google Scholar]
- 12. Tao S., Wang S., Moghaddam S.J., Ooi A., Chapman E.. Oncogenic KRAS confers chemoresistance by upregulating Nrf2. Cancer Res. 2014; 74:7430–7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kong B., Erkan M., Kleeff J., Michalski C.W.. Overview on how oncogenic KRAS promotes pancreatic carcinogenesis by inducing low intracellular ROS levels. Front. Physiol. 2013; 4:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. DeNicola G.M., Karreth F.A., Humpton J., Gopinathan A., Wei C., Frese K., Mangal D., Yu K.H., Teo C.J., Calhoun E.S. et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011; 475:106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hezel A.F., Kimmelmann A.C., Stanger B.Z., Bardeesy N., Depinho R.A.. Genetic and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006; 20:1218–1249. [DOI] [PubMed] [Google Scholar]
- 16. Almoguera C., Shibata D., Forrester K., Martin J., Arnheim N., Perucho M.. Most human carcinomas of the exocrine pancreas contain mutant c-Ki-ras gene. Cell. 1988; 53:549–554. [DOI] [PubMed] [Google Scholar]
- 17. Yeh J.J., Der C.J.. Targeting signal transduction in pancreatic cancer treatment. Expert. Opin. Ther. Targets. 2007; 11:673–694. [DOI] [PubMed] [Google Scholar]
- 18. Collins M.A., Bednar F., Zhang Y., Brisset J.C., Galban S., Galban C.J., Rakshit S., Flannagan K.S., Adsay N.V., Pasca di Magliano M.. Oncogenic KRAS is required for both the initiation and maintenance of pancreatic cancer in mice. J. Clin. Invest. 2012; 122:639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pasca di Magliano M, Logsdon C.D.. Roles of KRAS in panceratic tumor development and progression. Gastroeneterology. 2013; 144:1220–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hingorani S.R., Petricoin E.F., Maitra A., Rajapakse V., King C., Jacobetz M.A., Ross S., Conrads T.P., Veenstra T.D., Hitt B.A. et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003; 4:437–450. [DOI] [PubMed] [Google Scholar]
- 21. Cadet J., Douki T., Ravanat J.L.. Oxidatively generated damage to the guanine moiety of DNA: mechanistic aspects and formation in cells. Acc. Chem. Res. 2008; 41:1075–1083. [DOI] [PubMed] [Google Scholar]
- 22. Kanvah S., Joseph J., Schuster G.B., Barnett R.N., Cleveland C.L., Landman U.. Oxidation of DNA: damage to nucleobases. Acc. Chem. Res. 2009; 43:280–287. [DOI] [PubMed] [Google Scholar]
- 23. Cogoi S., Xodo L.E.. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006; 34:2536–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cogoi S., Zorzet S., Rapozzi V., Géci I., Pedersen E.B., Xodo L.E.. MAZ-binding G4-decoy with locked nucleic acid and twisted intercalating nucleic acid modifications suppresses KRAS in pancreatic cancer cells and delays tumor growth in mice. Nucleic Acids Res. 2013; 41:4049–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cogoi S., Paramasivam M., Spolaore B., Xodo L.E.. Structural polymorphism within a regulatory element of the human KRAS promoter: formation of G4-DNA recognized by nuclear proteins. Nucleic Acids Res. 2008; 36:3765–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paramasivam M., Membrino A., Cogoi S., Fukuda H., Nakagama H., Xodo L.E.. Protein hnRNP A1 and its derivative Up1 unfold quadruplex DNA in the human KRAS promoter: implications for transcription. Nucleic Acids Res. 2009; 37:2841–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cogoi S., Rapozzi V., Cauci S., Xodo L.E.. Critical role of hnRNP A1 in activating KRAS transcription in pancreatic cancer cells: A molecular mechanism involving G4 DNA. Biochim. Biophys. Acta. 2016; 1861:1389–1398. [DOI] [PubMed] [Google Scholar]
- 28. Paramasivam M., Cogoi S., Xodo L.E.. Primer extension reactions as a tool to uncover folding motifs within complex G-rich sequences: analysis of the human KRAS NHE. Chem. Commun. (Camb). 2011; 47:4965–4967. [DOI] [PubMed] [Google Scholar]
- 29. Cogoi S., Xodo L.E.. G4 DNA in ras genes and its potential in cancer therapy. Biochim. Biophys. Acta. 2016; 1859:663–674. [DOI] [PubMed] [Google Scholar]
- 30. Bodepudi V., Shibutani S., Johnson F.. Synthesis of 2′-deoxy-7,8-dihydro-8-oxoguanosine and 2′-deoxy-7,8-dihydro-8-oxoadenosine and their incorporation into oligomeric DNA. Chem. Res. Toxicol. 1992; 5:608–617. [DOI] [PubMed] [Google Scholar]
- 31. Lin X., Tirichine L., Bowler C.. Protocol: chromatin immunoprecipitation (ChIP) methodology to investigate histone modifications in two model diatom species. Plant Methods. 2012; 8:48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Clark D.W., Phang T., Edwards M.G., Geraci M.W., Gillespie M.N.. Promoter G-quadruplex sequences are targets for base oxidation and strand cleavage during hypoxia-induced transcription. Free Radic. Biol. Med. 2012; 53:51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S.. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015; 33:877–881. [DOI] [PubMed] [Google Scholar]
- 34. Hänsel-Hertsch R., Beraldi D., Lensing S.V., Marsico G., Zyner K., Parry A., Di Antonio M., Pike J., Kimura H., Narita M. et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016; 48:1267–1272. [DOI] [PubMed] [Google Scholar]
- 35. Ding Y., Fleming A.M., Burrows C.J.. Sequencing the mouse genome for the oxidatively modified base 8-Oxo-7,8-dihydroguanine by OG-seq. J. Am. Chem. Soc. 2017; 139:2569–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morgan R.K., Batra H., Gaerig V.C., Hockings J., Brooks T.A.. Identification and characterization of a new G-quadruplex forming region within the KRAS promoter as a transcriptional regulator. Biochim. Biophys. Acta. 2016; 1859:235–245. [DOI] [PubMed] [Google Scholar]
- 37. Podbevsek P., Plavec J.. KRAS promoter oligonucleotide with decoy activity dimerazes into a unique topology consisting of two G-quadruplex units. Nucleic Acids Res. 2016; 44:917–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kerkour A., Marquevielle J., Ivashchenko S., Yatsunyk L.A., Mergny J.L., Salgado G.F.. High-resolution 3D NMR structure of the KRAS proto-oncogene promoter reveals key features of a G-quadruplex involved in transcriptional regulation. J. Biol. Chem. 2017; 292:8082–8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chu P.C., Yang M.C., Kulp S.K., Salunke S.B., Himmel L.E., Fang C.S., Jadhav A.M., Shan Y.S., Lee C.T., Lai M.D. et al. Regulation of oncogenic KRAS signaling via a novel KRAS-integrin-linked kinase-hnRNPA1 regulatory loop in human pancreatic cancer cells. Oncogene. 2016; 35:3897–3908. [DOI] [PubMed] [Google Scholar]
- 40. Szalai V.A., Singer M.J., Thorp H.H.. Site-specific probing of oxidative reactivity and telomerase function using 7,8-dihydro-8-oxoguanine in telomeric DNA. J Am. Chem. Soc. 2002; 124:1625–1631. [DOI] [PubMed] [Google Scholar]
- 41. Fleming A.M., Burrows C.J.. G-quadruplex folds of the human telomere sequence alter the site reactivity and reaction pathway of guanine oxidation compared to duplex DNA. Chem. Res. Toxicol. 2013; 26:593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vorlícková M., Tomasko M., Sagi A.J., Bednarova K., Sagi J.. 8-oxoguanine in a quadruplex of the human telomere DNA sequence. FEBS J. 2012; 279:29–39. [DOI] [PubMed] [Google Scholar]
- 43. Fleming A.M., Zhou J., Wallace S.S., Burrows C.J.. A role for the fifth G-track in G-quadruplex forming oncogene promoter sequences during oxidative stress: do these sparetires have an evolved function. ACS Cent. Sci. 2015; 1:226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. An N., Fleming A.M., Burrows C.J.. Human telomere G-quadruplexes with five repeats accommodate 8-Oxo-7,8-dihydroguanine by looping out the DNA damage. ACS Chem. Biol. 2016; 11:500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Beckett J., Burns J., Broxson C., Tornaletti S.. Spontaneous DNA lesions modulate DNA structural transitions occurring at nuclease hypersensitive element III of the human c-myc proto-oncogene. Biochemistry. 2012; 51:5257–5268. [DOI] [PubMed] [Google Scholar]
- 46. Tran P.L.T., Mergny J.L., Alberti P.. Stability of telomeric G-quadruplexes. Nucleic Acids Res. 2011; 39:3282–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mergny J.L., Phan A.T., Lacroix L.. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998; 435:74–78. [DOI] [PubMed] [Google Scholar]
- 48. Rosenquist T.A., Zharkov D.O., Grollman A.P.. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:7429–7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Roldán-Arjona T., Wei Y.F., Carter K.C., Klungland A., Anselmino C., Wang R.P., Augustus M., Lindahl T.. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:8016–8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Radicella J.P., Dherin C., Desmaze C., Fox M.S., Boiteux S.. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:8010–8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. David S.S., O'Shea V.L., Kundu S.. Base-excision repair of oxidative DNA damage. Nature. 2007; 447:941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Allgayer J., Kitsera N., Bartelt S., Epe B., Khobta A.. Widespread transcriptional gene inactivation initiated by a repair intermediate of 8-oxoguanine. Nucleic Acids Res. 2016; 44:7267–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tang X., Wang H., Fan L., Wu X., Xin A., Ren H., Wang X.J.. Luteolin inhibits Nrf2 leading to negative regulation of the Nrf2/ARE pathway and sensitization of human lung carcinoma A549 cells to therapeutic drugs. Free Radic. Biol. Med. 2011; 50:1599–1609. [DOI] [PubMed] [Google Scholar]
- 54. Chian S., Thapa R., Chi Z., Wang X.J., Tang X.. Luteolin inhibits the Nrf2 signaling pathway and tumor growth in vivo. Biochem. Biophys. Res. Commun. 2014; 447:602–608. [DOI] [PubMed] [Google Scholar]
- 55. Girard P.M., Guibourt N., Boiteux S.. The Ogg1 protein of Saccharomyces cerevisiae: a 7,8-dihydro-8-oxoguanine DNA glycosylase/AP lyase whose lysine 241 is a critical residue for catalytic activity. Nucleic Acids Res. 1997; 25:3204–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhou J., Fleming A.M., Averill A.M., Burrows C.J., Wallace S.S.. The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 2015; 43:4039–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Amasova O., Coulter R., Fresco J.R.. Self calalyzed site-specific depurination of guanine residues within gene sequences. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:4392–4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Parks C.L., Shenk T.. The serotonin 1a receptor gene contains a TATA-less promoter that responds to MAZ and Sp1. J. Biol. Chem. 1996; 271:4417–4430. [DOI] [PubMed] [Google Scholar]
- 59. Cogoi S., Shchekotikhin A.E., Xodo L.E.. HRAS is silenced by two neighboring G-quadruplexes and activated by MAZ, a zinc-finger transcription factor with DNA unfolding property. Nucleic Acids Res. 2014; 42:8379–8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jean-Philippe J., Paz S., Caputi M.. hnRNP A1: the Swiss Army Knife of Gene Expression. Int. J. Mol. Sci. 2013; 14:18999–19024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Furlan-Magaril M., Rincón-Arano H., Recillas-Targa F.. Sequential chromatin immunoprecipitation protocol: ChIP-reChIP. Methods Mol. Biol. 2009; 543:253–266. [DOI] [PubMed] [Google Scholar]
- 62. Moore S.P., Toomire K.J., Strauss P.R.. DNA modifications repaired by base excision repair are epigenetic. DNA Repair (Amst). 2013; 12:1152–1158. [DOI] [PubMed] [Google Scholar]
- 63. Sporn M.B., Liby K.T.. NRF2 and cancer: the good, the bad and the importance of context. Nat. Rev. Cancer. 2012; 12:564–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Allgayer J., Kitsera N., Bartelt S., Epe B., Khobta A.. Widespread transcriptional gene inactivation initiated by a repair intermediate of 8-oxoguanin. Nucleic Acids Res. 2016; 44:7267–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fleming A.M., Ding Y., Burrows C.J.. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:2604–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Broxson C., Hayner J.N., Beckett J., Bloom L.B., Tornaletti S.. Human AP endonuclease inefficiently removes abasic sites within G4 structures compared to duplex DNA. Nucleic Acids Res. 2014; 42:7708–7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pan L., Zhu B., Hao X., Vlahopoulos S.A., Hazra T.K., Hedge M.L., Rodak Z., Bacsi A., Brasier A.R., Ba X. et al. Oxidized guanine base lesions Function in 8-oxoguanine DNA glycosylase-1-mediated epigenetic regulation of nuclear factor kB-driven gene expression. J. Biol. Chem. 1016; 291:25553–25566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sun D., Hurley L.H.. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression. J. Med. Chem. 2009; 52:2863–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Brooks T.A., Hurley L.H.. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat. Rev. Cancer. 2009; 9:849–861. [DOI] [PubMed] [Google Scholar]
- 70. Manzini G., Yathindra N., Xodo L.E.. Evidence for intramolecularly folded i-DNA structures in biologically relevant CCC-repeat sequences. Nucleic Acids Res. 1994; 22:4634–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Miglietta G., Cogoi S., Pedersen E.B., Xodo L.E.. GC-elements controlling HRAS transcription form i-motif structures unfolded by heterogeneous ribonucleoprotein particle A1. Sci. Rep. 2015; 5:18097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kang H.J., Kendrick S., Hecht S.M., Hurley L.H.. The transcriptional complex between the BCL2 i-Motif and hnRNP LL is a molecular switch for control of gene expression that can be modulated by small molecules. J. Am. Chem. Soc. 2014; 136:4172–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Arnold A.R., Grodick M.A., Barton J.K.. DNA charge transport: from chemical principls to the cell. Cell Chem. Biol. 2016; 23:183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tell G., Quadrufoglio F., Tiribelli C, Kelley M.R.. The many functions of APE1/Ref-1: not only a DNA repair enzyme. Antioxid. Redox Signal. 2009; 11:601–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.