Abstract
Nuclear actin-binding proteins (ABPs) perform distinguishable functions compared with their cytoplasmic counterparts in extensive activities of living cells. In addition to the ability to regulate actin cytoskeleton dynamics, nuclear ABPs are associated with multiple nuclear biological processes, including chromatin remodeling, gene transcriptional regulation, DNA damage response, nucleocytoplasmic trafficking and nuclear structure maintenance. The nuclear translocation of ABPs is affected by numerous intracellular or extracellular stimuli, which may lead to developmental malformation, tumor initiation, tumor progression and metastasis. Abnormal expression of certain ABPs have been reported in different types of cancer. This review focuses on the newly identified roles of nuclear ABPs in the pathological processes associated with cancer.
Keywords: actin-binding protein, nuclear, actin, transcription, cancer
1. Introduction
Actin is one of the most abundant proteins identified in almost all eukaryotic cells, and it is a highly conserved protein during evolution (1). Actin-binding proteins (ABPs) refer to proteins that contain actin-binding domains that interact with actin. They can bind to actin monomers, actin polymers or both (2).
Early studies have focused on the biological features and physiological mechanisms of cytoplasmic actin. Therefore, ABPs were considered to be distributed only in the cytoplasm and associated with the organization of actin cytoskeleton (3). In the cytoplasm, actin is associated with numerous cellular activities, including sustaining cellular morphology, determining cellular organelle distribution, mediating intracellular transfer, endocytosis and exocytosis, cell division, cell migration and adhesion (4–8). Meanwhile, ABPs regulate actin cytoskeletal structure by modulating actin filament cross-linking into networks or depolymerizing into monomers, allowing actin to switch between the polymeric (F-actin form, filamentous actin) and monomeric state (G-actin form, globular actin) (2,9).
However, recent studies (10,11) indicate that a great number of actin and ABPs exist in the nucleus. Nuclear actin and nuclear ABPs exhibit nuclear-specific functions that are different from those in the cytoplasm. Although the precise biological mechanisms remain elusive, we are fortunate to uncover several observations (12). This review aims to present up-to-date discoveries of nuclear actin and nuclear ABPs in the field of cancer research.
2. Actin and actin-binding proteins (ABPs) in the nucleus
Studies in the recent decades provide a plethora of evidence that has broadened our horizon on the functions of nuclear actin and nuclear ABPs in the eukaryotic cell life (12–14). Since the existence of nuclear actin was confirmed, subsequent studies also established the presence of ABPs in the nucleus (12,14,15). The very first nuclear ABP was reported as early as 1987, henceforth, the rest of the ABP family in the nucleus has come to light comprising of proflilin, anillin, flightless I (Fli I), filamin α (FLNα), α-actinins, myosins, gelsolin and ezrin-radixin-moesin proteins (12,13,16,17). Although these ABPs are primarily in the cytoplasm, they can translocate into the nucleus under certain circumstances, for example, extracellular stimuli (stress), hormone stimulation and intracellular signaling (16).
In the nucleus, actin is associated with chromatin remodeling, DNA replication, DNA repair, gene transcriptional regulation, RNA processing, nuclear protein transportation and maintenance of nuclear structure, for instance, the nuclear envelope assembly (12,18–20). Nuclear ABPs are closely associated to nuclear actin and implicated in various nuclear activities. ABPs promote actin filament nucleation or sequestering, manipulate nuclear actin dynamics and determine the ratio of nuclear to cytoplasmic actin (13). Therefore, ABPs directly or indirectly associate with chromatin remodeling, DNA replication, transcription, DNA repair, nucleocytoplasmic transport and maintenance of nuclear structure integrity (12,13,21). Furthermore, nuclear actin is required by all three RNA polymerases in transcriptional activation (22–24). The study by Miyamoto and Gurdon (25) mentions the major function of nuclear actin and ABPs in transcriptional regulation and nuclear reprogramming.
In eukaryotic cells, the cytoplasm and nucleus are separated by the nuclear envelope, which is a double membrane barrier. Trafficking of proteins and other molecules between these two compartments occurs by passing through the nuclear pore complex. Additionally, cytoplasmic ABPs can interact with the nuclear receptor in the cytoplasm, form complexes and facilitate nuclear translocation. Therefore, ABPs mediate transcriptional activation of nuclear receptors. These nuclear receptors include the glucocorticoid and estrogen receptor, androgen receptor (AR), thyroid receptor and peroxisome proliferator-activated receptor-c (26–28). Furthermore, they are associated with transcriptional activation of multiple genes and are involved in a spectrum of functions, including cell proliferation, differentiation and apoptosis (21,29,30).
3. Nuclear ABPs in cancer cells
Numerous ABPs demonstrate the ability to shuttle between the cytoplasm and nucleus via different mechanisms. The question remains whether it is the shuttling of ABPs between different cellular compartments that is linked to pathological behaviors, including abnormal cell development and differentiation, carcinogenesis and metastasis. However, this needs to be clarified. The theory that ABPs in the nucleus affect the aforementioned pathological processes is intriguing. In the sections, the nuclear ABPs that are associated with chromatin remodeling, transcriptional regulation, DNA damage repair, protein nucleocytoplasmic shuttling and nuclear structure maintenance in human cancer cells are summarized in Table I and Fig. 1.
Table I.
Summary of nuclear functions of actin-binding proteins in human cancer.
| Nuclear function | ABP | Mechanism | Tumor | (Refs.) |
|---|---|---|---|---|
| Chromatin remodeling | Flightless I homolog | Recruits chromatin remodeling complex | Breast cancer | (31,32) |
| Transcriptional regulation | Villin | Interacts with transcriptional corepressor | Lung, gastric, colorectal, pancreatic, biliary, liver, renal, cervical and endometrioid cancer | (33–36) |
| α-actinin 4 | Interacts with nuclear receptor (act as a coactivator); interacts with transcriptional corepressor | Breast, prostate, ovarian, colorectal, pancreatic and esophageal cancer, and salivary gland carcinoma | (37–39,41–46) | |
| Filamin α | Interact with transcription factors | Colon, breast and prostate cancer | (47–49) | |
| Transgelin | Speculated to act as a transcriptional regulator | Colorectal cancer | (57,58) | |
| DNA damage response | Filamin α | Participates in double strand break repair (DSBR) | Breast cancer, certain melanomas | (61,63) |
| Nesprin-1 | Participates in the DNA damage response (DDR), DNA mismatch repair (MMR) | Liver cancer | (64) | |
| Protein nuclear translocation | Nesprin-2 | Participates in protein nuclear localization | Breast cancer | (65–67) |
| Nuclear structure maintenance | Nesprin-1 | Constitutes the linker of nucleoskeleton and cytoskeleton (LINC) complex | Lung and pancreatic cancer | (69) |
| Nesprin-2 | Constitutes LINC complex | Breast and colorectal cancer, head and neck squamous cell carcinoma | (70–72) |
ABP, actin-binding protein.
Figure 1.
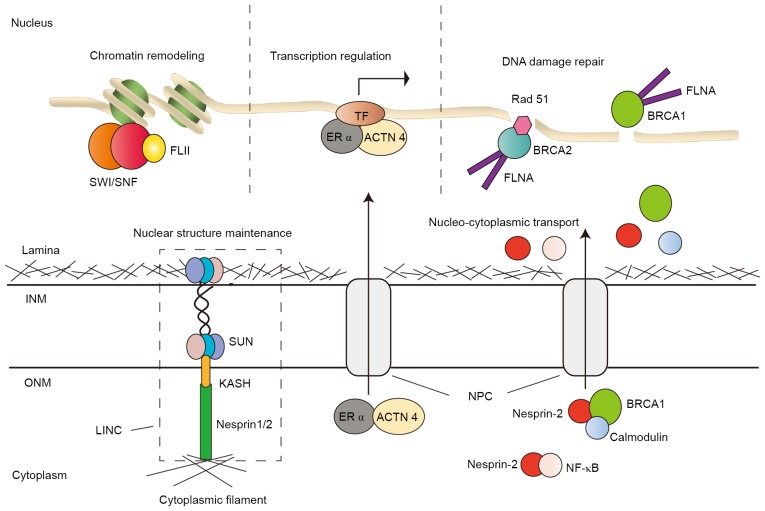
Illustrations of ABPs in the nucleus. ABPs are involved in chromatin remodeling, gene transcriptional regulation, DNA damage repair, nucleocytoplasmic transport and nuclear structure maintenance. ABP, actin-binding protein; INM, inner nuclear membrane; ONM, outer nuclear membrane; NPC, nuclear pore complex; LINC, linker of nucleoskeleton and cytoskeleton complex; SUN, SUN domain proteins; KASH, KASH domain; FLII, flightless I homolog; SWI/SNF, SWItch/Sucrose Non-Fermentable chromatin remodeling complex; ACTN 4, α-actinin 4; ERα, estrogen receptor α; TF, transcription factor; FLNα, filamin α; NF-κB, nuclear factor κ-light-chain enhancer of activated B-cells; BRCA2, breast cancer type 2; BRCA1, breast cancer type 1.
Fli I homolog (FLII), with a C-terminal gelsolin-like actin-binding domain, is a member of the gelsolin protein superfamily. FLII interacts with BAF53, a subunit of the SWItch/Sucrose Non-Fermentable chromatin-remodeling complex, and recruits the latter to the promoter and enhancer regions of the trefoil factor 1 gene, an estrogen receptor (ERα) target gene (31) (Fig. 1). FLII also regulates chromatin accessibility for the binding of RNA polymerase II and other transcriptional coactivators to the promoter and enhancer of other ERα target genes, including growth regulation by estrogen in breast cancer 1, continuous traumatic stress disorder and MYC in MCF-7 breast cancer cells. In addition, FLII promotes the hormone-dependent growth of breast cancer cells (32).
Villin is a tissue-specific ABP predominantly expressed in the epithelium, including the gastrointestinal tract and digestive organs. Overexpression of villin is reported in tissues of Barrett's metaplasia, gastric and colorectal adenocarcinoma (33,34). Furthermore, villin is distributed in the cytoplasm and nucleus and cytoplamic-nuclear transport of villin keeps the system dynamically stable. It migrates to the nucleus upon stimulation, including hypoxia and injury. Another postulation is that tyrosine phosphorylation of villin may prompt its gathering in the nucleus. A study reveals that villin in the nucleus can interact with ZBRK1 [also called zinc finger and breast cancer type 1 (BRCA1)-interaction protein], a transcriptional corepressor and also a ligand of the human Slug promoter, thus regulating the activity of Slug and gene expression (34).
The villin-ZBRK1 complex eliminates the corepressor effect of ZBRK1 and upregulates Slug expression. Slug is a crucial transcriptional regulator of epithelial-mesenchymal transition (EMT). Therefore, nuclear villin performs an important role in inducing EMT, which is considered essential in tumorigenesis (34), invasiveness and metastasis. A previous study demonstrates that severe combined immunodeficiency mice injected with xenografts derived from five colon cancer cell lines developed tumors in 21 days at a 100% frequency, and by staining with villin antibodies, the xenografts reveal strong nuclear accumulation of villin (34). Furthermore, villin expression is observed in gastric, colorectal, pancreatic, biliary, liver, renal, cervical, endometrioid, lung and other types of cancer (34–36), particularly with propensity for metastasis and poor prognosis.
α-actinin 4 (ACTN4), a member of the ABP family, is a regulator of gene transcription that is presumably mediated by nuclear hormone receptors. Although the majority of ACTN4 is located in the cytoplasm, the proportion of ACTN4 in the nucleus is unneglectable. ACTN4 binds to the nuclear receptors in a hormone-dependent manner. Furthermore, ACTN4 is a coactivator of the estrogen receptor α (ER-α) that regulates target gene transcription in MCF-7 breast cancer cells, subsequently promoting tumor cell proliferation (37). Overexpression of ACTN4 in the nucleus increases the expression of progesterone receptors and antigen related to ER (pS2), and target genes of ER-α (Fig. 1). Furthermore, it can interact with the pS2 promoter and potentiate estradiol (E2)-induced transcription. Additionally, it interacts with the AR and functions as a co-regulator of AR-mediated transcription (38).
Nuclear ACTN4 serves as a transcriptional coactivator for nuclear factor κ-light-chain enhancer of activated B-cell (NF-κB) (39). NF-κB activation in ERα-negative breast cancer promotes cancer cell proliferation (40). Furthermore, ACTN4 also interacts with histone deacetylase 7 (HDAC7), a transcriptional corepressor of myocyte enhancer factor 2 (MEF2), competitively inhibiting the repressing effect of HDAC7 and potentiating MEF2 transcription activity (41). ACTN4 interacts with HDAC7 by its C-terminal calmodulin (CaM) -like domain, and activates ERα transcription, which contributes to tumorigenesis in breast cancer (42). In addition, elevated levels of ACTN4 are widely identified in other malignancies, including colorectal, pancreatic, prostate and ovarian cancer, salivary gland carcinoma and esophageal cancer (38,42–46).
FLNα, also called ABP-280, is a scaffold protein. Filamin deficit is prevalent among carcinomas, including colon, prostate and breast cancer. It is verified that FLNα acts as a promoter in cancer metastasis and invasion in the cytoplasm, while it functions as a tumor suppressor in the nucleus (47). Furthermore, cytoplasmic FLNα interacts with a number of proteins, for example, β1-integrin, phosphatidylinositols and small GTPases, which facilitate cell adhesion and migration. Furthermore, nuclear FLNα interacts with transcription factors and the associate transcription machinery subsequently restrains cell migration and represses cell growth. A previous study demonstrates that nuclear FLNα prohibits ribosome RNA transcription by interacting with RNA polymerase I (48). In addition, nuclear FLNα binds to the AR and modulates the nuclear translocation of the latter, thus regulates AR-induced gene expression. AR is a steroid nuclear receptor and closely associated with prostate cancer. Nuclear FLNα inhibits AR target gene transcription, thus negatively regulating cancer development (49).
Transgelin is an ABP mostly distributed in the cytoplasm of fibroblasts and several epithelial cells (50). Its expression is often altered in human types of cancer (51–55). Previous research by this group (56) revealed that activated AKT and c-jun NH2-terminal kinases promote the expression of transgelin in the cytoplasm and contribute to colorectal cancer progression. Additionally, it is revealed that transgelin was located in the cytoplasm and nucleus of colorectal cancer cells (57). Overexpression of transgelin in human colon cancer cells affects the expression of ~250 other transcripts and enhances the metastatic behavior (58). Thus, it is speculated that transgelin is another transcriptional regulator within the ABP family.
The recovery of DNA damage is essential in the maintenance of genome integrity. The nuclear FLNα is associated with DNA damage repair (59). By contacting BRCA1 and breast cancer type 2 (BRCA2), nuclear FLNα is involved in the process of homologous recombinational and non-homologous DNA repair (60). Furthermore, FLNα interacts with BRCA1 with its extreme C-terminus and mediates BRCA1 and Rad51 foci formation following DNA damage (Fig. 1). In addition, the lack of FLNα may contribute to predisposition to breast cancer (61). In double strand break repair (DSBR), FLNα interacts with BRCA2 and subsequently forms the repair complex (Fig. 1). The deficiency in FLNα makes the cells susceptible to ionizing radiation and delays the recovery from G2/M arrest, which may trigger the incidence of cancer (60–63). Measurement of FLNα expression reveals that FLNα is negative in several melanomas (63). In the absence of FLNα, cells impair to recover from DNA damage and incline to accumulate genetic mutations and initiate tumorigenesis.
Nesprin-1, also known as Enaptin, is a nuclear envelope protein, consisting of a C-terminal KASH domain, a long spectrin repeat region and an N-terminal F-actin binding domain. Novel observations indicate that Nesprin-1 is involved in the DNA damage response and DNA mismatch repair (MMR), thus maintaining genetic stability (64). The study indicates that Nesprin-1 interacts with the DNA MMR proteins, MSH2 and MSH6, and regulates the expression level and function of these proteins, which are associated to DSBR. Reduction of Nesprin-1 may lead to deficiency in correcting DNA damage, therefore triggering tumorigenesis and accelerating tumor progression. Consistent with this, Nesprin-1 expression significantly decreases in liver cancer and numerous other types of human cancer (64). Nesprin-2 is a nuclear membrane protein of the nuclear envelope spectrin-repeat (nesprin) family, which contains an actin-binding domain. Loss of Nesprin-2 is associated with less nuclear accumulation of c-Fos, mothers against decapentaplegic homolog (SMAD) 2, 3 and 4, holding back the course of nuclear translocation (65,66). Furthermore, the latest studies imply that Nesprin-2 is a prerequisite for the nuclear transport of certain proteins, for example, BRCA1 and NF-κB (Fig. 1). The nuclear localization of BRCA1 is regulated by a RAN-independent Ca2+/CaM mediated machinery in which Nesprin-2 is necessary (66). Additionally, abnormality in Nesprin-2 nuclear trafficking results in impaired nuclear translocation and mislocalization of BRCA1. Downregulation of Nesprin-2 is revealed in breast cancer tissue (67). Therefore, disturbance of nuclear translocation of certain proteins by the ABPs may be associated to a number of diseases, including cancer (66).
Nesprins, a family of nuclear envelope proteins, together with SUN domain proteins, form the core of the linker of nucleoskeleton and cytoskeleton (LINC) complex. As a key component of the LINC complex, nesprins tether nuclei to the cytoskeleton and are essential in the maintenance of the nuclear architecture (Fig. 1). Loss of nesprins leads to risk of nuclear structural instability, including nuclear shape, size and chromatin organization, which is implicated in tumorigenesis (68). Furthermore, nesprin-1 downregulation is reported in lung cancer, and synaptic nuclear envelope protein 1 gene mutation is observed in pancreatic cancer with metastasis (69).
Spectrin repeat containing, nuclear envelope protein 2 abnormality is revealed in breast and colorectal cancer, and head and neck squamous cell carcinoma (70–72). One possible mechanism is that nesprin downregulation modulates nuclear stiffness via the LINC complex, and increases nuclear malleability for cells to migrate through restricted tissue spaces (69).
4. Conclusion and perspectives
ABPs have been revealed in both the cytoplasm and nucleus of eukaryotic cells. These proteins share an actin-binding calponin homology domain, exhibit different functions and are involved in diverse activities in the two cellular compartments. Numerous studies observe a significant proportion of ABP shuttle between the cytoplasm and the nucleus. Cytoplasmic ABPs may translocate into the nucleus in response to the alternation of the extracellular microenvironment, including hypoxia, inflammation and injury. The subcellular localization of ABPs is associated with the onset and development of various pathogenesis and carcinogenesis processes. Recent observations (73–75) demonstrate that nuclear ABPs may be involved in chromatin remodeling, function as transcriptional regulators, are involved in the DNA damage response and DNA mismatch repair, mediate protein nuclear translocation and maintain nuclear structural stability. Altogether, the nuclear ABPs maintain the genomic integrity and reduce cellular oncogenic potential; whereas mutations and deficiencies of nuclear ABPs contribute to tumorigenesis and metastasis.
Although the comprehensive molecular mechanism of specific nuclear ABPs remains elusive, the understanding of the association between nuclear ABPs and relevant diseases is extended. The investigation of ABPs' nuclear function and their effects on cancer remains underway and lots of questions remain to be answered. These studies will help to identify novel therapeutic targets in fighting against cancer in the near future.
Acknowledgements
The National Natural Science Foundation of China (grant no. 81641179, YL) and the Natural Science Foundation of Guangdong Province (grant no. 2017A030313603, YL) supported the present study. Grant (2013) 163 from the Key Laboratory of Malignant Tumor Molecular Mechanism and Translational Medicine of Guangzhou Bureau of Science and Information Technology, and grant no. KLB09001 from the Key Laboratory of Malignant Tumor Gene Regulation and Target Therapy of Guangdong Higher Education Institutes also supported the present study.
References
- 1.Pollard TD, Cooper JA. Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu Rev Biochem. 1986;55:987–1035. doi: 10.1146/annurev.bi.55.070186.005011. [DOI] [PubMed] [Google Scholar]
- 2.dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, Nosworthy NJ. Actin binding proteins: Regulation of cytoskeletal microfilaments. Physiol Rev. 2003;83:433–473. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- 3.Hartwig JH, Tyler J, Stossel TP. Actin-binding protein promotes the bipolar and perpendicular branching of actin filaments. J Cell Biol. 1980;87:841–848. doi: 10.1083/jcb.87.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 5.Amann KJ, Pollard TD. Cellular regulation of actin network assembly. Curr Biol. 2000;10:R728–R730. doi: 10.1016/S0960-9822(00)00751-X. [DOI] [PubMed] [Google Scholar]
- 6.Vasioukhin V, Bauer C, Yin M, Fuchs E. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 2000;100:209–219. doi: 10.1016/S0092-8674(00)81559-7. [DOI] [PubMed] [Google Scholar]
- 7.Winder SJ, Ayscough KR. Actin-binding proteins. J Cell Sci. 2005;118:651–654. doi: 10.1242/jcs.01670. [DOI] [PubMed] [Google Scholar]
- 8.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weston L, Coutts AS, La Thangue NB. Actin nucleators in the nucleus: An emerging theme. J Cell Sci. 2012;125:3519–3527. doi: 10.1242/jcs.099523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fairley EA, Kendrick-Jones J, Ellis JA. The Emery-Dreifuss muscular dystrophy phenotype arises from aberrant targeting and binding of emerin at the inner nuclear membrane. J Cell Sci. 1999;112:2571–2582. doi: 10.1242/jcs.112.15.2571. [DOI] [PubMed] [Google Scholar]
- 11.Tse WT, Tang J, Jin O, Korsgren C, John KM, Kung AL, Gwynn B, Peters LL, Lux SE. A new spectrin, beta IV, has a major truncated isoform that associates with promyelocytic leukemia protein nuclear bodies and the nuclear matrix. J Biol Chem. 2001;276:23974–23985. doi: 10.1074/jbc.M009307200. [DOI] [PubMed] [Google Scholar]
- 12.Castano E, Philimonenko VV, Kahle M, Fukalová J, Kalendová A, Yildirim S, Dzijak R, Dingová-Krásna H, Hozák P. Actin complexes in the cell nucleus: New stones in an old field. Histochem Cell Biol. 2010;133:607–626. doi: 10.1007/s00418-010-0701-2. [DOI] [PubMed] [Google Scholar]
- 13.Kristo I, Bajusz I, Bajusz C, Borkuti P, Vilmos P. Actin, actin-binding proteins and actin-related proteins in the nucleus. Histochem Cell Biol. 2016;145:373–388. doi: 10.1007/s00418-015-1400-9. [DOI] [PubMed] [Google Scholar]
- 14.Hofmann WA. Cell and molecular biology of nuclear actin. Int Rev Cell Mol Biol. 2009;273:219–263. doi: 10.1016/S1937-6448(08)01806-6. [DOI] [PubMed] [Google Scholar]
- 15.de Lanerolle P, Serebryannyy L. Nuclear actin and myosins: Life without filaments. Nat Cell Biol. 2011;13:1282–1288. doi: 10.1038/ncb2364. [DOI] [PubMed] [Google Scholar]
- 16.Gettemans J, Van Impe K, Delanote V, Hubert T, Vandekerckhove J, De Corte V. Nuclear actin-binding proteins as modulators of gene transcription. Traffic. 2005;6:847–857. doi: 10.1111/j.1600-0854.2005.00326.x. [DOI] [PubMed] [Google Scholar]
- 17.Rando OJ, Zhao K, Crabtree GR. Searching for a function for nuclear actin. Trends Cell Biol. 2000;10:92–97. doi: 10.1016/S0962-8924(99)01713-4. [DOI] [PubMed] [Google Scholar]
- 18.Bettinger BT, Gilbert DM, Amberg DC. Actin up in the nucleus. Nat Rev Mol Cell Biol. 2004;5:410–415. doi: 10.1038/nrm1370. [DOI] [PubMed] [Google Scholar]
- 19.Blessing CA, Ugrinova GT, Goodson HV. Actin and ARPs: Action in the nucleus. Trends Cell Biol. 2004;14:435–442. doi: 10.1016/j.tcb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 20.Miralles F, Visa N. Actin in transcription and transcription regulation. Curr Opin Cell Biol. 2006;18:261–266. doi: 10.1016/j.ceb.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 21.Zheng B, Han M, Bernier M, Wen JK. Nuclear actin and actin-binding proteins in the regulation of transcription and gene expression. FEBS J. 2009;276:2669–2685. doi: 10.1111/j.1742-4658.2009.06986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Philimonenko VV, Zhao J, Iben S, Dingová H, Kyselá K, Kahle M, Zentgraf H, Hofmann WA, de Lanerolle P, Hozák P, Grummt I. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat Cell Biol. 2004;6:1165–1172. doi: 10.1038/ncb1190. [DOI] [PubMed] [Google Scholar]
- 23.Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, Kysela K, Goodrich JA, Lessard JL, Hope TJ, et al. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat Cell Biol. 2004;6:1094–1101. doi: 10.1038/ncb1182. [DOI] [PubMed] [Google Scholar]
- 24.Hu P, Wu S, Hernandez N. A role for beta-actin in RNA polymerase III transcription. Genes Dev. 2004;18:3010–3015. doi: 10.1101/gad.1250804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyamoto K, Gurdon JB. Transcriptional regulation and nuclear reprogramming: Roles of nuclear actin and actin-binding proteins. Cell Mol Life Sci. 2013;70:3289–3302. doi: 10.1007/s00018-012-1235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ting HJ, Yeh S, Nishimura K, Chang C. Supervillin associates with androgen receptor and modulates its transcriptional activity; Proc Natl Acad Sci USA; 2002; pp. 661–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishimura K, Ting HJ, Harada Y, Tokizane T, Nonomura N, Kang HY, Chang HC, Yeh S, Miyamoto H, Shin M, et al. Modulation of androgen receptor transactivation by gelsolin: A newly identified androgen receptor coregulator. Cancer Res. 2003;63:4888–4894. [PubMed] [Google Scholar]
- 28.Yang Z, Chang YJ, Miyamoto H, Ni J, Niu Y, Chen Z, Chen YL, Yao JL, di Sant'Agnese PA, Chang C. Transgelin functions as a suppressor via inhibition of ARA54-enhanced androgen receptor transactivation and prostate cancer cell growth. Mol Endocrinol. 2007;21:343–358. doi: 10.1210/me.2006-0104. [DOI] [PubMed] [Google Scholar]
- 29.Baek SH, Ohgi KA, Nelson CA, Welsbie D, Chen C, Sawyers CL, Rose DW, Rosenfeld MG. Ligand-specific allosteric regulation of coactivator functions of androgen receptor in prostate cancer cells; Proc Natl Acad Sci USA; 2006; pp. 3100–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang F, Liu XQ, Li H, Liang KN, Miner JN, Hong M, Kallel EA, van Oeveren A, Zhi L, Jiang T. Structure of the ligand-binding domain (LBD) of human androgen receptor in complex with a selective modulator LGD2226. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:1067–1071. doi: 10.1107/S1744309106039340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Won Jeong K, Chodankar R, Purcell DJ, Bittencourt D, Stallcup MR. Gene-specific patterns of coregulator requirements by estrogen receptor-alpha in breast cancer cells. Mol Endocrinol. 2012;26:955–966. doi: 10.1210/me.2012-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeong KW. Flightless I (Drosophila) homolog facilitates chromatin accessibility of the estrogen receptor α target genes in MCF-7 breast cancer cells. Biochem Biophys Res Commun. 2014;446:608–613. doi: 10.1016/j.bbrc.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 33.Khurana S. Structure and function of villin. In: Khurana S, editor. Aspects of the Cytoskeleton. Elsevier; New York: 2006. pp. 89–1159. [DOI] [Google Scholar]
- 34.Patnaik S, George SP, Pham E, Roy S, Singh K, Mariadason JM, Khurana S. By moonlighting in the nucleus, villin regulates epithelial plasticity. Mol Biol Cell. 2016;27:535–548. doi: 10.1091/mbc.E15-06-0453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shillingford NM, Calicchio ML, Teot LA, Boyd T, Kurek KC, Goldsmith JD, Bousvaros A, Perez-Atayde AR, Kozakewich HP. Villin immunohistochemistry is a reliable method for diagnosing microvillus inclusion disease. Am J Surg Pathol. 2015;39:245–250. doi: 10.1097/PAS.0000000000000355. [DOI] [PubMed] [Google Scholar]
- 36.Yang Z. The utility of villin and mammaglobin in the differential diagnosis between intrahepatic cholangiocarcinoma and breast cancer. Appl Immunohistochem Mol Morphol. 2015;23:19–25. doi: 10.1097/PAI.0000000000000035. [DOI] [PubMed] [Google Scholar]
- 37.Khurana S, Chakraborty S, Cheng X, Su YT, Kao HY. The actin-binding protein, actinin alpha 4 (ACTN4), is a nuclear receptor coactivator that promotes proliferation of MCF-7 breast cancer cells. J Biol Chem. 2011;286:1850–1859. doi: 10.1074/jbc.M110.162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jasavala R, Martinez H, Thumar J, Andaya A, Gingras AC, Eng JK, Aebersold R, Han DK, Wright ME. Identification of putative androgen receptor interaction protein modules: Cytoskeleton and endosomes modulate androgen receptor signaling in prostate cancer cells. Mol Cell Proteomics. 2007;6:252–271. doi: 10.1074/mcp.M600169-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Zhao X, Hsu KS, Lim JH, Bruggeman LA, Kao HY. α-Actinin 4 potentiates nuclear factor κ-light-chain-enhancer of activated B-cell (NF-κB) activity in podocytes independent of its cytoplasmic actin binding function. J Biol Chem. 2015;290:338–349. doi: 10.1074/jbc.M114.597260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biswas DK, Shi Q, Baily S, Strickland I, Ghosh S, Pardee AB, Iglehart JD. NF-kappa B activation in human breast cancer specimens and its role in cell proliferation and apoptosis; Proc Natl Acad Sci USA; 2004; pp. 10137–10142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chakraborty S, Reineke EL, Lam M, Li X, Liu Y, Gao C, Khurana S, Kao HY. Alpha-actinin 4 potentiates myocyte enhancer factor-2 transcription activity by antagonizing histone deacetylase 7. J Biol Chem. 2006;281:35070–35080. doi: 10.1074/jbc.M602474200. [DOI] [PubMed] [Google Scholar]
- 42.Hsu KS, Kao HY. Alpha-actinin 4 and tumorigenesis of breast cancer. Vitam Horm. 2013;93:323–351. doi: 10.1016/B978-0-12-416673-8.00005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hara T, Honda K, Shitashige M, Ono M, Matsuyama H, Naito K, Hirohashi S, Yamada T. Mass spectrometry analysis of the native protein complex containing actinin-4 in prostate cancer cells. Mol Cell Proteomics. 2007;6:479–491. doi: 10.1074/mcp.M600129-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto S, Tsuda H, Honda K, Kita T, Takano M, Tamai S, Inazawa J, Yamada T, Matsubara O. Actinin-4 expression in ovarian cancer: A novel prognostic indicator independent of clinical stage and histological type. Mod Pathol. 2007;20:1278–1285. doi: 10.1038/modpathol.3800966. [DOI] [PubMed] [Google Scholar]
- 45.Kikuchi S, Honda K, Tsuda H, Hiraoka N, Imoto I, Kosuge T, Umaki T, Onozato K, Shitashige M, Yamaguchi U, et al. Expression and gene amplification of actinin-4 in invasive ductal carcinoma of the pancreas. Clin Cancer Res. 2008;14:5348–5356. doi: 10.1158/1078-0432.CCR-08-0075. [DOI] [PubMed] [Google Scholar]
- 46.Honda K. The biological role of actinin-4 (ACTN4) in malignant phenotypes of cancer. Cell Biosci. 2015;5:41. doi: 10.1186/s13578-015-0031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Savoy RM, Ghosh PM. The dual role of filamin A in cancer: Can't live with (too much of) it, can't live without it. Endocr Relat Cancer. 2013;20:R341–R356. doi: 10.1530/ERC-13-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng W, Lopez-Camacho C, Tang JY, Mendoza-Villanueva D, Maya-Mendoza A, Jackson DA, Shore P. Cytoskeletal protein filamin A is a nucleolar protein that suppresses ribosomal RNA gene transcription; Proc Natl Acad Sci USA; 2012; pp. 1524–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loy CJ, Sim KS, Yong EL. Filamin-A fragment localizes to the nucleus to regulate androgen receptor and coactivator functions; Proc Natl Acad Sci USA; 2003; pp. 4562–4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lawson D, Harrison M, Shapland C. Fibroblast transgelin and smooth muscle SM22alpha are the same protein, the expression of which is down-regulated in many cell lines. Cell Motil Cytoskeleton. 1997;38:250–257. doi: 10.1002/(SICI)1097-0169(1997)38:3<250::AID-CM3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 51.Shields JM, Rogers-Graham K, Der CJ. Loss of transgelin in breast and colon tumors and in RIE-1 cells by Ras deregulation of gene expression through Raf-independent pathways. J Biol Chem. 2002;277:9790–9799. doi: 10.1074/jbc.M110086200. [DOI] [PubMed] [Google Scholar]
- 52.Sitek B, Lüttges J, Marcus K, Klöppel G, Schmiegel W, Meyer HE, Hahn SA, Stühler K. Application of fluorescence difference gel electrophoresis saturation labelling for the analysis of microdissected precursor lesions of pancreatic ductal adenocarcinoma. Proteomics. 2005;5:2665–2679. doi: 10.1002/pmic.200401298. [DOI] [PubMed] [Google Scholar]
- 53.Mikuriya K, Kuramitsu Y, Ryozawa S, Fujimoto M, Mori S, Oka M, Hamano K, Okita K, Sakaida I, Nakamura K. Expression of glycolytic enzymes is increased in pancreatic cancerous tissues as evidenced by proteomic profiling by two-dimensional electrophoresis and liquid chromatography-mass spectrometry/mass spectrometry. Int J Oncol. 2007;30:849–855. [PubMed] [Google Scholar]
- 54.Huang Q, Huang Q, Chen W, Wang L, Lin W, Lin J, Lin X. Identification of transgelin as a potential novel biomarker for gastric adenocarcinoma based on proteomics technology. J Cancer Res Clin Oncol. 2008;134:1219–1227. doi: 10.1007/s00432-008-0398-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun X, Zhang H, Luo L, Zhong K, Ma Y, Fan L, Fu D, Wan L. Comparative proteomic profiling identifies potential prognostic factors for human clear cell renal cell carcinoma. Oncol Rep. 2016;36:3131–3138. doi: 10.3892/or.2016.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou H, Zhang Y, Chen Q, Lin Y. AKT and JNK signaling pathways increase the metastatic potential of colorectal cancer cells by altering transgelin expression. Dig Dis Sci. 2016;61:1091–1097. doi: 10.1007/s10620-015-3985-1. [DOI] [PubMed] [Google Scholar]
- 57.Lin Y, Buckhaults PJ, Lee JR, Xiong H, Farrell C, Podolsky RH, Schade RR, Dynan WS. Association of the actin-binding protein transgelin with lymph node metastasis in human colorectal cancer. Neoplasia. 2009;11:864–873. doi: 10.1593/neo.09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou HM, Fang YY, Weinberger PM, Ding LL, Cowell JK, Hudson FZ, Ren M, Lee JR, Chen QK, Su H, et al. Transgelin increases metastatic potential of colorectal cancer cells in vivo and alters expression of genes involved in cell motility. BMC Cancer. 2016;16:55. doi: 10.1186/s12885-016-2105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuan Y, Shen Z. Interaction with BRCA2 suggests a role for filamin-1 (hsFLNa) in DNA damage response. J Biol Chem. 2001;276:48318–48324. doi: 10.1074/jbc.M102557200. [DOI] [PubMed] [Google Scholar]
- 60.Yue J, Huhn S, Shen Z. Complex roles of filamin-A mediated cytoskeleton network in cancer progression. Cell Biosci. 2013;3:7. doi: 10.1186/2045-3701-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Velkova A, Carvalho MA, Johnson JO, Tavtigian SV, Monteiro AN. Identification of Filamin A as a BRCA1-interacting protein required for efficient DNA repair. Cell cycle. 2010;9:1421–1433. doi: 10.4161/cc.9.7.11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meng X, Yuan Y, Maestas A, Shen Z. Recovery from DNA damage-induced G2 arrest requires actin-binding protein filamin-A/actin-binding protein 280. J Biol Chem. 2004;279:6098–6105. doi: 10.1074/jbc.M306794200. [DOI] [PubMed] [Google Scholar]
- 63.Yue J, Wang Q, Lu H, Brenneman M, Fan F, Shen Z. The cytoskeleton protein filamin-A is required for an efficient recombinational DNA double strand break repair. Cancer Res. 2009;69:7978–7985. doi: 10.1158/0008-5472.CAN-09-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sur I, Neumann S, Noegel AA. Nesprin-1 role in DNA damage response. Nucleus. 2014;5:173–191. doi: 10.4161/nucl.29023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rashmi RN, Eckes B, Glöckner G, Groth M, Neumann S, Gloy J, Sellin L, Walz G, Schneider M, Karakesisoglou I, et al. The nuclear envelope protein Nesprin-2 has roles in cell proliferation and differentiation during wound healing. Nucleus. 2012;3:172–186. doi: 10.4161/nucl.19090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kelkar P, Walter A, Papadopoulos S, Mroß C, Munck M, Peche VS, Noegel AA. Nesprin-2 mediated nuclear trafficking and its clinical implications. Nucleus. 2015;6:479–489. doi: 10.1080/19491034.2015.1128608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsumoto A, Hieda M, Yokoyama Y, Nishioka Y, Yoshidome K, Tsujimoto M, Matsuura N. Global loss of a nuclear lamina component, lamin A/C, and LINC complex components SUN1, SUN2 and nesprin-2 in breast cancer. Cancer Med. 2015;4:1547–1557. doi: 10.1002/cam4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neumann S, Noegel AA. Nesprins in cell stability and migration. Adv Exp Med Biol. 2014;773:491–504. doi: 10.1007/978-1-4899-8032-8_22. [DOI] [PubMed] [Google Scholar]
- 69.Cartwright S, Karakesisoglou I. Nesprins in health and disease. Semin Cell Dev Biol. 2014;29:169–179. doi: 10.1016/j.semcdb.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 70.Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 71.Chittenden TW, Howe EA, Culhane AC, Sultana R, Taylor JM, Holmes C, Quackenbush J. Functional classification analysis of somatically mutated genes in human breast and colorectal cancers. Genomics. 2008;91:508–511. doi: 10.1016/j.ygeno.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang P, Sridharan D, Lambert MW. Nuclear α spectrin differentially affects monoubiquitinated versus non-ubiquitinated FANCD2 function after DNA interstrand cross-link damage. J Cell Biochem. 2016;117:671–683. doi: 10.1002/jcb.25352. [DOI] [PubMed] [Google Scholar]
- 74.Almuzzaini B, Sarshad AA, Farrants AK, Percipalle P. Nuclear myosin 1 contributes to a chromatin landscape compatible with RNA polymerase II transcription activation. Bmc Biol. 2015;13:35. doi: 10.1186/s12915-015-0147-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Savoy RM, Chen L, Siddiqui S, Melgoza FU, Durbin-Johnson B, Drake C, Jathal MK, Bose S, Steele TM, Mooso BA, et al. Transcription of Nrdp1 by the androgen receptor is regulated by nuclear filamin A in prostate cancer. Endocr Relat Cancer. 2015;22:369–386. doi: 10.1530/ERC-15-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]