Abstract
The initiation of mRNA translation from start codons other than AUG was previously believed to be rare and of relatively low impact. More recently, evidence has suggested that as much as half of all translation initiation utilizes non-AUG start codons, codons that deviate from AUG by a single base. Furthermore, non-AUG start codons have been shown to be involved in regulation of expression and disease etiology. Yet the ability to gauge expression based on the sequence of a translation initiation site (start codon and its flanking bases) has been limited. Here we have performed a comprehensive analysis of translation initiation sites that utilize non-AUG start codons. By combining genetic-reporter, cell-sorting, and high-throughput sequencing technologies, we have analyzed the expression associated with all possible variants of the -4 to +4 positions of non-AUG translation initiation site motifs. This complete motif analysis revealed that 1) with the right sequence context, certain non-AUG start codons can generate expression comparable to that of AUG start codons, 2) sequence context affects each non-AUG start codon differently, and 3) initiation at non-AUG start codons is highly sensitive to changes in the flanking sequences. Complete motif analysis has the potential to be a key tool for experimental and diagnostic genomics.
INTRODUCTION
Codons other than the canonical AUG start codon are capable of initiating translation (1–3). Recent ribosomal profiling studies suggest that many translation initiation sites (TIS) utilize non-AUG start codons that differ from AUG by a single nucleotide (i.e., CUG, GUG, UUG, ACG, AAG, AGG, AUA, AUU, and AUC) (4–6). These additional TISs increase proteomic diversity (4). When one or more non-AUG start codons are positioned upstream of an AUG start codon, a single transcript may encode multiple protein isoforms that initiate at alternative non-AUG start codons or novel proteins (7–9). This alternative initiation can create isoforms with N-terminal extensions that have alternative localization properties and modified activity, as has been observed with PTEN, HCK, and c-Myc (10–12). In the case of the proto-oncogene c-Myc, which is misregulated in many cancers, the two isoforms have altered DNA binding capabilities and the ratio of their expression levels regulates their activity (11,13,14). In addition to increasing proteomic diversity through alternative initiation sites, some non-AUG start codons enable translation of upstream open reading frames (uORFs) that may regulate expression from the downstream protein coding sequences (4,15–17).
In eukaryotic cells, translation initiation typically follows a ribosomal scanning model. According to this model, the 40S ribosomal subunit, along with several initiation factors, binds to the 5′ cap of the mRNA and then begins scanning in the 3′ direction in search of a start codon (18,19). Once the 40S ribosomal subunit reaches a potential start codon it initiates protein synthesis if it recognizes the start codon. Alternatively, if the scanning ribosome does not recognize a potential start codon, it can bypass the potential start codon and continue scanning in search of the next start codon—a phenomenon known as leaky scanning (19,20). Here we use the term ‘efficiency’ to describe the level of translation initiation attributed to a TIS, i.e., the more likely that a ribosome is to initiate translation at a TIS, the more ‘efficient’ the TIS. Whether a potential start codon initiates translation depends on several factors including the nucleotide context—the sequence immediately surrounding the start codon (20,21). In this study, we investigated the importance of the four nucleotides before the start codon (-4 to -1 positions, where +1 is the first nucleotide of the start codon), the start codon (+1 to +3), and the nucleotide immediately following the start codon (+4 position).
Much of what is known about the effect of sequence context on TIS efficiency comes from the study of AUG start codons. Marilyn Kozak identified a highly efficient TIS sequence motif that is sometimes referred to as the ‘consensus Kozak’ sequence, i.e., CACCAUGG (20). She also showed that the -3 and the +4 positions are the most important for determining the efficiency of AUG start codons (20). In the past, we and others have often followed the general −3/+4 position guidelines first described by Kozak for AUG start codons to determine whether a non-AUG codon was in a good TIS context for efficient initiation. Yet behavior governing AUG-mediated initiation may not hold for non-AUG start codons. To address this issue, here we have analyzed the sequence requirements for all TIS sequences with non-AUG start codons.
With traditional methods, it can be quite an undertaking to study the sequence-phenotype relationship associated with every possible sequence of a genetic motif. To better understand how the TIS motif governs translation initiation efficiency, here we have utilized a high-throughput approach that we previously developed called FACS-seq (21). Using FACS-seq, we previously measured translation initiation efficiency from TIS sequences that utilized AUG start codons. Here we have used this approach to measure the efficiency of all nine non-AUG codons that differ from AUG by a single nucleotide while varying the TIS sequence from the -4 to the +4 position. Additionally, we demonstrate that our results are in line with independently generated ribosomal profiling data.
MATERIALS AND METHODS
FACS-seq
FACS-seq was performed as described previously (21). Briefly, we generated a library of genetic reporters using degenerate PCR primers and a highly efficient Gibson assembly reaction (22). PD-31 cells were stably transduced with the library of genetic reporters. Cells with fluorescence in the GFP channel greater than GFP negative cells were sorted by FACS into 20 equally populated gates based on the ratio of GFP to RFP expression. Cells that were below the GFP expression threshold, but expressed RFP, were sorted into another gate and considered to have no expression. The genomic DNA from the cells in each sorted population was isolated and the TIS sequences were extracted by PCR, purified by gel extraction, and barcoded by PCR. The DNA was then pooled and sequenced using an Illumina MiSeq. A full description of this method is available in Supplementary Data: Materials and Methods.
Calculating TIS efficiency from sequencing results
The TIS sequences and corresponding barcodes were extracted from the DNA sequencing results. The reads from each barcode were scaled such that the total reads for each barcode were proportional to the fraction of cells in each sorted population. These values were then used to calculate the median efficiency of each TIS sequence. Due to a finite number of DNA sequencing reads, some TIS sequences are not adequately represented and so their TIS efficiency could not be definitively measured. To correct for this, we used the ‘glmnet’ package in R (the programming language) to fit the data with a generalized linear model (GLM) (23). We subtracted the noise threshold (based on the GFP values of non-fluorescent cells) below which we could not accurately measure TIS efficiency. Dinucleotide interactions were included in the GLM because they were found to be important for modeling the efficiency of AUG start codons (21). For each codon, we used a 10-fold cross validated GLM with LASSO which accounted for dinucleotide interactions to fit the natural log of the median efficiencies for all of the TIS sequences that did not contain an AUG codon upstream of the start codon of interest (21,24). The natural log was used to describe the interaction between the TIS sequence and the ribosomal preinitiation complex as an association reaction at equilibrium. We used the values calculated by these GLMs for all non-AUG codons and the TIS efficiencies for AUG codons reported in Noderer et al. (21) for all analyses in this report.
Cell culture
Retroviral particles of the TIS reporters were produced by transiently transfecting HEK-293T cells with equal amounts of pCru5-GFP-IRES-mCherry-F2A-Puro DNA and a retrovirus packaging vector pCL-Eco (ecotropic pseudotyping for mouse cell lines) or pCL-Ampho (amphotropic pseudotyping for human cell lines) (25). The Calphos Mammalian Transfection kit was used to perform the transfections (Clontech Laboratories, Inc., Mountain View, CA). After 24 hours the media from the transfected cells was collected and filtered using a 0.4 μm-filter, and the virus was stored at -80°C until it was used.
FACS-seq was performed using PD-31 cells, an Abelson murine leukemia virus-transformed pre-B cell line (21,26). They were cultured in RPMI-1640 medium (Life Technologies) supplemented with 10% fetal bovine serum (FBS, Gemini Bio Products, Sacramento, CA), 2 mM glutamine, 1 mM sodium pyruvate, 0.05 mM 2-mercaptoethanol, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C with 5% CO2. 100 million cells were infected with the virus with 8 μg/mL polybrene (hexadimethrine bromide) added to the media. The infection frequency was 1%, measured by flow cytometry 2 days post-infection. 1.5 μg/mL puromycin was added 2 days post-infection to select for infected cells. Note that we did not observe any changes in TIS efficiency when puromycin was added.
Additional experiments were carried out in other cell lines and under different conditions. NIH-3T3 cells, HeLa cells, and HepG2 cells were cultured in DMEM medium (Life Technologies) with 10% FBS, 4.5 g/ml glucose, and 2 mM glutamine. PD-31 cells (standard conditions) and K562 cells were cultured in RPMI-1640 medium with 10% FBS, 2 mM glutamine, 1 mM sodium pyruvate, and 0.05 mM 2-mercaptoethanol. HCT-116 cells were cultured in McCoy's 5A medium (HyClone Laboratories, Logan, UT) with 10% FBS. MCF-10A cells were cultured in DMEM/F12 media (11330–032; Life Technologies) supplemented with horse serum (H1138; Sigma-Aldrich), 2 mM glutamine, EGF (SRP3027; Sigma-Aldrich), 10 μg/mL insulin (I1882; Sigma-Aldrich), 500 ng/mL hydrocortisone (H0888; Sigma-Aldrich), 100 ng/mL cholera toxin (C8052; Sigma-Aldrich), 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C and 5% (vol/vol) CO2. Full growth medium contained 5% (vol/vol) serum and 20 ng/mL EGF. All cells were cultured with 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C with 5% CO2.
Analysis of reporter expression in altered environmental conditions
The individual TIS reporters used to measure the efficiency of GFP TIS sequences used a GFP fusion protein that is stabilized by addition of trimethoprim, GFP-DHFR (27). The only exception is the GFP reporters that were used to compare cell lines, which used the monomeric enhanced GFP identical to the reporter used in the FACS-seq experiment. PD-31 cells infected with GFP-DHFR TIS reporters were treated with 3 μm trimethoprim for 72 hours before being analyzed by flow cytometry. Where indicated, 3 μM trimethoprim was added to PD-31 cells for 6 hours instead of 72 hours along with one of the following conditions: no additive (normal conditions), 1 mM spermidine added to the medium (increased polyamine levels), 300 nM thapsigragin added to the medium (endoplasmic reticulum stress), or incubated at 40°C (heat shock) prior to analysis by flow cytometry. There was also an additional sample that was used as a control to establish the baseline expression for each TIS reporter in the absence of trimethoprim. The baseline expression for each reporter was then subtracted from the expression measured for each environmental condition. Relative expression was scaled so that the sequence CACCAUGG under normal conditions had a value of 100.
Flow cytometry
An LSRII flow cytometer (BD Biosciences) was used for all analyses. GFP and RFP levels were quantified by measuring fluorescence intensities by flow cytometry. The relative efficiency of each TIS was gauged by computing the quotient GFP divided by RFP levels. Flow cytometry data were analyzed with FlowJo software (Tree Star).
mRNA secondary structure
mRNA secondary structure predictions were performed with NUPACK software (28). The pfunc command was used to calculate the mRNA folding energy (ΔG) of each sequence. The default RNA parameters were used (-material rna1995), dangle energies were included (-dangles all), and the temperature was set at 37°C.
Ribosomal profiling analysis
Translation initiation sites previously identified by ribosomal profiling were downloaded from links provided by Ingolia et al., Lee et al., and Fritsch et al. (4–6). The sequence of each transcript with at least one TIS in the 5′ leader sequence was downloaded from RefSeq and analyzed to identify all possible AUG and non-AUG start codons. TISs identified by ribosomal profiling were matched to the corresponding sequences in RefSeq. All of the potential start codons present in the 5′ leaders were then categorized based on TIS associated properties. The frequency of start codon utilization was calculated by dividing the number of TISs in each category that were observed in the ribosomal profiling study by the total number of AUG or non-AUG start codons present according to RefSeq. P-values were calculated using Fisher's exact test.
RESULTS
Motif analysis of translation initiation sites containing non-AUG start codons
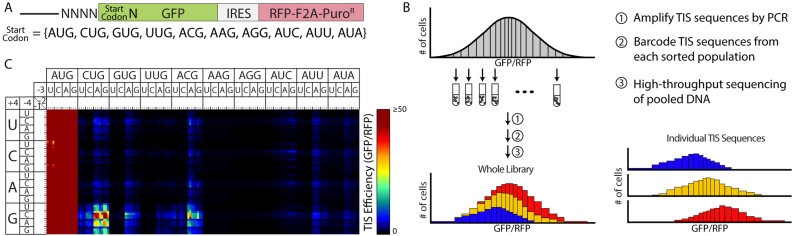
Our objective was to measure the translation efficiency associated with every non-AUG start codon (differing from AUG at a single position) in combination with every possible nucleotide sequence at the adjacent -4 to -1 and +4 positions. Translation efficiency was measured using a genetic reporter (Figure 1A) (26). The reporter encodes expression of green fluorescent protein (GFP) using a TIS sequence of interest. Red fluorescent protein (RFP) is expressed from the same transcript and used to normalize GFP expression, and as a result, calculate the relative translation initiation efficiency of the TIS sequence. Instead of separately generating a reporter construct for each TIS sequence of interest, we generated a single library containing all 10240 possible TIS sequences in the motif NNNN[Start Codon]N (where Start Codon represents AUG, CUG, GUG, UUG, ACG, AAG, AGG, AUA, AUU, or AUC and N represents A,C,G, or U), 9,216 of which utilized non-AUG codons. We then transduced a culture of PD-31 cells (mouse pre-B lymphocytes) with the reporter library and analyzed reporter expression by FACS-seq (Figure 1B) (21). Briefly, with the FACS-seq method we first sorted cells based on their level of reporter expression using FACS (fluorescence-activated cell sorting). Then high-throughput sequencing was used to count the number of copies of each TIS sequence in each sorted population. We were then able to associate reporter expression levels with each TIS sequence in the library.
Figure 1.
High-throughput analysis of TIS motifs utilizing non-AUG start codons. A) The TIS reporter used to measure translation initiation efficiency from every AUG and non-AUG start codon. The -4 to -1 and +4 positions were varied to create a library of all possible sequences at those positions (N = A, C, G, or U). RFP was expressed from the same transcript using an internal ribosome entry site (IRES) and served to normalize GFP expression. F2A is a peptide that allows multiple proteins to be expressed from a single open reading frame. PuroR is the puromycin resistance gene, which enabled selection of stably transduced cells. B) Summary of the FACS-seq method. A population of stably transduced cells is sorted into 20 equally populated gates based on TIS efficiency (GFP/RFP). The TIS sequences are then PCR-amplified and barcoded before being pooled and sequenced. FACS-seq histograms were then created for each TIS sequence based on the number of reads for each TIS in each gated population. The median efficiency values for each TIS sequence were then fit with a generalized linear model that accounted for important dinucleotide interactions. C) Heat map of the TIS efficiencies measured via FACS-seq. The labels of the nucleotides at the -2 and -1 positions follow the same pattern as the other positions: U, C, A, G. A TIS efficiency of 100 corresponds to the TIS sequence CACCAUGG.
To accurately measure protein expression from non-AUG TISs, we chose a FACS gating scheme that would increase the resolution of FACS-seq for TIS sequences that cause low to moderate levels of expression. This limited the resolution we could obtain for translation initiation levels of AUG start codons, which typically cause much higher levels of expression. Therefore, we used the values for AUG start codons reported in Noderer et al. (21) for data normalization and other analyses requiring AUG translation initiation efficiency values. Specifically, to compare data between these two data sets, we normalized TIS efficiency levels to the average efficiency of a subset of highly efficient AUG TIS sequences (N[A/G]NNAUGG) present in both data sets. Here, in reporting the efficiency of each TIS sequence, the efficiency values are scaled so that the efficiency of the TIS sequence CACCAUGG (listed as GCCACCAUGGC in Noderer et al.) equals 100 (Figure 1C, Supplementary Table S1).
A subset of non-AUG codons can initiate translation as efficiently as AUG start codons
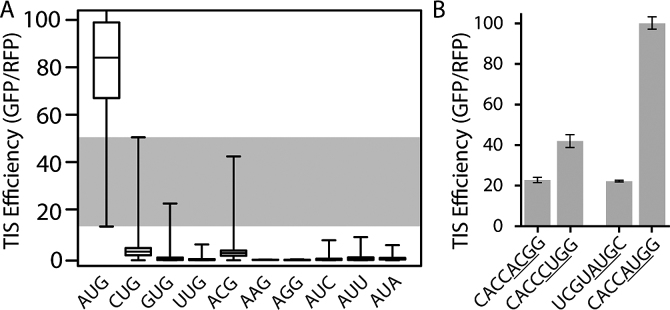
While most non-AUG TIS sequences have very low expression, from our FACS-seq analysis we observed that there was a small subset of flanking sequences that support high levels of translation initiation for several codons (Figure 1C). Within these highly efficient TIS sequences, CUG codons were capable of causing the most expression of all the non-AUG codons, followed by ACG and then GUG, with TIS efficiencies up to 50, 40, and 20 respectively (Figures 1C, 2A). This means that some CUG TISs can cause ∼50% as much protein expression as the highly efficient ‘consensus Kozak’ sequence (CACCAUGG), ∼5 to 10-fold higher than many of the previous reports (2,3). Such highly efficient non-AUG TIS sequences can generate more protein expression than some AUG codons that are in less efficient sequence contexts (Figure 2A, here ‘context’ refers to the nucleotide sequence flanking the start codon). AUU, AUA, AUC, and UUG are less efficient, but still produce detectable levels of expression with maximal TIS efficiencies between 5 and 10 (Figure 1C, 2A). Also, note that due to very low levels of expression we were unable to accurately detect and measure any sequence-dependence in the efficiency of AAG and AGG TIS sequences. This is in agreement with previous reports that found AAG and AGG were the least efficient start codons (2,29).
Figure 2.
A subset of non-AUG codons can be as efficient as AUG start codons. A) Box and whisker plot of TIS efficiencies for each start codon. The edges of the box represent the inner quartile range, the bar within the box represents the median, and the bars extending out of the rectangle extend to the maximum and minimum of the range of efficiencies for each codon. The shaded area represents the range of TIS efficiencies where AUG and non-AUG codons overlap. B) TIS efficiencies determined from individually deployed TIS reporter constructs via flow cytometry. Start codons are underlined. Error bars represent the standard deviation between experimental replicates (N = 3).
We sought to confirm that artifacts of the FACS-seq method did not cause the high levels of expression that we observed. We generated individual TIS reporter constructs (Figure 1A), each with a specified TIS sequence and an inducible GFP fusion protein that is stabilized by addition of trimethoprim (GFP-DHFR) (27). Then, we measured the efficiency of each TIS sequence individually using traditional flow cytometry after the addition of 3 μM trimethoprim for 72 hours. The flow cytometry results agreed with those measured by FACS-seq and showed that some non-AUG TIS sequences generated higher protein expression levels than AUG codons in inefficient contexts (Figure 2B).
Translation initiation efficiency is more dependent on sequence context for non-AUG start codons than AUG start codons
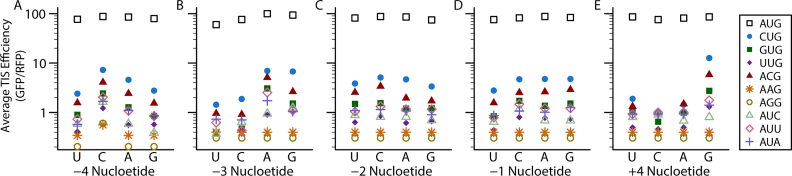
To further investigate how a small fraction of non-AUG start codons can act as efficient TISs, we analyzed the relative importance of sequence context for non-AUG and AUG start codons. For each nucleotide position, we calculated the average efficiency of the TIS sequences with each nucleotide at each position for each start codon (e.g. we calculated the average efficiency of all 256 TIS sequences that had a G in the -4 position and a CUG codon), for every combination of start codon and nucleotide at each position (Figure 3). According to the FACS-seq results, nucleotides in the -4 position caused the efficiency of non-AUG TISs to vary by greater than 100%, but the efficiency of AUG TISs varied by ∼10% (Figure 3A). When only AUG codons in a poor context (those with a C or U in both the -3 and +4 position) were considered, the presence of a C in the -4 position caused an average increase in efficiency of ∼10%. The efficiency of a small subset of the TISs (<5%) increased more than 25%, but none increased by more than 50%. Using individual TIS reporters, we confirmed that the -4 position had a larger impact on all non-AUG codons than on an AUG codon (except AAG and AGG, which had expression levels too close to the background for accurate assessment). We found that a C in the -4 position caused a ∼70% increase in expression from all non-AUG codons compared to a G in the same position (except for AAG and AGG). The same change did not affect the efficiency of an AUG codon (Supplementary Figure S1A). In contrast with our FACS-seq results, we also found that an A in the -4 position was similar to a C for both of the codons tested (Supplementary Figure S1B, C). Additionally, our FACS-seq results indicated the presence of either an A or G in the -3 position increases the efficiency of non-AUG and AUG TISs, but the enhancement observed for non-AUG codons was up to 6-fold greater than the enhancement observed for AUG codons (Figure 3B). Using individual TIS reporters we found that all non-AUG codons (excluding AAG and AGG) with an A in the -3 position were more efficient than those with a G, although the extent of this effect varied between codons and in some cases differed from the high-throughput FACS-seq results (Supplementary Figure S1D). For example, according to the FACS-seq results CUG codons with an A or a G in the -3 position should be equal, but the individual constructs show that CACCCUGG was 50% more efficient than CGCCCUGG. However, except for these cases, throughout this study our FACS-seq results generally were reproduced by flow cytometry experiments performed with individually generated reporter constructs. Altogether, our results demonstrate that the efficiency of non-AUG codons is more dependent on sequence context than the efficiency of AUG codons, both at nucleotide positions that have large impact on the efficiency of AUG TISs and those that do not.
Figure 3.
Non-AUG start codons are more dependent on their surrounding nucleotide context than AUG start codons. The average TIS efficiency of all TIS sequences with each codon and nucleotide in a specified position. A) -4 position. B) -3 position. C) -2 position. D) -1 position. E) +4 position.
G in the +4 position greatly enhances the translation initiation efficiency of CUG, ACG, and GUG codons
A major goal of this study was to identify how the effect of sequence context varies across start codons. We found that the effect of the -4 to -1 position was similar between non-AUG codons, except for AAG and AGG where expression was too low to be accurately measured (Figure 3A–D). In contrast, the +4 position affects each codon differently (Figure 3E). The codons that we found to be the most efficient non-AUG start codons (CUG, ACG, and GUG) experience the greatest increase in efficiency from a G in the +4 position (10x, 7x, 5x respectively). In contrast, the efficiency of an AUU start codon only increases ∼2-fold, similar to the effect of the -4 position. When the +4 nucleotide was not G, all of the non-AUG codons (excluding AAG and AGG, where expression was too low to be quantified accurately) had comparable, low efficiencies. In fact, when the +4 position is not a G, other codons such as ACG and even AUU were sometimes more efficient than a CUG codon in the same sequence context. We verified these results using individual TIS reporter constructs (Supplementary Figure S2).
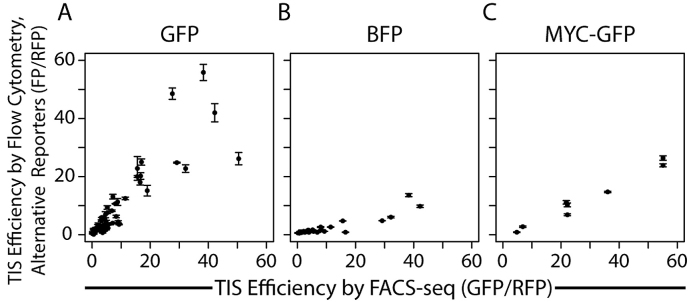
The relative translation initiation efficiency of non-AUG TISs is conserved between proteins
We next wanted to determine how broadly our results could be applied. As discussed above, the TIS efficiencies measured from individual TIS reporters with GFP generally matched the FACS-seq results (Figure 4A). However, we previously found that the efficiency of AUG start codons varied slightly depending on the sequence of the gene (21,26). Therefore, we wanted to determine whether our results were dependent on our choice of reporter gene. We replaced GFP with two other fluorescent reporters to determine if the efficiency of non-AUG TISs also varied between proteins. We replaced GFP with a blue fluorescent protein (mTagBFP, referred to as BFP), which has a different sequence and comes from a different organism. As an additional fluorescent reporter, we replaced the start codon of GFP with the first 669 nucleotides of the c-Myc transcript (NM_002467), including the first 144 nucleotides of the protein coding sequence, to generate a fusion protein (described in greater detail in a later section). The efficiencies of non-AUG TIS sequences measured with the BFP and MYC-GFP reporters increased linearly with the corresponding efficiencies measured by FACS-seq (Figure 4B, C). Although the efficiencies measured with each fluorescent reporter correlated linearly with the efficiencies measured by FACS-seq, the efficiencies for BFP and Myc-GFP were significantly lower than for GFP. The efficiencies measured with the BFP reporter were highly correlated, but lower than those measured with individual GFP reporters by a factor of ∼4 (Figure 4B, Supplementary Figure S3D) and those measured using the MYC-GFP reporter are ∼50% as efficient as measured by FACS-seq (Figure 4C). We also investigated the impact of cell type, protein degradation, and environmental conditions on TIS efficiency, but did not observe any significant effects (Supplementary Figure S3A-C). More details on this analysis can be found in the Supplementary Data: Results. Thus, while the general trends that we observed between TIS sequence and efficiency are conserved, absolute TIS efficiencies varied between proteins. This indicates that there are factors other than sequence context that significantly affect TIS efficiency.
Figure 4.
Efficiency of TISs utilizing non-AUG start codons varies linearly between proteins. The efficiencies of TIS sequences were measured individually by flow cytometry with several genetic reporters and were then compared to the efficiency predicted by FACS-seq. A) GFP reporter (R2 = 0.79). B) BFP reporter (R2 = 0.83). C) Myc-GFP reporter (the N-terminal of c-Myc fused to GFP, R2 = 0.97). Reporter constructs were the same as in Figure 1A with GFP substituted with the other reporter genes. All measured efficiencies were normalized to RFP expression. The TIS sequence CACCAUGG has a TIS efficiency of 100. Error bars represent the standard deviation between experimental replicates (N≥3).
Ribosomal profiling confirms a predictive role for TIS efficiency in start codon selection
We sought to determine whether the TIS efficiencies we measured by FACS-seq could be used to analyze gene expression throughout the genome accurately despite the variation in TIS efficiency between proteins. Several ribosomal profiling experiments have focused on identifying TISs in the genome (4–6). Each of these studies used a different small molecule drug (harringtonine, puromycin, or lactimidomycin) to stall ribosomes as they initiate translation. Then, they identified TISs in the genome by sequencing the RNA fragments that were protected from nucleases by stalled ribosomes.
We compared the publicly available TISs identified by ribosomal profiling to all of the non-AUG codons in the subset of RefSeq transcripts present in each study (30). Rather than attempt to calculate the efficiency of each TIS in the ribosomal profiling experiments, which can be difficult to interpret (31), we measured the frequency of start codon utilization—the fraction of non-AUG and AUG codons within the 5′ leader sequences of transcripts that were observed to cause translation initiation events in each ribosomal profiling study. We investigated the effect of upstream AUG start codons on the frequency of start codon utilization to validate this simplified analysis. We observed that as the number of AUG codons upstream of a potential start codon increased, the less likely the codon was to be utilized as a TIS (Supplementary Figure S4A). This is consistent with the ribosomal scanning model, which predicts that a fraction of the scanning ribosomes will be diverted by each upstream start codon.
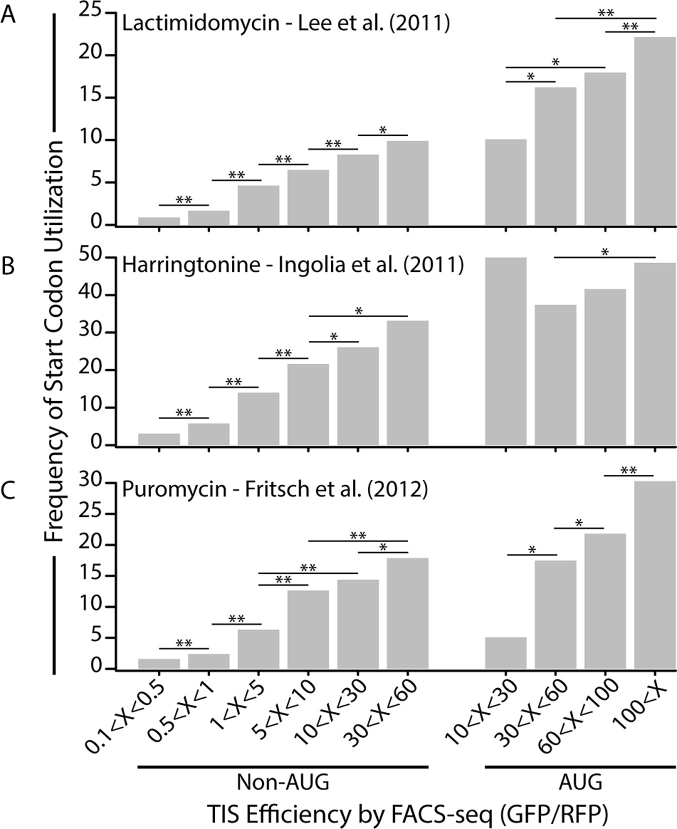
We found that the frequency of start codon utilization increased significantly with TIS efficiency, as measured by FACS-seq (Figure 5). Non-AUG codons with a TIS efficiency greater than 30 were ∼10 times more likely to have observable translation initiation than those with an efficiency less than 0.5. The frequency of start codon utilization also increased with TIS efficiency for AUG codons, albeit to a lesser degree. In the range of efficiencies where both AUG and non-AUG codons overlapped, the percent of non-AUG codons that caused translation initiation was similar to that of AUG codons. Specifically, in the data sets from Ingolia et al. and Fritsch et al., the frequency of start codon utilization for AUG codons was not significantly greater than non-AUG codons with similar TIS efficiencies (Figure 5B, C). In the data from Lee et al., the frequency of start codon utilization was ∼50% greater for AUG codons than for non-AUG codons with TIS efficiencies between 30 and 60 (Figure 5A). We also investigated the effect of the distance between the 5′ cap and the start codon, open reading frame length, and downstream mRNA secondary structure on frequency of start codon utilization. The details of this analysis can be found in Supplementary Data: Results. These results demonstrate that the TIS efficiencies measured by FACS-seq can be used as a predictor of the non-AUG codons that serve as TISs in the genome. Therefore, the efficient TIS sequences identified in this study can potentially be used to help predict and identify novel protein isoforms with N-terminal extensions and regulatory upstream open reading frames.
Figure 5.
Ribosomal profiling confirms a predictive role for TIS efficiency in start codon utilization in the genome. The frequency of start codon utilization, the percentage of potential TIS motifs that were observed to act as translation initiation sites, in the 5′ leader sequences of three independent ribosomal profiling experiments. Each study used a unique small molecule drug to stall initiating ribosomes. A) Lactimidomycin - Lee et al, 2011, B) Harringtonine - Ingolia et al, 2011, C) Puromycin - Fritsch et al, 2012. P-values were calculated using Fisher's exact test. * denotes a P-value < 0.05; ** denotes a P-value < 0.001.
Identification of genetic variants within the human genome that alter the efficiency of non-AUG start codons
Our FACS-seq analysis indicated that non-AUG codons were more sensitive to mutations in their TIS sequence than AUG start codons. Single base mutations in the TIS sequence of AUG codons can cause up to a 160% change in efficiency, but the median percent change caused by a mutation is only 12%. In contrast, a single-base mutation in the sequence context of a CUG codon causes a 70% change in efficiency on average and can cause up to a 30-fold change in expression (Dataset S1). We analyzed the National Center for Biotechnology Information's (NCBI) dbSNP database and the Catalog of Somatic Mutations in Cancer (COSMIC) to identify genetic variants that are located in the TIS sequence of annotated non-AUG start codons (annotated in the NCBI RefSeq database) and may have a biologically relevant effect on protein expression (Table 1). Several of the genetic variants alter or eliminate the start codon leading to drastic changes in protein expression as observed in HCK, a proto-oncogene that has two isoforms with distinct subcellular localizations and functions (12,32). Other variants do not alter the start codon but cause a greater than 5-fold change in protein expression according to our FACS-seq results, such as those found in the transcription factors FGF2 and MYC. Additionally, mutations that are outside of the -3 and +4 positions should be carefully considered because they can have a significant impact on the efficiency of non-AUG TISs. For example, our results indicate that the -4A→T mutation in DDX17 causes a 2.4-fold reduction in protein expression. Each of these types of genetic variation should be considered when investigating genes known to utilize non-AUG translation initiation.
Table 1. Genetic variants within the human genome alter the TIS efficiency of non-AUG start codons.
| Gene | Original DNA Sequence | Mutation | WT Efficiency (From FACS-seq) | Efficiency after Mutation (From FACS-seq) | Fold Change in Efficiency | Mutation IDs (dbSNP and COSMIC) |
|---|---|---|---|---|---|---|
| HCK | CGACCTGG | +3G deletion, +3G insertion | 45.76 | FS, FS | -, - | 755024531, COSM296913 |
| TEAD1 | CAAAATTG | +2T→C | 6.086 | NA | - | 748767790 |
| TEAD3 | CACAATAG | +1A→G | 3.537 | NA | - | COSM4153322 |
| MRVI1 | TGTCCTGA | +3G→T | 0.963 | NA | - | 766752155 |
| NR1I2 | AAACCTGG | +1C→T, +3G→A | 25.062 | 1.607, NA | 15.7, - | 770100550, 775173325 |
| PRPS1L1 | CAAGACGC | -4C→A, +2C→T, +3G→A/T | 3.048 | 2.07, 79.78, NA | 1.4, 26.6, - | 746684840, 73313931, 752999268 |
| VEGFA | CGCGCTGA | -2C→T | 3.6002 | 2.03 | 1.8 | 745949912 |
| BAG1 | GGGCCTGG | -3G→A | 10.674 | 8.54 | 1.25 | COSM7566952 |
| FGF2 | GAGGCTGG | +4G→A | 11.257 | 2.066 | 5.4 | COSM4620808 |
| PIM1 | GGCGCTGC | -3G→A, -1G→A, +1C→T | 1.05 | 1.49, 0.893, 0.433 | 1.4, 1.2, 2.4 | 113378883, 11550058, 192449585 |
| EIF4G2 | CAAAGTGG | +1G→A, +4G→A | 8.74 | 107.44, 3.38 | 12.3, 2.6 | COSM2110788, 111411775 |
| DDX17 | AAACCTGT | -4A→T, -1C→T | 4.585 | 1.922, 2.682 | 2.4, 1.7 | 200991904,753604497 |
| MYC | GACGCTGG | -3A→U*, -2C→G, -1G→A, +1C→G*, +4G→T | 17.00 | 3.67, 11.25, 17.00, 5.24, 3.04 | 4.6, 1.5, 1, 3.2, 5.6 | Wiman et al. (1984), 754742138, COSM4876635, Hartl et al. (1987), 778595707 |
| HMHB1 | AGAACTGG | -3G→C | 29.96 | 7.911 | 3.8 | 748541714 |
| RARB | AAGCCTGG | -1C→G, +1C→G | 13.923 | 18.317, 5.640 | 1.3, 2.5 | 570353505, 756610050 |
| TEAD4 | AGCCTTGG | -2C→A, -1C→T | 2.147 | 1.607, 1.59 | 1.3, 1.3 | 747738730, 769333846 |
| R3HCC1 | CACCCTGG | -2C→T | 42.206 | 47.58 | 1.1 | 754555262 |
Genetic variants listed in the NCBI's dbSNP database or COSMIC. Each variant is located in the TIS sequence of a non-AUG start codon that is annotated in the NCBI RefSeq database. * The mutation was identified in previous reports, but not listed in dbSNP or COSMIC. NA - The start codon was altered and the efficiency could not be calculated. FS - The variant resulted in a frameshift.
Mutations in the c-Myc non-AUG TIS can affect expression levels
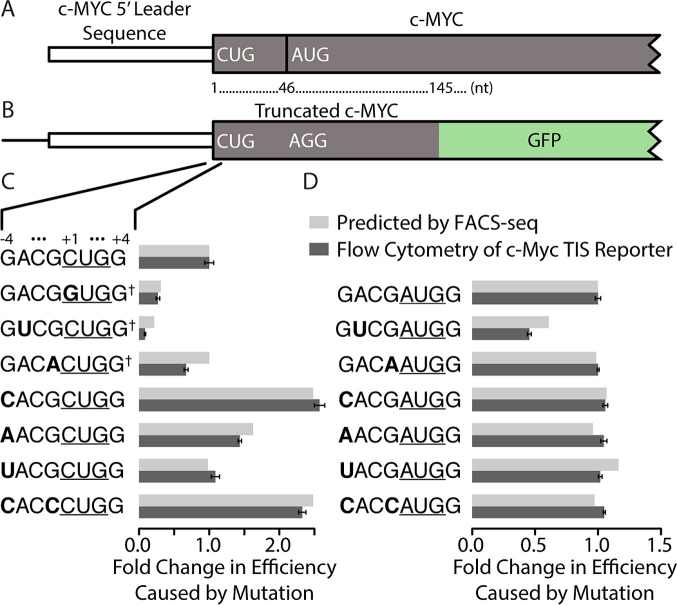
We further analyzed c-Myc, a transcription factor that regulates genes involved in cell growth, differentiation, and apoptosis (33,34), to demonstrate that the results from our FACS-seq could be used to predict changes in protein expression. c-Myc utilizes a non-AUG TIS and is misregulated in many carcinomas (11). In humans, there are two isoforms of c-Myc that are expressed from a single transcript (Figure 6A). The longer isoform initiates at a CUG codon while the shorter isoform initiates at an in-frame, downstream AUG codon (11). In some tumor samples, the N-terminally extended isoform is no longer expressed due to a loss of the first exon during chromosomal translocation (11). Interestingly, in other tumor samples the first exon is maintained, but mutations have been reported in the TIS of the N-terminal extension (35–37). Specifically, we expected a mutation in the -3 position, which converted an A into a U, and a mutation in the +1 position, which converted the CUG codon into a GUG codon, to significantly decrease protein expression levels, while a mutation in the -1 position would have only a moderate effect. To show that these mutations in the TIS may cause a decrease in the expression of the longer isoform, we constructed a reporter to measure expression from the TIS of the N-terminal extension (also used to produce results shown in Figure 4C). The downstream AUG was mutated to AGG to ensure the expression was associated with the TIS of interest. We then inserted the c-Myc 5′ leader and the first 144 nucleotides of the c-Myc coding sequence before GFP to create a fusion protein (Figure 6B).
Figure 6.
Single-base mutations in the TIS can alter protein expression of c-Myc. A) The wild type (WT) c-Myc transcript of Homo sapiens (NM_002467, not to scale). The CUG and AUG codons are the two native TISs and are separated by 45 nucleotides. B) The c-Myc-GFP reporter construct used to measure expression. The downstream AUG start codon was mutated to AGG, disabling expression of the truncated isoform. The 5′ leader sequence and the first 144 nucleotides of c-Myc were inserted into GFP-IRES-RFP reporter (Figure 1A). C, D) Expression of c-Myc-GFP measured by flow cytometry relative to expression from the WT TIS sequence with either CUG (C) or AUG (D) as the start codon. The observed relative expression is compared with the results predicted from the FACS-seq data presented in Figure 1C. Start codons are underlined and the bases that differ from the WT sequence are in bold. † following the TIS sequence denotes mutations that were previously documented in tumor samples. Error bars represent the standard deviation between experimental replicates (N = 3).
Using flow cytometry, we measured the change in expression caused by each mutation that was identified in a tumor sample. In agreement with our FACS-seq analysis using the native GFP reporter (Figure 1A), the mutations found in the TIS of c-Myc in tumor samples decreased expression (Figure 6C). The -3A→U mutation caused expression to be reduced by a factor of 10 and the +1C→G mutation reduced expression by a factor of 4. These mutations would alter the ratio between the two isoforms, which is known to affect the transcriptional activity of c-Myc and is disrupted in Burkitt's Lymphoma (14). In addition to the mutations that have been observed in tumor samples, we generated several reporters with mutations in the -4 position. Mutations at positions other than the -3 or +4 position significantly altered protein expression when the WT CUG codon was used (Figure 6C), but only had a small impact on AUG start codons (Figure 6D). This is consistent with previous reports that the nucleotides in positions other than the -3 and +4 positions had a larger impact on inefficient TIS sequences (20). These results demonstrate that the TIS efficiencies reported here can be used to predict the change in expression caused by mutations in TIS sequences and that mutations at positions other than the -3 and +4 positions can have a large impact on efficiency of non-AUG TISs.
DISCUSSION
The genetically engineered reporter is one of the fundamental tools of experimental molecular genetics. This tool allows us to determine a relationship between a genetic sequence of interest and a cellular outcome. Traditionally, genetic reporters are delivered to cells and then analyzed one at a time—a process that can be laborious and subject to experiment-to-experiment variation. This study utilized an emerging technology, FACS-seq, capable of processing a library of reporter constructs in parallel. Using this high-throughput approach to analyze a complete motif library should allow us to gauge the phenotypic behavior associated with all variants of a genetic motif (assuming it can be measured using a reporter construct). In our study, we measured the output associated with 9,216 novel motifs and we estimate that the FACS-seq method can process as many as 106 motif variants in a single experiment. Complete motif analysis generates a comprehensive database of motif sequence-phenotype relationships and can be a powerful tool in the research lab and the clinic.
In demonstrating the potential of complete motif analysis, we probed the sequence-phenotype relationship between motifs utilizing non-AUG start codons and subsequent translation initiation. Using FACS-seq, we measured the translation initiation efficiency of the 9 codons that differ from AUG by a single nucleotide plus all possible sequences surrounding the codon from the -4 to the -1 and the +4 positions. The TIS efficiencies of non-AUG start codons varied over a broad range, from undetectable levels of expression up to levels approximately half that of an AUG codon in the ‘consensus Kozak’ motif (CACCAUGG). These expression levels are higher than those reported in many previous studies (3,29,38), likely due to differences between reporters. We demonstrated that non-AUG codons are more dependent on their surrounding nucleotide sequence context than AUG codons. Base pairing between an AUG start codon and anticodon of the initiator tRNA along with interactions between the scanning ribosome and the nucleotides surrounding the start codon (e.g., the interaction between Arg55 of eukaryotic initiation factor 2α and the -3 position (39)) cause the preinitiation complex to shift from an open conformation to a closed conformation so that translation initiation can occur. Most preinitiation complexes undergo translation initiation when they encounter an AUG start codon whether it is an efficient or inefficient context because the strong interaction between the codon and anticodon provide enough energy to drive the conformational shift. However, mismatches between a non-AUG start codon and the anticodon reduce the binding energy from the codon and anticodon. Therefore, the contributions from interactions between the preinitiation complex and the ‘context nucleotides’ likely become more significant and necessary. We also showed that sequence context, specifically the +4 position, affects the efficiency of each non-AUG start codon differently. The observed differential effect of the +4 position demonstrates that sequence properties can have codon-specific effects on TIS efficiency. It is possible that there are other properties with codon-specific effects. Furthermore, differences in these properties between reporters may explain why some previous studies have identified GUG or ACG as the most efficient non-AUG start codon (2,3) while others are in agreement with our findings (29,38).
To further investigate if it was appropriate to apply our FACS-seq results to genome-wide analyses, we used publicly available ribosomal profiling data sets. By using small molecule drugs to stall ribosomes during translation initiation, ribosomal profiling experiments are able to identify TISs and other features of mRNA translation. These experiments generate large, detailed data sets that can be used to investigate mRNA translation at the genomic level. Publicly available ribosomal profiling data sets allowed us to use an independent and unrelated experimental technique to demonstrate that our results could be applied to the genome. We showed that non-AUG codons with greater efficiencies were more likely to cause translation initiation than those with lower efficiencies. Therefore, TIS efficiency is a predictor (but not the only predictor) of non-AUG codons that are utilized as TISs in the genome. In future ribosomal profiling studies, TIS efficiency could be used to identify the most likely start codon when there are multiple potential non-AUG start codons within the TIS footprint. This can help to corroborate ribosomal profiling data and distinguish between upstream open reading frames and N-terminal extensions by establishing the proper reading frame for each TIS. Additionally, in the future it will be important to better understand the other factors that influence which AUG and non-AUG start codons cause translation initiation. Understanding the effect of these factors in determining start codon selection will help us to predict all functional TISs and provide us with a more complete understanding of gene expression and proteome diversity.
In addition to helping to identify novel translation products, the TIS efficiencies that we have measured can be used to predict changes in expression levels caused by mutations near a start codon. Due to the increased sensitivity of non-AUG codons to sequence context, genes that utilize non-AUG start codons are much more susceptible to mutations in their TIS sequence. As demonstrated by our c-Myc genetic reporter experiments, mutations that would not alter the expression of proteins that use AUG start codons can greatly reduce or increase expression from non-AUG start codons. Those changes in expression levels could in turn impact cell behavior and human health, particularly when the gene is a proto-oncogene like c-Myc. Using our results as a guide, researchers and clinicians will be able to better identify potentially dangerous mutations that drive tumorigenesis or other diseases. For example, when DNA from a patient's tumor is sequenced, somatic mutations may be found near a non-AUG start codon associated with a gene suspected to have oncogenic potential. Without further information about a sequence-phenotype relationship, a disease-causing ‘driver’ mutation could be missed or a ‘passenger’ mutation might be improperly implicated. Instead, armed with a database of all motif sequence-phenotype relationships, we will have a greater ability to identify the mutations contributing to disease.
Supplementary Material
ACKNOWLEDGEMENTS
Flow cytometry measurements were performed at the Stanford Shared FACS Facility, and sorting was performed on an instrument at the Stanford Shared FACS Facility obtained using NIH S10 Shared Instrument Grant (S10RR025518–01). A.J.D. was supported by a NSF Graduate Research Fellowship.
Conflict of interest statement. None declared.
Footnotes
Present address: Clifford L Wang, Illumina, 499 Illinois St., San Francisco, CA 94158, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
REFERENCES
- 1. Tikole S., Sankararamakrishnan R.. A survey of mRNA sequences with a non-AUG start codon in RefSeq database. J. Biomol. Struct. Dyn. 2006; 24:33–41. [DOI] [PubMed] [Google Scholar]
- 2. Peabody D.S. Translation initiation at non-AUG triplets in mammalian cells. J. Biol. Chem. 1989; 264:5031–5035. [PubMed] [Google Scholar]
- 3. Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol. Cell. Biol. 1989; 9:5073–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ingolia N.T., Lareau L.F., Weissman J.S.. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011; 147:789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee S., Liu B., Lee S., Huang S.-X., Shen B., Qian S.-B.. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:14728–14729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fritsch C., Herrmann A., Nothnagel M., Szafranski K., Huse K., Schumann F., Schreiber S., Platzer M., Krawczak M., Hampe J. et al. Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res. 2012; 22:2208–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Touriol C., Bornes S., Bonnal S., Audigier S., Prats H., Prats A.-C., Vagner S.. Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol. Cell. 2003; 95:169–178. [DOI] [PubMed] [Google Scholar]
- 8. Ivanov I.P., Firth A.E., Michel A.M., Atkins J.F., Baranov P.V.. Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic. Acids. Res. 2011; 39:4220–4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Slavoff S.A., Mitchell A.J., Schwaid A.G., Cabili M.N., Ma J., Levin J.Z., Karger A.D., Budnik B.A., Rinn J.L., Saghatelian A.. Peptidomic discovery of short open reading frame–encoded peptides in human cells. Nat. Chem. Biol. 2013; 9:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hopkins B.D., Fine B., Steinbach N., Dendy M., Rapp Z., Shaw J., Pappas K., Yu J.S., Hodakoski C., Mense S. et al. A secreted PTEN phosphatase that enters cells to alter signaling and survival. Science. 2013; 341:399–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hann S.R., King M.W., Bentley D.L., Anderson C.W., Eisenman R.N.. A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt's lymphomas. Cell. 1988; 52:185–195. [DOI] [PubMed] [Google Scholar]
- 12. Carréno S., Caron E., Cougoule C., Emorine L.J., Maridonneau-Parini I.. p59Hck isoform induces F-actin reorganization to form protrusions of the plasma membrane in a Cdc42- and Rac-dependent manner. J. Biol. Chem. 2002; 277:21007–21016. [DOI] [PubMed] [Google Scholar]
- 13. Hann S.R., Dixit M., Sears R.C., Sealy L.. The alternatively initiated c-Myc proteins differentially regulate transcription through a noncanonical DNA-binding site. Genes Dev. 1994; 8:2441–2452. [DOI] [PubMed] [Google Scholar]
- 14. Batsche E., Crémisi C.. Opposite transcriptional activity between the wild-type c-myc gene coding for c-Myc1 and c-Myc2 proteins and c-Myc1 and c-Myc2 separately. Oncogene. 1999; 18:5662–5671. [DOI] [PubMed] [Google Scholar]
- 15. Ingolia N.T., Ghaemmaghami S., Newman J.R.S., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sarrazin S., Starck J., Gonnet C., Doubeikovski A., Melet F., Morle F.. Negative and translation termination-dependent positive control of FLI-1 protein synthesis by conserved overlapping 5′ upstream open reading frames in Fli-1 mRNA. Mol. Cell. Biol. 2000; 20:2959–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ivanov I.P., Loughran G., Atkins J.F.. uORFs with unusual translational start codons autoregulate expression of eukaryotic ornithine decarboxylase homologs. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:10079–10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA?. Cell. 1978; 15:1109–1123. [DOI] [PubMed] [Google Scholar]
- 19. Hinnebusch A.G., Lorsch J.R.. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb. Perspect. Biol. 2012; 4:a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986; 44:283–292. [DOI] [PubMed] [Google Scholar]
- 21. Noderer W.L., Flockhart R.J., Bhaduri A., Diaz de Arce A.J., Zhang J., Khavari P.A., Wang C.L.. Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol. Syst. Biol. 2014; 10:748–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gibson D.G., Smith H.O., Iii C.A.H., Venter J.C., Merryman C.. Chemical synthesis of the mouse mitochondrial genome. Nat. Methods. 2010; 7:901–903. [DOI] [PubMed] [Google Scholar]
- 23. Friedman J., Hastie T., Tibshirani R.. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010; 33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 24. Salis H.M., Mirsky E.A., Voigt C.A.. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009; 27:946–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Naviaux R.K., Costanzi E., Haas M., Verma I.M.. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 1996; 70:5701–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ferreira J.P., Overton K.W., Wang C.L.. Tuning gene expression with synthetic upstream open reading frames. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:11284–11289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Iwamoto M., Björklund T., Lundberg C., Kirik D., Wandless T.J.. A general chemical method to regulate protein stability in the mammalian central nervous system. Chem. Biol. 2010; 17:981–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zadeh J.N., Steenberg C.D., Bois J.S., Wolfe B.R., Pierce M.B., Khan A.R., Dirks R.M., Pierce N.A.. NUPACK: analysis and design of nucleic acid systems. J. Comput. Chem. 2011; 32:170–173. [DOI] [PubMed] [Google Scholar]
- 29. Ivanov I.P., Loughran G., Sachs M.S., Atkins J.F.. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1). Proc. Natl. Acad. Sci. U.S.A. 2010; 107:18056–18060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M. et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014; 42:D756–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Michel A.M., Andreev D.E., Baranov P.V.. Computational approach for calculating the probability of eukaryotic translation initiation from ribo-seq data that takes into account leaky scanning. BMC Bioinformatics. 2014; 15:380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carréno S., Gouze M.E., Schaak S., Emorine L.J., Maridonneau-Parini I.. Lack of palmitoylation redirects p59Hck from the plasma membrane to p61Hck-positive lysosomes. J. Biol. Chem. 2000; 275:36223–36229. [DOI] [PubMed] [Google Scholar]
- 33. Benassayag C., Montero L., Colombie N., Gallant P., Cribbs D., Morello D.. Human c-Myc isoforms differentially regulate cell growth and apoptosis in Drosophila melanogaster. Mol. Cell. Biol. 2005; 25:9897–9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hann S.R. Regulation and function of non-AUG-initiated proto-oncogenes. Biochimie. 1994; 76:880–886. [DOI] [PubMed] [Google Scholar]
- 35. Wiman K.G., Clarkson B., Hayday A.C., Saito H., Tonegawa S., Hayward W.S.. Activation of a translocated c-myc gene: role of structural alterations in the upstream region. Proc. Natl. Acad. Sci. U.S.A. 1984; 81:6798–6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hartl P., Lipp M.. Generation of a variant t(2;8) translocation of Burkitt's lymphoma by site-specific recombination via the kappa light-chain joining signals. Mol. Cell. Biol. 1987; 7:2037–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Forbes S.A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S. et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015; 43:D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Loughran G., Sachs M.S., Atkins J.F., Ivanov I.P.. Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res. 2012; 40:2898–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hussain T., Llácer J.L., Fernández I.S., Munoz A., Martin-Marcos P., Savva C.G., Lorsch J.R., Hinnebusch A.G., Ramakrishnan V.. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell. 2014; 159:597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.