Abstract
Nucleus accumbens-associated protein 1 (NAC1), a transcriptional co-regulator, is overexpressed in advanced prostate cancer. However, the NAC1-regulated transcriptome has not been completely explored. In the present study, the functional silencing of NAC1 blocked the migration of prostate cancer cells and suppress osteoclastogenesis. The present study also determined that NAC1 was overexpressed in the highly aggressive prostate cancer cell lines PC-3, DU-145 and LNCaP. NAC1 small interfering RNA treatment of DU-145 cells decreased cell migration, but interestingly had no significant effects on cell proliferation. Furthermore, microarray analysis showed that a group of genes may be associated with the development of prostate cancer after NAC1 knockdown, including interferon-β (IFNβ), which is reported to be involved in osteoclastogenesis, an important factor affecting bone metastasis. The mechanisms of NAC1 function were further explored by co-culture studies using PC-3 and RAW264.7 osteoclast precursor cells, which demonstrated that silencing NAC1 downregulated the genes associated with the activation of osteoclasts. Furthermore, it was revealed that NAC1 had the ability to affect the release of IFNβ into the extracellular environment. Together, these findings indicated that NAC1 promoted cell migration, and that NAC1 may have a key role in osteoclastogenesis.
Keywords: nucleus accumbens-associated protein 1, interferon β, prostate cancer, migration, osteoclastogenesis
Introduction
Prostate cancer is the second most frequently diagnosed cancer in men worldwide, and with an increasing incidence. It is also the fifth leading cause of cancer-related death worldwide, and >250,000 men die of prostate cancer each year (1,2). In prostate cancer patients, bone is the most common site of distant metastasis, which is the predominant cause of morbidity and mortality. In addition, bone destruction can increase the risk of death by >20% in patients with bone metastasis (3,4). However, at present, few drugs are able to delay the skeletal events (such as bone destruction or pain caused by bone metastasis), and the available treatments for bone pain are unsatisfactory (5). Therefore, the molecular mechanisms of bone metastasis from prostate cancer must be further explored, and more efficacious therapeutic targets are urgently required.
Nucleus accumbens-associated protein 1 (NAC1) belongs to a family of proteins with the Pox virus and zinc finger (POZ; also known as BTB) domain, a versatile protein-protein interaction motif in the N-terminus that mediates either homodimer or heterodimer formation (6–8). Moreover, NAC1 is a transcriptional co-factor that participates in self-renewal and maintenance of pluripotency in embryonic stem cells (9,10). Recent studies have revealed that NAC1 is markedly up-regulated in several types of human carcinomas, and is involved in the proliferation, autophagy, senescence, chemotherapy resistance, cytokinesis, migration and motility of cancer cells (11–16). An abundance of NAC1 protein seems to correlate with poorer prognosis in certain carcinomas in humans (17). However, the explicit mechanism of NAC1 in prostate cancer has not yet been reported. Given the significant and diverse roles of NAC1 in cancer biology, the purpose of the present study was to investigate the role of NAC1 in the development of prostate cancer and bone metastasis.
Materials and methods
Patients and sampling
The protocol utilized in this study was approved by the Central South University Institutional Review Board. For western blot analysis, eight pairs of prostate cancer and adjacent normal tissue specimens were collected from the patients who underwent surgery at the Third Xiangya Hospital of the Central South University in 2016. In addition, 84 paraffin-embedded samples of prostate cancer and 84 specimens of adjacent non-cancerous prostate tissue were collected between 2014 and 2016 for use in immunohistochemical assays. All tumors were histologically and clinically diagnosed by the Cancer Center of Central South University. For the use of these clinical materials, prior patient consent and approval from the institutional research ethics committee (approval nο: 2016-S064) were obtained.
Immunohistochemistry
Immunohistochemistry was performed on deparaffinized sections using a NAC1 antibody (Abcam, Cambridge, MA, USA) at a dilution of 1:100 and an UltraSensitive™ S-P hypersensitivity kit (Fuzhou Maixin Biotech Co., Ltd., China). Following antigen retrieval in a sodium citrate buffer, sections were incubated overnight at 4°C with the antibody. The slides for all samples were evaluated with a light microscope by two researchers. The antibody staining intensity was then analyzed using the H-SCORE system with modifications, as follows: H-SCORE=ΣPi(i), where i is the intensity of staining (0=undetectable, 1=weakly positive, 2=moderately positive, and 3=intensely positive).
Cell lines and culture conditions
The prostate cancer cell lines PC-3, DU-145 and LNCaP were kindly provided by the Cancer Center of Sun Yat-sen University, and RAW264.7 was kindly provided by the Central Laboratory of Central South University. Cells were cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) or DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal calf serum (Biological Industries, Cromwell, CT, USA) and 100 units/ml penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc.).
siRNA knockdown of NAC1 gene expression
Two small interfering RNAs (siRNAs) targeting NAC-1 were designed, with the sequences 5′-UGAUGUACACGUUGGUGCCUGUCACCA-3′ and 5′-UGUAGCAGAAGCUGAGGAUCUGCUG-3′ (16). The NAC-1 siRNAs and negative control siRNA (luciferase siRNA) were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). Cells were seeded into 6-well plates or 96-well plates and transfected with siRNAs using siRNA-Mate (Shanghai GenePharma Co., Ltd.). At 24 h after transfection, cells were harvested and prepared for assays.
Western blot analysis
Cells or tissues were lysed in RIPA Lysis Buffer supplemented with a cocktail of protease inhibitors (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China), followed by centrifugation at 14,000 × g for 10 min. After centrifugation, the cell lysates were collected and the protein concentrations measured. Proteins (20–30 µg) were resolved by SDS-PAGE, and then transferred to PVDF membranes (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The membranes were blocked with 5% BSA at room temperature for 3 h, and then incubated with primary antibodies in 3% BSA/TBST at 4°C overnight, followed by incubation with secondary antibodies (Auragene Bioscience Corp., Inc., Changsha, China) at room temperature for 1 h. The protein signals were detected by the ECL method. The following antibodies were used for western blotting: NAC1, GAPDH, β-actin, IFNβ, RANK, MMP9 and PKM2 antibodies, all purchased from Abcam.
MTT assay
DU-145 cell growth ability was measured by MTT assay. Briefly, cells subjected to different treatments were plated at a density of 3×103 cells per well in 96-well plates, and then incubated at 37°C in a humidified atmosphere containing 5% CO2/95% air. After 24, 48, 72 and 96 h of incubation, MTT reagent was added to each well, and the samples were incubated for another 4 h. The resultant formazan product was dissolved in DMSO and absorbance was read at 490 nm on a Victor3 Multi Label plate reader (PerkinElmer, Boston, MA, USA).
Colony formation assay
At 24 h after transfection, DU-145 cells (1.5×103 cells per well were plated in 6-well plates and incubated at 37°C in a humidified atmosphere containing 5% CO2/95% air for 14 days to allow for the formation of colonies. The colonies were washed twice with PBS, and the cells were then stained with 0.1% crystal violet for 20 min and washed with water before the colonies (each ≥50 cells) were counted.
Wound healing assay and Transwell assay
To study cell migration, DU-145 cells were cultured to confluence in 6-well plates, and a 2-mm-wide single, uninterrupted scratch in the cell layer was made across the diameter of each well. The culture plates were incubated with RPMI-1640 for 24 h and then washed with PBS, and photomicrographs were captured to assess the change in the size of the wound area.
Transwell assays were also performed to quantify the migration of DU-145 cells. Uncoated inserts (cat. no. 354578; BD Biosciences, San Jose, CA, USA) were used for the migration assays. Duplicate wells were assessed. DU-145 cells were seeded on top of the inserts in RPMI-1640 and cultured for 24 h, after which the cells that had migrated to the underside of the membrane were visualized. Cells were counted in five individual fields on each insert.
Microarray analysis
DU-145 cells were transfected with the two NAC1 siRNAs or a negative control siRNA. At 48 h after transfection, the cells were harvested for microarray analysis. An Arraystar Human messenger RNA (mRNA) Microarray v3.0 was used and run by the service provider. The array contained 30,215 protein-coding transcripts. A specific exon or splice junction probe which could identify an individual transcript accurately was used to represent each transcript. Positive probes for housekeeping genes and negative probes were also included in the array for hybridization quality control. The microarray hybridization and bioinformatics analysis were performed by KangChen Bio-tech (Shanghai, China).
RT-qPCR
Total RNA was extracted from the cells with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The relative expression levels of the mRNAs were determined by real-time RT-qPCR using a standard SYBR Green RT-PCR Kit (Takara Bio, Inc., Otsu, Japan). The specific primer pairs used in this study are listed in Table I. Averages of the Ct values from duplicate measurements were obtained. The relative expression levels of mRNAs were quantified using GraphPad Prism v5.0 software (GraphPad Software, Inc., San Diego, CA, USA) and the 2−ΔΔCt method.
Table I.
Oligonucleotide primers used for reverse transcription-quantitative polymerase chain reaction.
| Primer | Sequence |
|---|---|
| NAC1 F | 5′-CCAGACACTGCAGATGGAGA-3′ |
| NAC1 R | 5′-AAGCTGAGGATCTGCTGGAA-3′ |
| IFNβ F | 5′-GCTCTCCTGTTGTGCTTCTCCACT-3′ |
| IFNβ R | 5′-AGCTGCTTAATCTCCTCAGGGATG-3′ |
| IL-24 F | 5′-TTCTCTGGAGCCAGGTATC-3′ |
| IL-24 R | 5′-TAGAATTTCTGCATCCAGGT-3′ |
| NFATc1 F | 5′-GTCCCACCACCGAGCCCACTACG-3′ |
| NFATc1 R | 5′-GACCATCTTCTTCCCGCCCACGAC-3′ |
| S100A9 F | 5′-CACCCAGACACCCTGAACCA-3′ |
| S100A9 R | 5′-CCTCGAAGCTCAGCTGCTTG-3′ |
| GAB1 F | 5′-ATCAGAAACGCCAGCGAAGA-3′ |
| GAB1 R | 5′-TCAGATACCACAAAGCACCA-3′ |
| MMP9 F | 5′-TGTACCGCTATGGTTACACTCG-3′ |
| MMP9 R | 5′-GGCAGGGACAGTTGCTTCT-3′ |
| VEGFA F | 5′-GAGCCTTGCCTTGCTGCTCTA-3′ |
| VEGFA R | 5′-CACCAGGGTCTCGATTGGATG-3′ |
| RANK F | 5′-CATCGGGTTCCCATAAAG-3′ |
| RANK R | 5′-GAAGCAAATGTTGGCGTA-3′ |
| GAPDH F | 5′-TTGGTATCGTGGAAGGACTC-3′ |
| GAPDH R | 5′-ACAGTCTTCTGGGTGGCAGT-3′ |
NAC1, Nuclear accumbens-associated protein 1; IFNβ, interferon-β; IL-24, interleukin 24; NFATc1, nuclear factor of activated T-cells cytoplasmic 1; S100A9, S100 calcium-binding protein A9; GAB1, GRB 2-associated binding protein 1; MMP9, matrix metalloproteinase 9; VEGFA, vascular endothelial growth factor A; RANK, receptor activator of NF-kB; GAPDH, Glyceraldehyde 3-Phosphate Dehydrogenase.
Co-culture studies
Boyden chambers (3-µm hole inserts; Corning Incorporated, Corning, NY, USA) were used to assess the influence of PC-3 cells on osteoclasts using RAW 264.7 mouse monocytes. RAW 264.7 cells (1×105) were plated in 6-well plates for 24 h, then PC-3 cells (1×105) transfected with NAC1 siRNA or negative control siRNA were added into the upper chamber. RPMI-1640 supplemented with 10% fetal bovine serum was used to culture the PC-3 cells and DMEM supplemented with 10% fetal bovine serum was used to culture the RAW264.7 cells. IFNβ concentrations were detected by ELISA of the co-culture medium (as described below), and osteoclast formation was evaluated by RT-qPCR and western blot analyses of the total RNA and protein extracted from RAW 264.7 cells after culturing for 72 h.
Enzyme-linked immunosorbent assay (ELISA)
The protein level of IFNβ in the conditioned medium was assessed using human immunoassay ELISA kits (CUSABIO, Wuhan, China). The optical densities were measured at 450 nm and the detection limits were set in accordance with the log-log correlative coefficient of the standard curve.
Statistical analysis
Data were expressed as the mean ± SEM. Student's t-test (for comparison between two groups) was used to evaluate the numeric data. The χ2 test was used for comparisons of categorical data. A value of P<0.05 was considered to indicate a statistically significant difference.
Results
NAC1 expression is up-regulated in the progression of prostate cancer
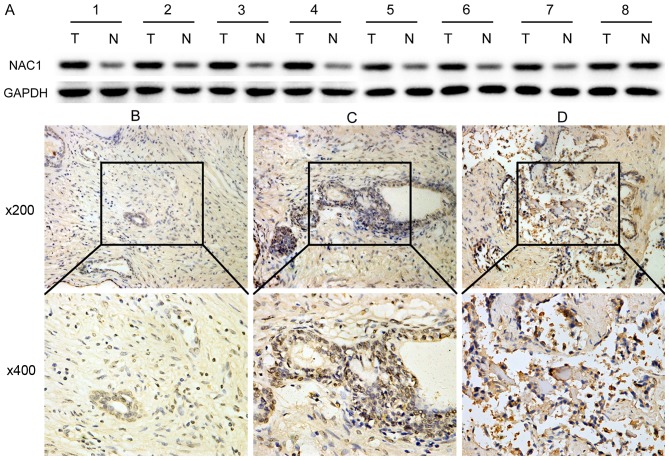
Initially, we examined the expression of NAC1 protein by western blotting in 8 prostate cancer and 8 adjacent normal tissues taken from the same patients. The results revealed that NAC1 expression was markedly higher in tumor tissues (Fig. 1A). To validate this result, immunohistochemistry was used to analyze the correlation of NAC1 expression with several clinicopathological characteristics in 84 prostate cancer and 84 non-cancerous tissue samples. In contrast to non-cancerous prostate tissues, prostate cancer demonstrated a higher NAC1 immunoreactivity, with 79% of the cases showing reactivity scores of 2+ or 3+ (χ2 test, P<0.001) (Fig. 1B-D and Table II). It also revealed that the NAC1 immunoreactivity was closely related to the clinical stage of prostate cancer (P<0.05). However, there were no significant associations with patient age, preoperative prostate-specific antigen level or Gleason score (Table II).
Figure 1.
NAC1 expression is up-regulated in prostate cancer. (A) Western blot analysis of eight paired prostate cancer and matched adjacent normal tissues. (B) Negative immunohistochemical staining of NAC1 in non-cancerous prostate tissues. The marked section in the 200× image was selected for further observation at 400x. (C) Positive staining of NAC1 graded as (+) in prostate cancer tissue. (D) Positive staining of NAC1 graded as (+++) in prostate cancer tissue.
Table II.
Correlation of NAC1 expression with clinicopathologic factors in prostate cancer.
| Immunointensity, n (%) | ||||
|---|---|---|---|---|
| Factors | Case no. | 0 or 1+ | 2+ or 3+ | P-value |
| Histologic type | <0.001 | |||
| Prostate cancer | 84 | 18 (21) | 66 (79) | |
| Normal prostate | 84 | 62 (74) | 22 (26) | |
| Patients' age | 0.09 | |||
| <60 y | 20 | 7 (35) | 13 (65) | |
| ≥60 y | 64 | 11 (17) | 53 (83) | |
| Clinical stages | 0.002 | |||
| ≤IIb | 38 | 14 (37) | 24 (63) | |
| ≥IIc | 46 | 4 (1) | 42 (99) | |
| Preoperative | 0.878 | |||
| PSA (ng/ml) | ||||
| ≤20 | 48 | 10 (21) | 38 (79) | |
| >20 | 36 | 8 (22) | 28 (78) | |
| Gleason score | 0.437 | |||
| ≤7 | 62 | 12 (19) | 50 (81) | |
| ≥8 | 22 | 6 (27) | 16 (73) | |
NAC1, nucleus accumbens-associated protein 1; PSA, prostate-specific antigen.
Silencing NAC1 expression decreases the migratory ability of prostate cancer cells
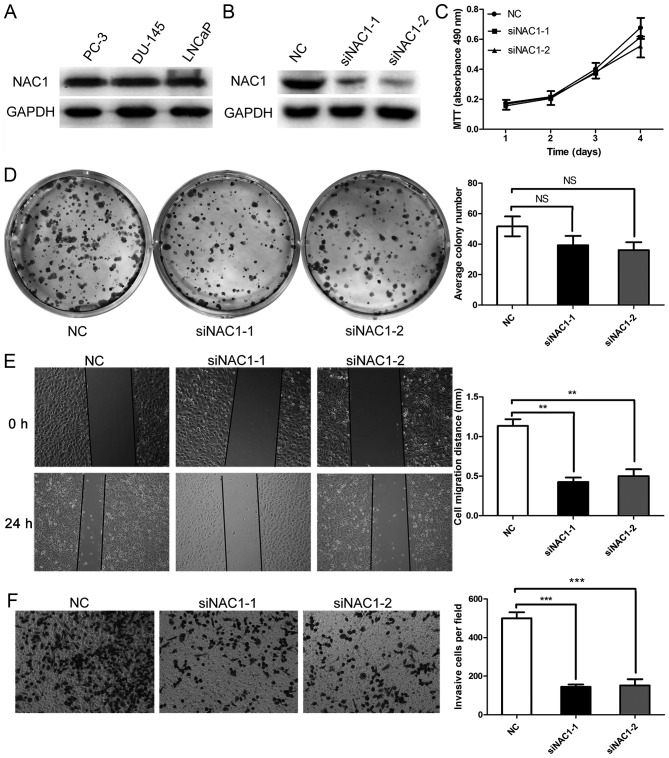
Since NAC1 was up-regulated in prostate cancer at advanced clinical stages, we next assessed the biological function of NAC1 in prostate cancer cells. NAC1 was expressed in the highly aggressive prostate cancer cell lines PC-3, DU-145 and LNCaP (Fig. 2A). DU-145 cells, which have the highest degree of malignancy, were selected for further study. Two NAC1 siRNAs (designated siNAC1-1 and siNAC1-2) were transfected into the cells. Treatment with siNAC1-1 or siNAC1-2 significantly reduced NAC1 protein expression compared with that in DU-145 cells transfected with negative control siRNA (NC) (Fig. 2B). However, siNAC1-1 and siNAC1-2 did not significantly reduce the numbers of DU145 cells compared with the NC group (Fig. 2C). To confirm the result, a colony formation assay was performed, which revealed a slight but non-significant difference between the NAC1 siRNA and NC groups (Fig. 2D).
Figure 2.
Silencing NAC1 expression decreased the migratory ability of prostate cancer cells. (A) Western blots showing expression of NAC1 in the highly aggressive prostate cancer cell lines PC-3, DU-145 and LNCaP. (B) Western blots showing a significant reduction of NAC1 protein in DU-145 cells transfected with NAC1 siRNAs (siNAC1-1 and siNAC1-2) compared with those transfected with negative control siRNA (NC). (C) MTT assay showing that siNAC1-1 and siNAC1-2 had no obvious effect on the cell number of DU-145 cells (P>0.05). (D) Detection of cell proliferation by plate colony formation assay in DU-145 cells transfected with NAC1 or NC siRNAs. Representative photographs show the DU-145 cell colonies in 6-well plates on the left. The cell colonies were scored visually and counted using a light microscope, as shown in the graph on the right; the siNAC1-1 and siNAC1-2 transfection groups exhibited no distinct difference in the number of colonies compared with the NC siRNA group (NS P>0.05). (E) Wound healing assay showed that NAC1 silencing affected the migration of prostate cancer DU-145 cells, as shown in the photographs on the left. The comparison of cell migration distance between siNAC1-1 or siNAC1-2 and NC siRNA transfection is shown on the right (**P<0.01). (F) Transwell migration analysis showed that NAC1 silencing affected the migration of prostate cancer cells, as shown in the photograph on the left (magnification, 200x). The quantitative analysis of the migratory ability of prostate cancer cells transfected with NAC1 or NC siRNAs is shown on the right (***P<0.001).
Subsequently, we investigated whether NAC1 played a causal role in the migration characteristics of prostate cancer cells by employing a wound-healing assay. A wound was scratched on the surface of DU-145 cells transfected with siNAC1-1, siNAC1-2 or NC siRNA, and the width of the gap was measured after 24 h. The cells in the siNAC-1 and siNAC1-2 groups migrated less than those in the NC group, as shown in Fig. 2E. Furthermore, Transwell assays demonstrated that down-regulation of NAC-1 by siNAC1 in DU-145 cells decreased the cell migration abilities compared with NC siRNA (Fig. 2F). These results revealed that silencing of NAC1 expression inhibited the migration of prostate cancer cells.
Identification of NAC1-regulated genes in prostate cancer cells
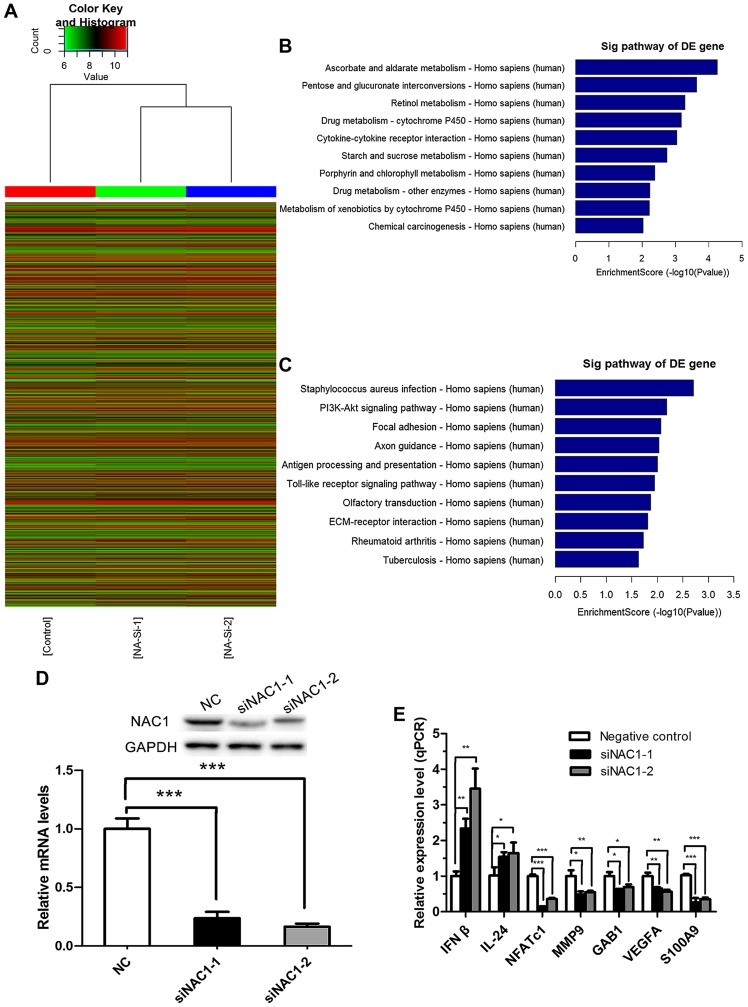
According to the preceding data, we performed a microarray analysis to identify the potential genes regulated by NAC1 in the DU-145 cell line. mRNA expression profiles were determined in DU145 cells transfected with NAC1 or NC siRNA. In total, 291 up-regulated mRNAs and 488 down-regulated mRNAs (with fold change |>2| and P<0.05) were identified consistently in the DU145 cells transfected with siNAC1-1 or siNAC1-2 compared with the NC siRNA-transfected cells (data not shown). The results of a Hierarchical Clustering analysis are shown in Fig. 3A, and the top ten pathways associated with the up-regulated and down-regulated genes are shown in Fig. 3B and C respectively.
Figure 3.
Profiling of NAC1-regulated genes in prostate cancer cells. (A) Hierarchical clustering of mRNA by the Z-score method. The results are displayed as a heat map, in which red indicates relatively high expression and green denotes relatively low expression. (B) Pathways corresponding to the up-regulated transcripts. (C) Pathways corresponding to down-regulated transcripts. (D) Western blots and RT-qPCR shown a significant reduction of NAC1 protein and mRNA in DU-145 cells transfected with NAC1 siRNAs compared with those transfected with NC (***P<0.001) (E). RT-qPCR was performed to validate the NAC1-regulated genes identified by microarray analysis in DU-145 cells transfected with siNAC1-1 or siNAC1-2 and NC. All candidate genes were validated (*P<0.05,**P<0.01,***P<0.001) by both siRNAs.
To validate the microarray results, qPCR was performed in cells transfected with the two independent NAC1 siRNAs, in order to assess the levels of seven genes identified as being associated with the biological process of cancer development based on the microarray analysis. All genes were validated by both siRNAs (P<0.05). NAC1 knockdown efficiency was verified by RT-qPCR and western blotting (Fig. 3D). Furthermore, we selected β-actin and PKM2 whose expression was unchanged in the microarray to verify the efficiency by western blotting (data not shown). The results revealed that NAC1 siRNAs did not affect the expression of β-actin and PKM2, but effectively down-regulated NAC1. As shown in Fig. 3E, the seven genes were IFNβ and IL-24, whose transcription was repressed by NAC1, and NFATc1, MMP9, GAB1, VEGFA and S100A9, whose transcription was promoted by NAC1.
NAC1 participates in osteoclastogenesis by negatively regulating IFNβ
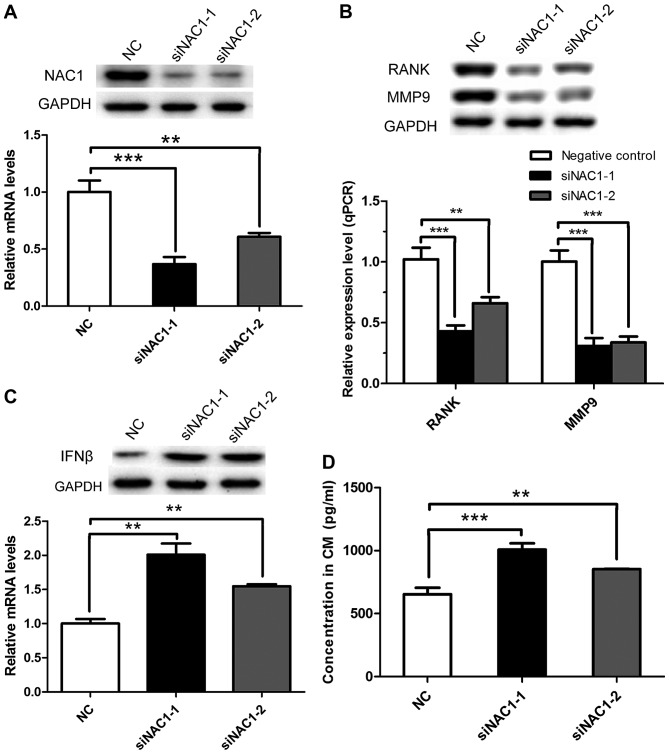
Interestingly, upon further exploration of the microarray data, a GO analysis of biological processes revealed that NAC1 negatively regulates IFNβ (data not shown). A previous study reported that IFNβ has the function of regulating osteoclastogenesis (18); therefore, we hypothesized that NAC1 may mediate osteoclastogenesis by negatively regulating IFNβ expression in prostate cancer. To verify this hypothesis, PC-3 cells, which are isolated from a bone metastatic site of prostate cancer, were employed for further study. PC-3 cells were transfected with the two NAC1 siRNAs or NC siRNA and after 24 h were seeded into the upper compartment of a Boyden chamber, while RAW 264.7 cells were cultured in the bottom chamber to allow exchange of secreted factors that generate osteoclasts. When NAC1 was silenced in PC-3 cells (Fig. 4A), the expression of genes associated with the activation of osteoclasts (such as RANK and MMP9) was significantly decreased in RAW 264.7 cells, as identified by RT-qPCR and western blot analyses (Fig. 4B). The mRNA and protein levels of IFNβ were increased in PC-3 cells transfected with NAC1 siRNAs compared with the NC siRNA (Fig. 4C). Furthermore, the protein level of IFNβ, detected by ELISA, was increased in the co-culture medium in the NAC1 siRNA-transfected group compared with the control group (Fig. 4D). This series of results confirmed that NAC1 participates in osteoclastogenesis through the negative regulation of IFNβ.
Figure 4.
NAC1 promotes prostate cancer bone metastasis in a manner dependent on the induction of IFNβ. (A) NAC1 knockdown efficiency was verified in PC-3 cells by RT-qPCR and western blotting. (B) Genes associated with activation of osteoclasts were identified in RAW 264.7 cells by RT-qPCR and western blotting (**P<0.01, ***P<0.001). (C) The mRNA and protein levels of IFNβ were verified in PC-3 cells by RT-qPCR and western blotting (**P<0.01). (D) ELISA was performed to validate the protein level of IFNβ in the co-culture medium The differences between the siNAC1-1 or siNAC1-2 transfection groups and the NC group in the Figure were statistically significant (**P<0.01, ***P<0.001).
Discussion
The key findings of the present study were that, as a cancer-associated gene, NAC1 promotes the migration of prostate cancer cells and participates in osteoclastogenesis. NAC1 is up-regulated at advanced clinical stages of prostate cancer, indicating that it may be useful biomarker for tracking the progression of prostate cancer. At present, the treatment of bone metastasis in patients with prostate cancer is unsatisfactory. As an upstream regulatory gene of osteoclastogenesis, NAC1 may represent a novel potential therapeutic target.
The results of previous studies have implicated NAC1 in cell proliferation and survival in certain gynecological tumors, such as ovarian cancer, endometrial cancer, cervical carcinomas and uterine sarcoma (11,17,19,20). However, interestingly, the results of the colony formation and MTT assays in the present study showed that NAC1 expression had no significant effects on cell proliferation in prostate cancer (Fig. 2C and D), which is similar to our previous studies in bladder urothelial carcinoma (In press). However, in the Transwell and wound healing assays (Fig. 2E and F), NAC1-silencing resulted in a decreased capacity for cell migration in the DU-145 cell line, indicating that NAC1 may be essential for metastasis in prostate cancer. Similarly, NAC1 has been shown to promote cell invasion and motility through FOXQ1 in ovarian cancer (16). Furthermore, NAC1 participate in multitudinous cancer-associated pathways like autophagy, apoptosis and epithelial mesenchymal transition in different cancer cells (21–23), indicating that NAC1 exert different effects in different cancer types. This may account for why NAC1 inhibition suppresses proliferation in some cancer cell types (11,19), but not in the prostate cancer, where it suppresses migration.
On the basis of comparing the transcripts present in NAC1-knockdown and control DU-145 prostate cancer cells by microarray analysis, we validated a set of target genes whose expression levels depend on NAC1. After NAC1 silencing, IFNβ and IL-24 were up-regulated, and NFATc1, S1009, GAB1, MMP9 and VEGFA were down-regulated (Fig. 3E). In these genes, IFNβ is particularly interesting, as previous studies have shown that IFNβ can negatively regulate osteoclastogenesis; this mechanism involves the secretion of IFNβ by osteocytes, which then acts on osteoclast precursors and down-regulates the protein level of c-Fos, a key factor in osteoclast formation (20,24,25). Combined with the GO analysis (data not shown), which demonstrated that NAC1 can negatively regulate IFNβ, we hypothesized that NAC1 may participate in osteoclastogenesis by reducing IFNβ secretion. To validate this hypothesis, PC-3 cells isolated from prostate cancer bone metastasis were co-cultured with RAW 264.7 cells. RANK and MMP9, which are associated with the activation of osteoclasts, were down-regulated in RAW 264.7 cells when NAC1 was significantly silenced in PC-3 cells (Fig. 4B). Furthermore, the protein level of IFNβ was increased in the culture medium of PC-3 cells (Fig. 4D). This finding is consistent with the supposition that NAC1 may play an important role in promoting osteoclastogenesis.
However, it should be noted that this study was only an in vitro examination. Therefore, further studies in vivo are needed to clarify the relationship between NAC1 and osteoclastogenesis. Moreover, other underlying mechanisms of prostate cancer migration and its downstream target genes must be further explored. For example, VEGFA, the most important factor among the angiogenic cytokines (26–28), was identified to be positively regulated by NAC1, suggesting that NAC1 may play an important role in promoting tumor angiogenesis in prostate cancer. Indeed, several NAC1-regulated genes identified in the microarray analysis, including IL-24, GAB1 and S100A9, are also involved in cell motility or invasion (29–31).
In conclusion, we have demonstrated that NAC1 promotes the migration of prostate cancer cells, and identified a significant correlation between NAC1 and osteoclastogenesis. These findings suggest that NAC1 may serve as a biomarker for the diagnosis of prostate cancer in the future, and that a NAC1-based molecular-targeting therapy is a potentially novel treatment for prostate cancer.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Rucci N, Angelucci A. Prostate cancer and bone: The elective affinities. Biomed Res Int. 2014;2014:167035. doi: 10.1155/2014/167035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saad F, Lipton A, Cook R, Chen YM, Smith M, Coleman R. Pathologic fractures correlate with reduced survival in patients with malignant bone disease. Cancer. 2007;110:1860–1867. doi: 10.1002/cncr.22991. [DOI] [PubMed] [Google Scholar]
- 5.Zustovich F, Fabiani F. Therapeutic opportunities for castration-resistant prostate cancer patients with bone metastases. Crit Rev Oncol Hematol. 2014;91:197–209. doi: 10.1016/j.critrevonc.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: A new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1995;6:1193–1198. [PubMed] [Google Scholar]
- 7.Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Privé GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bardwell VJ, Treisman R. The POZ domain: A conserved protein-protein interaction motif. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 10.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakayama K, Nakayama N, Davidson B, Sheu JJ, Jinawath N, Santillan A, Salani R, Bristow RE, Morin PJ, Kurman RJ, et al. A BTB/POZ protein, NAC-1, is related to tumor recurrence and is essential for tumor growth and survival; Proc Natl Acad Sci USA; 2006; pp. 18739–18744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jinawath N, Vasoontara C, Yap KL, Thiaville MM, Nakayama K, Wang TL, Shih IM. NAC-1, a potential stem cell pluripotency factor, contributes to paclitaxel resistance in ovarian cancer through inactivating Gadd45 pathway. Oncogene. 2009;28:1941–1948. doi: 10.1038/onc.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Cheng Y, Ren X, Hori T, Huber-Keener KJ, Zhang L, Yap KL, Liu D, Shantz L, Qin ZH, et al. Dysfunction of nucleus accumbens-1 activates cellular senescence and inhibits tumor cell proliferation and oncogenesis. Cancer Res. 2012;72:4262–4275. doi: 10.1158/0008-5472.CAN-12-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Cheng Y, Ren X, Zhang L, Yap KL, Wu H, Patel R, Liu D, Qin ZH, Shih IM, Yang JM. NAC1 modulates sensitivity of ovarian cancer cells to cisplatin by altering the HMGB1-mediated autophagic response. Oncogene. 2012;31:1055–1064. doi: 10.1038/onc.2011.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yap KL, Fraley SI, Thiaville MM, Jinawath N, Nakayama K, Wang J, Wang TL, Wirtz D, Shih IeM. NAC1 is an actin-binding protein that is essential for effective cytokinesis in cancer cells. Cancer Res. 2012;72:4085–4096. doi: 10.1158/1538-7445.AM2012-LB-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao M, Wu RC, Herlinger AL, Yap K, Kim JW, Wang TL, Shih IeM. Identification of the NAC1-regulated genes in ovarian cancer. Am J Pathol. 2014;184:133–140. doi: 10.1016/j.ajpath.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeasmin S, Nakayama K, Ishibashi M, Katagiri A, Iida K, Purwana IN, Nakayama N, Miyazaki K. Expression of the bric-a-brac tramtrack broad complex protein NAC-1 in cervical carcinomas seems to correlate with poorer prognosis. Clin Cancer Res. 2008;14:1686–1691. doi: 10.1158/1078-0432.CCR-07-4085. [DOI] [PubMed] [Google Scholar]
- 18.Takayanagi H, Kim S, Matsuo K, Suzuki H, Suzuki T, Sato K, Yokochi T, Oda H, Nakamura K, Ida N, et al. RANKL maintains bone homeostasis through c-Fos-dependent induction of interferon-beta. Nature. 2002;416:744–749. doi: 10.1038/416744a. [DOI] [PubMed] [Google Scholar]
- 19.Ishikawa M, Nakayama K, Yeasmin S, Katagiri A, Iida K, Nakayama N, Miyazaki K. NAC1, a potential stem cell pluripotency factor expression in normal endometrium, endometrial hyperplasia and endometrial carcinoma. Int J Oncol. 2010;36:1097–1103. doi: 10.3892/ijo_00000591. [DOI] [PubMed] [Google Scholar]
- 20.Rahman MT, Nakayama K, Ishikawa M, Rahman M, Katagiri H, Katagiri A, Ishibashi T, Iida K, Yamada T, Miyazaki K. NAC1, a BTB/POZ protein overexpressed in uterine sarcomas. Anticancer Res. 2012;32:3841–3845. [PubMed] [Google Scholar]
- 21.Zhang Y, Yang JW, Ren X, Yang JM. NAC1 and HMGB1 enter a partnership for manipulating autophagy. Autophagy. 2011;7:1557–1558. doi: 10.4161/auto.7.12.17910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ju T, Jin H, Ying R, Xie Q, Zhou C, Gao D. Overexpression of NAC1 confers drug resistance via HOXA9 in colorectal carcinoma cells. Mol Med Rep. 2017;16:3194–3200. doi: 10.3892/mmr.2017.6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faiola F, Yin N, Fidalgo M, Huang X, Saunders A, Ding J, Guallar D, Dang B, Wang J. NAC1 regulates somatic cell reprogramming by controlling Zeb1 and E-cadherin expression. Stem Cell Rep. 2017;9:913–926. doi: 10.1016/j.stemcr.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashida C, Ito J, Nakayachi M, Okayasu M, Ohyama Y, Hakeda Y, Sato T. Osteocytes produce interferon-β as a negative regulator of osteoclastogenesis. J Biol Chem. 2014;289:11545–11555. doi: 10.1074/jbc.M113.523811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grigoriadis AE, Wang ZQ, Cecchini MG, Hofstetter W, Felix R, Fleisch HA, Wagner EF. c-Fos: A key regulator of osteoclast-macrophage lineage determination and bone remodeling. Science. 1994;266:443–448. doi: 10.1126/science.7939685. [DOI] [PubMed] [Google Scholar]
- 26.Kieran MW, Kalluri R, Cho YJ. The VEGF pathway in cancer and disease: Responses, resistance, and the path forward. Cold Spring Harb Perspect Med. 2012;2:a006593. doi: 10.1101/cshperspect.a006593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potente M, Gerhardt H, Carmeliet P. Basic and therapeutic aspects of angiogenesis. Cell. 2011;146:873–887. doi: 10.1016/j.cell.2011.08.039. [DOI] [PubMed] [Google Scholar]
- 28.Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panneerselvam J, Jin J, Shanker M, Lauderdale J, Bates J, Wang Q, Zhao YD, Archibald SJ, Hubin TJ, Ramesh R. IL-24 inhibits lung cancer cell migration and invasion by disrupting the SDF-1/CXCR4 signaling axis. PLoS One. 2015;10:e0122439. doi: 10.1371/journal.pone.0122439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Song W, Shen W, Yang X, Sun W, Qu S, Shang R, Ma B, Pu M, Tao K, et al. MicroRNA-200a suppresses cell invasion and migration by directly targeting GAB1 in hepatocellular carcinoma. Oncol Res. 2017;25:1–10. doi: 10.3727/096504016X14685034103798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim SY, Yuzhalin AE, Gordon-Weeks AN, Muschel RJ. Tumor-infiltrating monocytes/macrophages promote tumor invasion and migration by upregulating S100A8 and S100A9 expression in cancer cells. Oncogene. 2016;35:5735–5745. doi: 10.1038/onc.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]