Abstract
Meiotic recombination is initiated by programmed formation of DNA double strand breaks (DSBs), which are mainly formed at recombination hotspots. Meiotic DSBs require multiple proteins including the conserved protein Spo11 and its cofactors, and are influenced by chromatin structure. For example, local chromatin around hotspots directly impacts DSB formation. Moreover, DSB is proposed to occur in a higher-order chromatin architecture termed ‘axis-loop’, in which many loops protrude from cohesin-enriched axis. However, still much remains unknown about how meiotic DSBs are generated in chromatin. Here, we show that the conserved histone H2A variant H2A.Z promotes meiotic DSB formation in fission yeast. Detailed investigation revealed that H2A.Z is neither enriched around hotspots nor axis sites, and that transcript levels of DSB-promoting factors were maintained without H2A.Z. Moreover, H2A.Z appeared to be dispensable for chromatin binding of meiotic cohesin. Instead, in H2A.Z-lacking mutants, multiple proteins involved in DSB formation, such as the fission yeast Spo11 homolog and its regulators, were less associated with chromatin. Remarkably, nuclei were more compact in the absence of H2A.Z. Based on these, we propose that fission yeast H2A.Z promotes meiotic DSB formation partly through modulating chromosome architecture to enhance interaction between DSB-related proteins and cohesin-loaded chromatin.
INTRODUCTION
Meiotic homologous recombination plays essential roles in ensuring faithful chromosome segregation during the first meiotic division and conferring genetic diversity to gametes. This event is crucial for sexually reproducing organisms as its failure can result in aneuploid or even dead gametes. In order to avoid such dire consequences, meiotic recombination is strictly regulated throughout the whole process. In particular, the initiation of the recombination is tightly controlled, since it is triggered by programmed formation of DNA double strand breaks (DSBs), potentially deleterious DNA lesions. Recent studies have established that successful meiotic DSB formation relies on various factors, which are themselves meticulously regulated (1–4).
One of the most critical regulations is imposed on proteins that induce DSBs; DSBs are formed by the conserved Spo11 protein and its partner proteins (referred to as DSB proteins). These proteins, essential for DSB formation, are subject to various surveillance systems. For example, their expression is in many cases restricted only in meiosis through transcription, splicing or translation. In another example, activation of and interactions among them, which are both indispensable to DSB formation, are under control of other cellular events such as premeiotic DNA replication (5,6) or systems such as checkpoint systems (7,8). These instances argue that the DSB machinery is a central factor to ensure adequate DSB formation.
Chromatin structured DNA, the in vivo substrate of meiotic DNA breaks, is another pivotal factor for flawless execution of meiotic recombination (9,10). Notably, this recombination preferentially occurs at discrete chromosomal sites called hotspots. As chromatin is structured in multiple layers, it regulates the event at multiple levels. On one hand, local chromatin structure around hotspots can directly impact DSB formation. Several hotspot-associated traits such as, low nucleosome density and histone modifications have been reported in several organisms (11–17). How these traits lead to DSB generation has been understood in some cases (18,19), but it is noteworthy that DSBs are still observed even in the absence of these traits (13,16,20), implying that currently unidentified factors are involved. On the other hand, higher-order chromatin structure influences recombination initiation (21). Meiotic recombination is proposed to proceed in a uniquely organized chromosome architecture called the ‘axis-loop’; many loops are emanated from the axis, a cohesin-enriched proteinaceous structure and a precursor of the lateral element of the synaptonemal complex (SC). This structure has been extensively studied in budding yeast, and axis components (e.g. cohesin subunits and other proteins such as Hop1 and Red1) promote DSB formation and DSB proteins are located in axis sites (22). Importantly axis association of DSB-regulating proteins has been reported in several species (23), and therefore, this structure appears to be universally important for successful DSB formation. It should be, however, pointed out that many points are left unknown about how the axis-loop structure contributes to the event. In sum, roles of chromatin in meiotic recombination are yet to be fully understood, but high conservation of chromatin structure is expected to give us clues to comprehensively understand the in vivo mechanism of DSB formation.
A likely candidate for such chromatin-related factors is histone variants, among which is the evolutionally conserved histone H2A variant histone H2A.Z. It plays versatile roles in DNA transactions including transcription, DNA repair, anti-gene silencing and chromosome segregation by posing various and intricate effects on chromatin dynamics (24,25). At the single nucleosome level, H2A.Z-containing nucleosomes have different stability than H2A-containing ones (26). When incorporated into nucleosome arrays, H2A.Z promotes intramolecular interactions but inhibits intermolecular interactions among multiple arrays (27). These observations can suggest that H2A.Z may influence meiotic recombination. Supporting this idea, recent studies on female Arabidopsis have reported that Arp6, which is essential for H2A.Z deposition onto chromatin, controls meiosis and transcription of meiotic genes (28,29). More importantly, another study on the same but male organism has shown that H2A.Z localizes at a group of hotspots probably to regulate formation and repair of meiotic DSBs (17). However, how Arabidopsis H2A.Z controls recombination and whether the involvement is conserved in other species are still left elusive. Moreover, reverse genetics is sometimes not easy to apply for H2A.Z, since it is essential in metazoans, and is encoded by multiple genes in Arabidopsis (30).
In light of the situation, fission yeast Schizosaccharomyces pombe is suitable to study H2A.Z since the protein is encoded by the single non-essential gene pht1+. Additionally, fission yeast is one of the most popular model organisms to study meiotic recombination, and much knowledge has been accumulated. In this organism, DSB formation strictly requires Rec12 (the homolog of Spo11) along with its partner proteins (DSB proteins, namely Rec6, Rec7, Rec14, Rec15, Rec24 and Mde2). To break DNA, these DSB proteins interact with each other in a way that Mde2 bridges two ternary complexes, the catalytic DSBC (DSB catalytic core) complex composed of Rec12, Rec6 and Rec14 and the regulatory Seven-Fifteen-Twenty four (SFT) complex composed of Rec7 (Seven), Rec15 (Fifteen) and Rec24 (Twenty four) (8,31). Furthermore, previous works have unveiled an intimate connection between chromatin and recombination initiation. For instance, hotspots are associated with acetylated histone H3 lysine 9 (H3K9ac) and chromatin modifying factors activate DSB formation (11,16,32), suggesting that open local chromatin around hotspots at least partly facilitates initiation of recombination. In addition, higher-order chromatin structure also influences meiotic recombination, as a meiosis-specific architecture called linear element (LinE) has been shown to participate in recombination regulation (33). Remarkably, multiple factors related to LinE, such as cohesin, Rec10 (budding yeast Red1), Rec25, Rec27, Mug20 and Hop1, promote DSB formation (34–38). Also, previous studies have shown that many proteins involved in meiotic recombination colocalize with LinE (39). These observations imply that LinE is similar to the budding yeast axial element in terms of interacting proteins and function, although fission yeast lacks the canonical SC.
Here, we show that H2A.Z is required for the wild-type (wt) level of DSB formation in fission yeast. Our analyses also found that pht1-deleted cells are proficient at incorporating the meiotic cohesin subunit Rec8 into LinE sites, but exhibit reduced chromatin-binding of Rec10, Rec12 and Rec15. Moreover, microscopic observation unveiled that nucleus lengths are shorter in the absence of H2A.Z, suggesting that H2A.Z would alleviate chromosome compaction. Based on these findings, we speculate that fission yeast H2A.Z regulates initiation of meiotic recombination partly by modulating chromosome architecture to promote chromatin binding of proteins involved in DSB formation.
MATERIALS AND METHODS
Yeast strains
Strains used in this study are listed in Supplementary Table S1. pht1-deleted (pht1Δ) cells were constructed by replacing pht1+ open reading frame with a selection marker cassette. pht1–4KR cells (see below) were constructed by transforming a pht1Δ::ura4+ strain with pht1 DNA fragments containing appropriate mutations.
DSB detection
Chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) analyses
Chromatin immunoprecipitation-quantitative polymerase chain reaction (ChIP-qPCR) experiments were performed as described before (16,42). Cells were disrupted either by Multi-beads Shocker (Yasui Kikai) or Beads Crusher μT-12 (TAITEC). qPCR was performed with KAPA SYBR FAST qPCR Kit (KAPA Biosystems), and by either Fast Real-Time PCR system 7300 (Applied Biosystems) or Thermal Cycler Dice Real Time System II (Takara). Antibodies used were anti-FLAG M2 antibody (SIGMA), Living Colors Full-Length GFP Polyclonal Antibody (Clontech) and anti-H2A.Z (Abcam). Sequences of primers used for real-time PCR were described before (8,16), or are listed in Supplementary Table S2.
ChIP–chip and expression array analyses
ChIP–chip experiments were performed as in (16) with slight modifications. Briefly, DNA from whole-cell extracts and ChIP fractions were amplified, end-labeled and hybridized against GeneChip®S. pombe Tilling 1.0 FR Arrays (P/N 900647, Affymetrix). ChIP signals obtained with the anti-H2A.Z antibody were quantile normalized to those with DNA in whole-cell extract. Normalized signals were smoothed with a half window size of 250 bp and visualized. H2A.Z-binding sites were determined as described in (16).
The regions around the pht1 gene (chr2: 1503041–1504557), the ura4 gene (chr3: 114780–117575), 100 kb regions from each end of every chromosome, centromeres (chr1: 3752000–3792000; chr2: 1595000–1650000; chr3: 1060000–1150000) and mating-type loci (chr2: 2110000–2140000) and SPBPJ4664.02 gene (chr2: 688616–700531) were not analyzed in this study due to the difference in genotype and the low density of probes used in ChIP–chip data. Schizosaccharomyces pombe sequence information was according to the Wellcome Trust Sanger Institute (September 2004).
mRNA from h+/h− diploid cells at 1.5 and 3 h after meiosis induction was obtained by purifying total RNA with RQ1 DNase I (Promega) and Oligotex™-dT30 Super mRNA Purification Kit (TaKaRa). mRNA was converted to biotin-labeled cRNA by using Affymetrix One-Cycle cDNA synthesis kit and then hybridized to GeneChip® Yeast Genome 2.0 Array (Affymetrix). The GeneChips were scanned with GeneChip® Scanner 3000 G7 and GeneChip® Operating Software (GCOS1.4), and expression values were obtained by performing background correction, normalization and summarization with gcrma method (43) in the platform R (http://www.r-project.org) and bioconductor (https://www.bioconductor.org) (44). Transcription start sites of annotated genes were according to a previous study (45).
Microscopic observation
The nucleus lengths were measured in Rec10-GFP expressing cells undergoing horsetail movement. A set of 13 focus planes at 0.4 μm intervals were taken in every 30 s using a DeltaVision Elite microscope (GE Healthcare) with a 60× PlanApo NA 1.4 oil objective lens (Olympus). Lengths of the nuclei that were moving straight and most elongated were obtained using the Measure Distances tool in DeltaVision softWoRx (GE Healthcare). The distance between ade1 and ade8 was measured as described in (46).
Statistical analyses
Throughout the article, one-tailed Student’s t-test was applied and statistical significance is indicated as *P < 0.05, **P < 0.01 and ***P < 0.001. P-values for all analyses are listed in Supplementary Table S3.
RESULTS
Spore viability in pht1Δ cells
As an initial step to examine involvement of the histone H2A variant H2A.Z in meiosis, we measured spore viability in pht1+ and pht1Δ cells by counting colony formation efficiency of spores. The calculation indicated that >80% of spores form a colony in both cells (Supplementary Figure S1). We note, however, that pht1 deletion reduced spore viability slightly, but significantly, and that the reduction was accompanied by increase of asci containing abnormal number (i.e. 0, 1, 2 or 3) of viable spores (Supplementary Table S4). These defects could be attributable to many factors including chromosome mis-segregation or impaired DSB formation/repair, and thus suggest that H2A.Z plays a role in meiotic events.
DSB formation is compromised in pht1Δ cells
To test whether H2A.Z is involved in recombination initiation, we monitored meiotic DSBs in pht1+ cells and pht1Δ mutants. In the experiments described in the following paragraph, genomic DNA was isolated from pat1–114 rad50S haploid cells, which are convenient to analyze meiotic DSB formation, since the pat1–114 mutation enables synchronous meiosis (even in haploid cells) by temperature-shift and the rad50S mutation helps DSB detection by blocking DSB repair.
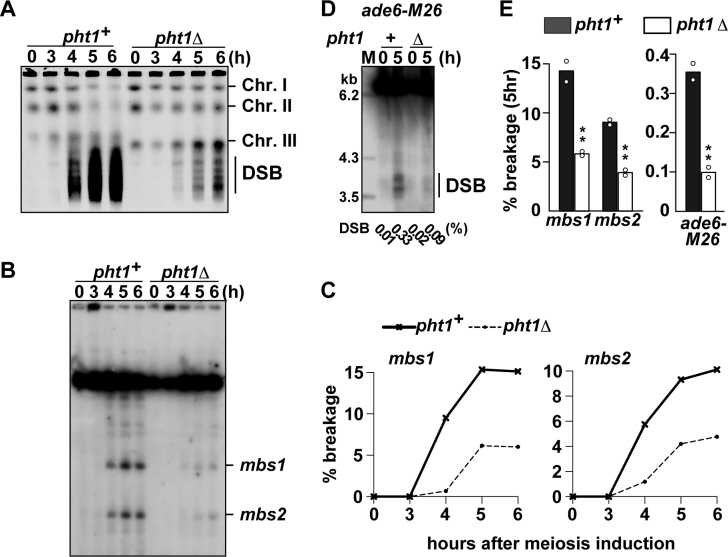
We first analyzed whole chromosomes by pulse field gel electrophoresis and ethidium bromide staining. In this analysis, the DSB frequency can be estimated by comparing signal intensities of three intact chromosomes, which do not suffer meiotic DSBs, and those of smeared fragments, which are the product of meiotic DSBs. As shown in later time-points of Figure 1A, pht1Δ cells showed weaker DSB signals and stronger intact chromosome signals than pht1+ cells, suggesting that H2A.Z facilitates DSB formation. This tendency was confirmed by Southern blotting analyses focused on several hotspots. mbs1 and mbs2, two strong hotspots in the fission yeast genome, were monitored for DSB amount for the first 6 h after meiosis induction (Figure 1B). Quantification of broken and unbroken DNA fragments demonstrated that DSB frequencies in pht1Δ cells are reduced to a third and a half of those in wt levels at mbs1 and mbs2, respectively (Figure 1C). Similar reduction was observed also at a weaker hotspot, ade6-M26 (Figure 1D). Therefore, at three hotspots we have tested, break formation was significantly and reproducibly impaired (Figure 1E). The decreased DSB was not caused by impaired progression of meiotic events, since premeiotic DNA replication and meiotic nuclear divisions were not compromised in pht1Δ cells (Supplementary Figure S2A and B).
Figure 1.
Effects of pht1 deletion on meiotic DSB formation. pat1–114 rad50S cells were induced into meiosis and were collected at indicated times after the induction. Genomic DNA was analyzed by pulse-field gel electrophoresis (A–C) or standard gel electrophoresis (D). (A) Formation of meiotic DSBs at the chromosome level. DNA was visualized with ethidium bromide. The positions of chromosomes I, II, III and smears resulting from meiotic DSBs are shown. (B) An example showing the meiotic DSBs formed at mbs1 and mbs2. DNA digested with Not I restriction endonuclease was analyzed by Southern blotting with the c227 probe recognizing the left end of the Not I J fragment. (C) Quantification of the DSBs at mbs1 and mbs2 shown in (B). These values were obtained by dividing the signal intensity of broken DNA fragments over the signal intensity of the unbroken Not I J fragment. (D) An example showing the meiotic DSBs formed at ade6-M26. DNA digested with Afl II restriction endonuclease was analyzed by Southern blotting as described in (41). Break frequencies obtained by quantification of the DSBs are indicated underneath the lanes. (E) Summary of DSB frequency quantification at hotspots shown in (B) and (D). Bar graphs are created based on mean breakage frequencies at 5 h after induction of two independent experiments (shown by open circles).
The above finding that H2A.Z promoted DSB formation was corroborated by testing DSB-associated events. The first one is pairing of homologous chromosomes. This event is followed and facilitated by DSB formation especially at chromosome arm regions (47), since broken DNA is repaired by homologous chromosomes as a template. To examine homologous pairing around DSB hotspots, the lacO repeat sequence was integrated near mbs1 and visualized by the LacI-GFP fusion protein. We found that mbs1 loci pair slowly in the absence of H2A.Z (Supplementary Figure S2C), consistent with the involvement of H2A.Z in DSB formation. We also tested production of Rec12-oligonucleotides (Rec12-oligo) complexes, which are byproducts of meiotic DSB processing and can be used to estimate total DSB formation (48). According to a previously described method (49), Rec12–oligo complexes immunoprecipitated from meiotic cell extract were radiolabeled and detected by autoradiography. Comparison between pht1+ and pht1Δ cells revealed that Rec12–oligo signals are reduced in the pht1Δ mutants (Supplementary Figure S2D), consistent with the observation that H2A.Z promotes DSB formation. These results reinforce the conclusion that histone H2A.Z promotes initiation of meiotic recombination in fission yeast.
Gene conversion and crossover frequencies in pht1Δ cells
We then tested whether lack of H2A.Z decreases meiotic recombination products as well. To this end, frequencies of intragenic (gene conversion) and intergenic recombination (crossover) were measured in cells with and without H2A.Z.
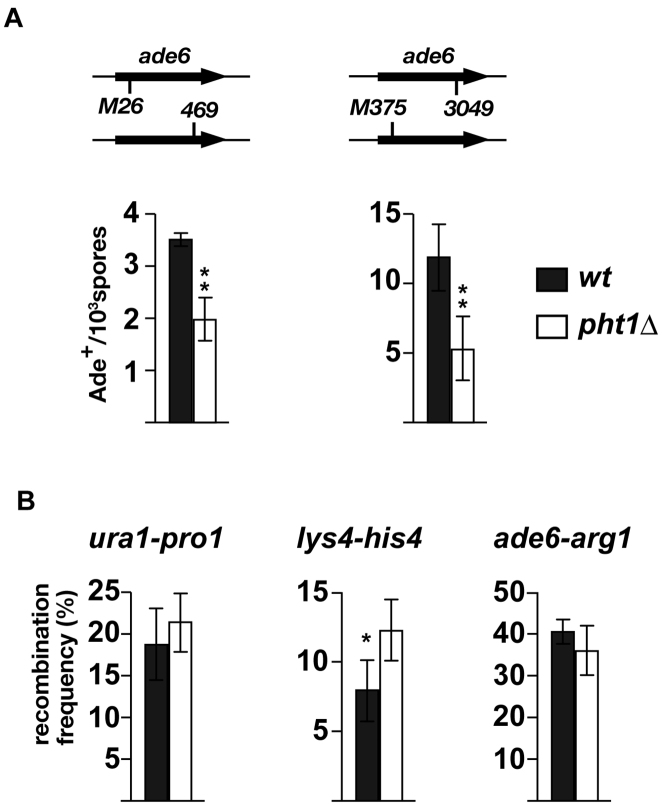
Gene conversion was examined at two hotspots located within the ade6 ORF, ade6-M26 and ade6–3049. Cells carrying either of hotspot alleles were crossed to those carrying an appropriate non-hotspot allele and produced spores were tested for adenine prototrophy. Those experiments indicated that, at both M26 and 3049 hotspots, deletion of pht1 reduced gene conversion frequency to about a half of wt levels (Figure 2A), indicating that histone H2A.Z is involved in promoting gene conversions at ade6 hotspots. Crossover frequencies in pht1+ and pht1Δ cells were compared at several intervals in the genome (one interval for each chromosome; Figure 2B). In contrast to gene conversion, pht1 deletion did not significantly affected genetic distance at ura1-pro1 and at ade6-arg1, but increased it at his4-lys4 at a statistically significant level. The biological significance of the upregulated crossover at his4-lys4 is currently unknown, but at all three intervals, pht1Δ cells maintained wt level of recombination. The stronger impacts of pht1 deletion on gene conversion than crossover could be accounted for by ‘crossover homeostasis’, which suggests that, in DSB-reduced circumstances, crossover levels are maintained at the expense of non-crossover (50,51). This hypothesis should be further verified by measuring gene conversion frequency at several other loci. Nevertheless, based on the significant reduction of gene conversion frequencies at ade6 hotspots in the absence of H2A.Z, we confirmed that H2A.Z is involved in meiotic recombination in fission yeast.
Figure 2.
Effects of pht1 deletion on meiotic recombination frequency. h+ cells and h− cells were crossed and resultant spores were tested for recombination by checking appropriate auxotrophies. Results are shown as the average of three independent experiments, and error bars indicate standard deviations. (A) Effects of pht1 deletion on gene conversion at the ade6-M26 and ade6–3049 hotspots. Recombination frequencies between ade6-M26 and ade6–469 (left), and between ade6–3049 and ade6-M375 (right) were measured. (B) Effects of pht1 deletion on crossover recombination. Recombination frequencies at the ura1-pro1 (left), lys4-his4 (middle) and ade6-arg1 (right) intervals were measured.
Neither hotspots nor axis sites are significantly enriched with H2A.Z
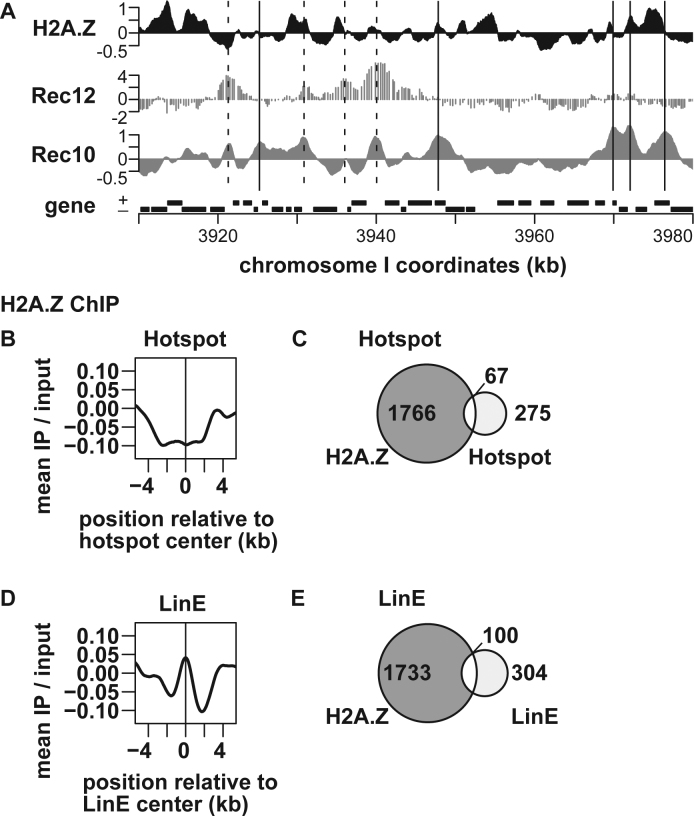
Having shown the involvement of H2A.Z in DSB formation, we sought to understand how H2A.Z regulates the event. One hypothesis is that the histone variant localizes around hotspots and/or axis sites and modifies surrounding chromatin to promote DSB formation. This possibility was tested by comprehensively examining the H2A.Z levels at hotspots and axis sites across the fission yeast genome. By combining ChIP using an anti-H2A.Z antibody and genome tiling array analysis (ChIP–chip), H2A.Z distribution was determined in pat1–114 haploid cells 4 h after meiotic induction (Figure 3A). Distribution of H2A.Z in meiotic cells, which we determined in this study and that in vegetatively growing cells, which a previous study determined, were similar to each other (Supplementary Figure S3).
Figure 3.
Genome-wide analysis of H2A.Z levels around recombination hotspots and LinE sites. pat1–114 cells were induced into meiosis and harvested at 4 h after meiosis induction. ChIP was performed using an anti-histone H2A.Z anitibody, and the resultant DNA was analyzed by genome tiling arrays. (A) An example of ChIP–chip data. The X-axis shows the chromosomal coordinates in kb and the Y-axis shows the log2 of signal strength. Locations of Rec12–DNA linkage sites (52) and Rec10-binding sites (8) are also shown for comparison. The vertical dotted and solid lines indicate hotspots and LinE sites, respectively. Genes are shown as filled boxes at the bottom of the figure. (B and D) Average distributions of H2A.Z around meiotic recombination hotspots (B) and LinE sites out of hotspots (D). The charts were created by a moving average method with a window size of 1 kb and a step size of 0.1 kb. The Y-axis shows the log2 of signal strength. (C and E) Venn diagrams showing the overlap between H2A.Z and hotspots (C), and between H2A.Z and LinE sites out of hotspots (E).
The H2A.Z levels were calculated at all hotspots that had been previously identified by ChIP–chip analysis of Rec12 covalently attached to broken DNA ends (52). As shown in Figure 3B, H2A.Z was not enriched, but was rather slightly depleted around the hotspot center. Consistently, the vast majority of hotspots were not colocalized with H2A.Z and vice versa (Figure 3C). The latter finding was confirmed with a more recent and higher-resolution hotspot map obtained by deep-sequencing oligonucleotides linked to Rec12 (53) (Supplementary Figure S4).
Also examined was association of H2A.Z with axis sites (LinE sites), which we had previously defined as Rec10-binding sites outside of DSB hotspots (8). We measured the enrichment value of H2A.Z at axis/LinE sites, but statistical analyses failed to find significant localization of the histone variant at these sites (Figure 3D). Further, H2A.Z-binding sites and LinE sites do not substantially coincide with each other either (Figure 3E). Taken together, genome-wide mapping of H2A.Z does not support the model that H2A.Z is localized at hotspots or LinE sites and regulates surrounding local chromatin structure to promote DSB formation. To confirm this conclusion, three hotspots and two LinE sites were analyzed by ChIP followed by quantitative PCR (ChIP-qPCR) of H2A.Z, which showed these sites were not particularly associated with high level of H2A.Z (Supplementary Figure S5).
pht1 deletion does not downregulate transcription of DSB-related genes
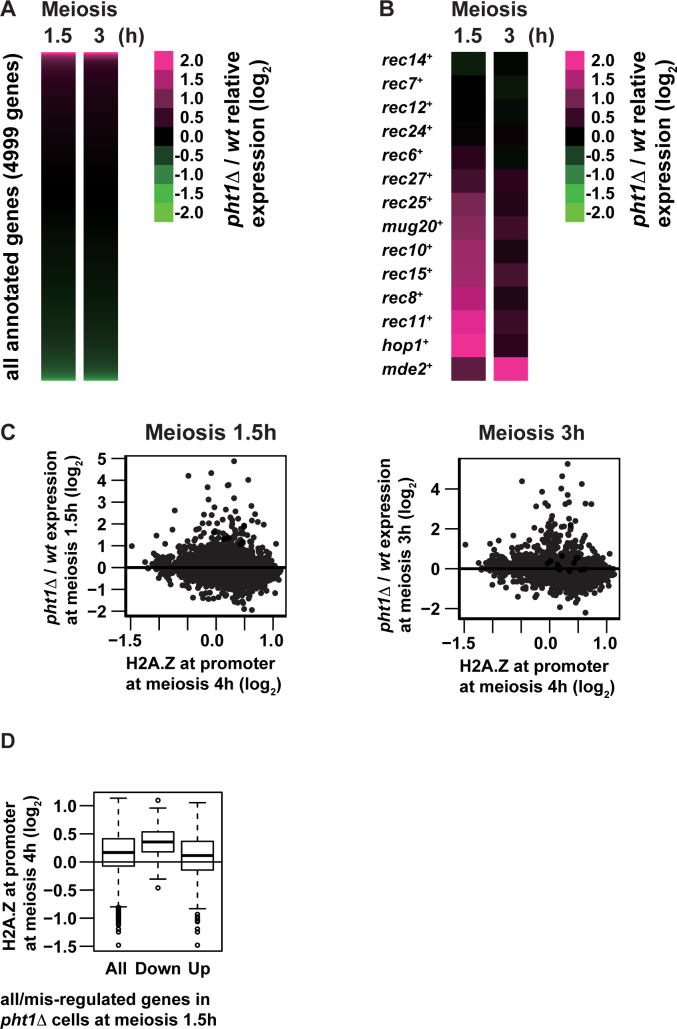
The absence of H2A.Z at hotspots and axis sites may mean that H2A.Z influences DSB formation at more global levels. From this point of view, we turned our attention to a well-known fact that H2A.Z is generally present adjacent to transcription start sites and controls transcription of a number of genes (54). Moreover, a subset of genes whose products promote DSB formation are transcriptionally induced after meiosis entry. Therefore, it is possible that H2A.Z activates transcription of genes required for recombination initiation to promote DSB formation. To verify this hypothesis, meiotic transcription profiles were compared in pht1+ and pht1Δ cells. In this experiment, meiosis was induced in h+/h− diploid cells by nitrogen withdrawal, instead of temperature-shift induction in pat1–114 cells, in order to eliminate indirect effects caused by high temperature. mRNA was purified from meiotic cells, and analyzed by gene expression microarrays displaying all annotated genes (4999 genes).
We observed that the transcriptional landscape is merely slightly affected by the absence of the pht1+ gene (Supplementary Table S5). At 1.5 h after meiosis induction, pht1 deletion significantly (i.e. >2-fold) increased mRNAs of 109 genes (2.2% of total genes), while it decreased 46 genes (0.92%) (Figure 4A, left). At 3 h, pht1 deletion resulted in higher and lower mRNA levels of 107 (2.1%) and 47 (0.94%) genes, respectively (Figure 4A, right).
Figure 4.
Transcriptome analyses of pht1+ cells and pht1Δ mutants. h+/h− diploid cells were induced into meiosis and harvested at 1.5 or 3 h after meiosis induction. mRNA was purified from meiotic pht1+ and pht1Δ cells and analyzed with microarray. (A and B) Heat maps of pht1Δ/pht1+ expression ratio (log2) for all annotated genes (4999 genes, A) and genes coding for DSB proteins and LinE components (B). Each row and column represent a gene, and hours after meiosis induction, respectively. The color scale shown on the right illustrates the log2 ratio of pht1Δ to pht1+ expression levels. Magenta and green colors represent high and low relative expression levels respectively. (C) Scatter plot of all annotated genes comparing H2A.Z levels at promoters (log2, X-axis) and pht1Δ/pht1+ expression ratio at 1.5 (left) and 3 (right) h after meiosis induction (log2Y-axis). H2A.Z levels at promoters were calculated by averaging H2A.Z ChIP–chip signals for 4 h after meiosis induction from 500 to 1 bp upstream of transcription start sites defined elsewhere (45). (D) Box-and-Whisker plots showing H2A.Z levels at promoters at 4 h after meiosis induction (log2) for all annotated genes (All), downregulated genes in pht1Δ cells (Down) and upregulated genes in pht1Δ cells (Up). H2A.Z levels at promoters were calculated as in (C).
Detailed investigation into the obtained profiles revealed two notable features. First, at 1.5 h after meiosis induction, mRNAs of several recombination-related genes were more abundant in pht1Δ mutants than in pht1+ cells (Figure 4B). Since this tendency was less overt at 3 h after induction (Figure 4B), lack of H2A.Z would accelerate transcriptional activation of recombination-related genes. Considering that meiotic DSB formation is a sophisticatedly controlled event, a subtle change in the transcription timing of involved genes could be detrimental to the event. Nevertheless, we were more interested in the other feature that mRNA levels of no genes coding for DSB proteins and LinE-related factors were less detected in pht1Δ cells at both time points (Figure 4B). Consistently, western blotting confirmed that Rec10, Rec15 and Hop1 protein levels are not altered by pht1 deletion (Supplementary Figure S6). Therefore, from our transcriptome analyses, we propose that decreased DSBs in the pht1Δ mutant would not be attributable to reduced transcript levels of DSB-promoting genes.
Lack of correlation between genomic sites bound by H2A.Z and transcriptionally altered genes by pht1 deletion
Because we observed the lack of correlation between H2A.Z-binding sites and hotspots/LinE sites (Figure 3), we wondered whether similar is the case with transcription: correlation between H2A.Z-binding sites and genes transcriptionally regulated by H2A.Z was gauged. This analysis should be performed on the same time point, but we could not chromatin-immunoprecipitate enough H2A.Z-bound DNA from cells used in expression array analyses. Therefore, results of H2A.Z ChIP–chip (Figure 3) and those of transcriptome experiments (Figure 4A and B) were compared. The comparison showed that there is no correlation between genes whose transcript levels were affected by the lack of H2A.Z and genes whose promoter regions were bound by H2A.Z (Figure 4C and D). To verify the uncorrelation, we revisited data on vegetative cells, which had been previously studied by other research groups and conducted similar analyses (55,56). The results supported our observation on meiotic cells such that no correlation was found between the two fractions (Supplementary Figure S7). These findings may indicate that the roles of H2A.Z in transcriptional control are more intricate than simply residing at a promoter to regulate the downstream gene. A possible interpretation could be that H2A.Z exerts its effects globally on overall chromosomes, rather than limitedly on local chromatin around H2A.Z-binding sites.
Lack of H2A.Z does not compromise chromatin binding of the Rec8 cohesin subunit
The results explained in the preceding section led us to consider a possibility that H2A.Z impacts meiotic DSB formation through higher-order chromosome structure, rather than through local chromatin around hotspots/LinE sites, to facilitate DSB formation. In this regard, LinE is a meiosis-specific higher-order chromatin structure in fission yeast. We therefore investigated its integrity in pht1Δ cells by examining chromatin binding of several LinE-associated factors.
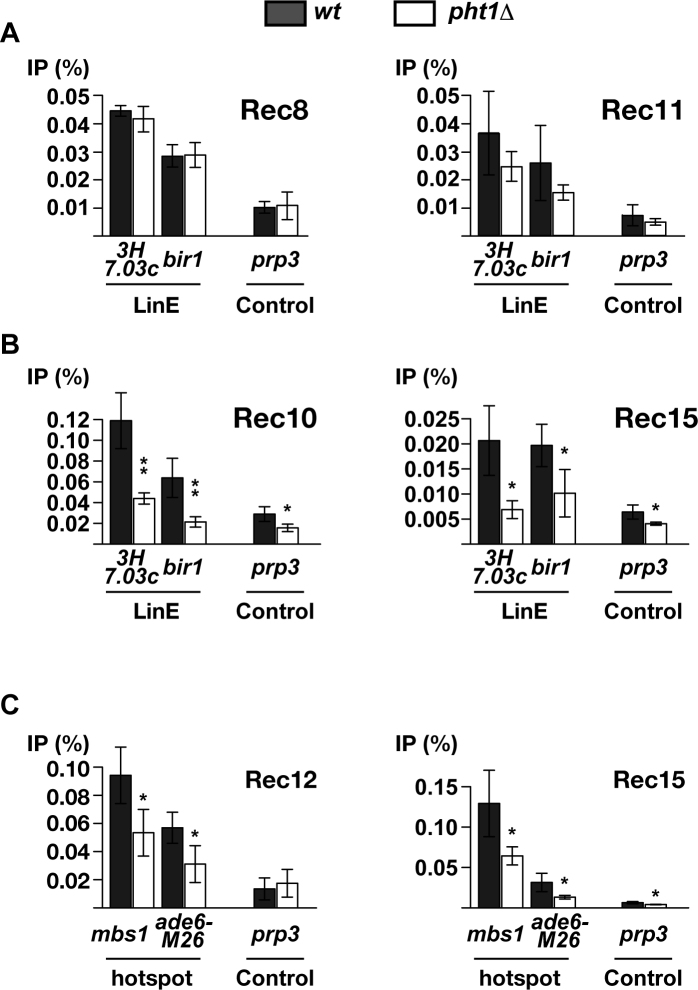
Cohesin stably binds to chromatin during premeiotic DNA synthesis, and is required for LinE assembly. Among several subunits, the kleisin subunit Rec8 and the SA subunit Rec11 have been shown to promote meiotic DSB formation (34). To assess their interaction with chromatin, ChIP of epitope-tagged proteins (Rec8-HA and Rec11-GFP) were performed using relevant antibodies, and DNA was analyzed by quantitative PCR directed at two previously identified LinE sites, namely bir1 and SPBC3H7.03c (8), as well as the control site prp3. The experiments were done in pat1–114 haploid cells that were crosslinked with formaldehyde 4 h after the meiosis induction. As shown in Figure 5A, Rec8 protein level at LinE sites were not affected at all by deletion of pht1. We observed a slight reduction of Rec11 binding at the sites in pht1Δ, but the difference from pht1+ cells was statistically insignificant. Accordingly, pht1Δ cells are likely to be proficient at incorporating meiotic cohesin, in particular Rec8, into appropriate chromosomal positions.
Figure 5.
ChIP-qPCR analyses of cohesin subunits and LinE/DSB proteins in pht1+ cells and pht1Δ mutants. pat1–114 cells expressing an epitope-tagged protein (Rec8-HA, Rec11-GFP, Rec10-FLAG or Rec15-FLAG) and pat1–114 rad50S cells expressing Rec12-FLAG were induced into meiosis and crosslinked with formaldehyde at 4 and 5 h after meiosis induction, respectively. Protein-associated DNA fragments were recovered from cell extracts by ChIP and analyzed by quantitative PCR. Results are shown as the average of multiple independent experiments (3× for Rec8, Rec10 and Rec15; 4× for Rec11 and Rec12) and error bars indicate standard deviations. (A) LinE binding of cohesin subunits in pht1+ and pht1Δ cells. Rec8-HA (left) and Rec11-GFP (right) proteins were assessed for binding to LinE sites (SPBC3H7.03c (3H7.03c) and bir1) as well as a control site (prp3). (B) LinE binding of Rec10 and Rec15 in pht1+ and pht1Δ cells. Rec10-FLAG (left) and Rec15-FLAG (right) proteins were analyzed as in (A). (C) Hotspot binding of DSB proteins in pht1+ and pht1Δ cells. Rec12-FLAG (left) and Rec15-FLAG (right) proteins were assessed for binding to hotspots (mbs1 and ade6-M26) as well as a control site (prp3).
Lack of H2A.Z compromises LinE development
Once bound to chromatin, cohesin subsequently recruits the LinE protein Rec10 (57,58) to promote assembly of LinE. As Rec10 physically interacts with other proteins involved in DSB formation, LinE development help these proteins bind to axis sites. For example, Rec15, one of the Rec12 partner proteins essential for DSB formation, is brought to axis by Rec10 (8). To evaluate LinE development in pht1Δ cells, Rec10 and Rec15 levels at LinE sites were examined by applying the above ChIP method to Rec10-FLAG or Rec15-FLAG expressing strains, respectively. The results demonstrated that lack of H2A.Z partially impaired both Rec10 (Figure 5B and Supplementary Figure S8A) and Rec15 (Figure 5B and Supplementary Figure S8B) levels at bir1, SPBC3H7.03c and five other axis sites.
Altogether, our results show that H2A.Z is required for accumulation of both Rec10 and Rec15 at axis sites. In addition, we also noticed that pht1 deletion reduces Rec10 level at two control sites and Rec15 level at one control site in a statistically significant manner. This tendency could be because compromised Rec10/15 binding to appropriate sites lowers experimental background as well, and/or H2A.Z influences weak non-specific chromatin binding of these factors. In any case, it is conceivable that LinE would be inefficiently developed, or defective in the pht1Δ mutant, and we speculate that reduced DSBs in the mutant can be ascribed to incomplete LinE development.
H2A.Z is required for efficient hotspot binding of DSB proteins
Several DSB proteins bind to hotspots, and such protein–hotspot interaction must be important for successful DSB formation. Although how they are recruited to hotspots is not fully understood, the compromised LinE binding of Rec10 and Rec15 prompted us to test whether hotspot-associated DSB proteins might be delocalized as well, by ChIP experiments.
Rec12, which breaks DNA, is mainly found at hotspots. To compare hotspot binding of Rec12 in pht1+ and pht1Δ cells, ChIP of Rec12-FLAG was carried out to analyze seven hotspots, including the well-characterized and -known ade6-M26 and mbs1 hotspots. The ChIP efficiency at all seven hotspots was reduced in pht1Δ mutants (Figure 5C and Supplementary Figure S8C), similar to the case of Rec10 at LinE sites. The same experiments were conducted with Rec15, which has been shown to bind to hotspots partly independently of Rec10 (8). Notably, Rec15-FLAG was less associated with five of seven hotspots in H2A.Z-lacking cells (Figure 5C and Supplementary Figure S8D). At the other two hotspots, Rec15 localization was either not affected, or mildly and insignificantly decreased, possibly implying a distinct hotspot binding mechanism than Rec12. These experiments argue that H2A.Z is also required for proper binding of Rec12 and its partner proteins to hotspots, leading us to infer that compromised interaction of DSB proteins with hotspots may be another reason for DSB reduction in pht1Δ cells.
Results shown in Figure 5 collectively suggest that H2A.Z is required for efficient chromatin binding of Rec10, Rec15 and Rec12, but not of cohesin subunits. Remarkably, this notion was ascertained by live observation of meiotic prophase nuclei, as signal intensity of Rec10-GFP, but not Rec8-GFP was reduced by pht1 deletion (Supplementary Figure S9). We therefore propose that one of the plausible roles of H2A.Z is to facilitate loading of proteins that are involved in DSB formation onto cohesin-bound chromatin.
H2A.Z antagonizes chromosome compaction
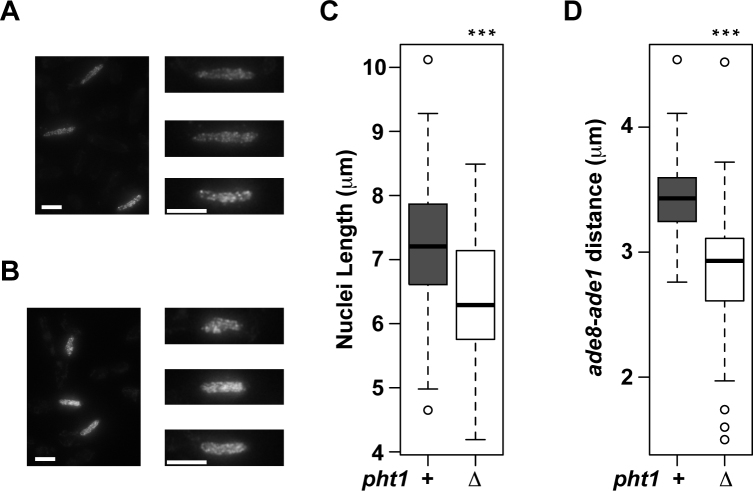
We wished to understand a possible reason of impaired chromatin binding of Rec10, Rec12 and Rec15 in the pht1 mutant. In this regard, it is known that Rec8 compacts meiotic chromosomes (46,59), and one of feasible models would be that H2A.Z may antagonize cohesin-mediated chromosome compaction to promote interaction of chromatin and DSB-related proteins. To test this model, we measured the length of horsetail nuclei, which were visualized by Rec10-GFP (Note that lack of Rec10 does not affect chromosome compaction (46)). Analyses of >80 nuclei for each genotype revealed that the nuclear length was longer in pht1+ cells than in pht1Δ cells (Figure 6A–C), suggesting that chromosomes in pht1Δ cells are more compact than pht1+ cells. Such compaction of pht1Δ nuclei was substantiated by measuring the distance between ade8 and ade1 loci in chromosome II; the distance was significantly shorter in the absence of H2A.Z (Figure 6D). These findings support the above hypothesis that H2A.Z may mitigate chromosome compaction to facilitate chromatin association of Rec10, Rec12 and Rec15, and assemble chromosomes favorable to DSB formation.
Figure 6.
Nucleus is more compact in pht1Δ cells than in pht1+ cells. (A–C) pht1+ and pht1Δ cells expressing Rec10-GFP were induced to enter meiosis by nitrogen withdrawal. Cells undergoing horsetail movement were selected and their nucleus lengths were measured. (A and B) (Left panels) A maximum-intensity projection image of pht1+ (A) and pht1Δ(B) cells. The scale bars are 5 μm. (Right panels) Nuclei shown in the left panels are magnified and aligned. The scale bars are 5 μm. Note that these images are maximum-intensity projections and do not reflect decreased Rec10-GFP signal intensity in pht1Δ cells (Supplementary Figure S9B), which was revealed by summing signal intensities of all focal panels. (C) A box plot summarizing the observation of 96 (pht1+) and 83 (pht1Δ) nuclei. (D) The distance between ade1 and ade8 loci in pht1+ and pht1Δ horsetail nuclei was measured as described before (46). A box plot summarizing the observation of 47 (pht1+) and 49 (pht1Δ) nuclei is shown.
DISCUSSION
This study showed that histone H2A.Z promotes initiation of meiotic recombination in fission yeast (Figures 1 and 2). Several follow up experiments were performed in order to understand its underlying mechanisms, and possible scenarios were examined.
Absence of H2A.Z around hotspots and axis sites
It is possible that H2A.Z occupies hotspots and/or axis sites to directly regulate the reaction, for example by decondensing surrounding chromatin or recruiting proteins involved in DSB formation. This model is reasonable as H2A.Z containing nucleosomes are, in some cases, labile and tend to create open chromatin that is preferable for recombination. It is consistent with, especially at hotspots, a previous study on male Arabidopsis. In this organism, H2A.Z is enriched around crossover hotspots and the H2A.Z-deposition mutant arp6 exhibits decreased number of Rad51 and DMC1 foci, indicating that DSB formation and processing are impaired (17).
Our comprehensive analysis in fission yeast, however, pointed to a contrasting landscape, where the H2A.Z level is not elevated around hotspots (Figure 3B and D). Such difference may imply distinct recombination initiation mechanisms between Arabidopsis and S. pombe. Alternatively, it may just reflect their different hotspot distribution patterns; hotspots in Arabidopsis are often localized near transcription start sites, which are enriched with H2A.Z, while those in fission yeast are not. In either case, fission yeast H2A.Z would have nominal influence on local chromatin around hotspots.
Likewise, our genome-wide analyses also found that LinE sites are not associated with H2A.Z either (Figure 3C and E). Therefore, although it could be possible that H2A.Z binds to hotspots/LinE sites very transiently, our present results do not support the model that H2A.Z stably resides at these sites to induce DSBs. They do not imply either that roles of H2A.Z are mediated by strong binding to hotspot-associated proteins such as Rec12 or LinE-associated proteins such as Rec10. Rather, we favor a view that it acts in a more global manner.
Possible roles of H2A.Z in DSB formation: controlling the meiotic transcription program
Because H2A.Z has been well known to regulate transcription of many genes, it is possible that pht1 deletion downregulates transcription of DSB-related genes, thereby inhibiting DSB formation. But, our microarray analysis demonstrated that pht1 deletion only mildly affects the transcription profile, as far as we tested (Figure 4A). More importantly, lack of H2A.Z did not reduce mRNAs of any genes that are known to promote DSB formation (Figure 4B) and abundance of several proteins involved in the event (Supplementary Figure S6). Based on these findings, we surmise that defective DSBs in pht1Δ cells would not be a consequence of impaired transcription of DSB-related genes.
However, our expression array results should be interpreted with two caveats. First, genes that are not currently perceived to promote the process may be transcriptionally compromised in pht1Δ cells. Second, lack of H2A.Z accelerated the transcription of several recombination-related genes (Figure 4B). Of these, the latter point is reminiscent of a previous study on female Arabidopsis showing that, in ovules, arp6 mutations increase expression of meiotic recombination genes and result in defective meiosis (29). It is hence conceivable that, in pht1Δ fission yeast cells, mis-regulated transcriptional activation of recombination factors could impair DSB formation. Further studies are necessary to address these points.
Possible roles of H2A.Z in DSB formation: chromatin binding of DSB/LinE proteins
Comparing H2A.Z ChIP–chip analyses and expression array analyses, we noticed that H2A.Z may have different targets for transcriptional regulation and for promoter binding (Figure 4C and D). One possible interpretation of this uncorrelation is that H2A.Z regulates transcription of various genes without binding to their promoters. If this is the case, it may be similar to the case of meiotic DSB formation in that H2A.Z controls chromosomal events (i.e. transcription and DSB formation) without occupying important chromosomal sites (i.e. promoters and hotspots/LinE sites). Such tendencies could be understood, if H2A.Z influences overall chromosome structure.
Indeed, H2A.Z is functionally linked to cohesin and condensin at least in vegetative cells (60–62), hinting the possibility that H2A.Z regulates DSB formation through these complexes. However, results shown in Figure 5A indicate that cohesin levels were not significantly altered in pht1Δ cells at two LinE sites examined and our preliminary experiments revealed that a condensin mutant produced the wt level of Rec12–oligo complexes (our unpublished observation). Therefore, although we have not comprehensively tested subunits of the complexes, roles of H2A.Z in DSB promotion may not be mediated by cohesin and condensin.
Instead, an alternative mechanism is suggested by our findings: unlike cohesin subunits, several LinE and DSB proteins were less associated with chromosomes (Figure 5B and C; Supplementary Figure S8). These observations point to a model that H2A.Z would help DSB-related proteins bind to cohesin-loaded chromosomes. Then how does H2A.Z promote interaction between these proteins and chromatin? A clue to solve this question may be the compaction of chromosomes. It has been reported that cohesin forms the core of chromosome axis and compacts meiotic chromosomes (46). Furthermore, during meiotic prophase, homologous chromosomes are at some sites juxtaposed to each other, possibly causing chromosome entanglement. In fact, our microscopic observation showed horsetail nuclei are longer in pht1+ cells than pht1Δ cells, indicating that H2A.Z makes nuclei less compact (Figure 6). Therefore, H2A.Z may counteract chromosome compaction and/or entanglement to deliver LinE proteins to cohesin-binding sites as well as DSB proteins to hotspots.
Such role may be similar to the one proposed for acetylated histones (63,64). In this regard, we observed that mutating four known acetylatable lysines of H2A.Z (55,60) to arginines (pht1–4KR) causes several defects (Supplementary Figure S10); pht1–4KR mutation compromised DSB formation at the chromosome level and gene conversion frequency at ade6-M26 as much as pht1Δ, and reduced Rec10 levels at LinE sites more severely than pht1Δ. These results may stem from decreased H2A.Z protein abundance in pht1–4KR (Supplementary Figure S10C) (62), and further investigation of H2A.Z acetylation will be interesting to test this point.
Moreover, our idea can be supported by several previous reports. For example, an in vitro study using nucleosome arrays has shown that H2A.Z inhibits the formation of highly condensed chromatin fibers (27). Another study on fission yeast has reported that acetylated H2A.Z prevents chromosome entanglement, although it is proposed in this case that H2A.Z exerts its effects by retaining condensin to chromosomes until mitotic anaphase (60). Taken altogether, we infer that (acetylated) H2A.Z may have an intrinsic property to alleviate compaction of overall chromosome structure, which may be required to assemble DSB-competent chromosomes. To our knowledge, this model has not been proposed or discussed in terms of meiotic DSB formation, and should be more thoroughly explored.
H2A.Z and meiotic recombination in other species
Considering the high conservation and versatility of the histone H2A.Z protein, it would be likely that other species engages H2A.Z in controlling meiotic DSB formation. Previous studies on various organisms allow us to infer a couple of possibilities.
First, as proposed in Arabidopsis, H2A.Z can reside around hotspots/axis sites and be directly involved in recombination initiation. This scenario may be the case with a number of species including budding yeast, birds, and dogs, since their hotspots colocalize with transcription start sites, which are enriched with H2A.Z. Second, H2A.Z may modulate chromatin architecture to assemble DSB-susceptible chromosomes, as this study proposes for fission yeast (Figures 5 and 6). This idea is worth testing, given that binding of recombination-related proteins to axis has been reported in multiple model organisms (22,23). In particular, for higher organisms that have larger chromosome size and large tracts of heterochromatin, it is imperative to prevent abnormal compaction of chromatin for successful loading of axis/DSB proteins onto meiotic chromosomes, and H2A.Z may be more strictly required.
As H2A.Z is one of the most important components of chromatin, studying how it functions in meiotic recombination is essential to understand its whole process. While how chromatin influences meiotic DSB formation varies among species (for example, local chromatin structure around hotspots), H2A.Z, a highly conserved protein, may play universal roles. Fission yeast is an excellent model to study H2A.Z, and knowledge obtained from this study as well as future ones can illuminate conserved aspects of meiotic recombination.
ACCESSION NUMBERS
The ChIP–chip data and expression data were submitted to Gene Expression omnibus (GEO) with accession number GSE81777.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Drs Gerald Smith, Yoshinori Watanabe and Takeshi Sakuno, NBRP and Mr Yuto Sumitani for strains, Ms Emi Takaya for technical help, and Drs Katsuhiko Shirahige and Yuki Kato for allowing us to use their experimental apparatus. We thank Drs Akira Shinohara, Takeshi Sakuno, Tomoichiro Miyoshi and Ryo Kariyazono for helpful advice, Dr Ed Luk for comments on the manuscript and Dr Matt Neale for sharing unpublished observation.
Footnotes
Present addresses:
Shintaro Yamada, Memorial Sloan Kettering Cancer Center, NY 10065, USA.
Kazuto Kugou, Kazusa DNA Research Institute, Chiba 292-0818, Japan.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
KAKENHI [26440186 to T.Y., 3114003, 212414946, 26291018 to K.O., JP17H01444, JP16H01309 to Y.H.]; Japan Society for the Promotion of Science Research Fellowship for Young Scientists [10J08626 to S.Y.]; MEXT (to K.O.); Japan Agency for Medical Research and Development (to K.O.). Funding for open access charge: KAKENHI [26440186].
Conflict of interest statement. None declared.
REFERENCES
- 1. de Massy B. Initiation of meiotic recombination: how and where? conservation and specificities among eukaryotes. Annu. Rev. Genet. 2013; 47:563–599. [DOI] [PubMed] [Google Scholar]
- 2. Lam I., Keeney S.. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb. Perspect. Biol. 2015; 7:a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Keeney S., Lange J., Mohibullah N.. Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annu. Rev. Genet. 2014; 48:187–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hunter N. Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol. 2015; 7:a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Murakami H., Keeney S.. Temporospatial coordination of meiotic DNA replication and recombination via DDK recruitment to replisomes. Cell. 2014; 158:861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu P.Y., Nurse P.. Replication origin selection regulates the distribution of meiotic recombination. Mol. Cell. 2014; 53:655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tonami Y., Murakami H., Shirahige K., Nakanishi M.. A checkpoint control linking meiotic S phase and recombination initiation in fission yeast. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:5797–5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miyoshi T., Ito M., Kugou K., Yamada S., Furuichi M., Oda A., Yamada T., Hirota K., Masai H., Ohta K.. A central coupler for recombination initiation linking chromosome architecture to S phase checkpoint. Mol. Cell. 2012; 47:722–733. [DOI] [PubMed] [Google Scholar]
- 9. Lichten M. Meiotic Chromatin: the Substrate for Recombination Initiation. 2008; Berlin: Springer. [Google Scholar]
- 10. Yamada T., Ohta K.. Initiation of meiotic recombination in chromatin structure. J. Biochem. 2013; 154:107–114. [DOI] [PubMed] [Google Scholar]
- 11. Yamada T., Mizuno K., Hirota K., Kon N., Wahls W.P., Hartsuiker E., Murofushi H., Shibata T., Ohta K.. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J. 2004; 23:1792–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pan J., Sasaki M., Kniewel R., Murakami H., Blitzblau H.G., Tischfield S.E., Zhu X., Neale M.J., Jasin M., Socci N.D. et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011; 144:719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Borde V., Robine N., Lin W., Bonfils S., Geli V., Nicolas A.. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009; 28:99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smagulova F., Gregoretti I.V., Brick K., Khil P., Camerini-Otero R.D., Petukhova G.V.. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011; 472:375–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Castro E., Soriano I., Marin L., Serrano R., Quintales L., Antequera F.. Nucleosomal organization of replication origins and meiotic recombination hotspots in fission yeast. EMBO J. 2012; 31:124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yamada S., Ohta K., Yamada T.. Acetylated Histone H3K9 is associated with meiotic recombination hotspots, and plays a role in recombination redundantly with other factors including the H3K4 methylase Set1 in fission yeast. Nucleic Acids Res. 2013; 41:3504–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Choi K., Zhao X., Kelly K.A., Venn O., Higgins J.D., Yelina N.E., Hardcastle T.J., Ziolkowski P.A., Copenhaver G.P., Franklin F.C. et al. Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat. Genet. 2013; 45:1327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Acquaviva L., Szekvolgyi L., Dichtl B., Dichtl B.S., de La Roche Saint Andre C., Nicolas A., Geli V.. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science. 2013; 339:215–218. [DOI] [PubMed] [Google Scholar]
- 19. Sommermeyer V., Beneut C., Chaplais E., Serrentino M.E., Borde V.. Spp1, a member of the Set1 complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol. Cell. 2013; 49:43–54. [DOI] [PubMed] [Google Scholar]
- 20. Brick K., Smagulova F., Khil P., Camerini-Otero R.D., Petukhova G.V.. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012; 485:642–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Borde V., de Massy B.. Programmed induction of DNA double strand breaks during meiosis: setting up communication between DNA and the chromosome structure. Curr. Opin. Genet. Dev. 2013; 23:147–155. [DOI] [PubMed] [Google Scholar]
- 22. Panizza S., Mendoza M.A., Berlinger M., Huang L., Nicolas A., Shirahige K., Klein F.. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell. 2011; 146:372–383. [DOI] [PubMed] [Google Scholar]
- 23. Kumar R., Bourbon H.M., de Massy B.. Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev. 2010; 24:1266–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Santisteban M.S., Kalashnikova T., Smith M.M.. Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000; 103:411–422. [DOI] [PubMed] [Google Scholar]
- 25. Kalocsay M., Hiller N.J., Jentsch S.. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol. Cell. 2009; 33:335–343. [DOI] [PubMed] [Google Scholar]
- 26. Zlatanova J., Thakar A.. H2A.Z: view from the top. Structure. 2008; 16:166–179. [DOI] [PubMed] [Google Scholar]
- 27. Fan J.Y., Gordon F., Luger K., Hansen J.C., Tremethick D.J.. The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat. Struct. Biol. 2002; 9:172–176. [DOI] [PubMed] [Google Scholar]
- 28. Rosa M., Von Harder M., Cigliano R.A., Schlogelhofer P., Mittelsten Scheid O.. The Arabidopsis SWR1 chromatin-remodeling complex is important for DNA repair, somatic recombination, and meiosis. Plant Cell. 2013; 25:1990–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qin Y., Zhao L., Skaggs M.I., Andreuzza S., Tsukamoto T., Panoli A., Wallace K.N., Smith S., Siddiqi I., Yang Z. et al. Actin-related protein6 regulates female meiosis by modulating meiotic gene expression in Arabidopsis. Plant Cell. 2014; 26:1612–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. March-Diaz R., Reyes J.C.. The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Mol. Plant. 2009; 2:565–577. [DOI] [PubMed] [Google Scholar]
- 31. Steiner S., Kohli J., Ludin K.. Functional interactions among members of the meiotic initiation complex in fission yeast. Curr. Genet. 2010; 56:237–249. [DOI] [PubMed] [Google Scholar]
- 32. Yamada S., Okamura M., Oda A., Murakami H., Ohta K., Yamada T.. Correlation of meiotic DSB formation and transcription initiation around fission Yeast recombination hotspots. Genetics. 2017; 206:801–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Loidl J. S. pombe linear elements: the modest cousins of synaptonemal complexes. Chromosoma. 2006; 115:260–271. [DOI] [PubMed] [Google Scholar]
- 34. Ellermeier C., Smith G.R.. Cohesins are required for meiotic DNA breakage and recombination in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:10952–10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Latypov V., Rothenberg M., Lorenz A., Octobre G., Csutak O., Lehmann E., Loidl J., Kohli J.. Roles of Hop1 and Mek1 in meiotic chromosome pairing and recombination partner choice in Schizosaccharomyces pombe. Mol. Cell. Biol. 2010; 30:1570–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Davis L., Rozalen A.E., Moreno S., Smith G.R., Martin-Castellanos C.. Rec25 and Rec27, novel linear-element components, link cohesin to meiotic DNA breakage and recombination. Curr. Biol. 2008; 18:849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Estreicher A., Lorenz A., Loidl J.. Mug20, a novel protein associated with linear elements in fission yeast meiosis. Curr. Genet. 2012; 58:119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fowler K.R., Gutierrez-Velasco S., Martin-Castellanos C., Smith G.R.. Protein determinants of meiotic DNA break hot spots. Mol. Cell. 2013; 49:983–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lorenz A., Estreicher A., Kohli J., Loidl J.. Meiotic recombination proteins localize to linear elements in Schizosaccharomyces pombe. Chromosoma. 2006; 115:330–340. [DOI] [PubMed] [Google Scholar]
- 40. Young J.A., Schreckhise R.W., Steiner W.W., Smith G.R.. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol. Cell. 2002; 9:253–263. [DOI] [PubMed] [Google Scholar]
- 41. Steiner W.W., Schreckhise R.W., Smith G.R.. Meiotic DNA breaks at the S. pombe recombination hot spot M26. Mol. Cell. 2002; 9:847–855. [DOI] [PubMed] [Google Scholar]
- 42. Oda A., Takemata N., Hirata Y., Miyoshi T., Suzuki Y., Sugano S., Ohta K.. Dynamic transition of transcription and chromatin landscape during fission yeast adaptation to glucose starvation. Genes Cells. 2015; 20:392–407. [DOI] [PubMed] [Google Scholar]
- 43. Wu Z., Irizarry R.A.. Preprocessing of oligonucleotide array data. Nat. Biotechnol. 2004; 22:656–658. [DOI] [PubMed] [Google Scholar]
- 44. Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004; 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rhind N., Chen Z., Yassour M., Thompson D.A., Haas B.J., Habib N., Wapinski I., Roy S., Lin M.F., Heiman D.I. et al. Comparative functional genomics of the fission yeasts. Science. 2011; 332:930–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ding D.Q., Sakurai N., Katou Y., Itoh T., Shirahige K., Haraguchi T., Hiraoka Y.. Meiotic cohesins modulate chromosome compaction during meiotic prophase in fission yeast. J. Cell Biol. 2006; 174:499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ding D.Q., Yamamoto A., Haraguchi T., Hiraoka Y.. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev. Cell. 2004; 6:329–341. [DOI] [PubMed] [Google Scholar]
- 48. Neale M.J., Keeney S.. End-labeling and analysis of Spo11-oligonucleotide complexes in Saccharomyces cerevisiae. Methods Mol. Biol. 2009; 557:183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Milman N., Higuchi E., Smith G.R.. Meiotic DNA double-strand break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes. Mol. Cell Biol. 2009; 29:5998–6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Martini E., Diaz R.L., Hunter N., Keeney S.. Crossover homeostasis in yeast meiosis. Cell. 2006; 126:285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kan F., Davidson M.K., Wahls W.P.. Meiotic recombination protein Rec12: functional conservation, crossover homeostasis and early crossover/non-crossover decision. Nucleic Acids Res. 2011; 39:1460–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hyppa R.W., Cromie G.A., Smith G.R.. Indistinguishable landscapes of meiotic DNA breaks in rad50+ and rad50S strains of fission yeast revealed by a novel rad50+ recombination intermediate. PLoS Genet. 2008; 4:e1000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fowler K.R., Sasaki M., Milman N., Keeney S., Smith G.R.. Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Res. 2014; 24:1650–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang H.Y., Roberts D.N., Cairns B.R.. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005; 123:219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Buchanan L., Durand-Dubief M., Roguev A., Sakalar C., Wilhelm B., Stralfors A., Shevchenko A., Aasland R., Shevchenko A., Ekwall K. et al. The Schizosaccharomyces pombe JmjC-protein, Msc1, prevents H2A.Z localization in centromeric and subtelomeric chromatin domains. PLoS Genet. 2009; 5:e1000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Anders A., Watt S., Bahler J., Sawin K.E.. Improved tools for efficient mapping of fission yeast genes: identification of microtubule nucleation modifier mod22-1 as an allele of chromatin-remodelling factor gene swr1. Yeast. 2008; 25:913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sakuno T., Watanabe Y.. Phosphorylation of cohesin Rec11/SA3 by casein kinase 1 promotes homologous recombination by assembling the meiotic chromosome axis. Dev. Cell. 2015; 32:220–230. [DOI] [PubMed] [Google Scholar]
- 58. Phadnis N., Cipak L., Polakova S., Hyppa R.W., Cipakova I., Anrather D., Karvaiova L., Mechtler K., Smith G.R., Gregan J.. Casein kinase 1 and phosphorylation of cohesin subunit Rec11 (SA3) promote meiotic recombination through linear element formation. PLoS Genet. 2015; 11:e1005225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ding D.Q., Matsuda A., Okamasa K., Nagahama Y., Haraguchi T., Hiraoka Y.. Meiotic cohesin-based chromosome structure is essential for homologous chromosome pairing in Schizosaccharomyces pombe. Chromosoma. 2016; 125:205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kim H.S., Vanoosthuyse V., Fillingham J., Roguev A., Watt S., Kislinger T., Treyer A., Carpenter L.R., Bennett C.S., Emili A. et al. An acetylated form of histone H2A.Z regulates chromosome architecture in Schizosaccharomyces pombe. Nat. Struct. Mol. Biol. 2009; 16:1286–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tada K., Susumu H., Sakuno T., Watanabe Y.. Condensin association with histone H2A shapes mitotic chromosomes. Nature. 2011; 474:477–483. [DOI] [PubMed] [Google Scholar]
- 62. Tapia-Alveal C., Lin S.J., Yeoh A., Jabado O.J., O’Connell M.J.. H2A.Z-dependent regulation of cohesin dynamics on chromosome arms. Mol. Cell. Biol. 2014; 34:2092–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shogren-Knaak M., Ishii H., Sun J.M., Pazin M.J., Davie J.R., Peterson C.L.. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006; 311:844–847. [DOI] [PubMed] [Google Scholar]
- 64. Ruan K., Yamamoto T.G., Asakawa H., Chikashige Y., Kimura H., Masukata H., Haraguchi T., Hiraoka Y.. Histone H4 acetylation required for chromatin decompaction during DNA replication. Sci. Rep. 2015; 5:12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.