Abstract
Enhancer of zeste homolog 2 (EZH2), the critical component of polycomb group protein family, has been demonstrated to be overexpressed in various types of human cancer, including hepatocellular carcinoma, breast, bladder and lung cancer. The mechanism of how EZH2 promotes oncogenesis has also been well studied. However, little is known about the role of EZH2 in colorectal cancer (CRC). The main purpose of the present study was to analyze the association between EZH2 expression and the clinicopathological features of CRC. Therefore, the mRNA and protein expression levels were analyzed in tumor tissues and adjacent non-cancerous tissues by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blot analysis. The expression of EZH2 was demonstrated to be significantly increased in tumor tissues compared with adjacent noncancerous tissues, according to the results of western blot analysis and RT-qPCR in the majority of cases. Patients with low EZH2 expression had a longer overall survival rate compared with those with high EZH2 expression. An analysis of the association between clinicopathological features and EZH2 expression indicated that high EZH2 expression was significantly associated with tumor stage, tumor size, histological differentiation and lymph node metastasis. Multivariate analysis demonstrated that high EZH2 expression was an independent predictor of overall survival. In conclusion, to the best of our knowledge, the data presented in the present study is the first to indicate that EZH2 is upregulated in CRC and may serve as a predictor of poor outcome for patients with CRC.
Keywords: enhancer of zeste homolog, polycomb group protein, colorectal cancer, prognosis, clustered regularly interspaced short palindromic repeats
Introduction
The morbidity of colorectal cancer (CRC) ranks third place in males and second place in females worldwide (1). In 2012, ~1.4 million new cases of CRC were diagnosed, with ~694,000 CRC-associated mortalities (2). In previous years, the incidence of CRC has continued to increase worldwide, particularly in developed countries, accounting for >65% of newly diagnosed cases each year (3). The risk factors of CRC include lifestyle, inherited genetic disorders, family history of colon cancer, exposure to radiation and other diseases, such as inflammatory bowel disease, obesity and diabetes (4). The treatment for CRC includes surgery, radiation therapy, chemotherapy and targeted therapy (4).
However, CRC remains one of the leading causes of cancer-associated mortality worldwide. The main challenge is that patients with CRC are not diagnosed at early stages, which results in a poor prognosis, with a 5-year survival rate of 50–59% (3). However, carcinoembryonic antigen and cancer antigen 19-9 have been widely used as biomarkers for CRC diagnosis and have also been shown to efficiently reduce the mortality rate of patients with CRC (5,6). Therefore, it is critical to identify the molecular markers, which are able to monitor or predict the progression and prognosis of patients with CRC and to investigate these potential biomarkers as therapeutic targets to improve the survival quality of patients.
Previous studies have highlighted the role of chromosome structure on the regulation of genome transcriptional status, which was frequently observed in diseases, particularly cancer (7,8). Enhancer of zeste homolog 2 (EZH2), the key component of polycomb repressive complex 2, has a crucial role in the regulation of cell proliferation and cell cycle through gene repression or histone H3 lysine 27 (H3K27) methylation (8). The methyltransferase activity of EZH2 lies in the catalytic domain of the C-terminal, which is also termed the SET domain (9). Mutations of the tyrosine 641 (Y641F, Y641N, Y641S, Y641C and Y641H) within the SET domain results in reduced methylation of unmethylated H3K27 but enhanced methylation of the dimethylated version of H3K27 (H3K27me2), and thus represses the expression of polycomb targets (10). An increasing body of evidence has demonstrated that a specific cell or its behavior mainly relies on the expression and repression of genes (11,12). There is evidence that exogenous expression of EZH2 in mice may lead to the development of myeloproliferative disorder (13). Another in vitro study that aimed to investigate the role of EZH2 in human breast epithelial cell lines demonstrated that expression of EZH2 causes neoplastic transformation of epithelial cells, which highlighted that EZH2 may perform an important role in cancer (14).
Additional studies have implicated the oncogenic role of EZH2 in cancer. The overexpression of EZH2 was shown to be associated with the poor prognosis of prostate cancer (15). In addition, overexpression of EZH2 in a number of types of human cancer, including hepatocellular carcinoma (16), breast cancer (17), bladder cancer (18) and melanoma (19), has been observed. Furthermore, several independent groups have revealed the mechanism of how EZH2 is involved in the development and progression of a variety of types of cancer (20). However, there is limited knowledge on the expression of EZH2 in CRC. In the present study, the expression status of EZH2 was analyzed in patients with CRC, and its association with the prognosis of CRC was also investigated.
Materials and methods
Cell lines
The noncancerous colon epithelial cell line (HCEC) and the two CRC cell lines (HCT-116 and SW480) used in a pre-experiment were purchased from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). The cells were cultured in complete Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) with 100 µg/ml streptomycin and 100 U/ml penicillin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and maintained in a humidified incubator (5% CO2, 37°C).
RNA interference
Knockdown of EZH2 in the CRC cell line was accomplished with small interfering RNA (siRNA) duplex as previously described (15). The EZH2 siRNA sequence (5′-AGUCUCAUGUACGCTGACUCUG-3′) was designed by Genepharm, Inc. (Sunnyvale, CA, USA) to target the 85–106 region of human EZH2. In vitro transient transfection was performed as described previously (21).
Generation of EZH2 tyrosine 641 mutation cell line using the CRISPR/Cas9 complex
To investigate the effect of EZH2 Y641 mutations on cell behavior, in vivo gene mutagenesis was performed on HCEC cells using CRISPR/Cas9-based technology. The guide RNA (gRNA) pairs and donor sequences were designed and synthesized by GenScript (Nanjing, China). The gRNA was cloned into pSpCas9n-BB (PX460) according manufacturer's recommendations and the sequences were as follows: gRNA-1, 5′-GGATTTGCATGCTTAGTAAC-3′; and gRNA-2, 5′-TGCAGAAGTCCAGGCTGAAA-3′. The donor sequences were presented in Table I and cloned into the EcoRV site of pUC57 (GenScript).
Table I.
Donor sequences used in this study.
| Mutation type | Sequence |
|---|---|
| EZH2 Y641F | ctgaacgatggtcattgcagaggaccaacaccaccaaaaggttttctgtaagacagagattcttatctgctgtataaggaaaacataatgttcatagccattctcagcagctttcacgttgactgaagctgtgtgcccaattactgccttagaacaaacaggtctgaggatttacagtgatagcttttgttttcattctgtagtctactttgtccccagtccattttcaccctccttttttgatgatgtgattgtgttttattctctagcatctattgctggcaccatctgacgtggcaggctgggggatttttatcaaagatcctgtgcagaaaaatgaattcatctcagaaTTGtgtggagaggtaaggcactgataacctgtattcaggtggcattgtatatactaactttactttattttagattgattttattaggtaagtctgtgggtttgattggaaatgaattgccataaactgccttttcagcctggacttctgcatgtttgtggatttgcatgcttagtaactggattgtgctgggcgcggtggccgactcctgcaatcccagcactttgggaggccgaggcaggtggattgcttgagctcaggagttggagaccagcatgggcaacatggcaagaccccattgctacaaaaaatgcaaaaattagccgggcgtggtggtgcatacttgtagtcccagctacttgggagc |
| EZH2 Y641N | ctgaacgatggtcattgcagaggaccaacaccaccaaaaggttttctgtaagacagagattcttatctgctgtataaggaaaacataatgttcatagccattctcagcagctttcacgttgactgaagctgtgtgcccaattactgccttagaacaaacaggtctgaggatttacagtgatagcttttgttttcattctgtagtctactttgtccccagtccattttcaccctccttttttgatgatgtgattgtgttttattctctagcatctattgctggcaccatctgacgtggcaggctgggggatttttatcaaagatcctgtgcagaaaaatgaattcatctcagaaAAGtgtggagaggtaaggcactgataacctgtattcaggtggcattgtatatactaactttactttattttagattgattttattaggtaagtctgtgggtttgattggaaatgaattgccataaactgccttttcagcctggacttctgcatgtttgtggatttgcatgcttagtaactggattgtgctgggcgcggtggccgactcctgcaatcccagcactttgggaggccgaggcaggtggattgcttgagctcaggagttggagaccagcatgggcaacatggcaagaccccattgctacaaaaaatgcaaaaattagccgggcgtggtggtgcatacttgtagtcccagctacttgggagc |
| EZH2 Y641S | ctgaacgatggtcattgcagaggaccaacaccaccaaaaggttttctgtaagacagagattcttatctgctgtataaggaaaacataatgttcatagccattctcagcagctttcacgttgactgaagctgtgtgcccaattactgccttagaacaaacaggtctgaggatttacagtgatagcttttgttttcattctgtagtctactttgtccccagtccattttcaccctccttttttgatgatgtgattgtgttttattctctagcatctattgctggcaccatctgacgtggcaggctgggggatttttatcaaagatcctgtgcagaaaaatgaattcatctcagaaAGUtgtggagaggtaaggcactgataacctgtattcaggtggcattgtatatactaactttactttattttagattgattttattaggtaagtctgtgggtttgattggaaatgaattgccataaactgccttttcagcctggacttctgcatgtttgtggatttgcatgcttagtaactggattgtgctgggcgcggtggccgactcctgcaatcccagcactttgggaggccgaggcaggtggattgcttgagctcaggagttggagaccagcatgggcaacatggcaagaccccattgctacaaaaaatgcaaaaattagccgggcgtggtggtgcatacttgtagtcccagctacttgggagc |
| EZH2Y641C | ctgaacgatggtcattgcagaggaccaacaccaccaaaaggttttctgtaagacagagattcttatctgctgtataaggaaaacataatgttcatagccattctcagcagctttcacgttgactgaagctgtgtgcccaattactgccttagaacaaacaggtctgaggatttacagtgatagcttttgttttcattctgtagtctactttgtccccagtccattttcaccctccttttttgatgatgtgattgtgttttattctctagcatctattgctggcaccatctgacgtggcaggctgggggatttttatcaaagatcctgtgcagaaaaatgaattcatctcagaaTGCtgtggagaggtaaggcactgataacctgtattcaggtggcattgtatatactaactttactttattttagattgattttattaggtaagtctgtgggtttgattggaaatgaattgccataaactgccttttcagcctggacttctgcatgtttgtggatttgcatgcttagtaactggattgtgctgggcgcggtggccgactcctgcaatcccagcactttgggaggccgaggcaggtggattgcttgagctcaggagttggagaccagcatgggcaacatggcaagaccccattgctacaaaaaatgcaaaaattagccgggcgtggtggtgcatacttgtagtcccagctacttgggagc |
| EZH2 Y641H | ctgaacgatggtcattgcagaggaccaacaccaccaaaaggttttctgtaagacagagattcttatctgctgtataaggaaaacataatgttcatagccattctcagcagctttcacgttgactgaagctgtgtgcccaattactgccttagaacaaacaggtctgaggatttacagtgatagcttttgttttcattctgtagtctactttgtccccagtccattttcaccctccttttttgatgatgtgattgtgttttattctctagcatctattgctggcaccatctgacgtggcaggctgggggatttttatcaaagatcctgtgcagaaaaatgaattcatctcagaaCACtgtggagaggtaaggcactgataacctgtattcaggtggcattgtatatactaactttactttattttagattgattttattaggtaagtctgtgggtttgattggaaatgaattgccataaactgccttttcagcctggacttctgcatgtttgtggatttgcatgcttagtaactggattgtgctgggcgcggtggccgactcctgcaatcccagcactttgggaggccgaggcaggtggattgcttgagctcaggagttggagaccagcatgggcaacatggcaagaccccattgctacaaaaaatgcaaaaattagccgggcgtggtggtgcatacttgtagtcccagctacttgggagc |
Bold letters indicate nuclotide sequence used to obtain the mutant type of EZH2.
To generate EZH2 tyrosine 641 mutant cell lines, two gRNA constructs along with the donor sequence construct were co-transfected into the HCEC cell line using Lipofectamine (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol (22). The transfected cells were cultured in the aforementioned conditions with the addition of ampicillin (Sigma-Aldrich; Merck KGaA) as a selective marker. Following continuous cultivation under the antibiotic selective pressure, several single colonies were successfully obtained in every independent experiment aimed to obtain tyrosine mutant cell lines. The successful introduction of tyrosine mutation in EZH2 gene was verified by Sanger Sequencing (GenScript).
Cell proliferation assay
The cell proliferation of wild-type cell lines (HCEC, HCT116 and SW480) and the mutant cell lines (HCEC/Y641F, HCEC/Y641N, HCEC/Y641S, HCEC/Y641C and HCEC/Y641H) were assessed by the widely used MTT assay. In brief, the cells in the logarithmic growth phase were harvested and the cell density was adjusted to ~5×104/ml. A total of 100 µl cell suspension for each cell line was seeded onto 96-well plates with a final cell density of ~5,000 cells/well and cultured in the described medium and atmosphere. MTT (20 µl; Sigma-Aldrich; Merck KGaA) was added after 24, 48 and 72 h of incubation, followed by 4 h incubation in the same conditions, and the supernatant was then removed by centrifugation (1,000 × g for 10 min at room temperature). A total of 150 µl DMSO was added to each well, and the plates were oscillated at a lower speed (100 rpm) until the crystals fully dissolved. The absorbance of each well was measured at a wavelength of 570 nm using Thermo Multiskan Spectrum (Thermo Fisher Scientific, Inc.). Each experiment was repeated three times under the same conditions.
Clinical patients
A total of 95 patients with CRC who underwent surgical treatment between March 2009 and June 2011 at Guangzhou First People's Hospital (Guangzhou, China) were enrolled in the present study. The cohort included 54 males and 41 females; age range, 39 to 76 years; average age of 57.3 years. In addition, none of the patients had received any anticancer treatments, including radiation, chemotherapy and surgery. The colorectal tissues were surgically removed from patients with CRC, and the matched non-cancerous tissues were obtained from the distal edge of the resection ≥5 cm from the tumor. The tissues were immediately frozen with liquid nitrogen and preserved at −80°C for further experiments. According to the tumor-node-metastasis (TNM) staging system (23), the patients enrolled in the present study were classified into stages I–IV. In addition, the histological grade was confirmed by microscopic examination and conducted by an independent pathologist. Other clinicopathological data, including tumor size, age, sex and distant metastasis were also collected (Table II), and the prognostic factors and disease progression were retrospectively collected. The present study was approved and monitored by the Ethics Committee of Guangzhou First People's Hospital, and written consent was obtained from all the recruited patients.
Table II.
Association between EZH2 expression and clinicopathological features of colorectal cancer.
| EZH2 expression | ||||
|---|---|---|---|---|
| Variable | No. of cases | High | Low | P-value |
| Sex | ||||
| Male | 54 | 40 | 14 | NS |
| Female | 41 | 26 | 15 | |
| Age, years | ||||
| ≥50 | 49 | 34 | 15 | NS |
| <50 | 46 | 32 | 12 | |
| Tumor size, cm | ||||
| ≥5 | 63 | 48 | 15 | 0.046 |
| <5 | 32 | 18 | 14 | |
| Lymph node metastasis | ||||
| Absent | 53 | 42 | 11 | 0.041 |
| Present | 42 | 24 | 18 | |
| Histological differentiation | ||||
| Well/moderate | 44 | 26 | 18 | 0.041 |
| Poor | 51 | 40 | 11 | |
| Tumor stage | ||||
| I–II | 59 | 46 | 13 | 0.021 |
| III–IV | 36 | 20 | 16 | |
NS, not significant; EZH2, enhancer of zeste homolog 2.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from cell lines and all 95 frozen tumor tissues was isolated using the miRNeasy Mini kit (Qiagen, Nordrhein-Westfalen, Germany) according to the detailed protocols provided by the manufacturer. The isolated mRNA was then treated with DNase (Invitrogen; Thermo Fisher Scientific, Inc.), and cDNA was synthesized using the Universal cDNA synthesis kit II (Exiqon A/S, Vedbaek, Denmark) according to the manufacturer's protocol.
RT-qPCR was performed on the ABI PRISM 7300 sequence detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.) using SYBR-Green PCR Master mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. To estimate the level of EZH2 mRNA expression, GAPDH expression level in the corresponding tissue was used an internal control. The primers were synthesized by Invitrogen (Thermo Fisher Scientific, Inc.), and the sequences were as follows: EZH2 sense, 5′-TTGTTGGCGGAAGCGTGTAAAATC-3′; and antisense, 5′-TCCCTAGTCCCGCGCAATGAGC-3′; GAPDH sense, 5′-TGAACGGGAAGCTCACTGG-3′; and antisense, 5′-TCCACCACCCTGTTGCTGTA-3′. The detailed PCR cycling conditions were as follows: Initial denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 1 min, and annealing/extension at 56°C for 1 min. All samples were performed in triplicate and normalized to internal controls. The fold-changes or relative EZH2 expression levels were calculated based on the 2−ΔΔCq method (24). The 75th percentile of EZH2 expression level was used as cut-off value to classified the patients into either the overexpressed or underexpressed group.
Western blot analysis
All 95 frozen tumor tissues and cellular protein samples were isolated using the method described in a previously published study (14) with slight modifications. Initially, the tissue or cell samples were homogenized with homogenization buffer [1 M Tris HCl pH 7.5, 1% Triton X-100, 1% Nonidet P-40 (NP-40), 10% SDS, 0.5% sodium deocycholate, 0.5 M EDTA, 10 µg/ml leupeptin, 10 µg/ml aprotinin and 1 M PMSF] and centrifuged at 10,000 × g for 30 min at 4°C. The protein concentration was determined using the standard Bradford method.
Total protein (50 µg) was loaded to each lane of a 10% SDS-PAGE gel and transferred to a nitrocellulose membrane. The membranes were then blocked with 5% fat-free milk in PBS for ~2 h at room temperature and incubated with antibodies against EZH2 (cat. no. 4905; dilution, 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA) and GAPDH (cat. no. 2118; dilution, 1:1,000; Cell Signaling Technology, Inc.) in PBS containing 5% milk for ~1 h at room temperature. The membranes were washed three times with PBS buffer and then incubated with horseradish peroxidase-conjugated secondary antibodies (cat. no. 7074; dilution, 1:500; Cell Signaling Technology, Inc.) for 1 h at room temperature. The specific band was developed using an enhanced chemiluminescence reagent (ECL reagent; NEN Life Science Products; PerkinElmer, Inc., Waltham, MA, USA) in a gel imaging system (ImageQuant 300/RT ECL; GE Healthcare, Chicago, IL, USA). The band was analyzed using an Image J 1.37 software (National Institutes of Health, Bethesda, MA, USA). A total of three independent experiments were performed.
Immunohistochemical analysis
To visualize the localization of EZH2 in tumor tissues obtained from patients with CRC, immunohistochemical staining was performed. The tissues were fixed in 4% (v/v) formalin at 4°C for 48 h and embedded in paraffin, and dewaxed in xylene and rehydrated in graded ethanol solutions (100, 95, 90, 80, 70 and 50%). Antigen retrieval was performed by immersing sections (5 µm) in 10 mM citrate buffer (pH 6.0) for between 15 and 20 min at 95°C, prior to incubation with 0.3% hydrogen peroxide for 15 min at room temperature to block endogenous peroxidase activity. The sections were incubated with 5% goat serum (CWBiotech, Beijing, China) to block the non-specific binding sites at 37°C for 30 min. The sections were then incubated overnight with the anti-EZH2 antibody (cat. no. 4905; dilution, 1:100; Cell Signaling Technology, Inc.) at 4°C. All sections were processed using the peroxidase-anti-peroxidase method. Subsequent to washing, the sections were counterstained with hematoxylin for 10 min at room temperature, dehydrated and mounted with a coverslip. The Olympus BX61-32S04 microscope was used to observe the stained tissues. The immunostaining scores for each section were evaluated in a blinded manner by two independent pathologists. The staining intensity (SI) was scored on a scale of 0 to 3: Negative (0), weak positive (1), moderately positive (2), and strongly positive (3). The percentage of positive cells (PP) was regarded as: None (0), <10% (1), 11–50% (2), 51–80% (3) and ≥80% (4). The product of SI and PP is the Immunoreactive score (0–12). A score of 0–2 was regarded as low, 3–12 as positive.
Statistical analysis
Statistical analysis was performed with SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA). The values are presented as the mean ± standard deviation, and P<0.05 was considered to indicate a statistically significant difference. One-way analysis of variance (ANOVA) followed by Tukey's multiple comparisons test was used for the comparison of means. The association between EZH2 expression and the clinicopathological characteristics was analyzed using χ2 test. Survival analysis was performed using the Kaplan-Meier log-rank test. Univariate and multivariate analyses were performed using Cox proportional hazard models.
Results
EZH2 is overexpressed in CRC cell lines
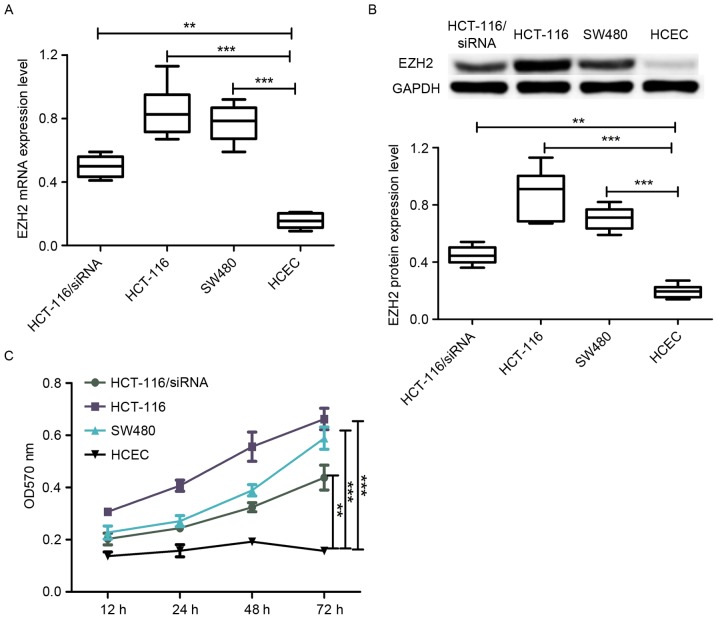
To date, little is known about the role of EZH2 in the development and progression of CRC, therefore, the levels of EZH2 mRNA and protein expression were analyzed in CRC cell lines. A total of two CRC cell lines (HCT-116 and SW480) and one normal colon epithelial cell line (HCEC) were selected in the present study. RT-qPCR was performed to estimate the level of EZH2 mRNA expression in these cell lines. As shown in Fig. 1A, the level of EZH2 expression in the HCT-116 and SW480 cell lines was significantly higher compared with the HCEC cell line. In addition, the level of protein expression in the aforementioned cell lines harvested in the same conditions was analyzed. As with the results obtained from RT-qPCR, the protein expression level of EZH2 in HCT-116 and SW480 cells was higher compared with HCEC cells (Fig. 1B). The observation that EZH2 expression in CRC cell lines was much higher compared with the normal colon epithelial cell line (HCEC) indicates that EZH2 may perform an important role in the development of CRC.
Figure 1.
EZH2 expression status in CRC cell lines and normal colon colorectal cell line. (A) Reverse transcription-quantitative polymerase chain reaction analysis of the EZH2 mRNA expression level in CRC HCT-116 and SW480 cell lines, normal colon colorectal cell line and siRNA-transfected HCT-116 cell line. (B) Western blot analysis of the protein expression of EZH2 in CRC cell lines HCT-116 and SW480, normal colon colorectal cell line HCEC and siRNA-transfected HCT-116 cell line. (C) MTT assay to determine the cell proliferation rate of CRC cell lines HCT-116 and SW480, normal colon colorectal cell line HCEC and siRNA-transfected HCT-116 cell line. ***P<0.001, **P<0.01. NS, not significant; EZH2, enhancer of zeste homolog 2; CRC, colorectal cancer; siRNA, small interfering RNA; OD, optical density.
EZH2 overexpression promotes cell proliferation
To further investigate the role of EZH2 overexpression on the proliferation of CRC cell lines and normal colon epithelial cell line, the cell proliferation rate of HCEC, HCT-116 and SW480 was analyzed using the well-known MTT assay. The results are depicted in Fig. 1C, where the cell proliferation rate in the CRC HCT-116 and SW480 cell lines was markedly higher compared with the HCEC cell line, which showed that EZH2 overexpression promoted cell proliferation. In addition, the cell proliferation rate of HCT-116 was higher compared with SW480 (Fig. 1C), which is consistent with comparisons of EZH2 expression level between HCT-116 and SW480 cells.
In addition, the level of EZH2 mRNA and protein expression was analyzed in the EZH2-specific siRNA-transfected HCT-116 cell line. The results revealed that EZH2 expression was downregulated in the siRNA-transfected HCT-116 cell line compared with the normal HCT-116 cell line (Fig. 1A and B). Importantly, the cell proliferation rate of the siRNA-transfected HCT-116 cell line was lower compared with the wild-type HCT-116 cell line. Taken together (Fig. 1C), EZH2 overexpression was able to promote cell proliferation and the degree of promotion was observed to be associated with the level of EZH2 expression.
Effects of EZH2 tyrosine 641 mutations on cell proliferation
Multiple studies have demonstrated that EZH2 is easy to mutate, particularly in tyrosine 641 (8,10). Notably, mutations in this specific site will affect the activity of EZH2, either gain of function or loss of function, and thus bring a discrepancy effect on the downstream target genes. In the present study, mutagenesis was performed on the tyrosine 641 of EZH2 using the CRISPR/Cas9-based gene editing technology to introduce Y641F, Y641N, Y641S, Y641C and Y641H mutations to EZH2. The gRNA pairs and donor sequences were designed according to the instructions in previous published studies. The HCEC cell line was co-transfected with The gRNA and donor constructs using Lipofectamine and the successful introduction of mutation was verified by genome sequencing (data not shown). The cell lines containing the desired mutations were termed HCEC/Y641F, HCEC/Y641N, HCEC/Y641S, HCEC/Y641C and HCEC/Y641H.
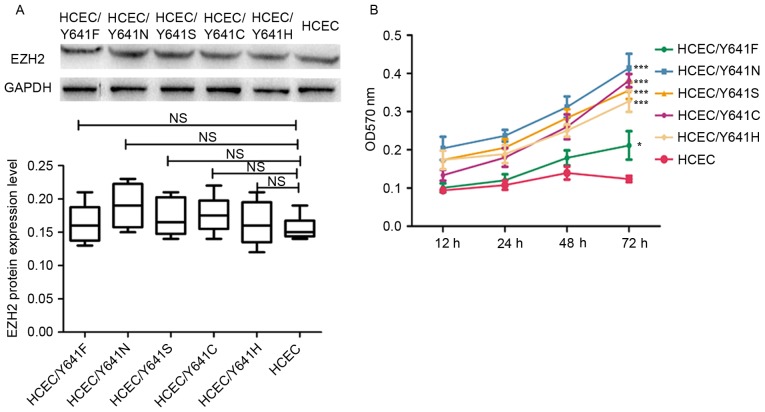
The protein expression of the mutant EZH2 in the HCEC cell line was then analyzed by western blot analysis. The results presented in Fig. 2A demonstrated that the expression level of EZH2 variants was almost the same as wild-type EZH2, which indicates that the expression of EZH2 was not affected by the point mutation. The cell proliferation rate of HCEC cells with the wild-type EZH2 or mutated EZH2 was then analyzed. According to the results shown in Fig. 2B, the cell lines with EZH2 mutations had a significantly higher cell proliferation rate compared with the wild-type HCEC cell line (P<0.05), which is consistent with previous findings (9). The present study therefore provided more support that EZH2 may have an important role in the malignancy of colorectal cell and is a potential treatment target for CRC.
Figure 2.
Expression of EZH2 variants in HCEC cell lines. (A) Expression of EZH2 variants in HCEC cell lines. (B) MTT assay to determine the cell proliferation rate of HCEC cell lines containing EZH2 variants. The significance was measured by comparing the HCEC cell lines containing EZH2 variants with wild-type HCEC cell line. ***P<0.001, *P<0.05. NS, not significant; EZH2, enhancer of zeste homolog 2; OD, optical density.
Overexpression of EZH2 in patients with CRC
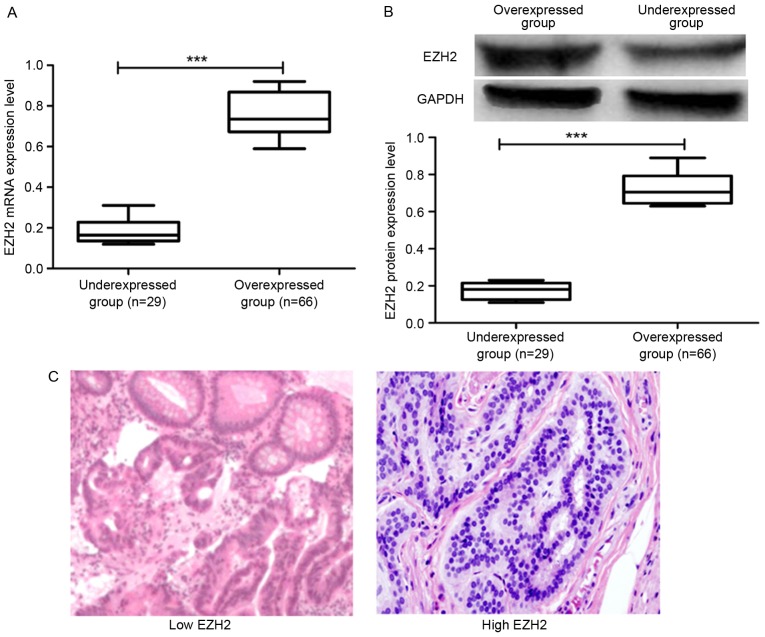
In addition to analyzing the EZH2 expression in CRC cell lines, the expression of EZH2 was examined in patients with CRC. In total, 95 pairs of tumor tissues and adjacent non-cancerous tissues were obtained from the patients with CRC involved in the present study and the expression pattern of EZH2 in all the collected tissues was analyzed. Initially, RT-qPCR was performed to analyze the level of EZH2 expression in CRC tissues. It was observed that 66 of 95 (69.47%) of the cancer tissue samples exhibited high EZH2 expression and 29 of 95 (30.53%) demonstrated low expression (Fig. 3A). Patients with CRC were then classified into overexpressed and under-expressed groups according to the results obtained from RT-qPCR. Furthermore, the protein expression level of EZH2 in the archived tumor tissues was analyzed by western blot analysis. The results presented in Fig. 3B demonstrated that the protein expression level of EZH2 in the overexpressed group was higher compared with the under-expressed group, which was consistent with the mRNA expression results obtained from RT-qPCR.
Figure 3.
EZH2 expression status in patients with CRC. (A) EZH2 mRNA expression in patients with CRC. Patients with high EZH2 expression were classified into overexpressed group and those with low expression were into under-expressed group (cut-off value, 0.39). (B) EZH2 protein expression in patients with CRC. (C) Immunohistochemical analysis of EZH2 expression in patients with CRC. Images were captured at ×200 magnification. ***P<0.001. NS, not significant; CRC, colorectal cancer; EZH2, enhancer of zeste homolog 2.
Immunohistochemical staining was performed to analyze the expression of EZH2 in the CRC tissues in addition to RT-qPCR and western blot analysis. The results shown in Fig. 3C demonstrated that the level of EZH2 expression as detected by immunohistochemical staining in the samples from the overexpressed group was higher compared with the under-expressed group. These results demonstrated that EZH2 was highly expressed in patients with CRC.
Clinical significance of EZH2 expression in CRC
The association between EZH2 expression in patients with CRC and the different clinicopathological characteristics was analyzed (Table II). High EZH2 expression was significantly associated with tumor stage (P=0.021), tumor size (P=0.046), histological differentiation (P=0.041) and lymph node metastasis (P=0.041), respectively. The overall survival (OS) rate of the patients from overexpressed and under-expressed groups was examined using the log-rank test. As a result, a statistically significant difference was observed between these two groups (P=0.016; Fig. 4). The 5-year OS rates for the overexpressed group and under-expressed group were 32.08 and 50.68%, respectively, indicating that the patients with low EZH2 expression had a longer survival expectation compared with the patients with high expression.
Figure 4.
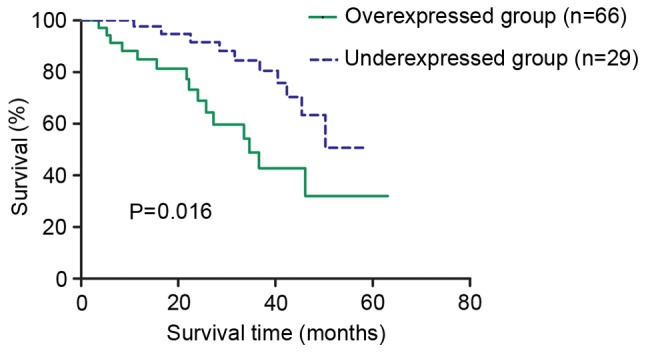
Overall survival rate assessed by Kaplan-Meier curves. Patients with CRC with EZH2 overexpression presented with shorter 5-year survival rates compared with those with EZH2 under-expression. CRC, colorectal cancer; EZH2, enhancer of zeste homolog 2.
EZH2 overexpression is associated with poor prognosis in CRC
Univariate analysis and multivariate analysis were performed to examine the association of clinicopathological characteristics, including EZH2 expression, age, sex, tumor size, lymph node metastases, histological differentiation, TNM stage with the survival of patients with CRC. It was concluded that tumor size, tumor stage, lymph node metastases, histological differentiation and EZH2 expression level could be reconsidered as independent prognosis factors for patients with CRC (Table III). The present study demonstrates the importance of EZH2 expression in the development and progression of CRC.
Table III.
Univariate analysis and multivariate analyses of overall survival in patients with colorectal cancer.
| A, Univariate analysis | |||
|---|---|---|---|
| Variables | HR | 95% CI | P-value |
| EZH2 expression | 2.872 | 1.204–6.349 | 0.009 |
| Age | 2.142 | 0.936–4.898 | 0.071 |
| Sex | 1.983 | 0.851–4.619 | 0.113 |
| Tumor size | 2.290 | 1.019–5.148 | 0.045 |
| Lymph node metastases | 2.297 | 1.022–5.164 | 0.044 |
| Histological differentiation | 2.448 | 1.107–5.414 | 0.027 |
| Tumor stage | 2.717 | 1.209–6.108 | 0.016 |
| B, Multivariate analysis | |||
| Variables | HR | 95% CI | P-value |
| EZH2 expression | 2.625 | 1.204–5.722 | 0.015 |
| Age | – | – | – |
| Sex | – | – | – |
| Tumor size | 2.499 | 1.128–5.537 | 0.024 |
| Lymph node metastases | 2.485 | 1.122–5.505 | 0.025 |
| Histological differentiation | 2.551 | 1.168–5.573 | 0.019 |
| Tumor stage | 2.737 | 1.270–5.900 | 0.010 |
HR, hazard ratio, CI, confidence interval; EZH2, enhancer of zeste homolog 2.
Discussion
An increasing number of studies have indicated that the chromosome structure is a key factor to regulate the development and progression of various types of human cancer (10). EZH2 has long been recognized as the most important participant to induce histone H3 lysine 27 (H3K27) methylation (7). Clinical study data has shown the oncogenic role of EZH2 in several types of human cancer including breast cancer and ovarian cancer (14,25). Notably, an in vitro experiment involving the EZH2 inhibitor in prostate cancel models demonstrated that EZH2 inhibitor was able to significantly increase death of mouse and human prostate cancer cells (26). Furthermore, in a preclinical study aimed to investigate the treatment function of EZH2 inhibitor on non-small cell lung cancer, researchers found the EZH2 inhibitor had differing effects depending on the mutations present (27). Although the development of EZH2 inhibitor is at an early stage, it provides a new potential strategy for cancer treatment (28,29).
However, little is known about the role of EZH2 expression in CRC and, to the best of our knowledge, this is the first study reported to investigate the expression status of EZH2 in CRC and also its role in the development and progression of CRC. The expression status of EZH2 in CRC and normal colon epithelial cell lines was examined with siRNA transfection. It was observed that the expression level of EZH2 was significantly higher in CRC cell lines compared with the normal colon epithelial cell line. The EZH2 expression in the siRNA-transfected HCT-116 was lower compared with the cell line without transfection. In addition, the cell proliferation rate of the CRC cell lines and normal colon epithelial cell line used in the present study was analyzed. Furthermore, the cell proliferation rate of CRC cell lines was higher compared with the normal colon epithelial cell line and the siRNA-transfected CRC cell line, which indicated that EZH2 overexpression may promote cell proliferation. Furthermore, the specified mutation in the tyrosine 641 of EZH2 was introduced using CRISPR/Cas9-based genome editing system. Consistent with the previous results (10), it was shown that the EZH2 Y641F, Y641N, Y641S, Y641C and Y641H mutations may increase the activity of EZH2. These in vitro results indicated that EZH2 may perform an important role in the development and progression of CRC.
Finally, the expression of EZH2 was investigated in patients with CRC and its role on the prognosis of patients with CRC was examined. Initially, it was verified that high EZH2 expression was observed in patients with CRC through examining the EZH2 expression in tumor tissues obtained from the patients with CRC using RT-qPCR, western blot analysis and immunohistochemistry approaches. These findings were consistent with the results obtained from analysis of EZH2 expression in CRC cell lines and normal colon epithelial cell line. Furthermore, the association of EZH2 expression with other clinical characteristics collected from the patients with CRC was analyzed and the results indicated EZH2 overexpression was associated with tumor stage, tumor size, histological differentiation and lymph node metastasis, which suggest that EZH2 overexpression is associated with tumor progression. In addition, the 5-year survival rate comparisons between the patients with CRC with high EZH2 expression and those with low expression revealed that the patients with high EZH2 expression had a shorter survival estimate, which provide further support that EZH2 overexpression is associated with poor outcomes in patients with CRC. Multivariate analyses revealed that EZH2 overexpression was an independent factor for prediction of prognosis of patients with CRC. Additionally, increased EZH2 expression, tumor size, tumor stage, lymph node metastases and histological differentiation are also considered to be independent factors for poor prognosis of CRC.
In conclusion, the in vitro and in vivo studies in the present study demonstrated that EZH2 was overexpressed in patients with CRC. The overexpression of EZH2 was an independent biomarker for the poor outcome of patients with CRC. The results indicated that EZH2 has potential value as a therapeutic target in patients with CRC in the future. However, more in vitro and in vivo studies are required to identify the downstream target genes in CRC to improve the understanding of the biological role of EZH2 in CRC.
Acknowledgements
The present study was funded by the National Nature Science Foundation of China (grant no. 81272556).
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Stewart BW, Wild CP. World Cancer Report 2014. IARC Publications; 2014. Colorectal cancer; pp. 392–402. [Google Scholar]
- 4.Laiyemo AO, Doubeni C, Pinsky PF, Doria Rose VP, Bresalier R, Lamerato LE, Crawford ED, Kvale P, Fouad M, Hickey T, et al. Race and colorectal cancer disparities: Health-care utilization vs different cancer susceptibilities. J Natl Cancer Inst. 2010;102:538–546. doi: 10.1093/jnci/djq068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Hees F, Saini SD, Lansdorp-Vogelaar I, Vijan S, Meester RG, de Koning HJ, Zauber AG, van Ballegooijen M. Personalizing colonoscopy screening for elderly individuals based on screening history, cancer risk, and comorbidity status could increase cost effectiveness. Gastroenterology. 2015;149:1425–1437. doi: 10.1053/j.gastro.2015.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma W, Yu Q, Jiang J, Du X, Huang L, Zhao L, Zhou QI. miR-517a is an independent prognostic marker and contributes to cell migration and invasion in human colorectal cancer. Oncol Lett. 2016;11:2583–2589. doi: 10.3892/ol.2016.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tonini T, D'Andrilli G, Fucito A, Gaspa L, Bagella L. Importance of Ezh2 polycomb protein in tumorigenesis process interfering with the pathway of growth suppressive key elements. J Cell Physiol. 2008;214:295–300. doi: 10.1002/jcp.21241. [DOI] [PubMed] [Google Scholar]
- 8.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan J, Ng SB, Tay JL, Lin B, Koh TL, Tan J, Selvarajan V, Liu SC, Bi C, Wang S, et al. EZH2 overexpression in natural killer/T-cell lymphoma confers growth advantage independently of histone methyltransferase activity. Blood. 2013;121:4512–4520. doi: 10.1182/blood-2012-08-450494. [DOI] [PubMed] [Google Scholar]
- 10.Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, Copeland RA. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas; Proc Natl Acad Sci USA; 2010; pp. 20980–20985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrera-Merchan A, Arranz L, Ligos JM, de Molina A, Dominguez O, Gonzalez S. Ectopic expression of the histone methyltransferase Ezh2 in haematopoietic stem cells causes myeloproliferative disease. Nat Commun. 2012;3:623. doi: 10.1038/ncomms1623. [DOI] [PubMed] [Google Scholar]
- 14.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells; Proc Natl Acad Sci USA; 2003; pp. 11606–11611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 16.Sudo T, Utsunomiya T, Mimori K, Nagahara H, Ogawa K, Inoue H, Wakiyama S, Fujita H, Shirouzu K, Mori M. Clinicopathological significance of EZH2 mRNA expression in patients with hepatocellular carcinoma. Br J Cancer. 2005;92:1754–1758. doi: 10.1038/sj.bjc.6602531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bachmann IM, Halvorsen OJ, Collett K, Stefansson IM, Straume O, Haukaas SA, Salvesen HB, Otte AP, Akslen LA. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 18.Weikert S, Christoph F, Köllermann J, Müller M, Schrader M, Miller K, Krause H. Expression levels of the EZH2 polycomb transcriptional repressor correlate with aggressiveness and invasive potential of bladder carcinomas. Int J Mol Med. 2005;16:349–353. [PubMed] [Google Scholar]
- 19.Zingg D, Debbache J, Schaefer SM, Tuncer E, Frommel SC, Cheng P, Arenas-Ramirez N, Haeusel J, Zhang Y, Bonalli M. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumor suppressors. Nat Commun. 2015;6:6051. doi: 10.1038/ncomms7051. [DOI] [PubMed] [Google Scholar]
- 20.Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, Woodward WA, Hsu JM, Hortobagyi GN, Hung MC. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-β-catenin signaling. Cancer Cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landen CN, Jr, Chavez-Reyes A, Bucana C, Schmandt R, Deavers MT, Lopez-Berestein G, Sood AK. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65:6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 22.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wittekind C, Compton CC, Greene FL, Sobin LH. TNM residual tumor classification revisited. Cancer. 2002;94:2511–2516. doi: 10.1002/cncr.10492. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Kuang Y, Lu F, Guo J, Xu H, Wang Q, Xu C, Zeng L, Yi S. Histone demethylase KDM2B upregulates histone methyltransferase EZH2 expression and contributes to the progression of ovarian cancer in vitro and in vivo. Onco Targets Ther. 2017;10:3131–3144. doi: 10.2147/OTT.S134784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirk JS, Sachaarchuch K, Dalimov Z, Lasorsa E, Ku S, Ramakrishnan S, Hu Q, Azabdaftari G, Wang J, Pili R, Ellis L. Top2a identifies and provides epigenetic rationale for novel combination therapeutic strategies for aggressive prostate cancer. Oncotarget. 2015;6:3136–3146. doi: 10.18632/oncotarget.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fillmore CM, Xu C, Desai PT, Berry JM, Rowbotham SP, Lin YJ, Zhang H, Marquez VE, Hammerman PS, Wong KK, Kim CF. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature. 2015;520:239–242. doi: 10.1038/nature14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, III, Diaz E, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 29.Qi W, Chan H, Teng L, Li L, Chuai S, Zhang R, Zeng J, Li M, Fan H, Lin Y, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation; Proc Natl Acad Sci USA; 2012; pp. 21360–21365. [DOI] [PMC free article] [PubMed] [Google Scholar]