Abstract
Colorectal cancer (CRC) is a common digestive tract tumor. Cancer tissues and healthy tissues were extracted from patients with CRC who were treated at our hospital. Targetscan and PicTar were used to identify microRNAs (miRNAs/miRs) that may interact with phosphatase and tensin homolog deleted on chromosome ten (PTEN). Dual luciferase reporter assay was applied to detect whether the 3′-untranslated region (UTR) of PTEN was targeted by miR-106a. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) showed that there was significantly higher miR-106a expression level in cancer tissue compared with in healthy tissue. The expression level of miR-106a in NCM640, SW620 and HT29 cell lines was detected by RT-qPCR, and HT29 cells showed the highest miR-106a level. HT29 cells were used for the present study, separated into control, miR-NC antagomiR and miR-106a antagomiR group. HT29 cell characteristics were tested. The results demonstrated that in the miR-106a antagomiR group, there was a lower cell proliferation and higher cell apoptosis rate compared with the control and miR-NC antagomiR groups. miR-106a was verified to target PTEN 3′-UTR in HT29 cells. In comparison with control and miR-NC antagomiR groups, the protein level of PTEN was increased and phosphatidylinositol-4,5-bisphosphate 3-kinase/protein kinase B was decreased following miR-766 antagomiR administration. The findings propose that miR-106a may serve a therapeutic target for the treatment of CRC.
Keywords: mir-106a, colon cancer, PTEN/PI3K/AKT signaling pathway
Introduction
Colorectal cancer (CRC), the third leading cause of cancer-related mortality and morbidity globally, is a common digestive tract tumor (1). Epigenetic variations, for instance, hyper-methylation of tumor suppressor genes, were associated with the development and progression of CRC (2). After years of researches, unfortunately, the molecular mechanisms of CRC metastasis have not been completely understood yet.
microRNAs (miRNAs), a cluster of non-coding RNA molecules that were at the length of about 19–24 nucleotides, could suppress gene expression via inducing target mRNA degradation and/or blocking translation (3) through base-pairing to bind to the 3′ untranslated region (UTR) of target mRNA (4,5). Up to now, more than 300 miRNAs have been identified from various organisms (6–9). miRNAs modulate numerous cell behaviors, for instance, cell differentiation, proliferation, apoptosis and metastasis (10–12).
Phosphatase and tensin homolog deleted on chromosome ten (PTEN) is a tumor suppressor gene located at 10q23, and plays a pivotal role in the pathogenesis of various human cancers. Evidences demonstrated the involvement of PTEN deactivation in tumorigenesis, whose loss increased the activity of PI3K/AKT and was correlated with cell proliferation, migration, invasion and apoptosis (13,14). Moreover, another study showed that PTEN, whose loss was positively correlated with malignant progression including tumor size and TNM advanced stage, played a crucial role at early/late stages of CRC (15); meanwhile, restoration of PTEN was reported to reduce the ratio of metastases in an orthotopic model of CRC (16). Current study aimed to investigate whether there are miRNAs that could target PTEN and regulate the progression of CRC, thus providing a therapeutic target for CRC. In our study, dual luciferase reporter assay was applied for the verification of interaction between miR-106a and PTEN 3′ UTR which was predicted by targetscan and PicTar. qRT-PCR was adopted to determine miR-106a expression level in tissues and cell lines. HT29 cells were used for the present study, separated into control group, miR-NC antagomiR group and miR-106a antagomiR group. HT29 cell proliferation and apoptosis were tested by 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and FCM assay, respectively. Western blot was used for the detection of PTEN, p-PI3K/p-AKT protein level.
Materials and methods
Clinical samples
Forty tumor tissues and the adjacent tissues were separated from CRC patients who underwent surgery in our hospital. Tissues were kept in liquid nitrogen as quick as possible. Informed consent was acquired from all the patients that participated in our research. Our study was approved by the ethics committee of Central Hospital of Chengde (Chengde, China).
Cell culture
The normal human colon epithelial cells (NCM640) and immortalized human colon epithelial cell lines including SW620 and HT29 were grown in RPMI-1640 medium (Hyclone; GE Healthcare, Logan, UT, USA) containing 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific Inc.) and 1% antibiotics at 37°C in a humidified chamber with 95% air and 5% CO2. Cells that expressed the highest miR-106a level was chosen for the following study.
Plasmid transfection
HT29 cells which showed the highest miR-106a level were used for the following experiments, and seeded into 24-well plates at the concentration of 1×105 cells/well. miR-106a antagomiR was synthesized by GenePharma (Shanghai, China). The antagomiR, a single-stranded RNA analogue was complementary to mature miR-106a (5′-CAAAGUGCUAACAGUGCAGGUAG-3′). A mismatched miR-negative control (NC) antagomiR (5′-UUGUACUACACAAAAGUACUG-3′) was also synthesized. Transfection of HT29 cells with miR-106a antagomiR or miR-NC antagomiR was conducted by Lipofectamine 2000 transfection reagent (Thermo Fisher, Inc., Waltham, MA, USA) in accordance with manufacturer's instruction.
HT29 cells were separated into 3 different groups including control group, miR-NC antagomiR group and miR-106a antagomiR group. Forty-eight hours after the transfection of plasmid, HT29 cells were collected.
Dual-luciferase reporter assay
The sequences of 3′UTR PTEN were amplified by the following primers (PTEN forward, 5′-cac aac tcg agT GGC AAT AGG ACA TTG TGTCA-3′ and reverse, 5′-aag gat ccA ACA ACA AGC AGT GAC AGCG-3′), then digested with BamHI/XhoI and constructed with pLuc vector. Downstream of the firefly luciferase stop codon was cloned in the pLuc control vector (Promega Corporation, Madison, WI, USA). HT29 cells were seeded into 48-well plates and incubated for 24 h, thereafter, co-transfected 1 mg 3′UTR-PTEN or 3′ UTR mut-PTEN with the control oligonucleotide (80 nM) or miR-106a antagomiR (80 nM) by Lipofectamine 2000 reagents (Invitrogen; Thermo Fisher Scientific Inc.) according to the manufacturer's protocol. Forty-eight h after transfection, luciferase activity in each group was determined with the Dual Luciferase Reporter Assay System (Promega Corporation).
MTT assay
HT29 cells (1×105 cells/ml) were seeded onto 12-well plates and incubated for 24 h, then incubated with 20 µl of MTT solution (5 mg/ml in PBS) for another 1 h. Followed by the removal of culture medium, converted purple formazan dye from MTT was solubilized in dimethyl sulfoxide (DMSO; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Finally, optical densities from all the samples were measured at 595 nm by a mocroplate reader (BioRad, Hercules, CA, USA).
Cell apoptosis analysis
A total of 1×105 HT29 cells were collected by centrifugation at the speed of 3,000 rpm/min for 5 min and washed three times with PBS. Cells were re-suspended in RPMI medium with 1% bovine serum albumin (BSA) and 10% FBS, mixed with Annexin V-enhanced green fluorescent protein (FITC) and propidium iodide (PI), and then incubated for 20 min at room temperature in the dark. Assay results were determined with flow cytometry.
Western blot
Cells were seeded into 6-well plate at the concentration of 1×105 cells/ml/plate and grown at 37°C incubator in a 5% CO2 atmosphere for 24 h. Cells were washed with cold PBS twice, scraped with radioimmunoprecipitation buffer (RIPA; Beyotime, Shanghai, China), then subjected to western blot analysis. Primary antibody reacted at 4°C overnight and the second antibody reacted at room temperature for 2 h. At room temperature, after washing the samples with Tris Buffered Saline with Tween-20 (TBST) three times with each time for 10 min. At last, bands were detected with ECL kit (Beyotime). Bands were quantified by Odyssey infrared imaging (LICOR, Lincoln, NE, USA) and GAPDH acted as an internal reference.
Quantitative real time polymerase chain reaction
Total RNA was extracted from HT29 cells with RNAiso Plus (Takara, Bio, Inc., Otsu, Japan). To assess the expression level of miR-106a, a stemloop reverse transcription-polymerase chain reaction (RT-PCR) was carried out. The relative expression level of miR-106a was calculated via the comparative cycle threshold (Ct) method and normalized to U6. Data were processed using the 2−ΔΔCT method.
Statistical analysis
Student's t test and one-way ANOVA were carried out to assess statistical significance. Results were presented as mean ± standard deviation (SD). P<0.05 was considered to indicate a statistically significant difference.
Results
miRNA that could target PTEN
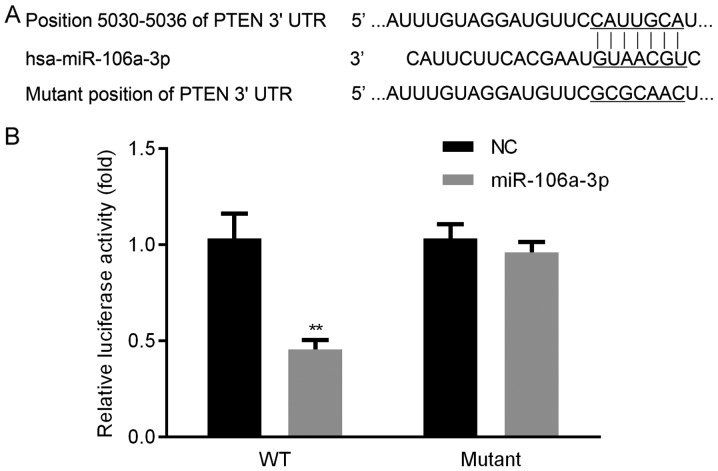
To explore the molecule by which PTEN was targeted, we used two algorithms (PicTar and Target Scan). Seed sequence of miR-106a was identified in 3′ UTR of PTEN, the wild-type and mutant seed sequences between PTEN and miR-106a were displayed (Fig. 1A). We found that, luciferase activity in WT group was obviously lower, whereas, there was no significant change of luciferase activity in mutant group (Fig. 1B). These results proved that PTEN could be targeted by miR-106a.
Figure 1.
miR-106a targets PTEN. Seed sequence of miR-106a was identified in 3′ UTR of PTEN. The mutant seed sequence of PTEN 3′ UTR was also displayed (A). Luciferase activity in WT group was lower, while no significant change was discovered in mutant group (B). **P<0.01 miR-106a-3P vs. NC. PTEN, phosphatase and tensin homolog deleted on chromosome ten; 3′ UTR, 3′-untranslated region; WT, wild-type; NC, negative control; miR, microRNA.
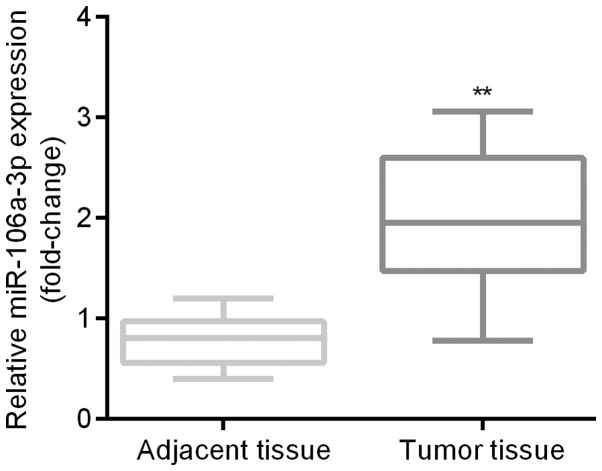
Expression level of miR-106a in patients' tissues
qRT-PCR was applied for the examination of expression differences of miR-106a between patients' tumor tissues and the adjacent tissues. Results indicated that miR-106a was significantly higher in patients' tumor tissues than in the adjacent tissues (Fig. 2).
Figure 2.
Higher expression level of miR-106a in patients' tumor tissues. qRT-PCR indicated that miR-106a was significantly higher in patients' tumor tissues than in the adjacent tissues. **P<0.01 tumor vs. Adjacent. miR, microRNA.
Correlations between miR-106a-3p or PTEN and CRC patients' characteristics
PTEN and CRC gene expression levels in patients' tumor tissues and the adjacent tissues were tested by qRT-PCR as presented in Table I. There were 15 male patients and 25 female patients in our study, with 19 patients aged <60 years old and 21 patients aged ≥60 years old; there were 18 patients whose tumor size was ≥5 cm and 22 patients whose tumor size was <5 cm; well-intermediately differentiation was found in 18 patients and poor differentiation was found in 22 patients; while metastasis was found in 17 patients. As for sex and age, there was no significant difference of miR-106a-3p or PTEN. Significant differences were discovered in tumor size, histological size and metastasis.
Table I.
The correlations between miR-106a-3p or PTEN and colorectal cancer patients' characteristics (mean ± SD).
| Factor | Case | miR-106a-3p | P-value | PTEN | P-value |
|---|---|---|---|---|---|
| Sex | 0.32 | 0.30 | |||
| Male | 15 | 2.58±0.34 | 2.12±0.51 | ||
| Female | 25 | 2.45±0.39 | 1.96±0.42 | ||
| Age (years) | 0.18 | 0.58 | |||
| <60 | 19 | 2.45±0.68 | 1.86±0.48 | ||
| ≥60 | 21 | 2.76±0.73 | 1.95±0.53 | ||
| Tumor size (cm) | 0.01 | 0.05 | |||
| ≥5 | 18 | 2.81±0.42 | 1.79±0.62 | ||
| <5 | 22 | 2.21±0.67 | 2.22±0.68 | ||
| Histological grade | 0.02 | 0.01 | |||
| Well-intermediately differentiation | 18 | 2.65±0.54 | 1.82±0.52 | ||
| Poor differentiation | 22 | 2.26±0.46 | 2.25±0.38 | ||
| Metastasis | 0.01 | 0.02 | |||
| No | 23 | 2.28±0.63 | 2.19±0.51 | ||
| Yes | 17 | 2.76±0.48 | 1.81±0.47 |
miR, microRNA; PTEN, phosphatase and tensin homolog deleted on chromosome ten; SD, standard deviation.
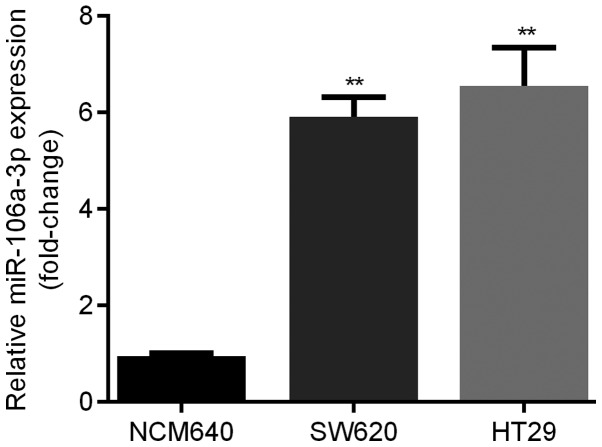
Expression level of miR-106a in NCM640, SW620 and HT29
qRT-PCR was applied for the examination of expression levels of miR-106a in 2 different human CRC cell lines compared with NCM640. We found that relative expression level of miR-106a was significantly elevated in SW620 and HT29 cell lines (about 6-fold). Moreover, HT29 cells showed the highest miR-106a level (Fig. 3).
Figure 3.
miR-106a was upregulated in CRC cell lines. qRT-PCR indicated that compared with NCM640 cells, miR-106a was obviously elevated in SW620 and HT29 cell lines, and HT29 cells showed the highest miR-106a level. **represents P<0.01 SW620 and HT29 vs. NCM640. miR, microRNA; CRC, colorectal cancer.
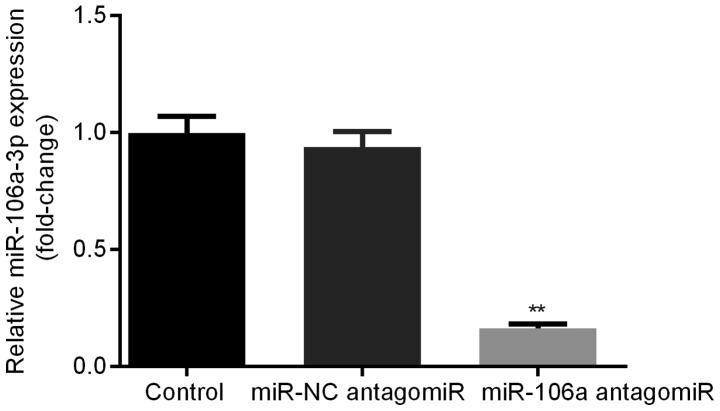
Expression level of miR-106a in HT29 cells after transfection
HT29 cells were transfected with miR-NC antagomiR or miR-106a antagomiR. Relative miR-106a expression levels were detected by qRT-PCR. We found that, there was no significant difference of miR-106a expression level between control group and miR-NC antagomiR group, however, miR-106a was obviously decreased in miR-106a antagomiR group compared with other two groups. This result verified the successful transfection of miR-106a antagomiR into HT29 cells (Fig. 4). Consequently, the transfected HT29 cells were used for the conduction of the experiments in our study.
Figure 4.
Successful transfection of miR-106a antagomiR in HT29 cells. qRT-PCR results indicated that, in HT29 cells, there was no significant difference of miR-106a level between control group and miR-NC antagomiR group, however, miR-106a was obviously decreased compared with other two groups. **represents P<0.01 antagomiR vs. Control and NC. miR, microRNA; NC, negative control.
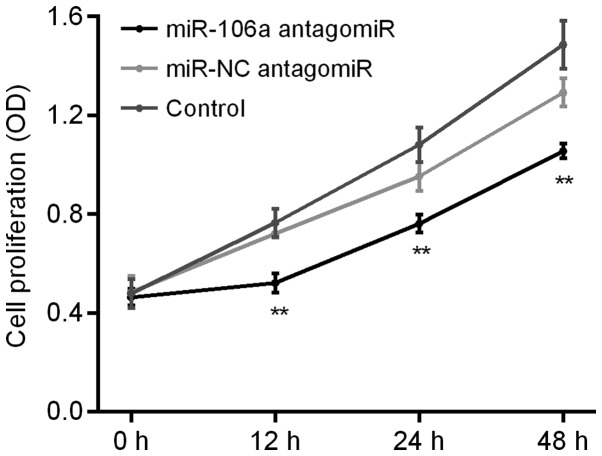
Influence of miR-106a antagomiR on HT29 cell proliferation
We carried out MTT assay to detect the influence of miR-106a antagomiR on the cell proliferation of HT29 cells. Results showed that, there was no significant difference of HT29 cell proliferation between control group and miR-NC antagomiR group, however, which was obviously decreased by miR-106a antagomiR (Fig. 5).
Figure 5.
miR-106a antagomiR inhibited HT29 cell proliferation. MTT assay showed that, there was no significant difference of HT29 cell proliferation between control group and miR-NC antagomiR group, which was obviously decreased by miR-106a antagomiR. **represents P<0.01 antagomiR vs. Control and NC. miR, microRNA; NC, negative control; MTT assay, 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay.
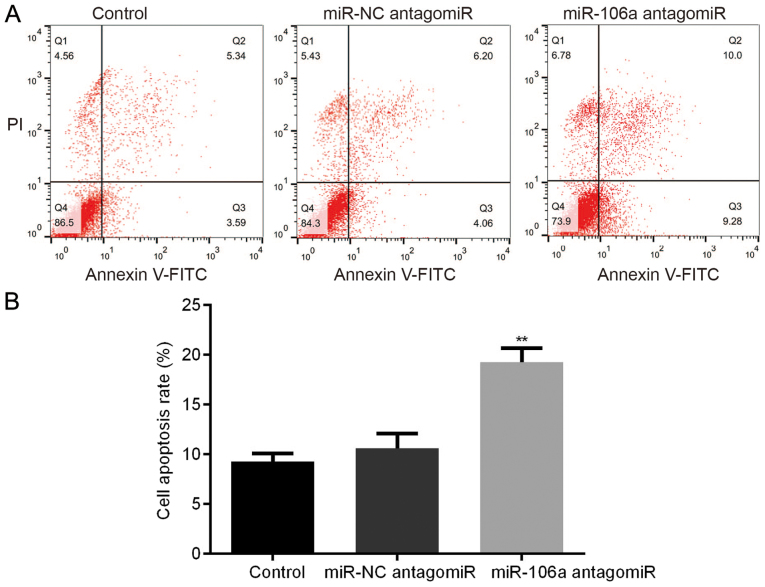
Influence of miR-106a antagomiR on HT29 cell apoptosis
We carried out FCM assay to detect the influence of miR-106a antagomiR on the cell apoptosis of HT29 cells. Results showed that, there was no significant difference of HT29 cell apoptosis between control group and miR-NC antagomiR group, however, miR-106a antagomiR treatment led to the highest cell apoptosis rate among the 3 groups (Fig. 6).
Figure 6.
miR-106a antagomiR induced HT29 cell apoptosis. FCM showed that, there was no significant difference of HT29 cell apoptosis between control group and miR-NC antagomiR group, which was obviously increased by miR-106a antagomiR (A and B). **represents P<0.01 antagomiR vs. Control and NC. miR, microRNA; NC, negative control.
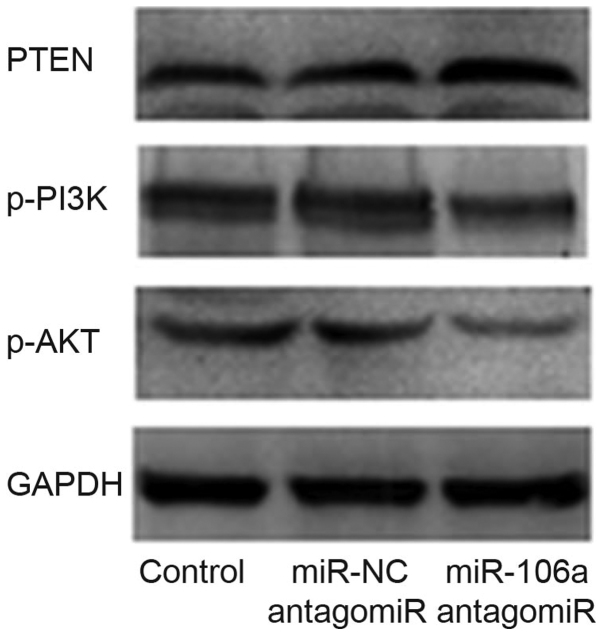
Influence of miR-106a antagomiR on protein levels of PTEN, p-PI3K and p-AKT
We performed western blot to detect the influence of miR-106a antagomiR on the expression level of PTEN, p-PI3K and p-AKT. Results showed that, there was no significant difference of the protein level between control group and miR-NC antagomiR group, however, p-PI3K/p-AKT protein levels were obviously decreased and PTEN protein level was obviously increased by the treatment of miR-106a antagomiR (Fig. 7).
Figure 7.
miR-106a antagomiR upregulated PTEN protein level and downregulated p-PI3K/p-AKT protein levels. Western blot analysis showed that, there was no significant difference of the protein levels between control group and miR-NC antagomiR group, however, p-PI3K/p-AKT expression levels were obviously decreased and PTEN expression level was obviously increased by miR-106a antagomiR. miR, microRNA; NC, negative control; PTEN, phosphatase and tensin homolog deleted on chromosome ten.
Discussion
CRC is a common digestive tract tumor (1). miRNAs were found to regulate cell differentiation, proliferation, apoptosis and metastasis (10–12).
Loss of PTEN was positively correlated with malignant progression of CRC (15), restoration of PTEN reduced the ratio of metastases of CRC (16). Current study aimed to investigate whether there were miRNAs that targeted PTEN and regulated the progression of CRC, thus providing a therapeutic target for CRC.
To explore the molecule by which PTEN was targeted, we used two algorithms (PicTar and TargetScan). Seed sequence of miR-106a was identified in 3′ UTR of PTEN (Fig. 1A). The interaction between miR-106a and PTEN were confirmed by dual luciferase activity (Fig. 1B). qRT-PCR indicated that, in patients' tumor tissues miR-106a was obviously upregulated compared to the adjacent tissues (Fig. 2), which was in consistent with previous studies that reported high expression of miR-106a in cancer tissues of CRC patients (17,18); meanwhile, compared to NCM640 cells, miR-106a was significantly elevated in SW620 and HT29 cell lines (Fig. 3), which was in line with a previous study (19). These results together suggested the oncogenetic role of miR-106a in CRC patients and in SW620/HT29 cells.
Successful transfection of miR-106a antagomiR into HT29 cells was confirmed by the lowest miR-106a level in the group treated with miR-106a antagomiR among the 3 groups (Fig. 4). Consequently, HT29 cells successfully transfected with miR-106a antagomiR were used for carrying out the following experiments.
As for the influences of miR-106a on HT29 cell proliferation and apoptosis, MTT assay and FCM assay were carried out, respectively. Results showed that, there was no significant difference of HT29 cell proliferation between control group and miR-NC antagomiR group, however, which was significantly decreased by the treatment of miR-106a antagomiR (Fig. 5). Otherwise, there was no significant difference of HT29 cell apoptosis between control group and miR-NC antagomiR group, which was notably increased by miR-106a antagomiR (Fig. 6). Whereas, the signaling pathways that were responsible for the changes remained to be investigated.
Activation of PI3K phosphorylated and activated AKT, thus localizing AKT to the plasma membrane; PI3K/AKT, an intracellular signaling pathway, was directly associated with cellular proliferation, cancer and longevity (20). And there was a natural inhibitor that named PTEN to inhibit the activation of PI3K/AKT signaling pathway, thus limiting cell proliferation and cancer progression; furthermore, knocking out of PTEN was shown to elevate the mass of brain on account of the corresponding unregulated proliferation (21). Administration of PTEN inhibitor was found to temporarily and safely impact the activation of PI3K/AKT signaling pathway thus influencing cell survival (22) and proliferation (23).
Consequently, we carried out western blot and found that, there was no significant difference of the protein level between control group and miR-NC antagomiR group, however, p-PI3K/p-AKT were obviously decreased and PTEN was obviously increased by the treatment of miR-106a antagomiR (Fig. 7).
Taken together, miR-106a antagomiR upregulated PTEN protein level and HT29 cell apoptosis, downregulated p-PI3K/p-AKT protein levels and HT29 cell proliferation via targeting 3′ UTR of PTEN, which suggested the potential role of miR-106a as a therapeutic target for the treatment of CRC.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer; Proc Natl Acad Sci USA; 1999; pp. 8681–8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Pasquinelli AE, Ruvkun G. Control of developmental timing by microRNAs and their targets. Annu Rev Cell Dev Biol. 2002;18:495–513. doi: 10.1146/annurev.cellbio.18.012502.105832. [DOI] [PubMed] [Google Scholar]
- 5.Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. doi: 10.1016/S1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 6.Dostie J, Mourelatos Z, Yang M, Sharma A, Dreyfuss G. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA. 2003;9:180–186. doi: 10.1261/rna.2141503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 8.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 9.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. Micro-RNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Croce CM, Calin GA. miRNAs, cancer and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 11.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 12.Ventura A, Jacks T. MicroRNAs and cancer: Short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bermúdez Brito M, Goulielmaki E, Papakonstanti EA. Focus on PTEN regulation. Front Oncol. 2015;5:166. doi: 10.3389/fonc.2015.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miao Y, Zheng W, Li N, Su Z, Zhao L, Zhou H, Jia L. Micro-RNA-130b targets PTEN to mediate drug resistance and proliferation of breast cancer cells via the PI3K/Akt signaling pathway. Sci Rep. 2017;7:41942. doi: 10.1038/srep41942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waniczek D, Śnietura M, Mlynarczyk-Liszka J, Piglowski W, Kopeć A, Lange D, Rudzki M, Arendt J. PTEN expression profiles in colorectal adenocarcinoma and its precancerous lesions. Pol J Pathol. 2013;64:15–20. doi: 10.5114/pjp.2013.34598. [DOI] [PubMed] [Google Scholar]
- 16.Chowdhury S, Ongchin M, Wan G, Sharratt E, Brattain MG, Rajput A. Restoration of PTEN activity decreases metastases in an orthotopic model of colon cancer. J Surg Res. 2013;184:755–760. doi: 10.1016/j.jss.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diaz R, Silva J, Garcia JM, Lorenzo Y, Garcia V, Peña C, Rodríguez R, Muñoz C, García F, Bonilla F, Domínguez G. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosomes Cancer. 2008;47:794–802. doi: 10.1002/gcc.20580. [DOI] [PubMed] [Google Scholar]
- 19.Feng B, Dong TT, Wang LL, Zhou HM, Zhao HC, Dong F, Zheng MH. Colorectal cancer migration and invasion initiated by microRNA-106a. PLoS One. 2012;7:e43452. doi: 10.1371/journal.pone.0043452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King D, Yeomanson D, Bryant HE. PI3King the Lock: targeting the PI3K/Akt/mTOR pathway as a novel therapeutic strategy in neuroblastoma. J Pediatr Hematol Oncol. 2015;37:245–251. doi: 10.1097/MPH.0000000000000329. [DOI] [PubMed] [Google Scholar]
- 21.Rafalski VA, Brunet A. Energy metabolism in adult neural stem cell fate. Prog Neurobiol. 2011;93:182–203. doi: 10.1016/j.pneurobio.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 22.Lai JP, Dalton JT, Knoell DL. Phosphatase and tensin homologue deleted on chromosome ten (PTEN) as a molecular target in lung epithelial wound repair. Br J Pharmacol. 2007;152:1172–1184. doi: 10.1038/sj.bjp.0707501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wyatt LA, Filbin MT, Keirstead HS. PTEN inhibition enhances neurite outgrowth in human embryonic stem cell-derived neuronal progenitor cells. J Comp Neurol. 2014;522:2741–2755. doi: 10.1002/cne.23580. [DOI] [PubMed] [Google Scholar]