Abstract
Controlling translation initiation is an efficient way to regulate gene expression at the post-transcriptional level. However, current knowledge regarding regulatory proteins and their modes of controlling translation initiation is still limited. In this study, we employed tandem affinity purification and mass spectrometry to screen for unknown proteins associated with the translation initiation machinery. Ubiquitin specific peptidase 9, X-linked (USP9X), was identified as a novel binding partner, that interacts with the eukaryotic translation initiation factor 4B (eIF4B) in a mRNA-independent manner. USP9X-deficient cells presented significantly impaired nascent protein synthesis, cap-dependent translation initiation and cellular proliferation. USP9X can selectively alter the translation of pro-oncogenic mRNAs, such as c-Myc and XIAP. Moreover, we found that eIF4A1, which is primarily ubiquitinated at Lys-369, is the substrate of USP9X. USP9X dysfunction increases the ubiquitination of eIF4A1 and enhances its degradation. Our results provide evidence that USP9X is a novel regulator of the translation initiation process via deubiquitination of eIF4A1, which offers new insight in understanding the pivotal role of USP9X in human malignancies and neurodevelopmental disorders.
INTRODUCTION
Regulating gene expression at the translational level allows cells to quickly and efficiently respond to extracellular and intracellular stimuli, and plays a key role in a number of biological processes, including cell growth, proliferation, differentiation, and survival (1,2). Translational control primarily occurs at the initiation stage, as that is the rate-limiting step for protein synthesis of most mRNAs (3). In eukaryotic cells, canonical translation initiation begins with the recruitment of the Met-tRNAi-40S ribosomal complex to the vicinity of the mRNA 5′ cap by the coordinated action of at least 11 eukaryotic initiation factors (eIFs) (3,4). Eukaryotic initiation factor 4F (eIF4F), a heterotrimeric protein complex, composes of a cap-binding subunit (eIF4E) that directly binds to the 5′cap structure of mRNA, a scaffolding protein (eIF4G), and a DEAD-box RNA helicase (eIF4A1), that unwinds the structured regions of the RNA and promotes mRNA scanning until the initiation complex locates the AUG start codon. A critical enzymatic step during translational initiation is the helicase activity mediated by eIF4A, which is stimulated by eIF4G, eIF4B or eIF4H (5–7). In addition to stimulating the helicase activity of eIF4A, eIF4B also directly interacts with RNA molecules (8,9), as well as the poly(A)-binding protein (PABP) (10), eIF3 (11) and the 40S ribosomal subunit (11,12), facilitating the recruitment of ribosomes to mRNA (13) and increasing cap-dependent translation initiation both in vitro and in vivo.
Regulating eukaryotic translation initiation appears to be primarily mediated through either altered levels or post-translational modifications of the initiation factors via several signal transduction pathways. For example, eIF4B phosphorylation is tightly regulated by multiple signaling pathways such as mTOR/PI3K and MAPK (14–16). Similarly, eIF4E regulation is also mediated by distinct MAP kinase pathways, which affects tumorigenesis (17–19). In addition to phosphorylation, ubiquitination, which involves the covalent attachment of ubiquitin to target proteins to regulate their half-lives, subcellular localization, activity and conformation, appears to be another important post-translational modification that helps fine-tune protein synthesis (20). Ubiquitination is a reversible process, and deubiquitinating enzymes (DUBs) cleave ubiquitin moieties from their substrates, to tightly control ubiquitination levels (21). It has been revealed that many ribosomal proteins, among both the small and large subunits (22,23), as well as several eIFs, including eIF5A (24), Paip1 (25), Paip2 (26), 4E-BP1 (27) and eIF2B epsilon (28) are ubiquitinated. Moreover, the cap-binding protein eIF4E is reported to be primarily ubiquitinated at Lys-159 and ubiquitination of eIF4E reduces its ability to bind eIF4G (29). Despite existing evidence of the involvement of ubiquitination in the translation initiation process, to the best of our knowledge, the role of DUBs in this process remains relatively unexplored.
In this study, we employed tandem affinity purification (TAP) to identify potential associating partners of eIFs. Ubiquitin-specific protease 9, X-linked (USP9X), a highly conserved and substrate-specific DUB (30), was identified and confirmed as a novel eIF4B-associated partner. We further confirmed that USP9X is a component of the translation initiation complex that regulates both global and individual mRNA translation, as well as cell proliferation. Furthermore, we identified that eIF4A1 is one of the substrates of USP9X in the translation initiation complex.
MATERIALS AND METHODS
Reagents and antibodies
WP1130 were purchased from Selleck Chemicals LLC. DMDA-pateamine A (D-PatA) was kindly made available to us by Dr Daniel Romo (Department of Chemistry, Texas A&M University, College Station, TX, USA). Sodium arsenite, cycloheximide, puromycin, IgG agarose, streptavidin-linked agarose and anti-FLAG M2 agarose were purchased from Sigma Aldrich. Anti-eIF4B monoclonal, anti-USP9X, and anti-eIF3c, anti-eIF4AI/II, GAPDH antibodies were purchased from Santa Cruz Biotechnology. Anti-c-Myc was from Abways Technology. Anti-XIAP antibody was obtained from BD Biosciences. Rabbit anti-human eIF4B polyclonal antibody was kindly provided by Dr John W.B. Hershey at UC Davis. Rabbit polyclonal Dcp1a antibody was kindly provided by Dr. Jens Lyke-Andersen at UC San Diego. Anti-Bcl-2 antibody was from Abcam Inc. anti-β-actin, β-tubulin and secondary antibodies conjugated with HRP were purchased from Kang-Cheng Biotech. Secondary antibodies conjugated with fluorescence were from Jackson ImmunoResearch Labs.
Plasmids
DNA plasmids were kindly provided by: FRT-USP9X-WT and FRT-USP9X-C1566A from Dr Dario R. Alessi at University of Dundee (31); The bicistronic reporter pRΔDE•HCVF (32) from Dr. Peter Sarnow at Stanford University; pcDNA3-Flag-eIF4B from Dr John W.B. Hershey at UC Davis. pcDNA3.1-c-TAP was constructed by inserting the streptavidin domain and a Protein A binding peptide separated by a TEV cleavage site into the pcDNA3.1(–) vector. pcDNA3.1-c-TAP-eIF4B was constructed by cloning the human eIF4B gene into the Not I and Bam HI sites of pcDNA3.1-c-TAP. The primers used to amplify the eIF4B gene are 5′-TCGAGCGGCCGCTATGGATTACAAGGATGACG and 5′-TACGGATCCTTCGGCATAATCTTCTCC. The sgRNA plasmid targeting USP9X sequence was cloned by annealing the following two complementary oligonucleotides (5′-CACCGGTTGATCATGTCATCCAACT-3′ and 5′- AAACAGTTGGATGACATGATCAACC-3′), phosphorylating the resulting double-stranded DNA product using T4 Quick Ligase (Thermo Fisher) and ligating this dsDNA into a BsmBI-digested lenti-CRISPR plasmid (Addgene ID: 49535) backbone.
Cell culture and transfection
The HEK293T, MCF-7, HeLa cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin in a 37°C incubator with 5% CO2. Transfection of the plasmids in HEK293T was performed using PEI transfection reagent.
Tandem affinity purification (TAP)
TAP was performed as previous described (33). Briefly, either an TAP-tagged target construct or an TAP plasmid alone was transiently expressed in HEK293T cells. After 24 h, the cells were lysed in TMK lysis buffer (10 mM Tris–HCl (pH 7.4), 100 mM KCl, 5 mM MgCl2, 1% Trition-100, 2 mM DTT and protease inhibitor cocktail), and the lysates were subsequently applied to first round of affinity purification by binding to rabbit IgG agarose. The beads were then cleaved by TEV protease in SBP binding buffer (10 mM Tris–HCl (pH 7.4), 100 mM NaCl, 0.2% NP-40). Next, the supernatants were subjected to a second round of affinity purification by binding to streptavidin-linked agarose, followed by subsequent elution with biotin and separation via SDS-PAGE. The identities of the proteins were obtained by excising the bands from the gel and performing subsequent tandem mass spectrometry.
Mass spectrometry LC–MS/MS
LC–MS/MS analysis of tryptic peptides were performed on a LTQ linear ion trap mass spectrometer (Thermo Fisher Scientific Inc.) (in the eIF4B interactome experiment) or a Obitrap Fusion mass spectrometer (Thermo Fisher Scientific Inc.) interfaced with an EASY-nLC 1000 System (Thermo Fisher Scientific Inc.) (in the eIF4A ubiquitination analysis). LC–MS/MS data were analyzed by Mascot (v2.3, Matrix Science Ltd., London, UK). Peak lists were generated by Proteome Discoverer software (v1.4, Thermo Fisher Scientific Inc). Precursor mass tolerance for Mascot analysis was set at ± 10 ppm, and fragment mass tolerance was set at ± 0.5 Da. Mascot emPAI (exponentially modified protein abundance index) score was used for label free protein quantification as previously described (34).
Co-immunoprecipitaion and immunoblotting
Cells in a 6-cm dish format were washed in PBS and lysed in 0.5 ml NP-40 lysis buffer (50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 4 mM EDTA, 0.5% NP-40) supplemented with protease inhibitors (Aprotinin, Leupeptin, Pepstatin A, and PMSF) and serine/threonine phosphatase inhibitors (Na3VO4 and NaF). Lysates were pre-cleared with 20 μl of protein-G Sepharose. Precleared supernatants containing 500 μg total protein were precipitated with the indicated antibodies and protein G sepharose beads at 4°C for 2 h, and immunoprecipitates were analyzed by immunoblotting. For immunoblotting, the whole cell lysates or immunoprecipitates were separated by SDS-PAGE, transferred onto NC membranes, blocked by 5% non-fat dry milk in PBST, probed with specific primary antibodies overnight at 4°C.The membranes were washed with PBST, and then incubated in HRP-conjugated secondary antibody for 1 h at room temperature. After the membranes were washed with PBST, the proteins were finally visualized by fluorography using an enhanced chemiluminescence system.
Pulldown assay using m7GTP-Sepharose
HEK293T cells were washed in PBS and lysed in lysis buffer (10 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 mM dithiothreitol, 10% glycerol and 0.5% NP-40) supplemented with protease inhibitors (Aprotinin, Leupeptin, Pepstatin A, and PMSF) and serine/threonine phosphatase inhibitors (Na3VO4 and NaF). For each sample, 0.5 mg of total lysates were incubated with 50 μl 7-methyl-GTP Sepharose 4B beads (GE Healthcare) for 2 h at 4°C. For competition analysis, 0.25 mM of excess free 7-methyl-GTP or 0.25 mM GTP were added, separately. Then, the resins were washed five times with lysis buffer, and the bound proteins attached to the washed Sepharose resins were eluted by 1 × SDS sample buffer and subjected to SDS-PAGE followed by immunoblotting with indicated antibodies.
Immunofluorescence and confocal microscopic analysis
Cells were grown on glass slides and treated as indicated. To terminate the reactions, the slides were quickly washed with PBS followed by fixing in 4% paraformaldehyde, then permeabilized in 0.1% Triton X-100. After blocking, the samples were incubated with anti-USP9X antibody (1:100) and anti-eIF4B antibody (1:100) in PBS containing 3% BSA at 4°C overnight. They were then stained with Cy3-conjugated anti-mouse secondary antibody and Alexa Fluo-488 conjugated anti-rabbit secondary antibody at a dilution of 1:200 in PBS containing 3% BSA at 37°C for 30 min. Following extensive washing, the cells with mounted on slides using a mounting media of SlowFade Gold antifade reagent (Invitrogen) added to reduce fading. Fluorescence was visualized under a confocal fluorescence microscopy (Leica, Mannheim, Germany).
Analysis of polysomes by sucrose gradient sedimentation
Polysome profiling were performed as previously described (35) with some modifications. Prior to analysis, cells were plated in a 10-cm dish, and incubated with 100 μg of CHX for 5 min. For the puromycin sensitivity experiments, the cells were incubated with 100 μg/ml puromycin for 3 min. Then, the cells were harvested and lysed in 0.5 ml TMK lysis buffer (10 mM Tris–HCl (pH 7.4), 5 mM MgCl2, 100 mM KCl, 1% Triton X-100, 2 mM DTT and 100 U/ml RNasin in DEPC water). After centrifugation at 10 000 × g for 10 min at 4°C, the supernatant was layered onto 11 ml of 15–45% linear sucrose gradients containing 20 mM HEPES (pH 7.4), 100 mM KCl, 5 mM MgCl2, and 2 mM DTT. Ultracentrifugation at 35 000 rpm for 3.5 h at 4°C was performed in an SW41 rotor. Following centrifugation of the sucrose gradients, the RNA content was measured across the gradient recording the absorption of samples at 254 nm using an in-line spectrophotometer. Polysome-to-monosome (P/M) ratios were quantitated by calculating the area under the curve corresponding to the polysome peaks divided by the area under the curve for the monosome peak.
Establishment and identification of USP9X−/− cells
0.5 μg of sgUSP9X plasmid was transfected into HEK293T cells (2 × 105 cells). Transfected cells were then cultured in medium containing 1.0 μg/ml puromycin for 2 days for selection. Surviving cells were trypsinized and diluted in medium for colony formation. Single colonies were selected, and each colony was passaged and genotyped. Genomic DNA was extracted from cell lines using a TIANamp Genomic DNA Kit (TIANGEN). A region of Exon 2 of the USP9X gene was amplified with genomic DNA-specific primers. Correct size of PCR products was checked by agarose gel electrophoresis and sent for DNA sequencing. USP9X protein expression levels in each colony were further examined using immunoblotting.
Measurement of protein synthesis
We used two different labeling methods to quantify newly synthesized proteins: L-azidohomoalanine (AHA) or [35S] metabolic labeling. L-AHA labeling was performed as follows: 1.5 × 105 of either HEK293T control cells or USP9X−/− cells were plated on 24-well plates and cultured overnight, after which the cells were rinsed twice with warm PBS and the medium was replaced with 450 μl methionine-free DMEM to which 50 μM of the methionine analogue L-AHA was added. After a 1.5 h incubation with L-AHA, the medium was removed, and the cells were washed three times with PBS. Then, the cells were lysed and subjected to the click reaction for 45 min in the dark according to manufacturer’ protocols (Click-chemistry LLC). After separation by SDS-PAGE, the labeled samples were visualized and scanned on a Typhoon FLA 9500 imager. Finally, the gel was stained with G250 Coomassie blue to determine protein loading. The [35S] metabolic labeling assay was performed as previously described (35).
Luciferase assay
HEK293T cells were seeded in 24-well plates and co-transfected with each reporter plasmid together with a different vector and/or a pGL4.74 Renilla luciferase-expressing vector for normalization. At 24 h after transfection, the cells were harvested in passive lysis buffer. Dual-luciferase assays were performed using a dual-luciferase reporter assay system according to the manufacturer's instructions (Promega). An EnVision Multilabel Plate Reader (PerkinElmer) was used to detect the luciferase activity. All values are presented as the mean ± standard error, and were calculated from the results of three independent transfections.
RNA interference
The sequences of siRNA (synthesized by Genepharma Corp., Shanghai, China) were the following, siUSP9X#1 sense: r(GCTAGTATTTAGCCCAAAT)dTdT, siUSP9X#1 antisense: r(AUUUGGGCUAAAUACUAGC)dTdG; siUSP9X#2 sense: r(GCCUGCAGUGGAAAGUGUA)dTdT, siUSP9X#2 antisense: r(UACACUUUC CACUGCAGGC)dGdG; siCtrl sense: r(UUCUCCGAACGUGUCACGU)dTdT, siCtrl antisense: r(ACGUGACACGUUCGGAGAA)dTdT. Cells were seeded in 6-well culture dish and transfected at approximately 50% confluent. Transfection was performed with Opti-MEM reduced serum medium, using RNAi Max transfection reagent, according to the manufacturer's instructions (Invitrogen Life Technologies, CA, USA). Seventy two hours after transfection, cells were either lysed for immunoblotting analysis or used in the assays of cell proliferation, colony formation, or in vivo translation.
RNA isolation and real-time PCR
All RNA samples are freshly obtained from cells treated as indicated. Total RNA was isolated using TRIzol (Invitrogen Life Technologies, CA, USA) following the manufacturer's guidelines. Concentrations and purity of RNA is determined by measuring the absorption of ultraviolet lights using a NanoDrop spectrophotometer. Then, 1 μg of total RNA were reverse-transcribed with ReverTra Ace-reverse transcription (RT) kit (Takara, JP) at 37°C for 15 min. DNase is added in the reverse transcription system. One-tenth of the RT reaction product was incubated in the real-time PCR reaction mixture (Takara, JP). CDNAs were stored at -20°C. The primers for real-time PCR were the following: Human GAPDH forward primer: TCGACAGTCAGCCGCATCTTCTTT, reversed primer: ACCAAATCCGTTGACTCCGACCTT, Human c-Myc forward primer: CGTCTCCACACATCAGCACAA, reversed primer: CACTGTCCAACTTGACCCTCTTG, Human XIAP forward primer: TGTTTCAGCATCAACACTGGCACG, reversed primer: TGCATGACAACTAAAGCACCGCAC, Human eIF4A1 forward primer: AAGGCGTCATCGAGAGTAACT, reversed primer: ATGTGGCCGTTTTCCCAGTC. Real-time PCR analysis was performed in triplicate for each sample at least three independent experiments.
Cell proliferation and colony formation assays
Cell proliferation was determined using the AlamarBlue assay (Invitrogen Inc.) according to the manufacturer's protocol. Each experiment was independently repeated three times. To determine the ability of the cells to form colonies after knocking out USP9X, we conducted colony formation assays by seeding 3000 cells in a six-well plate and cultivating for 10 days. Afterwards, the cells were stained with 0.01% (w/v) crystal violet and the colonies were counted using ImageJ analysis software.
CHX pulse-chase analysis
HEK293T WT cells or USP9X−/− cells were plated on 24-well plates, and then exposed to 50 μg/ml cycloheximide (CHX). Cells were harvested at different time points after CHX treatment, and cell extracts were immunoblotted with an anti-c-Myc, XIAP, eIF4A1, GAPDH or anti-β-Tubulin antibody.
Statistical analysis
Statistical analysis of the data was conducted using Student's t-test. P values <0.05 were considered statistically significant.
RESULTS
Identification of USP9X as a novel eIF4B-interacting protein
We employed tandem affinity purification with TAP-tagged human eIF4B coding sequence as bait to identify novel proteins associated with translation initiation (33). TAP-tagged eIF4B, TAP-tagged UNRIP (Unr-interacting protein, an unrelated control protein), or TAP vector alone (negative control) were ectopically expressed in HEK293T cells. Proteins associated with TAP-eIF4B were isolated using two-step TAP purification, separated by SDS-PAGE, and subjected to mass spectrometry for protein identification (Figure 1A). Proteins that were enriched in eIF4B precipitations are listed in Supplemental Table S1.
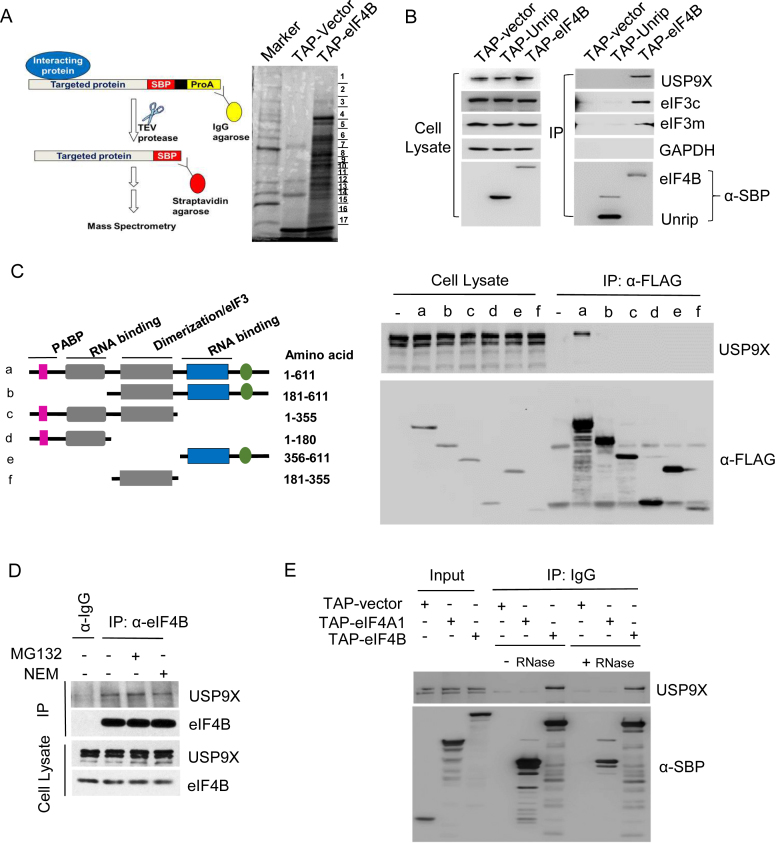
Figure 1.
Interactions of USP9X and eIF4B. (A) Schematic of the tandem affinity purification technique. Either the TAP-empty vector or TAP-eIF4B was transiently expressed in HEK293T cells, and tandem purification was performed as described in ‘Materials and Methods’. Then the purified fractions were separated by denaturing SDS-PAGE followed by silver staining. The indicated bands were excised, subjected to tryptic digestion and analyzed using mass spectrometry. (B) Immunoblotting analysis of the TAP-eIF4B affinity-purified fractions. The elutes after TEV protease treatment (first Step of TAP) were separated on gels using SDS-PAGE and analyzed by immunoblotting with antibodies targeting USP9X, eIF3c, eIF3m and GAPDH (negative control). The TAP-UNRIP construct was used as a non-specific control. (C) Co-immunoprecipitation of USP9X with a variety of eIF4B deletion mutants. left, Schematic representation of the FLAG-tagged eIF4B deletion mutant constructs used in this study. Color bars indicate the motifs involved in protein–protein interactions (interacting partner indicated), dimerization (DRYG) or RNA binding (RRM and ARM). right, HEK293T cells were transfected with vectors encoding eIF4B full-length or each deletion mutant of eIF4B, and whole cell lysates were immunoprecipitated with anti-FLAG beads, and immunoblotted with antibodies targeting USP9X. (D) Co-immunoprecipitation of USP9X with eIF4B from HEK293T cell lysates. HEK293T cells were treated with DMSO, MG132 or NEM for 6 h, and lysed. The cell lysates were then incubated with either an anti-eIF4B antibody or normal rabbit IgG antibody (negative control), and the precipitates were detected with anti-USP9X or anti-eIF4B antibodies. (E) The interaction of eIF4B with USP9X in the presence or absence of RNase. The elutes after TEV protease treatment (first step of TAP purification) were incubated in the presence or absence of 40 U/μl RNase for 120 min, and then analyzed by immunoblotting using USP9X antibodies. TAP-Vector and TAP-eIF4A1 were used as controls.
As expected, most of the identified proteins are known components of the translational apparatus, including all 13 subunits of the translation initiation factor eIF3 (eIF3a-m), small subunit ribosomal proteins (RPS), and other mRNA processing factors (e.g., DDX3 and poly A-binding protein (PABPC1)). Remarkably, USP9X, which has neither been reported as an interacting protein of eIF4B nor implicated in protein synthesis, was found in the high molecular weight fraction. To confirm the mass spectrometry data, eluted proteins after IgG affinity purification and TEV cleavage were subjected to immunoblotting. We observed a clear signal of USP9X in TAP-eIF4B precipitations, as well as other well-characterized eIF4B associating proteins including eIF3c and eIF3m (Figure 1B). GAPDH (negative control) was not detected in either of the precipitations. Next, we mapped the domains of eIF4B that are recognized by USP9X. eIF4B can be structurally divided into four functional domains: a PABP binding site, two RNA binding domains, and a DRYG-repeats region that mediates its interaction with eIF3 and promotes dimerization (36). Various fragments of FLAG-tagged eIF4B (Figure 1C, left) were generated and overexpressed in HEK293T cells. USP9X co-immunoprecipitated with the full-length eIF4B (Figure 1C, lane a), whereas all the remaining deletion mutants failed to interact with USP9X (Figure 1C, lanes b–f), suggesting that the full-length structure of eIF4B is required for its association with USP9X.
To determine whether the interaction between eIF4B and USP9X occurs at physiological level, we immunoprecipitated eIF4B from HEK293T cell lysates. As shown in Figure 1D, endogenous eIF4B was found to be coimmunoprecipitated with USP9X. Treatment with either MG132, a protease inhibitor, or N-ethylmaleimide (NEM), a deubiquitinase inhibitor, had no effect on the interaction between USP9X and eIF4B. Moreover, to rule out any RNA-mediated interactions, RNase A was added to the cell lysate during immunoprecipitation. As shown in Figure 1E, the interaction between USP9X and eIF4B was barely affected by RNase A treatment, ruling out the possibility that RNA molecules mediate this interaction.
USP9X co-localizes and co-sediments with translation initiation proteins
To further verify the association of USP9X with translation initiation complex, we next performed a pull-down assay using m7GTP-Sepharose beads to confirm the interaction between capped mRNA and USP9X. As shown in Figure 2A, the m7GTP-Sepharose was able to pull down USP9X from HEK293T cell lysates along with cap binding protein eIF4E, as well as other eIFs, such as eIF4G, eIF4A, and eIF4B. In addition, the cap analogue (m7GTP), but not GTP, interfered with the association between USP9X and m7GTP-Sepharose, which underscores the specificity of the association between USP9X and 5′ end capped mRNPs.
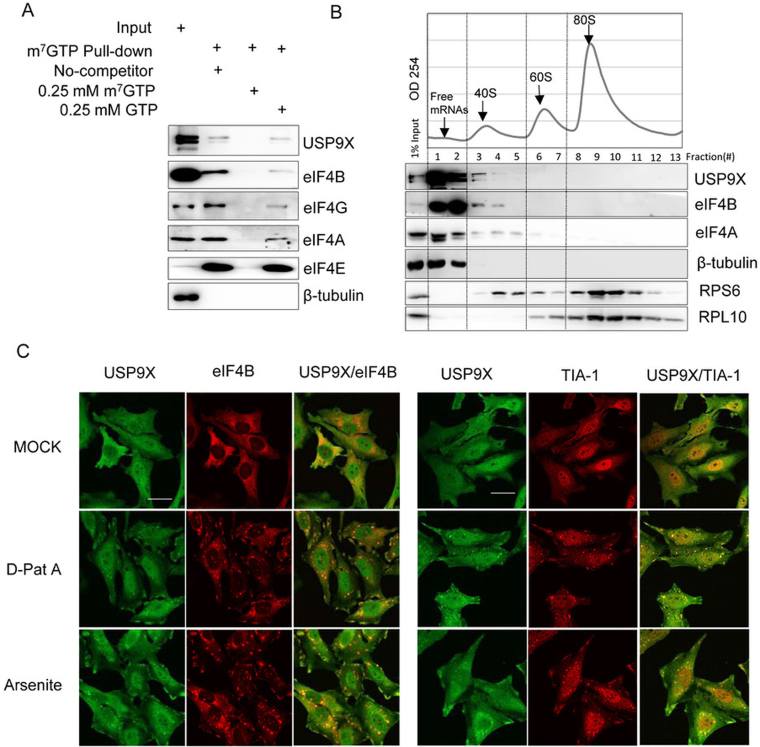
Figure 2.
USP9X co-localized and co-sedimented with the translation initiation complex. (A) HEK293T cell extracts were incubated with m7GTP-Sepharose beads in the presence or absence of 0.25 mM m7GTP or 0.25 mM GTP as indicated, and the eluted proteins were then analyzed using immunoblotting with the indicated antibodies. (B) Polyribosomal distribution of USP9X, eIF4B and eIF4A in HEK293T cells. Approximately 7 × 106 HEK239T cells were harvested and centrifuged over a 10%-35% sucrose gradient, and then RNA across the gradient were detected using a continuous 254-nm monitoring system as shown. The distribution of USP9X, eIF4B, eIF4A, β-Tubulin, RPS6 and RPL10 across the gradient was examined using immunoblotting. (C) Subcellular localization of USP9X upon either inhibition of translation initiation or oxidative stress. HeLa cells were treated upon with DMSO, 0.1 μM D-Pat A or 0.3 mM sodium arsenite for 1.5 h, and the localization of USP9X, eIF4B and the stress granule marker protein TIA-1 was monitored using immunofluorescence. Scale bar, 10 μm.
Furthermore, polysome profiling assays by separating the extract of HEK293T cells using sucrose gradient ultracentrifugation were performed. As shown in Figure 2B, USP9X, was abundant in the upper portion of the gradient corresponding to ribosome-free lysate (fraction 1–2), and trailed into the fractions containing ribosomal 40S subunits (fractions 3–5), whereas it was barely detected in the heavier 60S and 80S fractions (fractions 6–13). In contrast, the ribosome-irrelevant cytosol protein β-Tubulin was only detected in the ribosome-free fractions (fractions 1–2), but was scarcely able to be detected in the 40S, 60S or 80S fractions. The distribution of USP9X was highly similar to that of eIF4B, which is consistent with their newly established interaction. When cells were pre-treated with puromycin, which is an agent that leads premature termination of translating ribosomes and results in polysome disassembly, no significant differences were observed in the distribution of USP9X between the untreated and puromycin-treated cells (Supplemental Figure S1A). This result indicates that the co-sedimentation of USP9X with eIF4A and eIF4B is independent of the enforced disassembly of the translation elongation complex.
In eukaryotic cells, inhibiting translation initiation often leads to the formation of stress granules (SGs), which contain multiple translation initiation components (e.g. eIF4B, eIF4A, eIF4G) and RPS together with untranslating mRNA (37). Then, we further examined the subcellular localization of USP9X when translation initiation was inhibited. As shown in Figure 2C, USP9X localized to both the cytoplasm and nucleus of HeLa cells under normal conditions. However, upon exposure to either the oxidative stressor sodium arsenite or the eIF4A-specific inhibitor D-PatA, USP9X colocalized with eIF4B in the SGs, as confirmed by staining with an SG-specific marker TIA-1. Interestingly, endogenous USP9X was barely detected in processing bodies, another cytoplasmic granule-processing body that is involved in mRNA decay and can be identified via hDcp1a staining after stimulation with either sodium arsenite or D-PatA stimulation (Supplemental Figure S1B). This result showed that USP9X was also able to converge on stress granules along with canonical translation initiation factor such as eIF4A and eIF4B under stress conditions.
USP9X modulates nascent protein synthesis
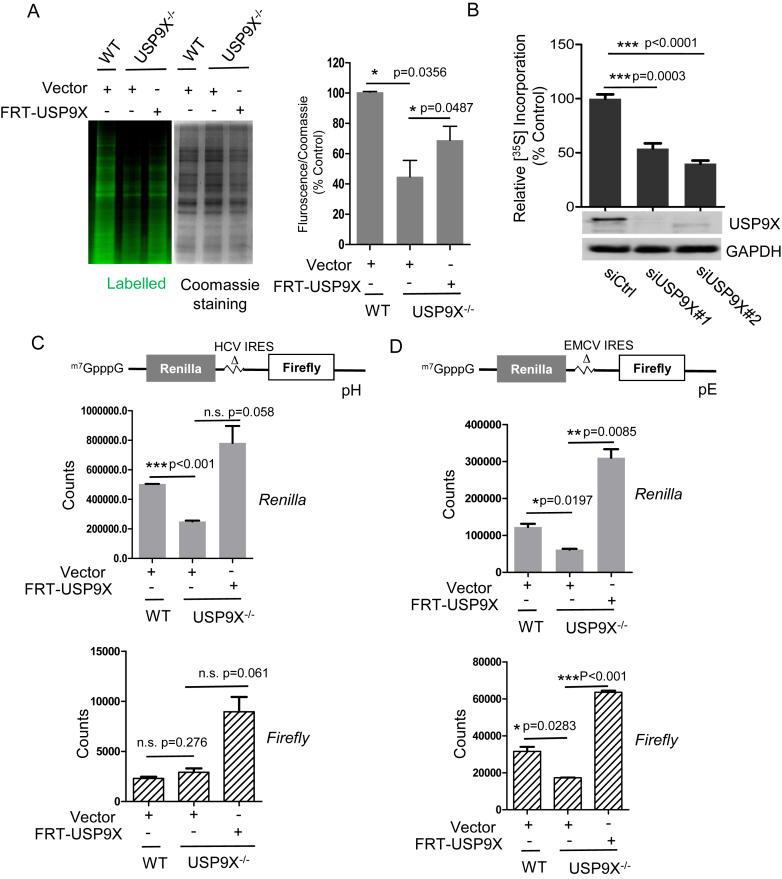
To assess the biological function of USP9X regarding translation, the effects of USP9X depletion or inhibition on global translation were evaluated. First, we generated a stable USP9X knockout (USP9X−/−) cell line using the CRISPR-Cas9 genome editing technique (Supplemental Figure S2A). Using the l-azidohomoalanine (AHA) metabolic labeling assay, we observed that nascent protein synthesis in USP9X−/− cells was significantly decreased to 44.7 ± 11.2% of the levels observed in HEK293T wild-type (WT) cells. Meanwhile, Re-expression of exogenous USP9X can partially rescue the translation impairment (Figure 3A). To further confirm it and rule out the off-target effect, we also performed 35S-methionine metabolic labeling to measure amino acid incorporation in human MCF-7 cells. Consistent with the observations in the knockout cells, silencing USP9X in MCF-7 cells resulted in a significant decrease in 35S-methionine incorporation relative to cells transfected with control siRNA (Figure 3B). These results suggested that USP9X participates in the regulation of nascent protein synthesis in mammalian cells.
Figure 3.
USP9X regulates the global translation and cap-dependent translation initiation. (A) HEK293T WT and USP9X−/- cells, were transfected with FRT-USP9X or FRT-Vector plasmid for 48 h, then incubated in methionine-free DMEM with l-azidohomoalanine (L-AHA) for 90 min, after which the cells were washed, lysed, labeled with a fluorescent alkyne, and subjected to SDS-PAGE. The gel was first scanned for fluorescence using a Typhoon FLA950, and stained with Coomassie blue. (left) Representative images of one replicate was shown. (right) The intensity of the signal in each lane was measured using ImageJ software, and the relative ratio of fluorescence intensity to Coomassie blue staining intensity is shown. *P < 0.05, n = 3 biological replicates. (B) MCF-7 cells were transfected with targeted siRNAs (siUSP9X#1, siUSP9X#2) or siCtrl for 48 h, then were incubated with 35S-labeled methionine/cysteine for 1 h, lysed, and subjected to SDS-PAGE. The incorporation of 35S into newly synthesized proteins was measured and normalized against the total cellular protein levels. ***P < 0.001, n = 3 (C, D) WT or USP9X−/− cells were co-transfected with the 0.1 μg bicistronic reporter pRΔDE·HCVF (pH) or pRΔDE·EMCVF (pE), and 0.4 μg vector or FRT-USP9X for 72 h, after which cells were lysed, and subjected to a dual-luciferase assay. The counts of Renilla and Firefly of each sample were measured by Envision and shown in the graph. n = 3 biological replicates.
To further study the effect of USP9X on global translation, we compared the polysome profiling of USP9X−/− to WT cells. As shown in Supplemental Figure S2D, an appreciable decrease of polysome-to-monosome (P/M) ratio was observed in USP9X−/− cells compared to WT cells. The perturbance of the polysome profiles was similar when USP9X expression was reduced by siRNA in MCF-7 cells (Supplemental Figure S2E).
USP9X regulates cap-dependent translational initiation efficiency
In mammalian cells, at least two different mechanisms of translation initiation exist: 5′ cap-dependent translation initiation and internal ribosomal entry site (IRES) mediated initiation. To confirm which initiation mode is affected by USP9X, we assessed the role of USP9X in both initiations, by taking advantage of bicistronic reporter constructs (32). The upstream reporter (Renilla luciferase) of this bicistronic mRNA is translated through a cap-dependent mechanism, whereas the downstream reporter (Firefly luciferase) is under the control of internal initiation from the hepatitis C virus (HCV)-IRES or (EMCV)-IRES (38). The construct was transiently transfected into either WT or USP9X−/− cells. As shown in Figure 3C and 3D, USP9X depletion caused a significant decrease of Renilla signals by approximately 50% in both cap-FF-HCV-Ren and cap-FF-EMCV-Ren system. Meanwhile, re-expression of USP9X was able to restore the impaired cap-dependent translation initiation activity in USP9X−/− cells. By contrast, HCV IRES-mediated translation of Firefly cistron, which is independent of eIF4F (39), was almost unaffected by lack of USP9X (Figure 3C). Interestingly, EMCV IRES mediated initiation, which requires canonical eIFs including eIF4A/4G, eIF4B, eIF2 and so on (39), was also regulated by the expression of USP9X (Figure 3D). These results indicated that USP9X exerted a direct effect on cap-dependent translation initiation.
USP9X regulates cells proliferation rates
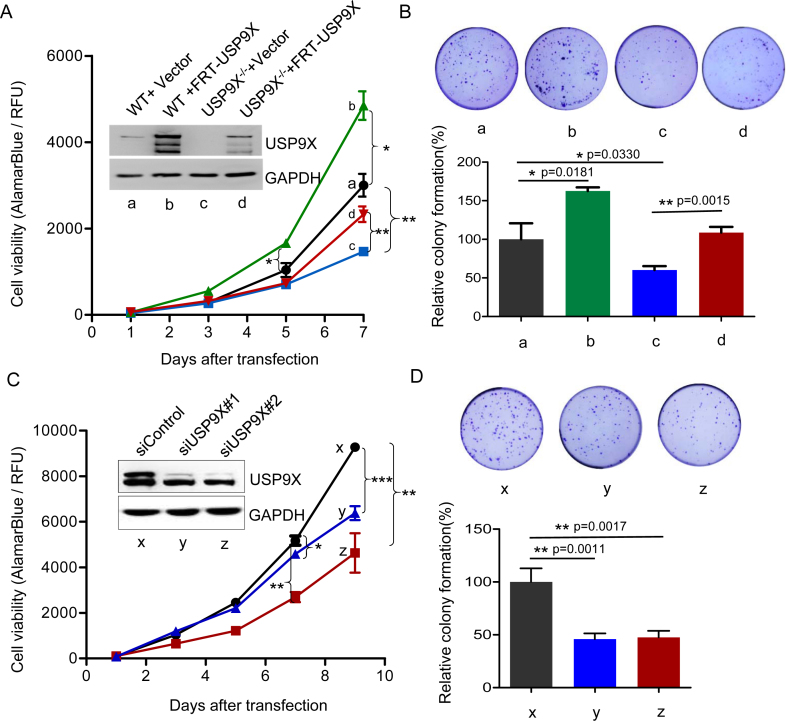
Dysregulation of translational control is believed to play an important role in cell growth, proliferation and survival (40). Therefore, we compared cell growth and the colony-forming ability of HEK293T WT and USP9X−/− cells. Consistent with the observed reduction in translation, USP9X−/− cells exhibited decreased proliferation and produced fewer colonies than WT cells (Figure 4A). When FRT-tagged USP9X was introduced into USP9X−/− cells, the cell proliferation rate nearly recovered to that of WT cells, whereas overexpression of USP9X in WT cells slightly promoted cell proliferation as well. The difference in cell proliferation became even more pronounced in the 14-day colony formation assays (Figure 4B). Consistent with these observations, silencing USP9X in MCF-7 cells led to significantly suppressed cell proliferation and colony formation (See also in Figure 4C and 4D).
Figure 4.
USP9X regulates cell proliferation rate. (A) HEK293T and USP9X−/− cells were transfected with either empty vector or FRT-USP9X for 48 h, then equal numbers of above cells were plated on day 0, and cells proliferation were assessed on day 1, day 3, day 5 and day 7. *P < 0.05. (B) Colony formation assays. HEK293T WT and USP9X−/− cells were seeded in a 24-well plate and transfected with either empty vector or FRT-USP9X for 48 h, after which a total of 3000 cells per condition were plated into a 35mm dish, and the number of colonies formed was counted and imaged 14 days later. (upper) Representative images of one replicate. (lower) Relative colonies numbers were normalized to HEK293T WT cells. **P < 0.01, *P < 0.05, n = 3 biological replicates. (C) MCF-7 cells were transfected with two siRNAs targeting USP9X or control siRNA, and then subjected to cell proliferation assay as described above. Cells proliferation on day 1, day 3, day 5, day 7, and day 9 after transfection were assessed. *P < 0.05 Western blot analysis of USP9X was also performed using cells lysates. (D) The growth of MCF-7 cells of (C) were evaluated by colony formation assays as described as (B). (upper) Representative images of one replicate. (lower) Relative colonies numbers were normalized to MCF-7 cells transfected with control siRNA. **P < 0.01, n = 6
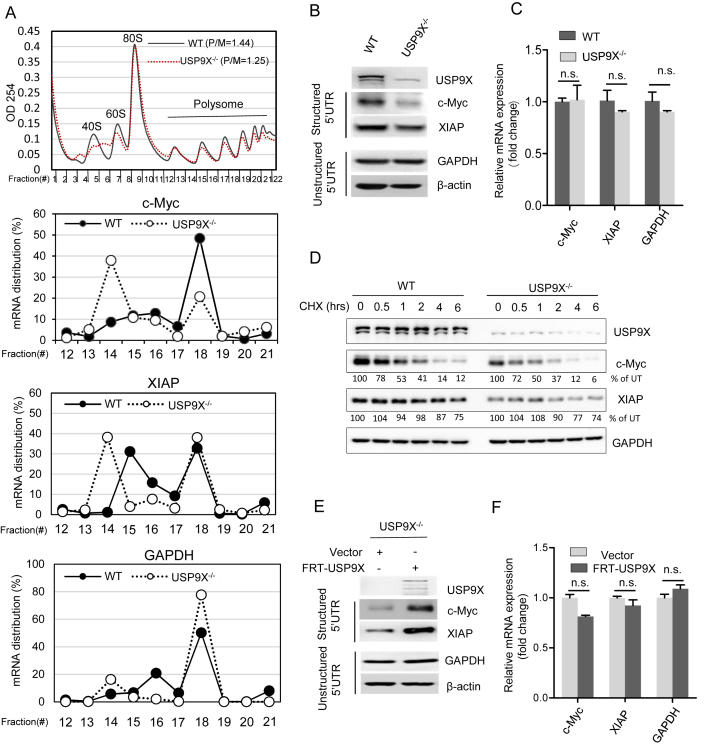
USP9X selectively alters the translational efficiency of specific mRNAs
Changes in translation initiation efficiency could cause either a change in overall protein synthesis or altered translation of specific mRNAs with structural elements within their 5′-UTRs. Therefore, the translation efficiency of mRNAs with complex 5′UTRs, including c-Myc (ΔG 5′UTR = –233 kcal/mol), and XIAP (ΔG 5′UTR = –63 kcal/mol), was examined. We performed sucrose density fractionation to evaluate the polysome association of the c-Myc, XIAP and GAPDH mRNA in HEK293T WT and USP9X−/− cells. We found that in USP9X−/- cells, the c-Myc and XIAP mRNA clearly shifted from the heavier polysome fractions to the lighter fractions, which is indicative of translation suppression. In contrast, USP9X depletion had a minimal effect on the polysome profiles of GAPDH (ΔG 5′UTR = –22 kcal/mol) (Figure 5A). In addition, loss of USP9X caused a substantial reduction in the protein expression of c-Myc and XIAP, whereas the protein levels of GAPDH and β-actin (ΔG 5′UTR = −16 kcal/mol), whose 5′UTR are relatively unstructured, remained unchanged (Figure 5B). Concurrently, the mRNA levels of c-Myc and XIAP in WT and USP9X−/− cells were barely altered (Figure 5C). In order to eliminate degradation influence on these 5′UTR structured genes, we tested the protein stability of c-Myc and XIAP by blocking protein synthesis with CHX treatment. There were no significant alterations in the c-Myc and XIAP degradation rate when comparing HEK293T WT to USP9X−/− cells (Figure 5D). Moreover, rescuing USP9X expression in USP9X−/− cells significantly increased the protein levels of c-Myc and XIAP with a minimal effect on their corresponding mRNA levels (Figure 5E and F). These results indicate that USP9X preferentially regulates the translation of individual mRNAs such as the well-characterized oncogene c-Myc, as well as XIAP.
Figure 5.
USP9X regulates the translation of specific mRNAs. (A) Polysome-shift quantitative PCR (qPCR) analysis of c-Myc, XIAP and GAPDH mRNA expression in the polysome fractions. mRNA distribution is presented as the percentage of mRNA in each fraction compared with the total mRNA in all the fractions. (B) Immunoblotting analysis of c-Myc, XIAP, GAPDH and β-actin protein expression in HEK293T WT and USP9X−/− cell lysates. (C) qPCR analysis of c-Myc, XIAP and GAPDH mRNA expression in WT and USP9X−/− cells normalized to 18S rRNA. n.s., P > 0.05, n = 3 biological replicates. (D) In HEK293T WT or USP9X−/− cells, a cycloheximide (CHX) pulse-chase experiment was performed to test the degradation rate of c-Myc and XIAP proteins by adding by 50 μg/ml CHX for the indicated length of time, after which cell lysates were collected and subjected to immunoblotting using the indicated antibodies. The intensity of c-Myc and XIAP were analysed by ImageJ software and normalized to untreated group. (E) Either FRT-USP9X or control vector was transfected into USP9X−/− cells for 48 h, after which cell lysates were collected and subjected to immunoblotting using the indicated antibodies. (F) Relative mRNA expression analysis of XIAP, c-Myc, and GAPDH (normalized to 18S rRNA) in the cells described in (D). n.s., P > 0.05, n = 3 biological replicates.
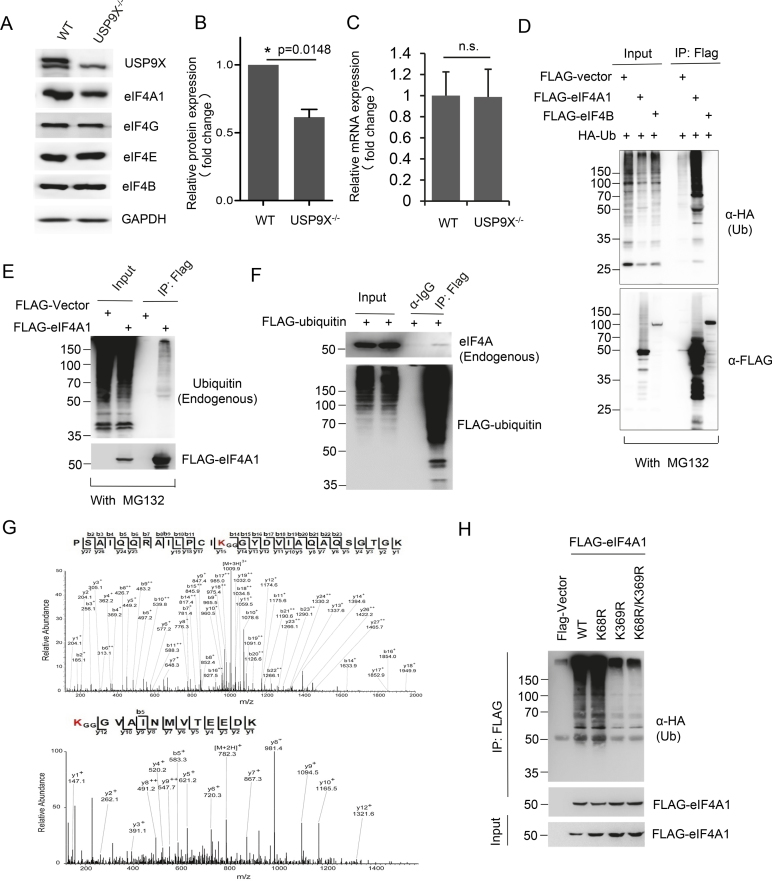
eIF4A1 is primarily ubiquitinated at Lys-369 in vivo
Because the previous results indicated that deubiquitinase activity of USP9X is critical for its function, we aimed to further look for its potential substrate within the translation pre-initiation complex. We analyzed the expression alteration of individual eIF4F complex component, including eIF4A1, eIF4B, eIF4E and eIF4G. Loss of USP9X in HEK293T cells decreased the expression of eIF4A1 by about 50%, while the protein levels of eIF4B, eIF4E and eIF4G were minimally affected by USP9X depletion (Figure 6A and B). In contrast, mRNA level of eIF4A1 in USP9X−/− cells was almost unchanged in comparison with that in WT cells (Figure 6C). In addition, we examined whether eIF4A1 and eIF4B were ubiquitinated in vivo. HEK293T cells were co-transfected with plasmids that expressed HA-ubiquitin and either FLAG-eIF4B or FLAG-eIF4A1, followed by treatment with proteasome inhibitor MG132. We observed extensive poly-ubiquitination of eIF4A1, primarily at molecular weights above the expected size, whereas minimal HA signal was detected in the precipitates of cells expressing FLAG-eIF4B (Figure 6D). Furthermore, endogenous ubiquitination of eIF4A1 was found through detecting in immunoprecipitated FLAG-eIF4A1 or endogenous eIF4A in FLAG-ubiquitin pull-downs (Figure 6E and F). Moreover, immunoprecipitated FLAG-eIF4A1 sample was subjected to mass spectrometry, which reveals two potential ubiquitination residues in eIF4A-Lys 68 and Lys 369 (Figure 6G). Remarkably, eIF4A1 mutants with either a Lys-369-to-Arg (K369R) substitution or both Lys-369-to-Arg (K369R) and Lys-68-to-Arg (K68R) substitution exhibited substantially reduced ubiquitin conjugation than either wild-type eIF4A1 or the K68R mutant (Figure 6H). This result suggested that Lys369 is likely the major site for eIF4A1 ubiquitination.
Figure 6.
eIF4A1 is ubiquitinated primarily at Lys-369. (A) Immunoblotting analysis of eIF4A1, eIF4B, eIF4E, eIF4G and GAPDH in HEK293T WT and USP9X−/− cells lysates. (B) Statistical analysis of endogenous eIF4A1 expression in HEK293T WT and USP9X−/− cells, P = 0.0148, n = 3 biological replicates. (C) qPCR analysis of eIF4A1 mRNA expression in WT and USP9X−/− cells (normalized to GAPDH), n.s., P > 0.05, n = 3 biological replicates. (D) HEK293T cells were co-transfected with HA-ubiquitin and FLAG-eIF4A1, FLAG-eIF4B, or FLAG-vector control plasmid and then treated with 2.5 μM MG132 for 12 h. Cells were then lysed, and the lysates were immunoprecipitated with anti-FLAG M2 beads and immunoblotted with an HA antibody. (E) HEK293T cells were transfected with either FLAG-eIF4A1 or vector control plasmid. FLAG-eIF4A1 was immunoprecipitated with anti-FLAG M2 beads, and immunoblotted with antibodies targeting endogenous ubiquitin to detect ubiquitinated eIF4A1. (F) HEK293T cells were transfected with FLAG-ubiquitin which was then immunoprecipitated using anti-FLAG M2 beads under denatured conditions and immunoblotted with antibodies targeting endogenous eIF4A1. (G) Mass spectra analysis of ubiquitinated eIF4A1 residues. (H) HEK293T cells were co-transfected with plasmids expressing HA-ubiquitin and FLAG-eIF4A1, the K68R mutant, the K369R mutant, or the K68R/K369R double mutant. eIF4A1 ubiquitination was analyzed by immunoprecipitating FLAG-eIF4A1 and blotting using an antibody targeting HA.
eIF4A1 is a novel substrate of USP9X in vivo
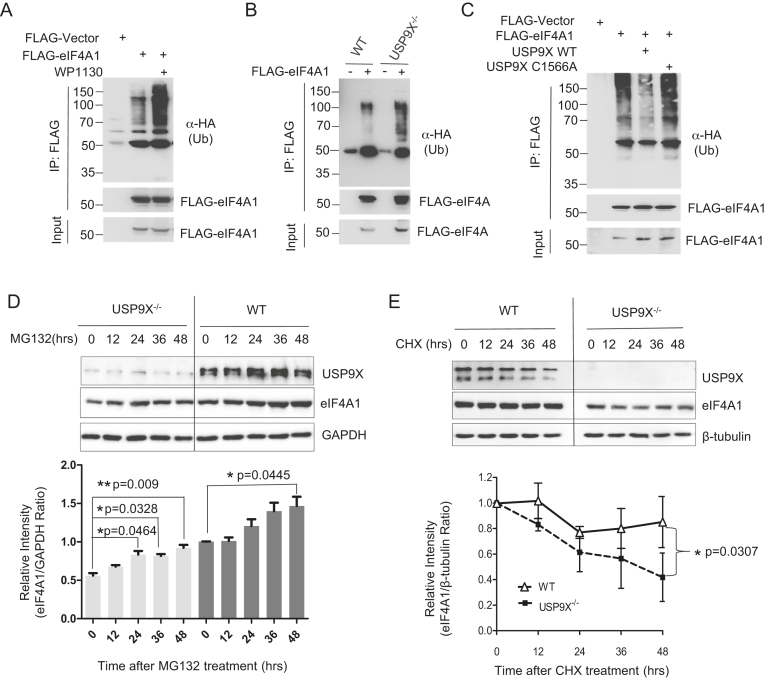
Then we sought to identify whether USP9X is responsible for the deubiquitination of eIF4A1. In HEK293T cells, FLAG- eIF4A1 and HA-ubiquitin were co-expressed in the presence or absence of the DUB inhibitor WP1130. We found that WP1130 significantly increased the ubiquitination of eIF4A1 (Figure 7A). Consistent with this, lack of USP9X in USP9X−/- cells also enhanced eIF4A1 ubiquitination relative to WT cells (Figure 7B). Moreover, overexpression of wild-type USP9X in 293T cells was able to partially deubiquitinate eIF4A1, whereas catalytically inactive USP9X C1566A mutant failed to decrease its ubiquitination levels (Figure 7C). These data suggested USP9X is one of the DUBs responsible for eIF4A1 deubiquitination.
Figure 7.
USP9X deubiquitinates eIF4A1 and is required for eIF4A1 stability. (A) HEK293T cells were co-transfected with plasmids expressing HA-ubiquitin and FLAG-eIF4A, treated in the presence or absence of 2.5 μM WP1130 for 12 h, then cell lysates were immunoprecipitated by anti-Flag M2 beads and blotted with HA antibody. (B) HEK293T WT or USP9X−/- cells were co-transfected with plasmids expressing HA-ubiquitin and FLAG-eIF4A or control FLAG-vector, and polyubiquitylated eIF4A1 protein was detected. (C) HEK293T cells were co-transfected with plasmids expressing HA-Ub and FLAG-eIF4A1 together with either a vector control, USP9X, or the USP9X C1566A mutant plasmid. Cells were treated with 2.5 μM MG132 for 12 h before cell lysates were immunoprecipitated to pull down FLAG-eIF4A1 protein, and the polyubiquitylated eIF4A1 protein was detected by anti-HA antibody. (D) HEK293T WT or USP9X−/− cells were treated with 1.25 μM MG132 for indicated time. Representative result of three independent experiment was shown. The intensity of the results was quantified and analysed. n = 3 biological replicates *P < 0.05, ** P < 0.01. (E) In HEK293T WT or USP9X−/− cells, a cycloheximide (CHX) pulse-chase experiment with endogenous eIF4A protein degradation was performed by adding by 50 μg/ml CHX for the indicated time. Representative result of three independent experiment was shown. The intensity of the results was quantified and analysed. *P = 0.0307, n = 3 biological replicates.
USP9X is required for eIF4A1 stabilization and protects it from proteasome- dependent degradation
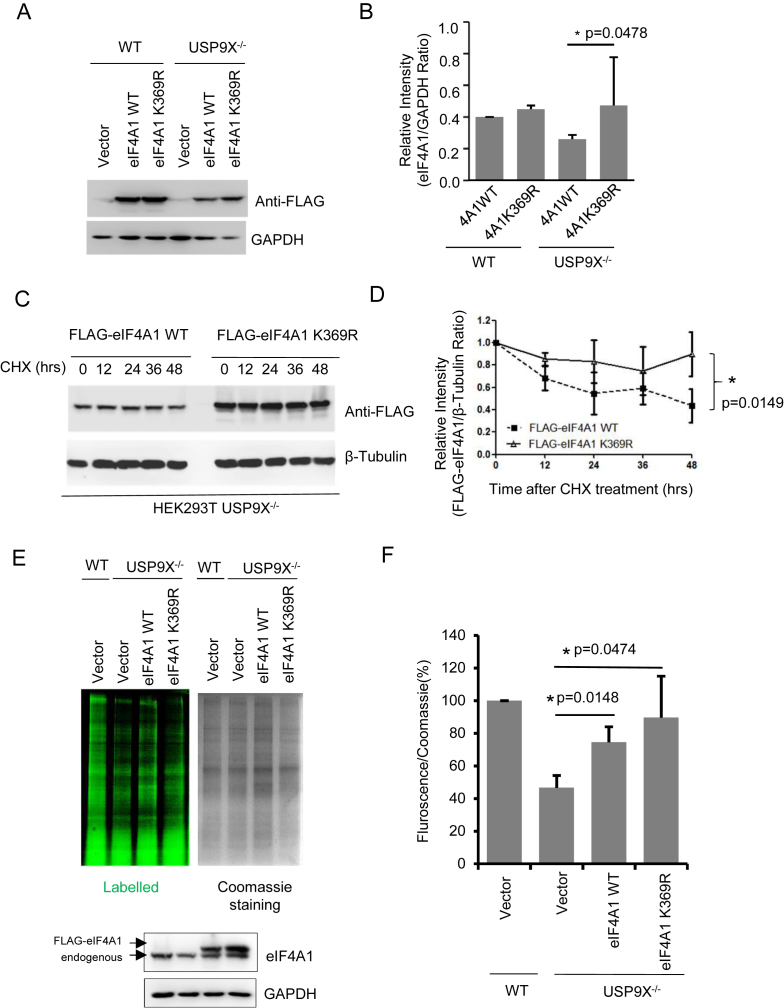
Next, we explored the role of USP9X in eIF4A1 ubiquitination and stability. When USP9X−/− and WT cells were treated with proteasome inhibitor MG132, eIF4A1 protein levels were increased in a time-dependent manner (Figure 7D). Especially in the USP9X−/− cells, a 48-h treatment significantly increased (P < 0.05) the expression of eIF4A1. This result demonstrated the turnover of eIF4A1 might be controlled by the ubiquitin-proteasome pathway. Hence, we compared the endogenous eIF4A1 degradation rate in WT and USP9X−/− cells through a CHX pulse-chase experiment. Depletion of USP9X in HEK293T cells accelerated eIF4A1 protein degradation, and eventually impaired endogenous eIF4A1 expression (Figure 7E). Furthermore, when the same amounts of wild-type FLAG-eIF4A1 or K369R mutant were exogenously expressed in WT or USP9X−/−cells, K369R mutant, which cannot be ubiquitinated, was expressed at relatively higher protein levels (Figure 8A and B), especially in USP9X−/−cells. Also, as shown in Figure 8C and D, the degradation rate of FLAG-eIF4A1 K369R mutant appears to be much lower in relative to wild-type FLAG-eIF4A1. These data suggested that the deubiquitination of eIF4A1 by USP9X is required for eIF4A1 protein stability.
Figure 8.
USP9X modulates protein translation through the regulation of eIF4A1 turnover. (A, B) FLAG-eIF4A1 WT or K369R construct was transfected into HEK293T WT or USP9X−/−cells, then FLAG-tagged proteins expression was determined by Western blot using anti-FLAG antibody. Western blot data were quantified using ImageJ software. *P = 0.0478, n = 3 biological replicates. (C) FLAG-eIF4A1 WT or K369R construct was transfected into HEK293T USP9X−/− cells, CHX pulse-chase experiment with FLAG-eIF4A protein degradation was performed by adding by 50 μg/ml CHX for the indicated time. Representative result of three independent experiment was shown. (D) The intensity of the results from (C) were quantified and analysed. n = 3 biological replicates. (E) HEK293T WT cells were transfected with 0.4 μg FLAG-vector and USP9X−/– cells were transfected with 0.4 μg FLAG-Vector, FLAG-eIF4A1 WT or FLAG-eIF4A1 K369R mutant plasmid for 48 h, then incubated in methionine-free DMEM with L-AHA for 90 min, after which the cells were washed, lysed, labeled with a fluorescent alkyne, and subjected to SDS-PAGE. The gel was first scanned for fluorescence using a Typhoon FLA950, and stained with Coomassie blue. Representative image of one replicate was shown. FLAG-eIF4A1 and endogenous eIF4A1 protein expression were analysed by immunoblotting using an eIF4A1 antibody. (F) The intensity of the signal in each lane from (E) was measured using Image J software, and the relative ratio of fluorescence intensity to Coomassie blue staining intensity is shown. *P < 0.05 n = 3 biological replicates.
To further confirm the loss of eIF4A1 is responsible for USP9X’s function in translation regulation, FLAG-eIF4A1 WT and K369R construct was exogenously over-expressed into USP9X−/−cells, and nascent protein synthesis was examined. As shown in Figure 8E and 8F, in USP9X−/− cells, ectopic expression of eIF4A1 WT and K389R mutant increased the nascent protein synthesis from 46.6% ± 7.5% of HEK293T control cells, to 74.5 ± 9.5%, and 89.6 ± 25.4% of WT cells, respectively. This result suggested that USP9X modulated protein translation mostly if not exclusively through the regulation of eIF4A1 turnover.
DISCUSSION
In this study, all the data suggest the role of USP9X as a novel binding partner for the translational preinitiation complex, which modulates the ubiquitination level of eIF4A1, thereby affecting cap-dependent translation initiation. Although a dozen USP9X substrates have been identified, including AF-6 (41,42), β-Catenin (43), Smad4 (44), Survivin (45), ERG (46) and MCL1 (47), this is the first evidence that USP9X is involved in the regulation of translation initiation. In addition, we first showed that eIF4A1, a key enzyme in translation initiation, is subject to ubiquitination and a novel substrate of USP9X. We have cloned over 20 of translation initiation factors including eIF4A1, eIF4E, eIF4B, eIF5A, eIF2α/β, eIF3m, eIF3l and some ribosomal small subunits for screening the substrates of USP9X (data not shown). Some of them indeed can be ubiquitinated, but we only confirmed that eIF4A1 is the substrate of USP9X. Further investigation would be needed to demonstrate whether other eIFs are also involved in the translation regulation by USP9X. Interestingly, compared to cap-dependent translation initiation, HCV IRES- and EMCV IRES- mediated translation initiation have different requirements for eIFs, which could give us a hint for screening potential target substrates. For example, EMCV-IRES mediated translation has no requirement for PABP, eIF4E and large N-terminal and C-terminal fragments of eIF4G (39), however, it was significantly reduced in the absence of USP9X (Figure 3D). This result makes PABP and eIF4E much less likely the direct substrate of USP9X. Of note, although eIF4B does not appear to be USP9X’s substrate, it may still play an important role in this process, most likely as a scaffolding protein connecting USP9X and eIF4A. This would be consistent with the interaction between eIF4B and USP9X.
We also demonstrated the requirement of USP9X in the maintenance of eIF4A1 homeostasis. It was previously shown that expression and activity of eIF4A1 directly contributes to aberrant global protein synthesis and an altered translational landscape, closely associated with translational control of a set of oncoproteins, such as c-myc, c-myb and NOTCH1 (40,48). Hence the tightly regulation and control of eIF4A1 expression by USP9X in this study might explain its critical biological function in modulating global protein synthesis via eIF4A1-dependent translation. In fact, we observed that completely depletion of USP9X is not lethal, and global translation is attenuated by approximately 50%, which is accordance with the decreased level of total eIF4A1 protein.
In this study, Lys-369 was identified as a major eIF4A1 ubiquitination site that is involved in proteasome-dependent degradation of eIF4A1. K369 is on the surface of C-terminal domain of eIF4A1 protein, which facilitates its conjunction to polyubiquitin chain. At same time, K369 is adjacent but not within the interacting domain of eIF4A with eIF4G, making it less likely to impede the complex formation of eIF4F. This is supported by our coimmunoprecipitation experiment using Flag-tagged eIF4A1 WT and K369A mutant where no difference in the amount of eIF4G that were pulled down by either WT or K369A mutant eIF4A (data not shown).
USP9X has been implicated in crucial roles in developmental disorders (49,50) and neurodegenerative diseases (30,51). For example, fat facets (faf), the Drosophila homolog of the Usp9x gene, is essential for early embryonic synaptic development in flies (30). USP9X enhances hippocampal axon growth (52) and synaptic plasticity (53), as well as the self-renewal of neural progenitors (54). Mutations in Usp9x are closely associated with X-linked intellectual disability (XLID) and appear to disrupt neuronal cell migration and growth (49). Despite the pivotal role of USP9X in neuronal development, the mechanism is still unclear. It has been well established that translational control is critical for synaptic activity as well as learning and memory (55), and exaggerated cap-dependent translation by eIF4E overexpression causes synaptic and behavioral aberrations associated with autism spectrum disorders (56,57). Therefore, we recommend future studies on whether the ability of USP9X to control translation initiation is correlated to its pivotal role in the nervous system.
Dysregulated translational control is also believed to play an important role in oncogenic transformation. Genes encoding canonical initiation factors (e.g. eIF3, eIF4G, eIF4E, eIF4A1, eIF4B, eIF5A2 and eIF4H) have been widely reported to be aberrantly expressed in a variety of human cancers and to contribute to tumorigenesis (2,58,59). Recent studies have demonstrated elevated expression and increased activity of USP9X, correlating with tumor progression and poor prognosis in a number of cancers including multiple myeloma (47), esophageal squamous cell carcinoma (60), non-small cell lung cancer (61), ERG-positive prostate tumor (46), colon cancer (62), breast cancer (63), cervical carcinoma (64), follicular lymphoma (47) as well as B-cell acute lymphoblastic leukaemia (65). Consistent with previous reports, the results disclosed herein support the idea that USP9X acts as a positive regulator of global translation as well as oncogenes such as c-Myc. This study, together with aforementioned observations, strongly suggested that USP9X is a pro-oncogene and may it present a promising therapeutic target in neoplasms associated with USP9X and eIF4A1 overexpression.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr Daniel Romo, Dr John W.B. Hershey, Dr Dario R. Alessi and Dr Peter Sarnow for providing the reagents described in the ‘Materials and Methods’. We would also like to recognize Dr. Tilman Schneider-Poetsch for his careful and critical reading of our manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
National Natural Science Foundation of China [31270830, 21572038 to Y.D.; 81372768 to X.N.]; Fund of the State Key Laboratory of Bioorganic and Natural Products Chemistry; Fund of the State Key Laboratory of Drug Research, Chinese Academy of Science [SIMM1601KF-08]. Funding for open access charge: National Natural Science Foundation of China [31270830, 21572038 to Y.D.; 81372768 to X.N.]; Fund of the State Key Laboratory of Bioorganic and Natural Products Chemistry; Fund of the State Key Laboratory of Drug Research, Chinese Academy of Science [SIMM1601KF-08].
Conflict of interest statement. None declared.
REFERENCES
- 1. Sonenberg N., Hinnebusch A.G.. New modes of translational control in development, behavior, and disease. Mol. Cell. 2007; 28:721–729. [DOI] [PubMed] [Google Scholar]
- 2. Ruggero D. Translational control in cancer etiology. Cold Spring Harb. Perspect. Biol. 2013; 5:a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sonenberg N., Hinnebusch A.G.. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009; 136:731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hinnebusch A.G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014; 83:779–812. [DOI] [PubMed] [Google Scholar]
- 5. Rogers G.W. Jr, Richter N.J., Lima W.F., Merrick W.C.. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 2001; 276:30914–30922. [DOI] [PubMed] [Google Scholar]
- 6. Harms U., Andreou A.Z., Gubaev A., Klostermeier D.. eIF4B, eIF4G and RNA regulate eIF4A activity in translation initiation by modulating the eIF4A conformational cycle. Nucleic Acids Res. 2014; 42:7911–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ozes A.R., Feoktistova K., Avanzino B.C., Fraser C.S.. Duplex unwinding and ATPase activities of the DEAD-box helicase eIF4A are coupled by eIF4G and eIF4B. J. Mol. Biol. 2011; 412:674–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Methot N., Pause A., Hershey J.W., Sonenberg N.. The translation initiation factor eIF-4B contains an RNA-binding region that is distinct and independent from its ribonucleoprotein consensus sequence. Mol. Cell. Biol. 1994; 14:2307–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Methot N., Pickett G., Keene J.D., Sonenberg N.. In vitro RNA selection identifies RNA ligands that specifically bind to eukaryotic translation initiation factor 4B: the role of the RNA remotif. RNA. 1996; 2:38–50. [PMC free article] [PubMed] [Google Scholar]
- 10. Bushell M., Wood W., Carpenter G., Pain V.M., Morley S.J., Clemens M.J.. Disruption of the interaction of mammalian protein synthesis eukaryotic initiation factor 4B with the poly(A)-binding protein by caspase- and viral protease-mediated cleavages. J. Biol. Chem. 2001; 276:23922–23928. [DOI] [PubMed] [Google Scholar]
- 11. Methot N., Song M.S., Sonenberg N.. A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol. Cell. Biol. 1996; 16:5328–5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rozovsky N., Butterworth A.C., Moore M.J.. Interactions between eIF4AI and its accessory factors eIF4B and eIF4H. RNA. 2008; 14:2136–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dmitriev S.E., Terenin I.M., Dunaevsky Y.E., Merrick W.C., Shatsky I.N.. Assembly of 48S translation initiation complexes from purified components with mRNAs that have some base pairing within their 5′ untranslated regions. Mol. Cell. Biol. 2003; 23:8925–8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shahbazian D., Roux P.P., Mieulet V., Cohen M.S., Raught B., Taunton J., Hershey J.W., Blenis J., Pende M., Sonenberg N.. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J. 2006; 25:2781–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Raught B., Peiretti F., Gingras A.C., Livingstone M., Shahbazian D., Mayeur G.L., Polakiewicz R.D., Sonenberg N., Hershey J.W.. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J. 2004; 23:1761–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Gorp A.G., van der Vos K.E., Brenkman A.B., Bremer A., van den Broek N., Zwartkruis F., Hershey J.W., Burgering B.M., Calkhoven C.F., Coffer P.J.. AGC kinases regulate phosphorylation and activation of eukaryotic translation initiation factor 4B. Oncogene. 2009; 28:95–106. [DOI] [PubMed] [Google Scholar]
- 17. Robichaud N., del Rincon S.V., Huor B., Alain T., Petruccelli L.A., Hearnden J., Goncalves C., Grotegut S., Spruck C.H., Furic L. et al. Phosphorylation of eIF4E promotes EMT and metastasis via translational control of SNAIL and MMP-3. Oncogene. 2015; 34:2032–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Furic L., Rong L., Larsson O., Koumakpayi I.H., Yoshida K., Brueschke A., Petroulakis E., Robichaud N., Pollak M., Gaboury L.A. et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. PNAS. 2010; 107:14134–14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang X., Flynn A., Waskiewicz A.J., Webb B.L., Vries R.G., Baines I.A., Cooper J.A., Proud C.G.. The phosphorylation of eukaryotic initiation factor eIF4E in response to phorbol esters, cell stresses, and cytokines is mediated by distinct MAP kinase pathways. J. Biol. Chem. 1998; 273:9373–9377. [DOI] [PubMed] [Google Scholar]
- 20. Pickart C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001; 70:503–533. [DOI] [PubMed] [Google Scholar]
- 21. Huo L., Dang Y., Feng R., Lv J., Li F.. Focal hepatic 11C-acetate activity on PET/CT scan due to lymphoid hyperplasia. Clin. Nucl. Med. 2015; 40:278–281. [DOI] [PubMed] [Google Scholar]
- 22. Silva G.M., Finley D., Vogel C.. K63 polyubiquitination is a new modulator of the oxidative stress response. Nat. Struct. Mol. Biol. 2015; 22:116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Higgins R., Gendron J.M., Rising L., Mak R., Webb K., Kaiser S.E., Zuzow N., Riviere P., Yang B., Fenech E. et al. The unfolded protein response triggers site-specific regulatory ubiquitylation of 40S ribosomal proteins. Mol Cell. 2015; 59:35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jin B.F., He K., Wang H.X., Wang J., Zhou T., Lan Y., Hu M.R., Wei K.H., Yang S.C., Shen B.F. et al. Proteomic analysis of ubiquitin-proteasome effects: insight into the function of eukaryotic initiation factor 5A. Oncogene. 2003; 22:4819–4830. [DOI] [PubMed] [Google Scholar]
- 25. Lv Y., Zhang K., Gao H.. Paip1, an effective stimulator of translation initiation, is targeted by WWP2 for ubiquitination and degradation. Mol. Cell. Biol. 2014; 34:4513–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yoshida M., Yoshida K., Kozlov G., Lim N.S., De Crescenzo G., Pang Z., Berlanga J.J., Kahvejian A., Gehring K., Wing S.S. et al. Poly(A) binding protein (PABP) homeostasis is mediated by the stability of its inhibitor, Paip2. EMBO J. 2006; 25:1934–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Elia A., Constantinou C., Clemens M.J.. Effects of protein phosphorylation on ubiquitination and stability of the translational inhibitor protein 4E-BP1. Oncogene. 2008; 27:811–822. [DOI] [PubMed] [Google Scholar]
- 28. Tuckow A.P., Kazi A.A., Kimball S.R., Jefferson L.S.. Identification of ubiquitin-modified lysine residues and novel phosphorylation sites on eukaryotic initiation factor 2B epsilon. Biochem. Biophys. Res. Commun. 2013; 436:41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murata T., Shimotohno K.. Ubiquitination and proteasome-dependent degradation of human eukaryotic translation initiation factor 4E. J. Biol. Chem. 2006; 281:20788–20800. [DOI] [PubMed] [Google Scholar]
- 30. Murtaza M., Jolly L.A., Gecz J., Wood S.A.. La FAM fatale: USP9X in development and disease. Cell. Mol. Life Sci: CMLS. 2015; 72:2075–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Al-Hakim A.K., Zagorska A., Chapman L., Deak M., Peggie M., Alessi D.R.. Control of AMPK-related kinases by USP9X and atypical Lys(29)/Lys(33)-linked polyubiquitin chains. Biochem. J. 2008; 411:249–260. [DOI] [PubMed] [Google Scholar]
- 32. Thompson S.R., Sarnow P.. Enterovirus 71 contains a type I IRES element that functions when eukaryotic initiation factor eIF4G is cleaved. Virology. 2003; 315:259–266. [DOI] [PubMed] [Google Scholar]
- 33. Burckstummer T., Bennett K.L., Preradovic A., Schutze G., Hantschel O., Superti-Furga G., Bauch A.. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods. 2006; 3:1013–1019. [DOI] [PubMed] [Google Scholar]
- 34. Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M.. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics: MCP. 2005; 4:1265–1272. [DOI] [PubMed] [Google Scholar]
- 35. Low W.K., Dang Y., Schneider-Poetsch T., Shi Z., Choi N.S., Merrick W.C., Romo D., Liu J.O.. Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol. Cell. 2005; 20:709–722. [DOI] [PubMed] [Google Scholar]
- 36. Shahbazian D., Parsyan A., Petroulakis E., Hershey J., Sonenberg N.. eIF4B controls survival and proliferation and is regulated by proto-oncogenic signaling pathways. Cell Cycle. 2010; 9:4106–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Parker R., Sheth U.. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007; 25:635–646. [DOI] [PubMed] [Google Scholar]
- 38. Sonenberg N. Measures and countermeasures in the modulation of initiation factor activities by viruses. New Biol. 1990; 2:402–409. [PubMed] [Google Scholar]
- 39. Hellen C.U., Sarnow P.. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001; 15:1593–1612. [DOI] [PubMed] [Google Scholar]
- 40. Wolfe A.L., Singh K., Zhong Y., Drewe P., Rajasekhar V.K., Sanghvi V.R., Mavrakis K.J., Jiang M., Roderick J.E., Van der Meulen J. et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014; 513:65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kanai-Azuma M., Mattick J.S., Kaibuchi K., Wood S.A.. Co-localization of FAM and AF-6, the mammalian homologues of Drosophila faf and canoe, in mouse eye development. Mech Dev. 2000; 91:383–386. [DOI] [PubMed] [Google Scholar]
- 42. Taya S., Yamamoto T., Kano K., Kawano Y., Iwamatsu A., Tsuchiya T., Tanaka K., Kanai-Azuma M., Wood S.A., Mattick J.S. et al. The Ras target AF-6 is a substrate of the fam deubiquitinating enzyme. J. Cell Biol. 1998; 142:1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Taya S., Yamamoto T., Kanai-Azuma M., Wood S.A., Kaibuchi K.. The deubiquitinating enzyme Fam interacts with and stabilizes beta-catenin. Genes Cells. 1999; 4:757–767. [DOI] [PubMed] [Google Scholar]
- 44. Dupont S., Mamidi A., Cordenonsi M., Montagner M., Zacchigna L., Adorno M., Martello G., Stinchfield M.J., Soligo S., Morsut L. et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009; 136:123–135. [DOI] [PubMed] [Google Scholar]
- 45. Vong Q.P., Cao K., Li H.Y., Iglesias P.A., Zheng Y.. Chromosome alignment and segregation regulated by ubiquitination of survivin. Science. 2005; 310:1499–1504. [DOI] [PubMed] [Google Scholar]
- 46. Wang S., Kollipara R.K., Srivastava N., Li R., Ravindranathan P., Hernandez E., Freeman E., Humphries C.G., Kapur P., Lotan Y. et al. Ablation of the oncogenic transcription factor ERG by deubiquitinase inhibition in prostate cancer. PNAS. 2014; 111:4251–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schwickart M., Huang X., Lill J.R., Liu J., Ferrando R., French D.M., Maecker H., O’Rourke K., Bazan F., Eastham-Anderson J. et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010; 463:103–107. [DOI] [PubMed] [Google Scholar]
- 48. Sen N.D., Zhou F., Ingolia N.T., Hinnebusch A.G.. Genome-wide analysis of translational efficiency reveals distinct but overlapping functions of yeast DEAD-box RNA helicases Ded1 and eIF4A. Genome Res. 2015; 25:1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Homan C.C., Kumar R., Nguyen L.S., Haan E., Raymond F.L., Abidi F., Raynaud M., Schwartz C.E., Wood S.A., Gecz J. et al. Mutations in USP9X are associated with X-linked intellectual disability and disrupt neuronal cell migration and growth. Am. J. Hum. Genet. 2014; 94:470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Paemka L., Mahajan V.B., Ehaideb S.N., Skeie J.M., Tan M.C., Wu S., Cox A.J., Sowers L.P., Gecz J., Jolly L. et al. Seizures are regulated by ubiquitin-specific peptidase 9 X-linked (USP9X), a de-ubiquitinase. PLoS Genet. 2015; 11:e1005022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang X., Zhou J.Y., Chin M.H., Schepmoes A.A., Petyuk V.A., Weitz K.K., Petritis B.O., Monroe M.E., Camp D.G., Wood S.A. et al. Region-specific protein abundance changes in the brain of MPTP-induced Parkinson's disease mouse model. J. Proteome Res. 2010; 9:1496–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tan M., Jolly L.A., Murtaza M., Camonis J., Wood S.A.. Usp9x regulates axon specification and growth. Int. J. Dev. Neurosci. 2015; 47:21. [Google Scholar]
- 53. Chen H., Polo S., Di Fiore P.P., De Camilli P.V.. Rapid Ca2+-dependent decrease of protein ubiquitination at synapses. PNAS. 2003; 100:14908–14913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jolly L.A., Taylor V., Wood S.A.. USP9X enhances the polarity and self-renewal of embryonic stem cell-derived neural progenitors. Mol. Biol. Cell. 2009; 20:2015–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Costa-Mattioli M., Sossin W.S., Klann E., Sonenberg N.. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009; 61:10–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Santini E., Huynh T.N., MacAskill A.F., Carter A.G., Pierre P., Ruggero D., Kaphzan H., Klann E.. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013; 493:411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gkogkas C.G., Khoutorsky A., Ran I., Rampakakis E., Nevarko T., Weatherill D.B., Vasuta C., Yee S., Truitt M., Dallaire P. et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature. 2013; 493:371–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vaysse C., Philippe C., Martineau Y., Quelen C., Hieblot C., Renaud C., Nicaise Y., Desquesnes A., Pannese M., Filleron T. et al. Key contribution of eIF4H-mediated translational control in tumor promotion. Oncotarget. 2015; 6:39924–39940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Modelska A., Turro E., Russell R., Beaton J., Sbarrato T., Spriggs K., Miller J., Graf S., Provenzano E., Blows F. et al. The malignant phenotype in breast cancer is driven by eIF4A1-mediated changes in the translational landscape. Cell Death Dis. 2015; 6:e1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Peng J., Hu Q., Liu W., He X., Cui L., Chen X., Yang M., Liu H., Wei W., Liu S. et al. USP9X expression correlates with tumor progression and poor prognosis in esophageal squamous cell carcinoma. Diagn. Pathol. 2013; 8:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang Y., Liu Y., Yang B., Cao H., Yang C.X., Ouyang W., Zhang S.M., Yang G.F., Zhou F.X., Zhou Y.F. et al. Elevated expression of USP9X correlates with poor prognosis in human non-small cell lung cancer. J. Thorac. Dis. 2015; 7:672–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Peddaboina C., Jupiter D., Fletcher S., Yap J.L., Rai A., Tobin R.P., Jiang W., Rascoe P., Rogers M.K., Smythe W.R. et al. The downregulation of Mcl-1 via USP9X inhibition sensitizes solid tumors to Bcl-xl inhibition. BMC Cancer. 2012; 12:541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Deng S., Zhou H., Xiong R., Lu Y., Yan D., Xing T., Dong L., Tang E., Yang H.. Over-expression of genes and proteins of ubiquitin specific peptidases (USPs) and proteasome subunits (PSs) in breast cancer tissue observed by the methods of RFDD-PCR and proteomics. Breast Cancer Res. Treat. 2007; 104:21–30. [DOI] [PubMed] [Google Scholar]
- 64. Rolen U., Kobzeva V., Gasparjan N., Ovaa H., Winberg G., Kisseljov F., Masucci M.G.. Activity profiling of deubiquitinating enzymes in cervical carcinoma biopsies and cell lines. Mol. Carcinog. 2006; 45:260–269. [DOI] [PubMed] [Google Scholar]
- 65. Zhou M., Wang T., Lai H., Zhao X., Yu Q., Zhou J., Yang Y.. Targeting of the deubiquitinase USP9X attenuates B-cell acute lymphoblastic leukemia cell survival and overcomes glucocorticoid resistance. Biochem. Biophys. Res. Commun. 2015; 459:333–339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.