Abstract
Different types of cancer exhibit distinct gene expression profiles. The present study aimed to identify a specific gene dysregulated in hepatocellular carcinoma (HCC) that was essential for cancer progression. The whole transcriptomes of primary HCC tissue samples were analyzed with microarrays. The most significantly differentially expressed gene was identified, specifically karyopherin subunit-α 2 (KPNA2), and an analysis using the Oncomine online tool was performed with data from The Cancer Genome Atlas to predict associated genes in HCC. Reverse transcription-quantitative polymerase chain reaction was performed to confirm the gene expression levels of KPNA2, and the RNA interference knockdown of KPNA2 was performed to identify the effect on putative downstream target genes. A proliferation assay and flow cytometry analysis was used to assess the function of KPNA2 in the regulation of the cell cycle. The results demonstrated that KPNA2 expression was significantly upregulated in HCC tumor tissues compared with liver tissues and was associated with cyclin B2 (CCNB2) and cyclin-dependent kinase 1 (CDK1) expression. KPNA2 expression was identified a novel marker to predict the outcome of patients. In addition, KPNA2 knockdown downregulated CCNB2 and CDK1, inhibited cell proliferation and induced cell cycle arrest in the G2/M phase. In conclusion, it was demonstrated that KPNA2 may promote tumor cell proliferation by increasing the expression of CCNB2/CDK1. KPNA2 could be a target for therapeutic intervention in HCC.
Keywords: karyopherin subunit-α 2, cyclin B2/cyclin-dependent kinase 1, hepatocellular carcinoma, cell cycle
Introduction
Hepatocellular carcinoma (HCC) is the most common type of liver cancer which results in ~80% of mortalities associated with hepatic cancer (1). Surgical resection is the principal strategy for treatment of HCC. The results of chemotherapy are often unsatisfactory as liver cancer typically develops resistance to chemotherapy, which may be due to a number of reasons; for example, the rapid metabolism of liver cells may quickly inactivate chemotherapy drugs administered to patients with HCC (2). In addition, the factors which enable the progression of HCC remain unidentified, and non-targeted drugs are less effective at preventing cancer recurrence and metastasis (3).
Karyopherin subunit-α 2 (KPNA2) is a nuclear transporter, allowing signal communication between the nucleus and cytoplasm (4). KPNA2 is required for cell survival (5); it may be mutated in cancer to enable disease progression (6). KPNA2 has been associated with the regulation of carcinogenesis, proliferation and recurrence in various cancer types, as previously reviewed (7). KPNA2 contributes to the relocation of DNA damage response proteins and has been associated with the prognosis of breast cancer (4). KPNA2 recognizes cargo with the nuclear location sequence (NLS) and may affect cancer cell progression by interacting with a number of transcriptional factors, including c-Myc and p53 (8). Additionally, the upregulation of KPNA2 increases the expression of octamer-binding transcription factor gene 4 (OCT4), which is associated with the stem-like properties and dedifferentiation of cancer cells (9). However, the association between KPNA2 and cancer progression has yet to be characterized and requires further study.
In the present study, it was identified that KPNA2 may serve a function in the progression of HCC. The knockdown of KPNA2 expression was associated with the downregulation of cancer-associated genes in HCC cells, and high KPNA2 expression was associated with a significantly reduced overall and disease-free survival time in patients with HCC.
Materials and methods
Tumor samples
The present study was approved by the Ethics Committee of the Third Affiliated Hospital of Kunming Medical University (Kunming, China) and written informed consent was obtained from all patients. The patients had not received radiation or chemotherapy prior to surgery. HCC tissues were obtained from 6 patients (3 males and 3 females), aged between 55 and 65 years old (mean age, 62 years old) with non-metastatic disease during surgical resection from the Department of Oncology of the Third Affiliated Hospital of Kunming Medical University between September 2013 and May 2014. Adjacent liver tissues were used as controls. All tissues were frozen with dry ice. RNA was extracted from the samples with TRIzol (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocol. The gene expression profile was analyzed using GeneChip® Human Transcriptome Array 2.0 microarrays (Affymetrix; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The raw data were analyzed by using the affy package (version 1.55.0; Affymetrix; Thermo Fisher Scientific, Inc.) in R (10). The robust multi-array average method was employed to corrects probe intensity values and the microarray data was then quantile-normalized (10) for the following comparisons. The normalized microarray data were analyzed using Multiple Experiment Viewer (version 4.9.0, http://mev.tm4.org/#/welcome).
Cell culture
The human HCC cell line Huh7 was purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). The cells were cultured in Dulbecco's Modified Eagle's Medium (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 5% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.). Cells were maintained in 37°C in an atmosphere containing 5% CO2 throughout the study.
RNA interference
Short interfering (si)RNA was synthesized by Jima Biotechnology Co., Ltd. (Shanghai, China); the si-KPNA2-1 sequence is UUACGAUUAUGUGGUCGACGG, si-KPNA2-2 is UUGUUUGGUUCCGACACCAUC and si-CTR is a commercial small RNA target with no sequence on known human gene. A total of 20 pmol siRNA was transfected using 3 µl RNAiMAX reagent/well (cat. no. 13778-100; Thermo Fisher Scientific, Inc.) in 24-well plates (cells grown up to 70% confluence). Further analysis, as subsequently described, was performed at 48 h after transfection.
MTT assay
A CellTiter 96® non-radioactive cell proliferation assay (cat. no. G4000; Promega Corporation, Madison, WI, USA) was used to analyze cell proliferation, according to the manufacturer's protocol. Briefly, 8,000 Huh7 cells were seeded in 96-well plates. Alterations in cell confluence were observed at 24 and 48 h, followed by the addition of 15 µl Dye Solution to each well. Following a further incubation of 2–3 h, 100 µl Solubilizing Solution/Stop mix was added to each well and the absorbance was determined at wavelength of 570 nm on a 96-well plate reader.
Reverse transcription-quantitative polymerase chain reaction
RNA was extracted from Huh7 cells using TRIzol, and reverse transcription was performed using the PrimeScript 1st strand cDNA Synthesis kit (cat. no. 6110A; Takara Bio, Inc., Otsu, Japan), according to the manufacturer's protocols. The SsoFast EvaGreen Supermix with Low ROX kit (cat. no. 1572-5211; Bio-Rad Laboratories, Inc., Hercules, CA, USA) was applied to quantify the mRNA expression level, according to the manufacturer's protocol. The primers used in qPCR were as follow: KPNA2 forward, CTGCAGGAAAACCGCAACAA and reverse, CCTGGCAGCTTGAGTAGCTT; CDK1 forward, TTTCTTTCGCGCTCTAGCCA and reverse, CAATCGGGTAGCCCGTAGAC; CCNB2 forward, GGCTGGTACAAGTCCAC-TCC and reverse, CTTCTTCCGGGAAACTGGCT; β-actin forward, GTCATTCC-AAATATGAGATGCGT and reverse, GCTATCACCTCCCCTGTGTG. Thermocycling protocol included pre-denatured at 95°C for 3 min, denaturation at 95°C for 15 sec, annealing at 60°C for 15 sec and extension at 72°C for 1 min for 40 cycles. The 2−ΔΔCq method was used for quantification of gene relative expression (11).
Cell cycle analysis
After cell transfection with siRNA for 24 h, the cells were dissociated using 0.25% trypsin in a 37°C cell incubator for 5 min. The detached cells were washed with PBS once and then fixed with cold 70% ethanol for 30 min at room temperature. Briefly, the cells were washed twice with PBS. The cells were treated with a final concentration of 5 µg/ml ribonuclease for 10 min at room temperature. Propidium iodide (PI) was added (10 µg/ml) and the cells were stained for 15 min at room temperature. The cells were analyzed using FACSCanto (BD Biosciences, Inc., San Jose, CA, USA). A wavelength of 605 nm was selected to determine the fluorescence of PI.
Bioinformatic data mining
Cancer Genome Atlas (TCGA) data was used. TCGA raw data was obtained and analyzed using the cBioportal (http://www.cbioportal.org/) and Oncomine (https://www.oncomine.org/) online tools. A total of 440 mRNA expression profiles for HCC (440 adjacent liver tissues were used as control tissues) were selected. The KPNA2 gene was input, with the cut-off for high/low expression level set at >1 standard deviation. To evaluate the hazard ratio of KPNA2, the OncoLnc (http://www.oncolnc.org/) was used to investigate the Cox coefficients of KPNA2 in HCC patients.
Statistical analysis
Pearson and Spearman correlation analysis were respectively performed to investigate the correlation between the KPNA2 and CDK1 as well as CCNB2. Cox's proportional hazards model was used for survival rate analysis. Kaplan-Meier estimator curves were plotted for survival analysis and a log-rank test was performed to determine the statistical significance of differences in survival between groups. GraphPad prism 7 (GraphPad Software, Inc., La Jolla, CA, USA) was used for data analysis. Data are presented as the mean ± standard error of the mean. Student's t-test was used to compare data between groups.
Results
KPNA2 mRNA expression is deregulated in hepatocellular carcinoma tissues and associated with cell regulators
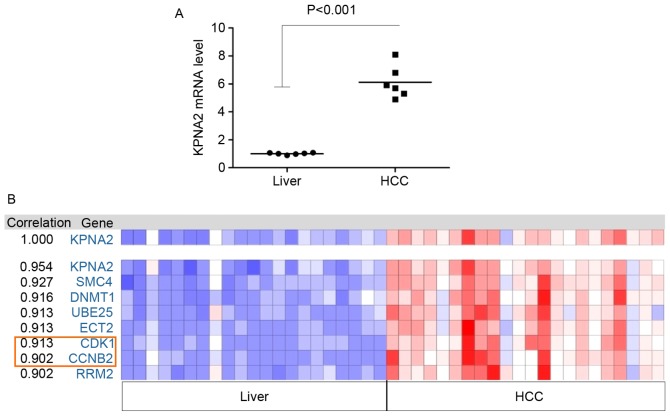
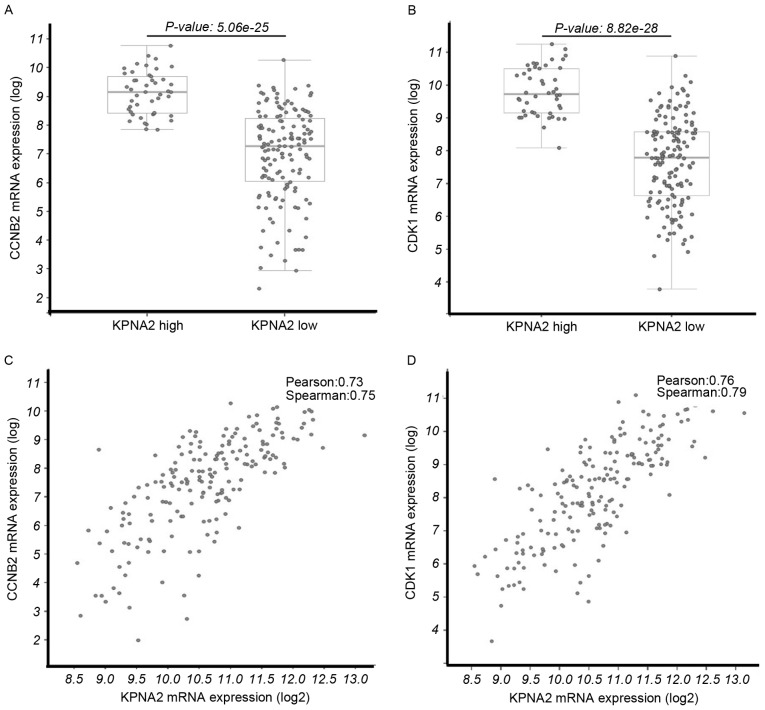
A total of 6 samples from patients with HCC were analyzed using microarrays. The mRNA expression of certain genes was upregulated compared with adjacent liver tissues in all tumor tissue samples; KPNA2 was the most significantly upregulated of all the differentially expressed genes (P<0.001; Fig. 1A). Therefore, the function of KPNA2 in the progression of malignant HCC was selected for further study. The Oncomine online tool was used to identify the correlation between the expression of KPNA2 and other identified genes from the TCGA tumor mRNA microarray data. In addition, the correlation in expression between KPNA2 and cell cycle-associated genes, including the cyclin family and CDKs, was analyzed. The results demonstrated that the expression of CCNB2 and CDK1 was correlated with KPNA2 (Fig. 1B). The mRNA expression levels of CCNB2 (P=5.06×10−25; Fig. 2A) and CDK1 (P=8.82×10−28; Fig. 2B) were significantly upregulated in the KPNA2 mRNA high expression group and the expression levels of CDK1 and CCNB2 were positively associated with the KPNA mRNA expression level (Fig. 2C and D).
Figure 1.
KPNA2 is upregulated in HCC tissues and correlated with CCNB2 and CDK1 expression. (A) A total of 6 HCC and non-tumor liver tissues were analyzed using microarrays, and the relative mRNA expression of KPNA2 was determined. (B) Analysis of KPNA2-associated genes with the Oncomine co-expression analysis tool. The mRNA expression levels of a number of genes, including CCNB2 and CDK1, were significantly correlated with KPNA2 expression. KPNA2, karyopherin subunit-α 2; HCC, hepatocellular carcinoma; CCNB2, cyclin B2; CDK1, cyclin-dependent kinase 1.
Figure 2.
Association between the mRNA expression levels of CNNB2/CDK1 and KPNA2 in HCC expression profiles from the Cancer Genome Atlas database, analyzed with the cBioportal enrichment tool. (A) CCNB2 and (B) CDK1 mRNA expression levels in the high and low KPNA2 groups. The mRNA expression levels of CCNB2 and CDK1 were upregulated in the high KPNA2 group. The correlation between the expression of KPNA2 with (C) CCNB2 and (D) CDK1. CCNB2 and CDK1 mRNA expression levels were significantly correlated with KPNA2 mRNA expression. CNNB2, cyclin B2; CDK1, cyclin-dependent kinase 1; KPNA2, karyopherin subunit-α 2.
Knockdown of KPNA2 affects the expression levels of CDK1 and cyclin B2, induces G2/M cell cycle arrest and inhibits cell proliferation
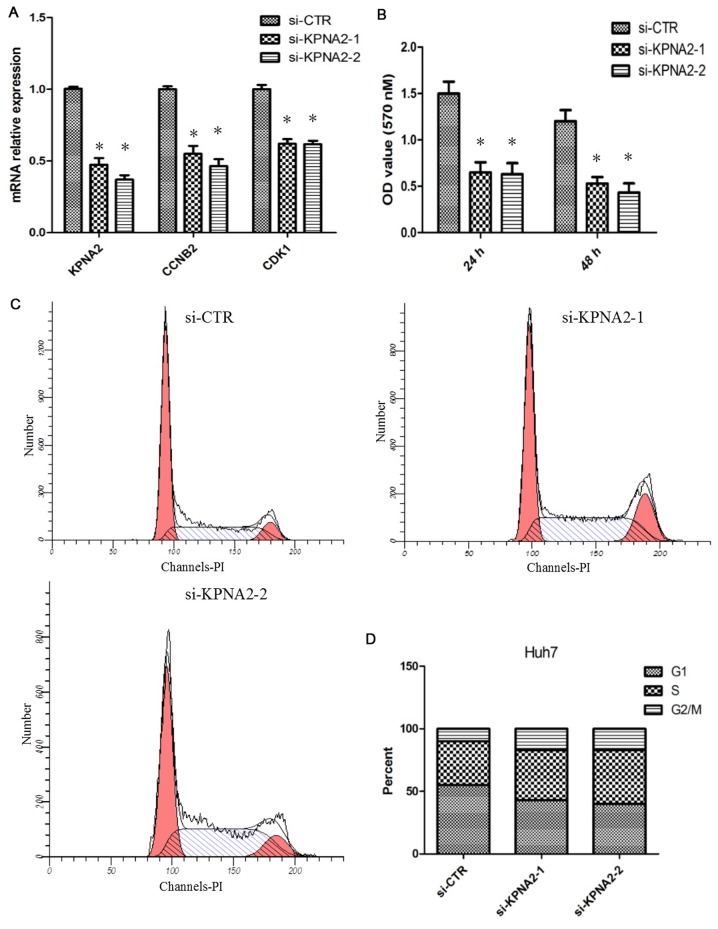
To identify the function of KPNA2 in hepatic tumor cell proliferation, siRNA was used to knockdown KPNA2 expression in Huh7 cells. The efficiency of the knockdown of KPNA2 in Huh7 cells was analyzed with RT-qPCR. It was demonstrated that the siRNA significantly reduced the KPNA2 mRNA level; the expression level of CCNB2 and CDK1 mRNA was simultaneously decreased (P<0.05; Fig. 3A). An MTT assay was performed to determine the effect of KPNA2 knockdown on Huh7 cell proliferation. The result demonstrated that KPNA2 knockdown significantly decreased the proliferative abilities of cells (P<0.05; Fig. 3B). As CDK1 and CCNB2 are associated with the regulation of the G2/M transition in the cell cycle, the effect of KPNA2 knockdown on cell cycle distribution was investigated using flow cytometry. The results suggested that knockdown of KPNA2 resulted in G2/M arrest in Huh7 cells (Fig. 3C and D).
Figure 3.
KPNA2 silencing inhibited HCC cell proliferation. (A) siRNA was used to knockdown KPNA2 in Huh7 cells, and the mRNA expression levels of KPNA2, CCNB2 and CDK1 were determined using reverse transcription-quantitative polymerase chain reaction. KPNA2 knockdown was confirmed; the mRNA expression levels of CCNB2 and CDK1 were also reduced. (B) Cell proliferation was determined using an MTT assay following KPNA2 knockdown in Huh7 cells. The knockdown of KPNA2 reduced cell proliferation. (C) Flow cytometry was applied to determine the cell cycle distribution in Huh7 cells following KPNA2 interference. (D) Quantification of (C). Knockdown of KPNA2 prompted HCC cell arrest in the G2/M phase. *P<0.05. KPNA2, karyopherin subunit-α 2; si, small interfering; CNNB2, cyclin B2; CDK1, cyclin-dependent kinase 1; CTR, control; PI, propidium iodide.
Expression of KPNA2 in tumor tissues is negatively associated with the prognosis
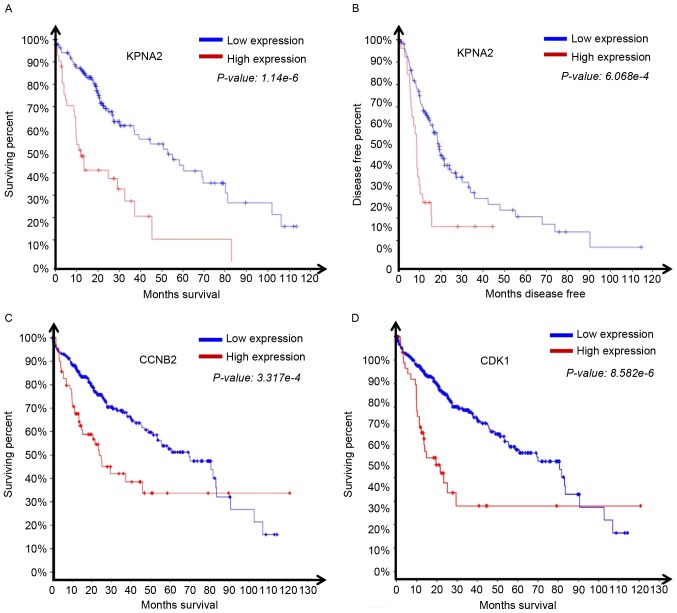
The survival data associated with HCC expression profiles from TCGA was analyzed. The results demonstrated that patients with high KPNA2 expression exhibited significantly reduced overall (1.14×10−6; Fig. 4A) and disease-free survival time (6.07×10−4; Fig. 4B) compared with the patients with low KPNA2 expression. We submitted in OncoLnc (http://www.oncolnc.org). The hazard ratio for high KPNA2 expression in HCC was 0.617. It was then assessed whether the expression of the KPNA2-associated genes CDK1 and CCNB2 affected overall survival time. The overall survival time of patients exhibiting a relatively high level of CCNB2 (P=3.32×10−4; Fig. 4C) or CDK1 (P=8.58×10−6; Fig. 4D) was significantly reduced. The results revealed a role for KPNA2, CDK1 and CCNB2 expression in the progression of HCC.
Figure 4.
KPNA2 expression is negatively associated with patient overall and disease-free survival time. (A) Overall survival curve for patients with high and low KPNA2 expression. (B) Disease-free survival curve for patients with high and low KPNA2 expression. (C) Overall survival curve for patients with high and low CCNB2 expression. (D) Overall survival curve for patients with high and low CDK1 expression. All figure sections were generated using cBioportal. KPNA2, karyopherin-α; CNNB2, cyclin B2; CDK1, cyclin-dependent kinase 1.
Discussion
HCC occurs worldwide, and the diversity in the genetic backgrounds of patients with HCC may cause difficulty in identifying the genes that promote the progression of HCC (12). In the present study, tumor tissues were selected from 6 patients with HCC and analysis was performed using microarrays; it was identified that KPNA2 was the most significantly upregulated gene. Therefore, it was hypothesized that KPNA2 may promote cancer progression.
TCGA is a public cancer research database containing high-throughput genome sequencing and transcriptome data to enable the study of cancer with bioinformatics tools. TCGA data was selected using the cBioportal and Oncomine online tools (13). To identify the genes associated with the KPNA2 gene in a wider HCC population. In the present study, the expression level of genes in the cyclin B family were demonstrated to be associated with the expression of KPNA2.
Unrestricted cell growth and proliferation is a hallmark of cancer cells (14). In a normal cell, cell proliferation is precisely controlled by cell cyclins and CDKs (15). In the present study, CCNB2 and CDK1 were relatively abundant in tumor tissues and their expression was significantly correlated with KPNA2 expression. CDK1, also known as CDC2, binds cyclin B1 and B2 to promote G2/M transition (16). The dimerization of CCNB2 with CDK1 is an essential component of the cell cycle regulatory machinery (17). A previous study demonstrated that KPNA2 affected cancer progression by regulating DNA damage response protein subcellular location (4). The CDK1 amino acid sequence was analyzed and an NLS was identified (18), which may be recognized by KPNA2 to result in CDK1 nuclear transportation. Further study is required to characterize the process of CCNB2/CDK1 transcriptional activation, the effect on cell cycle progression and how this is associated with KPNA2.
To validate an interaction between KPNA2, CCNB2 and CDK1 in HCC, Huh7 cells were selected. KPNA2 expression was silenced with siRNA. It was demonstrated that KPNA interference directly decreased the expression of cyclin B2 and CDK1 at the mRNA. In addition, it was revealed that cell proliferation was inhibited by KPNA interference. Therefore, the prediction of genes associated with genes, including KPNA2, using the cBioportal tool may be reliable, as the prediction result could be experimentally verified. Additionally, the present study aimed to determine whether KPNA2 was associated with the clinical prognosis for patients with HCC. Significant differences in overall and disease-free survival time were identified between patients with high and low KPNA2 expression; patients with high KPNA2 expression exhibited a relatively poor prognosis.
In conclusion, the results of the present study revealed that high KPNA2 expression is a risk factor in HCC and that KPNA2 regulated the cell cycle checkpoint-associated proteins cyclin B2 and CDK1. Therefore, KPNA2 may be a novel therapeutic target in HCC.
Acknowledgements
The authors thank Dr Xi Lan Shi (Department of Pharmacology, Kunming Medical University) for his critical comments on data analysis and presentation.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Housman G, Byler S, Heerboth S, Lapinska K, Longacre M, Snyder N, Sarkar S. Drug resistance in cancer: An overview. Cancers (Basel) 2014;6:1769–1792. doi: 10.3390/cancers6031769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sukowati CH, Rosso N, Crocè LS, Tiribelli C. Hepatic cancer stem cells and drug resistance: Relevance in targeted therapies for hepatocellular carcinoma. World J Hepatol. 2010;2:114–126. doi: 10.4254/wjh.v2.i3.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alshareeda AT, Negm OH, Green AR, Nolan CC, Tighe P, Albarakati N, Sultana R, Madhusudan S, Ellis IO, Rakha EA. KPNA2 is a nuclear export protein that contributes to aberrant localisation of key proteins and poor prognosis of breast cancer. Br J Cancer. 2015;112:1929–1937. doi: 10.1038/bjc.2015.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang L, Wang HY, Li JD, Wang JH, Zhou Y, Luo RZ, Yun JP, Zhang Y, Jia WH, Zheng M. KPNA2 promotes cell proliferation and tumorigenicity in epithelial ovarian carcinoma through upregulation of c-Myc and downregulation of FOXO3a. Cell Death Dis. 2013;4:e745. doi: 10.1038/cddis.2013.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teng SC, Wu KJ, Tseng SF, Wong CW, Kao L. Importin KPNA2, NBS1, DNA repair and tumorigenesis. J Mol Histol. 2006;37:293–299. doi: 10.1007/s10735-006-9032-y. [DOI] [PubMed] [Google Scholar]
- 7.Christiansen A, Dyrskjøt L. The functional role of the novel biomarker karyopherin α 2 (KPNA2) in cancer. Cancer Lett. 2013;331:18–23. doi: 10.1016/j.canlet.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim IS, Kim DH, Han SM, Chin MU, Nam HJ, Cho HP, Choi SY, Song BJ, Kim ER, Bae YS, Moon YH. Truncated form of importin alpha identified in breast cancer cell inhibits nuclear import of p53. J Biol Chem. 2000;275:23139–23145. doi: 10.1074/jbc.M909256199. [DOI] [PubMed] [Google Scholar]
- 9.Li XL, Jia LL, Shi MM, Li X, Li ZH, Li HF, Wang EH, Jia XS. Downregulation of KPNA2 in non-small-cell lung cancer is associated with Oct4 expression. J Transl Med. 2013;11:232. doi: 10.1186/1479-5876-11-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Maass T, Sfakianakis I, Staib F, Krupp M, Galle PR, Teufel A. Microarray-based gene expression analysis of hepatocellular carcinoma. Curr Genomics. 2010;11:261–268. doi: 10.2174/138920210791233063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: A changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 16.Dorée M, Hunt T. From Cdc2 to Cdk1: When did the cell cycle kinase join its cyclin partner? J Cell Sci. 2002;115:2461–2464. doi: 10.1242/jcs.115.12.2461. [DOI] [PubMed] [Google Scholar]
- 17.Huang Y, Sramkoski RM, Jacobberger JW. The kinetics of G2 and M transitions regulated by B cyclins. PLoS One. 2013;8:e80861. doi: 10.1371/journal.pone.0080861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs; Proc Natl Acad Sci USA; 2009; pp. 10171–10176. [DOI] [PMC free article] [PubMed] [Google Scholar]