Abstract
Studies of the repair pathways associated with DNA double strand breaks (DSBs) are numerous, and provide evidence for cell-cycle specific regulation of homologous recombination (HR) by the regulation of its associated proteins. Laser microirradiation is a well-established method to examine in vitro kinetics of repair and allows for live-imaging of DSB repair from the moment of induction. Here we apply this method to whole, live organisms, introducing an effective system to analyze exogenous, microirradiation-induced breaks in the Caenorhabditis elegans germline. Through this method we observed the sequential kinetics of the recruitment of ssDNA binding proteins RPA-1 and RAD-51 in vivo. We analyze these kinetics throughout different regions of the germline, and thus throughout a range of developmental stages of mitotic and meiotic nuclei. Our analysis demonstrates a largely conserved timing of recruitment of ssDNA binding proteins to DSBs throughout the germline, with a delay of RAD-51 recruitment at mid-pachytene nuclei. Microirradiated nuclei are viable and undergo a slow kinetics of resolution. We observe RPA-1 and RAD-51 colocalization for hours post-microirradiation throughout the germline, suggesting that there are mixed RPA-1/RAD-51 filaments. Finally, through live imaging analysis we observed RAD-51 foci movement with low frequency of coalescence.
INTRODUCTION
DNA double strand breaks (DSBs) are DNA lesions that when not repaired can lead to cell death or the loss of genetic information. Repair of DSBs occurs by either homology driven pathways or pathways that do not require sequence homology, such as non-homologous end joining (NHEJ) (1). The homology-dependent pathways include homologous recombination (HR), which branches into several sub-pathways, including double strand break repair (DSBR) and synthesis dependent strand annealing (SDSA) (Figure 1A). A common and early step in both these pathways is the resection of DSBs by endo- and exo-nucleases to form ssDNA used for a homology search. The success of this search is crucial for the repair of DSBs and is mediated by a large number of proteins. Key proteins that play a role in the early steps of the HR pathways are the ssDNA-binding proteins Rad51 and the Rpa protein complex. The Rpa complex (Rpa1, 2 and 3) binds DNA immediately after the ssDNA is formed (2). This binding protects the ssDNA from degradation and prevents the formation of secondary structures. These functions of Rpa are a prerequisite for the binding of Rad51, which then completely, or partially, displaces Rpa. This results in the formation of an ssDNA-Rad51 microfilament capable of a homology search. The invasion of the microfilament into the homologous sequence is followed either by DNA synthesis and strand rejection (via SDSA) or a double Holliday junction formation (via DSBR) (3). These pathways can lead to gene conversion that is (DSBR), or is not (DSBR and SDSA), accompanied by a crossover.
Figure 1.
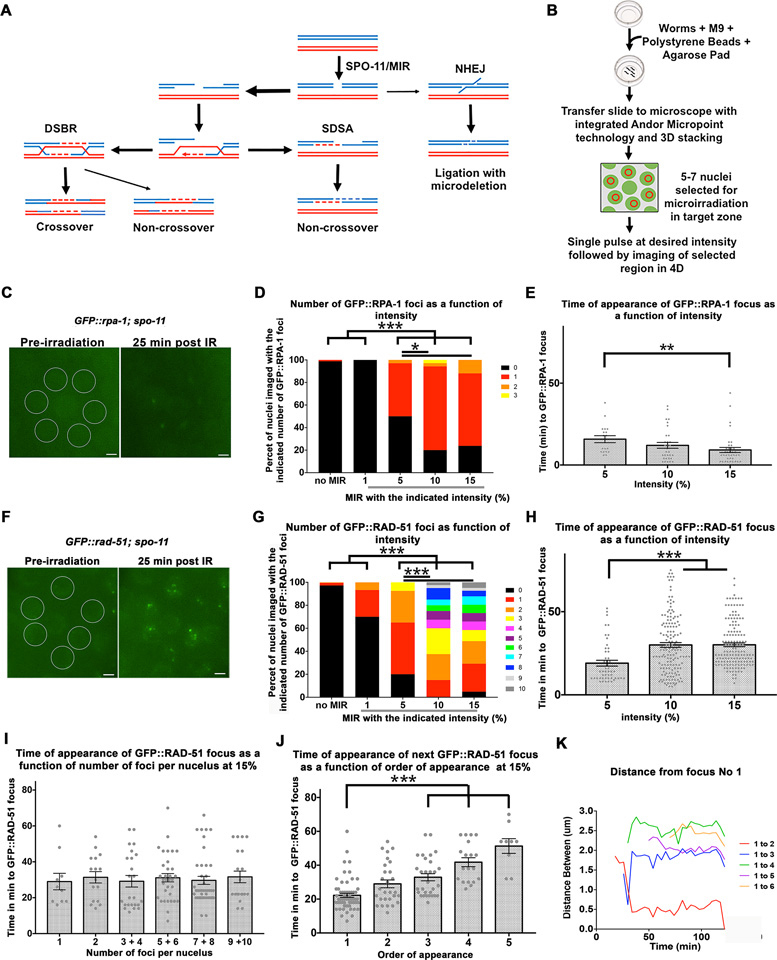
Laser microirradiation of C. elegans germline nuclei leads to GFP::RPA-1 and GFP::RAD-51 foci appearance. (A) Schematic representation of pathways for the repair of DSBs; meiotic DSBs are induced by SPO-11 (wild type cells) or MIR (in this manuscript) and then repaired by the double stand break repair pathway (DSB) or synthesis dependent strand annealing pathway (SDSA). Non homologous end joining can also repair DSBs, but it not a favored pathway in meiosis. (B) Schematic representation of microirradiation experimental design. (C) An example of nuclei selected for microirradiation prior to microirradiation (left image) and the same nuclei with visible GFP::RPA-1 foci 25 min post-microirradiation (right image). Scale bar, 2 μm. (D) Different laser pulse intensities (1–15%) were applied to MP nuclei and the number of GFP::RPA-1 foci per nucleus was counted for each intensity setting. n nuclei for 0 = 171, 1% = 30, 5% = 34, 10% = 35, 15% = 42. (E) The same foci from the experiment described in D were also quantified according to the time (in minutes) it took them to appear for each level of intensity. 1% intensity was intentionally left out given that this intensity did not produce any foci. Each data point represents a focus generated. n foci for 5% = 18, 10% = 31, 15% = 37. (F) An example of nuclei selected for microirradiation prior to microirradiation (left image) and the same nuclei with visible GFP::RAD-51 foci 25 min post-microirradiation (right image). Scale bar, 2 μm. (G) Different pulse intensities (1–15%) were applied to mid-pachytene nuclei and the number of GFP::RAD-51 foci per nucleus was counted for each intensity. n nuclei for 0 = 220, 1% = 30, 5% = 40, 10% = 40, 15% = 41. (H) The same foci from the experiment described in G were also quantified according to time of appearance (in minutes) for 5, 10, and 15%. 1% not included given that this intensity only produced foci in 2 nuclei. Each data point represents a focus generated. n foci for 5% = 49, 10% = 152, 15% = 149. (I) Time of appearance of foci in nuclei with a particular number of foci. Each data point represents a nucleus with a focus (or foci). n foci for nucleus with one focus = 10, two foci = 16, 3 and 4 foci = 24, 5 and 6 foci = 33, 7 and 8 foci = 37, 9 and 10 foci = 29. All values are insignificant from each other (Kruskal-Wallis test P = 0.42). (J) Time of appearance of the subsequent foci plotted as a function of the order of their appearance. Each data point represents a focus generated. n foci first focus = 62, second focus = 30, third focus = 28, fourth focus = 19, fifth focus = 9. For simplicity of representation p values only for the first focus comparisons are represented. (K) Movement of foci within one particular MP nucleus was quantified by measuring the distance (in μm) between each focus and the focus designated focus number one. Each data point indicates a measurement taken at that time point for the specific pairwise distance indicated by the graph legend. Data point in purple shows example of coalescing foci with focus #1. Bars in all graphs represent the mean ± SEM. * is for 0.01 < P < 0.05, ** is for 0.0001 < P < 0.01, *** is for P < 0.0001. For exact P-values, mean, SD and SEM values see Tables S1–S5 in Supplementary Material.
Mitotic DSBs are unintentional byproducts caused by replication errors or by the action of exogenous agents (1). Meiosis-specific DSBs are programmed and induced by the enzymatic activity of the topoisomerase-like protein Spo11 (3). Meiotic and mitotic DSBs can be processed by the HR pathway, but with one main difference: meiotic DSBs require the removal of Spo11 from DSBs by nucleases that also take part in resection (4). Studies of germline DSBR focus on Spo11-generated breaks, since they are the most prevalent form of DSBs in the germline. DSBR of other forms of DSBs in the germline have been investigated by ionizing radiation, such as gamma irradiation [e.g. (5)]. This tool was useful for identifying genes that play a role in DSBR, because mutants for HR pathway genes are sensitive to irradiation. However, due to the way gamma irradiation is delivered, this method cannot be used for recording early events of DSBR.
Laser microirradiation is a well-demonstrated method for the induction of DSBs, which allows the recording of DSB repair protein recruitment by live-imaging from the moment of DSB formation (6). Many methods and technologies are used for laser microirradiation. Some techniques require pre-sensitization of the cells by growth in BrdU. It was shown that a UV-spectrum laser can cause DSBs (7–9). Some UVA laser microirradiation systems were optimized to be used for the formation of DSBs and single strand breaks without the need for cell pre-sensitizing. High and medium laser intensities can induce DSBs, while low laser intensities (<4% of total power/∼3μJ) can induce single strand breaks, oxidation, or interstrand cross-links (10–12).
Laser microirradiation has the advantage of creating DSBs without the need to genetically engineer cells, as required by methods that rely on generating DSBs by site-specific endonucleases. Yet laser microirradiation is able to provide results similar to those obtained with other systems. Rpa1 and Rad51 are promptly recruited after DSB formation. Immunofluorescence analysis post-microirradiation revealed that Rpa1 is observed as early as 5–10 min post-microirradiation of human and yeast cells (13) and persists for 6 h after microirradiation (14,15). Faster kinetics of RPA recruitment were observed in yeast (16). Rad51 appears within 10–15 min post microirradiation and reaches saturation at 2–3 h in human and hamster cells lines (17,18). Rad51 foci repair kinetics are slow, as Rad51 foci can be seen even 24 h post-microirradiation of hamster cells lines (18). Despite the frequent use of laser microirradiation in single-cell systems this method has not been used for DSB formation in an intact multicellular organism.
The repair of DSBs is regulated throughout the cell cycle [review (19,20)]. Cells in G1 do not have sister chromatids available for HR, and therefore employ NHEJ as the main DSB repair pathway. S and G2 cells have a replicating or replicated genome and therefore can employ HR pathways. Many DNA repair proteins, including Rpa and Rad51, are the target of cell-cycle regulation (19). Some of the regulation is indirect, as the loading of RPA and RAD51 requires resection, and resection is a hub for cell cycle regulation (20). Others have described a more direct regulation: Rpa2 is phosphorylated by CDKs and its hyper-phosphorylation is critical for Rad51 recruitment and HR in response to DSBs created by replication stress (21,22). The oscillation in activity of Rpa and Rad51 throughout the cell cycle may also be an outcome of controlling their mediators. For example, Rad52, a protein required for the loading of Rad51, is recruited to the DSB in a cell-cycle regulated manner (23,24). Live imaging of laser microirradiated cells can also reveal cell-cycle regulation of DSBR proteins. As expected from their role in HR, Rad51 and Rpa1 recruitment to microirradiated DNA in hamster and human tissue culture cells is reduced in G1 compared to S/G2 (18,25).
Meiotic DSBs are committed to the HR pathway. However, in some situations NHEJ can repair meiotic DSBs, suggesting HR/NHEJ regulation is also imposed during the meiotic program (26–28). Meiotic prophase I is an extended phase of the meiotic program (ranging from hours to weeks depending on the organism), during which many events occur at the level of chromosome structure and dynamics (29). Meiotic prophase I cells regulate events in DSBR with meiotic progression in a manner analogous to mitotic cell-cycle regulation. In yeast meiotic prophase I cells both Rpa2 phosphorylation and phosphorylation of Rad51 mediators regulates crossover patterns (30–33).
In Caenorhabditis elegans meiotic and mitotic DSBR pathways have been well-documented, although much of the research into HR pathways is focused on the germline (34). This stems mainly from the ability to analyze meiotic events in a single germline in which nuclei are organized in a temporo-spatial pattern. Here, we use laser microirradiation to examine RPA-1 and RAD-51 recruitment to DSBs in the C. elegans germline. This represents the first method for timed induction of DSBs in living C. elegans throughout the germline. We show that this method is effective for DSB formation throughout the germline and can be used to record the recruitment of ssDNA binding proteins to the region of DNA damage. Through this analysis, we identify for the first time the kinetics of recruitment of RAD-51 and RPA-1 to DSBs in C. elegans in different regions of the germline. We show that these kinetics are largely constant throughout the germline, except in mid-prophase, which shows delayed RAD-51 recruitment.
MATERIALS AND METHODS
Worm strains and culture
Worms were grown and maintained on nematode growth media (NGM) plates seeded with Escherichia coli (OP50) and raised at 20°C. The following strains were used in experiments: N2 bristol, oxIs279 [pie-1p::GFP::H2B + unc-119(+)] II; spo-11 (ok79) IV / nT1 [qls51] (IV, V), spo-11 (ok79) IV / nT1 [qls51] (IV, V); gtIs2368 [pie-1p::GFP(lap)::rpa-1 + unc-119(+)], GFP::rad-51 spo-11(iow16) IV/nT1[qls 51] (IV, V), rad-51(ok2218) spo-11(iow14) IV/nT1[qls 51] (IV, V); gtIs2368 [pie-1p::GFP(lap)::rpa-1 + unc-119(+)]
oxIs279 [pie-1p::GFP::H2B + unc-119(+)] II; spo-11 (ok79) IV / nT1 [qls51] (IV, V) was created by crossing oxIs279 [pie-1p::GFP::H2B + unc-119(+)] and spo-11 (ok79) IV / nT1 [qls51] (IV, V).
rad-51(ok2218) spo-11(iow14) IV/nT1[qls 51] (IV, V); gtIs2368 [pie-1p::GFP(lap)::rpa-1 + unc-119(+)] was created first by making a spo-11 mutant (iow14, see CRISPR-Cas9) in a rad-51(ok2218)/nT1[qls 51] (IV, V) background, and then crossing that line to pie-1p::GFP(lap)::rpa-1.
spo-11 (ok79) IV / nT1 [qls51] (IV, V); gtIs2368 [pie-1p::GFP(lap)::rpa-1 + unc-119(+)]) was created by crossing gtIs2368 [pie-1p::GFP(lap)::rpa-1 + unc-119(+)]. ltIs37 [pie-1p::mCherry::his-58 + unc-119(+)] and spo-11 (ok79) IV / nT1 [qls51] (IV, V).
The pie-1p::GFP(lap)::rpa-1 of spo-11 (ok79) IV / nT1 [qls51] (IV, V); gtIs2368 [pie-1p::GFP(lap)::rpa-1 + unc-119(+)] was occasionally silenced, to induce de-silencing the strain was grown for 1–2 generations at 25°C and then grown for a generation at 20°C before analysis.
smIs34 [ced-1p::ced-1::GFP + rol-6(su1006)]; GFP::rad-51 spo-11(iow16) IV/nT1[qls 51] (IV, V).
meIs8 [pie-1p::GFP::cosa-1 + unc-119(+)] II; spo-11 (ok79) IV / nT1 [qls51] (IV, V) ojIs9 [zyg-12(all)::GFP + unc-119(+)].
CRISPR-Cas9
GFP::rad-51 IV/nT1[qls 51] (IV, V) was created with CRISPR-Cas9 to add a GFP tag to rad-51 using pDD282 as in (35). We decided to create an N- terminal tagged RAD-51, as N- terminal tagged RAD-51 were known to be more functional compared to C-terminal tags in other species (see supplementary Table S2 of (36)). GFP::RAD-51 does not complement a null RAD-51, but also does not exert a dominant negative effect (GFP::RAD-51/+ are fertile). Heterozygotes or homozygotes for GFP::RAD-51 show no defects in pairing or synapsis (Supplementary Figure S1A and B). GFP::RAD-51 accumulate in similar kinetics as GFP::RAD-51/+ and GFP::RAD-51 homozygotes, but persist in GFP::RAD-51 up to diakinesis (Supplementary Figure S1C). Six bivalents are observed in GFP::RAD-51/+ mutants, as found in wild type, but GFP::RAD-51 homozygotes show misshapen bivalents that frequently associate with each other, indicative of DSB repair defects (Supplementary Figure S1D). GFP::RAD-51 foci colocalize with untagged RAD-51 foci (anti-RAD-51 antibody). spo-11 is tightly linked to rad-51, therefore a new spo-11 partial 1034bp deletion mutation (iow16) was created in the GFP::rad-51 IV/nT1[qls 51] (IV, V) strain using a single crRNA that begins just before the N terminal coding region and selected using dyp-10 coconversion (37) to generate GFP::rad-51 spo-11(iow16) IV/nT1[qls 51] (IV, V). GFP::rad-51 spo-11(iow16) shows significantly reduced foci numbers compared to GFP::rad-51 as well as showing 12 DAPI-stained bodies in diakinesis. Some GFP::RAD-51 foci are found in the absence of spo-11, mainly in PMT. GFP::rad-51 spo-11(iow16) have 12 DAPI bodies in diakinesis, indicating that crossovers cannot be formed in this mutant.
The spo-11 mutation (iow14) in rad-51(ok2218) spo-11(iow14) IV/nT1[qls 51] (IV, V); gtIs2368 [pie-1p::GFP(lap)::rpa-1 + unc-119(+)] was made using the same techniques as above creating an insertion in this case. Loss of spo-11 function was assessed by DAPI body counts with all gonads showing 12 DAPI bodies indicating lack of crossovers.
Imaging
Live worms were imaged using MetaMorph Version 7.8.12.0 with 100×/1.4 NA oil Leica illuminated with 110LED. Intensity of light source was set to 5% and exposure times were 500 ms for the GFP channel, and 20 ms for D&C. Nuclei were located based on fluorescence of either GFP tagged Histone H2B, gtIs2368 [pie-1p::GFP(lap)::rpa-1 + unc-119(+)], or (SSM264 GFP::rad-51) spo-11 /nT1 [qls51] (IV, V). Row position in the PMT was determined by counting from the distal tip, and TZ was indicated by both crescent shaped nuclei, disruption in the organization of nuclei, and the beginning of the hollow lumen center of the gonad as PMT has nuclei throughout the center while pachytene nuclei are arranged on the surface. Mid-pachytene was determined to be midway between the start of transition zone and the start of Diplotene, and late pachytene were nuclei immediately preceding diplotene (all LP experiments used the last seven rows in pachytene).
Time series projects of microirradiated live worms were created using Metamorph Multidimensional Acquisition. Images were aquired at 2 min periods, imaging a 9 μm range centered on the z-plane coordinate of microirradiation at 1 μm intervals resulting in 10 image stacks per time point. The program was initiated and the time course was paused after the first stack of each target region was imaged. The stage was moved to each target region and microirradiated nucleus, this took approximately 10 seconds per gonad; then the program was resumed resulting in ≤2 min from microirradiation to the first post-irradiation image. Position of the center Z stack was sometimes adjusted during recording so that affected nuclei stayed in the image region (and did not move out of imaging focus) if the gonad shifted during recording. This could be done between images taken so as not to change the time points. The program ran for 46 time points.
Imaged stacks from live imaging were processed in MetaMorph to create a maximum projection for each time point. Nuclei were scored for foci in FIJI-Image J. Foci were counted in microirradiated nuclei and all nuclei immediately adjacent were scored as negative controls. The size of foci were also recorded as some foci in microirradiated nuclei were abnormally large. Foci that exceeded a diameter of 0.6 μm at the thickest point were recorded as a focus cluster. The likely death of the worm during live imaging was assumed at the time points when foci stopped moving within nuclei and auto-fluorescence of intestines diminished as well as small movements in the gonad completely ceased (while the worms were completely immobile, gonads would often pulse slightly internally), at which time nuclei were no longer scored. All strains had some loss (or bleaching) of fluorescence progressively during live imaging as live worms were exposed to more excitation light. This was especially true for the relatively weaker fluorescence of pie-1p::GFP(lap)::rpa-1 strains. Nuclei were not scored when fluorescence diminished to the point when foci count could no longer be assessed in a gonad.
As well as having some background foci, GFP::rad-51 spo-11(iow16) IV/nT1[qls 51] (IV, V) on rare occasion would have a bit of background fluorescence during live imaging that could not be distinguished as foci, but appeared as light heterogeneous coloration in the nuclei that increased in contrast during the experiments when background fluorescence diminished. Heterogeneous coloration was not counted and only clear, relatively bright foci were scored.
Each live imaging nucleus was scored for number of foci at every time point with the total foci appearance calculated by number of foci minus foci before microirradiation (i.e., new foci). In most cases there were zero foci present before microirradiation but in GFP::rad-51 spo-11(iow16) IV/nT1[qls 51] (IV, V), nuclei with foci were sometimes unavoidable, especially in the PMT and TZ.
Fixed slides were imaged using the DeltaVision wide-field fluorescence microscope system (Applied Precision) with 100×/1.4 NA oil Olympus and images were deconvolved with softWoRx software (Applied Precision). Length of gonad and location of foci were recorded using cell width counts from the distal tip cell. Images were analyzed using softWoRx Explorer 1.3.0 software (Applied Precision). Foci were distinguished as a focus cluster if their diameter exceeded 0.6 μm.
Microirradiation
Nuclei in the appropriate region of the germline were located and the Z-plane was focused on the center of the target nuclei before microirradiation. Target nuclei were selected at the center of each nucleus and microirradiated using a single pulse with Andor micropoint 365nm pulsed laser, and attenuated in MetaMorph to specified percentage of the total 80 μJ output. Live worms were microirradiated and recorded targeting the mid-pachytene nuclei at one pulse per nucleus for a total of 5–7 nuclei per gonad as imaging allowed. The manually driven optical attenuator was at max permissiveness and the motorized optical attenuator was adjusted and displayed as a percentage of full power. For RAD-51 and RPA-1 live imaging in Figures 1–3 live worms were microirradiated and recorded as in the indicated intensity one pulse per nucleus and foci counts for each nucleus at every time point was recorded. The data is presented in Figures 1–3 contains the first 85 min of recording. For the experiments in Figure 4 and Tables 1–3 live worms were microirradiated at 15% pulse intensity at one pulse per nucleus at the targeted gonad region in two adjacent rows. Multiple targeted worms were recovered and each experimental group (each row in the tables) contained data collected from at least four germlines. An example for the variability between worms is seen in Supplementary Figure S2. The amount of nuclei targeted was set per gonad region to allow for the number of nuclei in a row and are as follows: PMT (row 5) 8 nuclei, TZ 10 nuclei, MP 8 nuclei, LP 6 nuclei. Worms were recovered immediately after microirradiation and dissected at the appropriate time intervals.
Figure 3.
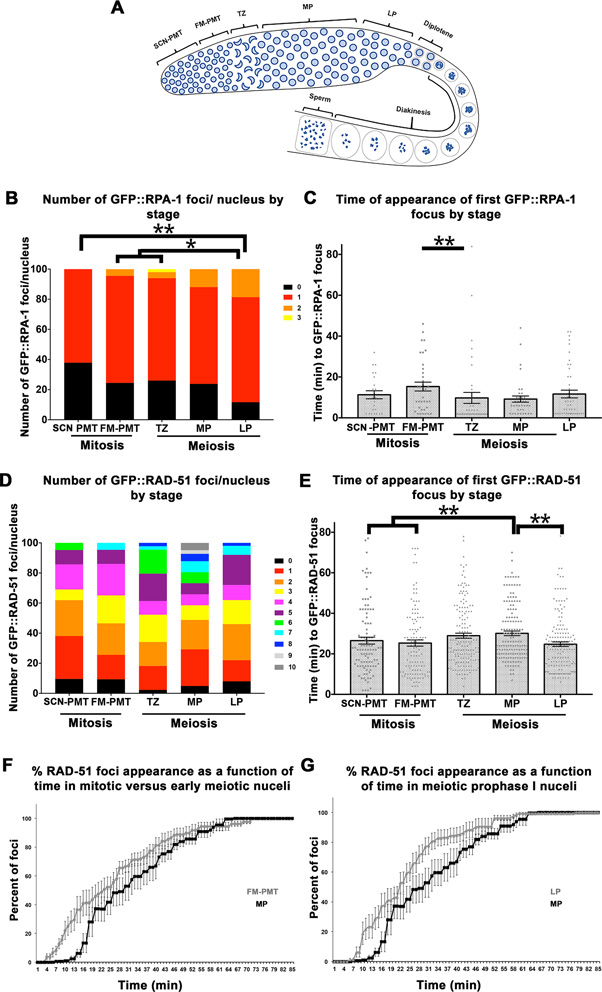
Differences in number and time of appearance of GFP::RPA-1 and GFP::RAD-51 foci between mitotic and meiotic cells. (A) Cartoon representation of the stages of meiosis in the C. elegans germline. Nuclei in mitosis (FM-PMT [row 5], SCN-PMT[rows 9–11]) and meiosis (TZ, MP, LP) were microirradiated and compared in terms of number of foci per nucleus (B) and time of appearance (C) in the GFP::RPA-1, spo-11. (B) n nuclei SCN-PMT = 34, FM-PMT = 45, TZ = 50, MP = 42, LP = 43. (C) Each data point in (C) represents a focus, when bars are the mean ± SEM. Only P < 0.01 are shown. n foci SCN-PMT = 23, FM-PMT = 36, TZ = 41, MP = 37, LP = 46.The same experiment described in B and C was applied to the GFP::RAD-51, spo-11. (D) n nuclei SCN-PMT = 42, FM-PMT = 43, TZ = 44, MP = 41, LP = 50. (E) Each data point in (E) represents a focus, when bars are the mean ± SEM. Only P < 0.01 are shown. n foci SCN-PMT = 100, FM-PMT = 118, TZ = 158, MP = 149, LP = 155. (F and G) The percent of GFP::RAD-51 foci appearance was plotted as a function of time for mitotic (FM-PMT) versus MP nuclei (F) and for meiotic prophase I nuclei MP and LP (G). Meiotic prophase I MP and LP. Mean of % foci at each time point in each gonad ± SEM. * is for 0.01 < P < 0.05, ** is for 0.0001 < P < 0.01. For exact P-values, mean, SD and SEM values see Supplementary Tables S8–S10 in Supplementary figures.
Figure 4.
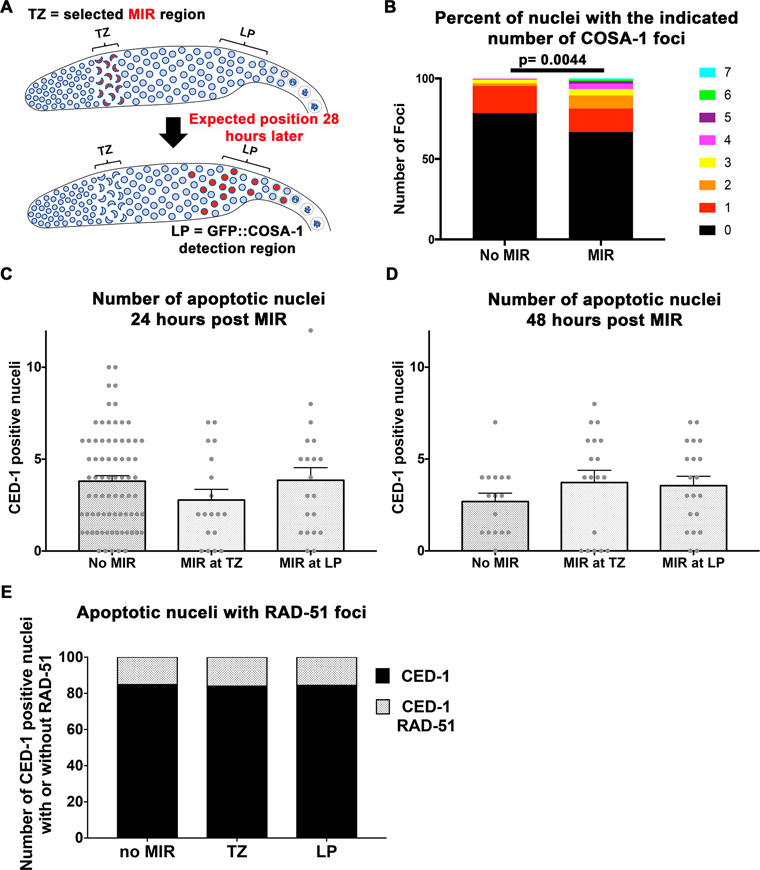
The effect of microirradiation on crossover formation and apoptosis. (A) Cartoon representation of the design of the GFP::COSA-1 experiment. (B) COSA-1 foci numbers with and without irradiation n = 130, 125 respectively. (C) The number of CED-1 positive cells a function of microirradiation in TZ or LP 24 h post treatment. No significant values are observed. n gonads no MIR = 76, TZ MIR = 18, LP MIR = 20. (D) same as (A), but at 48 h treatment. No significant values are observed. n gonads no MIR = 16, TZ MIR = 18, LP MIR = 20. (E) Fraction of RAD-51 positive nuclei among nuclei with CED-1. No significant values are observed. n gonads no MIR = 284, TZ MIR = 50, LP MIR = 77.
Table 1. GFP::RPA-1, spo-11.
| Region | Hours post- microirradiation | Nuclei with RPA-1 foci clusters | Total number of RPA-1 foci clusters | Total numbers of microirradiated nuclei | Fraction of nuclei with foci clusters/microirradiated nuclei | RPA-1 foci clusters/nucleus with a cluster |
|---|---|---|---|---|---|---|
| PMT | 2 | 33 | 39 | 48 | 68.8 | 1.2 |
| PMT | 24 | 19 | 33 | 40 | 47.5 | 1.7 |
| PMT | 48 | 16 | 19 | 112 | 14.3 | 1.2 |
| TZ | 2 | 20 | 27 | 50 | 40.0 | 1.4 |
| TZ | 24 | 25 | 47 | 40 | 62.5 | 1.9 |
| TZ | 48 | 0 | 0 | 40 | NA | NA |
| MP | 2 | 21 | 26 | 40 | 52.5 | 1.2 |
| MP | 24 | 18 | 27 | 56 | 32.1 | 1.5 |
| MP | 48 | 0 | 0 | 32 | NA | NA |
| LP | 2 | 40 | 45 | 60 | 66.7 | 1.1 |
| LP | 24 | 0 | 0 | 24 | 0.0 | NA |
| LP | 48 | 0 | 0 | 24 | 0.0 | NA |
Number of nuclei with foci, total number of foci, and total number of microirradiated nuclei per gonadal region for worms that were microirradiated and then dissected at three different time points post-microirradiation. Immunofluorescence was performed with an anti-GFP antibody. ‘Fraction of nuclei with Foci clusters/microirradiated nuclei’ indicates how many nuclei showed foci out of the total number of nuclei that were microirradiated. ‘RPA-1 foci clusters/nucleus with a cluster’ indicates the average number of foci clusters per nucleus.
Table 3. Colocalization of RPA-1 and RAD-51 clusters.
| Numbers of foci | % of RPA-1 clusters | % of RAD-51 clusters | ||||||
|---|---|---|---|---|---|---|---|---|
| Region | Hours post-microirradiation | Mostly RPA-1 | Mostly RAD-51 | Colocalized | Mostly RPA-1 | With RAD-51 | Mostly RAD-51 | With RPA-1 |
| PMT | 2 | 6 | 13 | 33 | 15 | 85 | 28 | 72 |
| 24 | 2 | 20 | 31 | 6 | 94 | 39 | 61 | |
| 48 | 2 | 7 | 17 | 11 | 89 | 29 | 71 | |
| TZ | 2 | 15 | 12 | 12 | 56 | 44 | 50 | 50 |
| 24 | 5 | 3 | 42 | 11 | 89 | 7 | 93 | |
| MP | 2 | 16 | 16 | 10 | 62 | 38 | 62 | 38 |
| 24 | 6 | 11 | 21 | 22 | 78 | 34 | 66 | |
| LP | 2 | 2 | 11 | 43 | 4 | 96 | 20 | 80 |
Data indicating the hours post-microirradiation for 4 different stages, and subsequent measurements of RPA-1, RAD-51, or both colocalized foci in dissected, fixed, and stained worms. Immunofluorescence was performed by the combined use of both an anti-GFP antibody and an anti-RAD-51 antibody. The data is further divided into the percentage of RPA-1 clusters that were either present mostly alone or with RAD-51 cluster, as well as RAD-51 cluster that were present mostly alone or with RPA-1 cluster.
Live image slides
Worms were positioned for live imaging and microirradiation using both 1’x3’x.04’ slides with the worms on top of a 3% agarose pad made with M9, or using a live imaging petri dish (35 mm petri dish with 14 mm Microwell No. 1.5 coverglass MatTek corporation). The worms were placed in M9 and Polybead 0.1-μm polystyrene beads (#00876; Polysciences) and a coverslip was placed on top, or when using an imaging petri dish, a thin circle of 10% agarose made with M9 was placed on top of the worms in M9 and polystyrene beads. Parafilm was used to seal the petri dish and lid. To recover worms after microirradiation, the entire slide was submerged in M9 and the coverslip gently pried off, then M9 was emptied in to a petri dish and worms were recovered with a glass pipet. To recover worms from the imaging petri dishes, M9 was added to the petri dish and the agarose pad was gently lifted to release worms before recovery with a glass pipet. When recovered, microirradiated worms were placed on NGM plates seeded with OP50 until time of dissection. A minimum of five worms were analyzed for each experimental group ( = each bar).
Immunofluorescent staining
Worms were dissected to extract their gonads which were fixed and stained as in (38) and are as follows: RAD-51 (Rabbit to RAD-51 1:10, 000; ModEncod), GFP (Chicken to GFP 1:500; Abcam), HIM-8 (Rabbit to HIM-8 1:1, 000 Novus) SYP-1 (Goat to SYP-1 1:500). Secondary antibodies used were α-rabbit Fluor 488 (1:500; Alexa), α-rabbit TRITC (1:200; Jackson Immuno Research 711-025-152), α-chicken Fluor 488 (Donkey to Chicken 1:500; Molecular Probes), α-chicken 488 (1:500; Jackson Immuno Research 103-545-155). All gonads were stained for 10 min with 1:2,000 dilution of 5-mg/ml DAPI in PBS-Tween.
Distance measurements
FIJI-ImageJ straight line measurement tool was used to measure distances in μm between foci within a mid-pachytene nucleus. The first focus that appeared was designated the primary focus and all measurements of subsequent foci were to this first focus. Each focus was numbered in order of their appearance (two appeared second, three appeared third etc.). Measurements began as soon as more than one focus was present in the nucleus and continued every four minutes for the duration of the experiment.
Analysis of fixed samples
RAD-51 formed large foci, which we defined as clusters, since they contained what appeared to be multiple RAD-51 foci (both by shape and overall size). The diameter of a RAD-51 cluster was 1.0 ± 0.3 μm, which is more than double the size of a meiotic focus (0.4 ± 0.1 μm). We were not able to individually count foci within this cluster in a reliable manner, as we did with our live-imaging data, and therefore the fixed sample analysis refers to foci clusters as opposed to individual foci as a mark of laser microirradiation. A control experiment in which non-microirradiated worms were subjected to the same regimen lead to gonads without any RAD-51 or RPA clusters (also see material and methods). We did see some small RAD-51 foci in our negative control and therefore small foci were not counted as positive in our microirradiation fixed sample experiments. Due to this background we believe that we are under estimating the number of foci in fixed samples, compared to live-imaging. In agreement, our dissected samples had about half of the expected numbers of RAD-51 positive nuclei observed, while we had higher targeting rates when the readout was by live-imaging. However, by this methodology we can be confidant that each RPA-1 or RAD-51 cluster in our dissected germlines was a result of laser microirradiation.
RNA interference
Frozen RNAi cultures were streaked on LB plates containing ampicillin and tetracycline, grown overnight and cultured the following morning in ampicillin LB for 6 h, then grown overnight in the dark at room temperature on IPTG plates. Worms for rpa-1(RNAi) and rad-51(RNAi) were bleached and embryos placed on OP-50 NGM plates and allowed to grow to L4s before being placed on prepared RNAi plates for 24 h, and microirradiated. rpa-1(RNAi) resulted in small gonads as an indication for successful RNAi.
Statistics
Kruskal-Wallis test was performed to determine if any of the groups are significantly different than the others. If determined significant, then pair-wise comparisons were made using Mann–Whitney U test to identify the significant pairs, which is presented in the supplementary data or in the text. Statistical analysis was performed using Prism 7 GraphPad software, which was also used for the graphic representation of the data in the figures.
Apoptosis assay
Live imaging slides were made and microirradiation was performed as described above, except the strain CED-1::GFP; GFP::RAD-51; spo-11 was used. The bend region of each gonadal arm was imaged 24 h, and 48 h post-microirradiation, with worms being recovered from the live imaging slides after each imaging session. Controls went through the same treatment regarding plating and recovery, but were not subjected to microirradiation, only imaging of the bend region 24 h, and 48 h later. The number of CED-1::GFP engulfed nuclei in the bend region were counted at each time point for all experimental conditions.
COSA-1 assay
Worms homozygous for GFP::COSA-1, GFP::ZYG-12 and spo-11 deletion were subjected to irradiation as indicated above, except all TZ nuclei were targeted. GFP::RPA -1 or GFP::RAD-51 could not be used for this experiment since both, as GFP::COSA-1, localize as foci following microirradiation. ZYG-12 assisted by DIC was used to identify TZ nuclei (nuclear patch appearance). Worms were recovered and dissected ∼28 h post-microirradiation. The late pachytene region of each germline was scored, and the number of GFP::COSA-1 foci in each nucleus was counted. Controls were worms of the same genotype and age that were not subjected to microirradiation.
RESULTS
A system for the analysis of RPA-1 and RAD-51 recruitment to DSBs generated by laser microirradiation in C. elegans
We hypothesized that the recruitment of DSBR proteins to DSBs would be regulated throughout the germline, which contains both mitotic and meiotic prophase I nuclei. To test this hypothesis, we developed a system in which recruitment of ssDNA binding proteins involved in DSBR can be followed by live-imaging of germline nuclei. We adopted the well-established laser microirradiation techniques used for tissue culture studies to our in vivo system (UV range laser using the Andor MicroPoint system, see Materials and Methods). Using this methodology, we generated DSBs in different regions of the germline and assessed if DSB numbers and timing of recruitment to the DSBs were similar in each region analyzed. For the experiments described here we immobilized C. elegans young adults under agarose pads. In these conditions, DSBs were created in a system in which protein recruitment to the breaks could be followed in vivo in the context of a whole, intact, living organism. The microirradiated nuclei were followed by live-imaging (Supplementary Figures S2 and S3, and Figures 1–3) until the death of the organism (1.5–4 h into imaging). Alternatively, the organism was moved to petri dishes immediately after microirradiation for variable amounts of times before analysis by immunofluorescence experiments (Figures 4 and 5, Supplementary Figures S1 and S4, and Tables 1–3). In these studies only laser induced DSBs were studied; all microirradiation experiments were performed in a genetic background (spo-11 null) with no meiotic DSBs and the analysis in live worms was done in strains that contain either RPA-1 [(39)] or RAD-51 fused to GFP [this study]. All P values reported below for live imaging experiments are for Mann-Whitney U test, and for fixed samples Fisher's exact test (more detail in Materials and Methods). An outline of the experimental design is presented in Figure 1B.
Figure 5.
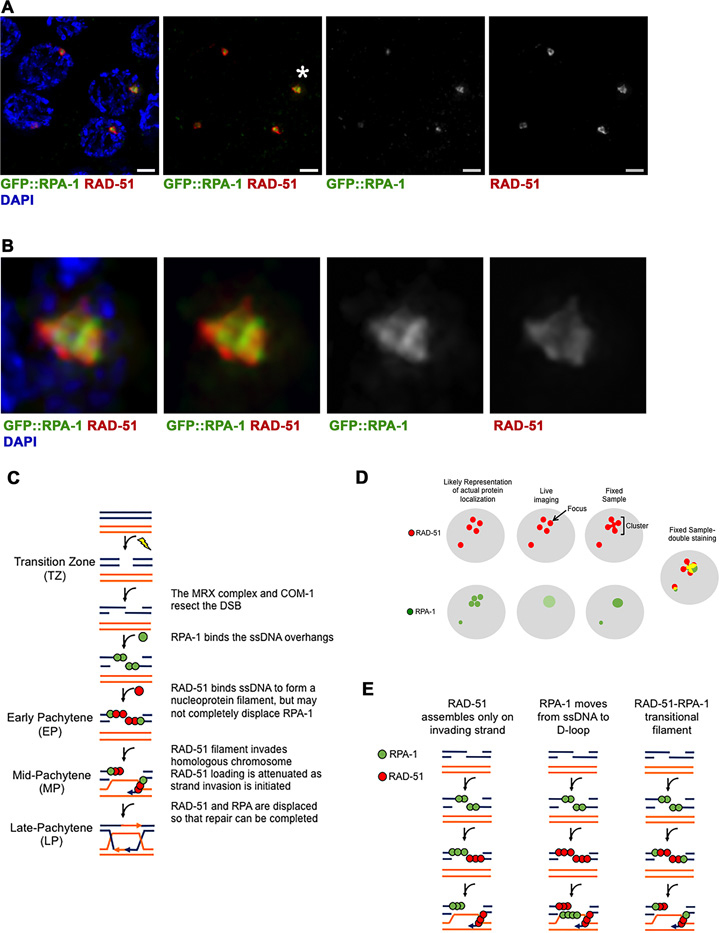
RPA-1 and RAD-51 colocalize on laser microirradiation DSBs. (A) GFP::RPA-1 strain was microirradiated and stained with both anti-GFP(green), anti-RAD-51(red) antibody and DAPI (blue) to visualize chromosomes. Four representative MP nuclei are shown. Three of the nuclei show colocalization between RPA-1 and RAD-51. The fourth nucleus (bottom left) shows RAD-51 cluster with very faint RPA-1 (defined as ‘mostly RAD-51’ in Table 3). Scale bar 2 μm. (B) Zoomed in images of the foci indicated by the asterisks in A. (C) Model depicting the process of DSB repair as it occurs in the different stages of the C. elegans germline. The model is canonical until Early Pachytene, where RAD-51 may not completely displace RPA-1 on the ssDNA overhang. RAD-51 may also be attenuated in Mid-Pachytene by the initiation of strand invasion. (D) Model depicting how individual RAD-51 and RPA-1 foci may appear as clusters upon fixed sample staining, with comparison to how similar foci appear in live imaging. The live imaging representation for RPA-1 is a lighter green, meant to demonstrate the weak signal of the GFP::RPA-1 strain. (E) Model depicting the three most likely explanations for the appearance of a mixed RPA-RAD-51 filament. The first two columns illustrate the possibility of RPA-1 and RAD-51 occupying different strands around the DSB at the same time, while the third column depicts how a mixed filament may look. RPA-1 is depicted in green, RAD-51 in red, for all sections.
RPA-1 is recruited to foci in microirradiated nuclei of the germline
RPA is an ssDNA binding protein that is the first to processively bind resected DSBs and was shown to localize to SPO-11 generated DSBs in the germline (27). As a first test of our system, we generated DSBs using variable amounts of laser power in GFP::rpa-1; spo-11 germline nuclei [a strain expressing RPA-1 fused to GFP and containing a deletion for spo-11]. We first calibrated the intensity by microirradiating nuclei at mid-pachytene (MP) with various amounts of energy (for n values and statistics, see Supplementary Table S1). High intensities frequently led to nuclear ablation and mechanical damage to the worm, but at lower intensities we observed DSBs forming, as evident by the generation of GFP::RPA-1 foci only in nuclei exposed to microirradiation (Figure 1C and Supplementary Movie S1). No GFP::RPA-1 foci appeared at non-microirradiated nuclei adjacent to the microirradiated nuclei (n nuclei = 171). This indicates that microirradiation is able to target nuclei autonomously, without affecting other nuclei in the germline syncytium. At a laser intensity of 1% (0.8 μJ) no foci were observed. The numbers of RPA-1 foci at intensity levels of 5% (4 μJ), 10% (8 μJ) and 15% (12 μJ) ranged from 0.5 to 0.9 foci/nucleus and were statistically different from no microirradiation (P < 0.0001) and 1% intensity (P < 0.05) (Figure 1D and Supplementary Table S1). While most nuclei had one focus (% nuclei with one focus: 5% = 47, 10% = 74 and 15% = 64), we also observed some nuclei with zero or two foci. The mean time of appearance of an RPA-1 focus was similar between the three intensity levels with most foci (5% = 71, 10% = 86 and 15% = 100) appearing in the first 20 min following microirradiation (Figure 1E and Supplementary Table S2).
RAD-51 is recruited to DSBs following RPA-1
We next assayed RAD-51 recruitment under the same conditions, using a spo-11; GFP::rad-51 strain. GFP::RAD-51 can not complement a null rad-51 mutant, but exhibits no early meiotic defects (see Materials and Methods and Supplementary Figure S1). GFP::RAD-51 foci appeared following microirradiation at all intensities tested (Figure 1G–H, Supplementary Movie S2; for n values and statistics, see Supplementary Table S3). However, at 1% only 9 (30%) of the microirradiated nuclei had foci, while at higher intensities 80–100% of the nuclei had foci (Figure 1G and Supplementary Table S3). There was no significant difference between 10% and 15% intensity in terms of mean number of RAD-51 foci, but other pairwise comparisons were significant (Supplementary Table S3). The large majority of the foci can be attributed to microirradiation, as adjacent non-microirradiated nuclei rarely showed new foci [15% 3.6±2.8 foci/nucleus (n = 41) versus non-microirradiated 0.01 ± 0.1 foci/nucleus (n = 87), P<0.0001]. We also verified that the bottom layer of germline nuclei is not affected by microirradiation (Supplementary Movie S3). The mean number of GFP::RAD-51 foci was three to five times higher than RPA-1 foci (Figure 1D and G 1.2 ± 0.9, 3.8 ± 2.5 and 3.6 ± 2.8 foci/nucleus in 5, 10 and 15% respectively). This may stem from under-detection of RPA-1 foci (see Discussion).
The current model of DSBR proposes that RAD-51 recruitment requires prior binding of RPA to the ssDNA. In agreement with this model, the mean time of RAD-51 foci formation was almost double that of RPA-1 [e.g. for 15%: 30.1 min (n = 149) versus 9.2 min (n = 37), P < 0.0001]. The mean time of appearance of RAD-51 focus in 10% and 15% intensities was slower when compared to the 5% intensities; however, in all conditions most foci appeared in the first 50 min following microirradiation (5% = 98, 10% = 87 and 15% = 89 and Figure 1H and Supplementary Table S4). Since there were overall more RAD-51 foci than RPA-1 foci in our experiments, we tested whether the apparent delay in RAD-51 foci appearance was biased by the late foci appearance. This was not the case, as the mean time of appearance of the first RAD-51 focus was larger compared to that of the first RPA-1 focus [Figure 1E compared to 1H, 22.8 min (n = 39) versus 6.3 min (n = 32), P < 0.0001]. Likewise, in nuclei with only one focus, the mean time of appearance of this focus was greater for RAD-51 compared to RPA-1 [29 min (n = 10) versus 6.4 min (n = 27), P < 0.0001]. There was no difference in the mean time of recruitment of RAD-51 foci between nuclei with different numbers of foci (Figure 1I). This indicates that RAD-51 foci appearance follows RPA recruitment, regardless of the number of foci in a microirradiated nucleus.
RAD-51 foci move and infrequently coalesce
Having multiple RAD-51 foci formed following microirradiation permitted the examination of the timing of recruitment as a function of the order of recruitment. We performed this analysis for the 15% intensity data. While the first focus is recruited in 23 min following microirradiation, the subsequent foci are recruited faster (Figure 1J and Supplementary Table S5).
We also analyzed the distance between GFP::RAD-51 foci over time in PMT, TZ and MP microirradiated nuclei. We found that foci change relative distance, indicating movement in the nucleus, but, for the most, they do not coalesce (% of two foci merging to one: FM-PMT 11.5%, n = 26, TZ 5.3%, n = 19, MP 12%, n = 25). Foci that initiated close to each other, remained in close proximity (Figure 1K and more examples in Supplementary Figure S3C). This observation was found regardless of the gonadal region tested. This may indicate that RAD-51 foci can move towards each other, but this is not a frequent phenomenon in the time frame examined.
RAD-51 is recruited to microirradiated foci in an RPA-1-dependent manner
RPA-1 binds resected ssDNA, which allows for the recruitment of RAD-51. If indeed the foci we observed are indicative of binding of RPA-1 and RAD-51 to ssDNA, it is expected that RAD-51 foci will be dependent on the presence of RPA-1, but not vice versa. We tested the genetic requirement for RPA-1 and RAD-51 recruitment by microirradiating nuclei depleted by RNAi of rad-51 or rpa-1, respectively (see Material and Methods). All microirradiation experiments below were performed with 15% intensity (for n values and statistics, see Supplementary Tables S6 and S7). We chose this intensity to avoid nicking as opposed to breaking which has been shown at lower intensities in other systems (and since 10% and 15% intensities were statistically indistinguishable for most parameters).
We analyzed the RPA-1 foci appearance in the rad-51 null mutant. Data obtained from spo-11; GFP::rpa-1; rad-51 was not different from that of spo-11; GFP::rpa-1 mutants (Figure 2A and B), both for number (P = 0.1) and time (P = 0.6) of appearance of foci. Similar results were obtained using rad-51(RNAi) (Figure 2C and D). These data indicate that RPA-1 forms foci independently of RAD-51. We then tested for the opposite relationship with RNAi for rpa-1 in the spo-11; GFP::rad-51 background. Feeding of spo-11; GFP::rad-51 with bacteria with empty vector control (pL4440) lead to similar results (P = 0.73 MW) to those observed above with no RNAi bacteria: 3.4 ± 1.9 GFP::RAD-51 foci appeared following microirradiation. However spo-11; GFP::rad-51 rpa-1(RNAi) germlines showed only 0.7 ± 1.3 foci/nucleus (P < 0.0001), indicating that RAD-51 recruitment is RPA dependent, as expected if the foci observed are indicative of RAD-51 bound to ssDNA (Figure 2E and F).
Figure 2.
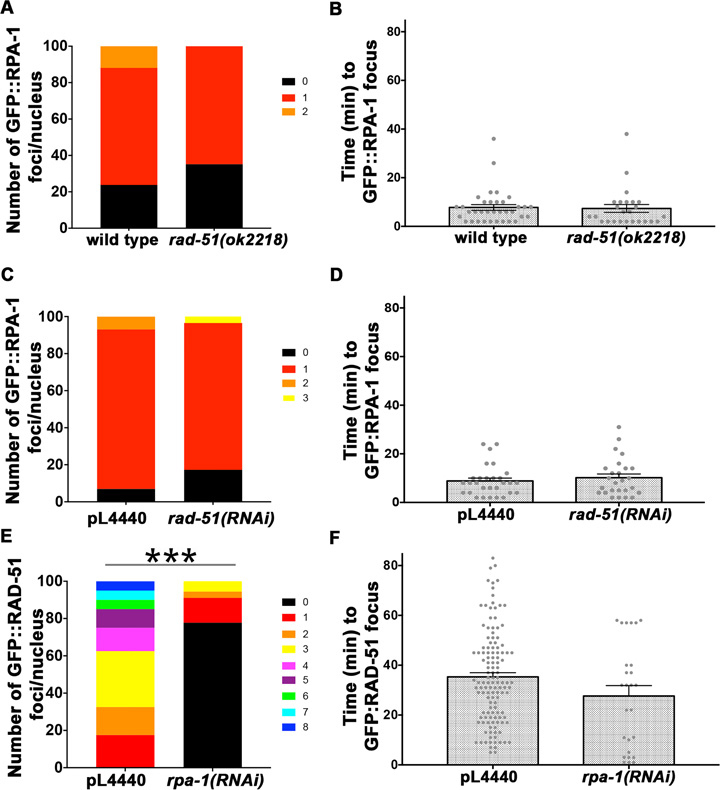
GFP::RAD-51 loading depends on RPA-1. Number of foci per nucleus (A, C, E) and time of appearance (B, D, F) for GFP::RPA-1 (A–D) or GFP::RAD-51 (E–F) in both a wild type and a rad-51 homozygous sterile deletion (ok2218) strain (A–B) or pL4440 and rad-51(RNAi) (C–D) or rpa-1(RNAi) (E–F). Each data point in (B, D, F) represents a focus generated. Bars represent the mean ± SEM. n nuclei and foci for wild type = 42, 35, and rad-51mutants = 37, 25, n nuclei and foci for pL4440 = 29, 29 and rad-51(RNAi) mutants = 29, 26, n nuclei and foci for pL4440 = 40, 128 and rpa-1(RNAi) mutants = 30, 26. * is for *** is for P < 0.0001. For exact P-values, mean, SD and SEM values see Supplementary Tables S6 and S7 in Supplementary figures.
RPA-1 is recruited in a similar manner in different regions of the germline
To test if RPA-1 recruitment is affected by the stage in which the nuclei are found, we microirradiated nuclei in five different regions of spo-11; GFP::rpa-1 germlines. C. elegans germline nuclei are arranged in rows in which their relative position reflects their mitotic or meiotic stage (Figure 3A). The distal region of the germline, the pre-meiotic tip (PMT), is composed of mitotically proliferating nuclei. Nuclei move from the distal tip into the transition zone, where they enter meiosis [∼row 16–20, this study, (40)]. The transition zone (TZ) contains nuclei in the leptotene-zygotene stage, where nuclei display clustered chromosome morphology. This clustered chromosomal organization can also be seen by the diffuse nuclear signal of GFP::RAD-51 or GFP::RPA-1. Nuclei exit leptotene-zygotene and enter pachytene when chromosomes disperse throughout the nuclear periphery. The pachytene stage is relatively long and is followed by a brief diplotene stage. The last meiotic prophase I stage, diakinesis, involves condensation of the chromosomes and formation of oocytes.
Meiotic DSBs are normally created in TZ, processed in MP and repaired as crossovers and non-crossovers at late pachytene (LP). However, in our system, meiotic DSBs were prevented by a spo-11 deletion and DSBs were induced by laser microirradiation. The rapid movement of chromosomes in diakinesis did not allow DSB targeting by laser microirradiation, however earlier stages could be targeted (see Materials and Methods). The three meiotic prophase I regions we tested were the TZ, MP and LP nuclei. The PMT contains mostly S and G2 phases of the cell cycle (41–43). Nuclei in rows 1 to 6–8 are part of the stem-cell niche, while the remainder of the PMT contains nuclei undergoing the last division prior to meiotic commitment (40). Therefore we decided to examine these two regions separately. We designated the two PMT regions based on this classification: microirradiation at rows 5–7 as stem cell niche PMT (SCN-PMT) and rows 10–12 as final mitosis (FM-PMT).
In all regions examined, RPA-1 foci appeared following microirradiation, as found in our MP data. The number of foci per nucleus ranged from 0.6 to 1.1 foci/nucleus, but the differences were significant with P < 0.01 only for LP (1.1 foci/nucleus) compared to SCN-PMT (Figure 3B and Supplementary Table S8). The timing of RPA-1 focus appearance was significant with P < 0.01, only for the FM-PMT and TZ comparison (Figure 3C and Supplementary Table S9).
RAD-51 is recruited slower in MP germline nuclei
We performed a similar analysis for RAD-51 using the spo-11; GFP::RAD-51 strain. As expected, RAD-51 foci appeared following microirradiation in all regions examined. There was no difference in the mean number of foci per nucleus in each gonadal region (Kruskal-Wallis test, P = 0.07). However, when timing of focus appearance was analyzed, MP nuclei showed delayed kinetics of RAD-51 foci appearance compared to all regions except TZ (P < 0.01 Figure 3E and Supplementary Table S10). Similar observations were made when data was pooled per germline and analyzed as a percent of total foci appearing at each time point (Figure 3F and G). These data indicate that RAD-51 foci are recruited with delayed kinetics to DSBs in MP nuclei, compared to other gonadal regions.
RPA-1 and RAD-51 foci resolution occurs more than 24 h following microirradiation
If laser microirradiation leads to nuclear ablation, we would expect microirradiated nuclei to be quickly eliminated from the germline. Live imaging of worms is restricted to no >4 h post-microirradiation. Therefore, to perform long-term analysis, we microirradiated GFP::rpa-1; spo-11 nuclei, and GFP::H2B spo-11 strains, rescued the worms and fixed dissected germlines 2, 24, 48 and 72 h post-microirradiation. RPA-1 was examined by the use of an anti-GFP antibody, while detection of RAD-51 was performed using an anti-RAD-51 antibody. RAD-51 foci formed clusters of multiple RAD-51 foci in close proximity (also see Materials and Methods). These clusters appear in live imaging as single foci, but as connected foci in fixed samples (Figure 5A, B and D).
Each nucleus contained on average slightly more than one RAD-51 or RPA-1 cluster/focus (Tables 1 and 2). Both RPA-1 and RAD-51 localization persisted for at least 24 h following microirradiation (Tables 1 and 2), indicating that microirradiated nuclei can maintain viability for at least a day. No nuclei with RPA-1 or RAD-51 staining were identified 48 and 72 h, respectively, post-microirradiation in meiotic zone. Similar results were found, in all regions microirradiated [SCN-PMT(PMT), TZ or MP]. As time passes, nuclei move in the germline (from distal to proximal); based on RAD-51 staining, PMT (SCN-PMT) microirradiated nuclei were found at MP 24 h post-microirradiation and at LP 48 h post-microirradiation (Supplementary Figure S4). The rate of nuclear movement in the germline has been suggested to be about one row per hour (43,44). In agreement with this, and as an indication of the viability of microirradiated nuclei, microirradiated PMT and TZ nuclei moved at the expected rates (∼0.85 rows/h). LP microirradiated nuclei with RPA-1/RAD-51 staining were mostly absent from samples obtained 24 h post-microirradiation, while MP microirradiated nuclei had reduced RAD-51/RPA-1 staining at 24 h and absent staining in samples obtained 48 h post-microirradiation. We conclude that RPA-1/RAD-51 foci disappeared more quickly in MP and LP compared to PMT and TZ microirradiated nuclei (also see Discussion).
Table 2. GFP::H2B, spo-11 & GFP::RPA-1, spo-11.
| Region | Hours post- microirradiation | Nuclei with RAD-51 foci clusters | Total number of RAD-51 foci clusters | Total numbers of microirradiated nuclei | Fraction of nuclei with foci clusters/microirradiated nuclei | RAD-51 foci clusters/nucleus with a cluster |
|---|---|---|---|---|---|---|
| PMT | 2 | 65 | 80 | 88 | 73.9 | 1.2 |
| PMT | 24 | 40 | 58 | 64 | 62.5 | 1.5 |
| PMT | 48 | 33 | 43 | 144 | 22.9 | 1.3 |
| PMT | 72 | 0 | 0 | 32 | 0.0 | NA |
| TZ | 2 | 36 | 49 | 90 | 40.0 | 1.4 |
| TZ | 24 | 44 | 71 | 70 | 62.9 | 1.6 |
| TZ | 48 | 7 | 8 | 80 | 8.8 | 1.1 |
| TZ | 72 | 0 | 0 | 40 | 0.0 | NA |
| MP | 2 | 57 | 71 | 96 | 59.4 | 1.2 |
| MP | 24 | 43 | 65 | 96 | 44.8 | 1.5 |
| MP | 48 | 0 | 0 | 80 | 0.0 | NA |
| MP | 72 | 0 | 0 | 56 | 0.0 | NA |
| LP | 2 | 64 | 76 | 78 | 82.1 | 1.2 |
| LP | 24 | 2 | 2 | 54 | 3.7 | 1.0 |
| LP | 48 | 0 | 0 | 30 | 0.0 | NA |
| LP | 72 | 0 | 0 | 24 | 0.0 | NA |
Number of RAD-51 nuclei with a foci cluster, total number of foci clusters, and total number of microirradiated nuclei per gonadal region for worms that were microirradiated and then dissected at 4 different time points post-microirradiation. Immunofluorescence was performed with an anti-RAD-51 antibody. ‘Fraction of nuclei with foci clusters/microirradiated nuclei’ indicates how many nuclei showed foci clusters out of the total number of nuclei that were microirradiated. ‘RAD-51 foci cluster/nucleus with a cluster’ indicates the average number of foci clusters per nucleus.
Microirradiated chromosomes can be repaired to form crossovers marked by COSA-1
If laser microirradiation leads to DSBs, their repair is expected to lead to formation of crossovers. The site of crossovers is marked by the protein COSA-1, that forms one focus per bivalents in wild type in late pachytene nuclei (45). COSA-1 foci are mostly, but not completely, eliminated in spo-11 mutants (45).To examine crossover formation, we microirradiated GFP::COSA-1 germline of spo-11 null mutants at TZ and examined the localization pattern of COSA-1 ∼28 h post-microirradiation (Figure 4A), when most nuclei are expected to reach late pachytene and RAD-51 foci are in the process of disappearing (Supplementary Figure S4). We observed a significant increase in the numbers of COSA-1 foci in microirradiated germlines (Figure 4B). These numbers are not representing a complete recovery of crossover defects, as will be discussed below.
Microirradiation does not lead to an increase in germline apoptosis
The reduction in numbers of nuclei with RAD-51 foci in LP could be due to accelerated apoptosis following microirradiation. γ-irradiation leads to a 5–10-fold increase in the number of apoptotic nuclei in the germline, which is found 24–48 h post-microirradiation (for example; (46–48)). We measured the levels of apoptosis using a ced-1::GFP reporter gene that marks engulfed nuclei to apoptosis. The numbers of apoptotic nuclei marked by CED-1 was not different between non-microirradiated nuclei and microirradiated nuclei 24 or 48 h post-microirradiation (Figure 4C and D). Non-microirradiated gonads showed very rare appearance of nuclei with RAD-51 foci (0.9/late pachytene; Figure 4E). These foci are formed by low-level of naturally occurring DNA damage (49). These spontaneous DNA damage RAD-51 positive nuclei are different from RAD-51 positive nuclei following microirradiation that are likely a result of microirradiation induced DNA damage. Despite these differences, the number of CED-1 positive nuclei contained RAD-51 foci, was not different between non-microirradiated and microirradiated nuclei (15.1%, 16% and 15.6% for non-microirradiated TZ and LP microirradiation nuclei respectively). This data indicates that microirradiation does not cause massive apoptosis (as generated by γ-irradiation) and that nuclei that are microirradiated have the same chance of being eliminated as nuclei suffering from ‘natural’ DNA damage. However, our data cannot completely exclude that microirradiated nuclei are selectively eliminated by apoptosis.
RPA-1 and RAD-51 colocalize at DNA damage sites for hours
It is well-established that RAD-51 displaces RPA-1 on ssDNA, but it is not clear if the whole filament is replaced by RAD-51 or if these two ssDNA binding proteins can cohabitate the same filament forming a transitional filament. In the first 30 min post-microirradiation, nuclei were more likely to contain RPA-1 than RAD-51 foci, but following that both RPA-1 and RAD-51 foci persisted for at least 24 h and disappeared around the same time. To test if these foci co-localize, we stained for endogenous RAD-51 in a GFP::RPA-1 spo-11(null) strain at 2, 24, and 48 h post-microirradiation. We found that most nuclei contained colocalized RPA-1 and RAD-51, regardless of the stage in which they were microirradiated or time of analysis post-microirradiation (Table 3). Some nuclei contain more intense RPA-1 or RAD-51 signal (‘Mostly RPA-1’ and ‘Mostly RAD-51’ categories in Table 3, respectively), but most had signals with similar intensity co-localizing with each other. The shape of the RPA-1 and RAD-51 cluster was not identical, suggesting that the proteins localize closely but not necessarily at the exact same position (Figure 5).
DISCUSSION
Our work establishes an experimental system for DSB induction in the germline of C. elegans, a living, intact organism. We show that RPA-1 and RAD-51 form foci following microirradiation, with an average appearance time indicative of sequential recruitment. For the first time we have a measurement of the kinetics of recruitment of RPA-1 and RAD-51 in C. elegans and an observation of the process in different regions of the germline. Our analysis indicates that the kinetics of appearance of RPA-1 and RAD-51 foci are largely uniform, with two exceptions that will be discussed below. These data suggests that germline nuclei of different stages are unique also in terms of regulation of the DSBR pathway.
Laser microirradiation is a valid method for live-imaging the cellular response to DSBs
Ionizing radiation, such as γ-irradiation has been used for many years as a tool for the study of DSBR in somatic cells of many biological systems, as well as the germline of C. elegans. However, there are several caveats for analysis of DSBR using ionizing radiation. First, many DSBs are induced simultaneously which complicates the quantitative analysis of HR protein recruitment to DSBs. Second, the exposure to many DSBs simultaneously may not reflect a situation analogous to in vivo exposure to natural DNA damage, which generates fewer DSBs. Third, exposing a sample to ionizing radiation requires specialized equipment which makes live-imaging analysis not possible for the early events of HR recruitment. Samples are typically processed not earlier than 30 min following γ-irradiation. Laser microirradiation can circumvent all of these problems by creating a break (or a few localized breaks) using the same microscope in which their repair is imaged. This allows for the imaging of recruitment of proteins to the DSB immediately after a DSB is formed.
We have shown that DSBs created by laser microirradiation can be imaged close to time zero in live, intact worms. This expands the analysis of DSBR beyond tissue culture to whole animals. We have shown that the recruitment of proteins to microirradiated germline DSBs follows the common model of DSBR: RAD-51 recruitment follows that of RPA-1, and RAD-51 recruitment requires RPA-1 loading but not vice-versa. We have also demonstrated that the DSBs are not toxic per se to the germline since nuclei can maintain viability for over a day post-microirradiation. Thus, the breaks that are caused by laser microirradiation in C. elegans follow the basic criteria of DSBs used for the study of DSBR in other systems.
γ-irradiation results in a 5–10-fold increase in the number of apoptotic nuclei 24 and 48 h post-irradiation (47,48). This may induce a stress response and other complications to the system. We have shown that microirradiation in our conditions (6 nuclei/worm), does not lead to increased apoptosis. Our experiments cannot exclude the possibility that microirradiated nuclei are selectively targeted for apoptosis, while others are not, so overall apoptosis levels remain the same. However, the overall lack of increase in apoptosis indicates that microirradiation is a less intrusive method of inducing DSBs compared to γ-irradiation.
Finally, we have shown that DSBs generated by microirradiation can be repaired by forming crossovers. If all DSBs were converted to crossovers and all crossovers could be visualized we would expect a larger increase in 1 and 2 foci category. We propose that we did not observe as many COSA-1 foci as expected due to technical reasons. COSA-1 foci formation is restricted only to the late pachytene region of the germline (45). The inability of microirradiation to fully rescue crossover numbers may result from the variable movement rate of nuclei in the germline. Despite the fact that nuclei move at an average of about 1 row per hour, movement rate is highly variable between nuclei (43). Therefore, nuclei that show COSA-1 foci 28 h post-microirradiation, are few among others that already passed or have not yet reached late pachytene, the region in which COSA-1 foci are visualized.
Limitations in comparing laser microirradiation foci and natural foci
We believe that our experimental approach can model DSBR of exogenous DSBs. This model may not be directly applied to understanding DSBR of meiotic DSBs because meiotic DSBs require the removal of a covalently bound SPO-11 protein. The kinetics of RPA-1 recruitment to microirradiated DSBs are likely faster than SPO-11 induced DSBs, since they do not require SPO-11 removal. To date there is no system for synchronous induction of SPO-11-mediated breaks in C. elegans that allows a direct comparison between the repair of exogenous versus SPO-11 breaks throughout the germline. Once RPA-1 loads, meiotic and laser-induced DSBs should be similar in nature. Therefore the timing of RAD-51 recruitment would likely not vary between meiotic and exogenous breaks. We therefore propose that the mean recruitment time of RAD-51 in the germline is ∼14 min after RPA-1 loading.
In the live-imaging studies RAD-51 foci appeared in 4-fold greater numbers compared to those of RPA-1. We believe that this is due to under-detection of RPA-1 foci in our system. The GFP::rpa-1 strain was generated by integration of an extra-chromosomal array, while the GFP::rad-51 strain is a CRISPR/Cas9 mediated single-copy insertion. The former is prone to silencing, while the latter is known to lead to expression similar to endogenous levels. GFP::RPA-1 foci appeared dimmer compared to GFP::RAD-51 foci (See also Figure 1C compared to 1F). In the future, generation of a CRISPR/Cas9 tagged RPA-1 can be used to verify RPA-1 foci numbers and to observe in vivo colocalization of RPA-1 and RAD-51.
Lastly, in our system we used a spo-11 null allele, so we can guarantee that the foci observed are likely due to breaks generated by microirradiation. We cannot completely exclude the possibility that in the absence of spo-11 kinetics of RPA-1 and/or RAD-51 recruitment are different than what is observed for wild type nuclei that contain SPO-11. In C. elegans, unlike what is found in yeast or mouse models, spo-11 null mutants only affect DSB formation and have no effect on any other key meiotic processes such a pairing or synapsis (50). Therefore, it is likely that any effects on the kinetics of RPA and/or RAD-51 recruitment will not be due to any effects on pairing or synapsis.
The timing of RPA-1 and RAD-51 recruitment to microirradiated DSBs in C. elegans germline is similar to that of mammalian tissue culture cells
The analysis of RPA-1 and RAD-51 localization to meiotic DSBs throughout the C. elegans germline was previously determined (27,28). However, this type of analysis cannot define the order of protein recruitment to DSBs, since both are present in TZ and SPO-11 DSB formation is relatively asynchronous (TZ takes hours). Using microirradiation, we were able to record the timing of RPA-1 and RAD-51 foci and the sequential loading of RPA-1 and RAD-51. Our studies indicate that in C. elegans both proteins are recruited within minutes to the DSB. Laser microirradiation experiments using the mammalian Rpa1 and Rad51 were done mainly on fixed tissue culture samples; this analysis revealed that Rpa1 is recruited within 5 min post-microirradiation (13). Other studies observed longer recruitment times, but this could be due to absence of sampling earlier time points (14,15,51,52). In yeast, RPA-1 recruitment is very fast, starting at approximately 50 seconds and peaking at ∼20 min post-microirradiation (16). We observed 27% of RPA-1 foci appear within 2 min of microirradiation and 86% of the foci appear in the first 30 min (MP), which is in the range of what is found in single-cell systems. Rad51 is recruited within the first 10–20 min in microirradiated tissue culture cells (17,18,51,53) and the number of foci peaks at 2–3 h post-microirradiation. In our system, 7% of RAD-51 foci are recruited within the first 15 min and 99% of the foci are formed in the first 60 min (MP). These data taken together point to more similarities than differences between the analysis of the C. elegans germline to mitotic single cell systems.
The timing of later stages of DSB repair, as monitored by the unloading of DSBR proteins, are also similar when C. elegans germline and mitotic single-cell systems are compared. In cell culture, Rpa1 persists 6 h after microirradiation (14,15); Rad51 foci are removed with slower kinetics and can be seen even 24 h post-microirradiation (18,51). In C. elegans, RAD-51 foci were observed even 48 h post-microirradiation of PMT nuclei. Foci formed by microirradiation of later stage (MP/LP) nuclei disappeared more quickly, yet remained for at least a day. This could be explained if repair of germline DSBs takes close to two days and that nuclei in MP/LP do not have the time to repair DSBs and therefore are eliminated by apoptosis. Alternatively, DSBR kinetics downstream of RAD-51 loading may be faster in MP/LP compared with PMT/TZ, leading to quick repair of DSBs in MP/LP microirradiated nuclei.
The germline is somewhat heterogeneous in terms of RPA-1 and RAD-51 recruitment
It is well established that repair pathway choice between HR and NHEJ is cell-cycle regulated in somatic cells. This is also supported by studies with DSBs created by laser microirradiation (18,25). Some of repair pathway choice occurs by inhibiting resection and down regulating RAD-51 loading in the G1 phase of the cell cycle (23,24). In C. elegans meiosis HR is imposed by efficient DSB resection (26,28). We have shown that the rate of RAD-51 recruitment varies in meiotic prophase I, suggesting another mode of regulation of HR.
Since HR is the predominant pathway in meiosis, we did not expect to find complete inhibition of RPA-1 and RAD-51 loading in any prophase I nuclei. We detected a small effect on RPA-1 loading in FM-PMT nuclei compared to TZ, which have more foci with slow loading kinetics. This may suggest that some FM-PMT nuclei have either delayed RPA loading or slower resection. RAD-51 loading in MP showed a more consistent pattern and was delayed 4–5 min compared to other stages. These MP nuclei also exhibited faster RPA-1 loading, leading to 20 min between RPA-1 and RAD-51 mean loading time. This suggests that MP nuclei take longer to load RAD-51. MP is the stage in which wild type nuclei show the highest number of RAD-51 foci and where strand invasion is thought to occur. Our studies imply that RAD-51 loading may be attenuated when strand invasion takes place (Figure 5C). This may serve as a mechanism to down regulate HR at this meiotic stage. Why later stages (LP) do not show this attenuation may be explained by the need to promote recombination of the last lingering DSBs before the apoptotic program eliminates such nuclei.
RPA-1 is not completely displaced by RAD-51 in the C. elegans germline
Colocalization of RPA-1 and RAD-51 on SPO-11 breaks has not been tested in the C. elegans germline. Two hours post laser microirradiation RPA-1 and RAD-51 showed high levels of colocalization. These levels remained high over time, until RPA-1 and RAD-51 disappeared from the DNA at a similar time point. These data are consistent with the timing of RPA-1 and RAD-51 appearance as largely overlapping. Studies of laser microirradiation in other systems did not involve double staining of Rpa and Rad51, but based on the timing of recruitment similar conclusions can be made. Despite being localized and restricted, a single pulse of microirradiation likely leads to several proximal breaks. Thus, it is possible that colocalization reflects different ssDNA molecules, some coated with RPA and some with RAD-51. Since RAD-51 has a conserved role in strand invasion, the RAD-51 filaments have to contain the invading side of the processed DSB (Figure 5E, left). This model suggests that ssDNA that is not involved in strand invasion does not acquire RAD-51, which contradicts the existing model of DSBR. Alternatively, ssDNA may be coated by RPA at different time points during DSBR, but on different types of intermediates that are adjacent (i.e. D-loop versus invading ssDNA, Figure 5E, middle). This model, however, predicts RPA-1 foci appearing, disappearing and reappearing as time progresses, which we did not observe in our live-imaging experiments (e.g. all of RPA-1 foci appeared in the first 20 min at MP). A third model suggests that our colocalization is a manifestation of mixed ssDNA RPA-RAD-51 transitional filament that contains one terminal RAD-51 domain and one internal RPA domain (Figure 5E, right). RPA-RAD-51 may cohabitate the same filament, at least in a transitional state. This model is consistent with our data, as well as ChIP data from yeast (54). The mixed filament may occur when resection exposes longer ssDNA than what can be completely coated by the pool of nuclear RAD-51.
Foci movement
Some studies proposed that part of the regulation of DSBR involves movement and coalescing of individual DSBs to a single location (55). Following individual RAD-51 foci for the first few hours of live-imaging after microirradiation at PMT, TZ and MP revealed their dynamic nature. However, we rarely (5–12% of foci) found them coalescing over time and their relative position remained fairly constant. The presence of similar levels of foci coalescence in nuclei that form the synaptonemal complex partially (TZ), fully (MP), or do not form the synaptonemal complex (PMT) indicates that the constraints placed by the synaptonemal complex are not affecting coalescence. Movement was present in nuclei that are known to show rapid chromosomal movement associated with chromosome pairing (TZ) or not (PMT, MP), yet coalescing did not increase in these nuclei. This data indicates that germline DSBs move, but most foci do not move to one location in the first 1–2 h following their appearance.
In conclusion, we propose that multiple DSBs are formed locally by laser microirradiation, but most foci remain constrained in one cellular location due to their presence on the same chromosome. Few foci may be on different chromosomes, and thus may drift apart, The numbers of RAD-51 foci induced by microirradiation is comparable to that observed in normal meiotic cells and they can be live-imaged immediately following DSB generation. These advantages may promote the use of laser microirradiation as a method of studying DSB repair proteins in C. elegans and other multi-cellular organisms.
Supplementary Material
ACKNOWLEDGEMENTS
Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). We are grateful to R.E. Malone, A. Malkova, M.S. Wold and members of the Smolikove laboratory for critical reading of this manuscript. We are also grateful for Yizhi Yin who did preliminary work establishing the system that is not included in this paper, and Matthew Wheat who helped with cloning.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
National Institutes of Health (NIH) [R01GM112657 to S.S.]. Funding for open access charge: NIH [R01GM112657].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ceccaldi R., Rondinelli B., D’Andrea A.D.. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016; 26:52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen R., Wold M.S.. Replication protein A: single-stranded DNA’s first responder: dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. Bioessays. 2014; 36:1156–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sung P., Klein H.. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006; 7:739–750. [DOI] [PubMed] [Google Scholar]
- 4. Keeney S. Spo11 and the formation of DNA double-strand breaks in meiosis. Genome Dyn. Stab. 2008; 2:81–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim H.-M., Colaiácovo M.P.. DNA damage sensitivity assays in Caenorhabditis elegans. Biol. Protoc. 2015; 5:e1487. [PMC free article] [PubMed] [Google Scholar]
- 6. Saha J., Wang S.Y., Davis A.J.. Examining DNA Double-Strand Break Repair in a Cell Cycle-Dependent Manner. Methods Enzymol. 2017; 591:97–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rogakou E.P., Boon C., Redon C., Bonner W.M.. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999; 146:905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lukas C., Melander F., Stucki M., Falck J., Bekker-Jensen S., Goldberg M., Lerenthal Y., Jackson S.P., Bartek J., Lukas J.. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 2004; 23:2674–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kong X., Mohanty S.K., Stephens J., Heale J.T., Gomez-Godinez V., Shi L.Z., Kim J.S., Yokomori K., Berns M.W.. Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells. Nucleic Acids Res. 2009; 37:e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Popuri V., Ramamoorthy M., Tadokoro T., Singh D.K., Karmakar P., Croteau D.L., Bohr V.A.. Recruitment and retention dynamics of RECQL5 at DNA double strand break sites. DNA Repair. 2012; 11:624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Iyama T., Wilson D.M.. Elements that regulate the DNA damage response of proteins defective in Cockayne Syndrome. J. Mol. Biol. 2016; 428:62–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ramamoorthy M., May A., Tadokoro T., Popuri V., Seidman M.M., Croteau D.L., Bohr V.A.. The RecQ helicase RECQL5 participates in psoralen-induced interstrand cross-link repair. Carcinogenesis. 2013; 34:2218–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang F., Shi J., Chen S.-H., Bian C., Yu X.. The PIN domain of EXO1 recognizes poly(ADP-ribose) in DNA damage response. Nucleic Acids Res. 2015; 43:10782–10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yuan J., Ghosal G., Chen J.. The HARP-like domain-containing protein AH2/ZRANB3 binds to PCNA and participates in cellular response to replication stress. Mol. Cell. 2012; 47:410–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ghosal G., Leung J.W.-C., Nair B.C., Fong K.-W., Chen J.. Proliferating cell nuclear antigen (PCNA)-binding protein C1orf124 is a regulator of translesion synthesis. J. Biol. Chem. 2012; 287:34225–34233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guarino E., Cojoc G., García-Ulloa A., Tolić I.M., Kearsey S.E.. Real-time imaging of DNA damage in yeast cells using ultra-short near-infrared pulsed laser irradiation. PLoS ONE. 2014; 9:e113325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tashiro S., Walter J., Shinohara A., Kamada N., Cremer T.. Rad51 accumulation at sites of DNA damage and in postreplicative chromatin. J. Cell Biol. 2000; 150:283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harper J.V., Reynolds P., Leatherbarrow E.L., Botchway S.W., Parker A.W., O’Neill P.. Induction of persistent double strand breaks following multiphoton irradiation of cycling and G1-arrested mammalian cells-replication-induced double strand breaks. Photochem. Photobiol. 2008; 84:1506–1514. [DOI] [PubMed] [Google Scholar]
- 19. Mathiasen D.P., Lisby M.. Cell cycle regulation of homologous recombination in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014; 38:172–184. [DOI] [PubMed] [Google Scholar]
- 20. Langerak P., Russell P.. Regulatory networks integrating cell cycle control with DNA damage checkpoints and double-strand break repair. Philos. Trans. R. Soc. B: Biol. Sci. 2011; 366:3562–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shi W., Feng Z., Zhang J., Gonzalez-Suarez I., Vanderwaal R.P., Wu X., Powell S.N., Roti Roti J.L., Gonzalo S., Zhang J.. The role of RPA2 phosphorylation in homologous recombination in response to replication arrest. Carcinogenesis. 2010; 31:994–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Anantha R.W., Vassin V.M., Borowiec J.A.. Sequential and synergistic modification of human RPA stimulates chromosomal DNA repair. J. Biol. Chem. 2007; 282:35910–35923. [DOI] [PubMed] [Google Scholar]
- 23. Lisby M., Rothstein R., Mortensen U.H.. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:8276–8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Antúnez de Mayolo A., Lisby M., Erdeniz N., Thybo T., Mortensen U.H., Rothstein R.. Multiple start codons and phosphorylation result in discrete Rad52 protein species. Nucleic Acids Res. 2006; 34:2587–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bekker-Jensen S. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 2006; 173:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lemmens B.B.L.G., Johnson N.M., Tijsterman M.. COM-1 promotes homologous recombination during Caenorhabditis elegans meiosis by antagonizing Ku-mediated non-homologous end joining. PLoS Genet. 2013; 9:e1003276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martin J.S., Winkelmann N., Petalcorin M.I.R., McIlwraith M.J., Boulton S.J.. RAD-51-dependent and -independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-strand break repair. Mol. Cell. Biol. 2005; 25:3127–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yin Y., Smolikove S.. Impaired resection of meiotic double-strand breaks channels repair to nonhomologous end joining in Caenorhabditis elegans. Mol. Cell. Biol. 2013; 33:2732–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reichman R., Alleva B., Smolikove S.. Prophase I: preparing chromosomes for segregation in the developing oocyte. Results Probl. Cell Differ. 2017; 59:125–173. [DOI] [PubMed] [Google Scholar]
- 30. Callender T.L., Laureau R., Wan L., Chen X., Sandhu R., Laljee S., Zhou S., Suhandynata R.T., Prugar E., Gaines W.A. et al. Mek1 down regulates Rad51 activity during yeast meiosis by phosphorylation of Hed1. PLoS Genet. 2016; 12:e1006226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Niu H., Wan L., Busygina V., Kwon Y., Allen J.A., Li X., Kunz R.C., Kubota K., Wang B., Sung P. et al. Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol. Cell. 2009; 36:393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brush G.S., Clifford D.M., Marinco S.M., Bartrand A.J.. Replication protein A is sequentially phosphorylated during meiosis. Nucleic Acids Res. 2001; 29:4808–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bartrand A.J., Iyasu D., Marinco S.M., Brush G.S.. Evidence of meiotic crossover control in Saccharomyces cerevisiae through Mec1-mediated phosphorylation of replication protein A. Genetics. 2006; 172:27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lui D.Y., Colaiácovo M.P.. Meiotic development in Caenorhabditis elegans. Adv. Exp. Med. Biol. 2013; 757:133–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dickinson D.J., Pani A.M., Heppert J.K., Higgins C.D., Goldstein B.. Streamlined genome engineering with a self-excising drug selection cassette. Genetics. 2015; 200:1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Da Ines O., Degroote F., Goubely C., Amiard S., Gallego M.E., White C.I.. Meiotic recombination in Arabidopsis is catalysed by DMC1, with RAD51 playing a supporting role. PLoS Genet. 2013; 9:e1003787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Arribere J.A., Bell R.T., Fu B.X.H., Artiles K.L., Hartman P.S., Fire A.Z.. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 2014; 198:837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Colaiácovo M.P., MacQueen A.J., Martinez-Perez E., McDonald K., Adamo A., La Volpe A., Villeneuve A.M.. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell. 2003; 5:463–474. [DOI] [PubMed] [Google Scholar]
- 39. Sonneville R., Querenet M., Craig A., Gartner A., Blow J.J.. The dynamics of replication licensing in live Caenorhabditis elegans embryos. J. Cell Biol. 2012; 196:233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rosu S., Cohen-Fix O.. Live-imaging analysis of germ cell proliferation in the C. elegans adult supports a stochastic model for stem cell proliferation. Dev. Biol. 2017; 423:93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Seidel H.S., Kimble J.. Cell-cycle quiescence maintains Caenorhabditis elegans germline stem cells independent of GLP-1/Notch. eLife. 2015; 4:3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fox P.M., Vought V.E., Hanazawa M., Lee M.-H., Maine E.M., Schedl T.. Cyclin E and CDK-2 regulate proliferative cell fate and cell cycle progression in the C. elegans germline. Development. 2011; 138:2223–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Crittenden S.L., Leonhard K.A., Byrd D.T., Kimble J.. Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol. Biol. Cell. 2006; 17:3051–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jaramillo-Lambert A., Ellefson M., Villeneuve A.M., Engebrecht J.. Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev. Biol. 2007; 308:206–221. [DOI] [PubMed] [Google Scholar]
- 45. Yokoo R., Zawadzki K.A., Nabeshima K., Drake M., Arur S., Villeneuve A.M.. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell. 2012; 149:75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang S., Tang M., Pei B., Xiao X., Wang J., Hang H., Wu L.. Cadmium-induced germline apoptosis in Caenorhabditis elegans: the roles of HUS1, p53, and MAPK signaling pathways. Toxicol. Sci. 2008; 102:345–351. [DOI] [PubMed] [Google Scholar]
- 47. Yang M., Sun J., Sun X., Shen Q., Gao Z., Yang C.. Caenorhabditis elegans protein arginine methyltransferase PRMT-5 negatively regulates DNA damage-induced apoptosis. PLoS Genet. 2009; 5:e1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schumacher B., Hofmann K., Boulton S.J., Gartner A.. The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr. Biol. 2001; 11:1722–1727. [DOI] [PubMed] [Google Scholar]
- 49. Pattabiraman D., Roelens B., Woglar A., Villeneuve A.M.. Meiotic recombination modulates the structure and dynamics of the synaptonemal complex during C. elegans meiosis. PLoS Genet. 2017; 13:e1006670–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dernburg A.F., McDonald K., Moulder G., Barstead R., Dresser M., Villeneuve A.M.. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998; 94:387–398. [DOI] [PubMed] [Google Scholar]
- 51. Kim J.-S., Krasieva T.B., Kurumizaka H., Chen D.J., Taylor A.M.R., Yokomori K.. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J. Cell Biol. 2005; 170:341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mu Y., Lou J., Srivastava M., Zhao B., Feng X.-H., Liu T., Chen J., Huang J.. SLFN11 inhibits checkpoint maintenance and homologous recombination repair. EMBO Rep. 2016; 17:94–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Coker H., Brockdorff N.. SMCHD1 accumulates at DNA damage sites and facilitates the repair of DNA double-strand breaks. J. Cell Sci. 2014; 127:1869–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang X., Haber J.E.. Role of Saccharomyces single-stranded DNA-binding protein RPA in the strand invasion step of double-strand break repair. PLoS Biol. 2004; 2:E21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Marnef A., Legube G.. Organizing DNA repair in the nucleus: DSBs hit the road. Curr. Opin. Cell Biol. 2017; 46:1–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


