Abstract
Actin-related protein 2/3 complex subunit 4 (ARPC4) acts as an actin nucleator in actin cytoskeleton branching and contributes to cell migration. ARPC4 has previously been demonstrated to be abnormally expressed in various colorectal carcinoma cell lines, particularly SW620 cells. The present study explored the biological action and the possible mechanisms underlying the function of ARPC4 in the progression of carcinoma. The proliferation and migration of SW620 cells transfected with ARPC4-specific short interfering (si)RNAs were assessed using western blot, cell counting, flow cytometry and transwell assays. SW620 cells exhibited the highest ARPC4 expression of the cell lines investigated, and siRNA538 was the most effective of the siRNAs considered. The results of the present study demonstrated that ARPC4-silencing exhibited a significant effect on the capacity of cells for migration, but did not affect their proliferative ability. ARPC4-silencing inhibited human SW620 cell migration, but not proliferation, in vitro, suggesting that ARPC4 may be a putative therapeutic target for colorectal carcinoma.
Keywords: actin-related protein 2/3 complex subunit 4, colorectal carcinoma, migration, proliferation
Introduction
Colorectal cancer is a major cause for morbidity and mortality and one of the most common malignant tumours of the digestive system (~9.7% of cases), thus posing a serious threat to human physical and mental health (1). The most hazardous characteristics of colorectal cancer are invasion and metastasis. Although basic and clinical cancer research has made notable progress in recent years, the survival rate for the majority of patients with cancer has not significantly improved, with metastasis and relapse following treatment being the cause for >90% of cancer-associated mortality (2). The current clinical treatment for tumour metastasis exhibits numerous adverse effects, and there is a lack of systematic and comprehensive knowledge regarding the biological mechanisms underlying tumour metastasis. Therefore, prediction, early diagnosis and effective interventions to prevent tumour metastasis to distal tissues or organs are limited. In order to improve the effects of therapeutics on malignant tumours, the molecular mechanisms underlying tumour invasion and metastasis should be investigated (3–5).
Previously, research into metastasis focused on the actin cytoskeleton and increases in actin-related protein (ARP) expression during metastasis. The ARP2/3 complex is an actin-assembly nucleating agent that promotes the nucleation of microfilaments and serves a significant function in a number of physiological activities, including cell migration (6,7). Cell migration is essential to normal biological processes, such as tissue repair and regeneration; however, abnormally activated cell movement is associated with various diseases and may eventually lead to the development of fatal metastatic tumours. Indeed, metastatic capacity is considered a cancer cell marker (8–10).
A previous study demonstrated that ARP subunits are abnormally expressed in tumours, and immunohistochemical experiments indicated that abnormally increased levels of ARPC4 were present in colorectal cancer tissues (1). Investigating the association between ARPC4 and tumours may, therefore, provide valuable insights for the development of strategies for early cancer diagnosis and gene therapy. Molecular and cell biology studies previously demonstrated abnormally increased ARPC4 expression levels in various colorectal cancer cell lines; however, the association between the ARPC4 gene and the occurrence and development of colorectal cancer has not yet been fully elucidated. In the present study, further explorative research was conducted to identify novel targets for colorectal cancer gene therapy.
Materials and methods
Tissue samples
Colorectal carcinoma and adjacent normal colon tissue were obtained from a female patient at the age of 67 in August 2015 by resection at The Third People's Hospital of Chengdu (Chengdu, China). Written informed consent was obtained from the patient and ethical approval was granted by the Medical Ethics Committee of The Third People's Hospital of Chengdu. Immunohistochemical staining images were analysed with Imaris 8.01 (Bitplane AG, Zurich, Switzerland).
Immunohistochemical analysis
Detection of ARPC4 protein levels was performed as follows: A paraffin section of the tumour sample 6–8-µm thick was dewaxed with xylene I and II for 5 min at room temperature, then incubated with descending ethanol series (100, 95, 90, 80 and 70%) for 3–5 min. Slices were rinsed twice with distilled water and then use PBS to rinse two to three times with PBS. H2O2 solution (3%) was used to block the endogenous peroxidase activity for 10 min at room temperature, followed by high-pressure antigen repair. It was sealed using 10% goat serum (cat no. ab7481; Abcam, Cambridge, UK) and the primary monoclonal rabbit anti-ARPC4 antibody (dilution, 1:100; cat no. ab217065; Abcam) was added, then incubated at 4°C overnight and washed three times with PBS. The biotin-labelled mouse anti-rabbit IgG secondary antibody (dilution, 1:100; cat no. ab6728; Abcam) was added and incubated for 30 min at room temperature, then washed three times with PBS, developed using DAB (93 µl ddH2O + 3 µl 0.05% DAB1 solution + 2 µl 0.05% DAB2 solution + 2 µl 0.05% DAB3 solution) and re-dyed using 2% haematoxylin at 4°C for 10 min. It was then sealed by neutral gum and imaged under a microscope (BX-42; Olympus Corporation, Tokyo, Japan).
Using light microscopy at magnification, ×400, cells from 3 fields of view were analysed and the average number of positive cells was noted. According to the following scoring standard, the number of positive stained cells was calculated and divided into the ARPC4 low-expression group (<10% positive cells) or the ARPC4 high-expression group (>10% positive cells). This was calculated by the following formula: Number of positively stained cells/total number of cells × 100.
Cell culture conditions and reagents
The human colorectal cancer SW620, HT-29, HCT116 and SW480 cell lines were obtained from the Institute of Biochemistry and Cell Biology (Shanghai, China) and maintained by the laboratory within the Medical Biology Department, Sichuan University (Chengdu, China). SW620, HT-29, HCT116 and SW480 cell lines were cultured with high-glucose Dulbecco's modified Eagle's medium (DMEM; HyClone; GE Healthcare Life Sciences, Logan, UT, USA) containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and penicillin-streptomycin (10,000 U/ml; 1:100; Gibco; Thermo Fisher Scientific, Inc.) in a Heracell™ VIOS 160i CO2 incubator (Thermo Fisher Scientific, Inc.), and were maintained in a 5% CO2 atmosphere at 37°C. all the cells were replaced with new medium every 2 days and passaged when the confluence reached ~90% by using 0.25% trypsin (Gibco; Thermo Fisher Scientific, Inc.).
Cell culture conditions and reagents
SW620 cells were maintained at 37°C in an atmosphere containing 5% CO2 in Dulbecco's modified Eagle's medium (DMEM; HyClone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% foetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Lipofectamine® 2000 was purchased from Life Sciences (Thermo Fisher Scientific, Inc.). Primary antibodies against β-actin (cat no. ab8227) and ARPC4 were purchased from Abcam (Cambridge, UK). Primary antibodies for vimentin (cat. no. 10366-1-AP), E-cadherin (cat. no. 20874-1-AP) and PCNA (cat. no. 24,036-1-AP) were supplied by ProteinTech Group, Inc. (Chicago, IL, USA). Secondary antibodies against vimentin (cat. no. sc-2370), E-cadherin (cat. no. sc-2030) and PCNA (cat. no. sc-2995) were purchased from Santa Cruz Biotechnology Inc. (Dallas, TX, USA).
Western blot assay
HT-29, HCT-116, SW480, SW620, SW-1116 or transfected and null SW620 colorectal cancer cells were harvested 48 h following transfection, during the logarithmic growth phase, and total cellular protein was extracted. All cells were washed three times with PBS, centrifuged at 1,000 × g for 5 min at room temperature, and re-suspended in radioimmunoprecipitation assay lysis (Beijing Solarbio Science & Technology, Beijing China) and extraction buffer containing 1 mM phenylmethylsulfonyl fluoride protease inhibitor (Beijing Solarbio Science & Technology) on ice for 30 min to ensure completely lysis. A total volume of 5 ml 5X loading buffer [(250 mM Tris-HCL (pH 6.8), 10% (w/v) SDS, 0.5% (w/v) BPB, 50% (v/v) glycerin and 5% (w/v) β-mercaptoethanol)] was added and boiled at 100°C for 5–10 min. then centrifuged at 12,000 × g for 15 min at 4°C to collect the supernatant. The total protein concentration was detected using the bicinchoninic acid method (BCA) (Beyotime Institute of Biotechnology, Haimen, China). A total of 40 µg protein for each sample was uploaded and separated by SDS-PAGE, with a 5% stacking gel and 12% separating gel, and transferred to polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA). The membranes were then blocked with TBS containing 0.05% Tween-20 (TBST) and 5% non-fat milk for 1 h at room temperature. Polyclonal rabbit anti-β actin (dilution, 1:200), rabbit monoclonal anti-ARPC4 (dilution, 1:200), polyclonal rabbit anti-vimentin antibody (dilution, 1:200), rabbit polyclonal anti-E-cadherin antibody (dilution, 1:200) and PCNA rabbit polyclonal antibody (dilution, 1:200) antibodies were added to incubate at 4°C overnight. Following washing three times for 10 min each with TBST, the membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit (dilution, 1:5,000; cat no. A8275; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and peroxidase-conjugated goat anti-rabbit (dilution, 1:5,000; cat no. A0418; Sigma-Aldrich; Merck KGaA), goat anti-rabbit IgG-HRP secondary antibody (dilution, 1:5,000; cat no. sc-2370; Santa Cruz Biotechnology, Inc.), goat anti-rabbit IgG-HRP secondary antibody (dilution, 1:5,000; cat no. sc-2030; Santa Cruz Biotechnology, Inc.) and chicken anti-rabbit IgG-HRP secondary antibody (dilution, 1:5,000; cat no. sc-2955; Santa Cruz Biotechnology, Inc.) for 1 h at room temperature. Subsequent to washing with TBST, the membranes were developed using an enhanced chemiluminescence western blot detection system (Merck KGaA). Quantity One 4.62 (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and Image J software (version 2.1; National Institutes of Health, Bethesda, MD, USA) was used analyze the results and calculate the expression of ARPC4 and β-actin. β-actin was used as a reference protein.
ARPC4-siRNA transfection
Cells were seeded in 6-well plates (3.5×105 cells per well). A negative control (NC) group was transfected with non-silencing siRNA of the same length as ARPC4-siRNA. A null group was incubated under normal conditions (DMEM containing 10% FBS) without siRNA transfection. Transfection was conducted using Lipofectamine® 2000 according to the manufacturer's protocol. The GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) was used to select the RNA interference target area for the ARPC4 gene, which was used for the design and synthesis of the three siRNA sequences and the NC (Shanghai GenePharma Co., Ltd., Shanghai, China). The siRNAs were designated as siRNA496, siRNA538 and siRNA679, according to the corresponding target sequence cDNA-initiation site, and BLAST (https://blast.ncbi.nlm.nih.gov/) queries were conducted to rule out homology with other genes (Table I).
Table I.
Oligonucleotide sequences of the siRNAs against actin-related protein 2/3 complex subunit 4.
| siRNA name | Sense | Antisense |
|---|---|---|
| Negative control | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
| siRNA 496 | GAGCAGAGAACUUCUUUAUTT | UAAAGAAGUUCUCUGCUCTT |
| siRNA 538 | GGUAUGAUAUCAGCUUUCUTT | AGAAAGCUGAUAUCAUACCTT |
| siRNA 687 | GCUGAAGAGUUCCUUAAGATT | UCUUAAGGAACUCUUCAGCTT |
siRNA, short interfering RNA.
Cell viability assays
Cells in the logarithmic growth phase immediately following transfection were seeded in 96-well plates (1×103 cells per well), with three wells allocated to each experimental group. The absorbance in each well was measured at 0, 1, 2, 3 and 4 days, using Cell Counting kit-8 (EnoGene Biotech Co., Ltd., Nanjing, China) according to the manufacturer's protocol. Absorbance was measured at 450 nm using a plate reader, and growth curves were generated from the resulting data.
Flow cytometry analysis of cell cycle distribution
A total of 48 h following transfection, cells from each experimental group were fixed overnight at 4°C with cold 70% ethanol prior to being incubated with RNase A at 37°C for 30 min. The cells were stained with propidium iodide for 30 min in the dark at room temperature, and cell cycle analysis was conducted using a FACSCanto II flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) and ModFit LT software 4.1 (Verity Software House, Inc., Topsham, ME, USA).
Cell migration assays
Cell migration assays were performed using transwell chamber dishes (6.5 mm) with 8.0-µm pore polycarbonate membrane inserts (EDM Millipore, Billerica, MA, USA). Serum-free cell suspensions (200 µl, 105 cells) were seeded in the upper compartment, while 450 µl DMEM containing 30% FBS was placed in the lower chamber. The cells were incubated for 48 h. The upper membrane surface was wiped to remove cells, and the migrated cells on the lower surface were fixed with 4% methanol at room temperature for 30 min and stained with 0.1% (g/ml) crystal violet for 30 min at room temperature. Using a light microscope, cells in three random fields of vision (magnification, ×400; DP-50; Olympus Corporation, Tokyo, Japan) were counted for each sample and means were reported.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA) and SPSS 16.0 statistical software (SPSS, Inc., Chicago, IL, USA). All data are presented as the mean ± standard deviation from 3 independent experiments. Student's t-test was used when comparing between two groups and one-way analysis of variance with Tukey's post-hoc test was used when comparing more than two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Immunohistochemical analysis of ARPC4 expression in human carcinoma-associated antigen-containing tissues and adjacent non-tumour colorectal tissues
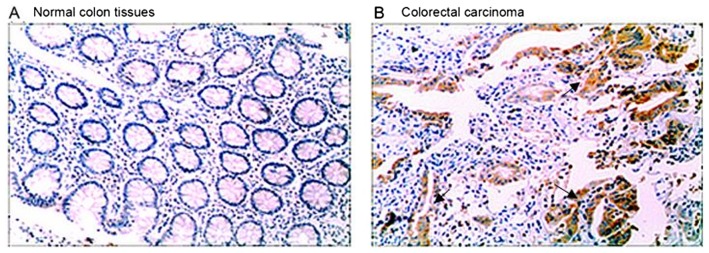
ARPC4 expression in colorectal carcinoma and normal colon tissues was assessed by immunohistochemistry. High levels of ARPC4 staining were observed in tumour tissues, whereas the normal tissues were negative for ARPC4 (Fig. 1).
Figure 1.
Immunohistochemical analysis of ARPC4 expression in (A) a colorectal carcinoma and (B) an adjacent normal colon tissue sample. Relatively high ARPC4 staining was observed in the tumour tissue, whereas the normal tissue was negative for ARPC4. ARPC4, actin-related protein 2/3 complex subunit 4.
Analysis of ARPC4 expression in colorectal cancer cells by western blot
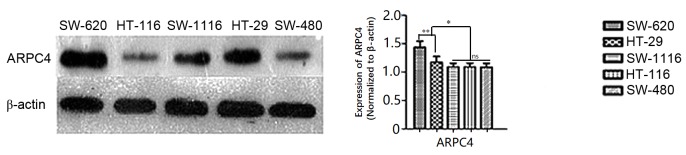
Western blot results revealed that ARPC4 was expressed at high levels in SW620 and HT-29 cell lines relative to other cell lines (Fig. 2), with the highest expression observed in SW620 cells (Fig. 2). For this reason, the SW620 cell line was selected for subsequent experiments.
Figure 2.
Representative image of relative ARPC4 expression levels in colorectal cancer cell lines were investigated using western blot analysis. ARPC4, actin-related protein 2/3 complex subunit 4. SW620 and HT-29 cell line groups demonstrated a relative higher ARPC4 expression levels when compare with the other groups (*P<0.05 vs. HT-116, SW1116, SW480 groups). However, the ARPC4 expression in SW620 group is much more apparent compared with the HT-29 group. (**P<0.01 vs. HT-29 group). The differences were not significant among HT-116, SW1116, SW480 groups. *P<0.05; **P<0.01; ns, not significant.
ARPC4-siRNA538 exhibited the strongest silencing effect
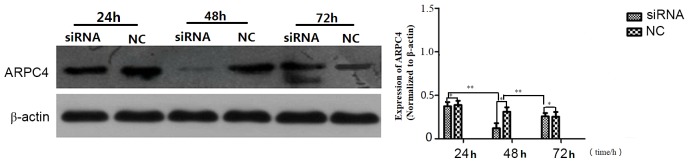
Western blot analysis was carried out to determine which of the ARPC4-siRNAs yielded the most effective silencing. As presented in (Fig. 3), siRNA538 resulted in the highest level of ARPC4 inhibition. and was, therefore, used in subsequent experiments. The inhibition rate was significantly decreased at all time points, but the optimal transfection time was 48 h. (Fig. 4).
Figure 3.
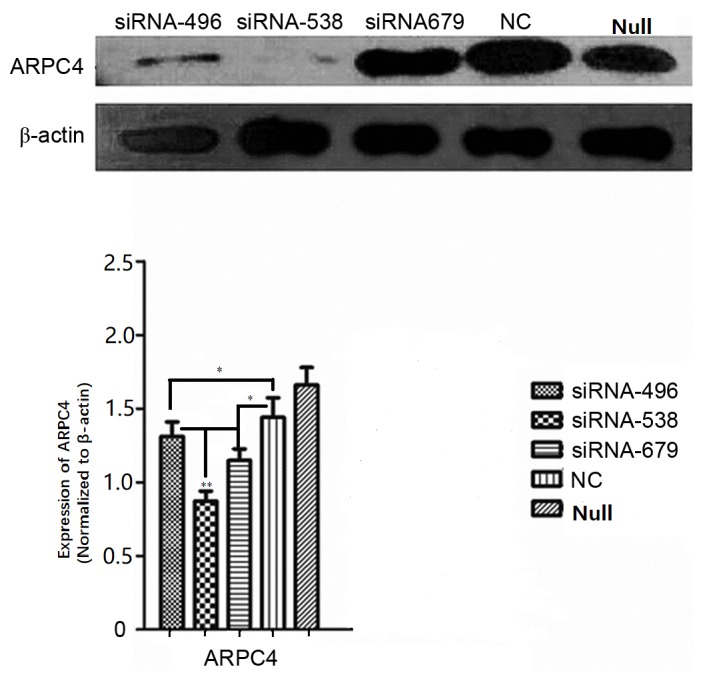
Comparison of silencing efficiencies for three ARPC4-siRNA oligonucleotide sequences. Comparison of ARPC4 protein expression levels among siRNA, NC and Null groups in SW620 cells after transfection with three ARPC4-siRNA oligonucleotide sequences and NC. Following transfection with siRNA and NC, silencing efficiencies between siRNA and NC groups were significant (*P<0.05 vs. NC). siRNA538 resulted in the highest level of ARPC4 inhibition when compared with the other siRNA groups (**P<0.01 vs. siRNA496/siRNA679). ARPC4, actin-related protein 2/3 complex subunit 4; siRNA, short interfering RNA; NC, nonspecific siRNA; siRNA496, short interfering RNA with oligonucleotide sequences number 496; siRNA538, short interfering RNA with oligonucleotide sequences number 538; siRNA679, short interfering RNA with oligonucleotide sequences number 679; NC, nonspecific siRNA; null, control group; *P<0.05; **P<0.01.
Figure 4.
ARPC4 protein expression in SW620 cells following transfection with ARPC4-siRNA538. Comparison of ARPC4 protein expression silencing efficiencies at time point 24, 48 and 72 h after transfected with siRNA538 and NC. The groups include: 24 h group (siRNA538/NC); 48 h group (siRNA538/NC); 72 h group (siRNA538/NC). The protein expression differences between siRNA538 and NC were statistically significant at each time point (*P<0.05 vs. NC). The siRNA538 silencing efficiencies at 48 h group were much more significant than at the other time points, the differences were statistically significant (**P<0.01 vs. 24/72 h group). siRNA538, small interfering target RNA with oligonucleotide sequences number 538; NC, nonspecific siRNA; *P<0.05; **P<0.01.
Effect of ARPC4 on viability
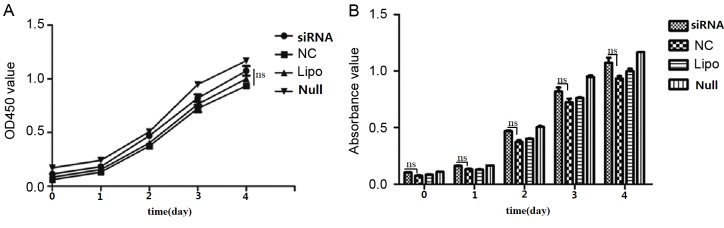
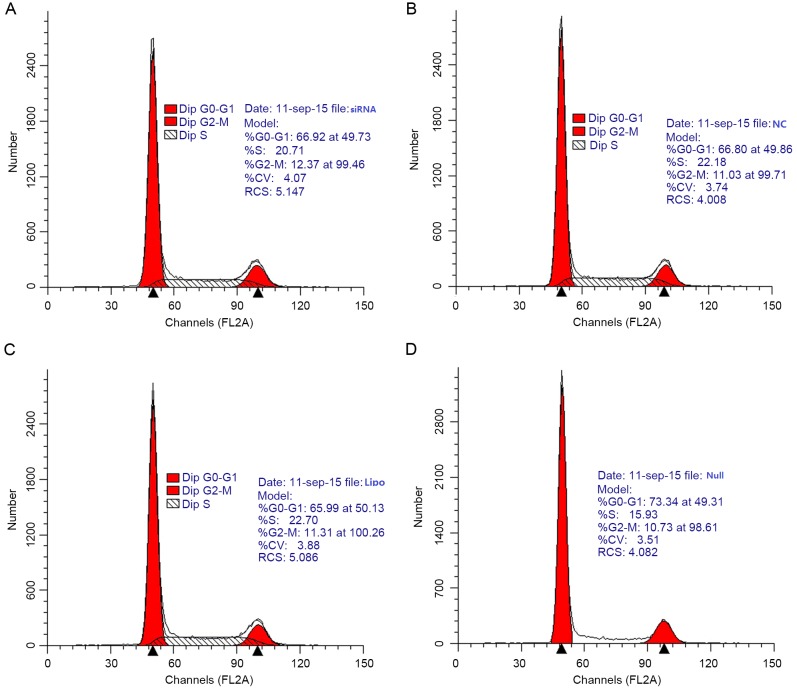
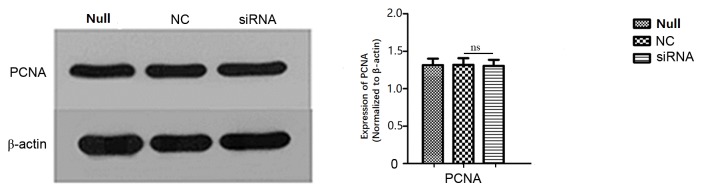
As determined from cell growth curves, cell viability rates did not differ significantly among the siRNA, NC, Lipo and Null groups (Fig. 5). Cell cycle analysis by flow cytometry yielded the following results for the siRNA group vs. the NC group: G0/G1 phase, 66.92 vs. 66.80%; S phase, 20.71 vs. 22.18%; and G2/M phase, 12.37 vs. 11.03% (Fig. 6). Differences in the G0/G1, S, and G2/M phase distributions between the groups were not statistically significant. Furthermore, PCNA expression, as determined by western blot, did not differ significantly between groups (Fig. 7).
Figure 5.
Growth curves for cells subsequent to actin-related protein 2/3 complex subunit 4 siRNA transfection. (A) OD450 value of each group. (B) Mean absorbance of each group. The relative proliferation rates at 0, 1, 2, 3 and 4 days subsequent to transfection in SW620 cells show no significant differences between the siRNA group and NC group (ns vs. NC). OD, optical density; siRNA, small interfering target RNA; NC, nonspecific siRNA; Lipo, Lipofectamine-only group; Null, control group; ns, not significant.
Figure 6.
Cell cycle distribution analysis by flow cytometry. (A) siRNA group; (B) NC group; (C) Lipo group and (D) Null group 24 h after transfection of SW620 cells. Differences in the G0/G1, S and G2/M phase distribution were not statistically significant between the siRNA group and NC group (ns vs. NC). siRNA, small interfering target RNA; Null, control group; NC, nonspecific siRNA; Lipo, Lipofectamine-only group; ns, not significant.
Figure 7.
Analysis of PCNA expression by western blotting subsequent to transfection with actin-related protein 2/3 complex subunit 4 siRNA. PCNA expression differences in siRNA and NC groups of SW620 cells after transfected 48 h were not statistically significant (ns vs. NC). PCNA, proliferating cell nuclear antigen; siRNA, small interfering target RNA; Null, control group; NC, nonspecific siRNA; ns, not significant.
ARPC4 promotes SW620 cell migration
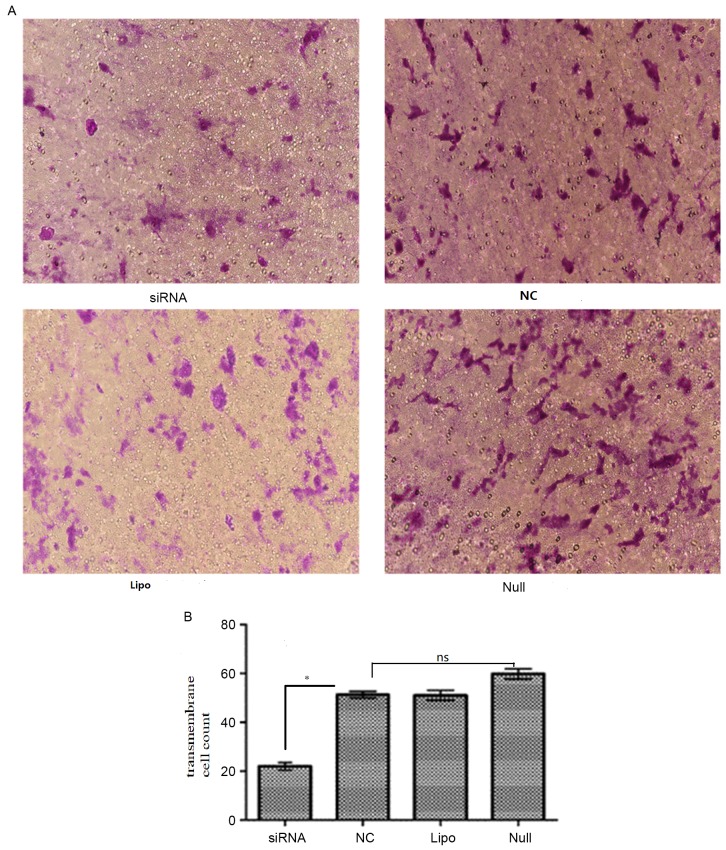
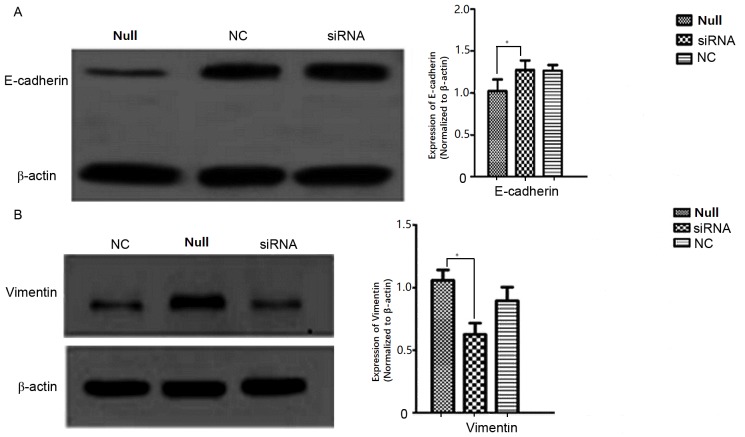
To examine the effect of ARPC4 on SW620 cell migration, Transwell and western blot analyses were conducted. In the transwell assay, the number of migrated cells in siRNA group were significantly decreased compared with that observed in the NC group (transmembrane cell count, siRNA vs. NC: 20.4±1.14 vs. 60.6±2.07; P<0.05; Fig. 8), while the differences among NC/Null/Lipo groups were not significant (transmembrane cell count 60.6±2.07, 50.0±1.58 and 50.8±1.59; P>0.05). Western blotting results revealed that E-cadherin (Fig. 9A) and vimentin (Fig. 9B) expression differed significantly between the siRNA-transfected cells and the Null SW620 cells, respectively. It was therefore concluded that cells with ARPC4 knockdown exhibited a decreased metastatic capacity compared to Null/NC cells. NC, nonspecific siRNA; Null, control group; siRNA, small interfering target RNA; ns; not significant.
Figure 8.
Transwell assay analysis of cell migration. (A) Representative images (magnification, ×400) and (B) quantification of the Transwell cell migration assay. Comparing the transmembrane cell count in siRNA, NC, Lipo and Null groups of SW620 cells after transfected 48 h. The number of migrated cells in siRNA group were significantly decreased compared with that observed in the NC group (*P<0.05 vs. NC), while the differences were not statistically significant among Null group, Lipo group and NC group (ns vs. Null/Lipo). *P<0.05. NC, negative control; Lipo, Lipofectamine-only group; siRNA, short interfering RNA group; ns, not significant.
Figure 9.
Analysis of E-cadherin and vimentin expression by western blot. (A) E-cadherin protein expression in Null, NC and siRNA groups of SW620 cells after transfected 48 h. the protein expression show significant differences between siRNA group and Null group (*P<0.05 vs. Null), while the differences were not statistically significant between Null group and NC group. (B) vimentin protein expression in NC, Null and siRNA groups of SW620 cells after transfected 48 h. The protein expression level show significant differences between siRNA group and Null group (*P<0.05 vs. Null), while the differences were not statistically significant between Null group and NC group. NC, nonspecific siRNA; Null, control group; siRNA, small interfering target RNA.
Discussion
While high levels of ARPC4 expression have been identified in colorectal cancer tissues, the association between the ARPC4 gene and the occurrence and development of colorectal cancer has not yet been elucidated (5).
Therefore, the present study explored the underlying molecular mechanisms of the function of ARPC4 in the progression of colorectal cancer and revealed that ARPC4 may serve a crucial function in colorectal cancer cell migration.
The findings of the present study indicated that although ARPC4-siRNA538 transfection did not influence cell viability, the invasiveness of cells transfected with siRNA538 was significantly diminished. The actin cytoskeleton formed by monomeric globular actin serves an essential function in several cellular processes, including division, migration, adhesion, and endocytosis. A number of these functions involve contact with the plasma membrane to allow the actin network outside of the cell to respond to extracellular signals. The aforementioned processes result from actin cytoskeleton rearrangement, which involves numerous regulatory factors, including the ARP2/3 complex, which is an evolutionary conserved 220-kDa complex comprised of ARP2, ARP3, and five affiliated proteins (ARPC1-5) (6–10).
The ARP2/3 complex is an important component of the cytoskeleton that promotes the nucleation of new microfilaments and functions in the maintenance of cell shape, motility, and cytokinesis. ARPC4 and ARPC2 constitute the centre of the complex, whereas ARPC4 was previously demonstrated to serve an important function in the biological function of ARP2/3 in pancreatic cancer (11–14). ARPC4, the expression of which is abnormally high in colorectal cancer cell lines, regulates the actin nucleation process in cells, forms fusion proteins with the products of the downstream genes and influences the migration of pancreatic cancer cells (15,16).
PCNA is a 36 kDa protein that is only identified in the nuclei of normal proliferative and tumour cells. PCNA is associated with cell DNA synthesis and serves an important function in the initiation of cell proliferation (17). Tumour cells exhibit strong proliferative activity; PCNA may be used as an evaluation index of the cell proliferation state. In order to further determine the influence of ARPC4 on SW620 colorectal cancer cell proliferation in the present study, the PCNA protein expression level was investigated; its expression was not significantly different between groups.
The expression of E-cadherin was markedly increased, whereas the expression of vimentin was decreased in ARPC4-silenced cells compared with the control cells. E-cadherin is considered to be a tumour invasion and metastasis suppressor gene, and belongs to the calcium-dependent cadherin family. The expression of E-cadherin, which maintains the stability of the connection between normal cells, is negatively correlated with the occurrence of the epithelial-mesenchymal transition (EMT) and tumourigenesis. E-cadherin is connected to the cytoskeleton by its interaction with catenin to inhibit the proliferation of tumour cells and the production of matrix metalloproteinases by the host cell (18–20).
Additionally, E-cadherin prevents the degradation of various proteins of the matrix and basement membrane surrounding the tumour cells, thereby inhibiting tumour cell degradation of the matrix and basement membrane barriers (21–23). Invasion of tumour cells is regulated by tumour-matrix interactions. Expression of the ARP2/3 complex is associated with stromal cells in colorectal cancer, and therefore ARP2/3 expression enhances the motility between stromal cells and tumour cells, thereby providing a more suitable environment for invasion by these two cell types (24). Vimentin, however, is considered an interstitial cell marker, the expression of which correlates positively with the occurrence of EMT and with tumour oncogenesis. Vimentin is the dominant central fibre in mesenchymal cells and participates in the maintenance of cell integrity. Decreased E-cadherin expression is associated with elevated vimentin expression, and waveform protein expression may interfere with cell adhesion mediated by E-cadherin (25–27). Therefore, the results of the present study suggested that ARPC4 may enhance the expression of vimentin, whereas it may inhibit the expression of E-cadherin, and that the expression of ARPC4 may therefore have decreased cell adhesion to promote migration in tumour cells, thus serving a function in tumour development.
In summary, the use of RNA interference may effectively suppress human ARPC4 expression in colorectal cancer SW620 cells, thereby inhibiting cell migration. These results suggested that specific targeting of ARPC4 may represent a potential treatment for colorectal cancer. Although previous studies demonstrated that ARPC4 influences the migration of pancreatic and colorectal cancer cells (15,16), further experiments should be undertaken to define the function of ARPC4 during the initiation of colorectal cancer. Future research will include determination of the mechanism by which ARPC4 expression influences the biological behaviour of tumour cells. This information will enable the elucidation of novel targets for the treatment of colorectal cancer.
Acknowledgements
The present study was supported by the Scientific Project of Sichuan Province (grant no. 16ZC1671) and the Program Science and Technology Bureau of Chengdu China (grant no. 2015-HM01-00141-SF).
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2839–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Inra JA, Syngal S. Colorectal cancer in young adults. Dig Dis Sci. 2015;60:722–733. doi: 10.1007/s10620-014-3464-0. [DOI] [PubMed] [Google Scholar]
- 3.Gross K, Brand MI. Common Surgical Diseases. New York: Springer; 2014. Genetic predisposition to colorectal cancer; pp. 189–191. [Google Scholar]
- 4.August DA, Ottow RT, Sugarbaker PH. Clinical perspective of human colorectal cancer metastasis. Cancer Metastasis Rev. 1984;3:303–324. doi: 10.1007/BF00051457. [DOI] [PubMed] [Google Scholar]
- 5.Belvitch P, Brown ME, Brinley BN, Letsiou E, Rizzo AN, Garcia JGN, Dudek SM. The ARP 2/3 complex mediates endothelial barrier function and recovery. Pulm Circ. 2017;7:200–210. doi: 10.1086/690307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamaguchi H, Condeelis J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim Biophys Acta. 2007;1773:642–652. doi: 10.1016/j.bbamcr.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Clainche C, Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- 8.Firat-Karalar EN, Welch MD. New mechanisms and functions of actin nucleation. Curr Opin Cell Biol. 2011;23:4–13. doi: 10.1016/j.ceb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goley ED, Welch MD. The ARP2/3 complex: An actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 10.Bailly M, Ichetovkin I, Grant W, Zebda N, Machesky LM, Segall JE, Condeelis J. The F-actin side binding activity of the Arp2/3 complex is essential for actin nucleation and lamellipod extension. CurrBiol. 2001;11:620–625. doi: 10.1016/S0960-9822(01)00152-X. [DOI] [PubMed] [Google Scholar]
- 11.Rauhala HE, Teppoi S, Niemelä S, Kallioniemi A. Silencing of the ARP2/3 complex disturbs pancreatic cancer cell migration. Anticancer Res. 2013;33:45–52. [PubMed] [Google Scholar]
- 12.Welch MD, DePace AH, Verma S, Iwamatsu A, Mitchison TJ. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol. 1997;138:375–384. doi: 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson RC, Turbedsky K, Kaiser DA, Marchand JB, Higgs HN, Choe S, Pollard TD. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 14.Higgs HN, Pollard TD. Regulation of actin filament network formation through ARP2/3 complex: Activation by a diverse array of proteins. Ann Rev Biochem. 2001;70:649–676. doi: 10.1146/annurev.biochem.70.1.649. [DOI] [PubMed] [Google Scholar]
- 15.Otsubo T, Iwaya K, Mukai Y, Mizokami Y, Serizawa H, Matsuoka T, Mukai K. Involvement of Arp2/3 complex in the process of colorectal carcinogenesis. Mod Pathol. 2004;17:461–467. doi: 10.1038/modpathol.3800062. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh A, Tousif S, Bhattacharya D, Samuchiwal SK, Bhalla K, Tharad M, Kumar S, Prakash P, Kumar P, Das G, Ranganathan A. Expression of the ARPC4 subunit of human Arp2/3 severely affects Mycobacterium tuberculosis growth and suppresses immunogenic response in murine macrophages. PLoS One. 2013;8:e69949. doi: 10.1371/journal.pone.0069949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong Y, Zhang H, Beach D. D type cyclins associate with multiple protein kinases and the DNA replication and repair factor PCNA. Cell. 1992;71:505–514. doi: 10.1016/0092-8674(92)90518-H. [DOI] [PubMed] [Google Scholar]
- 18.Goodlad RA. Quantification of epithelial cell proliferation, cell dynamics and cell kinetics in vivo. Wiley Interdiscip Rev Dev Biol. 2017;6 doi: 10.1002/wdev.274. [DOI] [PubMed] [Google Scholar]
- 19.Xie Y, Li P, Gao Y, Gu L, Chen L, Fan Y, Zhang F, Zhang X. Reduced E-cadherin expression is correlated with poor prognosis in patients with bladder cancer: A systematic review and meta-analysis. Oncotarget. 2017;8:62489–62499. doi: 10.18632/oncotarget.19934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao H, Xu E, Liu H, Wan L, Lai M. Epithelial-mesenchymal transition in colorectal cancer metastasis: A system review. Pathol Res Pract. 2015;211:557–69. doi: 10.1016/j.prp.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 21.Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol. 2009;1:a003129. doi: 10.1101/cshperspect.a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abu Taha A, Schnittler HJ. Dynamics between actin and the VE-cadherin/catenin complex: Novel aspects of the ARP2/3 complex in regulation of endothelial junctions. Cell Adh Migr. 2014;8:125–135. doi: 10.4161/cam.28243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen A, Beetham H, Black MA, Priya R, Telford BJ, Guest J, Wiggins GA, Godwin TD, Yap AS, Guilford PJ. E-cadherin loss alters cytoskeletal organization and adhesion in non-malignant breast cells but is insufficient to induce an epithelial-mesenchymal transition. BMC Cancer. 2014;14:552. doi: 10.1186/1471-2407-14-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kovacs EM, Goodwin M, Ali RG, Paterson AD, Yap AS. Cadherin-directed actin assembly: E-cadherin physically associates with the Arp2/3 complex to direct actin assembly in nascent adhesive contacts. Curr Biol. 2002;12:379–382. doi: 10.1016/S0960-9822(02)00661-9. [DOI] [PubMed] [Google Scholar]
- 25.Nijkamp MM, Span PN, Hoogsteen IJ, van der Kogel AJ, Kaanders JH, Bussink J. Expression of E-cadherin and vimentin correlates with metastasis formation in head and neck squamous cell carcinoma patients. Radiother Oncol. 2011;99:344–348. doi: 10.1016/j.radonc.2011.05.066. [DOI] [PubMed] [Google Scholar]
- 26.Canel M, Serrels A, Frame MC, Brunton VG. E-cadherin-integrin crosstalk in cancer invasion and metastasis. J Cell Sci. 2013;126:393–401. doi: 10.1242/jcs.100115. [DOI] [PubMed] [Google Scholar]
- 27.Kuefer R, Hofer MD, Gschwend JE, Pienta KJ, Sanda MG, Chinnaiyan AM, Rubin MA, Day ML. The role of an 80 kDa fragment of E-cadherin in the metastatic progression of prostate cancer. Clin Cancer Res. 2003;9:6447–6452. [PubMed] [Google Scholar]