Abstract
Cytoplasmic polyadenylation element-binding protein-4 (CPEB4) is involved in several biological processes that are associated with cancer progression. However, it remains unknown whether CPEB4 expression levels are associated with head and neck squamous cell carcinoma (HNSCC). The aim of the present study was to explore the potential function of CPEB4 in HNSCC. The expression of CPEB4 was analyzed in HNSCC from six Gene Expression Omnibus (GEO) datasets. Immunohistochemical staining was conducted to examine CPEB4 protein levels in an HNSCC tissue microarray (TMA). According to the GEO dataset analyses, CPEB4 gene expression was downregulated in HNSCC compared with normal samples (P<0.05). Notably, a statistical difference was observed between different tumor grades (P<0.05). Furthermore, the methylation of the CPEB4 gene in HNSCC was significantly increased compared with that observed in normal samples (P<0.01). The outcome from the TMA demonstrated that CPEB4 protein expression in human HNSCC tumors was significantly decreased compared with normal samples (P<0.05). In addition, the expression of CPEB4 protein was negatively associated with histological grades of HNSCC (P<0.05). The results from the present study suggested that CPEB4 may function as a tumor suppressor gene in HNSCC, which identifies the potential value of CPEB4 in predicting prognosis of HNSCC. Hypermethylation of the CPEB4 gene may be responsible for the downregulation of CPEB4 expression in HNSCC and result in tumorigenesis.
Keywords: cytoplasmic polyadenylation element-binding protein-4, head and neck squamous cell carcinoma, Gene Expression Omnibus dataset, tissue microassay, hypermethylation
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common type of cancer worldwide, representing 90% of all head and neck cancers (1–3). Despite improvements in therapeutic interventions over the last 20 years, in 2010 the 5-year survival rate was ~50% (4,5). According to the literature, from 1975–2010, the morbidity and mortality of patients with HNSCC remained at a high level, with >650,000 novel HNSCC cases diagnosed annually worldwide (6). To date, a series of biomarkers associated with HNSCC including p16, p53, epidermal growth factor receptor and vascular endothelial growth factor have been identified (7), which have proven to be beneficial in directing diagnosis, prognosis and therapy for this disease. However, these are insufficient to accurately define the pathogenesis of HNSCC. Therefore, there is an urgent requirement to explore and identify novel molecular biomarkers that are associated with HNSCC, as potential therapeutic targets.
Cytoplasmic polyadenylation element-binding protein (CPEB) is a type of sequence-specific highly conserved RNA-binding protein (8,9), which regulates the translational activation and cytoplasmic polyadenylation of target mRNAs (10). The CPEB family of proteins all confer a similar structure, including highly variable N-termini and relatively conservative C-termini, consisting of two RNA recognition motifs and a zinc finger domain essential for RNA binding (11–13). The family of CPEBs, which are widely expressed in vertebrates, are composed of four family members (CPEB1-CPEB4), of which CPEB1 differs from CPEB2-CPEB4 in terms of binding specificity and regulatory domains (11,14,15). Several previous studies have demonstrated that CPEBs are associated with various biological processes, including cell cycle progression (16), development (17,18), cellular senescence (19) and malignant tumor progression (20). The direct link between the aberrant expression of CPEBs and tumorigenesis has been previously observed in glioblastomas and pancreatic ductal adenocarcinomas (PDA) (11).
CPEB4 belongs to the CPEB family, and functions to directly mediate translation and polyadenylation (11). A previous study reported that CPEB4 was abundantly expressed in glioblastomas and PDA, and influenced the acceleration of tumor proliferation, vascularization and invasion (21). Several previous studies researched the association between CPEB4 and various types of cancer, and further validated the crucial involvement of CPEB4 in tumorigenesis. Taken together, these results highlighted the probability that CPEB4-mediated abnormal regulation of downstream target gene expression may be a common mechanism in malignant tumors (21,22). To the best of our knowledge, the biological functions and clinical significance of CPEB4 expression in HNSCC has not been previously reported. The aim of the present study was to investigate the potential function of CPEB4 in the tumorigenesis of HNSCC, and identify the potential underlying molecular mechanisms involved.
Materials and methods
Datasets retrieved
The Gene Expression Omnibus (GEO) database, established and maintained by the National Center for Biotechnology Information, is an international public functional genomics data repository (23). A total of six microarray datasets were downloaded from the GEO repository for data analysis, including GSE33205 (24), GSE59102 (25), GSE58911 (26), GSE51985 (27), GSE39366 (28) and GSE25093 (29). The CPEB4 expression data series originated from tumor tissues or adjacent non-neoplastic tissues of human HNSCC, benign lesions of the head and neck, and normal tissues from non-HNSCC. In the six datasets, the association between CPEB4 expression and the occurrence of HNSCC was analyzed independently.
Tissue microarray (TMA)
The TMA was obtained from US Biomax, Inc. (Rockville, MD, USA; cat no. HN803b). The TMA was composed of 7 nasal carcinoma, 31 laryngeal carcinoma, 32 tongue carcinoma and 10 normal tongue tissues, from subjects ranging between 18 and 90 years of age, with a median age of 53.4 years. The samples were obtained from 18 women and 62 men. Detailed information of the TMA is summarized in Table I. The present study was approved by the appropriate Ethical Committees of Renmin Hospital of Wuhan University.
Table I.
Characteristics of the head and neck squamous cell carcinoma tissue microarray.
| Patient characteristic | n (%) |
|---|---|
| Localization | |
| Tongue | 42 (52.5) |
| Larynx | 31 (38.6) |
| Nose | 7 (8.9) |
| T-stage | |
| T1 | 5 (8.3) |
| T2 | 30 (50.0) |
| T3 | 17 (28.3) |
| T4 | 8 (13.3) |
| N-stage | |
| N0 | 42 (68.9) |
| N1 | 16 (26.2) |
| N2 | 3 (5.0) |
| Grade | |
| I | 12 (16.7) |
| II | 36 (50.0) |
| III | 24 (33.3) |
| NAT | 2 (2.5) |
| Normal | 9 (11.2) |
NAT, no grade data was provided.
Immunohistochemical (IHC) staining
IHC analysis of CPEB4 protein expression in the TMA was performed, using the two-step staining method. TMA slides were dried for 2 h at 60°C, dewaxed in pure xylene at room temperature three times for 15 min, and rehydrated through a graded series of alcohols. Antigen retrieval was performed on TMA slides, which were incubated in sodium citrate buffer (pH 6.0) at 100°C for 15 min in the pressure cooker. Slides were immersed in 0.3% hydrogen peroxide at room temperature for 10 min to block endogenous peroxidase activity. Non-specific binding was blocked with normal goat serum (50 µl) (cat. no. KIT-9706; Fuzhou Maixin Biotech, Co., Ltd., Fuzhou, China) at room temperature for 10 min. Subsequently, the slides were incubated with primary antibodies against CPEB4 (1:200; cat. no. HPA038394; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) overnight at 4°C. TMA slides were rinsed twice in PBS at room temperature for 5 min each time, prior to incubation with the secondary antibody (poly-horseradish peroxidase-conjugated goat anti-rabbit IgG; cat. no. KIT-9706; 1:200, Maixin Biotech, Co., Ltd, Fuzhou, China) for 30 min at room temperature. TMA sections were stained with 3,3-diaminobenzidine at room temperature for 5 min to detect the antigen, and the cell nucleus was counterstained with Mayer's hematoxylin at room temperature for 3 min. Slides were dehydrated with an ascending series of alcohols, prior to mounting.
Assessment of IHC staining
The majority of the CPEB4 protein is located in the cytoplasm, but enters the cell nucleus in response to the lack of oxygen and glucose (30). The TMA slides were scored by two independent investigators using a two-index scoring system, which considers the staining intensity and proportion of tumor cells stained. A 4-point intensity scoring system was graded as follows: Grade 0, negative staining; grade 1, weak staining, light yellow; grade 2, moderate staining, yellow-brown; and grade 3, strong staining, brown. According to the percentage of positive cancer cells, the proportion score was divided into four levels: 0–25% positive tumor cells, 26–50% positive tumor cells, 51–75% positive tumor cells and 76–100% positive tumor cells. Finally, the total scores from each stained area were calculated as a composite expression score (CES; range 0–12) for further statistical analysis, using the following formula; CES=intensity × proportion. The CES for tumors was defined as negative (score=0), weak positive (score=1–4), positive (score=5–8) and strong positive (score=9–12).
Statistical analysis
Statistical evaluation was performed using SPSS (version 16.0; SPSS, Inc., Chicago, IL, USA). All gene expression data downloaded from GEO were inputted into Microsoft Excel. Analysis of variance was used to compare differences among the groups, with a Tukey's multiple comparison test performed following ANOVA. An unpaired Student's t-test was used for comparisons between two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
CPEB4 gene expression in HNSCC and normal tissues
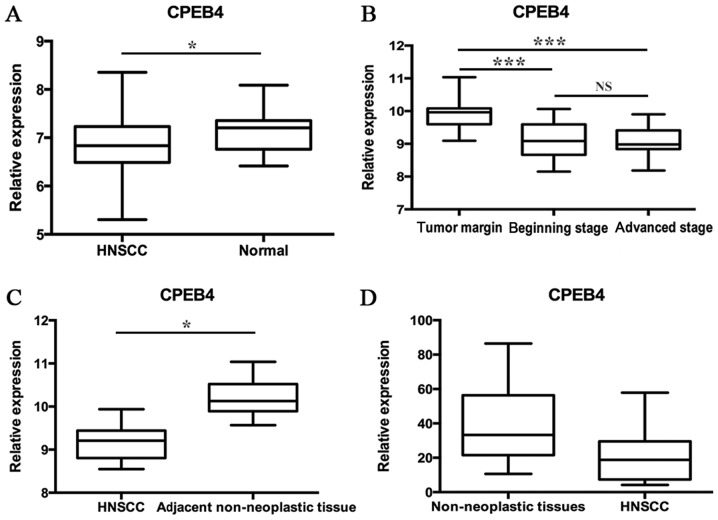
The GSE33205 data consisted of 44 HNSCC tumor samples and 25 normal mucosal samples, of which the normal samples were taken from uvulopalatopharyngoplasty. The statistical analyses were made between tumors and normal tissues (P<0.05; Fig. 1A). The GSE59102 data included 29 tumor specimens and 13 margin samples, which were derived from patients suffering surgical procedures of laryngeal squamous cell carcinoma (LSCC). Results indicated that the CPEB4 gene expression level was lower in early- and advanced-stage LSCC tumor tissues compared with the tumor margin (P<0.0001), whereas no significant difference between early- and advanced-stage LSCC tumor tissues was identified (Fig. 1B). The GSE58911 data consisted of 15 paired normal and tumor samples obtained from patients who were diagnosed with HNSCC (oropharynx, hypopharynx and larynx). Prior to therapy, the samples taken from a site at a distance from the tumor tissues and from the tumor site were used for normal samples and tumor samples, respectively. The statistical analyses were made between normal and tumor samples (P<0.01; Fig. 1C). The GSE51985 data originated from 10 patients undergoing surgery for LSCC. The cancer tissues were compared with corresponding adjacent non-neoplastic tissues. The statistical analyses were made between cancer tissues and the corresponding adjacent non-neoplastic tissues (P=0.0577; Fig. 1D). These data indicated that, in the majority of cases, CPEB4 expression was significantly downregulated in HNSCC samples compared with normal corresponding tissue samples.
Figure 1.
Analysis of CPEB4 expression levels in HNSCC and normal tissues from GEO datasets (A) Expression level of CPEB4 in HNSCC compared with normal controls from uvulopalatopharyngoplasty (GSE 33205). (B) Expression level of CPEB4 in HNSCC compared with normal samples from tumor margin (GSE 59102). (C) Expression level of CPEB4 in HNSCC compared with normal tissues (GSE 58911). (D) Expression level of CPEB4 in HNSCC compared with normal tissues (GSE 51985). *P<0.01, with comparisons indicated by lines, determined using analysis of variance and Tukey's post hoc test. CPEB4, cytoplasmic polyadenylation element-binding protein-4; HNSCC, head and neck squamous cell carcinoma.
Associations between CPEB4 gene expression and pathological grading
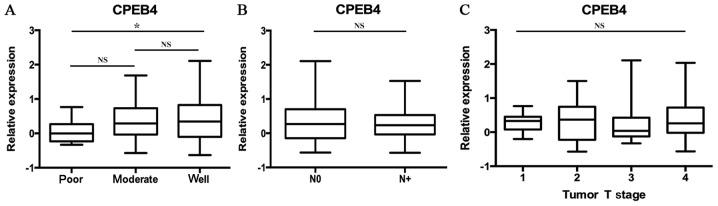
The data of GSE39366 included a total of 138 HNSCC specimens. According to tumor differentiation, these samples were divided into three grades: Poorly, moderately and well-differentiated. The statistical analyses were made between these three grades (P<0.05; Fig. 2A). The results indicated that decreased CPEB4 expression was associated with increasing pathological grading. Statistically significant differences were observed for CPEB4 expression between poor and well tumor grades (P<0.05). The CPEB4 gene expression level was lower in poorly differentiated HNSCC tissues compared with well-differentiated HNSCC tissues (P<0.05), whereas no significant difference in CPEB4 expression between distinct tumor grades was identified.
Figure 2.
Analysis of CPEB4 expression between grades, N-stages and T-stages from the GEO database. (A) CPEB4 expression level in each grade of HNSCC (GSE 39366). (B) CPEB4 expression level in each N-stage of HNSCC (GSE 39366). (C) CPEB4 expression level in each T-stage of HNSCC (GSE 39366). *P<0.05 with comparisons indicated by lines, determined using analysis of variance and Tukey's post hoc test (A and C) or Student's t-test (B). CPEB4, cytoplasmic polyadenylation element-binding protein-4; HNSCC, head and neck squamous cell carcinoma.
Associations between CPEB4 gene expression and N-stage
Statistical analyses of the expression of the CPEB4 gene were made between different N-stages of HNSCC, from the data originating from the dataset GSE39366 (P>0.05; Fig. 2B). No statistically significant associations were identified between CPEB4 gene expression and N-stages of HNSCC.
Associations between CPEB4 gene expression and T-stage
Statistical analyses of CPEB4 gene expression were made between different T-stages of HNSCC, using the data obtained from the dataset GSE39366 (P>0.05; Fig. 2C). No statistically significant associations were identified between CPEB4 gene expression and T-stages of HNSCC.
Analysis of CPEB4 gene methylation in HNSCC and normal tissues
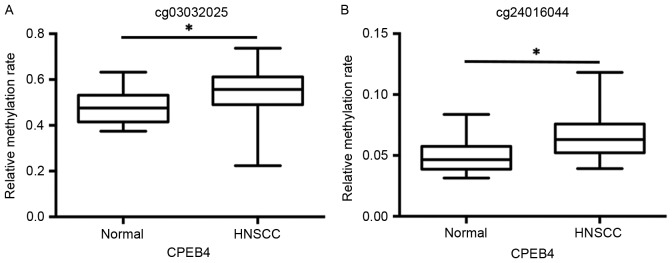
The data from the GSE25093 dataset was collected from 91 fresh-frozen HNSCC tumor tissues and 18 fresh-frozen normal samples drawn from the larynx, pharynx and oral cavity. The data from the cg03032025 and cg24016044 datasets revealed that CPEB4 methylation was significantly increased in HNSCC tumor samples and compared with normal tissue samples (P<0.01; Fig. 3A and B).
Figure 3.
Analysis of the methylation levels of CPEB4 in HNSCC and normal controls from GEO database. (A) Methylation levels of the CPEB4 gene in HNSCC compared with normal tissues (cg03032025). (B) Methylation level of CPEB4 in HNSCC compared with normal tissues (cg24016044). *P<0.01 with comparisons indicated by lines, determined using Student's t-test. CPEB4, cytoplasmic polyadenylation element-binding protein-4; HNSCC, head and neck squamous cell carcinoma.
CPEB4 protein expression in the HNSCC TMA
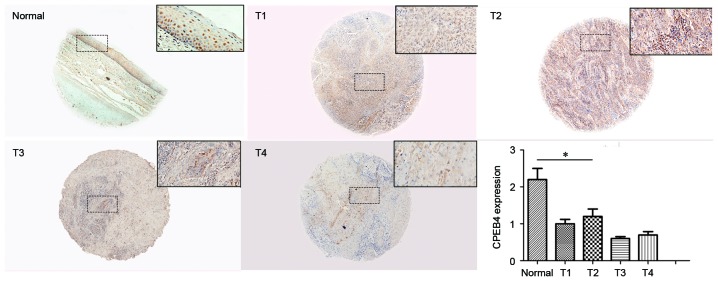
To further verify CPEB4 expression in HNSCC, IHC staining was performed to examine CPEB4 protein in the TMA. Each TMA section was analyzed by two individual index parameters, i.e., proportion and intensity, which were transformed into a CES for assessment. CPEB4 protein expression in HNSCC tissue and normal tissue is presented in Fig. 4. The immunostaining indicated that CPEB4 protein was significantly decreased in tumor tissues compared with normal tissues (P<0.05). This provides further evidence that CPEB4 was frequently downregulated in HNSCC, which is comparable with the outcome of CPEB4 gene expression, obtained from the GEO dataset.
Figure 4.
CPEB4 protein levels in normal and HNSCC tissues with different T-stages. CPEB4 protein expression in each T-stage was detected using immunohistochemical staining. No significant differences in CPEB4 protein expression were detected between different T-stages of HNSCC in the tissue microarray. *P>0.05 vs. normal. Analysis of variance was used for the comparison among four T-stages. Student's t-test was used for comparisons between normal tissues and tumor tissues. CPEB4, cytoplasmic polyadenylation element-binding protein-4; HNSCC, head and neck squamous cell carcinoma. Magnification, ×40.
Differences in CPEB4 protein between different tumor grades in the TMA
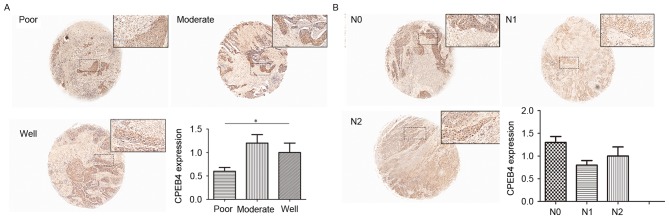
The range of tumor grades (poor to well) represents increasing deterioration of histological differentiation, ranging from well differentiated to poorly differentiated. CES scores were calculated for each tumor grade for statistical analysis. The results from the present study demonstrated that CES for poorly differentiated tumors was significantly decreased compared with that of moderately differentiated tumors (P<0.05; Fig. 5A). Furthermore, moderately differentiated tumors had a decreased CES score compared with that of the well differentiated tumors (P<0.05; Fig. 5A). These data demonstrated that a significant decrease in expression of CPEB4 protein was associated with the poorest tumor differentiation.
Figure 5.
CPEB4 protein levels in different grades and N-stages of HNSCC tissues. (A) CPEB4 protein expression in each tumor grade was detected using immunohistochemical staining. CPEB4 protein expression was decreased with increasing tumor grade. (B) CPEB4 protein expression in each N-stage was detected using immunohistochemical staining. No significant differences in CPEB4 protein expression were detected between different N-stages of HNSCC in tissue microarray. *P>0.05 with comparisons indicated by lines, determined using analysis of variance. CPEB4, cytoplasmic polyadenylation element-binding protein-4; HNSCC, head and neck squamous cell carcinoma. Magnification, ×40.
Differences in CPEB4 protein expression between different T-stages in TMA
CES scores for each T-stage were calculated for statistical analysis. No significant differences for CPEB4 protein were observed between different T-stages (P>0.05; Fig. 4), which demonstrated that no significant associations between CPEB4 and T-stage of HNSCC were identified. The outcome was comparable with that for the analysis from the GEO dataset.
Differences in CPEB4 protein between different N-stages in TMA
The same method as that for T-stage was used for N-stage. However, there was no significant difference in CPEB4 protein expression between N-stages (P>0.05; Fig. 5B), indicating that CPEB4 was not significantly associated with the N-stage of HNSCC. This result was consistent with the conclusion from the GEO dataset.
Discussion
Ortiz-Zapater et al (21) initially demonstrated the direct link between CPEB4 expression and cancer etiology, and suggested that overexpression may be a common mechanism for regulating the reprogramming of gene expression involved in cancer progression. Subsequently, a number of studies have been performed that demonstrated that the aberrant expression of CPEB4 is significantly associated with the clinical prognosis of patients with malignant tumors (22,31–33).
In the present study, CPEB4 expression was investigated between various types of cancer and corresponding normal tissues. Notably in previous studies, the expression of CPEB4 protein was increased in several malignant tumors, including metastatic prostate cancer (34), colorectal cancer (CRC) (31), astrocytic tumors (32), invasive ductal breast carcinoma (IDC) (22) and glioma (33). In contrast, another study reported the opposite outcome in that CPEB4 protein was downregulated in hepatocellular carcinoma and non-small cell lung cancer (35). However, to date, there is limited knowledge concerning the association between CPEB4 abnormalities and HNSCC. In order to investigate CPEB4 gene expression levels in HNSCC, four independent GEO datasets, including GSE33205, GSE59102, GSE58911 and GSE51985, were analyzed. The analysis of CPEB4 gene expression between cancer and normal samples identified statistical differences from three datasets, in which P-values were confirmed to be <0.05. The results of the present study confirmed that CPEB4 expression was downregulated in the majority of cases of HNSCC compared with normal tissues, based on the results obtained from GEO datasets.
In addition, an HNSCC TMA was used to examine CPEB4 protein expression to further verify the gene expression results from GEO datasets. The results from the present study confirmed that CPEB4 protein expression was significantly decreased in cancer tissues compared with normal corresponding tissues, consistent with the outcomes from the GEO datasets. Taken together, the results of the present study confirmed that CPEB4 was consistently downregulated in HNSCC tissues, which may be associated with the progression of tumorigenesis. Therefore, CPEB4 may function as a tumor suppressor gene in HNSCC.
According to previous studies, there has been controversy surrounding CPEB4 expression levels in various malignant tumors (22,31,35,36). Therefore, we speculate that the discrepancies between the results of the present study and those of previous studies may be due to the different pathological patterns of cancers. For example, CPEB4 expression was upregulated in adenoma but downregulated in solid tumors. In different cancer cells, CPEB4 binds to various downstream genes and regulates the reprogramming of these, acting either as a cancer suppressor gene or an oncogene and affecting tumor development in several ways (22,31,32,34).
Several previous studies have been performed to investigate the associations between CPEB4 and clinicopathological parameters, including age, sex, T-stage and N-stage (22,31,32). The positive association between CPEB4 expression and T-stage (tumor size) or N-stage (lymph node status) of cancer was demonstrated in astrocytic tumors, IDC and CRC (22,31,32). However, the statistical analysis from the GSE39366 dataset revealed that CPEB4 expression was not significantly associated with T-stages or N-stages in HNSCC. Furthermore, the data from the TMA which explored the association between CPEB4 protein expression and T/N-stages, revealed an outcome comparable with that from the GEO dataset. Therefore, it was concluded that there was no association between CPEB4 and T/N-stages in HNSCC. The inconsistent conclusion between the results from the present study and those of previous studies may be caused by several factors, including tumor microenvironment, location, histological origin and histological type.
The association between CPEB4 and histological grading has been identified in several previous studies, indicating that CPEB4 expression was positively associated with differing tumor grades (22,32,33). In the present study, the analysis of the GEO datasets suggested that downregulated CPEB4 expression further decreased in association with increasing histological grades of HNSCC. The differences in CPEB4 protein between different tumor grades were statistically significant. In addition, it was observed that CPEB4 protein levels were lower in high-grade HNSCC compared with that in low-grade HNSCC in TMA. Furthermore, the significant differences between different pathological grades supported our previous conclusions from the GEO dataset. In summary, it was confirmed that CPEB4 expression was negatively correlated with pathological grading, and CPEB4 may represent a valuable marker for HNSCC prognosis.
Currently, the molecular mechanisms underlying the contribution of CPEB4 to cancer progression have not been elucidated. According to previous studies, CPEB4 targets specific genes that are associated with tumorigenesis. For example, CPEB4 us able to regulate the activation of genes including Ras-associated molecules, cyclins, cell signaling components, apoptosis-related molecules, chromatin remodeling proteins, metabolic enzymes, and genes involved in migration and metastasis, indicating a significant effect on tumor development in several capacities (21,37).
There are three suggested mechanisms responsible for CPEB4-mediated malignant formation in cancer. CPEB4 may be critically involved in regulating several downstream signaling pathways relevant to proliferation, apoptosis and the cell cycle (31,34). Zhong et al (31) reported that knockdown of CPEB4 in CRC cells may contribute to the downregulation of B-cell lymphoma (Bcl) extra-large expression and upregulation of Bcl-2-associated X protein, apoptosis regulation, resulting in the promotion of cell apoptosis and the inhibition of cell proliferation. Xu and Liu (34) demonstrated that CPEB4 accelerated the development of metastasis and invasion through the transforming growth factor-β signaling pathway. In addition, the expression of vital genes involved in tumorigenesis were modulated by CPEB4. In pancreatic carcinoma, enhanced CPEB4 expression accelerated the progression of tumor malignancy by increasing the expression level of tissue plasminogen activator mRNA which is an essential component for PDA (21). Furthermore, CPEB4-induced translational control of vimentin may be responsible for the development of astrocytic tumors (32). Finally, CPEB4 may act as a downstream target gene of other oncogenes or cancer suppressor genes, and be involved in promoting invasion and metastasis of tumor cells. MicroRNA-203, a tumor suppressor, directly targets CPEB4 and negatively regulates CPEB4 expression to influence the apoptosis signaling pathway in CRC (31). In addition, microRNA-550a binds to the 3′-untranslated region of the CPEB4 gene to negatively regulate CPEB4 expression, leading to accelerated migration and invasion of liver cancer cells (35). Furthermore, the downregulation of CPEB4 expression was induced by microRNA-1246 to facilitate migration and invasion of tumor cells in non-small cell lung carcinoma (36). It may be speculated that microRNA-mediated CPEB4 regulation frequently exists in tumor progression.
However, to the best of our knowledge, the present study is the first to report on the potential action of CPEB4 in the tumor progression of HNSCC. The results of the present study implicated an important function for CPEB4 in HNSCC tumorigenesis, and further investigation is urgently required. In the present study, the evidence from the GEO dataset analysis demonstrated that the methylation status of the CPEB4 gene was significantly higher in HNSCC compared with normal samples. This is indicative of an association between the hypermethylation of CPEB4 and the downregulation of CPEB4 in HNSCC. Thus, is it hypothesized that CPEB4 hypermethylation may be involved in the tumorigenesis of HNSCC by downregulating CPEB4 gene expression. This result warrants further investigation in a follow-up study to confirm the underlying mechanisms involved.
In conclusion, CPEB4 gene expression was significantly downregulated in HNSCC, which indicates that CPEB4 may act as a tumor suppressor gene in HNSCC. Considering that CPEB4 gene expression was associated with pathological grading, this indicates the potential value of CPEB4 in directing prognosis for HNSCC. Hypermethylation of the CPEB4 gene may be responsible for the downregulation of CPEB4 gene expression in HNSCC, and results in cancer tumorigenesis. However, further consideration must be given to the potential of CPEB4 as a therapeutic target for patients with HNSCC.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 81372880) and the Natural Science Foundation of Hubei Province (grant no. 2012FFA045).
References
- 1.Howren MB, Christensen AJ, Karnell LH, Funk GF. Psychological factors associated with head and neck cancer treatment and survivorship: Evidence and opportunities for behavioral medicine. J Consult Clin Psychol. 2013;81:299–317. doi: 10.1037/a0029940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khan Z, Khan AA, Prasad GB, Khan N, Tiwari RP, Bisen PS. Growth inhibition and chemo-radiosensitization of head and neck squamous cell carcinoma (HNSCC) by survivin-siRNA lentivirus. Radiother Oncol. 2016;118:359–368. doi: 10.1016/j.radonc.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Döbrossy L. Epidemiology of head and neck cancer: Magnitude of the problem. Cancer Metastasis Rev. 2005;24:9–17. doi: 10.1007/s10555-005-5044-4. [DOI] [PubMed] [Google Scholar]
- 4.Boeckx C, Op de Beeck K, Wouters A, Deschoolmeester V, Limame R, Zwaenepoel K, Specenier P, Pauwels P, Vermorken JB, Peeters M, et al. Overcoming cetuximab resistance in HNSCC: The role of AURKB and DUSP proteins. Cancer Lett. 2014;354:365–377. doi: 10.1016/j.canlet.2014.08.039. [DOI] [PubMed] [Google Scholar]
- 5.Molinolo AA, Amornphimoltham P, Squarize CH, Castilho RM, Patel V, Gutkind JS. Dysregulated molecular networks in head and neck carcinogenesis. Oral Oncol. 2009;45:324–334. doi: 10.1016/j.oraloncology.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kulasinghe A, Perry C, Jovanovic L, Nelson C, Punyadeera C. Circulating tumour cells in metastatic head and neck cancers. Int J Cancer. 2015;136:2515–2523. doi: 10.1002/ijc.29108. [DOI] [PubMed] [Google Scholar]
- 7.Thomas GR, Nadiminti H, Regalado J. Molecular predictors of clinical outcome in patients with head and neck squamous cell carcinoma. Int J Exp Pathol. 2005;86:347–363. doi: 10.1111/j.0959-9673.2005.00447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burns DM, D'Ambrogio A, Nottrott S, Richter JD. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature. 2011;473:105–108. doi: 10.1038/nature09908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendez R, Richter JD. Translational control by CPEB: A means to the end. Nat Rev Mol Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- 10.Radford HE, Meijer HA, de Moor CH. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim Biophys Acta. 2008;1779:217–229. doi: 10.1016/j.bbagrm.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernández-Miranda G, Méndez R. The CPEB-family of proteins, translational control in senescence and cancer. Ageing Res Rev. 2012;11:460–472. doi: 10.1016/j.arr.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Hake LE, Mendez R, Richter JD. Specificity of RNA binding by CPEB: Requirement for RNA recognition motifs and a novel zinc finger. Mol Cell Biol. 1998;18:685–693. doi: 10.1128/MCB.18.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hake LE, Richter JD. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during xenopus oocyte maturation. Cell. 1994;79:617–627. doi: 10.1016/0092-8674(94)90547-9. [DOI] [PubMed] [Google Scholar]
- 14.Theis M, Si K, Kandel ER. Two previously undescribed members of the mouse CPEB family of genes and their inducible expression in the principal cell layers of the hippocampus; Proc Natl Acad Sci USA; 2003; pp. 9602–9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang YS, Kan MC, Lin CL, Richter JD. CPEB3 and CPEB4 in neurons: Analysis of RNA-binding specificity and translational control of AMPA receptor GluR2 mRNA. EMBO J. 2006;25:4865–4876. doi: 10.1038/sj.emboj.7601322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tay J, Richter JD. Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev Cell. 2001;1:201–213. doi: 10.1016/S1534-5807(01)00025-9. [DOI] [PubMed] [Google Scholar]
- 17.Hafer N, Xu S, Bhat KM, Schedl P. The drosophila CPEB protein Orb2 has a novel expression pattern and is important for asymmetric cell division and nervous system function. Genetics. 2011;189:907–921. doi: 10.1534/genetics.110.123646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang XP, Cooper NG. Characterization of the transcripts and protein isoforms for cytoplasmic polyadenylation element binding protein-3 (CPEB3) in the mouse retina. BMC Mol Biol. 2009;10:109. doi: 10.1186/1471-2199-10-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groisman I, Ivshina M, Marin V, Kennedy NJ, Davis RJ, Richter JD. Control of cellular senescence by CPEB. Genes Dev. 2006;20:2701–2712. doi: 10.1101/gad.1438906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen CN, Ketabi Z, Rosenstierne MW, Palle C, Boesen HC, Norrild B. Expression of CPEB, GAPDH and U6snRNA in cervical and ovarian tissue during cancer development. APMIS. 2009;117:53–59. doi: 10.1111/j.1600-0463.2008.00015.x. [DOI] [PubMed] [Google Scholar]
- 21.Ortiz-Zapater E, Pineda D, Martínez-Bosch N, Fernández-Miranda G, Iglesias M, Alameda F, Moreno M, Eliscovich C, Eyras E, Real FX, et al. Key contribution of CPEB4-mediated translational control to cancer progression. Nat Med. 2011;18:83–90. doi: 10.1038/nm.2540. [DOI] [PubMed] [Google Scholar]
- 22.Sun HT, Wen X, Han T, Liu ZH, Li SB, Wang JG, Liu XP. Expression of CPEB4 in invasive ductal breast carcinoma and its prognostic significance. Onco Targets Ther. 2015;8:3499–3506. doi: 10.2147/OTT.S87587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41(Database Issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun W, Gaykalova DA, Ochs MF, Mambo E, Arnaoutakis D, Liu Y, Loyo M, Agrawal N, Howard J, Li R, et al. Activation of the NOTCH pathway in head and neck cancer. Cancer Res. 2014;74:1091–1104. doi: 10.1158/0008-5472.CAN-13-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Barros E Lima, Bueno R, Ramão A, Pinheiro DG, Alves CP, Kannen V, Jungbluth AA, de Araújo LF, Muys BR, Fonseca AS, Plaça JR, et al. HOX genes: Potential candidates for the progression of laryngeal squamous cell carcinoma. Tumour Biol. 2016;37:15087–15096. doi: 10.1007/s13277-016-5356-8. [DOI] [PubMed] [Google Scholar]
- 26.Lobert S, Graichen ME, Hamilton RD, Pitman KT, Garrett MR, Hicks C, Koganti T. Prognostic biomarkers for HNSCC using quantitative real-time PCR and microarray analysis: β-tubulin isotypes and the p53 interactome. Cytoskeleton (Hoboken) 2014;71:628–637. doi: 10.1002/cm.21195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Regla-Nava JA, Nieto-Torres JL, Jimenez-Guardeño JM, Fernandez-Delgado R, Fett C, Castaño-Rodríguez C, Perlman S, Enjuanes L, DeDiego ML. Severe acute respiratory syndrome coronaviruses with mutations in the E protein are attenuated and promising vaccine candidates. J Virol. 2015;89:3870–3887. doi: 10.1128/JVI.03566-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walter V, Yin X, Wilkerson MD, Cabanski CR, Zhao N, Du Y, Ang MK, Hayward MC, Salazar AH, Hoadley KA, et al. Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PLoS One. 2013;8:e56823. doi: 10.1371/journal.pone.0056823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poage GM, Houseman EA, Christensen BC, Butler RA, Avissar-Whiting M, McClean MD, Waterboer T, Pawlita M, Marsit CJ, Kelsey KT. Global hypomethylation identifies Loci targeted for hypermethylation in head and neck cancer. Clin Cancer Res. 2011;17:3579–3589. doi: 10.1158/1078-0432.CCR-11-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kan MC, Oruganty-Das A, Cooper-Morgan A, Jin G, Swanger SA, Bassell GJ, Florman H, van Leyen K, Richter JD. CPEB4 is a cell survival protein retained in the nucleus upon ischemia or endoplasmic reticulum calcium depletion. Mol Cell Biol. 2010;30:5658–5671. doi: 10.1128/MCB.00716-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong X, Xiao Y, Chen C, Wei X, Hu C, Ling X, Liu X. MicroRNA-203-mediated posttranscriptional deregulation of CPEB4 contributes to colorectal cancer progression. Biochem Biophys Res Commun. 2015;466:206–213. doi: 10.1016/j.bbrc.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Chen W, Hu Z, Li XZ, Li JL, Xu XK, Li HG, Liu Y, Liu BH, Jia WH, Li FC. CPEB4 interacts with Vimentin and involves in progressive features and poor prognosis of patients with astrocytic tumors. Tumour Biol. 2016;37:5075–5087. doi: 10.1007/s13277-015-3975-0. [DOI] [PubMed] [Google Scholar]
- 33.Hu W, Yang Y, Xi S, Sai K, Su D, Zhang X, Lin S, Zeng J. Expression of CPEB4 in human glioma and its correlations with prognosis. Medicine (Baltimore) 2015;94:e979. doi: 10.1097/MD.0000000000000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu H, Liu B. CPEB4 is a candidate biomarker for defining metastatic cancers and directing personalized therapies. Med Hypotheses. 2013;81:875–877. doi: 10.1016/j.mehy.2013.08.030. [DOI] [PubMed] [Google Scholar]
- 35.Tian Q, Liang L, Ding J, Zha R, Shi H, Wang Q, Huang S, Guo W, Ge C, Chen T, et al. MicroRNA-550a acts as a pro-metastatic gene and directly targets cytoplasmic polyadenylation element-binding protein 4 in hepatocellular carcinoma. PLoS One. 2012;7:e48958. doi: 10.1371/journal.pone.0048958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang W, Li H, Luo R. The microRNA-1246 promotes metastasis in non-small cell lung cancer by targeting cytoplasmic polyadenylation element-binding protein 4. Diagn Pathol. 2015;10:127. doi: 10.1186/s13000-015-0366-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.D'Ambrogio A, Nagaoka K, Richter JD. Translational control of cell growth and malignancy by the CPEBs. Nat Rev Cancer. 2013;13:283–290. doi: 10.1038/nrc3485. [DOI] [PubMed] [Google Scholar]