Abstract
Cancer prevention using natural micronutrition on epigenetic mechanisms primarily revolves around plant extracts. However, the role of macronutrition, including animal peptides, on epigenetic modification in cancer has been elusive. In traditional Chinese medicine, the soft-shelled turtle has a long-history of being a functional food that strengthens immunity through unknown mechanisms. The present study aimed to investigate the impact of soft-shelled turtle peptide on microRNA (miRNA) expression in gastric cancer (GC) cells and to analyze the potential anticancer mechanisms for GC. Affymetrix GeneChip miRNA 3.0 Array and quantitative polymerase chain reaction were used to detect the miRNA expression profile in human GC AGS cells treated with the soft-shelled turtle peptide. The results demonstrated that 101 miRNAs (49 upregulated miRNAs and 52 downregulated miRNAs) were significantly differentially expressed in the AGS cells following soft-shelled turtle peptide treatment. Several tumor suppressor miRNAs were upregulated markedly, including miRNA-375, let-7d, miRNA-429, miRNA-148a/148b and miRNA-34a. Pathway analysis indicated that soft-shelled turtle peptide may function with anticancer properties through the Hippo signaling pathway and the forkhead box O signaling pathway. Therefore, these results demonstrated that soft-shelled turtle peptide has the capacity to influence cancer-related pathways through the regulation of miRNA expression in GC cells.
Keywords: soft-shelled turtle peptide, microRNA profiling, gastric cancer cells, macronutrition, nutri-epigenetics
Introduction
Cancers result from abnormal cell growth, which is caused by abnormalities in the genome and the epigenome (1). Alterations of epigenetic modifications are always reversible and can be influenced by external factors, including diet or environmental exposure (2). Given that epigenetic dysregulation occurs early during tumorigenesis, epigenetic modifications have been confirmed as potential targets for cancer prevention or cancer therapy (3). Nutri-epigenetics, which revolves around the impact of micronutritions and macronutritions on epigenetic mechanisms, has renewed the study of traditional methods in the epigenetic field. Epigenetic modifications in response to functional foods consist of DNA methylation, histone modifications and the effect of non-coding RNAs. However, it has been confirmed that a few functional foods not only play a role in health promotion, but also promote anticancer activity for several tumors. Epigallocatechin-3-gallate (EGCG) extracted from green tea inhibits DNA methylation and increases histone acetylation to recover silenced tumor suppressor genes in cancer cells (4,5). Isothiocyanates extracted from cruciferous vegetables suppress the metastasis potential in lung cancer cells by inducing apoptosis and cell cycle arrest, and inhibiting deubiquitinating enzymes associated with tumorigenesis to exert anticancer effects (6,7).
The occurrence of gastric adenocarcinoma is a stepwise process that may follow several years or decades of gastritis, intestinal metaplasia and dysplasia to malignancy with epigenetic alterations (3). Gradual epigenetic dysregulations are strongly associated with the tumorigenesis of certain inflammation-associated cancer types, particularly gastric cancer (GC). Hence, more and more natural anticancer components are being studied. The search for effective and safe natural anticancer factors, and mechanisms to reverse or counteract cancer-associated epigenetic alterations for GC prevention and interventions are required.
To date, nutraceuticals for cancer chemoprevention have primarily been derived from plants, for example, curcumin from ginger plants (8), allicin from garlic (9), resveratrol from grapes and EGCG from green tea (10,11). However, the influence of animal-derived macronutrition, including proteins or peptides, on cancer epigenetic mechanisms remains uninvestigated. Despite the fact that the consumption of red meat is undeniably associated with cancer risk (12–14), molecular mechanisms of animal peptides on epigenetic regulation have not been fully elucidated. The soft-shelled turtle is commonly consumed in China. According to excavations from the Hemudu site (Zhejiang, China), the history of eating soft-shelled turtles in Asia dates back to the Hemudu culture ~6,000 years ago in the neolithic period. The soft-shelled turtle is also used in traditional Chinese medicine where it is believed to strengthen immunity among other benefits (15). Recently, with the development of turtle aquaculture, soft-shelled turtles have been processed into various types of health products, including products in capsule and liquid forms, and are used to improve the prognosis of cancer patients undergoing radiotherapy and chemotherapy, according to the beliefs of traditional Chinese medicine (16). To the best of our knowledge, for the first time, the present study investigates the impact of soft-shelled turtle peptides on epigenetic mechanisms in GC cells. A microarray was used to detect the expression profile of microRNA (miRNA) in the human GC AGS cell line treated with soft-shelled turtle peptide. The target genes of the soft-shelled turtle peptide-specific miRNAs and corresponding pathways were further analyzed to determine the potential anticancer properties of the soft-shelled turtle peptide.
Materials and methods
Materials
The peptides extracted from soft-shelled turtles were offered by Zhejiang Agricultural Group Co., Ltd. (Hangzhou, China) and stored in the form of a dry powder in a brown glass bottle at room temperature. Every unit of the soft-shelled turtle peptide powder consisted of a polypeptide (40.68%), acid soluble protein (47.97%), free amino acids (7.29%) and hydroxyproline (4.06%). The polypeptide was the major functional component and the treatment concentration was dependent on the actual polypeptide content. The peptide powder was dissolved in cell culture medium at the required concentration immediately prior to use.
Cell culture and cell treatment
The human GC AGS cell line was purchased from American Type Culture Collection (ATCC; Manassas, VA, USA) and cultured in F12 medium supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in T-75 flasks (Corning Inc., Corning, NY, USA) at 37°C in a humidified atmosphere supplied with 5% CO2. The AGS cells were treated during the exponential phase of growth. The experimental group of cells were incubated with the soft-shelled turtle peptide dissolved in F12 medium (100 mg/l), while the control group of cells were incubated with F12 medium only. The experimental group and the control group were incubated for 72 h, and the medium was renewed every 24 h. After 72 h, the cells were collected by trypsinization.
RNA extraction, labeling and miRNA expression profiling analysis
The total RNA of AGS cells with or without soft-shelled turtle peptide treatment was extracted and purified using the mirVana™ miRNA Isolation kit without phenol (Ambion; Thermo Fisher Scientific, Inc.), following the manufacturers instructions, and checked for an RNA integrity number to inspect RNA integration using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). The genome-wide miRNA expression profiling was detected by the Affymetrix platform with Genechip miRNA 3.0 array (Affymetrix; Thermo Fisher Scientific, Inc.) based on the Sanger miRBase version 17. This chip contained 19,913 probe sets, including 1,789 mature miRNAs, 1,693 precursor miRNAs and 2,336 small nucleolar RNAs and small Cajal body-specific RNAs. After total RNA quality inspection, total RNAs of each sample were tailed with poly A and labeled with biotin by the FlashTag™ Biotin HSR RNA Labeling kit (Affymetrix; Thermo Fisher Scientific, Inc.) according to the manufacturers instructions. Hybridization of bio-labeled RNA samples was performed according to the manufacturers instructions for the Affymetrix GeneChip miRNA 3.0 Array with GeneChip Hybridization Wash and Stain kit, and GeneChip Hybridization Oven 645 (all from Affymetrix; Thermo Fisher Scientific, Inc.). After washing and staining with GeneChip Fluidics Station 450, the arrays were scanned by the GeneChip Scanner 3000 (both from Affymetrix; Thermo Fisher Scientific, Inc.).
Statistical analysis
Differentially expressed miRNAs were evaluated by the ratio of fluorescence between the control and the soft-shelled turtle peptide treated sample. Fold-change of ≥2 and ≤0.5, used for threshold values, were regarded as upregulation and downregulation, respectively. Hierarchical clustering analysis was performed to display the discrepant miRNA expression profile in two samples through red and green color blocks via Gene Cluster (version 3.0) and Java TreeView software program (bonsai.hgc.jp/~mdehoon/software/cluster/software.htm). The potential target genes of differentially expressed miRNAs in the treated cells were predicted using five databases, including TARGETMINER (www.isical.ac.in/~bioinfo_miu/targetminer20.htm), miRDB (mirdb.org), microRNA (www.mirbase.org/), TarBase (diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index) and RNA22 (cm.jefferson.edu/rna22/Interactive). Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (https://david.ncifcrf.gov/; DAVID Bioinformatics Resources 6.8) were performed to demonstrate corresponding biological processes and the regulatory network in which specific miRNA-genes may have participated (17,18).
Validation of miRNA array by reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Differentially expressed miRNAs were selected and validated according to RT-qPCR analysis with Platinum Quantitative PCR SuperMix-UDG w/ROX (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA were extracted from AGS cells with or without soft-shelled turtle peptide treatment. PCR was performed using a Applied Biosystems 7500 (Foster City, CA, USA). Amplification was performed at 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 34 sec. U6 was selected as an internal reference to normalize the miRNA expression levels, and each sample was validated in triplicate. The specific primer sequences of U6 is as follow: 5′-CGCAAGGATGACACGCAAATTC-3. The reverse transcription of poly-A tailed miRNAs using anchored Oligo dT primers for the candidate miRNAs are presented in Table I. The relative expression of differentially expressed miRNAs was evaluated according to the 2−∆∆Cq method (19).
Table I.
Reverse transcription of poly-A tailed miRNAs using anchored Oligo dT primers for the candidate miRNAs.
| microRNA | Name in mirBase | Sequence in mirBase | Length | Modified primer sequence | Primer length | |
|---|---|---|---|---|---|---|
| hsa-miR-4455 | >hsa-miR-4455 MIMAT0018977 | AGGGUGUGUGUGUUUUU | AGGGTGTGTGTGTTTTT | 17 | GCCAGGGTGTGTGTGTTTTT | 20 |
| hsa-miR-29a | >hsa-miR-29a-3p MIMAT0000086 | UAGCACCAUCUGAAAUCGGUUA | TAGCACCATCTGAAATCGGTTA | 22 | TAGCACCATCTGAAATCGGTTA | 22 |
| hsa-miR-378b | >hsa-miR-378b MIMAT0014999 | ACUGGACUUGGAGGCAGAA | ACTGGACTTGGAGGCAGAA | 19 | ACTGGACTTGGAGGCAGAA | 19 |
| hsa-miR-29b | >hsa-miR-29b-3p MIMAT0000100 | UAGCACCAUUUGAAAUCAGUGUU | TAGCACCATTTGAAATCAGTGTT | 23 | GGTAGCACCATTTGAAATCAG | 21 |
| hsa-miR-205 | >hsa-miR-205-5p MIMAT0000266 | UCCUUCAUUCCACCGGAGUCUG | TCCTTCATTCCACCGGAGTCTG | 22 | TCCTTCATTCCACCGGAGT | 19 |
| hsa-miR-100 | >hsa-miR-100-5p MIMAT0000098 | AACCCGUAGAUCCGAACUUGUG | AACCCGTAGATCCGAACTTGTG | 22 | AACCCGTAGATCCGAACTTGTG | 22 |
| hsa-miR-7 | >hsa-miR-7-5p MIMAT0000252 | UGGAAGACUAGUGAUUUUGUUGU | TGGAAGACTAGTGATTTTGTTGT | 23 | GCCTGGAAGACTAGTGATTTG | 21 |
| hsa-miR-375 | >hsa-miR-375 MIMAT0000728 | UUUGUUCGUUCGGCUCGCGUGA | TTTGTTCGTTCGGCTCGCGTGA | 22 | TGTTCGTTCGGCTCGCGTGAA | 21 |
| hsa-miR-4299 | >hsa-miR-4299 MIMAT0016851 | GCUGGUGACAUGAGAGGC | GCTGGTGACATGAGAGGC | 18 | TAGCTGGTGACATGAGAGG | 19 |
| hsa-miR-378h | >hsa-miR-378h MIMAT0018984 | ACUGGACUUGGUGUCAGAUGG | ACTGGACTTGGTGTCAGATGG | 21 | ACTGGACTTGGTGTCAGATGG | 21 |
Results
Alteration of miRNA expression profile in GC cells treated with soft-shelled turtle peptide
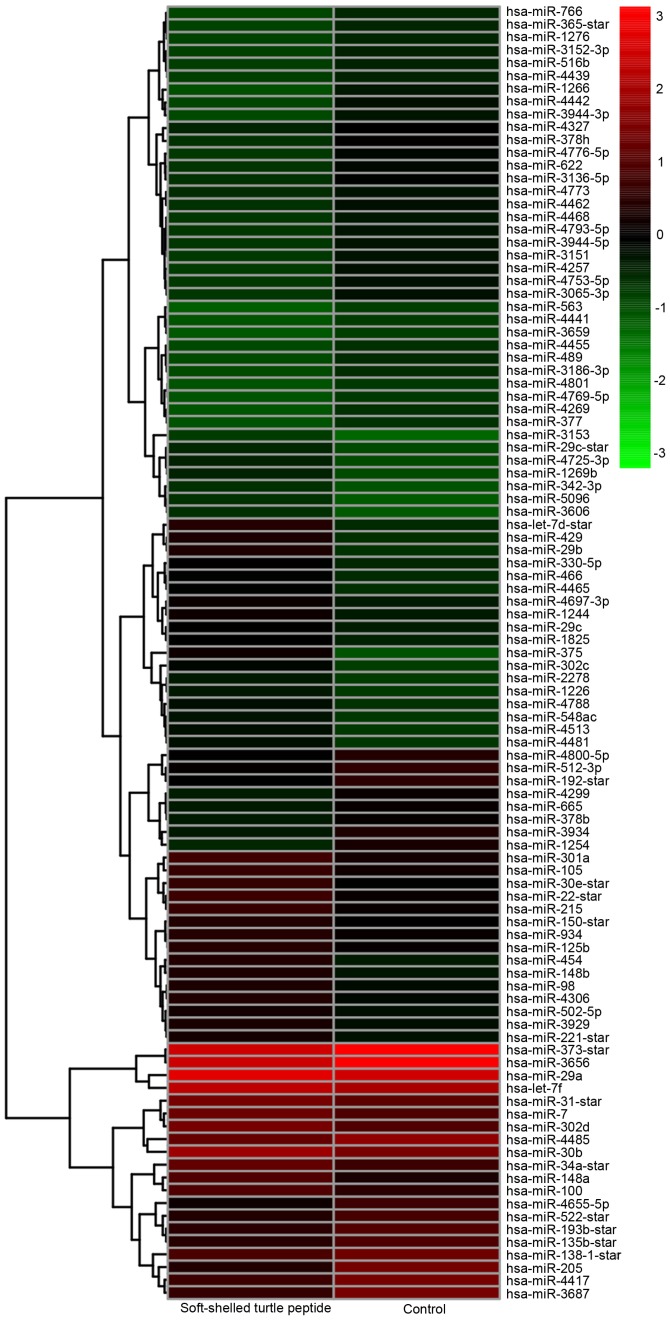
In order to evaluate the soft-shelled turtle peptide-mediated anticancer capabilities in GC cells, the AGS cells were treated with 100 mg/l soft-shelled turtle peptide for 72 h and the medium was renewed every 24 h. A total of 1,744 human miRNAs were screened to analyze the regulation of soft-shelled turtle peptide on miRNA expression in the AGS cells. Compared with the untreated cells, the miRNA expression of AGS cells with soft-shelled turtle peptide treatment varied markedly. According to the hierarchical cluster analysis, there were 101 differentially expressed miRNAs in the AGS cells treated with the soft-shelled turtle peptide (Fig. 1). Among them, 49 miRNAs were upregulated and 52 miRNAs were downregulated after treatment with the soft-shelled turtle peptide (Table II). Particularly, the expression of miRNA-375 was increased intensely by 8.32-fold and other upregulated miRNAs, including let-7d, miRNA-29b, miRNA-429, miRNA-454, miRNA-148a, miRNA-22, miRNA-30e, miRNA-302 and miRNA-148b, were altered markedly by >3-fold. Additionally, the expression of miRNA 205, miRNA-1254, miRNA-3687 and miRNA-1266 was significantly downregulated by <0.25-fold.
Figure 1.
Gene Cluster (version 3.0) and Java TreeView software programs were used to perform the hierarchical cluster analysis of differentially expressed miRNA microarray data in AGS cells treated with soft-shelled turtle peptides. A hierarchical cluster analysis of 101 miRNAs whose expression differed significantly between the soft-shelled turtle peptides-treated group (treatment for 72 h) and the untreated group (fold-change >2 or <0.5; P<0.05). Each row represents a miRNA and each column represents a sample. The miRNA expression levels are illustrated using a color key and histogram. The red and green colors denote high and low expression, respectively. miR/miRNA, microRNA.
Table II.
Differentially expressed miRNAs (upregulated >2-fold or downregulated <0.5-fold) in AGS cells following treatment with soft-shelled turtle peptides.
| miRNA | Fold-change |
|---|---|
| Upregulated | |
| hsa-miR-375 | 8.32 |
| hsa-let-7d-star | 6.05 |
| hsa-miR-29b | 5.82 |
| hsa-miR-429 | 5.40 |
| hsa-miR-454 | 4.10 |
| hsa-miR-148a | 3.66 |
| hsa-miR-22-star | 3.44 |
| hsa-miR-30e-star | 3.43 |
| hsa-miR-302c | 3.42 |
| hsa-miR-148b | 3.23 |
| hsa-miR-215 | 3.08 |
| hsa-miR-301a | 2.82 |
| hsa-miR-302d | 2.69 |
| hsa-miR-34a-star | 2.66 |
| hsa-miR-4306 | 2.62 |
| hsa-miR-100 | 2.59 |
| hsa-miR-330-5p | 2.54 |
| hsa-miR-98 | 2.52 |
| hsa-miR-4465 | 2.50 |
| hsa-miR-1244 | 2.49 |
| hsa-miR-30b | 2.46 |
| hsa-miR-4513 | 2.45 |
| hsa-miR-3929 | 2.44 |
| hsa-miR-105 | 2.43 |
| hsa-miR-221-star | 2.43 |
| hsa-miR-466 | 2.35 |
| hsa-miR-4481 | 2.35 |
| hsa-miR-3153 | 2.32 |
| hsa-miR-3606 | 2.32 |
| hsa-miR-5096 | 2.32 |
| hsa-miR-1269b | 2.32 |
| hsa-miR-4697-3p | 2.29 |
| hsa-miR-4725-3p | 2.29 |
| hsa-miR-7 | 2.21 |
| hsa-miR-1825 | 2.17 |
| hsa-miR-502-5p | 2.14 |
| hsa-miR-4788 | 2.13 |
| hsa-miR-31-star | 2.11 |
| hsa-miR-150-star | 2.06 |
| hsa-miR-29c | 2.06 |
| hsa-miR-29a | 2.05 |
| hsa-miR-29c-star | 2.04 |
| hsa-miR-342-3p | 2.04 |
| hsa-miR-1226 | 2.04 |
| hsa-miR-2278 | 2.04 |
| hsa-miR-548ac | 2.04 |
| hsa-let-7f | 2.04 |
| hsa-miR-125b | 2.02 |
| hsa-miR-934 | 2.01 |
| hsa-miR-4769-5p | 0.48 |
| hsa-miR-3656 | 0.49 |
| hsa-miR-138-1-star | 0.49 |
| hsa-miR-4773 | 0.49 |
| hsa-miR-4455 | 0.49 |
| Downregulated | |
| hsa-miR-205 | 0.17 |
| hsa-miR-1254 | 0.22 |
| hsa-miR-3687 | 0.23 |
| hsa-miR-1266 | 0.25 |
| hsa-miR-4442 | 0.27 |
| hsa-miR-378h | 0.27 |
| hsa-miR-3944-3p | 0.29 |
| hsa-miR-4776-5p | 0.29 |
| hsa-miR-4417 | 0.29 |
| hsa-miR-3934 | 0.30 |
| hsa-miR-3136-5p | 0.33 |
| hsa-miR-4257 | 0.33 |
| hsa-miR-4655-5p | 0.33 |
| hsa-miR-4753-5p | 0.36 |
| hsa-miR-192-star | 0.36 |
| hsa-miR-622 | 0.36 |
| hsa-miR-4299 | 0.38 |
| hsa-miR-4269 | 0.39 |
| hsa-miR-3065-3p | 0.39 |
| hsa-miR-4327 | 0.39 |
| hsa-miR-3151 | 0.39 |
| hsa-miR-512-3p | 0.39 |
| hsa-miR-3944-5p | 0.40 |
| hsa-miR-377 | 0.40 |
| hsa-miR-489 | 0.42 |
| hsa-miR-193b-star | 0.42 |
| hsa-miR-665 | 0.42 |
| hsa-miR-3152-3p | 0.42 |
| hsa-miR-4793-5p | 0.42 |
| hsa-miR-563 | 0.43 |
| hsa-miR-766 | 0.44 |
| hsa-miR-1276 | 0.44 |
| hsa-miR-4462 | 0.44 |
| hsa-miR-373-star | 0.44 |
| hsa-miR-4468 | 0.44 |
| hsa-miR-4485 | 0.45 |
| hsa-miR-365-star | 0.45 |
| hsa-miR-3186-3p | 0.45 |
| hsa-miR-378b | 0.45 |
| hsa-miR-135b-star | 0.45 |
| hsa-miR-4800-5p | 0.46 |
| hsa-miR-516b | 0.46 |
| hsa-miR-522-star | 0.48 |
| hsa-miR-4441 | 0.48 |
| hsa-miR-4439 | 0.48 |
| hsa-miR-3659 | 0.48 |
| hsa-miR-4801 | 0.48 |
Soft-shelled turtle peptides associated with miRNAs in previous studies of GC (20–48) were reviewed and are summarized in Table III. To further validate the microarray results, 10 miRNAs, including hsa-miR-4455, hsa-miR-29a, hsa-miR-378b, hsa-miR-29b, hsa-miR-205, hsa-miR-100, hsa-miR-7, hsa-miR-375, hsa-miR-4299 and hsa-miR-378h, were selected to be verified according to RT-qPCR analysis. Results of the RT-qPCR validation were similar to results of the microarray, indicating that the miRNA data was reliable (Fig. 2).
Table III.
Differentially expressed miRNAs induced by soft-shelled turtle peptides in GC.
| miRNA | Regulationa | Expression in the literatureb | Sourcec (ref.) |
|---|---|---|---|
| miR-375 | Upregulated | Downregulated | GC tissues, gastric epithelial with Hp infection (20) |
| GC cell lines: BGC823, MGC803, SGC7901, AGS, | |||
| N87 and MKN45 (21,22) | |||
| Plasma in GC patients (20) | |||
| miR-7d | Upregulated | Downregulated | Serum in GC patients with Hp infection (23) |
| miR-29b | Upregulated | Downregulated | GC tissues (24) |
| GC cell lines: SGC7901, AGS, MKN45, MGC803 and BGC823 (25) | |||
| miR-429 | Upregulated | Downregulated | GC tissues (26) |
| GC cell lines: BGC823, MGC803 and AGS (27–29) | |||
| miR-454 | Upregulated | / | / |
| miR-148a | Upregulated | Downregulated | GC tissues (20,30) |
| GC cell lines: AGS, BGC823, MKN1, MKN45 and MKN74 (30,31) | |||
| Plasma and serum in GC patients (20,32) | |||
| miR-22 | Upregulated | Downregulated | GC tissues (33–35) |
| GC cell lines: AGS, BGC823, HGC27 and SGC7901 (33–35) | |||
| miR-30e | Upregulated | Downregulated | GC tissues (36) |
| miR-148b | Upregulated | Downregulated | GC tissues (37) |
| GC cell lines: AGS, MGC803, SGC7901 and BGC823 (37) | |||
| miR-34a | Upregulated | Downregulated | GC tissues (38,39) |
| GC cell lines: MGC803, HGC27, NCI-N87 and SGC7901 (39,40) | |||
| Gastric MALT lymphoma tissues and B-cell lymphoma tissues (41) | |||
| miR-205 | Downregulated | Downregulated | GC tissues (42) |
| GC cell lines: AGS and NCI-N87 (42) | |||
| miR-1254 | Downregulated | / | / |
| miR-3687 | Downregulated | / | / |
| miR-1266 | Downregulated | Downregulated | GC tissues (43) |
| GC cell lines: BGC823, MKN28 and SGC7901 (43) | |||
| miR-192 | Downregulated | Upregulated | GC tissues (44,45) |
| GC cell lines: BGC823, SGC7901 and MKN45 (46) | |||
| Plasma in GC patients (46) | |||
| miR-377 | Downregulated | Upregulated | GC tissues (47) |
| GC cell lines: MKN45 (47) | |||
| miR-665 | Downregulated | Upregulated | Intestinal GC tissues (48) |
Regulation means the expression level induced by soft-shelled turtle peptides
literature represents the expression of miRNAs in previous gastric cancer-associated studies
source indicates the expression of miRNAs in different sources presented in the previous literature. miR/miRNA, microRNA; GC, gastric cancer; Hp, Helicobacter pylori; MALT, mucosa-associated lymphoid tissue; /, no relevant reference published.
Figure 2.
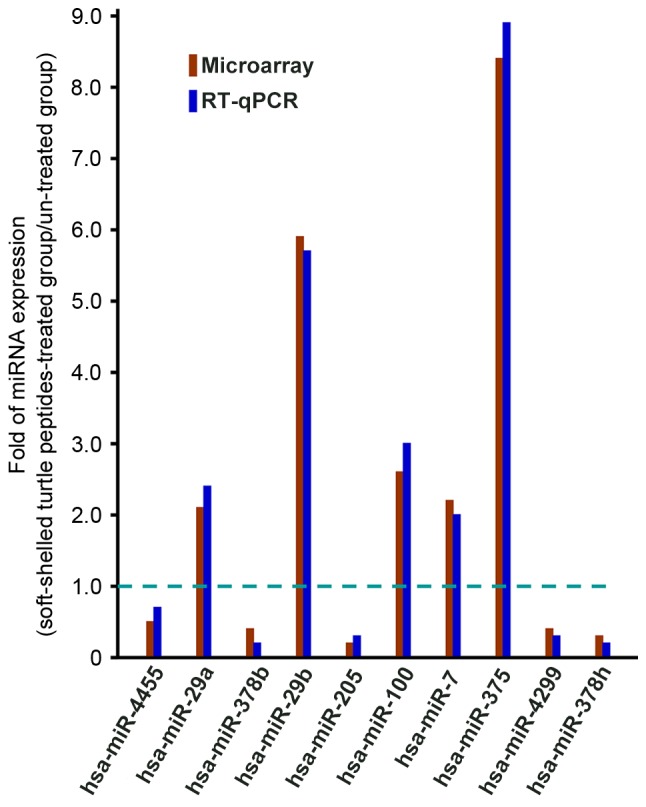
RT-qPCR validation of differentially expressed miRNAs that were screened by the microarray assay. Upregulated and downregulated miRNAs are indicated by bars above and below the dotted line respectively. The RT-qPCR results were representative of three independent experiments. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; miRNA, microRNA.
Target genes of differentially expressed miRNAs modified by soft-shelled turtle peptide
With the identification of differentially expressed miRNAs in AGS cells following treatment of the soft-shelled turtle peptide, the target genes of these miRNA were further investigated according to 5 online software programs, including TargetMiner, miRDB, microRNA, TarBase, and RNA-22 (Table IV). As presented in Table V, potential target genes were identified.
Table IV.
Predicted target genes of differentially expressed miRNAs from five miRNA databases.
| miRNA databases | ||||||
|---|---|---|---|---|---|---|
| miRNA | TargetMiner | miRDB | microRNA | TarBase | RNA22 | Total |
| Upregulated | ||||||
| hsa-miR-429 | 161 | 767 | 92 | 3,283 | 33 | 3,625 |
| hsa-miR-454 | 0 | 0 | 380 | 0 | 0 | 380 |
| hsa-miR-4306 | 3,194 | 1,689 | 402 | 3,560 | 714 | 6,716 |
| hsa-miR-98 | 2,685 | 0 | 209 | 2,426 | 0 | 4,504 |
| hsa-miR-4465 | 2,285 | 512 | 0 | 651 | 1,020 | 3,806 |
| hsa-miR-3929 | 2,763 | 484 | 0 | 3,363 | 3,169 | 7,130 |
| hsa-miR-466 | 72 | 741 | 115 | 3,225 | 745 | 3,917 |
| hsa-miR-4697-3p | 1,807 | 247 | 0 | 2,111 | 4,791 | 7,210 |
| hsa-miR-7 | 0 | 0 | 457 | 0 | 0 | 457 |
| hsa-miR-502-5p | 109 | 216 | 447 | 2,456 | 3,351 | 5,498 |
| hsa-miR-934 | 18 | 133 | 174 | 1,125 | 646 | 1,893 |
| Downregulated | ||||||
| hsa-miR-378h | 132 | 171 | 0 | 1,976 | 1,602 | 3,426 |
| hsa-miR-4417 | 1,942 | 218 | 0 | 2,175 | 8,113 | 9,630 |
| hsa-miR-3934 | 2,754 | 0 | 0 | 2,772 | 0 | 4,829 |
| hsa-miR-4655-5p | 278 | 21 | 0 | 221 | 7,511 | 7,659 |
| hsa-miR-4299 | 855 | 276 | 447 | 2,313 | 2,089 | 4,754 |
| hsa-miR-4327 | 1,405 | 190 | 396 | 1,280 | 7,696 | 9,158 |
| hsa-miR-512-3p | 1,078 | 383 | 498 | 3,066 | 2,153 | 5,475 |
| hsa-miR-665 | 966 | 612 | 1,747 | 3,158 | 6,901 | 9,266 |
| hsa-miR-4485 | 1,275 | 0 | 0 | 191 | 0 | 1,438 |
| hsa-miR-378b | 132 | 167 | 555 | 2,048 | 1,926 | 4,033 |
| hsa-miR-4800-5p | 152 | 83 | 0 | 1,296 | 1,900 | 3,093 |
| hsa-miR-522-5p | 2,001 | 206 | 0 | 460 | 1,864 | 4,057 |
Number of target genes predicted by five miRNA databases for each miRNA was different. miR/miRNA, microRNA.
Table V.
Potential target genes for miRNAs differentially expressed in response to the soft-shelled turtle peptides.
| miRNA | Target gene | Function of the encoded protein |
|---|---|---|
| hsa-miR-429 | CERS6 | Sphingolipid biosynthesis |
| MIB1 | Ubiquitination | |
| CBL | Regulation of tyrosine kinases | |
| HOOK1 | Early endosome trafficking | |
| ZNF148 | Regulation of cell growth, apoptosis | |
| hsa-miR-454 | SUN2 | Interaction with chromatin and the nuclear lamina |
| ADAM28 | Sperm maturation and immune suppression | |
| KDM2A | Cell proliferation, differentiation of mesenchymal stem cells derived from dental tissue | |
| hsa-miR-98 | COL4A2 | Assembled with COL4A1, interaction with growth factors and cell surface receptors |
| MAP4K3 | Cell growth, viability, cell death, body size and metabolism | |
| IL13 | Mediator of allergic inflammation | |
| IGF2BP1 | Embryo development, oncofetal RNA-binding protein | |
| hsa-miR-7 | S100A7A | Inflammation, tumorigenesis |
| EGFR | Cell growth, proliferation, differentiation | |
| SUCNR1 | Paired with succinate, cell metabolism | |
| ZEB2 | A key regulator of epithelial mesenchymal differentiation | |
| PIK3AP1 | Regulation of cell proliferation, survival, metabolism, cytoskeletal reorganization and membrane trafficking | |
| CD86 | Immune responses | |
| ADAM9 | Upregulation associated with tumorigenesis | |
| hsa-miR-502-5p | KIF1B | Axonal transport of synaptic vesicles and mitochondria, axon myelination and outgrowth |
| RNF14 | Mitochondrial transcription, immune function | |
| GRAP2 | Calcium influx and cytokine production | |
| hsa-miR-378h | CBL | Regulation of tyrosine kinases |
| PTCH1 | A key regulator of cell proliferation, cell surveillance, embryonic development, adult tissue homeostasis and stem cell quiescence | |
| SULF1 | Blocking the binding of growth factors and receptors, regulation of cell growth and differentiation | |
| TRAF3 | Regulation of signaling through B-lymphocyte receptors, inhibition of B-cell homoeostatic survival | |
| CELF2 | Regulation of mRNA editing, stability and translation | |
| hsa-miR-665 | SRF | A transcription factor for coupling actin dynamics and signaling pathways to gene expression |
| SMAD7 | An antagonist of TGF-β signaling | |
| BCL7B | Negatively regulating the Wnt-signaling pathway and positively regulating the apoptotic pathway | |
| ELK1 | Regulation of proto-oncogene c-fos | |
| FGF9 | Epithelial-mesenchymal interaction | |
| hsa-miR-4417 | PCDH17 | Functioning as a tumor suppressor |
| FOXO1 | Stopping cell cycle progression, promotes apoptosis, and negatively regulating angiogenesis | |
| IGFBP5 | Inducing extracellular matrix production and deposition | |
| SH2B3 | A negative regulator of multiple cytokine signaling pathways | |
| RAB6A | Regulating intracellular vesicle transport |
miR/miRNA, microRNA.
GO analysis and pathway analysis
To further evaluate the role of soft-shelled turtle peptide influenced miRNAs in physiological functions and pathological processes in GC, interactions between miRNAs and associated target genes were predicted according to GO analysis and pathway analysis. The GO cellular component analysis demonstrated that target genes were mainly clustered into the extracellular region, extracellular space, keratin filament and nucleus categories (Fig. 3). The GO molecular function analysis indicated that the bulk of the target genes participated in olfactory receptor activity and G-protein coupled receptor activity (Fig. 3). In addition, the GO biological process analysis demonstrated that these target genes were closely associated with the detection of chemical stimuli involved in the sensory perception of smell and the G-protein coupled receptor signaling pathway (Fig. 3). However, the pathway analysis indicated that the target genes were associated with the thyroid hormone signaling pathway, the Hippo signaling pathway, the forkhead box O (FoxO) signaling pathway, the AMP-activated protein kinase signaling pathway, the mechanistic target of rapamycin signaling pathway and transcriptional misregulation in cancer, among others (Fig. 4).
Figure 3.
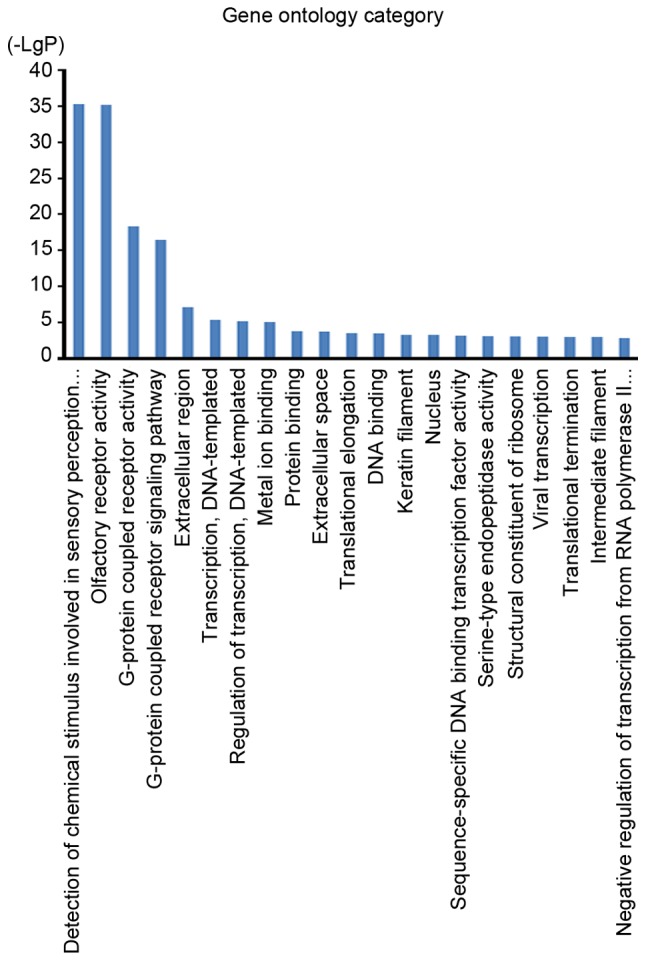
GO analysis for target genes of differentially expressed miRNAs. To investigate the biological functions of the differentially expressed miRNAs, public online databases were used to obtain miRNA-regulated putative target genes. A GO function analysis of miRNA targets was performed to obtain the significantly over-represented GO terms (P<0.01). GO category for target genes indicated that several important GO terms were possibly associated with cell activation, including intracellular signal transduction, protein phosphorylation, metabolic process and the positive regulation of genes. GO, Gene Ontology; miRNA, microRNA.
Figure 4.
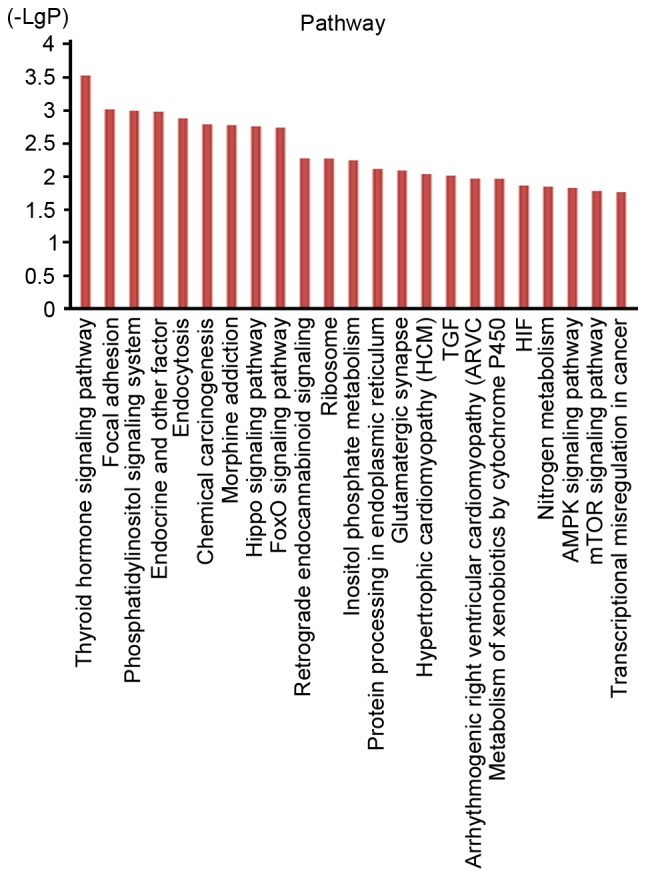
Pathway analysis for target genes of differentially expressed miRNAs. The target genes of soft-shelled turtle peptides related miRNAs were predicted with Kyoto Encyclopedia of Genes and Genomes pathway analysis. miRNA, microRNA; FoxO, forkhead box O; TGF, transforming growth factor; HIF, hypoxia-inducible factor; AMPK, AMP-activated protein kinase; mTOR, mechanistic target of rapamycin.
Discussion
Dietary components function as cancer chemoprevention agents principally through improving the nutritional supplement, enhancing immunity or directly blocking cancer cells (49). Soft-shelled turtle derived proteins can alleviate the side effects of cancer patients undergoing chemotherapy and radiotherapy, including myelosuppression (16). In the present study, the primary focus was upon the direct influence of soft-shelled turtle extract on the miRNA expression profile in human GC AGS cells.
From the results of the miRNA array, a total of 101 differentially expressed miRNAs, including 49 upregulated miRNAs and 52 downregulated miRNAs, were identified. Among the 49 miRNAs with increased expression, the expression of miRNA-375 was altered markedly, with 8.32-fold upregulation. The expression of miRNA-375 has been identified to be frequently downregulated in various types of cancer, including GC, head and neck squamous cell carcinoma, esophageal cancer, pancreatic ductal adenocarcinoma and hepatocellular carcinoma (21,50–53). In GC, miRNA-375 may function as a tumor suppressor. Overexpression of miRNA-375 could inhibit the proliferation of GC cells by targeting Janus kinase 2 (21). Moreover, Snail, a metastasis-associated transcription factor, directly binds to the putative promoter of miR-375 (54). Chang et al (55) also identified that the expression of miRNA-375 was dependent on Helicobacter pylori infection in GC; the miRNA-375 expression level in H. pylori-positive GC tissues was lower than that in the H. pylori-negative GC tissues. In the present study, the expression of miRNA-375 was upregulated to the greatest extent following treatment with the soft-shelled turtle peptide. This indicated that the soft-shelled turtle peptide may have the capacity to block the proliferation and metastasis of GC cells by increasing the expression of miRNA-375. Other significantly upregulated miRNAs, including let-7d, miRNA-29b, miRNA-429, miRNA-22, miRNA-148a/148b and miRNA-34a, played an anticancer role in GC. miRNA-429 acts as a tumor suppressor and has been identified to be downregulated in certain cancer types, including GC (26), clear cell renal cell carcinoma (56) and glioblastoma multiforme (57). It has been demonstrated that miRNA-429 induces the apoptosis of glioblastoma cells via B-cell lymphoma 2 (Bcl-2), while the suppression of miRNA-429 promotes Bcl-2-mediated cancer cell survival (57). Other studies indicated that c-Myc was a critical target gene of miRNA-429, and that miRNA-429 significantly downregulated the expression of c-myc to control the growth of GC cells (26). Additionally, miRNA-429 was demonstrated to not only inhibit GC cell growth, but to regulate GC cell invasiveness through zinc finger E-box-binding homeobox protein (58). miRNA-148a and miRNA-148b play important roles, functioning as tumor suppressors in GC. However, downregulation of miRNA-148a contributes to GC lymph node-metastasis and progression, while upregulation of miRNA-148b suppresses GC cell growth. These two miRNAs are therefore potential therapeutic target candidates (37,59). However, in the study, it was revealed that after treatment with soft-shelled turtle peptide, these suppressor function-associated miRNAs whose expression was restrained in GC cells were significantly restored to an upregulated state. This indicated that the soft-shelled turtle peptide not only improves immunity to contend against cancer cells indirectly in vivo, but that it also has the capacity to block GC cell growth directly through regulation of associated miRNAs and target genes. Furthermore, certain miRNAs, including miRNA-375, could be detected in plasma and may become potential biomarkers for the dynamic monitoring and evaluation of cancer prognosis (20,60).
Olfactory receptor neurons express olfactory receptor gene to detect odor molecules and transport the action potential to the olfactory bulb of the brain. Olfactory receptors are G protein-coupled receptors that are mainly expressed in the olfactory epithelium to detect odorants (61). However, besides olfactory tissues, non-olfactory tissues have also recently been demonstrated to exhibit the expression of olfactory signaling components (olfactory receptor, olfactory G-protein, adenylate cyclase III and olfactory marker protein) (62,63). The role of olfactory signaling components in non-olfactory tissues has not been clarified. Nevertheless, it has been demonstrated that olfactory receptors are not only involved in monitoring extracellular chemical cues, but that they are also frequently overexpressed in various types of cancer cells (64,65). In the present study, the GO analysis results indicated that target genes of soft-shelled turtle peptide-specific miRNAs were involved in a number of essential biological processes. Moreover, two biological processes, namely detection of chemical stimuli involved in the sensory perception of smell and the G-protein coupled receptor signaling pathway, were closely associated with the target genes. The role of olfactory receptor-associated events in GC has not been reported. In pancreatic cancer cells, the involvement of PI3 kinase γ-dependent signaling pathway in the promotion of tumor cell invasiveness could be triggered by olfactory receptor stimulation (64). Hence, the tight association between olfactory receptors and soft-shelled turtle peptide-specific miRNA-gene indicated that soft-shelled turtle peptide may influence the abilities of growth, metastasis and invasion in cancer cells through the regulation of olfactory receptor-associated events.
In the present study, KEGG analysis revealed that the target genes were associated with several cancer-related pathways. Particularly, the Hippo signaling pathway and the FoxO signaling pathway, which function as tumor suppressors, may be regulated by soft-shelled turtle peptide. The major functions of the Hippo signaling pathway have been defined as restricting cell growth and modulating cell proliferation and differentiation in developing organs (66,67). Transcriptional coactivator YAP1/telomere length regulator Taz1 (YAP1/TAZ) are the most important effectors and are negatively regulated by the Hippo pathway through phosphorylation-dependent and -independent mechanisms. The upregulation of YAP1 and TAZ induces epithelial-mesenchymal transition and increases drug resistance in cancer cells (68–70). Deregulation of the Hippo signaling pathway is involved in the initiation, progression and metastasis of a number of cancer types (71–73), including GC (74). Hence, the Hippo pathway has been speculated to be a drug target inhibitor of YAP1 and TAZ in cancer therapy (75,76). Additionally, miRNA-375 has been identified to target the Hippo pathway effector YAP to inhibit the proliferation and invasion of liver cancer cells (77). Therefore, we hypothesize that soft-shelled turtle peptide may exert anticancer functions through the miRNA-375-Hippo pathway. FoxO factors not only play an anticancer role in various types of tumors (78,79), but also contribute to extreme longevity and life span (80). miRNA-22 forms a regulatory loop to fine-tune the dynamics of the phosphatase and tensin homolog/AKT/FoxO1 pathway (81). miRNA-30d induces apoptosis and is regulated by the AKT/FoxO pathway in renal cell carcinoma, and is formed as the AKT/FoxO/miR-30d/metastasis adhesion protein signaling transduction pathway (82).
In conclusion, the microarray results of the present study indicated that the soft-shelled turtle peptide has the potential function to influence cancer-related miRNAs and pathways. However, the anticancer properties of soft-shelled turtle peptide in GC require further validation in vitro and in vivo.
Acknowledgements
This study was supported in part by Special Grants from the Zhejiang Agricultural Group Co., Ltd., Scientific Research Grants in Traditional Chinese Medicine, Zhejiang Province of China (no. 2012ZB018), Research Grants from the National Natural Science Foundation of China (nos. 81372332 and 81572822) and Key Projects from the Zhejiang Provincial Natural Science Foundation of China (no. LZ13H160002 and LZ18H160002), and was partly sponsored by Zhejiang Provincial Program for the Cultivation of High-level Innovative Health talents (no. Zjwjw2014-108) and the Major Training Personnel from Zhejiang Provincial Program for the Training and Development Project for 151 talents (no. Zjhrss2014-150).
References
- 1.Toyota M, Suzuki H, Yamamoto E, Yamano H, Imai K, Shinomura Y. Integrated analysis of genetic and epigenetic alterations in cancer. Epigenomics. 2009;1:291–299. doi: 10.2217/epi.09.20. [DOI] [PubMed] [Google Scholar]
- 2.Gerhauser C. Cancer chemoprevention and nutriepigenetics: State of the art and future challenges. Top Curr Chem. 2013;329:73–132. doi: 10.1007/128_2012_360. [DOI] [PubMed] [Google Scholar]
- 3.Lu XX, Yu JL, Ying LS, Han J, Wang S, Yu QM, Wang XB, Fang XH, Ling ZQ. Stepwise cumulation of RUNX3 methylation mediated by Helicobacter pylori infection contributes to gastric carcinoma progression. Cancer. 2012;118:5507–5517. doi: 10.1002/cncr.27604. [DOI] [PubMed] [Google Scholar]
- 4.Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, Welsh W, Yang CS. Tea polyphenol (−)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563–7570. [PubMed] [Google Scholar]
- 5.Nandakumar V, Vaid M, Katiyar SK. (−)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis. 2011;32:537–544. doi: 10.1093/carcin/bgq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan H, Zhu Y, Liu B, Wu H, Li Y, Wu X, Zhou Q, Xu K. Mitogen-activated protein kinase mediates the apoptosis of highly metastatic human non-small cell lung cancer cells induced by isothiocyanates. Br J Nutr. 2011;106:1779–1791. doi: 10.1017/S0007114511002315. [DOI] [PubMed] [Google Scholar]
- 7.Lawson AP, Long MJ, Coffey RT, Qian Y, Weerapana E, El Oualid F, Hedstrom L. Naturally occurring isothiocyanates exert anticancer effects by inhibiting deubiquitinating enzymes. Cancer Res. 2015;75:5130–5142. doi: 10.1158/0008-5472.CAN-15-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan F, Ding Y, Zhang Y, Zhou Y, Li M, Wang C. Curcumin suppresses proliferation and migration of MDA-MB-231 breast cancer cells through autophagy-dependent Akt degradation. PLoS One. 2016;11:e0146553. doi: 10.1371/journal.pone.0146553. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Chhabria SV, Akbarsha MA, Li AP, Kharkar PS, Desai KB. In situ allicin generation using targeted alliinase delivery for inhibition of MIA PaCa-2 cells via epigenetic changes, oxidative stress and cyclin-dependent kinase inhibitor (CDKI) expression. Apoptosis. 2015;20:1388–1409. doi: 10.1007/s10495-015-1159-4. [DOI] [PubMed] [Google Scholar]
- 10.Sinha D, Sarkar N, Biswas J, Bishayee A. Resveratrol for breast cancer prevention and therapy: Preclinical evidence and molecular mechanisms. Semin Cancer Biol. 2016;40-41:1–232. doi: 10.1016/j.semcancer.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Park JS, Khoi PN, Joo YE, Lee YH, Lang SA, Stoeltzing O, Jung YD. EGCG inhibits recepteur dorigine nantais expression by suppressing Egr-1 in gastric cancer cells. Int J Oncol. 2013;42:1120–1126. doi: 10.3892/ijo.2013.1775. [DOI] [PubMed] [Google Scholar]
- 12.Wang H, Iwasaki M, Haiman CA, Kono S, Wilkens LR, Keku TO, Berndt SI, Tsugane S, Le Marchand L. Interaction between red meat intake and NAT2 genotype in increasing the risk of colorectal cancer in Japanese and African Americans. PLoS One. 2015;10:e0144955. doi: 10.1371/journal.pone.0144955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taunk P, Hecht E, Stolzenberg-Solomon R. Are meat and heme iron intake associated with pancreatic cancer? Results from the NIH-AARP Diet and Health Cohort. Int J Cancer. 2015;138:2172–2189. doi: 10.1002/ijc.29964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lippi G, Mattiuzzi C, Cervellin G. Meat consumption and cancer risk: A critical review of published meta-analyses. Crit Rev Oncol Hematol. 2016;97:1–14. doi: 10.1016/j.critrevonc.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Zhao CG, Shen YM. Anti-cancer effect and edible methods of soft-shelled turtle. China Agricultural Publishing House; Beijing: 2008. [Google Scholar]
- 16.Fu JJ, Tan SL, Li YG, Lv H, Zhu WF, Liu HN. Adjuvant effects of snapping turtle co-peptide (STCP) on radiotherapy for cancer. J Jiangxi Uni Trad Chin Med. 2015;27:68–71. (In Chinese) [Google Scholar]
- 17.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Juzėnas S, Saltenienė V, Kupcinskas J, Link A, Kiudelis G, Jonaitis L, Jarmalaite S, Kupcinskas L, Malfertheiner P, Skieceviciene J. Analysis of deregulated microRNAs and their target genes in gastric cancer. PLoS One. 2015;10:e0132327. doi: 10.1371/journal.pone.0132327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding L, Xu Y, Zhang W, Deng Y, Si M, Du Y, Yao H, Liu X, Ke Y, Si J, Zhou T. miR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010;20:784–793. doi: 10.1038/cr.2010.79. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Xing R, Zhang X, Dong W, Zhang J, Yan Z, Li W, Cui J, Lu Y. miR-375 targets the p53 gene to regulate cellular response to ionizing radiation and etoposide in gastric cancer cells. DNA Repair (Amst) 2013;12:741–750. doi: 10.1016/j.dnarep.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Shiotani A, Murao T, Kimura Y, Matsumoto H, Kamada T, Kusunoki H, Inoue K, Uedo N, Iishi H, Haruma K. Identification of serum miRNAs as novel non-invasive biomarkers for detection of high risk for early gastric cancer. Br J Cancer. 2013;109:2323–2330. doi: 10.1038/bjc.2013.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui H, Wang L, Gong P, Zhao C, Zhang S, Zhang K, Zhou R, Zhao Z, Fan H. Deregulation between miR-29b/c and DNMT3A is associated with epigenetic silencing of the CDH1 gene, affecting cell migration and invasion in gastric cancer. PLoS One. 2015;10:e0123926. doi: 10.1371/journal.pone.0123926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lang N, Liu M, Tang QL, Chen X, Liu Z, Bi F. Effects of microRNA-29 family members on proliferation and invasion of gastric cancer cell lines. Chin J Cancer. 2010;29:603–610. doi: 10.5732/cjc.009.10597. [DOI] [PubMed] [Google Scholar]
- 26.Sun T, Wang C, Xing J, Wu D. miR-429 modulates the expression of c-myc in human gastric carcinoma cells. Eur J Cancer. 2011;47:2552–2559. doi: 10.1016/j.ejca.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 27.Liu D, Xia P, Diao D, Cheng Y, Zhang H, Yuan D, Huang C, Dang C. miRNA-429 suppresses the growth of gastric cancer cells in vitro. J Biomed Res. 2012;26:389–393. doi: 10.7555/JBR.26.20120029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ning X, Shi Z, Liu X, Zhang A, Han L, Jiang K, Kang C, Zhang Q. DNMT1 and EZH2 mediated methylation silences the microRNA-200b/a/429 gene and promotes tumor progression. Cancer Lett. 2015;359:198–205. doi: 10.1016/j.canlet.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Zhu P, Zhang J, Zhu J, Shi J, Zhu Q, Gao Y. MiR-429 induces gastric carcinoma cell apoptosis through Bcl-2. Cell Physiol Biochem. 2015;37:1572–1580. doi: 10.1159/000438524. [DOI] [PubMed] [Google Scholar]
- 30.Sakamoto N, Naito Y, Oue N, Sentani K, Uraoka N, Zarni Oo H, Yanagihara K, Aoyagi K, Sasaki H, Yasui W. MicroRNA-148a is downregulated in gastric cancer, targets MMP7, and indicates tumor invasiveness and poor prognosis. Cancer Sci. 2014;105:236–243. doi: 10.1111/cas.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuo J, Xia J, Ju F, Yan J, Zhu A, Jin S, Shan T, Zhou H. MicroRNA-148a can regulate runt-related transcription factor 3 gene expression via modulation of DNA methyltransferase 1 in gastric cancer. Mol Cells. 2013;35:313–319. doi: 10.1007/s10059-013-2314-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim SY, Jeon TY, Choi CI, Kim DH, Kim DH, Kim GH, Ryu DY, Lee BE, Kim HH. Validation of circulating miRNA biomarkers for predicting lymph node metastasis in gastric cancer. J Mol Diagn. 2013;15:661–669. doi: 10.1016/j.jmoldx.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 33.Zuo QF, Cao LY, Yu T, Gong L, Wang LN, Zhao YL, Xiao B, Zou QM. MicroRNA-22 inhibits tumor growth and metastasis in gastric cancer by directly targeting MMP14 and Snail. Cell Death Dis. 2015;6:e2000. doi: 10.1038/cddis.2014.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang Y, Liu X, Su B, Zhang Z, Zeng X, Lei Y, Shan J, Wu Y, Tang H, Su Q. microRNA-22 acts as a metastasis suppressor by targeting metadherin in gastric cancer. Mol Med Rep. 2015;11:454–460. doi: 10.3892/mmr.2014.2682. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Yu H, Lu X, Zhang P, Wang M, Hu Y. miR-22 suppresses the proliferation and invasion of gastric cancer cells by inhibiting CD151. Biochem Biophys Res Commun. 2014;445:175–179. doi: 10.1016/j.bbrc.2014.01.160. [DOI] [PubMed] [Google Scholar]
- 36.Sugihara H, Ishimoto T, Watanabe M, Sawayama H, Iwatsuki M, Baba Y, Komohara Y, Takeya M, Baba H. Identification of miR-30e* regulation of Bmi1 expression mediated by tumor-associated macrophages in gastrointestinal cancer. PLoS One. 2013;8:e81839. doi: 10.1371/journal.pone.0081839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song YX, Yue ZY, Wang ZN, Xu YY, Luo Y, Xu HM, Zhang X, Jiang L, Xing CZ, Zhang Y. MicroRNA-148b is frequently down-regulated in gastric cancer and acts as a tumor suppressor by inhibiting cell proliferation. Mol Cancer. 2011;10:1. doi: 10.1186/1476-4598-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang B, Huang J, Liu H, Guo W, Li G. miR-335 directly, while miR-34a indirectly modulate survivin expression and regulate growth, apoptosis, and invasion of gastric cancer cells. Tumour Biol. 2016;37:1771–1779. doi: 10.1007/s13277-015-3951-8. [DOI] [PubMed] [Google Scholar]
- 39.Peng Y, Guo JJ, Liu YM, Wu XL. MicroRNA-34A inhibits the growth, invasion and metastasis of gastric cancer by targeting PDGFR and MET expression. Biosci Rep. 2014;34:e00112. doi: 10.1042/BSR20140020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Cao W, Fan R, Wang L, Cheng S, Li H, Jiang J, Geng M, Jin Y, Wu Y. Expression and regulatory function of miRNA-34a in targeting survivin in gastric cancer cells. Tumour Biol. 2013;34:963–971. doi: 10.1007/s13277-012-0632-8. [DOI] [PubMed] [Google Scholar]
- 41.He M, Gao L, Zhang S, Tao L, Wang J, Yang J, Zhu M. Prognostic significance of miR-34a and its target proteins of FOXP1, p53, and BCL2 in gastric MALT lymphoma and DLBCL. Gastric Cancer. 2014;17:431–441. doi: 10.1007/s10120-013-0313-3. [DOI] [PubMed] [Google Scholar]
- 42.Yin WZ, Li F, Zhang L, Ren XP, Zhang N, Wen JF. Down-regulation of microRNA-205 promotes gastric cancer cell proliferation. Eur Rev Med Pharmacol Sci. 2014;18:1027–1032. [PubMed] [Google Scholar]
- 43.Chen L, Lü MH, Zhang D, Hao NB, Fan YH, Wu YY, Wang SM, Xie R, Fang DC, Zhang H, et al. miR-1207-5p and miR-1266 suppress gastric cancer growth and invasion by targeting telomerase reverse transcriptase. Cell Death Dis. 2014;5:e1034. doi: 10.1038/cddis.2013.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu YJ, Fan Y. miR-215/192 participates in gastric cancer progression. Clin Transl Oncol. 2015;17:34–40. doi: 10.1007/s12094-014-1194-6. [DOI] [PubMed] [Google Scholar]
- 45.Jin Z, Selaru FM, Cheng Y, Kan T, Agarwal R, Mori Y, Olaru AV, Yang J, David S, Hamilton JP, et al. MicroRNA-192 and −215 are upregulated in human gastric cancer in vivo and suppress ALCAM expression in vitro. Oncogene. 2011;30:1577–1585. doi: 10.1038/onc.2010.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Q, Ge X, Zhang Y, Xia H, Yuan D, Tang Q, Chen L, Pang X, Leng W, Bi F. Plasma miR-122 and miR-192 as potential novel biomarkers for the early detection of distant metastasis of gastric cancer. Oncol Rep. 2014;31:1863–1870. doi: 10.3892/or.2014.3004. [DOI] [PubMed] [Google Scholar]
- 47.Wen X, Wu JQ, Peng W, Feng JF, Tang JH. MicroRNA-377 predicts poor clinical outcome of gastric cancer and induces tumorigenesis by targeting multiple tumor-suppressor genes. Oncol Rep. 2015;34:203–210. doi: 10.3892/or.2015.3981. [DOI] [PubMed] [Google Scholar]
- 48.Chen J, Sun D, Chu H, Gong Z, Zhang C, Gong B, Li Y, Li N, Jiang L. Screening of differential microRNA expression in gastric signet ring cell carcinoma and gastric adenocarcinoma and target gene prediction. Oncol Rep. 2015;33:2963–2971. doi: 10.3892/or.2015.3935. [DOI] [PubMed] [Google Scholar]
- 49.Link A, Balaguer F, Goel A. Cancer chemoprevention by dietary polyphenols: Promising role for epigenetics. Biochem Pharmacol. 2010;80:1771–1792. doi: 10.1016/j.bcp.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Avissar M, Christensen BC, Kelsey KT, Marsit CJ. MicroRNA expression ratio is predictive of head and neck squamous cell carcinoma. Clin Cancer Res. 2009;15:2850–2855. doi: 10.1158/1078-0432.CCR-08-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mathé EA, Nguyen GH, Bowman ED, Zhao Y, Budhu A, Schetter AJ, Braun R, Reimers M, Kumamoto K, Hughes D, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: Associations with survival. Clin Cancer Res. 2009;15:6192–6200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhatti I, Lee A, James V, Hall RI, Lund JN, Tufarelli C, Lobo DN, Larvin M. Knockdown of microRNA-21 inhibits proliferation and increases cell death by targeting programmed cell death 4 (PDCD4) in pancreatic ductal adenocarcinoma. J Gastrointest Surg. 2011;15:199–208. doi: 10.1007/s11605-010-1381-x. [DOI] [PubMed] [Google Scholar]
- 53.Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, Zucman-Rossi J. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 54.Xu Y, Jin J, Liu Y, Huang Z, Deng Y, You T, Zhou T, Si J, Zhuo W. Snail-regulated MiR-375 inhibits migration and invasion of gastric cancer cells by targeting JAK2. PLoS One. 2014;9:e99516. doi: 10.1371/journal.pone.0099516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang H, Kim N, Park JH, Nam RH, Choi YJ, Lee HS, Yoon H, Shin CM, Park YS, Kim JM, Lee DH. Different microRNA expression levels in gastric cancer depending on Helicobacter pylori infection. Gut Liver. 2015;9:188–196. doi: 10.5009/gnl13371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen D, Li Y, Li Y, Jin L, Su Z, Yu Z, Yang S, Mao X, Lai Y. Tumor suppressive microRNA429 regulates cellular function by targeting VEGF in clear cell renal cell carcinoma. Mol Med Rep. 2016;13:1361–1366. doi: 10.3892/mmr.2015.4653. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Z, Zhou Q, Miao Y, Tian H, Li Y, Feng X, Song X. MiR-429 induces apoptosis of glioblastoma cell through Bcl-2. Tumour Biol. 2015 Oct 28; doi: 10.1007/s13277-015-4291-4. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 58.Liu W, An J, Li K, Hou H. miR-429 regulates gastric cancer cell invasiveness through ZEB proteins. Tumour Biol. 2015 Oct 15; doi: 10.1007/s13277-015-4094-7. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 59.Zheng B, Liang L, Wang C, Huang S, Cao X, Zha R, Liu L, Jia D, Tian Q, Wu J, et al. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin Cancer Res. 2011;17:7574–7583. doi: 10.1158/1078-0432.CCR-11-1714. [DOI] [PubMed] [Google Scholar]
- 60.Patel M, Verma A, Aslam I, Pringle H, Singh B. Novel plasma microRNA biomarkers for the identification of colitis-associated carcinoma. Lancet. 2015;385(Suppl 1):S78. doi: 10.1016/S0140-6736(15)60393-2. [DOI] [PubMed] [Google Scholar]
- 61.Touhara K, Sengoku S, Inaki K, Tsuboi A, Hirono J, Sato T, Sakano H, Haga T. Functional identification and reconstitution of an odorant receptor in single olfactory neurons; Proc Natl Acad Sci USA; 1999; pp. 4040–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feldmesser E, Olender T, Khen M, Yanai I, Ophir R, Lancet D. Widespread ectopic expression of olfactory receptor genes. BMC Genomics. 2006;7:121. doi: 10.1186/1471-2164-7-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang N, Kim H, Jae Y, Lee N, Ku CR, Margolis F, Lee EJ, Bahk YY, Kim MS, Koo J. Olfactory marker protein expression is an indicator of olfactory receptor-associated events in non-olfactory tissues. PLoS One. 2015;10:e0116097. doi: 10.1371/journal.pone.0116097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sanz G, Leray I, Dewaele A, Sobilo J, Lerondel S, Bouet S, Grébert D, Monnerie R, Pajot-Augy E, Mir LM. Promotion of cancer cell invasiveness and metastasis emergence caused by olfactory receptor stimulation. PLoS One. 2014;9:e85110. doi: 10.1371/journal.pone.0085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gelis L, Jovancevic N, Bechara FG, Neuhaus EM, Hatt H. Functional expression of olfactory receptors in human primary melanoma and melanoma metastasis. Exp Dermatol. 2017;26:569–576. doi: 10.1111/exd.13316. [DOI] [PubMed] [Google Scholar]
- 66.Zeng Q, Hong W. The emerging role of the Hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell. 2008;13:188–192. doi: 10.1016/j.ccr.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 67.Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao Y, Yang X. The Hippo pathway in chemotherapeutic drug resistance. Int J Cancer. 2015;137:2767–2773. doi: 10.1002/ijc.29293. [DOI] [PubMed] [Google Scholar]
- 69.Xie D, Cui J, Xia T, Jia Z, Wang L, Wei W, Zhu A, Gao Y, Xie K, Quan M. Hippo transducer TAZ promotes epithelial mesenchymal transition and supports pancreatic cancer progression. Oncotarget. 2015;6:35949–35963. doi: 10.18632/oncotarget.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Z, Wang Y, Zhu Y, Yuan C, Wang D, Zhang W, Qi B, Qiu J, Song X, Ye J, et al. The Hippo transducer TAZ promotes epithelial to mesenchymal transition and cancer stem cell maintenance in oral cancer. Mol Oncol. 2015;9:1091–1105. doi: 10.1016/j.molonc.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yagi H, Asanoma K, Ohgami T, Ichinoe A, Sonoda K, Kato K. GEP oncogene promotes cell proliferation through YAP activation in ovarian cancer. Oncogene. 2016;35:4471–4480. doi: 10.1038/onc.2015.505. [DOI] [PubMed] [Google Scholar]
- 72.Lamar JM, Stern P, Liu H, Schindler JW, Jiang ZG, Hynes RO. The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain; Proc Natl Acad Sci USA; 2012; pp. E2441–E2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang G, Lu X, Dey P, Deng P, Wu CC, Jiang S, Fang Z, Zhao K, Konaparthi R, Hua S, et al. Targeting YAP-dependent MDSC infiltration impairs tumor progression. Cancer Dis. 2016;6:80–95. doi: 10.1158/2159-8290.CD-15-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou GX, Li XY, Zhang Q, Zhao K, Zhang CP, Xue CH, Yang K, Tian ZB. Effects of the hippo signaling pathway in human gastric cancer. Asian Pac J Cancer Prev. 2013;14:5199–5205. doi: 10.7314/APJCP.2013.14.9.5199. [DOI] [PubMed] [Google Scholar]
- 75.Perra A, Kowalik MA, Ghiso E, Ledda-Columbano GM, Di Tommaso L, Angioni MM, Raschioni C, Testore E, Roncalli M, Giordano S, Columbano A. YAP activation is an early event and a potential therapeutic target in liver cancer development. J Hepatol. 2014;61:1088–1096. doi: 10.1016/j.jhep.2014.06.033. [DOI] [PubMed] [Google Scholar]
- 76.Santucci M, Vignudelli T, Ferrari S, Mor M, Scalvini L, Bolognesi ML, Uliassi E, Costi MP. The Hippo pathway and YAP/TAZ-TEAD protein-protein interaction as targets for regenerative medicine and cancer treatment. J Med Chem. 2015;58:4857–4873. doi: 10.1021/jm501615v. [DOI] [PubMed] [Google Scholar]
- 77.Liu AM, Poon RT, Luk JM. MicroRNA-375 targets Hippo-signaling effector YAP in liver cancer and inhibits tumor properties. Biochem Biophys Res Commun. 2010;394:623–627. doi: 10.1016/j.bbrc.2010.03.036. [DOI] [PubMed] [Google Scholar]
- 78.Guan H, Tan P, Xie L, Mi B, Fang Z, Li J, Yue J, Liao H, Li F. FOXO1 inhibits osteosarcoma oncogenesis via Wnt/β-catenin pathway suppression. Oncogenesis. 2015;4:e166. doi: 10.1038/oncsis.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bullock M. FOXO factors and breast cancer: Outfoxing endocrine resistance. Endocr Relat Cancer. 2016;23:R113–R130. doi: 10.1530/ERC-15-0461. [DOI] [PubMed] [Google Scholar]
- 80.Martins R, Lithgow GJ, Link W. Long live FOXO: Unraveling the role of FOXO proteins in aging and longevity. Aging Cell. 2016;15:196–207. doi: 10.1111/acel.12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bar N, Dikstein R. miR-22 forms a regulatory loop in PTEN/AKT pathway and modulates signaling kinetics. PloS One. 2010;5:e10859. doi: 10.1371/journal.pone.0010859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu C, Jin B, Chen L, Zhuo D, Zhang Z, Gong K, Mao Z. MiR-30d induces apoptosis and is regulated by the Akt/FOXO pathway in renal cell carcinoma. Cell Signal. 2013;25:1212–1221. doi: 10.1016/j.cellsig.2013.01.028. [DOI] [PubMed] [Google Scholar]