The crystal structure of a novel binuclear copper(II) complex with a dianionic Schiff base derived from 5-bromosalicylic aldehyde and cysteamine prepared by direct synthesis is reported.
Keywords: crystal structure, dinuclear copper(II) complex, Schiff base, 5-bromosalicylaldehyde, cysteamine (2-aminoethanthiol)
Abstract
The crystal structure of the title compound, [Cu2(C18H12Br2N4O2S2)2], consists of binuclear complex units which lie across inversion centres and are connected by weak Cu—O coordination bonds forming chains along the b axis. The CuII ion is five-coordinated by two N atoms and two O atoms of the chelating ligand and one symmetry-related O atom forming a square-pyramidal coordination geometry. In the crystal, short S⋯Br contacts connect neighbouring chains into a two-dimensional network parallel to (101).
Chemical context
Schiff bases and their metal complexes represent one of the most widely used classes of compound because of their synthetic flexibility and wide range of applications (Mitra et al., 1997 ▸; Bera et al., 1998 ▸; Prabhakaran et al., 2004 ▸). Such complexes having sulfur-containing ligands are of considerable interest because of their diverse coordination modes and bridging ability. The formation and cleavage of disulfide bonds are known to be important for the biological activity of several sulfur-containing peptides and proteins (Gilbert et al., 1999 ▸; Jacob et al., 2003 ▸).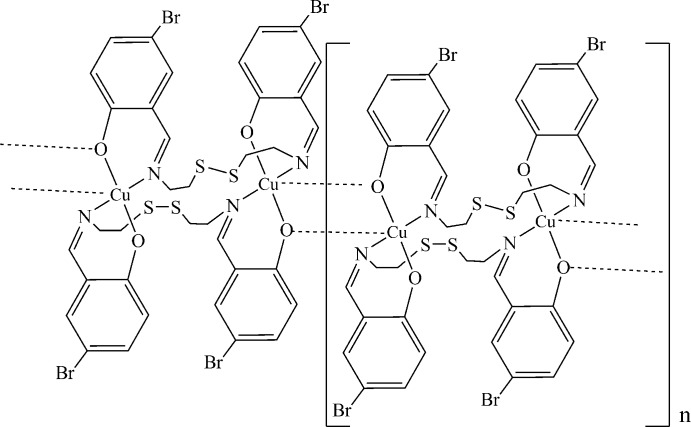
In this study we have continued our investigations in the field of direct synthesis, which is an efficient method to obtain novel polynuclear complexes (Babich & Kokozay, 1997 ▸; Vinogradova et al., 2001 ▸; Nesterova et al., 2008 ▸). The title compound was prepared by direct synthesis starting from zero-valent Cu with a Schiff-base ligand, the product of condensation between 5-bromsalicylaldehyde and cysteamine, formed in situ in a methanol/dimethylformamide (DMF) mixture.
Structural commentary
In the title compound, binuclear complex units lie across an inversion centre (Fig. 1 ▸). The coordination geometry around the CuII ion is comparable to that found in copper complexes reported earlier (CSD refcode FEDCIB; Dhar et al., 2005 ▸; Rusanova & Bederak, 2017 ▸). Despite the close structural similarity, neighboring centrosymmetric binuclear fragments are connected by additional weak Cu⋯O (2 − x, 1 − y, 2 − z) coordination bonds with the oxygen atoms of the ligand [2.520 (3) Å] and organized in chains along the b-axis direction (Fig. 2 ▸). Thus, each CuII ion is five-coordinated by two nitrogen atoms (N1, N2), two oxygen atoms (O1, O2) and one symmetry-related O atom [O1 (2 − x, 1 − y, 2 − z)], forming a distorted square-pyramidal geometry.
Figure 1.
The molecular structure of the binuclear complex unit of the title compound, showing 50% probability displacement ellipsoids. Unlabelled atoms are related to labelled ones by the symmetry operation (2 − x, −y, 2 − z).
Figure 2.
The crystal packing of the title compound viewed along the a axis. Short S⋯Br contacts are shown as dashed lines. H atoms are not shown.
The chelating fragments coordinated to the CuII ions are twisted relative to each other, as defined by the dihedral angle of 28.9 (2)° formed between the mean planes of atoms O1/N1/C1/C6/C7 and O2/N2/C8/C13/C14. The thiosulfonate moiety is not involved in any metal–ligand interactions.
The separation between the two symmetry-related CuII ions within the binuclear fragment is 5.2161 (11) Å and between neighboring fragments is 3.4458 (11) Å. In general, all bonding parameters and the dimensions of the angles in the title complex are in good agreement with those encountered in related complexes (Dhar et al., 2005 ▸; Zhang et al., 2010 ▸).
Supramolecular features
In the crystal, short S⋯Br(− + x,
+ x,  − y,
− y,  + z) contacts with a distance of 3.5108 (13) Å connect neighboring chains, forming a two-dimensional network parallel to (101) (Fig. 3 ▸).
+ z) contacts with a distance of 3.5108 (13) Å connect neighboring chains, forming a two-dimensional network parallel to (101) (Fig. 3 ▸).
Figure 3.
The crystal packing of the title compound viewed along the b axis. Short S⋯Br contacts are shown as dashed lines. H atoms are not shown.
In contrast to the previously reported complex (Rusanova & Bederak, 2017 ▸), there are no hydrogen-bond or π-π stacking interactions in the title complex. In terms of C—H⋯Br interactions, the intermolecular C16—H16B⋯Br2(x + 1, y, z) distance of 3.03 Å and C17—H17B⋯S1(− + x,
+ x,  − y, −
− y, − + z) distance of 2.95 Å are almost equal to the sum of the van der Waals radii for the atoms involved and may be worthy of note.
+ z) distance of 2.95 Å are almost equal to the sum of the van der Waals radii for the atoms involved and may be worthy of note.
Database survey
A search of the Cambridge Structural Database (Version 5.38; last update November 2016; Groom et al., 2016 ▸) for related complexes with an aminoethanethiol group gave 165 hits, including two closely related structures with a disulfide moiety, viz. bis[(μ2-sulfato)(6-salicylideneamino-3,4-dithiahexylammonium]copper(II) and bis(μ2-N,N′-(3,4-dithiahexane-1,6-diyl)bis(salicylideneiminato)-N,N′,O,O′)dicopper(II) (Dhar et al., 2004 ▸, 2005 ▸). The length of the S⋯Br contact in the title compound is in good agreement with those in related complexes (CSD refcodes WEMCAT and QELVIN; Salivon et al., 2006 ▸, 2007 ▸; CSD refcode PODDAO; Xia et al., 2008 ▸)
Synthesis and crystallization
A solution of KOH (0.12 g, 2 mmol) in a minimum amount of methanol was added to a solution of aminoethanethiol hydrochloride (0.23g, 2 mmol) in methanol (5 ml) and stirred on an ice bath for 10 min. The white precipitate of solid KCl was removed by filtration and 5-bromsalicylaldehyde (0.402 g, 2 mmol) in dimethylformamide (10 ml) were added to the filtrate and stirred magnetically for 50 min. Copper powder (0.064 g, 1 mmol) were added to the yellow solution of the Schiff base formed in situ, and the resulting deep green–brown solution was stirred magnetically and heated in air at 323–333 K for 2 h, resulting in a dark-green precipitate. Crystals suitable for crystallographic study were grown from a saturated solution in DMF after successive addition of CH2Cl2. The crystals were filtered off, washed with dry i-PrOH and finally dried at room temperature (yield: 20%).
The IR spectrum of the title compound (as KBr pellets) is consistent with the above structural data. In the range 4000–400 cm−1 it shows all characteristic functional groups peaks: ν(CH) due to aromatic =C—H stretching at 3000–3100cm−1, the aromatic ring vibrations in the 1600–1400 cm−1 region, weak S–S absorptions at 500–540 cm−1 as well absorbance at 1630 cm−1 assigned to the azomethine ν(C=N) group. Analysis calculated for C36H32Br4Cu2N4O4S4: C37.28, H 2.78, N 4.83, S 11.06%; found: C 37.32, H 3.01, N 4.70, S 11.10%.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 1 ▸. All hydrogen atoms were placed at calculated positions (C–H = 0.93–0.97 Å) and refined as riding with U iso(H) = 1.2U eq(C).
Table 1. Experimental details.
| Crystal data | |
| Chemical formula | [Cu2(C18H16Br2N2O2S2)2] |
| M r | 1159.61 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 296 |
| a, b, c (Å) | 12.3596 (5), 8.3442 (3), 19.5002 (7) |
| β (°) | 95.156 (2) |
| V (Å3) | 2002.94 (13) |
| Z | 2 |
| Radiation type | Mo Kα |
| μ (mm−1) | 5.31 |
| Crystal size (mm) | 0.45 × 0.10 × 0.06 |
| Data collection | |
| Diffractometer | Bruker SMART APEXII |
| Absorption correction | Multi-scan SADABS |
| T min, T max | 0.36, 0.74 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 18553, 3939, 2644 |
| R int | 0.075 |
| (sin θ/λ)max (Å−1) | 0.617 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.046, 0.098, 1.05 |
| No. of reflections | 3939 |
| No. of parameters | 244 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.82, −0.66 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989017017790/lh5862sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017017790/lh5862Isup2.hkl
CCDC reference: 1810837
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| [Cu2(C18H16Br2N2O2S2)2] | F(000) = 1140 |
| Mr = 1159.61 | Dx = 1.923 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 12.3596 (5) Å | Cell parameters from 2534 reflections |
| b = 8.3442 (3) Å | θ = 2.7–23.6° |
| c = 19.5002 (7) Å | µ = 5.31 mm−1 |
| β = 95.156 (2)° | T = 296 K |
| V = 2002.94 (13) Å3 | Needle, green |
| Z = 2 | 0.45 × 0.10 × 0.06 mm |
Data collection
| Bruker SMART APEXII diffractometer | 3939 independent reflections |
| Radiation source: sealed tube | 2644 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.075 |
| phi and ω scans | θmax = 26.0°, θmin = 1.9° |
| Absorption correction: multi-scan sadabs | h = −15→8 |
| Tmin = 0.36, Tmax = 0.74 | k = −10→10 |
| 18553 measured reflections | l = −23→24 |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.046 | H-atom parameters constrained |
| wR(F2) = 0.098 | w = 1/[σ2(Fo2) + (0.0426P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max = 0.004 |
| 3939 reflections | Δρmax = 0.82 e Å−3 |
| 244 parameters | Δρmin = −0.66 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| BR1 | 1.46222 (5) | 0.24549 (7) | 0.82836 (3) | 0.04443 (19) | |
| BR2 | 0.40450 (5) | 0.21977 (8) | 1.11957 (3) | 0.0456 (2) | |
| CU1 | 0.95419 (5) | 0.30506 (7) | 1.00084 (3) | 0.01799 (16) | |
| N1 | 1.0809 (3) | 0.1638 (4) | 1.03365 (18) | 0.0184 (9) | |
| N2 | 0.8169 (3) | 0.3643 (4) | 0.94366 (18) | 0.0174 (9) | |
| O1 | 1.0408 (2) | 0.4239 (3) | 0.94197 (15) | 0.0184 (7) | |
| O2 | 0.8768 (3) | 0.2123 (4) | 1.07014 (16) | 0.0239 (8) | |
| S1 | 1.10722 (11) | −0.01078 (14) | 1.23102 (6) | 0.0231 (3) | |
| S2 | 1.21861 (10) | −0.17845 (15) | 1.20755 (6) | 0.0229 (3) | |
| C1 | 1.1308 (4) | 0.3764 (5) | 0.9170 (2) | 0.0187 (11) | |
| C2 | 1.1648 (4) | 0.4542 (5) | 0.8585 (2) | 0.0241 (12) | |
| H2 | 1.120805 | 0.533137 | 0.836940 | 0.029* | |
| C3 | 1.2610 (4) | 0.4166 (6) | 0.8324 (2) | 0.0279 (13) | |
| H3 | 1.282029 | 0.470919 | 0.794135 | 0.033* | |
| C4 | 1.3278 (4) | 0.2961 (6) | 0.8634 (3) | 0.0265 (12) | |
| C5 | 1.2960 (4) | 0.2146 (6) | 0.9191 (3) | 0.0261 (12) | |
| H5 | 1.340017 | 0.133916 | 0.939219 | 0.031* | |
| C6 | 1.1973 (4) | 0.2517 (5) | 0.9464 (2) | 0.0205 (11) | |
| C7 | 1.1680 (4) | 0.1571 (5) | 1.0031 (2) | 0.0184 (11) | |
| H7 | 1.218760 | 0.081147 | 1.019849 | 0.022* | |
| C8 | 0.7732 (4) | 0.2183 (5) | 1.0778 (2) | 0.0193 (11) | |
| C9 | 0.7380 (4) | 0.1630 (6) | 1.1407 (2) | 0.0260 (12) | |
| H9 | 0.788849 | 0.123649 | 1.174609 | 0.031* | |
| C10 | 0.6303 (4) | 0.1663 (6) | 1.1528 (3) | 0.0247 (12) | |
| H10 | 0.608900 | 0.131899 | 1.194901 | 0.030* | |
| C11 | 0.5537 (4) | 0.2208 (6) | 1.1021 (3) | 0.0282 (12) | |
| C12 | 0.5833 (4) | 0.2746 (6) | 1.0406 (3) | 0.0287 (13) | |
| H12 | 0.530635 | 0.311494 | 1.007245 | 0.034* | |
| C13 | 0.6938 (4) | 0.2746 (5) | 1.0273 (2) | 0.0214 (11) | |
| C14 | 0.7216 (4) | 0.3439 (5) | 0.9635 (2) | 0.0206 (11) | |
| H14 | 0.663784 | 0.377719 | 0.933102 | 0.025* | |
| C15 | 1.0733 (4) | 0.0487 (5) | 1.0908 (2) | 0.0191 (11) | |
| H15A | 1.121681 | −0.040961 | 1.085098 | 0.023* | |
| H15B | 0.999770 | 0.007538 | 1.089495 | 0.023* | |
| C16 | 1.1034 (4) | 0.1278 (6) | 1.1595 (2) | 0.0222 (12) | |
| H16A | 1.051164 | 0.211612 | 1.166482 | 0.027* | |
| H16B | 1.174199 | 0.177672 | 1.158804 | 0.027* | |
| C17 | 0.8220 (4) | 0.4465 (5) | 0.8773 (2) | 0.0219 (11) | |
| H17A | 0.865362 | 0.542817 | 0.884674 | 0.026* | |
| H17B | 0.749153 | 0.478879 | 0.860140 | 0.026* | |
| C18 | 0.8697 (4) | 0.3449 (5) | 0.8229 (2) | 0.0212 (11) | |
| H18A | 0.939036 | 0.302111 | 0.841906 | 0.025* | |
| H18B | 0.883066 | 0.412405 | 0.784101 | 0.025* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| BR1 | 0.0376 (4) | 0.0474 (4) | 0.0522 (4) | −0.0041 (3) | 0.0256 (3) | −0.0010 (3) |
| BR2 | 0.0234 (3) | 0.0744 (5) | 0.0400 (4) | −0.0008 (3) | 0.0092 (3) | −0.0080 (3) |
| CU1 | 0.0192 (3) | 0.0175 (3) | 0.0170 (3) | 0.0009 (3) | 0.0002 (2) | 0.0026 (3) |
| N1 | 0.028 (2) | 0.017 (2) | 0.010 (2) | −0.0013 (18) | −0.0003 (18) | −0.0023 (16) |
| N2 | 0.024 (2) | 0.015 (2) | 0.013 (2) | 0.0012 (17) | −0.0001 (18) | −0.0034 (16) |
| O1 | 0.0209 (19) | 0.0170 (18) | 0.0174 (17) | 0.0007 (14) | 0.0019 (15) | 0.0013 (14) |
| O2 | 0.0197 (19) | 0.029 (2) | 0.0229 (18) | 0.0018 (15) | 0.0025 (15) | 0.0086 (15) |
| S1 | 0.0316 (7) | 0.0187 (7) | 0.0190 (7) | 0.0033 (6) | 0.0029 (6) | 0.0010 (5) |
| S2 | 0.0253 (7) | 0.0192 (7) | 0.0229 (7) | 0.0026 (6) | −0.0044 (6) | −0.0016 (6) |
| C1 | 0.022 (3) | 0.018 (3) | 0.015 (3) | −0.001 (2) | −0.002 (2) | −0.004 (2) |
| C2 | 0.037 (3) | 0.018 (3) | 0.018 (3) | −0.007 (2) | 0.004 (2) | 0.002 (2) |
| C3 | 0.040 (3) | 0.026 (3) | 0.020 (3) | −0.009 (3) | 0.010 (3) | −0.003 (2) |
| C4 | 0.029 (3) | 0.027 (3) | 0.025 (3) | −0.003 (2) | 0.010 (2) | −0.006 (2) |
| C5 | 0.027 (3) | 0.020 (3) | 0.031 (3) | −0.001 (2) | 0.005 (2) | −0.003 (2) |
| C6 | 0.020 (3) | 0.022 (3) | 0.020 (3) | −0.004 (2) | 0.003 (2) | −0.007 (2) |
| C7 | 0.017 (3) | 0.019 (3) | 0.018 (3) | 0.004 (2) | −0.002 (2) | 0.000 (2) |
| C8 | 0.021 (3) | 0.014 (3) | 0.024 (3) | 0.000 (2) | 0.004 (2) | −0.006 (2) |
| C9 | 0.029 (3) | 0.027 (3) | 0.022 (3) | 0.003 (2) | 0.001 (2) | 0.004 (2) |
| C10 | 0.023 (3) | 0.026 (3) | 0.026 (3) | −0.006 (2) | 0.008 (2) | −0.001 (2) |
| C11 | 0.024 (3) | 0.033 (3) | 0.029 (3) | −0.002 (2) | 0.009 (2) | −0.006 (2) |
| C12 | 0.021 (3) | 0.033 (3) | 0.030 (3) | 0.001 (2) | −0.003 (2) | −0.003 (2) |
| C13 | 0.021 (3) | 0.021 (3) | 0.022 (3) | −0.002 (2) | −0.002 (2) | −0.002 (2) |
| C14 | 0.018 (3) | 0.019 (3) | 0.023 (3) | 0.006 (2) | −0.005 (2) | −0.002 (2) |
| C15 | 0.014 (3) | 0.017 (3) | 0.026 (3) | 0.002 (2) | 0.002 (2) | 0.002 (2) |
| C16 | 0.026 (3) | 0.021 (3) | 0.019 (3) | 0.002 (2) | −0.002 (2) | 0.003 (2) |
| C17 | 0.029 (3) | 0.021 (3) | 0.015 (3) | 0.004 (2) | −0.004 (2) | 0.003 (2) |
| C18 | 0.024 (3) | 0.022 (3) | 0.018 (3) | −0.004 (2) | 0.001 (2) | 0.004 (2) |
Geometric parameters (Å, º)
| BR1—C4 | 1.900 (5) | C6—C7 | 1.433 (6) |
| BR2—C11 | 1.905 (5) | C7—H7 | 0.9300 |
| CU1—O2 | 1.890 (3) | C8—C13 | 1.408 (6) |
| CU1—O1 | 1.915 (3) | C8—C9 | 1.415 (6) |
| CU1—N2 | 2.006 (4) | C9—C10 | 1.372 (6) |
| CU1—N1 | 2.018 (4) | C9—H9 | 0.9300 |
| CU1—O1i | 2.520 (3) | C10—C11 | 1.382 (7) |
| N1—C7 | 1.277 (5) | C10—H10 | 0.9300 |
| N1—C15 | 1.480 (5) | C11—C12 | 1.362 (7) |
| N2—C14 | 1.284 (5) | C12—C13 | 1.413 (6) |
| N2—C17 | 1.471 (5) | C12—H12 | 0.9300 |
| O1—C1 | 1.315 (5) | C13—C14 | 1.441 (6) |
| O2—C8 | 1.304 (5) | C14—H14 | 0.9300 |
| S1—C16 | 1.808 (5) | C15—C16 | 1.511 (6) |
| S1—S2 | 2.0435 (17) | C15—H15A | 0.9700 |
| S2—C18ii | 1.832 (5) | C15—H15B | 0.9700 |
| C1—C2 | 1.410 (6) | C16—H16A | 0.9700 |
| C1—C6 | 1.415 (6) | C16—H16B | 0.9700 |
| C2—C3 | 1.371 (6) | C17—C18 | 1.517 (6) |
| C2—H2 | 0.9300 | C17—H17A | 0.9700 |
| C3—C4 | 1.403 (7) | C17—H17B | 0.9700 |
| C3—H3 | 0.9300 | C18—S2ii | 1.832 (5) |
| C4—C5 | 1.369 (7) | C18—H18A | 0.9700 |
| C5—C6 | 1.407 (6) | C18—H18B | 0.9700 |
| C5—H5 | 0.9300 | ||
| O2—CU1—O1 | 170.63 (14) | C13—C8—C9 | 117.8 (4) |
| O2—CU1—N2 | 92.36 (15) | C10—C9—C8 | 121.4 (5) |
| O1—CU1—N2 | 91.68 (14) | C10—C9—H9 | 119.3 |
| O2—CU1—N1 | 87.85 (14) | C8—C9—H9 | 119.3 |
| O1—CU1—N1 | 91.88 (14) | C9—C10—C11 | 119.7 (5) |
| N2—CU1—N1 | 156.01 (14) | C9—C10—H10 | 120.1 |
| O2—CU1—O1i | 92.59 (12) | C11—C10—H10 | 120.1 |
| O1—CU1—O1i | 78.91 (12) | C12—C11—C10 | 121.2 (5) |
| N2—CU1—O1i | 90.61 (12) | C12—C11—BR2 | 120.0 (4) |
| N1—CU1—O1i | 113.35 (13) | C10—C11—BR2 | 118.8 (4) |
| C7—N1—C15 | 115.8 (4) | C11—C12—C13 | 120.2 (5) |
| C7—N1—CU1 | 122.7 (3) | C11—C12—H12 | 119.9 |
| C15—N1—CU1 | 121.2 (3) | C13—C12—H12 | 119.9 |
| C14—N2—C17 | 116.0 (4) | C8—C13—C12 | 119.7 (4) |
| C14—N2—CU1 | 123.6 (3) | C8—C13—C14 | 122.2 (4) |
| C17—N2—CU1 | 120.2 (3) | C12—C13—C14 | 117.9 (4) |
| C1—O1—CU1 | 127.1 (3) | N2—C14—C13 | 127.6 (4) |
| C8—O2—CU1 | 129.2 (3) | N2—C14—H14 | 116.2 |
| C16—S1—S2 | 103.67 (16) | C13—C14—H14 | 116.2 |
| C18ii—S2—S1 | 101.41 (16) | N1—C15—C16 | 110.9 (4) |
| O1—C1—C2 | 119.0 (4) | N1—C15—H15A | 109.5 |
| O1—C1—C6 | 123.5 (4) | C16—C15—H15A | 109.5 |
| C2—C1—C6 | 117.5 (4) | N1—C15—H15B | 109.5 |
| C3—C2—C1 | 121.8 (5) | C16—C15—H15B | 109.5 |
| C3—C2—H2 | 119.1 | H15A—C15—H15B | 108.0 |
| C1—C2—H2 | 119.1 | C15—C16—S1 | 113.1 (3) |
| C2—C3—C4 | 120.0 (4) | C15—C16—H16A | 109.0 |
| C2—C3—H3 | 120.0 | S1—C16—H16A | 109.0 |
| C4—C3—H3 | 120.0 | C15—C16—H16B | 109.0 |
| C5—C4—C3 | 119.9 (5) | S1—C16—H16B | 109.0 |
| C5—C4—BR1 | 119.9 (4) | H16A—C16—H16B | 107.8 |
| C3—C4—BR1 | 120.2 (4) | N2—C17—C18 | 113.8 (4) |
| C4—C5—C6 | 120.7 (5) | N2—C17—H17A | 108.8 |
| C4—C5—H5 | 119.7 | C18—C17—H17A | 108.8 |
| C6—C5—H5 | 119.7 | N2—C17—H17B | 108.8 |
| C5—C6—C1 | 120.1 (4) | C18—C17—H17B | 108.8 |
| C5—C6—C7 | 117.3 (4) | H17A—C17—H17B | 107.7 |
| C1—C6—C7 | 122.7 (4) | C17—C18—S2ii | 113.1 (3) |
| N1—C7—C6 | 128.2 (4) | C17—C18—H18A | 109.0 |
| N1—C7—H7 | 115.9 | S2ii—C18—H18A | 109.0 |
| C6—C7—H7 | 115.9 | C17—C18—H18B | 109.0 |
| O2—C8—C13 | 124.2 (4) | S2ii—C18—H18B | 109.0 |
| O2—C8—C9 | 118.0 (4) | H18A—C18—H18B | 107.8 |
Symmetry codes: (i) −x+2, −y+1, −z+2; (ii) −x+2, −y, −z+2.
References
- Babich, O. A. & Kokozay, V. N. (1997). Polyhedron, 16, 1487–1490.
- Bera, P., Butcher, R. J. & Saha, N. (1998). Chem. Lett. 27, 559–560.
- Bruker (2008). APEX2, SMART, SAINT and SADABS. Bruker AXS, Inc., Madison, Wisconsin, USA.
- Dhar, S., Nethaji, M. & Chakravarty, A. R. (2004). Dalton Trans. pp. 4180–4184. [DOI] [PubMed]
- Dhar, S., Nethaji, M. & Chakravarty, A. R. (2005). Dalton Trans. pp. 344–348. [DOI] [PubMed]
- Gilbert, B. C., Silvester, S., Walton, P. H. & Whitwood, A. C. (1999). J. Chem. Soc. Perkin Trans. 2, pp. 1891–1895.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Jacob, C., Giles, G. L., Giles, N. M. & Sies, H. (2003). Angew. Chem. Int. Ed. 42, 4742–4758. [DOI] [PubMed]
- Mitra, A., Banerjee, T., Roychowdhury, P., Chaudhuri, S., Bera, P. & Saha, N. (1997). Polyhedron, 16, 3735–3742.
- Nesterova, O. V., Petrusenko, S. R., Kokozay, V. N., Skelton, B. W., Jezierska, J., Linert, W. & Ozarowski, A. (2008). Dalton Trans. pp. 1431–1436. [DOI] [PubMed]
- Prabhakaran, R., Geetha, A., Thilagavathi, M., Karvembu, R., Krishnan, V., Bertagnolli, H. & Natarajan, K. (2004). J. Inorg. Biochem. 98, 2131–2140. [DOI] [PubMed]
- Rusanova, J. A. & Bederak, D. (2017). Acta Cryst. E73, 1797–1800. [DOI] [PMC free article] [PubMed]
- Salivon, N. F., Filinchuk, Y. E. & Olijnyk, V. V. (2006). Z. Anorg. Allg. Chem. 632, 1610–1613.
- Salivon, N. F., Olijnik, V. V. & Shkurenko, A. A. (2007). Russ. J. Coord. Chem. 33, 908–913.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Vinogradova, E. A., Vassilyeva, O. Y., Kokozay, V. N., Squattrito, P. J., Reedijk, J., Van Albada, G. A., Linert, W., Tiwary, S. K. & Raithby, P. R. (2001). New J. Chem. 25, 949–953.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Xia, J.-H., Liu, Z. & Jin, L.-X. (2008). Chin. J. Inorg. Chem. 5, 823–826.
- Zhang, S.-H., Wang, Y., Feng, C. & Li, G. Z. (2010). J. Coord. Chem. 63, 3697–3705.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989017017790/lh5862sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017017790/lh5862Isup2.hkl
CCDC reference: 1810837
Additional supporting information: crystallographic information; 3D view; checkCIF report





