The crystal structure features two O—H⋯O hydrogen bonds, forming chains along the [010] direction.
Keywords: crystal structure; 6,7-dehydroroyleanone; Taxodium distichum (L.) Rich
Abstract
The title compound, 6,7-dehydroroyleanone, C20H26O3 [systematic name: (4bS)-3-hydroxy-2-isopropyl-4b,8,8-trimethyl-4b,5,6,7,8,8a-hexahydrophenanthrene-1,4-dione] was isolated from Taxodium distichum (L.) Rich. The compound crystallizes in the space group P21. The crystal structure features two O—H⋯O hydrogen bonds, forming chains along the [010] direction.
Chemical context
Taxodium distichum (L.) Rich. is a tree native to North America that can grow to 25 m in height (Ogunwande et al., 2007 ▸). Its leaves and seeds are used for the treatment of malaria and liver disease (Kupchan et al., 1968 ▸). Previous studies revealed that it contains multiple compounds such as diterpenes (Kusumoto et al., 2010 ▸), flavonoids (Zaghloul et al., 2008 ▸), proanthocyanidins (Stafford & Lester, 1986 ▸), lignins (Logan & Thomas, 1985 ▸), sterols and fatty acids (Geiger & de Groot-Pfleiderer, 1979 ▸). A detailed phytochemical investigation of a petroleum ether extract of the seeds of Taxodium distichum (L.) Rich. led to the isolation of the title compound 6,7-dehydroroyleanone. Herein we present the crystal structure of 6,7-dehydroroyleanone, which was undertaken in order to establish unambiguously the stereochemical features of this natural compound.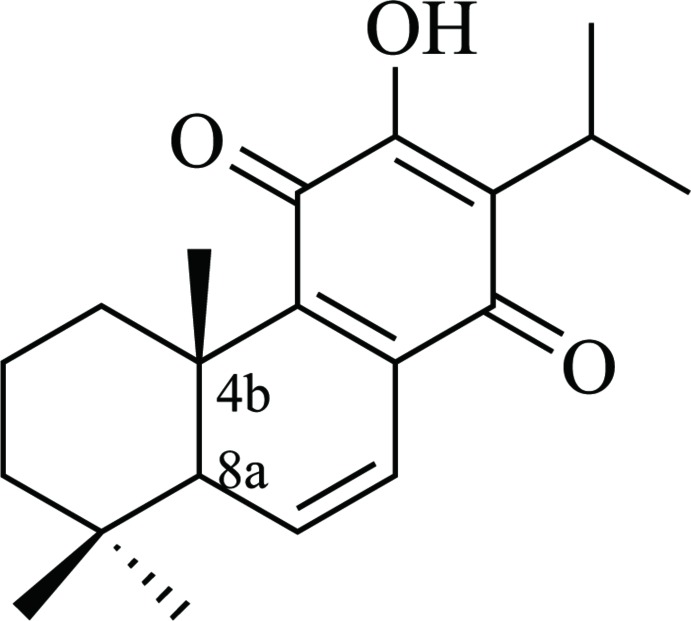
Structural commentary
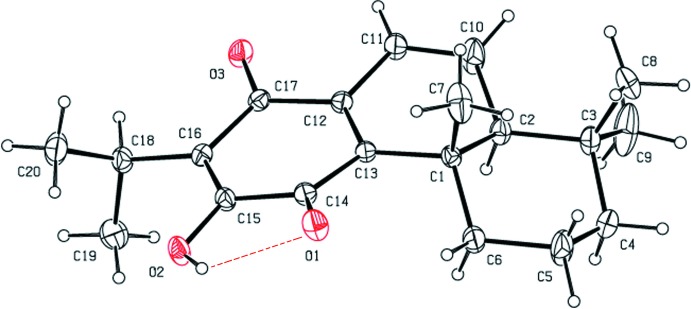
The molecular structure of the title compound is shown in Fig. 1 ▸. The title compound belongs to the class of abietane-type diterpenes and the structure contains two ketone groups at C14 and C17 and three double bonds located between atoms C10 and C11, C12 and C13, C15 and C16. The torsion angles C17—C12—C13—C1 [176.8 (2)°], C11—C12—C13—C14 [168.7 (3)°], C6—C1—C2—C10 [171.6 (3)°] and C13—C1—C2—C3 [−173.4 (3)°] describe the geometry at the junctions of the three rings. An intramolecular O2—H2A⋯O1 hydrogen bond (Table 1 ▸) stabilizes the molecular conformation.
Figure 1.
The molecular structure of the title compound, with the atom labelling and 50% probability displacement ellipsoids. The intramolecular O—H⋯O hydrogen bond (see Table 1 ▸) is shown as a red dashed line.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O2—H2A⋯O1 | 0.82 (6) | 2.03 (5) | 2.607 (3) | 128 (5) |
| O2—H2A⋯O3i | 0.82 (6) | 2.53 (6) | 3.160 (3) | 135 (5) |
| C11—H11⋯O1ii | 0.93 | 2.37 | 3.290 (3) | 173 (5) |
Symmetry codes: (i)  ; (ii)
; (ii) 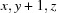 .
.
Supramolecular features
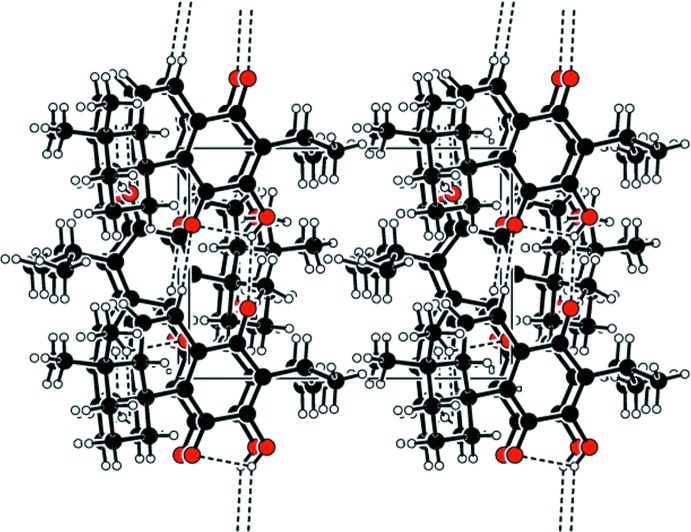
In the crystal, O2—H2A⋯O3i and C11—H11⋯O1i hydrogen bonds link the molecules, forming chains along [010] (Table 1 ▸ and Fig. 2 ▸).
Figure 2.
Part of the crystal structure of the title compound, with hydrogen bonds (see Table 1 ▸) shown as dashed lines.
Database survey
A search of the Cambridge Structural Database (CSD, Version 5.27, last update Feb 2017; Groom et al., 2016 ▸) yielded the compound royleanone (HACGUN01; Fun et al., 2011 ▸), which has a similar structure to the title compound but without the double bond between C10 and C11.
Synthesis and crystallization
The title compound was isolated from the seeds of Taxodium distichum (L.) Rich. collected in Xining, China, in April 2015 (SC0185). The air-dried seeds of Taxodium distichum (1.1 kg) were extracted with 95% EtOH and then partitioned successively with petroleum ether (PE), ethyl acetate (EtOAc) and n-butyl alcohol (n-BuOH) to give a PE extract (30 g), an EtOAc extract (50 g) and an n-BuOH extract (68 g). The PE extract (30 g) was subjected to normal-phase silica-gel column chromatography (300–400 mesh) with a gradient solvent system of petroleum ether–ethyl acetate (1:0-0:1, v/v, containing 0.1% formic acid) to give ten major fractions, denoted F1–F10. F7 (2.8 g) was sequentially subjected to Sephadex-LH20 gel column chromatography (CH2Cl2–MeOH, 3:1, v/v, containing 0.1% formic acid) to give four major fractions F7.1–F7.4. F7.3 was purified by semi-preparative HPLC (CNCH3/H2O, 20:80→100:0, 40 min, containing 0.1% formic acid in both phases) to give an orange solid, which was recrystallized from a solvent mix of CH2Cl2–MeOH (5:1) affording orange block-like crystals suitable for X-ray diffraction analysis. For the 1H and 13C NMR data of 6,7-dehydroroyleanone, see Chang et al. (2001 ▸).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. C-bound H atoms were positioned with idealized geometry and refined isotropically using a riding model with C—H = 0.94–0.99 Å and U iso(H) = 1.5U eq(C) for methyl H atoms and 1.2U eq(C) for all others. The OH hydrogen atom was refined freely with U iso(H) = 1.5U eq(O).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C20H26O3 |
| M r | 314.41 |
| Crystal system, space group | Monoclinic, P21 |
| Temperature (K) | 296 |
| a, b, c (Å) | 10.4348 (17), 7.6726 (13), 10.8210 (18) |
| β (°) | 97.773 (3) |
| V (Å3) | 858.4 (2) |
| Z | 2 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.08 |
| Crystal size (mm) | 0.3 × 0.2 × 0.2 |
| Data collection | |
| Diffractometer | Bruker P4 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 6718, 3416, 2980 |
| R int | 0.022 |
| (sin θ/λ)max (Å−1) | 0.625 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.047, 0.143, 1.08 |
| No. of reflections | 3416 |
| No. of parameters | 216 |
| No. of restraints | 1 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.27, −0.18 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989017017935/qm2118sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017017935/qm2118Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989017017935/qm2118Isup3.cdx
Supporting information file. DOI: 10.1107/S2056989017017935/qm2118Isup4.cml
CCDC reference: 1551127
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| C20H26O3 | F(000) = 340 |
| Mr = 314.41 | Dx = 1.216 Mg m−3 |
| Monoclinic, P21 | Mo Kα radiation, λ = 0.71073 Å |
| a = 10.4348 (17) Å | Cell parameters from 2696 reflections |
| b = 7.6726 (13) Å | θ = 2.6–27.3° |
| c = 10.8210 (18) Å | µ = 0.08 mm−1 |
| β = 97.773 (3)° | T = 296 K |
| V = 858.4 (2) Å3 | Block, orange |
| Z = 2 | 0.3 × 0.2 × 0.2 mm |
Data collection
| Bruker P4 diffractometer | θmax = 26.4°, θmin = 1.9° |
| φ and ω scans | h = −13→12 |
| 6718 measured reflections | k = −9→9 |
| 3416 independent reflections | l = −13→13 |
| 2980 reflections with I > 2σ(I) | 1 standard reflections every 300 reflections |
| Rint = 0.022 | intensity decay: 1% |
Refinement
| Refinement on F2 | 1 restraint |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.047 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.143 | w = 1/[σ2(Fo2) + (0.0865P)2 + 0.0799P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.08 | (Δ/σ)max = 0.003 |
| 3416 reflections | Δρmax = 0.27 e Å−3 |
| 216 parameters | Δρmin = −0.18 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.3123 (3) | 0.9175 (4) | 0.1442 (2) | 0.0325 (6) | |
| C2 | 0.4090 (3) | 1.0711 (4) | 0.1650 (3) | 0.0400 (7) | |
| H2 | 0.454767 | 1.067917 | 0.091644 | 0.048* | |
| C3 | 0.5186 (3) | 1.0545 (5) | 0.2774 (3) | 0.0471 (8) | |
| C4 | 0.5860 (4) | 0.8794 (7) | 0.2667 (4) | 0.0712 (13) | |
| H4A | 0.633484 | 0.882840 | 0.195663 | 0.085* | |
| H4B | 0.647861 | 0.861196 | 0.340907 | 0.085* | |
| C5 | 0.4925 (6) | 0.7270 (6) | 0.2515 (5) | 0.098 (2) | |
| H5A | 0.450167 | 0.716784 | 0.325687 | 0.117* | |
| H5B | 0.540153 | 0.620204 | 0.242845 | 0.117* | |
| C6 | 0.3896 (4) | 0.7501 (5) | 0.1370 (4) | 0.0696 (13) | |
| H6A | 0.431584 | 0.752332 | 0.062273 | 0.084* | |
| H6B | 0.331142 | 0.651222 | 0.130827 | 0.084* | |
| C7 | 0.2214 (4) | 0.9005 (8) | 0.2443 (3) | 0.0746 (14) | |
| H7A | 0.148629 | 0.829236 | 0.213057 | 0.112* | |
| H7B | 0.191762 | 1.014064 | 0.264675 | 0.112* | |
| H7C | 0.267276 | 0.847719 | 0.317782 | 0.112* | |
| C8 | 0.4756 (5) | 1.0761 (7) | 0.4025 (4) | 0.0797 (14) | |
| H8A | 0.427134 | 0.975447 | 0.421190 | 0.120* | |
| H8B | 0.422203 | 1.178020 | 0.402234 | 0.120* | |
| H8C | 0.549959 | 1.088847 | 0.464619 | 0.120* | |
| C9 | 0.6187 (5) | 1.1986 (9) | 0.2651 (6) | 0.106 (2) | |
| H9A | 0.581634 | 1.310139 | 0.279235 | 0.160* | |
| H9B | 0.643405 | 1.195624 | 0.182830 | 0.160* | |
| H9C | 0.693627 | 1.179893 | 0.325620 | 0.160* | |
| C10 | 0.3398 (5) | 1.2400 (6) | 0.1542 (5) | 0.0861 (17) | |
| H10 | 0.356742 | 1.324207 | 0.216089 | 0.103* | |
| C11 | 0.2507 (3) | 1.2703 (4) | 0.0517 (3) | 0.0480 (8) | |
| H11 | 0.222528 | 1.382398 | 0.029548 | 0.058* | |
| C12 | 0.2011 (3) | 1.1170 (4) | −0.0221 (3) | 0.0346 (6) | |
| C13 | 0.2247 (3) | 0.9533 (3) | 0.0209 (2) | 0.0312 (6) | |
| C14 | 0.1516 (3) | 0.8125 (4) | −0.0477 (3) | 0.0370 (6) | |
| C15 | 0.0868 (3) | 0.8459 (4) | −0.1776 (3) | 0.0376 (7) | |
| C16 | 0.0709 (3) | 1.0066 (4) | −0.2261 (3) | 0.0359 (6) | |
| C17 | 0.1215 (3) | 1.1519 (4) | −0.1454 (3) | 0.0346 (6) | |
| C18 | 0.0096 (3) | 1.0468 (5) | −0.3588 (3) | 0.0451 (7) | |
| H18 | 0.001388 | 1.173803 | −0.365603 | 0.054* | |
| C19 | 0.0958 (4) | 0.9899 (7) | −0.4517 (3) | 0.0680 (11) | |
| H19A | 0.177527 | 1.048720 | −0.434859 | 0.102* | |
| H19B | 0.055809 | 1.018277 | −0.534386 | 0.102* | |
| H19C | 0.109290 | 0.866270 | −0.445188 | 0.102* | |
| C20 | −0.1270 (4) | 0.9706 (7) | −0.3885 (4) | 0.0711 (12) | |
| H20A | −0.121950 | 0.845769 | −0.390726 | 0.107* | |
| H20B | −0.166517 | 1.012984 | −0.468077 | 0.107* | |
| H20C | −0.177917 | 1.005487 | −0.325198 | 0.107* | |
| O1 | 0.1376 (3) | 0.6667 (3) | −0.0048 (2) | 0.0561 (7) | |
| O2 | 0.0435 (3) | 0.7012 (3) | −0.2393 (2) | 0.0521 (6) | |
| H2A | 0.058 (5) | 0.628 (8) | −0.184 (5) | 0.078* | |
| O3 | 0.1021 (2) | 1.3039 (3) | −0.1790 (2) | 0.0497 (6) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0388 (14) | 0.0315 (15) | 0.0263 (12) | −0.0009 (12) | 0.0013 (10) | 0.0016 (10) |
| C2 | 0.0381 (14) | 0.0392 (17) | 0.0405 (14) | −0.0066 (13) | −0.0029 (11) | 0.0027 (13) |
| C3 | 0.0432 (16) | 0.0488 (19) | 0.0448 (16) | −0.0042 (15) | −0.0108 (13) | 0.0022 (15) |
| C4 | 0.060 (2) | 0.086 (3) | 0.060 (2) | 0.027 (2) | −0.0208 (18) | −0.012 (2) |
| C5 | 0.119 (4) | 0.047 (3) | 0.105 (4) | 0.026 (3) | −0.070 (3) | −0.010 (3) |
| C6 | 0.085 (3) | 0.043 (2) | 0.069 (2) | 0.022 (2) | −0.035 (2) | −0.0170 (18) |
| C7 | 0.056 (2) | 0.126 (4) | 0.0416 (19) | −0.030 (2) | 0.0079 (16) | 0.008 (2) |
| C8 | 0.082 (3) | 0.101 (4) | 0.050 (2) | 0.002 (3) | −0.0137 (19) | −0.031 (2) |
| C9 | 0.074 (3) | 0.113 (5) | 0.116 (4) | −0.049 (3) | −0.047 (3) | 0.047 (4) |
| C10 | 0.098 (3) | 0.034 (2) | 0.107 (4) | −0.002 (2) | −0.058 (3) | −0.010 (2) |
| C11 | 0.0561 (18) | 0.0279 (16) | 0.0545 (19) | −0.0007 (13) | −0.0124 (15) | −0.0005 (14) |
| C12 | 0.0351 (13) | 0.0292 (14) | 0.0383 (14) | −0.0001 (11) | 0.0009 (11) | 0.0001 (12) |
| C13 | 0.0334 (13) | 0.0278 (15) | 0.0318 (13) | −0.0006 (11) | 0.0022 (10) | 0.0009 (11) |
| C14 | 0.0433 (15) | 0.0285 (16) | 0.0377 (15) | −0.0001 (12) | 0.0003 (12) | 0.0015 (12) |
| C15 | 0.0410 (14) | 0.0318 (16) | 0.0375 (15) | −0.0029 (12) | −0.0038 (12) | −0.0037 (12) |
| C16 | 0.0352 (14) | 0.0364 (16) | 0.0348 (14) | 0.0040 (11) | 0.0000 (11) | 0.0021 (12) |
| C17 | 0.0321 (13) | 0.0309 (15) | 0.0403 (15) | 0.0017 (12) | 0.0026 (11) | 0.0016 (12) |
| C18 | 0.0541 (18) | 0.0399 (18) | 0.0372 (15) | 0.0030 (15) | −0.0087 (13) | 0.0022 (13) |
| C19 | 0.084 (3) | 0.083 (3) | 0.0374 (16) | 0.013 (2) | 0.0072 (17) | 0.0045 (19) |
| C20 | 0.058 (2) | 0.072 (3) | 0.074 (3) | 0.002 (2) | −0.025 (2) | −0.002 (2) |
| O1 | 0.0790 (16) | 0.0311 (12) | 0.0532 (14) | −0.0106 (11) | −0.0091 (12) | 0.0062 (10) |
| O2 | 0.0703 (15) | 0.0339 (13) | 0.0454 (12) | −0.0065 (11) | −0.0170 (11) | −0.0030 (10) |
| O3 | 0.0576 (13) | 0.0312 (12) | 0.0554 (14) | 0.0020 (10) | −0.0102 (11) | 0.0077 (10) |
Geometric parameters (Å, º)
| C1—C2 | 1.548 (4) | C9—H9C | 0.9600 |
| C1—C6 | 1.525 (5) | C10—H10 | 0.9300 |
| C1—C7 | 1.538 (4) | C10—C11 | 1.367 (5) |
| C1—C13 | 1.537 (4) | C11—H11 | 0.9300 |
| C2—H2 | 0.9800 | C11—C12 | 1.475 (4) |
| C2—C3 | 1.558 (4) | C12—C13 | 1.350 (4) |
| C2—C10 | 1.481 (6) | C12—C17 | 1.497 (4) |
| C3—C4 | 1.528 (6) | C13—C14 | 1.466 (4) |
| C3—C8 | 1.492 (5) | C14—C15 | 1.497 (4) |
| C3—C9 | 1.539 (6) | C14—O1 | 1.227 (4) |
| C4—H4A | 0.9700 | C15—C16 | 1.341 (4) |
| C4—H4B | 0.9700 | C15—O2 | 1.342 (4) |
| C4—C5 | 1.517 (8) | C16—C17 | 1.470 (4) |
| C5—H5A | 0.9700 | C16—C18 | 1.522 (4) |
| C5—H5B | 0.9700 | C17—O3 | 1.230 (4) |
| C5—C6 | 1.536 (5) | C18—H18 | 0.9800 |
| C6—H6A | 0.9700 | C18—C19 | 1.501 (5) |
| C6—H6B | 0.9700 | C18—C20 | 1.534 (5) |
| C7—H7A | 0.9600 | C19—H19A | 0.9600 |
| C7—H7B | 0.9600 | C19—H19B | 0.9600 |
| C7—H7C | 0.9600 | C19—H19C | 0.9600 |
| C8—H8A | 0.9600 | C20—H20A | 0.9600 |
| C8—H8B | 0.9600 | C20—H20B | 0.9600 |
| C8—H8C | 0.9600 | C20—H20C | 0.9600 |
| C9—H9A | 0.9600 | O2—H2A | 0.82 (6) |
| C9—H9B | 0.9600 | ||
| C6—C1—C2 | 108.0 (3) | C3—C9—H9B | 109.5 |
| C6—C1—C7 | 110.2 (4) | C3—C9—H9C | 109.5 |
| C6—C1—C13 | 111.3 (2) | H9A—C9—H9B | 109.5 |
| C7—C1—C2 | 114.5 (3) | H9A—C9—H9C | 109.5 |
| C13—C1—C2 | 106.8 (2) | H9B—C9—H9C | 109.5 |
| C13—C1—C7 | 106.0 (2) | C2—C10—H10 | 120.6 |
| C1—C2—H2 | 104.1 | C11—C10—C2 | 118.9 (4) |
| C1—C2—C3 | 116.5 (3) | C11—C10—H10 | 120.6 |
| C3—C2—H2 | 104.1 | C10—C11—H11 | 121.5 |
| C10—C2—C1 | 110.7 (3) | C10—C11—C12 | 117.0 (3) |
| C10—C2—H2 | 104.1 | C12—C11—H11 | 121.5 |
| C10—C2—C3 | 115.5 (3) | C11—C12—C17 | 116.8 (2) |
| C4—C3—C2 | 108.0 (3) | C13—C12—C11 | 121.4 (3) |
| C4—C3—C9 | 107.5 (4) | C13—C12—C17 | 121.8 (2) |
| C8—C3—C2 | 114.9 (3) | C12—C13—C1 | 121.7 (2) |
| C8—C3—C4 | 111.3 (4) | C12—C13—C14 | 116.9 (2) |
| C8—C3—C9 | 106.8 (4) | C14—C13—C1 | 121.0 (2) |
| C9—C3—C2 | 108.0 (3) | C13—C14—C15 | 119.2 (2) |
| C3—C4—H4A | 109.0 | O1—C14—C13 | 124.1 (3) |
| C3—C4—H4B | 109.0 | O1—C14—C15 | 116.8 (3) |
| H4A—C4—H4B | 107.8 | C16—C15—C14 | 122.8 (3) |
| C5—C4—C3 | 113.0 (3) | C16—C15—O2 | 123.4 (3) |
| C5—C4—H4A | 109.0 | O2—C15—C14 | 113.8 (3) |
| C5—C4—H4B | 109.0 | C15—C16—C17 | 116.7 (2) |
| C4—C5—H5A | 109.3 | C15—C16—C18 | 124.7 (3) |
| C4—C5—H5B | 109.3 | C17—C16—C18 | 118.6 (3) |
| C4—C5—C6 | 111.6 (4) | C16—C17—C12 | 120.4 (2) |
| H5A—C5—H5B | 108.0 | O3—C17—C12 | 118.8 (3) |
| C6—C5—H5A | 109.3 | O3—C17—C16 | 120.8 (3) |
| C6—C5—H5B | 109.3 | C16—C18—H18 | 107.1 |
| C1—C6—C5 | 111.9 (3) | C16—C18—C20 | 112.3 (3) |
| C1—C6—H6A | 109.2 | C19—C18—C16 | 111.0 (3) |
| C1—C6—H6B | 109.2 | C19—C18—H18 | 107.1 |
| C5—C6—H6A | 109.2 | C19—C18—C20 | 111.9 (3) |
| C5—C6—H6B | 109.2 | C20—C18—H18 | 107.1 |
| H6A—C6—H6B | 107.9 | C18—C19—H19A | 109.5 |
| C1—C7—H7A | 109.5 | C18—C19—H19B | 109.5 |
| C1—C7—H7B | 109.5 | C18—C19—H19C | 109.5 |
| C1—C7—H7C | 109.5 | H19A—C19—H19B | 109.5 |
| H7A—C7—H7B | 109.5 | H19A—C19—H19C | 109.5 |
| H7A—C7—H7C | 109.5 | H19B—C19—H19C | 109.5 |
| H7B—C7—H7C | 109.5 | C18—C20—H20A | 109.5 |
| C3—C8—H8A | 109.5 | C18—C20—H20B | 109.5 |
| C3—C8—H8B | 109.5 | C18—C20—H20C | 109.5 |
| C3—C8—H8C | 109.5 | H20A—C20—H20B | 109.5 |
| H8A—C8—H8B | 109.5 | H20A—C20—H20C | 109.5 |
| H8A—C8—H8C | 109.5 | H20B—C20—H20C | 109.5 |
| H8B—C8—H8C | 109.5 | C15—O2—H2A | 101 (4) |
| C3—C9—H9A | 109.5 | ||
| C1—C2—C3—C4 | 52.3 (4) | C11—C12—C13—C14 | 168.7 (3) |
| C1—C2—C3—C8 | −72.7 (4) | C17—C12—C13—C1 | 176.8 (2) |
| C1—C2—C3—C9 | 168.2 (4) | C17—C12—C13—C14 | −10.0 (4) |
| C1—C2—C10—C11 | −50.7 (6) | C11—C12—C17—C16 | 178.5 (3) |
| C1—C13—C14—C15 | −169.1 (2) | C11—C12—C17—O3 | 0.3 (4) |
| C1—C13—C14—O1 | 12.1 (4) | C12—C13—C14—C15 | 17.7 (4) |
| C2—C1—C6—C5 | 54.3 (5) | C12—C13—C14—O1 | −161.1 (3) |
| C2—C1—C13—C12 | −27.2 (3) | C13—C1—C2—C3 | −173.4 (3) |
| C2—C1—C13—C14 | 159.9 (2) | C13—C1—C2—C10 | 51.8 (4) |
| C2—C3—C4—C5 | −52.3 (4) | C13—C1—C6—C5 | 171.2 (4) |
| C2—C10—C11—C12 | 17.7 (7) | C13—C12—C17—C16 | −2.5 (4) |
| C3—C2—C10—C11 | 174.0 (4) | C13—C12—C17—O3 | 179.2 (3) |
| C3—C4—C5—C6 | 57.4 (6) | C13—C14—C15—C16 | −13.2 (4) |
| C4—C5—C6—C1 | −58.2 (6) | C13—C14—C15—O2 | 168.3 (3) |
| C6—C1—C2—C3 | −53.7 (4) | C14—C15—C16—C17 | 0.4 (4) |
| C6—C1—C2—C10 | 171.6 (3) | C14—C15—C16—C18 | 178.0 (3) |
| C6—C1—C13—C12 | −144.9 (3) | C15—C16—C17—C12 | 7.6 (4) |
| C6—C1—C13—C14 | 42.2 (4) | C15—C16—C17—O3 | −174.3 (3) |
| C7—C1—C2—C3 | 69.5 (4) | C15—C16—C18—C19 | −70.0 (4) |
| C7—C1—C2—C10 | −65.2 (4) | C15—C16—C18—C20 | 56.2 (4) |
| C7—C1—C6—C5 | −71.4 (5) | C17—C12—C13—C1 | 176.7 (2) |
| C7—C1—C13—C12 | 95.2 (4) | C17—C12—C13—C14 | −10.1 (4) |
| C7—C1—C13—C14 | −77.6 (4) | C17—C16—C18—C19 | 107.6 (3) |
| C8—C3—C4—C5 | 74.7 (4) | C17—C16—C18—C20 | −126.3 (3) |
| C9—C3—C4—C5 | −168.6 (4) | C18—C16—C17—C12 | −170.2 (2) |
| C10—C2—C3—C4 | −175.2 (4) | C18—C16—C17—O3 | 8.0 (4) |
| C10—C2—C3—C8 | 59.9 (5) | O1—C14—C15—C16 | 165.6 (3) |
| C10—C2—C3—C9 | −59.2 (5) | O1—C14—C15—O2 | −12.8 (4) |
| C10—C11—C12—C13 | 11.3 (5) | O2—C15—C16—C17 | 178.7 (3) |
| C10—C11—C12—C17 | −169.8 (4) | O2—C15—C16—C18 | −3.8 (5) |
| C11—C12—C13—C1 | −4.4 (4) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2A···O1 | 0.82 (6) | 2.03 (5) | 2.607 (3) | 128 (5) |
| O2—H2A···O3i | 0.82 (6) | 2.53 (6) | 3.160 (3) | 135 (5) |
| C11—H11···O1ii | 0.93 | 2.37 | 3.290 (3) | 173 (5) |
Symmetry codes: (i) x, y−1, z; (ii) x, y+1, z.
Funding Statement
This work was funded by Natural Science Foundation of Qinghai Province grant 2016-ZJ-908. National Natural Science Foundation of China grant grant 81573561.
References
- Bruker (2007). APEX2 and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Chang, J. M., Zhu, N. S. & Kasimu, R. N. (2001). Nat. Prod. Res. Dev, 13, 27–29.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Fun, H.-K., Chantrapromma, S., Salae, A. W., Razak, I. A. & Karalai, C. (2011). Acta Cryst. E67, o1032–o1033. [DOI] [PMC free article] [PubMed]
- Geiger, H. & de Groot-Pfleiderer, W. (1979). Phytochemistry, 18, 1709–1710.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Kupchan, S. M., Karim, A. & Marcks, C. (1968). J. Am. Chem. Soc. 90, 5923–5924. [DOI] [PubMed]
- Kusumoto, N., Ashitani, T., Murayama, T., Ogiyama, K. & Takahashi, K. (2010). J. Chem. Ecol. 36, 1381–1386. [DOI] [PubMed]
- Logan, K. J. & Thomas, B. A. (1985). New Phytol. 99, 571–585.
- Ogunwande, I. A., Olawore, N. O., Ogunmola, O. O., Walker, T. M., Schmidt, J. M. & Setzer, W. N. (2007). Pharm. Biol. 45, 106–110.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Stafford, H. A. & Lester, H. H. (1986). Am. J. Bot. 73, 1555–1562.
- Zaghloul, A. M., Gohar, A. A., Naiem, Z. A. A. M. & Bar, F. M. A. (2008). J. Biosci. 63, 355–360. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989017017935/qm2118sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017017935/qm2118Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989017017935/qm2118Isup3.cdx
Supporting information file. DOI: 10.1107/S2056989017017935/qm2118Isup4.cml
CCDC reference: 1551127
Additional supporting information: crystallographic information; 3D view; checkCIF report