The disubstituted urea molecule has a twisted conformation for each of the two molecules comprising the asymmetric unit. Intramolecular amine-N—H⋯N(imine) and hydroxy-O—H⋯O(methoxy) hydrogen bonds are noted. In the molecular packing, amide-N—H⋯O(amide), hydroxyl-O—H⋯N(imine) and phenylamine-N—H⋯O(methoxy) hydrogen bonding leads to layers in the ac plane.
Keywords: crystal structure, urea derivative, hydrogen bonding, Hirshfeld surface analysis
Abstract
Two independent molecules (A and B) comprise the asymmetric unit of the title compound, C18H21N3O3. The urea moiety is disubstituted with one amine being linked to a phenyl ring, which is twisted out of the plane of the CN2O urea core [dihedral angles = 25.57 (11) (A) and 29.13 (10)° (B)]. The second amine is connected to an imine (E conformation), which is linked in turn to an ethane bridge that links a disubstituted benzene ring. Intramolecular amine-N—H⋯N(imine) and hydroxyl-O—H⋯O(methoxy) hydrogen bonds close S(5) loops in each case. The molecules have twisted conformations with the dihedral angles between the outer rings being 38.64 (81) (A) and 48.55 (7)° (B). In the crystal, amide-N—H⋯O(amide) hydrogen bonds link the molecules A and B via an eight-membered {⋯HNCO}2 synthon. Further associations between molecules, leading to supramolecular layers in the ac plane, are hydrogen bonds of the type hydroxyl-O—H⋯N(imine) and phenylamine-N—H⋯O(methoxy). Connections between layers, leading to a three-dimensional architecture, comprise benzene-C—H⋯O(hydroxy) interactions. A detailed analysis of the calculated Hirshfeld surfaces shows molecules A and B participate in very similar intermolecular interactions and that any variations relate to conformational differences between the molecules.
Chemical context
Semicarbazones belong to the general class of molecules termed Schiff bases and are prepared from condensation of semicarbazides with aldehydes/ketones. They have attracted considerable attention due to their wide spectrum of biological activities, including anti-convulsant (Pandey & Srivastava, 2010 ▸), anti-tubercular (Sriram et al., 2004 ▸), anti-cancer (Ali et al., 2012 ▸) and anti-microbial (Beraldo & Gambino, 2004 ▸). Actually, they have been investigated extensively for their anti-convulsant properties with 4-(4-fluorophenoxy)benzaldehyde semicarbazone, in particular, attracting attention as a potent anti-epileptic drug over the past 15 years (Pandeya, 2012 ▸). Recently, the crystal structures of related chalcone-derived thiosemicarbazones and their transition metal complexes have been reported (Tan et al., 2015 ▸, 2017 ▸). In this contribution, aryl semicarbazide is introduced with vanillylacetone, which led to the formation of the title compound. Vanillylacetone is one of the active components of ginger and possesses strong anti-oxidant and chemopreventive properties (Kıyak et al., 2015 ▸). The structural elucidation of such compounds has not been extensively investigated. In order to redress this, herein the crystal and molecular structures of the title compound, (I), are described along with an analysis of the calculated Hirshfeld surface in order to ascertain more details of the supramolecular association operating in the crystal.
Structural commentary
Two independent molecules, A and B, comprise the asymmetric unit of (I) and these are shown in Fig. 1 ▸. Each molecule features a disubstituted urea molecule with one amine group connected to a phenyl ring and the other linked to a disubstituted imine group, with the longer side-chain carrying an ethane chain terminating with a disubstituted benzene ring. The four atoms comprising the urea core are strictly planar with an r.m.s. deviation of 0.0041 Å [0.0043 for the O4-molecule, molecule B]. The phenyl ring is inclined to this plane, forming a dihedral angle of 25.57 (11)° [29.13 (10)° for molecule B]. Intramolecular N—H⋯N hydrogen bonds are found within the urea residues, Table 1 ▸. A significant kink in the molecule occurs in the ethane bridge, as seen in the value of −157.88 (16)° for the C8—C9—C10—C11 torsion angle [C26—C27—C28—C29 = 162.93 (17)° for B]. As a result, the molecule is twisted with the terminal rings inclined to each other, forming a (C2–C7)/(C11–C16) dihedral angle of 38.64 (8)° [(C20–C25)/(C29–C34) = 48.55 (7)° for B]. The latter represents the major difference between molecules A and B, as illustrated in the overlay diagram shown in Fig. 2 ▸. In each of the disubstituted benzene rings, the hydroxyl-H atom is orientated to allow the formation of intramolecular O—H⋯O hydrogen bonds with the methoxy-O atom, Table 1 ▸. The conformation about the imine bond [N3=C8 = 1.281 (2) and N6=C26 = 1.276 (2) Å] is E in each molecule. Finally, each of the methoxy substituents is twisted out of the plane of the ring to which it is bonded [C18—O2—C13—C12 = 11.7 (3) and C36—O5—C31—C30 = −16.5 (3)°].
Figure 1.
The molecular structures of the two independent molecules comprising the asymmetric unit of (I), showing the atom-labelling scheme and displacement ellipsoids at the 70% probability level.
Table 1. Hydrogen-bond geometry (Å, °).
Cg1, Cg2 and Cg3 are the centroids of the C2–C7, C29–C34 and C20–C25 rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N⋯N3 | 0.86 (2) | 2.18 (2) | 2.635 (2) | 113 (2) |
| N4—H4N⋯N6 | 0.86 (2) | 2.23 (2) | 2.637 (2) | 109 (1) |
| O3—H3O⋯O2 | 0.84 (2) | 2.29 (3) | 2.660 (2) | 107 (2) |
| O6—H6O⋯O5 | 0.84 (2) | 2.28 (2) | 2.663 (2) | 108 (2) |
| O3—H3O⋯N6i | 0.84 (2) | 2.19 (2) | 2.994 (2) | 161 (2) |
| O6—H6O⋯N3ii | 0.84 (2) | 2.22 (2) | 3.013 (2) | 157 (2) |
| N2—H2N⋯O4iii | 0.88 (2) | 2.01 (2) | 2.873 (2) | 170 (2) |
| N4—H4N⋯O2i | 0.86 (2) | 2.54 (2) | 3.390 (2) | 167 (2) |
| N5—H5N⋯O1iv | 0.88 (2) | 2.04 (2) | 2.900 (2) | 169 (2) |
| C33—H33⋯O6v | 0.95 | 2.54 | 3.212 (2) | 128 |
| C15—H15⋯O3vi | 0.95 | 2.63 | 3.166 (2) | 113 |
| C33—H33⋯O6i | 0.95 | 2.54 | 3.212 (2) | 128 |
| C10—H10A⋯Cg1vii | 0.99 | 2.80 | 3.774 (2) | 168 |
| C18—H18A⋯Cg2ii | 0.98 | 2.66 | 3.603 (4) | 161 |
| C28—H28B⋯Cg3viii | 0.99 | 2.75 | 3.720 (2) | 166 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  ; (vi)
; (vi)  ; (vii)
; (vii)  ; (viii)
; (viii)  .
.
Figure 2.
Overlay diagram for (I), with the O1-molecule (red image) and O4-molecule (blue image) superimposed so that the urea residues are coincident.
Supramolecular features
Conventional O—H⋯N and N—H⋯O hydrogen bonding features significantly in the molecular packing of (I), Table 1 ▸, and this is highlighted in Fig. 3 ▸ a. The two molecules comprising the asymmetric unit associate via an eight-membered amide synthon, {⋯OCNH}2. The hydroxy-O—H groups at each end of the dimeric aggregate hydrogen bond to an imine-N atom of the other independent molecule. The hydroxyl-O3—H⋯N6(imine) interaction is incorporated within a 10-membered {⋯HOC2O⋯HNCNN} heterosynthon owing to the formation of a relatively weak phenylamine-N4—H⋯O2(methoxy) hydrogen bond. The putative phenylamine-N1—H⋯O5(methoxy) hydrogen bond is beyond the standard limits (Spek, 2009 ▸) as the H⋯O separation is 2.73 Å. As seen in Fig. 3 ▸ b, these hydrogen bonds extend laterally to from an array in (101). The most obvious connections between the supramolecular layers are of the type benzene-C—H⋯O(hydroxyl), which occur between centrosymmetrically related O6-benzene rings. A view of the unit-cell contents highlighting the stacking of layers is shown in Fig. 3 ▸ c. Other C—H⋯O and several C—H⋯π interactions occur in the crystal but within the layers stabilized by hydrogen bonding. These and other weak interactions are discussed in more detail in Analysis of the Hirshfeld surface (§4).
Figure 3.
The molecular packing in (I): (a) a detail of the supramolecular association sustained by O—H⋯N and N—H⋯O hydrogen bonding, shown as orange and blue dashed lines, respectively, (b) a view of the supramolecular layer in the ac plane, and (c) a view of the unit-cell contents shown in projection down the b axis. The C—H⋯O interactions are shown as green dashed lines, and one layer is highlighted in space-filling mode.
Analysis of the Hirshfeld surface
The Hirshfeld surface was calculated for the individual O1- and O4-molecules in (I), i.e. molecules A and B, and for overall (I) in accord with a recent report on a related molecule (Tan et al., 2017 ▸). These calculations provide additional information about the influence of weak intermolecular C—H⋯O and C—H⋯π interactions, Table 1 ▸, along with short interatomic H⋯H, C⋯H/H⋯C and O⋯H/H⋯O contacts, Table 2 ▸, on the molecular packing in the crystal.
Table 2. Summary of short interatomic contacts (Å) in (I).
| Contact | Distance | Symmetry operation |
|---|---|---|
| H3⋯H21 | 2.16 |
 − x, − − x, − + y, + y,  − z − z
|
| H28A⋯H35A | 2.24 |
 − x, − − x, − + y, + y,  − z − z
|
| O1⋯H22 | 2.50 |
 − x, − − x, − + y, + y,  − z − z
|
| O2⋯H27B | 2.56 | 1 − x, 1 − y, 1 − z |
| O3⋯H15 | 2.63 | −x, 2 − y, 1 − z |
| O3⋯C15 | 3.166 (2) | −x, 2 − y, 1 − z |
| O3⋯H23 | 2.58 | 1 − x, 2 − y, 1 − z |
| O4⋯H16 | 2.58 | 1 + x, −1 + y, z |
| O5⋯H9A | 2.46 | 1 − x, −y, 1 − z |
| C6⋯H10A | 2.63 | x, −1 + y, z |
| C15⋯H36C | 2.55 | 1 − x, 1 − y, 1 − z |
| C16⋯H36C | 2.80 | 1 − x, 1 − y, 1 − z |
| C24⋯H28B | 2.63 | x, 1 + y, z |
| C32⋯H18A | 2.76 | 1 − x, − y, 1 − z |
| C33⋯H18A | 2.62 | 1 − x, − y, 1 − z |
| C34⋯H18A | 2.78 | 1 − x, − y, 1 − z |
| C35⋯H18B | 2.71 |
 + x, + x,  − y, − − y, − + z + z
|
| C36⋯H9A | 2.80 | 1 − x, − y, 1 − z |
| C6⋯C6 | 3.210 (3) | 1 − x, − y, 1 − z |
| C24⋯C24 | 3.300 (3) | 2 − x, 1 − y, 1 − z |
The bright-red spots appearing near the hydroxyl-H3O and H6O, and imine-N3 and N6 atoms on the Hirshfeld surfaces mapped over d norm shown with labels ‘1’ and ‘2’ in Fig. 4 ▸ represent the donors and acceptors of intermolecular hydroxyl-O—H⋯N(imine) hydrogen bonds, respectively, Table 1 ▸. In the same way, the prominent red regions near the amide-H2N and H5N, and amide-O1 and O4 atoms, i.e. ‘3’ and ‘4’ in Fig. 4 ▸, indicate their participation in the intermolecular N—H⋯O hydrogen bonds between the symmetry-related independent molecules, Table 1 ▸. The donors and acceptors of comparatively weak intermolecular N—H⋯O and C—H⋯O interactions summarized in Table 1 ▸ are viewed as faint-red spots near the respective atoms on d norm-mapped Hirshfeld surfaces with labels ‘5–7’ in Fig. 4 ▸.
Figure 4.
Views of the Hirshfeld surface for (I) mapped over d norm in the ranges (a) −0.150 to +1.462 au for the O1-containing molecule and (b) −0.215 to + 1.462 au for the O4-molecule.
The presence of diminutive red spots viewed near phenyl atoms C6 in Fig. 4 ▸ a and C24 in Fig. 4 ▸ b, of the independent molecules, respectively, reflect short interatomic edge-to-edge C⋯C contacts, Table 2 ▸, although they contribute a very low contribution, i.e. 0.1%, to the Hirshfeld surface owing to the absence of π–π stacking between aromatic rings in the crystal, Table 3 ▸. The faint-red spots appearing near the labelled H10A, H18A, C28, C6, C33 and C24 atoms in the images of Fig. 4 ▸ represent their participation in short interatomic C⋯H/H⋯C contacts, Table 2 ▸, and confirm the influence of the intermolecular C—H⋯π interactions, Table 1 ▸, in the crystal. In addition to these short interatomic C⋯H/H⋯C contacts, the faint-red spots near the C15 O1, H9A and H18B atoms, Fig. 4 ▸ a, and O5, C35, H22 and H36C atoms, Fig. 4 ▸ b, indicate the contributions from the additional short interatomic C⋯H/H⋯C and O⋯H/H⋯O contacts, Table 2 ▸, to the molecular packing.
Table 3. Percentage contributions of interatomic contacts to the Hirshfeld surface for the O1-molecule, the O4-molecule and for overall (I).
| Contact | Percentage contribution | ||
|---|---|---|---|
| O1-molecule | O4-molecule | (I) | |
| H⋯H | 49.5 | 49.4 | 48.7 |
| O⋯H/H⋯O | 16.4 | 17.5 | 17.8 |
| N⋯H/H⋯N | 7.4 | 7.3 | 7.7 |
| C⋯H/H⋯C | 26.3 | 25.7 | 25.5 |
| C⋯C | 0.1 | 0.1 | 0.1 |
| O⋯O | 0.2 | 0.0 | 0.1 |
| C⋯O/O⋯C | 0.1 | 0.0 | 0.1 |
On the Hirshfeld surfaces mapped over the electrostatic potential for the independent molecules of (I), Fig. 5 ▸, the donors and acceptors of intermolecular interactions are represented with blue and red regions corresponding to positive and negative electrostatic potentials, respectively. The views of Hirshfeld surfaces about reference independent molecules of (I) mapped within the shape-index property, Fig. 6 ▸, highlight the short interatomic C⋯H/H⋯C and C—H⋯π/π⋯H—C contacts operating in the crystal.
Figure 5.
Views of the Hirshfeld surface for (I) mapped over the electrostatic potential in the range −0.103 to + 0.141 au for the (a) O1-containing molecule and (b) the O4-molecule. The red and blue regions represent negative and positive electrostatic potentials, respectively.
Figure 6.
Views of the shape-indexed Hirshfeld surfaces about reference molecules highlighting dominant short interatomic C—H/H—C and C—H⋯π/π⋯H—C interactions for the (a) O1-containing molecule and (b) the O4-molecule.
It is clear from the overall two-dimensional fingerprint plots for each independent molecule and for the entire asymmetric unit of (I) shown in Fig. 7 ▸ that the individual molecules have common features in their intermolecular O—H⋯N, N—H⋯O and C—H⋯π interactions. The small differences in the distribution of points in the fingerprint plots delineated into H⋯H, O⋯H/H⋯O, N⋯H/H⋯N and C⋯H/H⋯C contacts (McKinnon et al., 2007 ▸) in Fig. 7 ▸, are ascribed to the commented upon (§3) conformational differences, i.e. the twisting of the methoxy substituents on the respective benzene rings and the inclination of these benzene rings with respect to the ethane bridges.
Figure 7.
The full two-dimensional fingerprint plot and those delineated into H⋯H, O⋯H/H⋯O, N⋯H/H⋯N and C⋯H/H⋯C contacts for the (a) O1-containing molecule, (b) the O4-molecule and (c) (I).
The fingerprint plot delineated into H⋯H contacts for molecules A and B have almost the same percentage contribution to their respective Hirshfeld surfaces, Table 3 ▸, and the distinct distributions in the upper regions of the plots are due to the contributions from hydrogen atoms of their respective disubstituted benzene rings to the surfaces of molecules A and B. The single short peaks at d e + d i ∼ 2.1 Å in the delineated plots for both the molecules indicate the involvement of hydrogen atoms of both in short interatomic H⋯H contacts, Table 2 ▸. The intermolecular N—H⋯O and O—H⋯N hydrogen bonds in the crystal are characterized as the pairs of spikes with their tips at d e + d i ∼ 2.0 Å (inner region) and at ∼ 2.2 Å (outer region) in the fingerprint plots delineated into O⋯H/H⋯O and N⋯H/H⋯N contacts, respectively. The forceps-like distribution of points linked with the donor spike for molecule A and the acceptor spike for molecule B at d e + d i ∼ 2.5 Å in the fingerprint plots delineated into O⋯H/H⋯O contacts are due to weak intermolecular C—H⋯O interactions and the short interatomic contacts summarized in Table 2 ▸. The asymmetric forceps-like distribution of points with the tips at d e + d i ∼ 2.6 Å in the acceptor and donor regions of fingerprint plots delineated into C⋯H/H⋯C contacts for molecules A and B, respectively, represent the involvement of these atoms in the short interatomic C⋯H/H⋯C contacts, Table 2 ▸, whereas the intermolecular C—H⋯π interactions are viewed as the forceps-like tips at d e + d i ∼ 2.7 Å in the donor and acceptor regions of molecules A and B, respectively. The other C⋯O/O⋯C, O⋯O and C⋯C interatomic contacts summarized in Table 3 ▸, having only small contributions to the Hirshfeld surface, have negligible directional impact on the molecular packing.
Database survey
There are no direct precedents for the structure of (I) in the crystallographic literature (Groom et al., 2016 ▸). However, there are several precedents for the phenylsemicarbazone residue with the imine-carbon atom incorporated within an all-carbon ring (Groth, 1980 ▸; Hoek van den et al., 1980 ▸), as exemplified in the cyclodecane derivative (II) (Groth, 1980 ▸; Hoek van den et al., 1980 ▸), see Scheme 2 for the chemical diagram of (II). More exotic derivatives with cyclic residues at both ends of the semicarbazone core are also known (Behenna et al., 2011 ▸; Ma et al., 2014 ▸), as exemplified by (III) (Ma et al., 2014 ▸), Scheme 2.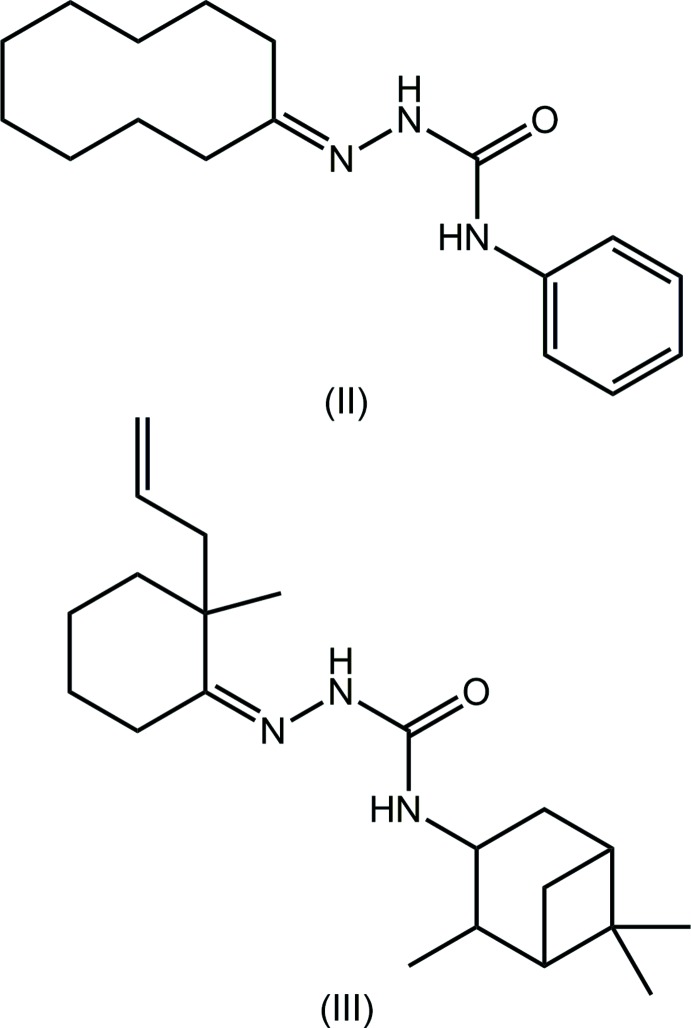
Synthesis and crystallization
Analytical grade reagents were used as procured without further purification. 4-Phenylsemicarbazide (1.51 g, 0.01 mol) and vanillylacetone (1.94 g, 0.01 mol) were dissolved separately in hot absolute ethanol (30 ml) and mixed with stirring. The reaction mixture was heated and stirred for 20 min., then stirred for another 30 min. at room temperature. The resulting white precipitate was filtered off, washed with cold absolute ethanol and dried in vacuo; yield: 75%. Light-yellow prisms of (I) were grown at room temperature from slow evaporation of mixed solvents of ethanol and acetonitrile (1:1; v/v 20 ml). IR (cm−1): 3201 ν(N—H), 1670 ν(C=N), 1213 ν(C—N), 1026 ν(C=O). MS m/z: 327.25 [M+1]+
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 4 ▸. The carbon-bound H atoms were placed in calculated positions (C—H = 0.95–0.99 Å) and were included in the refinement in the riding-model approximation, with U iso(H) set to 1.2–1.5U eq(C). The oxygen- and nitrogen-bound H atoms were located in a difference-Fourier map but were refined with distance restraints of O—H = 0.84±0.01 Å and N—H = 0.88±0.01 Å, and with U iso(H) set to 1.5U eq(O) and 1.2U eq(N), respectively. The maximum and minimum residual electron density peaks of 0.60 and 0.26 e Å−3, respectively, were located 0.95 and 0.75 Å from atoms H10A and H36A, respectively.
Table 4. Experimental details.
| Crystal data | |
| Chemical formula | C18H21N3O3 |
| M r | 327.37 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 100 |
| a, b, c (Å) | 16.5464 (4), 9.2184 (2), 22.3975 (4) |
| β (°) | 100.494 (2) |
| V (Å3) | 3359.18 (13) |
| Z | 8 |
| Radiation type | Cu Kα |
| μ (mm−1) | 0.73 |
| Crystal size (mm) | 0.25 × 0.16 × 0.06 |
| Data collection | |
| Diffractometer | Oxford Diffraction Xcaliber Eos Gemini |
| Absorption correction | Multi-scan (CrysAlis PRO; Agilent, 2011 ▸) |
| T min, T max | 0.917, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 23815, 6481, 5581 |
| R int | 0.019 |
| (sin θ/λ)max (Å−1) | 0.615 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.059, 0.180, 1.05 |
| No. of reflections | 6481 |
| No. of parameters | 455 |
| No. of restraints | 6 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.60, −0.26 |
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989017017273/hb7720sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017017273/hb7720Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989017017273/hb7720Isup3.cml
CCDC reference: 926756
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
We thank the staff of the University of Malaya’s X-ray diffraction laboratory for the data collection.
supplementary crystallographic information
Crystal data
| C18H21N3O3 | F(000) = 1392 |
| Mr = 327.37 | Dx = 1.295 Mg m−3 |
| Monoclinic, P21/n | Cu Kα radiation, λ = 1.5418 Å |
| a = 16.5464 (4) Å | Cell parameters from 9245 reflections |
| b = 9.2184 (2) Å | θ = 3.7–71.3° |
| c = 22.3975 (4) Å | µ = 0.73 mm−1 |
| β = 100.494 (2)° | T = 100 K |
| V = 3359.18 (13) Å3 | Prism (cut), light-yellow |
| Z = 8 | 0.25 × 0.16 × 0.06 mm |
Data collection
| Oxford Diffraction Xcaliber Eos Gemini diffractometer | 6481 independent reflections |
| Radiation source: fine-focus sealed tube | 5581 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.019 |
| Detector resolution: 16.1952 pixels mm-1 | θmax = 71.4°, θmin = 3.7° |
| ω scans | h = −20→20 |
| Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2011) | k = −11→11 |
| Tmin = 0.917, Tmax = 1.000 | l = −26→27 |
| 23815 measured reflections |
Refinement
| Refinement on F2 | 6 restraints |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.059 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.180 | w = 1/[σ2(Fo2) + (0.1131P)2 + 1.5134P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max < 0.001 |
| 6481 reflections | Δρmax = 0.60 e Å−3 |
| 455 parameters | Δρmin = −0.26 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.50355 (7) | 0.34300 (14) | 0.28183 (6) | 0.0292 (3) | |
| O2 | 0.22012 (9) | 0.87251 (16) | 0.56672 (6) | 0.0386 (4) | |
| O3 | 0.09139 (8) | 1.04987 (16) | 0.54412 (6) | 0.0350 (3) | |
| H3O | 0.1190 (15) | 1.034 (3) | 0.5787 (7) | 0.053* | |
| N1 | 0.40785 (9) | 0.29389 (16) | 0.34177 (6) | 0.0253 (3) | |
| H1N | 0.3694 (10) | 0.337 (2) | 0.3564 (9) | 0.030* | |
| N2 | 0.40724 (9) | 0.51144 (16) | 0.29066 (7) | 0.0256 (3) | |
| H2N | 0.4313 (12) | 0.5727 (19) | 0.2696 (8) | 0.031* | |
| N3 | 0.34334 (9) | 0.55389 (17) | 0.31934 (7) | 0.0284 (3) | |
| C1 | 0.44364 (10) | 0.37902 (19) | 0.30419 (7) | 0.0238 (3) | |
| C2 | 0.43316 (10) | 0.15426 (19) | 0.36316 (8) | 0.0259 (4) | |
| C3 | 0.47454 (13) | 0.0593 (2) | 0.33074 (9) | 0.0360 (4) | |
| H3 | 0.4884 | 0.0887 | 0.2932 | 0.043* | |
| C4 | 0.49550 (13) | −0.0785 (2) | 0.35355 (10) | 0.0407 (5) | |
| H4 | 0.5243 | −0.1425 | 0.3316 | 0.049* | |
| C5 | 0.47517 (13) | −0.1240 (2) | 0.40757 (10) | 0.0419 (5) | |
| H5 | 0.4894 | −0.2186 | 0.4227 | 0.050* | |
| C6 | 0.43353 (14) | −0.0291 (2) | 0.43949 (10) | 0.0442 (5) | |
| H6 | 0.4192 | −0.0594 | 0.4768 | 0.053* | |
| C7 | 0.41258 (13) | 0.1094 (2) | 0.41765 (9) | 0.0368 (4) | |
| H7 | 0.3842 | 0.1734 | 0.4400 | 0.044* | |
| C8 | 0.29887 (11) | 0.6607 (2) | 0.29621 (8) | 0.0297 (4) | |
| C9 | 0.23057 (11) | 0.7061 (2) | 0.32858 (9) | 0.0326 (4) | |
| H9A | 0.2323 | 0.6455 | 0.3653 | 0.039* | |
| H9B | 0.1769 | 0.6900 | 0.3016 | 0.039* | |
| C10 | 0.23837 (13) | 0.8663 (2) | 0.34707 (9) | 0.0355 (4) | |
| H10A | 0.2974 | 0.8905 | 0.3592 | 0.043* | |
| H10B | 0.2158 | 0.9266 | 0.3113 | 0.043* | |
| C11 | 0.19461 (12) | 0.9052 (2) | 0.39874 (8) | 0.0329 (4) | |
| C12 | 0.22808 (13) | 0.8620 (2) | 0.45796 (9) | 0.0367 (4) | |
| H12 | 0.2757 | 0.8024 | 0.4650 | 0.044* | |
| C13 | 0.19215 (12) | 0.9056 (2) | 0.50660 (8) | 0.0307 (4) | |
| C14 | 0.12349 (11) | 0.99627 (19) | 0.49664 (8) | 0.0261 (4) | |
| C15 | 0.08828 (11) | 1.0335 (2) | 0.43795 (9) | 0.0302 (4) | |
| H15 | 0.0400 | 1.0915 | 0.4307 | 0.036* | |
| C16 | 0.12319 (11) | 0.9866 (2) | 0.38942 (8) | 0.0322 (4) | |
| H16 | 0.0976 | 1.0108 | 0.3492 | 0.039* | |
| C17 | 0.31178 (11) | 0.7458 (2) | 0.24149 (8) | 0.0329 (4) | |
| H17A | 0.3595 | 0.8096 | 0.2526 | 0.049* | |
| H17B | 0.2628 | 0.8043 | 0.2266 | 0.049* | |
| H17C | 0.3214 | 0.6788 | 0.2095 | 0.049* | |
| C18 | 0.30011 (18) | 0.8105 (4) | 0.58127 (11) | 0.0742 (10) | |
| H18A | 0.2995 | 0.7133 | 0.5635 | 0.111* | |
| H18B | 0.3167 | 0.8039 | 0.6255 | 0.111* | |
| H18C | 0.3393 | 0.8718 | 0.5648 | 0.111* | |
| O4 | 0.99721 (7) | 0.19097 (14) | 0.27509 (6) | 0.0296 (3) | |
| O5 | 0.74121 (9) | −0.41262 (16) | 0.57259 (6) | 0.0389 (3) | |
| O6 | 0.59792 (8) | −0.55085 (16) | 0.54521 (6) | 0.0355 (3) | |
| H6O | 0.6277 (14) | −0.547 (3) | 0.5798 (7) | 0.053* | |
| N4 | 0.90978 (8) | 0.23778 (16) | 0.34148 (6) | 0.0245 (3) | |
| H4N | 0.8743 (10) | 0.198 (2) | 0.3602 (8) | 0.029* | |
| N5 | 0.90487 (9) | 0.02044 (16) | 0.28954 (7) | 0.0263 (3) | |
| H5N | 0.9267 (12) | −0.040 (2) | 0.2668 (8) | 0.032* | |
| N6 | 0.84546 (9) | −0.02354 (17) | 0.32229 (7) | 0.0288 (3) | |
| C19 | 0.94100 (10) | 0.15314 (18) | 0.30127 (7) | 0.0230 (3) | |
| C20 | 0.93614 (10) | 0.37987 (19) | 0.35945 (8) | 0.0251 (4) | |
| C21 | 0.96816 (12) | 0.4747 (2) | 0.32082 (8) | 0.0309 (4) | |
| H21 | 0.9735 | 0.4440 | 0.2812 | 0.037* | |
| C22 | 0.99218 (12) | 0.6136 (2) | 0.34046 (9) | 0.0350 (4) | |
| H22 | 1.0146 | 0.6768 | 0.3142 | 0.042* | |
| C23 | 0.98404 (12) | 0.6617 (2) | 0.39743 (9) | 0.0367 (4) | |
| H23 | 1.0006 | 0.7571 | 0.4104 | 0.044* | |
| C24 | 0.95139 (13) | 0.5686 (2) | 0.43530 (9) | 0.0384 (5) | |
| H24 | 0.9451 | 0.6008 | 0.4745 | 0.046* | |
| C25 | 0.92765 (12) | 0.4282 (2) | 0.41667 (8) | 0.0331 (4) | |
| H25 | 0.9056 | 0.3653 | 0.4432 | 0.040* | |
| C26 | 0.79860 (11) | −0.1278 (2) | 0.30028 (8) | 0.0292 (4) | |
| C27 | 0.73558 (11) | −0.1766 (2) | 0.33697 (9) | 0.0330 (4) | |
| H27A | 0.6797 | −0.1593 | 0.3134 | 0.040* | |
| H27B | 0.7417 | −0.1189 | 0.3748 | 0.040* | |
| C28 | 0.74571 (13) | −0.3380 (2) | 0.35290 (9) | 0.0364 (4) | |
| H28A | 0.7238 | −0.3958 | 0.3163 | 0.044* | |
| H28B | 0.8051 | −0.3600 | 0.3645 | 0.044* | |
| C29 | 0.70304 (12) | −0.3848 (2) | 0.40393 (9) | 0.0327 (4) | |
| C30 | 0.74324 (12) | −0.3697 (2) | 0.46427 (9) | 0.0342 (4) | |
| H30 | 0.7957 | −0.3245 | 0.4729 | 0.041* | |
| C31 | 0.70709 (12) | −0.4203 (2) | 0.51172 (8) | 0.0307 (4) | |
| C32 | 0.63080 (11) | −0.49006 (19) | 0.49940 (8) | 0.0265 (4) | |
| C33 | 0.58946 (11) | −0.5002 (2) | 0.44001 (9) | 0.0320 (4) | |
| H33 | 0.5367 | −0.5443 | 0.4314 | 0.038* | |
| C34 | 0.62477 (12) | −0.4461 (2) | 0.39286 (8) | 0.0326 (4) | |
| H34 | 0.5951 | −0.4510 | 0.3524 | 0.039* | |
| C35 | 0.80405 (11) | −0.2075 (2) | 0.24253 (8) | 0.0329 (4) | |
| H35A | 0.8160 | −0.1383 | 0.2120 | 0.049* | |
| H35B | 0.7516 | −0.2560 | 0.2274 | 0.049* | |
| H35C | 0.8481 | −0.2799 | 0.2506 | 0.049* | |
| C36 | 0.82787 (14) | −0.3844 (3) | 0.58750 (10) | 0.0481 (6) | |
| H36A | 0.8572 | −0.4519 | 0.5652 | 0.072* | |
| H36B | 0.8468 | −0.3976 | 0.6312 | 0.072* | |
| H36C | 0.8388 | −0.2845 | 0.5762 | 0.072* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0302 (6) | 0.0278 (6) | 0.0345 (7) | 0.0047 (5) | 0.0187 (5) | 0.0035 (5) |
| O2 | 0.0518 (8) | 0.0411 (8) | 0.0266 (7) | 0.0180 (6) | 0.0167 (6) | 0.0056 (5) |
| O3 | 0.0351 (7) | 0.0431 (8) | 0.0304 (7) | 0.0075 (6) | 0.0154 (5) | −0.0057 (6) |
| N1 | 0.0276 (7) | 0.0269 (7) | 0.0248 (7) | 0.0014 (6) | 0.0136 (6) | −0.0006 (6) |
| N2 | 0.0257 (7) | 0.0268 (7) | 0.0281 (7) | 0.0019 (6) | 0.0153 (6) | 0.0016 (6) |
| N3 | 0.0294 (7) | 0.0313 (8) | 0.0282 (8) | 0.0033 (6) | 0.0152 (6) | −0.0005 (6) |
| C1 | 0.0241 (8) | 0.0256 (8) | 0.0227 (8) | −0.0003 (6) | 0.0073 (6) | −0.0015 (6) |
| C2 | 0.0271 (8) | 0.0268 (9) | 0.0245 (8) | −0.0036 (7) | 0.0064 (6) | 0.0007 (7) |
| C3 | 0.0475 (11) | 0.0307 (10) | 0.0352 (10) | 0.0047 (8) | 0.0222 (9) | 0.0055 (8) |
| C4 | 0.0455 (11) | 0.0308 (10) | 0.0500 (12) | 0.0066 (8) | 0.0196 (9) | 0.0071 (9) |
| C5 | 0.0435 (11) | 0.0326 (10) | 0.0498 (12) | −0.0010 (8) | 0.0088 (9) | 0.0140 (9) |
| C6 | 0.0571 (13) | 0.0425 (12) | 0.0363 (11) | −0.0079 (10) | 0.0172 (9) | 0.0121 (9) |
| C7 | 0.0463 (11) | 0.0359 (10) | 0.0320 (10) | −0.0043 (8) | 0.0176 (8) | 0.0001 (8) |
| C8 | 0.0296 (9) | 0.0336 (9) | 0.0277 (9) | 0.0013 (7) | 0.0097 (7) | −0.0028 (7) |
| C9 | 0.0283 (9) | 0.0367 (10) | 0.0353 (10) | 0.0005 (7) | 0.0123 (7) | −0.0034 (8) |
| C10 | 0.0449 (11) | 0.0343 (10) | 0.0314 (10) | 0.0030 (8) | 0.0179 (8) | 0.0023 (8) |
| C11 | 0.0419 (10) | 0.0292 (9) | 0.0307 (9) | 0.0023 (8) | 0.0149 (8) | −0.0010 (7) |
| C12 | 0.0444 (11) | 0.0351 (10) | 0.0340 (10) | 0.0142 (8) | 0.0164 (8) | 0.0035 (8) |
| C13 | 0.0395 (10) | 0.0289 (9) | 0.0265 (9) | 0.0058 (7) | 0.0134 (7) | 0.0019 (7) |
| C14 | 0.0292 (9) | 0.0239 (8) | 0.0291 (9) | −0.0029 (6) | 0.0163 (7) | −0.0021 (6) |
| C15 | 0.0246 (8) | 0.0327 (9) | 0.0354 (10) | 0.0004 (7) | 0.0110 (7) | 0.0012 (7) |
| C16 | 0.0333 (9) | 0.0384 (10) | 0.0262 (9) | 0.0011 (8) | 0.0087 (7) | 0.0005 (7) |
| C17 | 0.0342 (9) | 0.0383 (10) | 0.0289 (9) | 0.0117 (8) | 0.0133 (7) | 0.0041 (7) |
| C18 | 0.0821 (19) | 0.111 (3) | 0.0325 (11) | 0.0655 (19) | 0.0182 (12) | 0.0211 (13) |
| O4 | 0.0312 (6) | 0.0265 (6) | 0.0362 (7) | −0.0032 (5) | 0.0200 (5) | −0.0031 (5) |
| O5 | 0.0459 (8) | 0.0460 (8) | 0.0266 (7) | −0.0145 (6) | 0.0118 (6) | −0.0018 (6) |
| O6 | 0.0317 (7) | 0.0471 (8) | 0.0312 (7) | −0.0050 (6) | 0.0152 (5) | 0.0059 (6) |
| N4 | 0.0245 (7) | 0.0272 (7) | 0.0244 (7) | −0.0009 (5) | 0.0116 (5) | 0.0008 (5) |
| N5 | 0.0271 (7) | 0.0279 (8) | 0.0279 (7) | −0.0029 (6) | 0.0159 (6) | −0.0030 (6) |
| N6 | 0.0296 (8) | 0.0313 (8) | 0.0296 (8) | −0.0042 (6) | 0.0166 (6) | −0.0001 (6) |
| C19 | 0.0223 (8) | 0.0250 (8) | 0.0230 (8) | 0.0008 (6) | 0.0075 (6) | 0.0015 (6) |
| C20 | 0.0223 (8) | 0.0286 (9) | 0.0249 (8) | 0.0030 (6) | 0.0057 (6) | −0.0027 (7) |
| C21 | 0.0371 (10) | 0.0287 (9) | 0.0297 (9) | −0.0013 (7) | 0.0140 (7) | −0.0046 (7) |
| C22 | 0.0376 (10) | 0.0287 (10) | 0.0421 (11) | −0.0032 (8) | 0.0162 (8) | −0.0037 (8) |
| C23 | 0.0379 (10) | 0.0301 (10) | 0.0431 (11) | 0.0004 (8) | 0.0104 (8) | −0.0121 (8) |
| C24 | 0.0458 (11) | 0.0394 (11) | 0.0313 (10) | 0.0059 (9) | 0.0103 (8) | −0.0108 (8) |
| C25 | 0.0384 (10) | 0.0346 (10) | 0.0286 (9) | 0.0039 (8) | 0.0125 (8) | −0.0005 (7) |
| C26 | 0.0275 (9) | 0.0306 (9) | 0.0311 (9) | −0.0004 (7) | 0.0098 (7) | 0.0036 (7) |
| C27 | 0.0293 (9) | 0.0332 (10) | 0.0395 (10) | −0.0004 (7) | 0.0144 (8) | 0.0038 (8) |
| C28 | 0.0450 (11) | 0.0340 (10) | 0.0345 (10) | 0.0026 (8) | 0.0186 (8) | 0.0026 (8) |
| C29 | 0.0403 (10) | 0.0281 (9) | 0.0326 (10) | 0.0010 (7) | 0.0143 (8) | 0.0029 (7) |
| C30 | 0.0373 (10) | 0.0318 (10) | 0.0361 (10) | −0.0073 (8) | 0.0131 (8) | −0.0007 (8) |
| C31 | 0.0375 (10) | 0.0296 (9) | 0.0270 (9) | −0.0041 (7) | 0.0111 (7) | −0.0026 (7) |
| C32 | 0.0288 (9) | 0.0249 (8) | 0.0299 (9) | 0.0034 (7) | 0.0165 (7) | 0.0012 (7) |
| C33 | 0.0241 (8) | 0.0377 (10) | 0.0357 (10) | 0.0035 (7) | 0.0095 (7) | 0.0007 (8) |
| C34 | 0.0341 (9) | 0.0365 (10) | 0.0280 (9) | 0.0043 (8) | 0.0077 (7) | 0.0021 (7) |
| C35 | 0.0330 (9) | 0.0400 (11) | 0.0267 (9) | −0.0114 (8) | 0.0076 (7) | −0.0018 (7) |
| C36 | 0.0474 (12) | 0.0608 (15) | 0.0358 (11) | −0.0236 (11) | 0.0064 (9) | −0.0054 (10) |
Geometric parameters (Å, º)
| O1—C1 | 1.235 (2) | O4—C19 | 1.237 (2) |
| O2—C13 | 1.376 (2) | O5—C31 | 1.379 (2) |
| O2—C18 | 1.424 (3) | O5—C36 | 1.436 (2) |
| O3—C14 | 1.365 (2) | O6—C32 | 1.366 (2) |
| O3—H3O | 0.838 (10) | O6—H6O | 0.840 (10) |
| N1—C1 | 1.363 (2) | N4—C19 | 1.362 (2) |
| N1—C2 | 1.410 (2) | N4—C20 | 1.416 (2) |
| N1—H1N | 0.862 (9) | N4—H4N | 0.865 (9) |
| N2—C1 | 1.371 (2) | N5—C19 | 1.366 (2) |
| N2—N3 | 1.3895 (19) | N5—N6 | 1.3899 (19) |
| N2—H2N | 0.876 (9) | N5—H5N | 0.878 (9) |
| N3—C8 | 1.281 (2) | N6—C26 | 1.276 (2) |
| C2—C7 | 1.388 (2) | C20—C25 | 1.388 (2) |
| C2—C3 | 1.394 (3) | C20—C21 | 1.400 (3) |
| C3—C4 | 1.390 (3) | C21—C22 | 1.389 (3) |
| C3—H3 | 0.9500 | C21—H21 | 0.9500 |
| C4—C5 | 1.379 (3) | C22—C23 | 1.380 (3) |
| C4—H4 | 0.9500 | C22—H22 | 0.9500 |
| C5—C6 | 1.389 (3) | C23—C24 | 1.384 (3) |
| C5—H5 | 0.9500 | C23—H23 | 0.9500 |
| C6—C7 | 1.388 (3) | C24—C25 | 1.394 (3) |
| C6—H6 | 0.9500 | C24—H24 | 0.9500 |
| C7—H7 | 0.9500 | C25—H25 | 0.9500 |
| C8—C17 | 1.503 (2) | C26—C35 | 1.504 (3) |
| C8—C9 | 1.509 (2) | C26—C27 | 1.509 (2) |
| C9—C10 | 1.533 (3) | C27—C28 | 1.532 (3) |
| C9—H9A | 0.9900 | C27—H27A | 0.9900 |
| C9—H9B | 0.9900 | C27—H27B | 0.9900 |
| C10—C11 | 1.516 (2) | C28—C29 | 1.512 (2) |
| C10—H10A | 0.9900 | C28—H28A | 0.9900 |
| C10—H10B | 0.9900 | C28—H28B | 0.9900 |
| C11—C16 | 1.383 (3) | C29—C34 | 1.393 (3) |
| C11—C12 | 1.399 (3) | C29—C30 | 1.400 (3) |
| C12—C13 | 1.392 (2) | C30—C31 | 1.392 (3) |
| C12—H12 | 0.9500 | C30—H30 | 0.9500 |
| C13—C14 | 1.395 (3) | C31—C32 | 1.399 (3) |
| C14—C15 | 1.381 (3) | C32—C33 | 1.384 (3) |
| C15—C16 | 1.389 (3) | C33—C34 | 1.389 (3) |
| C15—H15 | 0.9500 | C33—H33 | 0.9500 |
| C16—H16 | 0.9500 | C34—H34 | 0.9500 |
| C17—H17A | 0.9800 | C35—H35A | 0.9800 |
| C17—H17B | 0.9800 | C35—H35B | 0.9800 |
| C17—H17C | 0.9800 | C35—H35C | 0.9800 |
| C18—H18A | 0.9800 | C36—H36A | 0.9800 |
| C18—H18B | 0.9800 | C36—H36B | 0.9800 |
| C18—H18C | 0.9800 | C36—H36C | 0.9800 |
| C13—O2—C18 | 116.47 (15) | C31—O5—C36 | 116.80 (15) |
| C14—O3—H3O | 115 (2) | C32—O6—H6O | 115 (2) |
| C1—N1—C2 | 126.87 (14) | C19—N4—C20 | 125.70 (14) |
| C1—N1—H1N | 113.9 (15) | C19—N4—H4N | 116.7 (15) |
| C2—N1—H1N | 118.8 (15) | C20—N4—H4N | 117.3 (14) |
| C1—N2—N3 | 119.30 (14) | C19—N5—N6 | 119.11 (14) |
| C1—N2—H2N | 117.9 (14) | C19—N5—H5N | 118.1 (14) |
| N3—N2—H2N | 121.7 (14) | N6—N5—H5N | 121.7 (15) |
| C8—N3—N2 | 117.32 (15) | C26—N6—N5 | 117.01 (15) |
| O1—C1—N1 | 124.55 (16) | O4—C19—N4 | 124.09 (15) |
| O1—C1—N2 | 120.18 (15) | O4—C19—N5 | 120.13 (15) |
| N1—C1—N2 | 115.26 (14) | N4—C19—N5 | 115.77 (14) |
| C7—C2—C3 | 119.56 (17) | C25—C20—C21 | 118.93 (17) |
| C7—C2—N1 | 117.72 (16) | C25—C20—N4 | 118.80 (16) |
| C3—C2—N1 | 122.67 (15) | C21—C20—N4 | 122.24 (15) |
| C4—C3—C2 | 119.74 (17) | C22—C21—C20 | 119.91 (17) |
| C4—C3—H3 | 120.1 | C22—C21—H21 | 120.0 |
| C2—C3—H3 | 120.1 | C20—C21—H21 | 120.0 |
| C5—C4—C3 | 121.05 (19) | C23—C22—C21 | 121.23 (18) |
| C5—C4—H4 | 119.5 | C23—C22—H22 | 119.4 |
| C3—C4—H4 | 119.5 | C21—C22—H22 | 119.4 |
| C4—C5—C6 | 118.92 (19) | C22—C23—C24 | 118.82 (18) |
| C4—C5—H5 | 120.5 | C22—C23—H23 | 120.6 |
| C6—C5—H5 | 120.5 | C24—C23—H23 | 120.6 |
| C7—C6—C5 | 120.86 (19) | C23—C24—C25 | 120.83 (18) |
| C7—C6—H6 | 119.6 | C23—C24—H24 | 119.6 |
| C5—C6—H6 | 119.6 | C25—C24—H24 | 119.6 |
| C6—C7—C2 | 119.87 (19) | C20—C25—C24 | 120.26 (18) |
| C6—C7—H7 | 120.1 | C20—C25—H25 | 119.9 |
| C2—C7—H7 | 120.1 | C24—C25—H25 | 119.9 |
| N3—C8—C17 | 125.06 (16) | N6—C26—C35 | 124.91 (16) |
| N3—C8—C9 | 116.39 (16) | N6—C26—C27 | 116.46 (16) |
| C17—C8—C9 | 118.50 (16) | C35—C26—C27 | 118.58 (16) |
| C8—C9—C10 | 111.32 (16) | C26—C27—C28 | 111.04 (15) |
| C8—C9—H9A | 109.4 | C26—C27—H27A | 109.4 |
| C10—C9—H9A | 109.4 | C28—C27—H27A | 109.4 |
| C8—C9—H9B | 109.4 | C26—C27—H27B | 109.4 |
| C10—C9—H9B | 109.4 | C28—C27—H27B | 109.4 |
| H9A—C9—H9B | 108.0 | H27A—C27—H27B | 108.0 |
| C11—C10—C9 | 113.94 (16) | C29—C28—C27 | 114.03 (16) |
| C11—C10—H10A | 108.8 | C29—C28—H28A | 108.7 |
| C9—C10—H10A | 108.8 | C27—C28—H28A | 108.7 |
| C11—C10—H10B | 108.8 | C29—C28—H28B | 108.7 |
| C9—C10—H10B | 108.8 | C27—C28—H28B | 108.7 |
| H10A—C10—H10B | 107.7 | H28A—C28—H28B | 107.6 |
| C16—C11—C12 | 118.52 (17) | C34—C29—C30 | 118.35 (17) |
| C16—C11—C10 | 121.80 (17) | C34—C29—C28 | 121.90 (18) |
| C12—C11—C10 | 119.66 (17) | C30—C29—C28 | 119.74 (17) |
| C13—C12—C11 | 120.53 (18) | C31—C30—C29 | 120.59 (17) |
| C13—C12—H12 | 119.7 | C31—C30—H30 | 119.7 |
| C11—C12—H12 | 119.7 | C29—C30—H30 | 119.7 |
| O2—C13—C12 | 125.91 (17) | O5—C31—C30 | 125.60 (17) |
| O2—C13—C14 | 114.09 (15) | O5—C31—C32 | 114.27 (16) |
| C12—C13—C14 | 119.94 (17) | C30—C31—C32 | 120.10 (17) |
| O3—C14—C15 | 119.68 (16) | O6—C32—C33 | 119.97 (16) |
| O3—C14—C13 | 120.88 (16) | O6—C32—C31 | 120.60 (16) |
| C15—C14—C13 | 119.44 (16) | C33—C32—C31 | 119.41 (16) |
| C14—C15—C16 | 120.25 (17) | C32—C33—C34 | 120.25 (17) |
| C14—C15—H15 | 119.9 | C32—C33—H33 | 119.9 |
| C16—C15—H15 | 119.9 | C34—C33—H33 | 119.9 |
| C11—C16—C15 | 121.08 (17) | C33—C34—C29 | 121.09 (17) |
| C11—C16—H16 | 119.5 | C33—C34—H34 | 119.5 |
| C15—C16—H16 | 119.5 | C29—C34—H34 | 119.5 |
| C8—C17—H17A | 109.5 | C26—C35—H35A | 109.5 |
| C8—C17—H17B | 109.5 | C26—C35—H35B | 109.5 |
| H17A—C17—H17B | 109.5 | H35A—C35—H35B | 109.5 |
| C8—C17—H17C | 109.5 | C26—C35—H35C | 109.5 |
| H17A—C17—H17C | 109.5 | H35A—C35—H35C | 109.5 |
| H17B—C17—H17C | 109.5 | H35B—C35—H35C | 109.5 |
| O2—C18—H18A | 109.5 | O5—C36—H36A | 109.5 |
| O2—C18—H18B | 109.5 | O5—C36—H36B | 109.5 |
| H18A—C18—H18B | 109.5 | H36A—C36—H36B | 109.5 |
| O2—C18—H18C | 109.5 | O5—C36—H36C | 109.5 |
| H18A—C18—H18C | 109.5 | H36A—C36—H36C | 109.5 |
| H18B—C18—H18C | 109.5 | H36B—C36—H36C | 109.5 |
| C1—N2—N3—C8 | −164.49 (16) | C19—N5—N6—C26 | 162.35 (16) |
| C2—N1—C1—O1 | 2.0 (3) | C20—N4—C19—O4 | 0.0 (3) |
| C2—N1—C1—N2 | −179.42 (15) | C20—N4—C19—N5 | −178.54 (15) |
| N3—N2—C1—O1 | −175.81 (15) | N6—N5—C19—O4 | 175.95 (15) |
| N3—N2—C1—N1 | 5.6 (2) | N6—N5—C19—N4 | −5.5 (2) |
| C1—N1—C2—C7 | 154.49 (18) | C19—N4—C20—C25 | −151.84 (17) |
| C1—N1—C2—C3 | −28.1 (3) | C19—N4—C20—C21 | 30.1 (3) |
| C7—C2—C3—C4 | −0.7 (3) | C25—C20—C21—C22 | 1.1 (3) |
| N1—C2—C3—C4 | −178.00 (18) | N4—C20—C21—C22 | 179.19 (17) |
| C2—C3—C4—C5 | 0.8 (3) | C20—C21—C22—C23 | −1.0 (3) |
| C3—C4—C5—C6 | −0.4 (3) | C21—C22—C23—C24 | 0.1 (3) |
| C4—C5—C6—C7 | 0.0 (3) | C22—C23—C24—C25 | 0.5 (3) |
| C5—C6—C7—C2 | 0.2 (3) | C21—C20—C25—C24 | −0.5 (3) |
| C3—C2—C7—C6 | 0.2 (3) | N4—C20—C25—C24 | −178.64 (16) |
| N1—C2—C7—C6 | 177.68 (18) | C23—C24—C25—C20 | −0.3 (3) |
| N2—N3—C8—C17 | −1.6 (3) | N5—N6—C26—C35 | 1.1 (3) |
| N2—N3—C8—C9 | −179.23 (15) | N5—N6—C26—C27 | 178.73 (15) |
| N3—C8—C9—C10 | 123.12 (19) | N6—C26—C27—C28 | −122.47 (19) |
| C17—C8—C9—C10 | −54.7 (2) | C35—C26—C27—C28 | 55.3 (2) |
| C8—C9—C10—C11 | −157.88 (16) | C26—C27—C28—C29 | 162.93 (17) |
| C9—C10—C11—C16 | −108.0 (2) | C27—C28—C29—C34 | 95.8 (2) |
| C9—C10—C11—C12 | 73.9 (2) | C27—C28—C29—C30 | −85.6 (2) |
| C16—C11—C12—C13 | −2.8 (3) | C34—C29—C30—C31 | 2.6 (3) |
| C10—C11—C12—C13 | 175.28 (18) | C28—C29—C30—C31 | −176.05 (18) |
| C18—O2—C13—C12 | 11.7 (3) | C36—O5—C31—C30 | −16.5 (3) |
| C18—O2—C13—C14 | −165.4 (2) | C36—O5—C31—C32 | 161.41 (18) |
| C11—C12—C13—O2 | −178.72 (19) | C29—C30—C31—O5 | 179.33 (18) |
| C11—C12—C13—C14 | −1.7 (3) | C29—C30—C31—C32 | 1.5 (3) |
| O2—C13—C14—O3 | 2.7 (3) | O5—C31—C32—O6 | −3.6 (3) |
| C12—C13—C14—O3 | −174.61 (17) | C30—C31—C32—O6 | 174.50 (17) |
| O2—C13—C14—C15 | −178.04 (16) | O5—C31—C32—C33 | 177.97 (16) |
| C12—C13—C14—C15 | 4.6 (3) | C30—C31—C32—C33 | −4.0 (3) |
| O3—C14—C15—C16 | 176.25 (16) | O6—C32—C33—C34 | −176.20 (17) |
| C13—C14—C15—C16 | −3.0 (3) | C31—C32—C33—C34 | 2.3 (3) |
| C12—C11—C16—C15 | 4.5 (3) | C32—C33—C34—C29 | 1.9 (3) |
| C10—C11—C16—C15 | −173.55 (18) | C30—C29—C34—C33 | −4.4 (3) |
| C14—C15—C16—C11 | −1.6 (3) | C28—C29—C34—C33 | 174.28 (18) |
Hydrogen-bond geometry (Å, º)
Cg1, Cg2 and Cg3 are the centroids of the C2–C7, C29–C34 and C20–C25 rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···N3 | 0.86 (2) | 2.18 (2) | 2.635 (2) | 113 (2) |
| N4—H4N···N6 | 0.86 (2) | 2.23 (2) | 2.637 (2) | 109 (1) |
| O3—H3O···O2 | 0.84 (2) | 2.29 (3) | 2.660 (2) | 107 (2) |
| O6—H6O···O5 | 0.84 (2) | 2.28 (2) | 2.663 (2) | 108 (2) |
| O3—H3O···N6i | 0.84 (2) | 2.19 (2) | 2.994 (2) | 161 (2) |
| O6—H6O···N3ii | 0.84 (2) | 2.22 (2) | 3.013 (2) | 157 (2) |
| N2—H2N···O4iii | 0.88 (2) | 2.01 (2) | 2.873 (2) | 170 (2) |
| N4—H4N···O2i | 0.86 (2) | 2.54 (2) | 3.390 (2) | 167 (2) |
| N5—H5N···O1iv | 0.88 (2) | 2.04 (2) | 2.900 (2) | 169 (2) |
| C33—H33···O6v | 0.95 | 2.54 | 3.212 (2) | 128 |
| C15—H15···O3vi | 0.95 | 2.63 | 3.166 (2) | 113 |
| C33—H33···O6i | 0.95 | 2.54 | 3.212 (2) | 128 |
| C10—H10A···Cg1vii | 0.99 | 2.80 | 3.774 (2) | 168 |
| C18—H18A···Cg2ii | 0.98 | 2.66 | 3.603 (4) | 161 |
| C28—H28B···Cg3viii | 0.99 | 2.75 | 3.720 (2) | 166 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+1, −y, −z+1; (iii) −x+3/2, y+1/2, −z+1/2; (iv) −x+3/2, y−1/2, −z+1/2; (v) −x+1, −y−1, −z+1; (vi) −x, −y+2, −z; (vii) x, y+1, z; (viii) x, y−1, z.
Funding Statement
This work was funded by Sunway University grant INT-PRO-2017–096. Universiti Putra Malaysia grants RUGS 9199834 and RUGS 9174000. Kementerian Sains, Teknologi dan Inovasi grant 09–02–04–0752-EA001.
References
- Agilent (2011). CrysAlis PRO. Agilent Technologies, Yarnton, England.
- Ali, S. M. M., Azad, M. A. K., Jesmin, M., Ahsan, S., Rahman, M. M., Khanam, J. A., Islam, M. N. & Shahriar, S. M. S. (2012). Asian Pac. J. Trop. Biomed. 2, 438–442. [DOI] [PMC free article] [PubMed]
- Behenna, D. C., Mohr, J. T., Sherden, N. H., Marinescu, S. C., Harned, A. M., Tani, K., Seto, M., Ma, S., Novák, Z., Krout, M. R., McFadden, R. M., Roizen, J. L., Enquist, J. A. Jr, White, D. E., Levine, S. R., Petrova, K. V., Iwashita, A., Virgil, S. C. & Stoltz, B. M. (2011). Chem. Eur. J. 17, 14199–14223. [DOI] [PMC free article] [PubMed]
- Beraldo, H. & Gambino, D. (2004). Mini Rev. Med. Chem. 4, 31–39. [DOI] [PubMed]
- Brandenburg, K. (2006). DIAMOND. Crystal Impact GbR, Bonn, Germany.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Groth, P. (1980). Acta Chem. Scand. A34, 609–620.
- Hoek, W. G. M. van den, Bokkers, G., Krabbendam, H., Spek, A. L. & Kroon, J. (1980). Z. Kristallogr. 152, 215–225.
- Kıyak, B., Esenpınar, A. A. & Bulut, M. (2015). Polyhedron, 90, 183–196.
- Ma, S., Reeves, C. M., Craig, R. A. II & Stoltz, B. M. (2014). Tetrahedron, 70, 4208–4212. [DOI] [PMC free article] [PubMed]
- McKinnon, J. J., Jayatilaka, D. & Spackman, M. A. (2007). Chem. Commun. pp. 3814–3816. [DOI] [PubMed]
- Pandeya, S. N. (2012). Acta Pharm. 62, 263–286. [DOI] [PubMed]
- Pandey, S. & Srivastava, R. S. (2010). Lett. Drug. Des. Discov. 7, 694–706.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Sriram, D., Yogeeswari, P. & Thirumurugan, R. (2004). Bioorg. Med. Chem. Lett. 14, 3923–3924. [DOI] [PubMed]
- Tan, M. Y., Crouse, K. A., Ravoof, T. B. S. A., Jotani, M. M. & Tiekink, E. R. T. (2017). Acta Cryst. E73, 1001–1008. [DOI] [PMC free article] [PubMed]
- Tan, M. Y., Crouse, K. A., Ravoof, T. B. S. A. & Tiekink, E. R. T. (2015). Acta Cryst. E71, o1047–o1048. [DOI] [PMC free article] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989017017273/hb7720sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017017273/hb7720Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989017017273/hb7720Isup3.cml
CCDC reference: 926756
Additional supporting information: crystallographic information; 3D view; checkCIF report









