Abstract
Deregulation of microRNA (miR)-193b has been revealed to be associated with the proliferation of liver cells. However, the interaction between miR-193b and their targets inducing liver cancer remains largely unknown. The aim of the present study was to investigate the hypothesis that miR-193b affects the proliferation of liver cancer cells. In the present study, the overall survival of patients with liver cancer and low fold change of miR-193b was higher compared with that of patients with liver cancer patients and high fold change of miR-193b. The expression level of myeloid cell leukemia-1 (Mcl-1) in patients with liver cancer was lower compared with in the control group. The results of the present study demonstrated that downregulation of miR-193b suppressed the proliferation and induced apoptosis of liver cancer cells, and inhibited the Mcl-1 protein expression level in liver cancer cells. Upregulation of miR-193b increased cell proliferation and decreased apoptosis of liver cancer cells and promoted the expression level of Mcl-1 protein. The results of the present study demonstrated that the expression of miR-193b as a novel tumor suppressor serves an important role in the proliferation of liver cancer cells by mediating Mcl-1 expression.
Keywords: microRNA-193b, liver cancer, cell proliferation, myeloid cell leukemia-1
Introduction
Malignant tumors pose an increasing threat to humans and tumors are one of the most common causes of mortality (1). Tumors are a neoplasm formed by hypoplasia and prosoplasia of normal cells generated under long-term effects of initiating and promoting factors in the body, is a general and systematic disease with multi-factor participation and multi-step development (2). However, the pathogenesis of tumors is not yet clear. In the last two centuries, there have been two breakthroughs in the treatment of cancer. The first was in 1890 when Halsted proposed the concept of radical operation of cancer (3). The second was in the 1970s when chemotherapy was integrated into radial operation (adjuvant chemotherapy or neoadjuvant chemotherapy) (3). Subsequently, there was no marked progress in the treatment of cancer, until the appearance of molecular targeting treatment, which may represent a third breakthrough (4). Tumor biotherapy considers the apoptosis of tumor as a fundamental link, inducing tumor cell apoptosis by targeted control of expression level or function of key proteins or enzymes in the apoptotic signaling pathway. It has been a focus for the development of tumor molecular therapy (4).
Liver cancer was the second most common digestive system neoplasm and ~53% of novel cases of liver cancer worldwide occur in China with a survival rate of <5% in the last 5 years (5). The primary causes of liver cancer are the high intrahepatic transport rate and high postoperative recurrence rate (1,6). At present, the comprehensive treatment focusing on surgery is the first choice and most effective strategy for primary hepatic carcinoma. However, the overall therapeutic effect is not promising. Despite early stage liver cancer, following radical excision the transfer and recurrence rate after 5 years is 61.5% and for small hepatocellular carcinoma the rate is 43.5%. Therefore, investigation into effective treatment strategies is required to further improve the therapeutic options for the treatment of liver cancer.
Myeloid cell leukemia-1 (Mcl-1) is one of the inhibitor genes of apoptosis in the gene family of B cell lymphoma-2 (Bcl-2) (7). The Mcl-1 gene is a known important anti-apoptosis factor in hepatocellular carcinoma, which is expressed extensively in human normal tissues and also in tumor tissues and tumor cell lines (8). Mcl-1 is located at human chromosome 1q21, which is a variable region during neoplastic disease and its precancerous lesions (9). Coupled with the anti-apoptotic function of Mcl-1, it is estimated that the expression of Mcl-1 may be associated with tumorigenesis (7).
MicroRNA (miRNA) is a micromolecule and non-coding RNA with 19–23 nucleotides and combines with the mRNA 3′ untranslated region of the target gene to regulate and control the expression of the gene (10). Although miRNAs account for only 1% of the human gene group, they perform transcriptional-level control for the expression of key genes and therefore serve an important role in physiological processes (11). Previous studies have revealed that a series of miRNAs served an important role in the expression of cancer genes, therefore the abnormal expression levels of miRNA was identified in various types of cancer (12). During hepatocarcinogenesis and cancer development, miRNA is a key factor involved in the mediation of cancer (12). A previous study indicated that miRNA may be a molecular marker in the prediction and diagnosis of cancer and may have potential application value in the clinical treatment of liver cancer (13). However, the interaction between miR-193b and its targets inducing liver cancer remains largely unknown. Furthermore, the present study investigated the effect of miRNA-193b on the proliferation of liver cancer cells.
Materials and methods
Reagents, patients and tissue specimens
RPMI-1640 medium and fetal bovine serum (FBS) were purchased from Gibco; Thermo Fisher Scientific, Inc. (Waltham, MA, USA). A total of 50 patients with liver cancer and 50 control patients were recruited for the present study. The mean age of the 50 patients with liver cancer was 57.8±7.1 years and that of the 50 normal control patients was 56.1±8.5 years (Table I). There were 11 female and 39 male patients in the liver cancer group and 13 female and 37 male patients in the control group (Table I). Patients with liver cancer and complete clinical pathology data were studied and clinically staged using the 7th edition of Lauren type and stage (14). For the prognosis study, we selected subjects who underwent surgical treatment and were subjected to regular follow-up; ultimately, 357 patients with gastric cancer and complete clinical pathology data were studied. The clinical staging of gastric cancer used the 7th edition of Union for International Cancer Control tumor-node-metastasis staging (13) and Lauren typing (14) was used for the histological classification of gastric cancer.
Table I.
Patient characteristics.
| Liver cancer group (n=50) | Control group (n=50) | |
|---|---|---|
| Age, years | 57.8±7.1 | 56.1±8.5 |
| Sex | ||
| Female | 11 | 13 |
| Male | 39 | 37 |
| TNM staging | ||
| I | 7 | |
| II | 6 | |
| III–IV | 11 | |
| Liver cirrhosis | ||
| Positive | 19 | 0 |
| Negative | 5 | 30 |
TNM, tumor-node-metastasis.
Written informed consent was obtained from all patients prior to enrollment in the present study and the present study was approved by the Ethics Committee of The First Affiliated Hospital of the University of Southern China (Hengyang, China). All patients underwent resection of the tissue between March 2014 and May 2015 at The First Affiliated Hospital of University of Southern China.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from patient tissues and HepG2 cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. RNA template was reverse-transcribed using a PrimeScript RT Master Mix kit (Takara Biotechnology Co., Ltd., Dalian, China) for 30 min at 16°C, 30 min at 42°C, 5 min at 85°C and maintained at 4°C. The mRNA expression level of miR-193b was determined using RT-qPCR. Applied Biosystems 7900HT Fast Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) was employed using 2 µl total RNA with SYBR Green PCR master mix (Applied Biosystems; Thermo Fisher Scientific, Inc.). The following primers were used for RT-qPCR: miR-193b forward, 5′-AGGCAAGAUGCUGGCAUAGCU-3′ and reverse, 5′-CUAUGCCAGCAUCUUGCCUUU-3′; U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′. The PCR parameters were as follows: 95° for 45 sec, followed by 40 cycles of 95°C for 30 sec and 60°C for 45 sec. Analysis of relative gene expression data was performed using the 2−ΔΔCq method (15).
Cell culture and transfection
HepG2 human liver cancer cell line was purchased from the Cancer Research Institute of Medical College, University of Southern China (Hengyang, China) and were cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin in a humidified incubator at 37°C with 5% CO2 for between 2 and 3 days. HepG2 cells were seeded onto sterile 6-well flat-bottomed plates [(1–2) ×106 cells] and incubated overnight at 4°C. Subsequently, the cells were transfected with pre-miR-193b, anti-miR-193b and the control (50 nM) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C and the transfected cells were then incubated for 6 h, prior to removal and replacement of the medium, at 37°C. pre-miR-193b, 5′-AACUGGCCCUCAAAGUCCCGCU-3′ and 5′-CGGGACUUUGAGGGCCAGUUUU-3′; anti-miR-193b, 5′-AGCUAUGCCAGCAUCUUGCCU-3′ and 5′-AGCGGGACUUUGAGGGCCAGUU-3′; controls, 5′-GUACCUGACUAGUCGCAGATT-3′ and 5′-UCUGCGACUAGUCAGGUACTT-3′.
Immunohistochemistry
The tissues samples were fixed using 4% paraformaldehyde for 24 h at room temperature, embedded in paraffin and divided into 4 µm slices. The tissues samples were dehydrated with xylene and ethanol (100% ethanol for 5 min, 95% ethanol for 5 min, 80% ethanol for 5 min and 70% ethanol for 5 min; Nanjing Chemical Reagent Co., Ltd., Nanjing, China). Subsequently, the sections were incubated in a blocking solution of 10% normal goat serum and with rabbit anti-Mcl-1 (dilution, 1:200; catalog no. 94296; Cell Signaling Technology, Inc., Danvers, MA, USA) at 4°C overnight and subsequently incubated with anti-rabbit IgG (H+L) antibody (dilution, 1:500; catalog no. 8889; Cell Signaling Technology, Inc.) for 1 h at room temperature. The sections were incubated with streptavidin-biotin complex conjugated to horseradish peroxidase and diaminobenzene for 10 min at room temperature. Tissue samples were observed using a fluorescence microscope (LSM710; magnification, ×20; Carl Zeiss Microscopy GmbH, Jena, Germany).
Cell proliferation
The effect of miR-193b on the proliferation of liver cancer cells was determined using an MTT assay. HepG2 cells transfected with miR-193b were seeded onto sterile 96-well flat-bottomed plates (1×103 cells) and incubated overnight. Subsequently, 0.5 µmol/l MTT (20 µl) was added into the wells and incubated at 37°C with 5% CO2 for 4 h. Following this, 150 µl DMSO was added into each well. Cell proliferation was evaluated using a microplate reader (SpectraMax® M2, Molecular Devices, LLC, Sunnyvale, CA, USA) at 570 nm.
Determination of apoptosis using flow cytometry
The effect of miR-193b on the proliferation of liver cancer cells was determined using the MTT assay. HepG2 cells transfected with miR-193b were seeded onto sterile 6-well flat-bottomed plates (1–2×106 cells) and incubated overnight. The cells were stained with 10 µl annexin V-allophycocyanin (eBioscience, Inc., San Diego, CA, USA) and µl propidium iodide at room temperature in the dark for 30 min. Subsequently, apoptosis was evaluated using a FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Western blotting
HepG2 cells transfected with miR-193b were prepared using a lysis buffer containing 10 mg/l aprotinin and 10 mg/l leupeptin. Protein content was quantified using the Bradford dye-binding procedure. Proteins were separated by SDS-PAGE (10% gels) followed by electrotransfer onto polyvinylidene difluoride membranes. The membranes were blocked with 5% non-fat milk at room temperature for 1 h and incubated with anti-Mcl-1 (catalog no. sc-20679; 1:3,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), anti-caspase-3 (catalog no. sc-98785; 1:3,000; Santa Cruz Biotechnology, Inc.), Bax (catalog no. sc-6236; 1:5,000; Santa Cruz Biotechnology, Inc.) and β-actin (catalog no. sc-7210; 1:5,000; Santa Cruz Biotechnology, Inc.) overnight at 4°C. Membranes were then incubated with anti-rabbit IgG (H+L; catalog no. sc-2004; dilution, 1:2,000, Santa Cruz Biotechnology, Inc.) for 1 h at room temperature following washing with TBST for 15 min. Immunoblots were visualized by Odyssey V3.0 image scanning (n=3; LI-COR Biosciences, Lincoln, NE, USA).
Statistical analysis
All data are presented as the mean ± standard error and were analyzed using one-way analysis of variance followed by Tukey's multiple comparison test. Unconditional logistic regression analysis with odds ratios (OR) and 95% confidence intervals (95% CI) were used to estimate the association between the risk of liver cancer and miR-193b expression levels. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression level of miR-193b in patients with liver cancer
The expression level of miR-193b was evaluated in patients with liver cancer. The expression level of miR-193b in the normal group was significantly higher compared with in liver cancer exosomes and whole tissues (P<0.01; Fig. 1).
Figure 1.
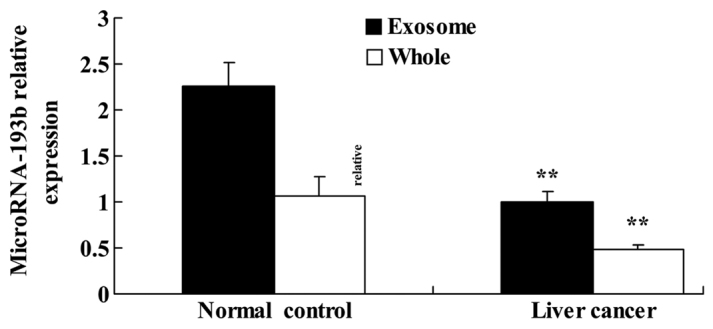
Expression level of microRNA-193b in patients with liver cancer. **P<0.01 compared with the control group.
Association between the clinicopathological features of liver cancer and the expression level of miR-193b
The association between clinicopathological features of liver cancer and the expression level of miR-193b were analyzed. Table II revealed that downregulated miR-193b was significantly associated with the depth of invasion, metastasis and tumor differentiation stage in patients with liver cancer.
Table II.
Expression of miR-193b in patients with patients with later-stage liver cancer.
| Variables | Crude OR (95% CI) | P-value |
|---|---|---|
| Depth of invasion | 0.002 | |
| T1+T2 | 1 | |
| T3+T4 | 4.01 (1.23–13.05) | |
| Metastasis | 0.08 | |
| Negative | 1 | |
| Positive | 3.77 (0.91–13.52) | |
| Tumor differentiation | 0.04 | |
| Moderate-well | 1 | |
| Poor | 3.82 (1.12–12.74) | |
| Lauren type | 0.04 | |
| Adenocarcinoma | 1 | |
| Diffusion | 4.02 (1.03–15.61) | |
| UICC stage | 0.001 | |
| I+II | 1 | |
| III+IV | 11.93 (2.21–39.89) |
miR, microRNA; OR, odds ratio; UICC, Union for International Cancer Control.
Association between the overall survival rate of patients with liver cancer and the expression level of miR-193b
In order to assess the association between the overall survival rate of patients with liver cancer and the expression level of miR-193b, the present study analyzed overall survival rate of all patients with liver cancer and their expression levels of miR-193b. As presented in Fig. 2, the overall survival rate of patients with liver cancer with lower miR-193b expression level is higher compared with that of patients with liver cancer and an increased miR-193b expression level.
Figure 2.
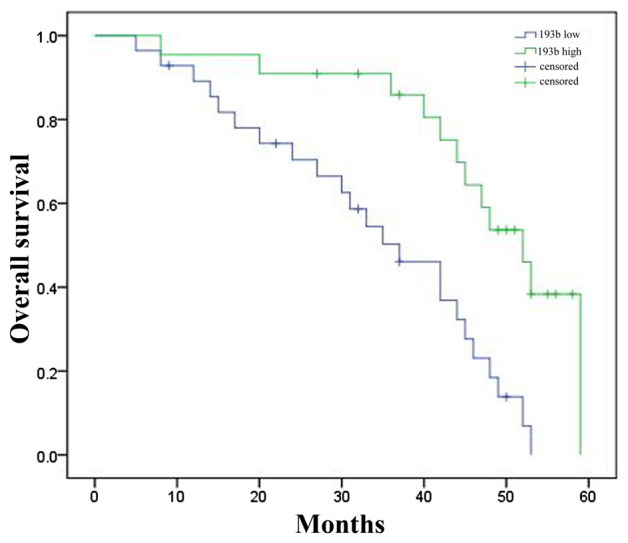
Association between the overall survival rate of patients with liver cancer and the expression level of microRNA-193b. 193b, microRNA-193b.
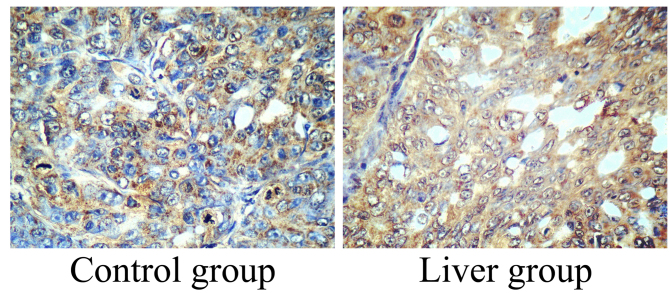
Expression level of Mcl-1 in patients with liver cancer
To determine the association between the expression of Mcl-1 and the incidence rate of liver cancer, the expression level of Mcl-1 was determined using immunohistochemistry. The expression level of Mcl-1 in liver cancer tissue samples was increased compared with in the control group (Fig. 3).
Figure 3.
Expression level of myeloid cell leukemia-1 in patients with liver cancer. Immunohistochemistry for myeloid cell leukemia sequence 1 expression analysis (magnification, ×20).
Effect of miR-193b on cell proliferation of HepG2 cells
The present study transfected anti-miR-193b plasmids into HepG2 cells and evaluated the change of cell proliferation. As presented at Fig. 4A, anti-miR-193b plasmids were transfected into HepG2 cells and significantly decreased the expression level of miR-193b in HepG2 cells (P<0.01). Additionally, downregulation of miR-193b significantly suppressed cell proliferation of HepG2 cells (P<0.01; Fig. 4B).
Figure 4.
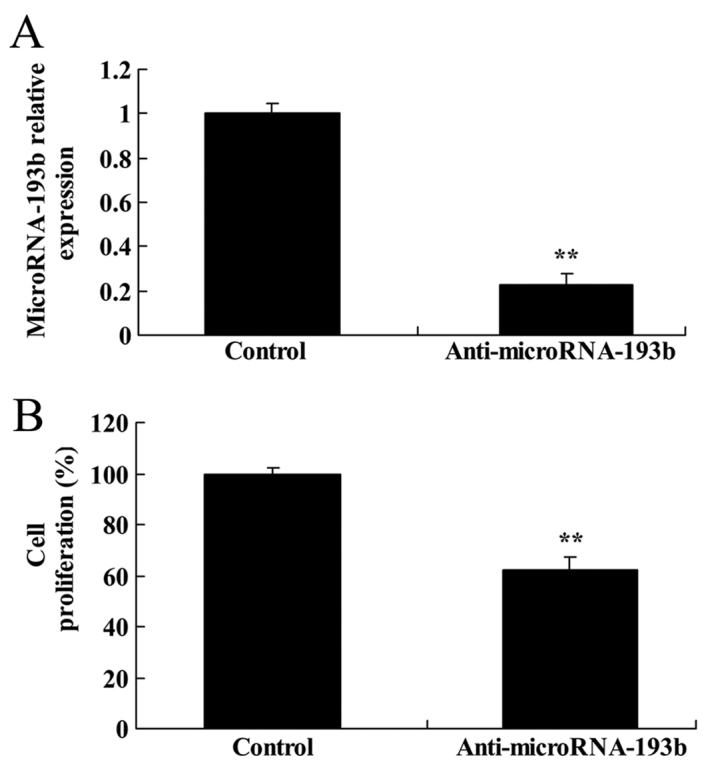
Effect of miR-193b expression on cell proliferation of HepG2 cells. (A) Expression level of miR-193b. (B) Effect of miR-193b on cell proliferation of HepG2 cells. **P<0.01 compared with the control group. miR, microRNA.
Effect of miR-193b on apoptosis of HepG2 cells
The present study determined the association between the downregulation of miR-193b on the rate of apoptosis of HepG2 cells. As presented in Fig. 5, there was a significant increase in the apoptotic rate of HepG2 cells with a downregulation of miR-193b compared with in the negative control group (P<0.01).
Figure 5.
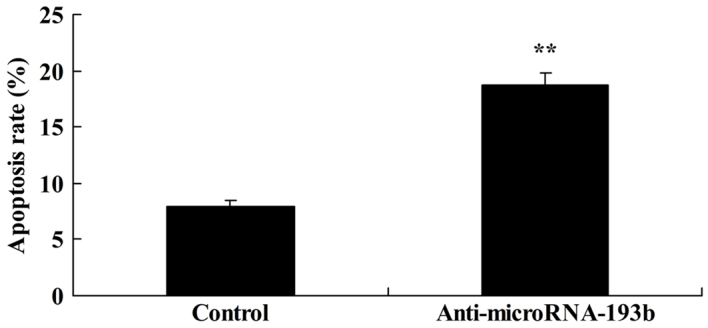
Effect of microRNA-193b expression on the rate of apoptosis of HepG2 cells. **P<0.01 compared with the control group.
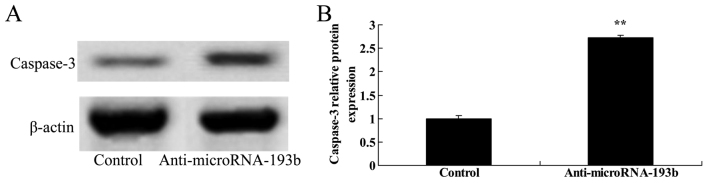
Effect of miR-193b on the caspase-3 protein expression level of HepG2 cells
The present study assessed the association between anti-miR-193b expression level on the caspase-3 protein expression level in HepG2 cells. Caspase-3 protein expression of HepG2 cells was significantly increased by anti-miR-193b transfected into cells compared with the negative control group (P<0.01; Fig. 6).
Figure 6.
Effect of miR-193b expression on caspase-3 protein expression level in HepG2 cells. (A) The effect of miR-193b on caspase-3 protein expression level using western blotting and (B) the statistical analysis of caspase-3 protein expression of HepG2 cells. β-actin was used as a loading control. **P<0.01 compared with the control group. miR, microRNA.
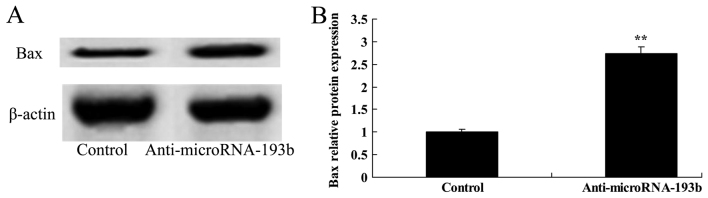
Effect of miR-193b on the Bax protein expression level of HepG2 cells
To characterize the molecular mechanism underlying apoptosis and anti-miR-193b regulation in human liver cancer, the Bax protein expression level was evaluated using western blotting. Western blotting demonstrated that Bax protein expression level of HepG2 cells transfected with anti-miR-193b was significantly increased compared with the control group (P<0.01; Fig. 7).
Figure 7.
Effect of miR-193b on Bax protein expression level in HepG2 cells. (A) Effect of miR-193b on Bax protein expression using western blotting and (B) quantification of Bax protein expression in HepG2 cells. β-actin was used as a loading control. **P<0.01 compared with the control group. miR, microRNA.
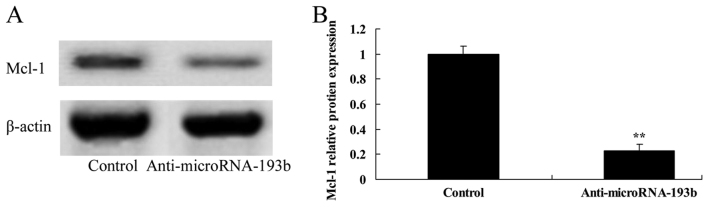
Effect of miR-193b on the Mcl-1 protein expression level in HepG2 cells
Initially, the present study assessed the association between the downregulation of miR-193b on the Mcl-1 protein expression level in HepG2 cells. Downregulation of miR-193b expression level significantly suppressed the Mcl-1 protein expression in HepG2 cells (P<0.01; Fig. 8).
Figure 8.
Effect of miR-193b on Mcl-1 protein expression level in HepG2 cells. (A) Effect of miR-193b on Mcl-1 protein expression level using western blotting and (B) quantification of Mcl-1 protein expression level in HepG2 cells. β-actin was used as a loading control. **P<0.01 compared with the control group. miR, microRNA; Mcl-1, myeloid cell leukemia-1.
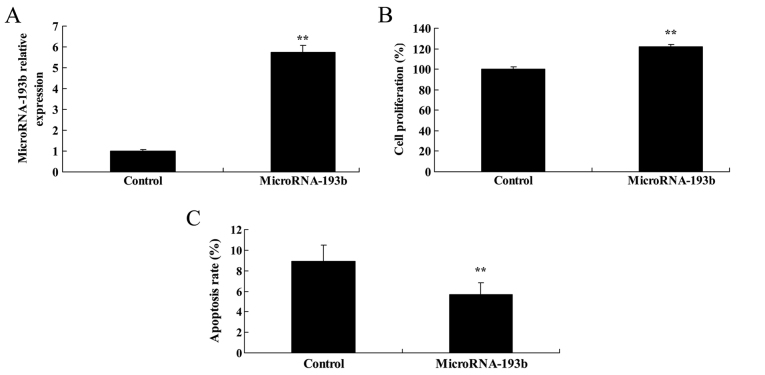
Effect of overregulation of miR-193b on cell proliferation and apoptosis of HepG2 cells
Subsequently, in order to determine whether overregulation of miR-193b affects cell proliferation and apoptosis of HepG2 cells, miR-193b plasmid was transfected into HepG2 cells. It was revealed that overregulation of miR-193b significantly increased the miR-193b expression level, promoted cell proliferation and suppressed cell apoptosis of HepG2 cells, as presented in Fig. 9 (P<0.01).
Figure 9.
Overregulation of miR-193b on cell proliferation and apoptosis of HepG2 cells. (A) Expression level of miR-193b, (B) effect of miR-193b on cell proliferation and (C) apoptosis rate of HepG2 cells. **P<0.01 compared with the control group. miR, microRNA.
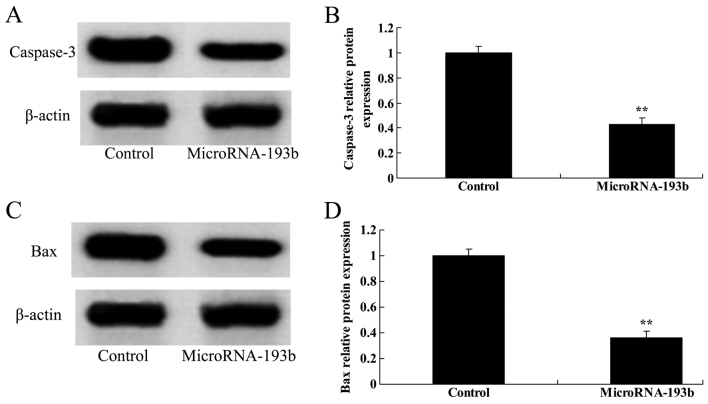
Overregulation of miR-193b on caspase-3 and Bax protein expression levels in HepG2 cells
The associations between the overregulation of miR-193b, caspase-3 and Bax protein expression levels in HepG2 cells were analyzed. Compared with their corresponding negative controls, overregulation of miR-193b significantly suppressed caspase-3 and Bax protein expression levels in HepG2 cells (P<0.01; Fig. 10).
Figure 10.
Overregulation of miR-193b on caspase-3 and Bax protein expression level in HepG2 cells. (A) Effect of miR-193b on the caspase-3 protein expression level using western blotting and (B) quantification of the caspase-3 protein expression level in HepG2 cells. (C) Effect of miR-193b on the Bax protein expression level using western blotting and (D) quantification of the Bax protein expression level in HepG2 cells. β-actin was used as a loading control. **P<0.01 compared with the control group. miR, microRNA.
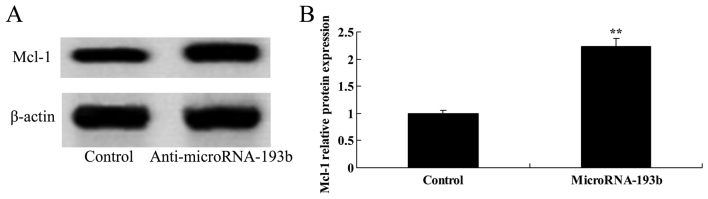
Overregulation of miR-193b on Mcl-1 protein expression levels in HepG2 cells
To further evaluate the associations between the overregulation of miR-193b and Mcl-1 protein expression levels of HepG2 cells, the Mcl-1 protein expression level was determined using western blotting. The results indicated that overregulation of miR-193b significantly suppressed the Mcl-1 protein expression level in HepG2 cells (P<0.01; Fig. 11).
Figure 11.
Overregulation of miR-193b on the Mcl-1 protein expression level in HepG2 cells. (A) The effect of miR-193b on Mcl-1 protein expression level using western blotting and (B) quantification of Mcl-1 protein expression levels in HepG2 cells. β-actin was used as a loading control. **P<0.01 compared with the control group. miR, microRNA; Mcl-1, myeloid cell leukemia-1.
Discussion
Clinically, the most common primary hepatic carcinoma is the primary hepatocellular carcinoma, which is a malignant tumor (16). Previous study data statistics have revealed that, worldwide, annual novel cases of liver cancer in 2008 accounted for ~4.1% of malignant tumors, which has exceeded 626,000 per year, ranking sixth among malignant tumors with a mortality rate of ~600,000 per year, and ranking third among cancer-associated mortalities (17). Furthermore, the liver cancer mortality rate in China was the highest worldwide in 2008 (18). The number of patients with liver cancer accounted for ~55% of those worldwide in 2010, ranking second among cancer-associated mortalities, with lung cancer being the most common (19). In the present study, 50 patients with liver cancer and 50 normal control patients were recruited. The results revealed that that the expression level of miR-193b in the normal group was significantly higher compared with that of the liver cancer exosome and whole tissue. Downregulated miR-193b was significantly associated with depth of invasion, metastasis, tumor differentiation, Lauren type and stage (14) in patients with liver cancer.
miRNA may interact with multiple cell regulatory genes, including regulatory factors of apoptosis and cell cycle, thereby serving a significant role in gene expression (20). Previous studies have revealed that the majority of miRNA have downregulated expression level in cancer cells. Thus, miRNA functions as a cancer suppressor gene in cells (21). A previous study demonstrated that miRNA had downregulated expression level in liver cancer and participated in the regulation and control of the signaling pathway of a series of cancer genes (22). Therefore, the preliminary study of miRNA has revealed the significance of miRNA as an important molecular marker in the prediction and diagnosis of hepatocellular carcinoma (23). In the present study, overall survival of patients with liver cancer and low miR-193b expression level is higher compared with that of patients with liver cancer and high miR-193b expression level. Kaukoniemi et al (24) demonstrated that miR-193b suppressed the proliferation of prostate cancer cells via cyclin D1.
Mcl-1 is a member of the Bcl-2 family, which is extensively expressed in normal human tissues, and also in human tumor tissues (25). Furthermore, a previous study revealed that it has a high expression level in tumors including lymphoma, leukemia, multiple myeloma, lung cancer and pancreatic cancer (26). Mcl-1 mainly participates in maintaining the stability of mitochondrial membrane of cells and inhibiting the release of cytochrome c, thereby promoting the survival of cells and preventing apoptosis (27). The subcellular localization of Mcl-1 is closely associated with cell proliferation (28). In exponential-phase cells, Mcl-1 is mainly located in the mitochondria; in stationary-phase cells, Mcl-1 is mainly located in the cell nucleus (8). A previous study demonstrated that Mcl-1 expression in hepatocellular carcinoma tissues was mostly located in the cytoplasm, indicating that hepatocellular carcinoma cells are in a rapid proliferation period, which corresponds to the biological characteristics of cancer (8). In addition, Mcl-1 may directly regulate and control cell differentiation and the cell cycle, combining Mcl-1 genes and proliferating cell nuclear antigen, which inhibits cells from entering the S phase, and an important regulatory factor in cell cycle transcription, E2F1, may inhibit the expression of Mcl-1 (29). The results of the present study demonstrated that downregulation of miR-193b suppressed cell proliferation and induced apoptosis of liver cancer cells and inhibited the expression level of Mcl-1 protein. Additionally, upregulation of miR-193b increased cell proliferation and decreased apoptosis of liver cancer cells and promoted the expression of Mcl-1 protein. Furthermore, Chen et al (30) revealed that miR-193b regulates Bcl-2 and Mcl-1 in melanoma.
To conclude, the results of the present study demonstrated that miR-193b is a novel suppressor for human liver cancer via inhibition of Mcl-1. The resulting expression of miR-193b regulated cell proliferation and apoptosis of human liver cancer. These results suggested that miR-193b and Mcl-1 account for the aggressive behavior of human liver cancer.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 81172575), Hunan Provincial Natural Science Committee and Hengyang City Government Unification Foundation of China (grant no. 12JJ9033), the Hunan Provincial Natural Science Foundation of China (grant no. 13JJ3079), National Natural Science Foundation (grant no. 81201331) and Experimental and Clinical Study of the New Chute Screw Growing Rod System in Correction of Children with Scoliosis (grant no. 2015JJ5003).
References
- 1.Xiong W, Qiu SY, Xu LY, Zhang CP, Yi Y, Wu Q, Huang LP, Liu SM, Wu B, et al. Effects of intermedin on dorsal root ganglia in the transmission of neuropathic pain in chronic constriction injury rats. Clin Exp Pharmacol Physiol. 2015;42:780–787. doi: 10.1111/1440-1681.12416. [DOI] [PubMed] [Google Scholar]
- 2.Du X, Cao Y, Xue P, Lin Z, Zeng Z, Xia Q. Protective effect of intermedin on myocardial cell in a rat model of severe acute pancreatitis. Cell Mol Biol Lett. 2011;16:462–476. doi: 10.2478/s11658-011-0020-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li P, Sun HJ, Han Y, Wang JJ, Zhang F, Tang CS, Zhou YB. Intermedin enhances sympathetic outflow via receptor-mediated cAMP/PKA signaling pathway in nucleus tractus solitarii of rats. Peptides. 2013;47:1–6. doi: 10.1016/j.peptides.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Choi JH, Comess KA, Xu C, Park J, Kim Y. Development of an interactive Coronary Doppler Vibrometry system for detection of coronary artery disease; Conf Proc IEEE Eng Med Biol Soc; 2011; pp. 7195–7198. [DOI] [PubMed] [Google Scholar]
- 5.Huang N, Wu J, Qiu W, Lyu Q, He J, Xie W, Xu N, Zhang Y. MiR-15a and miR-16 induce autophagy and enhance chemosensitivity of Camptothecin. Cancer Biol Ther. 2015;16:941–948. doi: 10.1080/15384047.2015.1040963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu L, Liu T, Xiao Y, Li X, Zhu Y, Zhao Y, Bao J, Wu C. Polygonatum odoratum lectin induces apoptosis and autophagy by regulation of microRNA-1290 and microRNA-15a-3p in human lung adenocarcinoma A549 cells. Int J Biol Macromol. 2016;85:217–226. doi: 10.1016/j.ijbiomac.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 7.Sabe AA, Elmadhun NY, Sadek AA, Chu LM, Bianchi C, Sellke FW. Differential effects of atorvastatin on autophagy in ischemic and nonischemic myocardium in Ossabaw swine with metabolic syndrome. J Thorac Cardiovasc Surg. 2014;148:3172–3178. doi: 10.1016/j.jtcvs.2014.07.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng J, Qiao L, Xu X, Zhai C, Zhao K, Ji X, Chen W. Lower AMP-activated protein kinase level is associated with the vulnerability of coronary atherosclerotic plaques by attenuating the expression of monocyte autophagy. Coron Artery Dis. 2015;26:322–327. doi: 10.1097/MCA.0000000000000243. [DOI] [PubMed] [Google Scholar]
- 9.De Jesus RA, Cechinel-Filho V, Oliveira AE, Schlemper V. Analysis of the antinociceptive properties of marrubiin isolated from Marrubium vulgare. Phytomedicine. 2000;7:111–115. doi: 10.1016/S0944-7113(00)80082-3. [DOI] [PubMed] [Google Scholar]
- 10.Wahdan-Alaswad RS, Cochrane DR, Spoelstra NS, Howe EN, Edgerton SM, Anderson SM, Thor AD, Richer JK. Metformin-induced killing of triple-negative breast cancer cells is mediated by reduction in fatty acid synthase via miRNA-193b. Horm Cancer. 2014;5:374–389. doi: 10.1007/s12672-014-0188-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blick C, Ramachandran A, McCormick R, Wigfield S, Cranston D, Catto J, Harris AL. Identification of a hypoxia-regulated miRNA signature in bladder cancer and a role for miR-145 in hypoxia-dependent apoptosis. Br J Cancer. 2015;113:634–644. doi: 10.1038/bjc.2015.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang S, Guo Y, Zhang C, Gao W, Wen S, Huangfu H, Wang B. Primary laryngeal cancer-derived miR-193b induces interleukin-10-expression monocytes. Cancer Invest. 2015;33:29–33. doi: 10.3109/07357907.2014.988344. [DOI] [PubMed] [Google Scholar]
- 13.Tessitore A, Cicciarelli G, Mastroiaco V, Filippo DV, Daria C, Daniela V, Mariafausta F, Davide V, Francesca Z, Edoardo A. Therapeutic use of micrornas in cancer. Anticancer Agents Med Chem. 2016;16:7–19. doi: 10.2174/1871520615666150824153358. [DOI] [PubMed] [Google Scholar]
- 14.Kemeny N, Brown K, Covey A, Kim T, Bhargava A, Brody L, Guilfoyle B, Haag NP, Karrasch M, Glasschroeder B, et al. Phase I, open-label, dose-escalating study of a genetically engineered herpes simplex virus, NV1020, in subjects with metastatic colorectal carcinoma to the liver. Hum Gene Ther. 2006;17:1214–1224. doi: 10.1089/hum.2006.17.1214. [DOI] [PubMed] [Google Scholar]
- 15.Shi Z, Zhou H, Lu L, Li X, Fu Z, Liu J, Kang Y, Wei Z, Pan B, Liu L, et al. The roles of microRNAs in spinal cord injury. Int J Neurosci. 2017;127:1104–115. doi: 10.1080/00207454.2017.1323208. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Wang W, Gong F, Fu S, Zhang Q, Hu J, Qi Y, Xie C, Zhang Y. Evaluation of intravenous immunoglobulin resistance and coronary artery lesions in relation to Th1/Th2 cytokine profiles in patients with Kawasaki disease. Arthritis Rheum. 2013;65:805–814. doi: 10.1002/art.37815. [DOI] [PubMed] [Google Scholar]
- 17.Yan GH, Wang M, Yiu KH, Lau CP, Zhi G, Lee SW, Siu CW, Tse HF. Subclinical left ventricular dysfunction revealed by circumferential 2D strain imaging in patients with coronary artery disease and fragmented QRS complex. Heart Rhythm. 2012;9:928–935. doi: 10.1016/j.hrthm.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 18.Li L, Zhao G. Downregulation of microRNA-218 relieves neuropathic pain by regulating suppressor of cytokine signaling 3. Int J Mol Med. 2016;37:851–858. doi: 10.3892/ijmm.2016.2455. [DOI] [PubMed] [Google Scholar]
- 19.Bi YF, Mao JY, Wang XL, Hou YZ, Lu YZ, Soh SB, Zhang BL. Clinical epidemiology survey of the traditional Chinese medicine etiology and syndrome differentiation of coronary artery disease: Study protocol of a multicenter trial. Zhong Xi Yi Jie He Xue Bao. 2012;10:619–627. doi: 10.3736/jcim20120604. [DOI] [PubMed] [Google Scholar]
- 20.Qian J, Xu B, Lansky AJ, Yang YJ, Qiao SB, Wu YJ, Chen J, Hu FH, Yang WX, Mintz GS, et al. First report of a novel abluminal groove filled biodegradable polymer rapamycin-eluting stent in de novo coronary artery disease: Results of the first in man FIREHAWK trial. Chin Med J (Engl) 2012;125:970–976. [PubMed] [Google Scholar]
- 21.Chen WR, Qian YA, Chen YD, Shi Y, Yin DW, Wang H, Zhu P, Liu HW, Sha Y. The effects of low vitamin D on coronary artery disease. Heart Lung Circ. 2014;23:314–319. doi: 10.1016/j.hlc.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 22.Hu CL, Li YB, Tang YH, Chen JB, Liu J, Tang QZ, Zhang QH, Huang CX. Effects of withdrawal of Xuezhikang, an extract of cholestin, on lipid profile and C-reactive protein: A short-term time course study in patients with coronary artery disease. Cardiovasc Drugs Ther. 2006;20:185–191. doi: 10.1007/s10557-006-7947-x. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Q, Xu B, Yang YJ, Zhang RY, Li JP, Qiao SB, Zhang JS, Hu J, Qin XW, Hong T, et al. Sirolimus-eluting cobalt alloyed stents in treating patients with coronary artery disease: Six-month angiographic and one-year clinical follow-up result. A prospective, historically controlled, multi-center clinical study. Chin Med J (Engl) 2007;120:533–538. [PubMed] [Google Scholar]
- 24.Kaukoniemi KM, Rauhala HE, Scaravilli M, Latonen L, Annala M, Vessella RL, Nykter M, Tammela TL, Visakorpi T. Epigenetically altered miR-193b targets cyclin D1 in prostate cancer. Cancer Med. 2015;4:1471–1425. doi: 10.1002/cam4.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burchill LJ, Lameijer H, Roos-Hesselink JW, Grewal J, Ruys TP, Kulikowski JD, Burchill LA, Oudijk MA, Wald RM, Colman JM, et al. Pregnancy risks in women with pre-existing coronary artery disease, or following acute coronary syndrome. Heart. 2015;101:525–529. doi: 10.1136/heartjnl-2014-306676. [DOI] [PubMed] [Google Scholar]
- 26.Zhao K, Xu XS, Meng X, Li YL, Li JF, Chen WQ. Autophagy of monocytes attenuates the vulnerability of coronary atherosclerotic plaques. Coron Artery Dis. 2013;24:651–656. doi: 10.1097/MCA.0000000000000035. [DOI] [PubMed] [Google Scholar]
- 27.Mostafalou S, Abdollahi M. Pesticides and human chronic diseases: Evidences, mechanisms and perspectives. Toxicol Appl Pharmacol. 2013;268:157–177. doi: 10.1016/j.taap.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 28.Shang FF, Xia QJ, Liu W, Xia L, Qian BJ, You L, He M, Yang JL, Wang TH. miR-434-3p and DNA hypomethylation co-regulate eIF5A1 to increase AChRs and to improve plasticity in SCT rat skeletal muscle. Sci Rep. 2016;6:22884. doi: 10.1038/srep22884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghavami S, Cunnington RH, Gupta S, Yeganeh B, Filomeno KL, Freed DH, Chen S, Klonisch T, Halayko AJ, Ambrose E, et al. Autophagy is a regulator of TGF-β1-induced fibrogenesis in primary human atrial myofibroblasts. Cell Death Dis. 2015;6:e1696. doi: 10.1038/cddis.2015.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, Zhang X, Lentz C, Abi-Daoud M, Paré GC, Yang X, Feilotter HE, Tron VA. miR-193b regulates Mcl-1 in melanoma. Am J Pathol. 2011;179:2162–2168. doi: 10.1016/j.ajpath.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]