Abstract
Esophageal squamous cell carcinoma (ESCC) is the most predominantly occurring type of esophageal cancer worldwide. Locally advanced ESCC patients are treated by neoadjuvant chemoradiation for tumor downstaging prior to tumor resection. Patients receiving this treatment have an increased expectation of cure via the following tumor resection and have better survival outcomes. However, not all patients respond well to chemoradiation and poor responders suffer from treatment-associated toxicity and complications without benefits. No method is currently available to predict patient chemoradiation response and to exclude poor responders from ineffective treatment. To address this clinical limitation, the present study aimed to identify non-invasive biomarkers for predicting patient chemoradiation response. Due to the features of microRNA (miRNA) in cancer diagnosis, prognosis and treatment response prediction, serum miRNA arrays were performed to identify potential miRNA(s) that may be used for chemoradiation response prediction in ESCC. Using an miRNA array to compare pre-treatment serum sample pools from 10 good responders and 10 poor responders, the present study identified miR-193b, miR-942 and miR-629* as candidate miRNAs for predicting chemoradiation response. Subsequent validation using reverse transcription-quantitative polymerase chain reaction confirmed that miR-193b, however not miR-942 and miR-629*, were significantly increased in sera from 24 good responders, compared with 23 poor responders. Further analyses using the receiver operating characteristic curve revealed a strong predictive power of serum miR-193b on discriminating good responders from poor responders to chemoradiation. In addition, a high serum level of miR-193b was significantly associated with better survival outcomes. Therefore, serum miR-193b may be considered a promising biomarker for predicting chemoradiation response and post-therapy survival of ESCC patients.
Keywords: esophageal squamous cell carcinoma, chemoradiation response, serum microRNAs, miR-193b, biomarker
Introduction
Esophageal cancer is the eighth most common cancer and the seventh leading cause of cancer-related death in men worldwide (1). It is composed of two major types, i.e. esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma. ESCC is the predominant type of esophageal cancer in Asian countries (2,3).
Tumor resection is the frontline treatment for ESCC patients. However, its benefit is limited to patients in early stage disease and those with resectable tumors. For patients with locally advanced tumors, they are alternatively treated with neoadjuvant chemoradiation for tumor down-staging before tumor resection. Based on the reports from our group and other research teams, such neoadjuvant treatment can enhance the chance of cure by tumor resection and therefore can profoundly increase patient outcomes (4–6). Despite that, patients exhibit a range of different responses towards chemoradiation from complete response, partial response to non-response. Till now, no clinical parameters or biomarkers are used to predict patient chemoradiation response and to identify non-responders to exclude them from treatment. Dismally, patients who do not respond to chemoradiation have to suffer from unnecessary and adverse side-effects of the treatment, such as drug-related toxicity and post-treatment complications. To address this clinical limitation, identification of biomarkers that can predict chemoradiation response can provide a guideline for selecting patients for chemoradiation, such that responders will be offered chemoradiation and non-responders will be excluded from such treatment.
MicroRNAs (miRNAs) are single-stranded, non-coding small RNA of 19 to 25 nucleotides. With their main functions on regulating gene expressions, they are capable of regulating diversified cellular functions and are involved in a range of physiological processes of growth and development (7). However, their deregulation can be commonly found in various diseased conditions including cancer of various types (8). MiRNAs involve in tumorigenesis by functioning both as tumor suppressors and oncogenes. A number of miRNAs have been found to have deregulated expressions in tumors when compared to adjacent non-tumor tissues. These observations have implicated for the potential uses of miRNAs for cancer prognosis and therapy (7,9).
Apart from the uses of miRNAs for cancer prognosis and therapy, recent studies have started to investigate the capability of miRNAs to predict patient responses towards different anti-cancer treatments including chemotherapy and targeting therapy for various cancers including ESCC (10–14). Collectively, these findings have consolidated the use of miRNAs for treatment response prediction. Due to the secretory nature of miRNAs and their small sizes making them resistant to degradation (15,16), current efforts have been focused on validating the use of circulatory miRNAs as non-invasive biomarkers for treatment response prediction for cancer patients (8,16,17).
The purpose of this study was to identify serum miRNAs that may serve as non-invasive biomarkers for treatment response prediction for ESCC patients. MiRNA array was employed in the analysis of serum samples from good responders vs. poor responders towards chemoradiation to identify candidate miRNAs with most differential difference in the serum levels between good responders and poor responders. The candidate miRNAs were further validated and analyzed for their potential use as predictive biomarkers for chemoradiation response of ESCC patients.
Materials and methods
Patient specimens
From 2000 to 2010, 47 locally advanced ESCC patients managed in Queen Mary Hospital, Hong Kong were included in this study. Age range of this patient cohort was 44 to 82 years old and the sex ratio was 44 (M) to 3 (F). All patients were treated with neoadjuvant cisplatin- and 5-fluorouracil-based chemoradiation followed by tumor resection. Chemoradiation was given concurrently. The chemotherapy regimen composed of cisplatin at 100 mg/m2 on day 1 and then day 22 and 5-fluorouracil at 500 mg/m2 per day from day 1 to day 5 and day 22 to day 26. Radiotherapy was given at a dosage of 40Gy at 2Gy per fraction (18). Serum specimens were prepared from blood samples collected from patients before treatment with chemoradiation. Patient chemoradiation responses were determined pathologically based on the percentage of viable tumor cells in the resected tissues collected from tumor resection (18). In this study, patients with 0% viable tumor cells were classified as good chemoradiation responders, while those with at least 50% viable tumor cells were regarded as poor responders. Informed consent was obtained for each patient recruited in this study. AJCC 6th edition was used for cancer staging. The use of clinical specimens for research was approved by the Institutional Review Board of The University of Hong Kong/Hospital Authority Hong Kong West Cluster (HKU/HA HKW IRB).
MiRNA array
Small RNAs from serum samples (10 samples from good responders and 10 samples from poor responders) were extracted using miRNeasy Serum/Plasma kit (Qiagen Inc., Hilden, Germany). RNA concentration and quality were determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). For both good responders and poor responders, pooled serum samples were prepared by mixing extracted RNAs from 5 samples, and that resulted in 2 serum sample pools for good responders and poor responders. The pooled RNA samples were then reverse transcribed to cDNA using TaqMan MicroRNA Reverse Transcription kit and Megaplex RT Primers (Applied Biosystems, Foster City, CA, USA) with the following thermal condition: 40 cycles of 16°C for 2 min; 42°C for 1 min; 50°C for 1 sec and 85°C for 5 min. TaqMan Low Density Array (TaqMan Array Human MicroRNA A+B Cards Set v3.0; Applied Biosystems) was employed to generate miRNA profiles from 754 different human miRNAs and U6, which was selected as an endogenous control. The reactions were performed in a 7900HT Fast Real-Time PCR System (Applied Biosystems), following the manufacturer protocol. Differentially expressed miRNAs were identified by comparing the miRNA profiles obtained from good responders and poor responders, and that miRNAs with at least 2.5-fold elevation in good responders vs. poor responders were subjected to further validation.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RNAs from serum specimens were prepared using miRNeasy Serum/Plasma kit (Qiagen). Concentration and quality of the extracted RNAs were determined as described above. To validate the miRNA array data, a TaqMan probe-based RT-qPCR system with the use of specific TaqMan MicroRNA Assays (Applied Biosystems) and TaqMan Fast Universal PCR Master Mix (Applied Biosystems) was performed on 24 good responders and 23 poor responders (including those used in the test cohort) (19–21). The reactions in duplicates were run on a 7900HT Fast Real-Time PCR System (Applied Biosystems) under the following thermal cycling condition: 2 min at 50°C; 10 min at 95°C and 45 cycles of 15 sec at 95°C and 1 min at 60°C. U6 was used as an endogenous control. Relative expressions for the examined miRNAs were calculated using the ∆∆Cq method (22).
Statistical analyses
GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA, USA) was used for statistical analyses. Quantitative data are described as the means with standard deviations (SD). Students t-test was used to compare expression level difference of each miRNA (expressed as ∆∆Cq value) between studied groups. To assess the capability of the studied miRNA for discriminating good responders and poor responders towards chemoradiation, receiver operating characteristic (ROC) curve and the area under the ROC curve (AUC) with 95% confidence interval (CI) were generated (19,23). Survival was calculated by the Kaplan-Meier method from the date of operation to the time of death or last follow-up date. Log-rank test was used to compare survival difference between groups. Clinical categorical data were analyzed using chi-squared test together with Pearson correlation. P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of candidate miRNAs for predicting patient chemoradiation responses
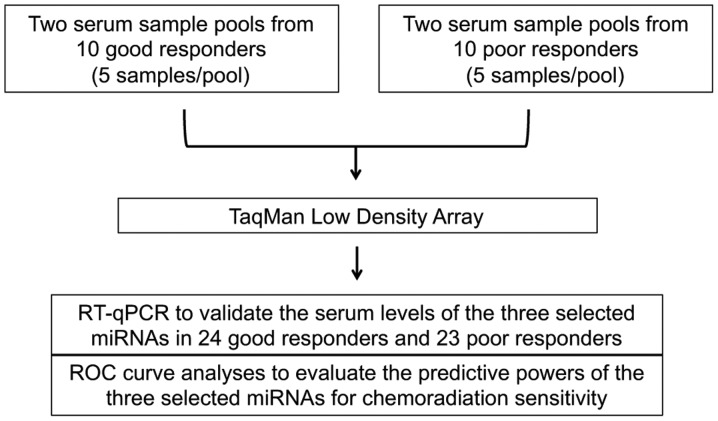
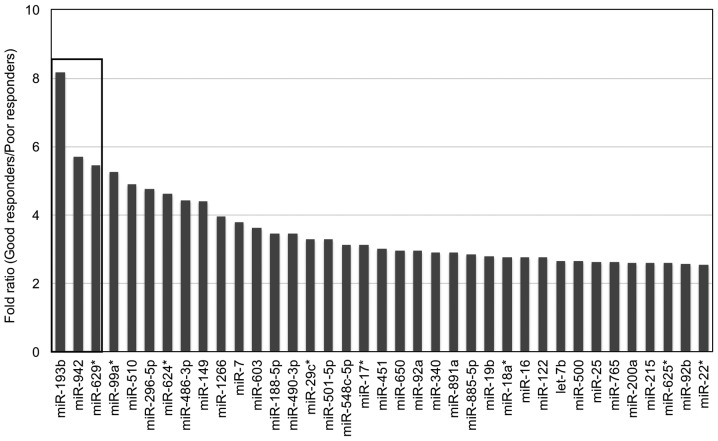
MiRNA array was performed on pooled serum samples from good responders and poor responders (Fig. 1). Of the 754 miRNAs examined, 37 miRNAs had at least 2.5-fold elevation in sera from good responders when compared to poor responders, while 23 of them (highlighted in grey) displayed consistent data between the two serum sample pools from the same group (Table I). Three serum miRNAs (miR-193b, miR-942 and miR-629*) with the most difference on the fold ratio between good responders and poor responders were selected as candidates for further validation (Fig. 2).
Figure 1.
Study design to identify serum biomarkers to predict chemoradiation responses of ESCC patients. Two serum sample pools from 10 good responders and 10 poor responders towards chemoradiation were subjected to TaqMan Low Density Array to identify miRNA(s) with differential serum levels between these two studied groups. Three miRNAs with most differential difference in their serum levels in good responder group when compared to poor responder group were selected for further validation by RT-qPCR. ROC curve analyses were performed on the three selected miRNAs to evaluate their predictive powers for chemoradiation sensitivity.
Table I.
Serum miRNAs with at least 2.5-fold elevation in good responders (GR) versus poor responders (PR).
| 1st array | 2nd array | 1st array | 2nd array | ||||
|---|---|---|---|---|---|---|---|
| miRNAs | GR1 | GR2 | PR1 | PR2 | Relative level (GR) | Relative level (PR) | Fold ratio (GR/PR) |
| miR-193ba | 34.83 | 35.17 | nd | nd | 0.3004 | 0.0368 | 8.1681 |
| miR-942a | 34.03 | 36.31 | nd | nd | 0.2544 | 0.0445 | 5.7160 |
| miR-629*a | 35.03 | 35.44 | nd | nd | 0.2432 | 0.0445 | 5.4642 |
| miR-99a* | 34.94 | nd | nd | 36.61 | 0.3132 | 0.0595 | 5.2598 |
| miR-510 | nd | 35.04 | 36.6 | nd | 0.2922 | 0.0597 | 4.8906 |
| miR-296-5p | 34.8 | 34.87 | 36.12 | nd | 0.3368 | 0.0706 | 4.7733 |
| miR-624*a | 34.97 | 35.98 | nd | nd | 0.2059 | 0.0445 | 4.6268 |
| miR-486-3pa | 34.17 | 34.78 | 36.32 | 37.56 | 0.4118 | 0.0928 | 4.4383 |
| miR-149 | 33.89 | 35.37 | 35.48 | nd | 0.3882 | 0.0881 | 4.4076 |
| miR-1266a | 35.29 | 36.1 | nd | nd | 0.1768 | 0.0445 | 3.9724 |
| miR-7a | 34.81 | 35.14 | 36.49 | 37.25 | 0.3057 | 0.0805 | 3.7974 |
| miR-603 | nd | 33.65 | nd | nd | 0.1615 | 0.0445 | 3.6301 |
| miR-188-5p | nd | 34.47 | nd | nd | 0.1276 | 0.0368 | 3.4702 |
| miR-490-3p | 34.17 | 30.59 | 32.61 | 35.67 | 1.8468 | 0.5340 | 3.4581 |
| miR-29c*a | 35.89 | 36.04 | nd | 38 | 0.1466 | 0.0445 | 3.2944 |
| miR-501-5p | 33.77 | 35.79 | nd | 34.93 | 0.3499 | 0.1066 | 3.2830 |
| miR-548c-5pa | 35.01 | 37.07 | nd | nd | 0.1392 | 0.0445 | 3.1275 |
| miR-17*a | 34.5 | 37.59 | nd | nd | 0.1387 | 0.0445 | 3.1167 |
| miR-451a | 20.23 | 21.1 | 22.48 | 21.98 | 6208.3751 | 2055.1101 | 3.0209 |
| miR-650a | 35.01 | 37.22 | nd | nd | 0.1321 | 0.0445 | 2.9690 |
| miR-92aa | 24.97 | 25.08 | 26.61 | 26.5 | 302.3341 | 102.5369 | 2.9485 |
| miR-340a | 37.07 | 35.9 | nd | nd | 0.1073 | 0.0368 | 2.9180 |
| miR-891a | nd | 34.98 | nd | nd | 0.1069 | 0.0368 | 2.9079 |
| miR-885-5p | 36.11 | 35.02 | 36.08 | nd | 0.2031 | 0.0715 | 2.8382 |
| miR-19ba | 25.61 | 26.35 | 27.29 | 27.57 | 155.9565 | 55.9087 | 2.7895 |
| miR-18a*a | 32.67 | 33.65 | 34.95 | 34.25 | 1.0755 | 0.3882 | 2.7702 |
| miR-16a | 23.72 | 24.28 | 25.55 | 25.33 | 615.2377 | 222.0899 | 2.7702 |
| miR-122a | 30.57 | 29.55 | 31.26 | 31.74 | 9.2215 | 3.3288 | 2.7702 |
| let-7ba | 28.18 | 28.66 | 29.64 | 29.96 | 28.7402 | 10.8153 | 2.6574 |
| miR-500a | 34.39 | 35.29 | 36.38 | 36.06 | 0.3356 | 0.1263 | 2.6574 |
| miR-25a | 28.08 | 28.8 | 29.98 | 29.64 | 28.3446 | 10.7406 | 2.6390 |
| miR-765 | 36.39 | 35.97 | nd | 37.09 | 0.1326 | 0.0504 | 2.6299 |
| miR-200a | nd | 35.79 | nd | 37.14 | 0.1737 | 0.0668 | 2.6027 |
| miR-215a | 32.69 | 33.68 | 33.91 | 35.16 | 1.0570 | 0.4061 | 2.6027 |
| miR-625* | 35.45 | 35.07 | 35.21 | nd | 0.2509 | 0.0967 | 2.5937 |
| miR-92b | 35.49 | 36.07 | nd | 36.23 | 0.1749 | 0.0679 | 2.5758 |
| miR-22*a | 34.05 | 33.68 | 34.72 | 35.66 | 0.6598 | 0.2579 | 2.5580 |
nd, not detectable.
miRNAs with consistent data between two arrays in the same group.
Figure 2.
Three miRNAs with most differential difference in serum levels between good responders and poor responders towards chemoradiation. MiRNA array result identified twenty-three miRNAs with consistent data between two serum sample pools in the same group and with at least 2.5-fold elevation in serum levels from good responders when compared to poor responders. MiR-193b, miR-942 and miR-629* were the three miRNAs with most differential difference in their serum levels between good responders and poor responders.
Validation of candidate miRNAs by RT-qPCR
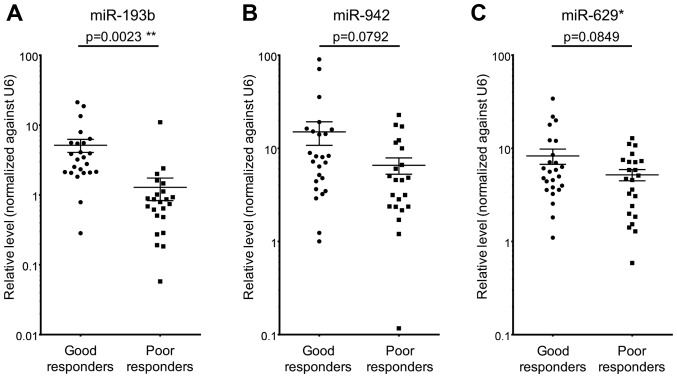
To validate the miRNA array data, RT-qPCR on serum miR-193b, miR-942 and miR-629* was performed on 24 good responders and 23 poor responders. The RT-qPCR results were consistent with those obtained by miRNA array, such that all three examined miRNAs demonstrated higher levels in sera from good responders when compared to poor responders (Fig. 3). However, only miR-193b achieved a statistically significant result (P<0.05) (Fig. 3A). These results suggested that high serum level of miR-193b could potentially be served as a biomarker to predict patient chemoradiation responses.
Figure 3.
Higher level of serum miR-193b in good responders when compared to poor responders towards chemoradiation. Among the three miRNAs with most differential difference in their serum levels between good responders and poor responders, only miR-193b (A), but not miR-942 (B) and miR-629* (C), showed statistically significant difference in its serum level between good responders and poor responders. Error bars represent the standard deviation; **P<0.01.
Serum miR-193b serves as a non-invasive predictive biomarker for patient chemoradiation response and survival outcome
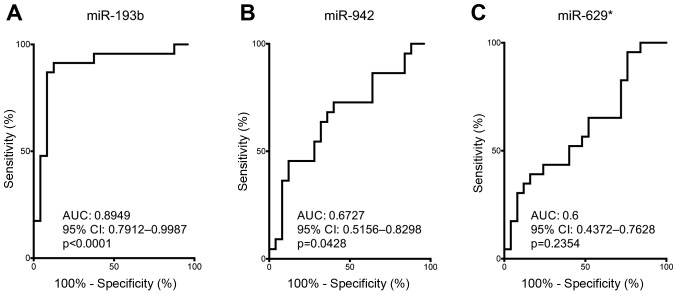
To examine the predictive power of serum miR-193b, miR-942 and miR-629* on distinguishing good responders and poor responders towards chemoradiation, ROC curves analysis was performed to obtain the AUC values and 95% CI. ROC curve data of serum miR-193b revealed a strong predictive power to distinguish good responders and poor responders (AUC: 0.8949, 95% CI: 0.7912–0.9987, P<0.0001) (Fig. 4A). Although ROC curve analysis supporting the use of serum miR-942 for distinguishing good responders and poor responders (AUC: 0.6727, 95% CI: 0.5156–0.8298, P=0.0428) (Fig. 4B), serum miR-942 failed to demonstrate significant difference between its level in good responders and poor responders (Fig. 3B). On the other hand, serum miR-629* failed to show sufficient discriminative power to distinguish good responders from poor responders (Fig. 4C). Based on the differential difference of serum miR-193b level in good responders vs. poor responders and the good discriminative power of serum miR-193b on distinguishing good responders from poor responders, our data have supported the potential use of miR-193b for predicting chemoradiation response.
Figure 4.
Serum miR-193b as the most powerful biomarker for predicting chemoradiation sensitivity for ESCC patients. ROC curve analyses were performed on miR-193b (A), miR-942 (B) and miR-629* (C) to evaluate their predictive power for chemoradiation sensitivity in ESCC patients. Both serum miR-193b and miR-942 showed a statistically significant difference.
As miR-193b is a potential predictor for chemoradiation response in ESCC patients, we also performed survival and clinical analyses by using the median value as a cut-off value to divide the patients into two groups with high and low serum level of miR-193b. Patients with high serum miR-193b level associated better survival (median survival=71.5 months) than those with low serum miR-193b level (median survival=16 months) (P=0.0099) (Fig. 5). High serum miR-193b level was inversely correlated with post-therapy pathological ypT stage (P<0.001), ypN stage (P=0.045), ypM stage (P=0.035) and overall ypTNM stage (P<0.001) (Table II). Moreover, high serum miR-193b level was associated with R0 category (no residual tumor after surgery) (P<0.001) and with pathological complete response (ypCR) (P<0.001) (Table II). Collectively, our data have suggested that high serum level of miR-193b can potentially serve as a non-invasive predictive biomarker for patient chemoradiation response and survival outcome.
Figure 5.
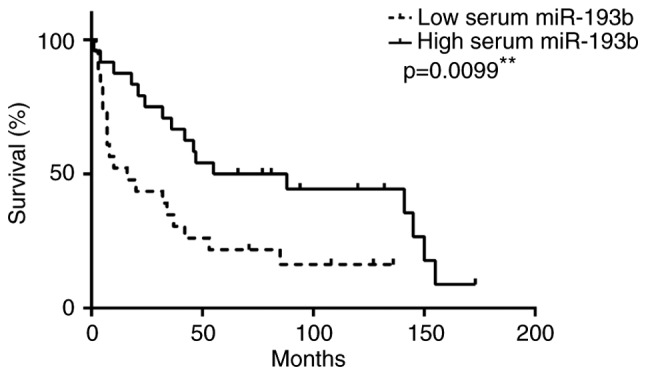
High serum level of miR-193b associated with better patient survivals. Kaplan-Meier survival curves for 47 ESCC patients were constructed. Log-rank test showed a statistically significant difference between low miR-193b level and high miR-193b level groups (P=0.0099). **P<0.01.
Table II.
Clinical correlation analyses of serum miR-193b level in ESCC patients treated with neoadjuvant chemoradiation.
| Serum miR-193b level | ||||
|---|---|---|---|---|
| Clinicopathological parameters | Patient number | High | Low | P-value |
| Age (years) | ||||
| ≤64.5 | 23 | 10 | 13 | 0.308 |
| >64.5 | 24 | 14 | 10 | |
| Gender | ||||
| Male | 44 | 24 | 20 | 0.067 |
| Female | 3 | 0 | 3 | |
| R category | ||||
| R0 | 34 | 24 | 10 | <0.001 |
| R1/R2 | 13 | 0 | 13 | |
| Pathological stage | ||||
| ypT stage | ||||
| ypT0 | 24 | 21 | 3 | <0.001 |
| ypT1/2 | 4 | 0 | 4 | |
| ypT3/4 | 19 | 3 | 16 | |
| ypN stage | ||||
| ypN0 | 33 | 20 | 13 | 0.045 |
| ypN1 | 14 | 4 | 10 | |
| ypM stage | ||||
| ypM0 | 40 | 23 | 17 | 0.035 |
| ypM1 | 7 | 1 | 6 | |
| ypTNM | ||||
| ypCRa | 24 | 21 | 3 | <0.001 |
| y-stage I/II | 9 | 1 | 8 | |
| y-stage III/IV | 14 | 2 | 12 | |
Pathological complete response.
Discussion
Neoadjuvant chemoradiation followed by tumor resection has become the standard treatment for locally advanced ESCC (24–26). However, patients respond differently towards such neoadjuvant treatment. On average, only 20–30% of patients demonstrate complete chemoradiation response. Poor responders, on the other hand, suffer unnecessary chemoradiation-related toxicity and complications without benefits. In view of this clinical situation, identification of reliable biomarker that can predict patient response before chemoradiation is important for selecting optimal treatment for patients.
For ESCC, previous studies have proposed the use of several biomarkers identified from ESCC tumors to predict patient chemoradiation response. A study has demonstrated the association of excision repair cross-complementing 1 (ERCC1) with cisplatin response in ESCC by revealing tumoral ERCC1 levels in patients with partial responses to chemoradiation were significantly lower than those in non-responsive patients. Besides, ESCC cell lines with lower ERCC1 level showed greater sensitivity to clinically relevant concentrations of cisplatin and its related drug oxaliplatin when compared to cell lines with higher ERCC1 level (27). In addition to this single molecule study, a number of studies have taken into accounts the use of more than one molecule for predicting chemoradiation responses. Using cDNA microarray to analyze pre-chemoradiation tumor biopsies from ESCC patients has identified a 32-gene classifier that can be used to predict patient responses towards chemoradiation (28). Another study by Wen et al has carried out gene expression analysis on pre-treatment ESCC biopsies from patients who have received neoadjuvant chemoradiation followed by surgery. Based on the gene expression analysis, they have developed a prediction model composing of three genes (MMP1, LIMCH1 and C1orf226) that can be used to predict pathological response towards chemoradiation with high accuracy (29). Apart from the above use of protein-coding genes for predicting chemoradiation responses of ESCC patients, research has also been focused on exploring the potential use of non-coding RNAs for this purpose. MiRNAs are non-coding RNAs with diversified roles on tumor progression and development, implicating their plausible uses for cancer classification and prognostication (7,30). By analyzing the pre-treatment tumor biopsies, a recent miRNA study has identified the combined use of four differentially expressed miRNAs (miR-145-5p, miR-152, miR-193b-3p and miR-376a-3p) for chemoradiation response prediction for ESCC patients (14).
When compared to the biomarkers derived from ESCC tumors, blood-based biomarkers are better predictors for chemotherapy/chemoradiation responses of patients as the procedure for blood collection is minimally invasive. Most studies on finding blood-based biomarkers for chemotherapy/chemoradiation response prediction put the focus on miRNAs because of their stability in human circulation (15,16). Kurashige et al have reported the use of serum miR-21 for indicating chemotherapy response by showing a significant reduction of its serum level in ESCC patients who are responsive to chemotherapy, but not in non-responders, after treatment (31). On the other hand, Tanaka et al have demonstrated serum level of miR-200c can be used for predicting chemotherapy responses for ESCC patients receiving chemotherapy (32). In the current study, we identified serum miR-193b by miRNA array, and validated it as a reliable predictor for chemoradiation responses for ESCC patients using TaqMan probe-based qPCR. Specifically, high serum level of miR-193b can indicate ESCC patients who are responsive to chemoradiation. Moreover, high serum level of miR-193b was significantly associated with better survival and inversely correlated with post-therapy pathological stages. Despite the advantageous feature of miRNAs, some studies have alternatively investigated the possible use of other circulatory protein or mRNA for predicting chemoradiation responses. Maher et al have utilized proteomics to analyze pre-treatment sera from esophageal cancer patients and have revealed significant elevation on the serum levels of complement C4a and C3a in poor responders towards chemoradiation (33). Moreover, a more recent study has also demonstrated the usefulness of circulating FAM84B mRNA and protein for predicting pathological chemoradiation responses (34). The above studies have supported the idea of using circulating biomarkers for predicting chemoradiation responses of ESCC patients.
In this study, we have reported high serum miR-193b level in ESCC patients responsive to chemoradiation, implicating the link between miR-193b and chemoradiation sensitivity. Similarly, Nyhan et al have reported the positive association between miR-193b and increased chemosensitivity of esophageal cancer cells, such that overexpression of miR-193b in chemoresistant esophageal cancer cells sensitized them towards chemotherapeutic drug treatment (35). Apart from the direct link between miR-193b with chemotherapy/chemoradiation sensitivity, this miRNA is proven to involve in other treatment-related sensitivities. In melanoma, over-expression of miR-193b in ABT-737-resistant tumor cells can restore their sensitivity towards ABT-737 (BH3 mimetic) treatment (36). In another scenario, over-expression of miR-193b in liver cancer cells can sensitize them towards treatment with molecular targeting drug sorafenib (37). Together, these prior studies including the current one depict the importance of miR-193b on determining the treatment sensitivity of tumor cells and implicate the potential of restoring miR-193b for treatment sensitization.
In summary, the present study has demonstrated high level of serum miR-193b as a promising biomarker for predicting chemoradiation responses and post-therapy survival of ESCC patients. Further validation on a larger and independent patient cohort is required for full assessment of its predictive power. Our findings can benefit the treatment selection process for locally advanced ESCC patients by predicting their sensitivities towards chemoradiation, such that only good responders will be offered this treatment.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 3.Hiyama T, Yoshihara M, Tanaka S, Chayama K. Genetic polymorphisms and esophageal cancer risk. Int J Cancer. 2007;121:1643–1658. doi: 10.1002/ijc.23044. [DOI] [PubMed] [Google Scholar]
- 4.Tong DK, Law S, Kwong DL, Wei WI, Ng RW, Wong KH. Current management of cervical esophageal cancer. World J Surg. 2011;35:600–607. doi: 10.1007/s00268-010-0876-7. [DOI] [PubMed] [Google Scholar]
- 5.Law S, Kwong DL, Kwok KF, Wong KH, Chu KM, Sham JS, Wong J. Improvement in treatment results and long-term survival of patients with esophageal cancer: Impact of chemoradiation and change in treatment strategy. Ann Surg. 2003;238:339–348. doi: 10.1097/01.sla.0000086545.45918.ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allum WH, Blazeby JM, Griffin SM, Cunningham D, Jankowski JA, Wong R, Association of Upper Gastrointestinal Surgeons of Great Britain and Ireland, the British Society of Gastroenterology and the British Association of Surgical Oncology Guidelines for the management of oesophageal and gastric cancer. Gut. 2011;60:1449–1472. doi: 10.1136/gut.2010.228254. [DOI] [PubMed] [Google Scholar]
- 7.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mostert B, Sieuwerts AM, Martens JW, Sleijfer S. Diagnostic applications of cell-free and circulating tumor cell-associated miRNAs in cancer patients. Expert Rev Mol Diagn. 2011;11:259–275. doi: 10.1586/erm.11.11. [DOI] [PubMed] [Google Scholar]
- 9.Garofalo M, Leva GD, Croce CM. MicroRNAs as anti-cancer therapy. Curr Pharm Des. 2014;20:5328–5335. doi: 10.2174/1381612820666140128211346. [DOI] [PubMed] [Google Scholar]
- 10.Tumilson CA, Lea RW, Alder JE, Shaw L. Circulating microRNA biomarkers for glioma and predicting response to therapy. Mol Neurobiol. 2014;50:545–558. doi: 10.1007/s12035-014-8679-8. [DOI] [PubMed] [Google Scholar]
- 11.Schwarzenbach H. The potential of circulating nucleic acids as components of companion diagnostics for predicting and monitoring chemotherapy response. Expert Rev Mol Diagn. 2015;15:267–275. doi: 10.1586/14737159.2015.980817. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, Xu Y, Liao X, Liao R, Zhang L, Niu K, Li T, Li D, Chen Z, Duan Y, Sun J. Plasma miRNAs in predicting radiosensitivity in non-small cell lung cancer. Tumour Biol. 2016;37:11927–11936. doi: 10.1007/s13277-016-5052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DAngelo E, Fassan M, Maretto I, Pucciarelli S, Zanon C, Digito M, Rugge M, Nitti D, Agostini M. Serum miR-125b is a non-invasive predictive biomarker of the pre-operative chemoradiotherapy responsiveness in patients with rectal adenocarcinoma. Oncotarget. 2016;7:28647–28657. doi: 10.18632/oncotarget.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wen J, Luo K, Liu H, Liu S, Lin G, Hu Y, Zhang X, Wang G, Chen Y, Chen Z, et al. MiRNA expression analysis of pretreatment biopsies predicts the pathological response of esophageal squamous cell carcinomas to neoadjuvant chemoradiotherapy. Ann Surg. 2016;263:942–948. doi: 10.1097/SLA.0000000000001489. [DOI] [PubMed] [Google Scholar]
- 15.Hoy AM, Buck AH. Extracellular small RNAs: What, where, why? Biochem Soc Trans. 2012;40:886–890. doi: 10.1042/BST20120019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarry J, Schadendorf D, Greenwood C, Spatz A, van Kempen LC. The validity of circulating microRNAs in oncology: Five years of challenges and contradictions. Mol Oncol. 2014;8:819–829. doi: 10.1016/j.molonc.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tong DK, Law S, Kwong DL, Chan KW, Lam AK, Wong KH. Histological regression of squamous esophageal carcinoma assessed by percentage of residual viable cells after neoadjuvant chemoradiation is an important prognostic factor. Ann Surg Oncol. 2010;17:2184–2192. doi: 10.1245/s10434-010-0995-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu AM, Yao TJ, Wang W, Wong KF, Lee NP, Fan ST, Poon RT, Gao C, Luk JM. Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: A retrospective cohort study. BMJ Open. 2012;2:e000825. doi: 10.1136/bmjopen-2012-000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu AM, Xu Z, Shek FH, Wong KF, Lee NP, Poon RT, Chen J, Luk JM. miR-122 targets pyruvate kinase M2 and affects metabolism of hepatocellular carcinoma. PLoS One. 2014;9:e86872. doi: 10.1371/journal.pone.0086872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsang FH, Au V, Lu WJ, Shek FH, Liu AM, Luk JM, Fan ST, Poon RT, Lee NP. Prognostic marker microRNA-125b inhibits tumorigenic properties of hepatocellular carcinoma cells via suppressing tumorigenic molecule eIF5A2. Dig Dis Sci. 2014;59:2477–2487. doi: 10.1007/s10620-014-3184-5. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Sun S, Poon RT, Lee NP, Yeung C, Chan KL, Ng IO, Day PJ, Luk JM. Proteomics of hepatocellular carcinoma: Serum vimentin as a surrogate marker for small tumors (<or=2 cm) J Proteome Res. 2010;9:1923–1930. doi: 10.1021/pr901085z. [DOI] [PubMed] [Google Scholar]
- 24.Berger AC, Farma J, Scott WJ, Freedman G, Weiner L, Cheng JD, Wang H, Goldberg M. Complete response to neoadjuvant chemoradiotherapy in esophageal carcinoma is associated with significantly improved survival. J Clin Oncol. 2005;23:4330–4337. doi: 10.1200/JCO.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 25.Stahl M, Stuschke M, Lehmann N, Meyer HJ, Walz MK, Seeber S, Klump B, Budach W, Teichmann R, Schmitt M, et al. Chemoradiation with and without surgery in patients with locally advanced squamous cell carcinoma of the esophagus. J Clin Oncol. 2005;23:2310–2317. doi: 10.1200/JCO.2005.00.034. [DOI] [PubMed] [Google Scholar]
- 26.Bonnetain F, Bouché O, Michel P, Mariette C, Conroy T, Pezet D, Roullet B, Seitz JF, Paillot B, Arveux P, et al. A comparative longitudinal quality of life study using the Spitzer quality of life index in a randomized multicenter phase III trial (FFCD 9102): Chemoradiation followed by surgery compared with chemoradiation alone in locally advanced squamous resectable thoracic esophageal cancer. Ann Oncol. 2006;17:827–834. doi: 10.1093/annonc/mdl033. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka K, Mohri Y, Ohi M, Yokoe T, Koike Y, Morimoto Y, Miki C, Tonouchi H, Kusunoki M. Excision-repair cross-complementing 1 predicts response to cisplatin-based neoadjuvant chemoradiotherapy in patients with esophageal squamous cell carcinoma. Mol Med Rep. 2009;2:903–909. doi: 10.3892/mmr_00000190. [DOI] [PubMed] [Google Scholar]
- 28.Duong C, Greenawalt DM, Kowalczyk A, Ciavarella ML, Raskutti G, Murray WK, Phillips WA, Thomas RJ. Pretreatment gene expression profiles can be used to predict response to neoadjuvant chemoradiotherapy in esophageal cancer. Ann Surg Oncol. 2007;14:3602–3609. doi: 10.1245/s10434-007-9550-1. [DOI] [PubMed] [Google Scholar]
- 29.Wen J, Yang H, Liu MZ, Luo KJ, Liu H, Hu Y, Zhang X, Lai RC, Lin T, Wang HY, Fu JH. Gene expression analysis of pretreatment biopsies predicts the pathological response of esophageal squamous cell carcinomas to neo-chemoradiotherapy. Ann Oncol. 2014;25:1769–1774. doi: 10.1093/annonc/mdu201. [DOI] [PubMed] [Google Scholar]
- 30.Hemmatzadeh M, Mohammadi H, Jadidi-Niaragh F, Asghari F, Yousefi M. The role of oncomirs in the pathogenesis and treatment of breast cancer. Biomed Pharmacother. 2016;78:129–139. doi: 10.1016/j.biopha.2016.01.026. [DOI] [PubMed] [Google Scholar]
- 31.Kurashige J, Kamohara H, Watanabe M, Tanaka Y, Kinoshita K, Saito S, Hiyoshi Y, Iwatsuki M, Baba Y, Baba H. Serum microRNA-21 is a novel biomarker in patients with esophageal squamous cell carcinoma. J Surg Oncol. 2012;106:188–192. doi: 10.1002/jso.23064. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka K, Miyata H, Yamasaki M, Sugimura K, Takahashi T, Kurokawa Y, Nakajima K, Takiguchi S, Mori M, Doki Y. Circulating miR-200c levels significantly predict response to chemotherapy and prognosis of patients undergoing neoadjuvant chemotherapy for esophageal cancer. Ann Surg Oncol. 2013;20(Suppl 3):S607–S615. doi: 10.1245/s10434-013-3093-4. [DOI] [PubMed] [Google Scholar]
- 33.Maher SG, McDowell DT, Collins BC, Muldoon C, Gallagher WM, Reynolds JV. Serum proteomic profiling reveals that pretreatment complement protein levels are predictive of esophageal cancer patient response to neoadjuvant chemoradiation. Ann Surg. 2011;254:809–817. doi: 10.1097/SLA.0b013e31823699f2. [DOI] [PubMed] [Google Scholar]
- 34.Hsu FM, Cheng JC, Chang YL, Lee JM, Koong AC, Chuang EY. Circulating mRNA profiling in esophageal squamous cell carcinoma identifies FAM84B as a biomarker in predicting pathological response to neoadjuvant chemoradiation. Sci Rep. 2015;5:10291. doi: 10.1038/srep10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nyhan MJ, ODonovan TR, Boersma AW, Wiemer EA, McKenna SL. MiR-193b promotes autophagy and non-apoptotic cell death in oesophageal cancer cells. BMC Cancer. 2016;16:101. doi: 10.1186/s12885-016-2123-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J, Zhang X, Lentz C, Abi-Daoud M, Paré GC, Yang X, Feilotter HE, Tron VA. miR-193b regulates Mcl-1 in melanoma. Am J Pathol. 2011;179:2162–2168. doi: 10.1016/j.ajpath.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mao K, Zhang J, He C, Xu K, Liu J, Sun J, Wu G, Tan C, Zeng Y, Wang J, Xiao Z. Restoration of miR-193b sensitizes Hepatitis B virus-associated hepatocellular carcinoma to sorafenib. Cancer Lett. 2014;352:245–252. doi: 10.1016/j.canlet.2014.07.004. [DOI] [PubMed] [Google Scholar]