Abstract
Homeobox A (HOXA) cluster genes, members of the HOX family, perform an important role in normal organ development. It has previously been reported that HOXA gene expression in various types of cancer is associated with poor patient outcomes. However, the role of HOXA genes, as well as their expression, in colorectal cancers (CRC) remains unknown. Therefore, the present study investigated HOXA gene expression in patients with CRC and revealed that HOXA9 expression was significantly increased in tumor tissues compared with non-tumor tissues. Additionally, the functional role of HOXA9 was assessed by knocking down the HOXA9 gene in CRC cells and by evaluating cell growth. Regarding gene expression, cases with positive HOXA9 expression (as detected by immunohistochemical staining) were significantly associated with higher TNM stage and positive lymph node metastasis, although no association was observed between increased HOXA9 levels and the rate of overall survival in the present cohort. Regarding the functional role, HOXA9 expression was demonstrated to be upregulated in patients with CRC and was associated with lymph node metastasis.
Keywords: homeobox A9, colorectal cancer, lymph node metastasis, immunohistochemistry
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in the United States (1). Over the past two decades, the lifetime risk of developing CRC has decreased in the USA, Europe and Japan (2). Screenings by fecal occult blood test or colonoscopy have been demonstrated to reduce the incidence of CRC and mortality (3). Early detection and treatment is known as the most effective measure to improve cancer mortality (4,5). However, despite recent advances in CRC treatment, once diagnosed, the overall prognosis remains poor (6,7). Treatment with several novel combination chemotherapies, including folinic acid (LV)/5-fluorouracil (5-FU)/oxaliplatin and LV/5-FU/irinotecan, exhibited high response rates and prolonged survival time in patients with CRC (8–11). However, patients with a positive response to chemotherapy remain as high as 50% of all patients with CRC (7). Additional molecular targets and biomarkers are required for improvements in chemotherapeutic effect and patient survival.
Homeobox A (HOXA) cluster genes, located on chromosome 7p15, serve a fundamental role in embryonic development (12). HOXA genes are required for the development of normal organs, including the central nervous system, axial skeleton, limbs, gut, hematopoietic and urogenital tract, and internal and external genitalia (13). Therefore, dysregulation of these genes may potentially lead to the development of malignant tumors. Numerous studies have reported that HOXA genes are altered in various types of cancer, including leukemia and skin, prostate, breast, ovarian, gastric and esophageal cancer, and those alterations are associated with patient survival or clinicopathological factors (14–19). In addition, several HOXA genes are also altered in CRC, and those alterations appear to be associated with poorer patient prognosis (20,21).
In the present study, HOXA gene expression was investigated using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) or immunohistochemical (IHC) staining in surgically resected CRC tissues, and the biological significance of HOXA genes was examined by in vitro experiments to clarify the function of HOXA genes in CRC.
Materials and methods
Clinical patient samples
A total of 231 patients with CRC, who had undergone surgical resection at Fukushima Medical University Hospital (Fukushima, Japan) between January 1991 and December 2007 were involved in the present study. Specimens from all 231 cases were used for IHC staining. The mRNA was extracted from tumor and adjacent non-tumor tissues in 40 of 231 cases for PCR analyses. Information regarding age, sex, tumor-node-metastasis (TNM) stage (22,23) and pathological diagnosis, including lymphatic and venous invasion, were retrospectively collected. The carcinomas at the time of primary tumor resection were staged according to the Union for International Cancer Control classification (22,23). The present study was approved by the Ethics Committee of the Fukushima Medical University (Fukushima, Japan; approval no. 1615). Written informed consent was obtained from all patients.
Cell line culture
The colon cancer cell lines (HCT116, LoVo, RKO, LS174T, Colo205, Colo201, SW620, LS180, SW837, SW480, HCT15, and SW48) used in the present study were originally obtained from the American Type Culture Collection (Manassas, VA, USA). RKO, LS174T and LS180 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (both from Thermo Fisher Scientific, Inc., Waltham, MA, USA). SW480, LoVo, HCT15, SW48, SW620 and HCT116 cells were cultured in RPMI-1640 (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) supplemented with 10% fetal bovine serum. The monolayer cells were maintained in a 37°C incubator with 5% CO2, observed regularly under a light microscope (×40) and subcultured when 80–90% confluence was reached.
RT-qPCR
Total RNA was extracted from tumor and non-tumor tissues from 20 patients and cells using TRIzol reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol as previously described (24–26). Complementary DNA (cDNA) was synthesized from 5 µg of the total RNA with a random hexamer using the SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific, Inc.) with the GeneAmp PCR system 9700 (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The thermocycling conditions maintained were as follows: Denaturation, 65°C for 5 min; annealing, 25°C for 10 min; elongation, 50°C for 50 min; and termination, 85°C for 5 min and 37°C for 20 min. Subsequently, the cDNA from CRC tissues were used for the measurement of expression of 11 HOXA family genes (HOXA1, HOXA2, HOXA3, HOXA4, HOXA5, HOXA6, HOXA7, HOXA9, HOXA10, HOXA11 and HOXA13). β-actin was used as an internal control. Primer sequences are shown in Table I. The PCR product was loaded onto a 2% agarose gel with 0.5 µg/ml ethidium bromide. Following electorophoresis, the product were visualized using an UV-transilluminator (E-Box-1000/20M; Cosmo Bio Co. Ltd., Tokyo, Japan).
Table I.
Primer sequences for reverse transcription-quantitative polymerase chain reaction analysis.
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) | Size, bp |
|---|---|---|---|
| HOXA1 | CCCATGGAGGAAGTGAGAAA | GGACCATGGGAGATGAGAGA | 489 |
| HOXA2 | AGTCTCGCCTTTAACCTAGCA | GGCCTCATACTGCTCTCAGG | 571 |
| HOXA3 | AATGCCAGCAACAACCCTAC | TGACCAGCGAATGCATAGAG | 535 |
| HOXA4 | CCCTGGATGAAGAAGATCCA | GAGGATCGCATCTTGGTGTT | 265 |
| HOXA5 | GTGAAGAAGCCCTGTTCTCG | AACGAGATTGAAGGGGGACT | 244 |
| HOXA6 | CGCGCAAATGAGTTCCTATT | GACCGAGTTGGACTGTTGGT | 199 |
| HOXA7 | GGCTTGCCTGCTACTAGTG | GAAGCTGGAAGCATCTCCAC | 326 |
| HOXA9 | CCACGCTTGACACTCACACT | TCGTCTTTTGCTCGGTCTTT | 382 |
| HOXA10 | GGGGGAAAAAGCCATATCAT | GGGAGAATTGTGGTGTGCTT | 671 |
| HOXA11 | GCTTGGAAGCTTCTGGTGAC | AATTGAGGACAGGCCAACAC | 557 |
| HOXA13 | CTGGAACGGCCAAATGTACT | AGAGATTCGTCGTGGCTGAT | 386 |
| β-actin | GCTCGTCGTCGACAACGGCTC | CAAACATGATCTGGGTCATCTTCTC | 353 |
HOX, homeobox.
PCR amplification using 4 µl of cDNA with the TaqMan® RT-qPCR kit (Thermo Fisher Scientific, Inc.) for HOXA family genes was performed with the 7500 Real-time PCR system (Thermo Fisher Scientific, Inc.) at 94°C for initial denaturation for 5 min, followed by 25–35 cycles at 94°C for 1 min, 55–60°C for 45 sec and 72°C for 45 sec. Probes for TaqMan gene expression assays [HOXA9 (assay ID. Hs00266821_m1) and β-actin (assay ID. Hs99999903_m1)] were purchased from Thermo Fisher Scientific, Inc., and β-actin was used as an internal control. The primer sequences are listed in Table I. Relative HOXA9 gene expression was calculated using the 2−ΔΔCq method (27), according to the supplier's protocol (Thermo Fisher Scientific, Inc.).
IHC staining and evaluation
IHC staining was performed on paraffin-embedded histological sections (thickness, 4 µm) that were fixed in 10% buffered formalin, using a polymer peroxidase method (Envision+/HRP; Dako; Agilent Technologies, Inc., Santa Clara, CA, USA) (24,25). Briefly, following deparaffinization with xylene and rehydration using a descending alcohol series, the tissue sections were treated with 0.3% hydrogen peroxide in methanol for 30 min at room temperature to block endogenous peroxidase activity. Following rinsing in PBS, the sections were incubated with anti-HOXA9 antibody (1:1,000; cat. no. NBP2-32356; Novus Biologicals, LLC, Littleton, CO, USA) at 4°C overnight. An additional wash in PBS was followed by treatment with ready-to-use peroxidase-labeled polymer conjugated to goat anti-rabbit immunoglobulins (cat. no. SM801; ENvision + kit; Dako; Agilent Technologies, Inc.) as the secondary antibody for 30 min at room temperature. The staining was visualized with diaminobenzidine, followed by counterstaining with hematoxylin.
Expression of these proteins was evaluated using optical microscopy (BX43; Olympus Corporation, Tokyo, Japan) as positive when the nucleus of the cancerous tissue was stained. The staining of each specimen was evaluated at ×40 or ×400 magnification by two investigators, Y.W. and K.S. (Fukushima Medical University School of Medicine, Fukushima, Japan), who were blinded to the sample name and the clinical outcomes. The rate of positive-stained cancer cells was evaluated in three randomly selected areas (size, 200×200 µm) from the tumor specimens. When the average positive tumor rate was >10%, the tumor was defined as being positively stained.
Knockdown experiment and cell counting
The knockdown experiment was performed using a small interfering RNA (siRNA) method using HOXA9 Stealth RNAi siRNA (HOXA9-siRNA; cat. no. HSS142497; Thermo Fisher Scientific, Inc.) and Stealth RNAi siRNA negative control (NC-siRNA; cat. no. 12935-300; Thermo Fisher Scientific, Inc.) using Lipofectamine RNAiMAX (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. At 1 day prior to transfection, SW48 was seeded at 2×106 cells/well into the wells of a 10 cm plate. Transfection with a final concentration of 20 nM siRNA oligo was performed when the cell confluence reached 30–50%. The cells were incubated at 37°C 48 h prior to harvest. The number of cells was subsequently counted after 24 and 48 h siRNA transfection using a Countess II FL Automated Cell Counter (Thermo Fisher Scientific, Inc.). All experiments were repeated three times.
Public database
HOXA9 gene aberrations were investigated using The Cancer Genome Atlas (TCGA) data portal (cBioPortal: http://www.cbioportal.org/).
Statistical analysis
Data are presented as the mean ± standard deviation. Statistical analyses were performed using GraphPad Prism 6 software (GraphPad Software, Inc., La Jolla, CA, USA). Wilcoxon signed-rank tests and unpaired Student's t-tests were performed to analyze the differences among groups. In addition, survival curves were generated using the Kaplan-Meier method and compared using the log rank test. A total of 226 patients were used in the Kaplan-Meier survival analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
HOXA9 expression is upregulated in CRC
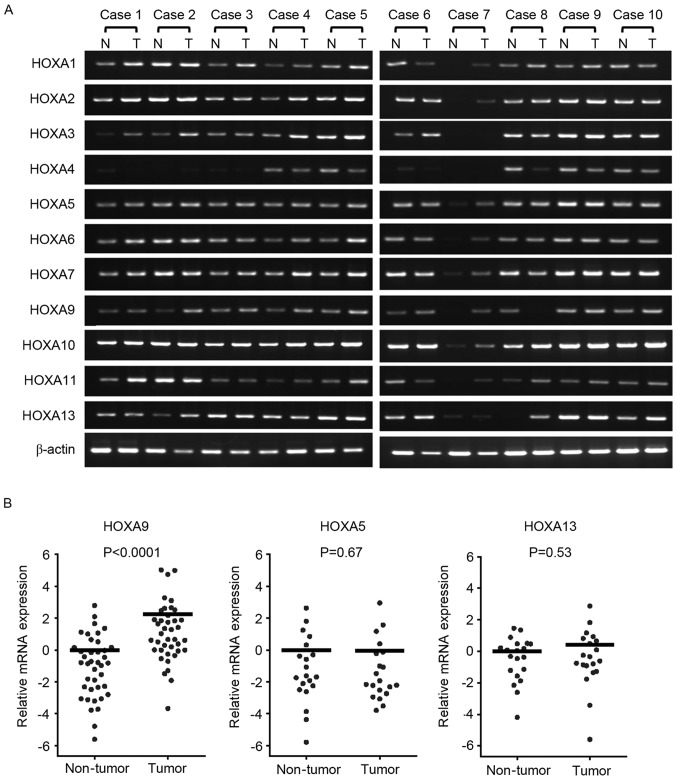
To investigate HOXA gene expression in CRC, the mRNA expression of each cluster gene was investigated in 10 representative cases by RT-qPCR (Fig. 1A). Although all HOXA genes were detected in tumor and adjacent non-tumor tissue samples, HOXA9 expression appeared to have increased in the tumor tissues compared with the non-tumor tissues as detected by PCR and gel electrophoresis. To investigate whether there is a difference in expression levels, RT-PCR was performed, and it was revealed that HOXA9 mRNA expression was increased in the tumor tissues by 4.71 fold compared with the non-tumor tissues (P<0.0001; Wilcoxon matched pairs test; Fig. 1B).
Figure 1.
mRNA expression levels of HOXA cluster genes in patients with CRC. (A) The mRNA expression of HOXA genes in patients with CRC. A total of 10 representative cases of tumor and non-tumor tissues were analyzed by RT-qPCR. β-actin was used as a loading control. (B) Differences in HOXA9 expression between tumor (n=40) and non-tumor (n=40) tissues, and HOXA5 and HOXA13 expression levels between tumor (n=20) and non-tumor (n=20) tissues as determined by qPCR and Wilcoxon matched pairs test. A log2 scale has been used for the y-axis. The mRNA expression in tumor tissues was normalized to the expression in non-tumor tissues. The horizontal bars indicate mean expression value. T, tumor; N, non-tumor; HOX, homeobox; CRC, colorectal cancer; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Upregulated HOXA9 expression is associated with poorer patient outcomes
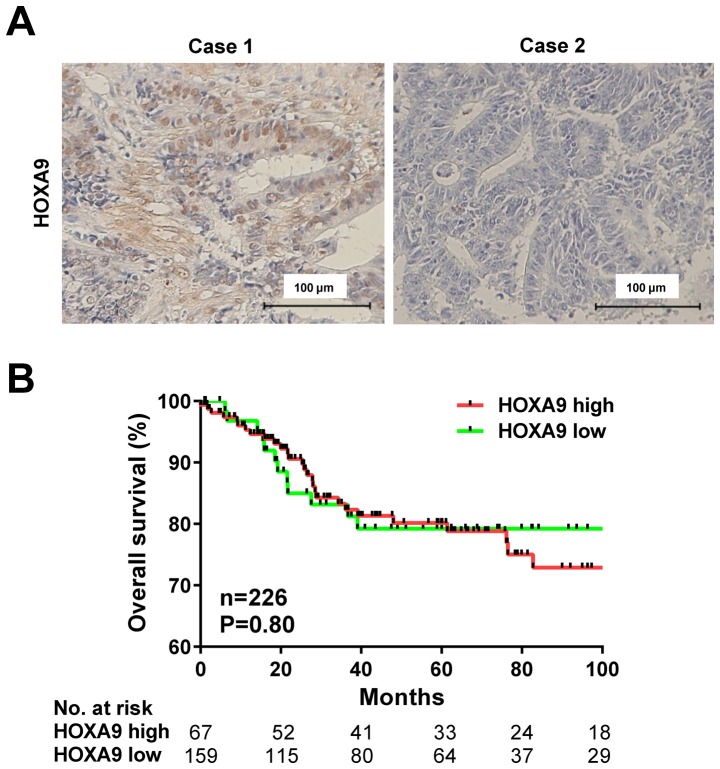
HOXA9 protein expression was evaluated using IHC staining in 231 patients with CRC (Fig. 2A). HOXA9 expression was detected in the nucleus of the CRC cells, and positive expression was observed in 161 cases (69.7%), and negative expression was detected in 70 cases (30.3%).
Figure 2.
HOXA9 expression and survival analysis of patients with CRC. (A) Immunohistochemical staining of HOXA9 in patients with CRC. Representative images showing positive (case 1) and negative (case 2) staining for HOXA9. Scale bar, 100 µm. (B) Kaplan-Meier survival analysis of 226 available cases in the present cohort stratified by HOXA9 expression in tumor specimens. HOX, homeobox; CRC, colorectal cancer.
The association between HOXA9 expression and clinicopathological factors was analyzed in patients with CRC (Table II). The rate of positive expression of HOXA9 was significantly associated with higher TNM stage (P=0.03) and positive lymph node metastasis (P=0.02). However, HOXA9 expression was not associated with age, gender, tumor location, histology, depth of invasion, venous invasion or liver metastasis. Furthermore, Kaplan-Meier analysis demonstrated no association between increased HOXA9 levels and overall survival rate (P=0.80; Fig. 2B).
Table II.
Association between clinicopathological factors and HOXA9 protein expression as determined by immunohistochemistry.
| HOXA9 IHC, n | ||||
|---|---|---|---|---|
| Characteristics | Total | Positive | Negative | P-value |
| Total | 231 | 161 | 70 | |
| Age | 1.00 | |||
| ≥60 years | 174 | 121 | 53 | |
| <60 years | 57 | 40 | 17 | |
| Gender | 0.77 | |||
| Male | 136 | 96 | 40 | |
| Female | 95 | 65 | 30 | |
| Stage | 0.03 | |||
| 0 | 5 | 4 | 1 | |
| I | 37 | 25 | 12 | |
| II | 85 | 52 | 33 | |
| III | 66 | 52 | 14 | |
| IV | 38 | 28 | 10 | |
| Tumor location | 0.65 | |||
| Colon | 165 | 113 | 52 | |
| Rectum | 66 | 48 | 18 | |
| Histology | 0.82 | |||
| tub | 203 | 142 | 61 | |
| por | 28 | 19 | 9 | |
| Depth | 0.28 | |||
| T1 | 28 | 19 | 9 | |
| T2 | 27 | 22 | 5 | |
| T3 | 159 | 106 | 53 | |
| T4 | 17 | 14 | 3 | |
| Lymphatic invasion | 1.00 | |||
| Absent | 48 | 34 | 14 | |
| Present | 183 | 127 | 56 | |
| Venous invasion | 0.60 | |||
| Absent | 50 | 33 | 17 | |
| Present | 181 | 128 | 53 | |
| Lymph node metastasis | 0.02 | |||
| Negative | 135 | 86 | 49 | |
| Positive | 96 | 75 | 21 | |
| Liver metastasis | 1.00 | |||
| Negative | 200 | 139 | 61 | |
| Positive | 31 | 22 | 9 | |
P-values were calculated using Fisher's exact test or a χ2 test, where appropriate. T, tumor; HOX, homeobox; tub, tubular adenocarcinoma; por, poorly differentiated adenocarcinoma.
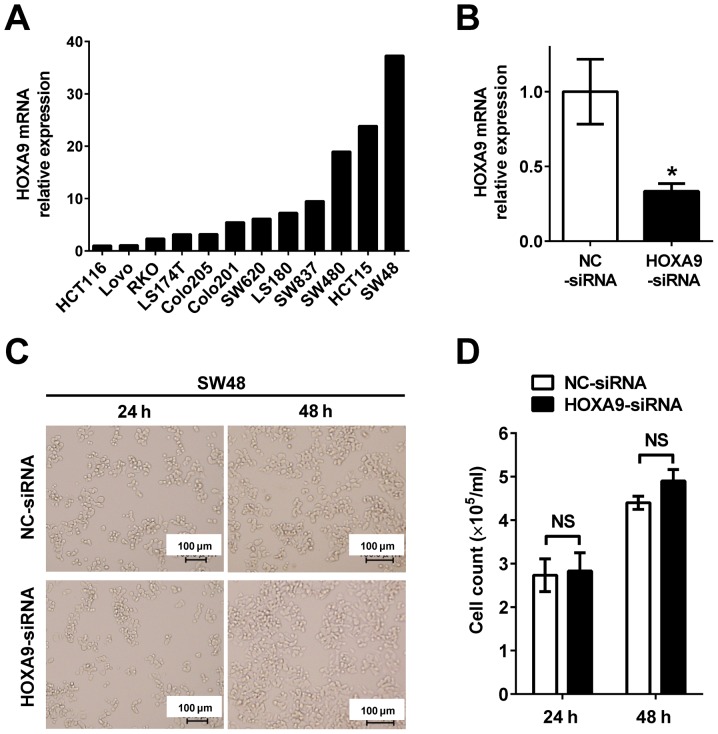
Knockdown of HOXA9 has no effect on growth in colon cancer cells
To evaluate the role of HOXA9 in colon cancer progression, gene knockdown of HOXA9 was performed to investigate cell proliferation. HOXA9 expression in 12 colon cancer cell lines (HCT116, LoVo, RKO, LS174T, Colo205, Colo201, SW620, LS180, SW837, SW480, HCT15 and SW48) was evaluated to select the appropriate cell line for subsequent experiments (Fig. 3A). As HOXA9 was most significantly expressed in SW48 cells compared with all other cells, they were chosen for subsequent knockdown experiments. HOXA9 was subsequently knocked down by transfection with siRNA oligonucleotide in the SW48 cells (HOXA9-siRNA) (Fig. 3B). No morphological changes were observed in the HOXA9 knocked down cells (Fig. 3C). Furthermore, cell proliferation was not decreased between HOXA9 knocked down cells and the negative control (Fig. 3D).
Figure 3.
In vitro analysis of the effects of HOXA9 knockdown. (A) RT-qPCR analysis of HOXA9 expression in colon cancer cell lines. Relative HOXA9 mRNA expression levels to HCT116 (HCT116=1) are shown. The expression of targets genes was normalized to β-actin expression. (B) RT-qPCR analysis of HOXA9 expression in SW48 cells transfected with HOXA9 siRNA and the negative control siRNA. Data are presented as the mean ± standard deviation. *P<0.05. (C) Images of SW48 cells in the NC-siRNA and HOXA9-siRNA group, 24 and 48 h following transfection. Scale bar, 100 µm. (D) Cell counts of SW48 cells in the NC-siRNA and HOXA9-siRNA group, 24 and 48 h following transfection. Data are presented as the mean ± standard deviation. NS, not significant; HOX, homeobox; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; siRNA, small interfering RNA; NC, negative control.
Discussion
In the present study, RT-qPCR analysis indicated that HOXA9 mRNA was upregulated in CRC tumor tissues compared with non-tumor tissues. Consistent with the results of the present study, expression of HOXA9 has been reported to be significantly increased in esophageal squamous cell cancer (17) and lung cancer (28) compared with normal non-tumor tissues. However, HOXA9 expression was decreased in hepatocellular carcinoma, as well as ovary, lung and bladder cancer tissues (29–32). Such suppression was reported to be due to hypermethylation of the HOXA9 promoter CpG island (29–32).
Another known regulator of HOXA9 mRNA expression is microRNA (miR) (33). It has been reported that HOXA9 is directly suppressed by upregulated miR-196b expression in non-small cell lung cancer cells (33). Although one study has reported that miR-196b is downregulated in cervical cancer (34), another study has suggested that HOXA9 is also downregulated in cervical cancer (35). These results indicate that the expression and aberration mechanisms of HOXA9 are tumor-specific.
The authors of the present study investigated whether HOXA9 gene aberrations attributed to high HOXA9 expression using the publicly available cBioPortal. HOXA9 gene amplification resulting in high HOXA9 expression was observed in 10/287 (3.5%) cases of stomach adenocarcinoma (36), 4/109 (3.7%) cases of pancreatic cancer (37) and 12/184 (6.5%) cases of esophageal cancer (TCGA provisional). However, HOXA9 gene amplification resulting in high HOXA9 expression was only detected in 1/212 (0.5%) cases in CRC (38). These findings indicated that gene amplification may not be involved in the increase of HOXA9 mRNA transcript levels in CRC. The underlying mechanism for the increase of HOXA9 mRNA in CRC tumors is yet to be elucidated. Notably, a similar upregulation of mRNA in human tumors has also been reported with other members of the HOX gene family (12,13,39). However, the main mechanism involved in this process also remains unknown (12,13,39).
The present study investigated HOXA9 protein expression in CRC tumor tissues and found that ~70% of patients with CRC exhibited positive staining for HOXA9. Furthermore, the increase in HOXA9 expression was significantly associated with positive lymph node metastasis in the present cohort. Consistent with the present results, a number of studies have demonstrated the involvement of HOX genes in lymph node metastasis (13,20,39). HOXC6 expression was upregulated in lymph node metastasis-positive oral squamous cell carcinoma compared with lymph node metastasis-negative oral squamous cell carcinoma (40). By contrast, HOXB7 expression was downregulated in breast cancer with lymph node metastasis compared with lymph node-negative breast cancer (41). Notably, no association between HOXA9 expression and patient survival was observed in the present cohort. This may be due to differences in tumor stage, as the population comprised primarily stage II and III patients.
In conclusion, to the best of our knowledge, this is the first study to report that HOXA9 expression is upregulated in CRC and is associated with lymph node metastasis. Additional studies are required to reveal the role of HOXA9 and its viability as a candidate therapeutic target or biomarker.
Acknowledgements
The present study was supported by the Japan Society for the Promotion of Science KAKENHI (grant no. 15k10143).
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Katanoda K, Hori M, Matsuda T, Shibata A, Nishino Y, Hattori M, Soda M, Ioka A, Sobue T, Nishimoto H. An updated report on the trends in cancer incidence and mortality in Japan, 1958–2013. Jpn J Clin Oncol. 2015;45:390–401. doi: 10.1093/jjco/hyv002. [DOI] [PubMed] [Google Scholar]
- 3.Mayer RJ, Venook AP, Schilsky RL. Progress against GI cancer during the American Society of Clinical Oncology's first 50 years. J Clin Oncol. 2014;32:1521–1530. doi: 10.1200/JCO.2014.55.4121. [DOI] [PubMed] [Google Scholar]
- 4.Brenner H, Kretschmann J, Stock C, Hoffmeister M. Expected long-term impact of screening endoscopy on colorectal cancer incidence: A modelling study. Oncotarget. 2016;7:48168–48179. doi: 10.18632/oncotarget.10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenner H, Chang-Claude J, Seiler CM, Hoffmeister M. Long-term risk of colorectal cancer after negative colonoscopy. J Clin Oncol. 2011;29:3761–3767. doi: 10.1200/JCO.2011.35.9307. [DOI] [PubMed] [Google Scholar]
- 6.Venook AP, Weiser MR, Tepper JE. Colorectal cancer: All hands on deck. Am Soc Clin Oncol Educ Book. 2014:1–89. doi: 10.14694/EdBook_AM.2014.34.83. [DOI] [PubMed] [Google Scholar]
- 7.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 8.de Gramont A, Figer A, Seymour M, Homerin M, Hmissi A, Cassidy J, Boni C, Cortes-Funes H, Cervantes A, Freyer G, et al. Leucovorin and fluorouracil with or without oxaliplatin as first-line treatment in advanced colorectal cancer. J Clin Oncol. 2000;18:2938–2947. doi: 10.1200/JCO.2000.18.16.2938. [DOI] [PubMed] [Google Scholar]
- 9.Douillard JY, Cunningham D, Roth AD, Navarro M, James RD, Karasek P, Jandik P, Iveson T, Carmichael J, Alakl M, et al. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: A multicentre randomised trial. Lancet. 2000;355:1041–1047. doi: 10.1016/S0140-6736(00)02034-1. [DOI] [PubMed] [Google Scholar]
- 10.Goldberg RM, Sargent DJ, Morton RF, Fuchs CS, Ramanathan RK, Williamson SK, Findlay BP, Pitot HC, Alberts SR. A randomized controlled trial of fluorouracil plus leucovorin, irinotecan, and oxaliplatin combinations in patients with previously untreated metastatic colorectal cancer. J Clin Oncol. 2004;22:23–30. doi: 10.1200/JCO.2004.09.046. [DOI] [PubMed] [Google Scholar]
- 11.Tournigand C, André T, Achille E, Lledo G, Flesh M, Mery-Mignard D, Quinaux E, Couteau C, Buyse M, Ganem G, et al. FOLFIRI followed by FOLFOX6 or the reverse sequence in advanced colorectal cancer: A randomized GERCOR study. J Clin Oncol. 2004;22:229–237. doi: 10.1200/JCO.2004.05.113. [DOI] [PubMed] [Google Scholar]
- 12.Pezzani L, Milani D, Manzoni F, Baccarin M, Silipigni R, Guerneri S, Esposito S. HOXA genes cluster: Clinical implications of the smallest deletion. Ital J Pediatr. 2015;41:31. doi: 10.1186/s13052-015-0137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinonez SC, Innis JW. Human HOX gene disorders. Mol Genet Metab. 2014;111:4–15. doi: 10.1016/j.ymgme.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Novak P, Jensen T, Oshiro MM, Wozniak RJ, Nouzova M, Watts GS, Klimecki WT, Kim C, Futscher BW. Epigenetic inactivation of the HOXA gene cluster in breast cancer. Cancer Res. 2006;66:10664–10670. doi: 10.1158/0008-5472.CAN-06-2761. [DOI] [PubMed] [Google Scholar]
- 15.Strathdee G, Holyoake TL, Sim A, Parker A, Oscier DG, Melo JV, Meyer S, Eden T, Dickinson AM, Mountford JC, et al. Inactivation of HOXA genes by hypermethylation in myeloid and lymphoid malignancy is frequent and associated with poor prognosis. Clin Cancer Res. 2007;13:5048–5055. doi: 10.1158/1078-0432.CCR-07-0919. [DOI] [PubMed] [Google Scholar]
- 16.Bach C, Buhl S, Mueller D, García-Cuéllar MP, Maethner E, Slany RK. Leukemogenic transformation by HOXA cluster genes. Blood. 2010;115:2910–2918. doi: 10.1182/blood-2009-04-216606. [DOI] [PubMed] [Google Scholar]
- 17.Chen KN, Gu ZD, Ke Y, Li JY, Shi XT, Xu GW. Expression of 11 HOX genes is deregulated in esophageal squamous cell carcinoma. Clin Cancer Res. 2005;11:1044–1049. [PubMed] [Google Scholar]
- 18.Widschwendter M, Apostolidou S, Jones AA, Fourkala EO, Arora R, Pearce CL, Frasco MA, Ayhan A, Zikan M, Cibula D, et al. HOXA methylation in normal endometrium from premenopausal women is associated with the presence of ovarian cancer: A proof of principle study. Int J Cancer. 2009;125:2214–2218. doi: 10.1002/ijc.24599. [DOI] [PubMed] [Google Scholar]
- 19.Orlovsky K, Kalinkovich A, Rozovskaia T, Shezen E, Itkin T, Alder H, Ozer HG, Carramusa L, Avigdor A, Volinia S, et al. Down-regulation of homeobox genes MEIS1 and HOXA in MLL-rearranged acute leukemia impairs engraftment and reduces proliferation; Proc Natl Acad Sci USA; 2011; pp. 7956–7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanai M, Hamada J, Takada M, Asano T, Murakawa K, Takahashi Y, Murai T, Tada M, Miyamoto M, Kondo S, Moriuchi T. Aberrant expressions of HOX genes in colorectal and hepatocellular carcinomas. Oncol Rep. 2010;23:843–851. [PubMed] [Google Scholar]
- 21.Bhatlekar S, Addya S, Salunek M, Orr CR, Surrey S, McKenzie S, Fields JZ, Boman BM. Identification of a developmental gene expression signature, including HOX genes, for the normal human colonic crypt stem cell niche: Overexpression of the signature parallels stem cell overpopulation during colon tumorigenesis. Stem Cells Dev. 2014;23:167–179. doi: 10.1089/scd.2013.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gospodarowicz MK, Wittekind Ch, Sobin LH, editors. Union for International Cancer Control (UICC), corp-author TNM Classification of Malignant Tumors. 7th. Wiley-Blackwell; Oxford: 2009. [Google Scholar]
- 23.Sobin LH, Compton CC. TNM seventh edition: What's new, what's changed: Communication from the International Union Against Cancer and the American Joint Committee on Cancer. Cancer. 2010;116:5336–5339. doi: 10.1002/cncr.25537. [DOI] [PubMed] [Google Scholar]
- 24.Okano M, Kumamoto K, Saito M, Onozawa H, Saito K, Abe N, Ohtake T, Takenoshita S. Upregulated Annexin A1 promotes cellular invasion in triple-negative breast cancer. Oncol Rep. 2015;33:1064–1070. doi: 10.3892/or.2015.3720. [DOI] [PubMed] [Google Scholar]
- 25.Tachibana K, Saito M, Imai JI, Ito E, Yanagisawa Y, Honma R, Saito K, Ando J, Momma T, Ohki S, et al. Clinicopathological examination of dipeptidase 1 expression in colorectal cancer. Biomed Rep. 2017;6:423–428. doi: 10.3892/br.2017.870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saito M, Shiraishi K, Matsumoto K, Schetter AJ, Ogata-Kawata H, Tsuchiya N, Kunitoh H, Nokihara H, Watanabe S, Tsuta K, et al. A three-microRNA signature predicts responses to platinum-based doublet chemotherapy in patients with lung adenocarcinoma. Clin Cancer Res. 2014;20:4784–4793. doi: 10.1158/1078-0432.CCR-14-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Calvo R, West J, Franklin W, Erickson P, Bemis L, Li E, Helfrich B, Bunn P, Roche J, Brambilla E, et al. Altered HOX and WNT7A expression in human lung cancer; Proc Natl Acad Sci USA; 2000; pp. 12776–12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin SH, Kim BH, Jang JJ, Suh KS, Kang GH. Identification of novel methylation markers in hepatocellular carcinoma using a methylation array. J Korean Med Sci. 2010;25:1152–1159. doi: 10.3346/jkms.2010.25.8.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beachy SH, Onozawa M, Silverman D, Chung YJ, Rivera MM, Aplan PD. Isolated Hoxa9 overexpression predisposes to the development of lymphoid but not myeloid leukemia. Exp Hematol. 2013;41:518–529.e5. doi: 10.1016/j.exphem.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorsteinsdottir U, Mamo A, Kroon E, Jerome L, Bijl J, Lawrence HJ, Humphries K, Sauvageau G. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood. 2002;99:121–129. doi: 10.1182/blood.V99.1.121. [DOI] [PubMed] [Google Scholar]
- 32.Aine M, Sjödahl G, Eriksson P, Veerla S, Lindgren D, Ringnér M, Höglund M. Integrative epigenomic analysis of differential DNA methylation in urothelial carcinoma. Genome Med. 2015;7:23. doi: 10.1186/s13073-015-0144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu SL, Lee DC, Sohn HA, Lee SY, Jeon HS, Lee JH, Park CG, Lee HY, Yeom YI, Son JW, et al. Homeobox A9 directly targeted by miR-196b regulates aggressiveness through nuclear Factor-kappa B activity in non-small cell lung cancer cells. Mol Carcinog. 2016;55:1915–1926. doi: 10.1002/mc.22439. [DOI] [PubMed] [Google Scholar]
- 34.How C, Hui AB, Alajez NM, Shi W, Boutros PC, Clarke BA, Yan R, Pintilie M, Fyles A, Hedley DW, et al. MicroRNA-196b regulates the homeobox B7-vascular endothelial growth factor axis in cervical cancer. PLoS One. 2013;8:e67846. doi: 10.1371/journal.pone.0067846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alvarado-Ruiz L, Martinez-Silva MG, Torres-Reyes LA, Pina-Sanchez P, Ortiz-Lazareno P, Bravo-Cuellar A, Aguilar-Lemarroy A, Jave-Suarez LF. HOXA9 is underexpressed in cervical cancer cells and its restoration decreases proliferation, migration and expression of epithelial-to-mesenchymal transition genes. Asian Pac J Cancer Prev. 2016;17:1037–1047. doi: 10.7314/APJCP.2016.17.3.1037. [DOI] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Research Network, corp-author. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, Mollaee M, Wagner KU, Koduru P, Yopp A, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cancer Genome Atlas Network, corp-author. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhatlekar S, Fields JZ, Boman BM. HOX genes and their role in the development of human cancers. J Mol Med (Berl) 2014;92:811–823. doi: 10.1007/s00109-014-1181-y. [DOI] [PubMed] [Google Scholar]
- 40.Hassan NM, Hamada J, Murai T, Seino A, Takahashi Y, Tada M, Zhang X, Kashiwazaki H, Yamazaki Y, Inoue N, Moriuchi T. Aberrant expression of HOX genes in oral dysplasia and squamous cell carcinoma tissues. Oncol Res. 2006;16:217–224. doi: 10.3727/000000006783981080. [DOI] [PubMed] [Google Scholar]
- 41.Makiyama K, Hamada J, Takada M, Murakawa K, Takahashi Y, Tada M, Tamoto E, Shindo G, Matsunaga A, Teramoto K, et al. Aberrant expression of HOX genes in human invasive breast carcinoma. Oncol Rep. 2005;13:673–679. [PubMed] [Google Scholar]