Abstract
The hallmark of Gram-negative bacteria and organelles such as mitochondria and chloroplasts is the presence of an outer membrane (OM). In bacteria such as Escherichia coli, the OM is a unique asymmetric lipid bilayer with lipopolysaccharide (LPS) in the outer leaflet. Integral, transmembrane proteins assume a β-barrel structure (OMPs) and their assembly is catalyzed by the heteropentomeric Bam complex containing the OMP BamA and four lipoproteins, BamB-E. How the Bam complex assembles a great diversity of OMPs into a membrane without an obvious energy source is a particularly challenging problem because folding intermediates are predicted to be unstable in either an aqueous or a hydrophobic environment. Two models have been put forward, the budding model based largely on structural data, and the BamA-assisted model based on genetic and biochemical studies. Here we offer a critical discussion of the pros and cons of each.
Keywords: envelope biogenesis, outer membrane protein, LptD, lateral gate, protein folding
1. Introduction
The defining feature of Gram-negative bacteria such as Escherichia coli is the presence of an outer membrane (OM) (Figure 1). The OM is an essential organelle that acts as a selective permeability barrier protecting the cell from harmful chemicals, including detergents and antibiotics (66). The OM is an asymmetric bilayer with phospholipids in the inner leaflet and lipopolysaccharide (LPS) in the outer leaflet. Like other biological membranes, the hydrophobic core of the OM, composed of phospholipids and the lipid A part of LPS, prevents penetration of the polar solutes. However, unlike other biological membranes, the OM also excludes hydrophobic compounds and this function is mediated by the carbohydrate chains of LPS. These sugar chains not only create a polar mesh around the cell limiting accessibility of the hydrophobic compounds to the lipid core but also enable strong lateral interactions between phosphorylated sugar residues that are bridged with divalent cations to seal the OM.
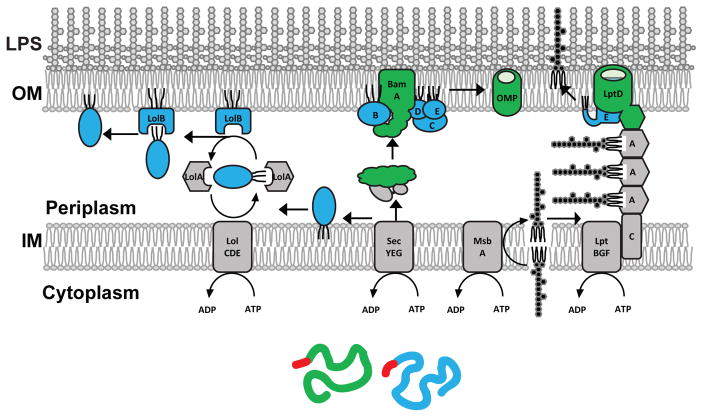
Figure 1. The structure of Gram-negative envelope and the pathways required for OM biogenesis.
The envelope of Gram-negative bacteria contains two membranes. The OM in an asymmetric bilayer with LPS in its outer leaflet. LPS is synthesized on the cytoplasmic side of the IM and translocated across the IM by MsbA. LPS is extracted from the IM by LptBFG. LPS is then translocated across the periplasm by being pushes by the following LPS molecule through the periplasmic LptA bridge to the OM translocon, LptD/LptE, which inserts LPS directly into the outer leaflet (see the recent reviews (62; 85). OM proteins, such as β-barrel OMPs (green) or lipoproteins (blue) are synthesized on ribosomes and contain a signal sequence (red) that targets them to the Sec complex, which translocates them across the IM. Unfolded OMPs are bound by periplasmic chaperones which escort them to the Bam complex. The Bam complex assembles OMPs into the OM (see the recent review (73). Lipoproteins undergo lipid modifications and are extracted from the IM by the LolCDE and passed to the periplasmic chaperone LolA that delivers them to the OM acceptor protein LolB, which inserts lipoproteins into the inner leaflet of the OM (see the recent review (64).
Nearly all of the integral OM proteins are β-barrel proteins, also known as OMPs. Some OMPs function as passive diffusion channels (porins), others are specific nutrient importers, only two are known to play an essential role in OM biogenesis. The peripheral OM proteins are lipoproteins anchored to the OM by covalently attached N-terminal lipid tails. A few lipoproteins play essential roles in OM biogenesis or peptidoglycan synthesis, others sense envelope stress, but the function of most of the OM lipoproteins remains unknown.
OM biogenesis is complex (Fig. 1). Biosynthesis of each of the components and their transport to and insertion into the OM has to be coordinated with the cell growth and accomplished without compromising the barrier function of the OM. Moreover, the cell must overcome the obstacles of translocating the hydrophobic components of the OM across the cytoplasmic or inner membrane (IM) and transporting them across the aqueous periplasmic space between the OM and the IM. While transport across the IM, and, in the case of lipoproteins and LPS, release from the IM is energy –dependent, transport across the periplasm and insertion into the OM presents special challenges as there is no ATP outside the cytoplasm and no useful ion gradient across the OM. For LPS, this transport is achieved in a process analogous to a PEZ candy dispenser, in which an LPS molecule, newly released from the IM using the energy of ATP, pushes the previous molecules across the periplasmic bridge of the Lpt complex and through the OM translocon onto the cell surface (69) (Fig. 1). Proteins other the other hand, traverse the periplasm bound to soluble chaperones, LolA in the case of lipoproteins (97), or a network of somewhat redundant chaperones such a SurA, Skp, DegP and FkpA for OMPs (84; 88). Chaperones deliver their substrates to the OM acceptors, LolB (61) and the β-barrel assembly machine, the Bam complex, which facilitate their insertion into the OM (32; 87; 94) (Fig. 1). Although the LPS, lipoprotein and OMP assembly pathways are distinct, they display interdependence, as the LptD/LptE transclocon and the Bam complex contain both β-barrel and lipoproteins (Fig. 1).
Although the Lol pathway, which delivers Lipoproteins to the OM has been well-studied, the final topology of lipoproteins cannot be easily predicted; they can face either the periplasm or the extracellular milieu or they can adopt a transmembrane topology and this final step in lipoprotein biogenesis is the least characterized (53). In contrast, the topology of β-barrels can be easily predicted due to the β-strands with their alternating hydrophobic and polar amino acids (30). These β-strands are arranged in antiparallel fashion into a β-sheet, which is wrapped to form a cylinder by establishing hydrogen bonds between the first and the last β-strands and this interaction is referred as the seam. In the final structure, polar amino acids line barrel interior, hydrophobic amino acids face the barrel exterior and interact with the OM lipid core, and the N- and C-termini face the periplasm. Although all OMPs feature such a cylindrical structure, they are quite diverse in their size and domain architecture. The size of β-barrels vary between 8 (OmpX, the smallest known (92)) and 26 (LptD, the largest known, (18; 74)) β-strands. Some OMPs can multimerize into dimers (PldA (89)) or trimers (OmpF/C, LamB, (3; 83; 98)) or form a single composite barrel made up of three subunits (TolC(54)) or as many as nine subunites (CsgG). Some OMPs feature large surface exposed loops and additional periplasmic (e.g. OmpA (41; 70)), or lumen-occluding plug domains (TonB-dependent transporters, e.g. FhuA (23)); some exist in complexes with lipoproteins (LptD with LptE (12) and OmpA/C/F with RcsF (51)). Despite the diversity, all OMPs are assembled by the Bam complex, highlighting the remarkable versatility of this cellular machine.
2. Overall architecture of the Bam complex
The Bam complex consists of five proteins (87; 94) (Fig. 2A). BamA is itself a β-barrel but also contains five periplasmic POTRA (polypeptide transport–associated) domains, which scaffold four lipoproteins, B-E. BamA and BamD are the essential core components and are conserved throughout bacteria (59; 94). BamB, C, and E are present in some but not all species, suggesting they play an accessory role in the β-barrel assembly pathway. The crystal structure of all individual components and their combinations, including the crystal and cryo-EM structures of the full pentameric complex are now available (1; 2; 17; 21; 31; 35; 37; 42–44; 49; 65; 68; 82). Despite the recent advances in the structural biology of the Bam complex, the mechanism of the β-barrel assembly pathway remains largely unknown.
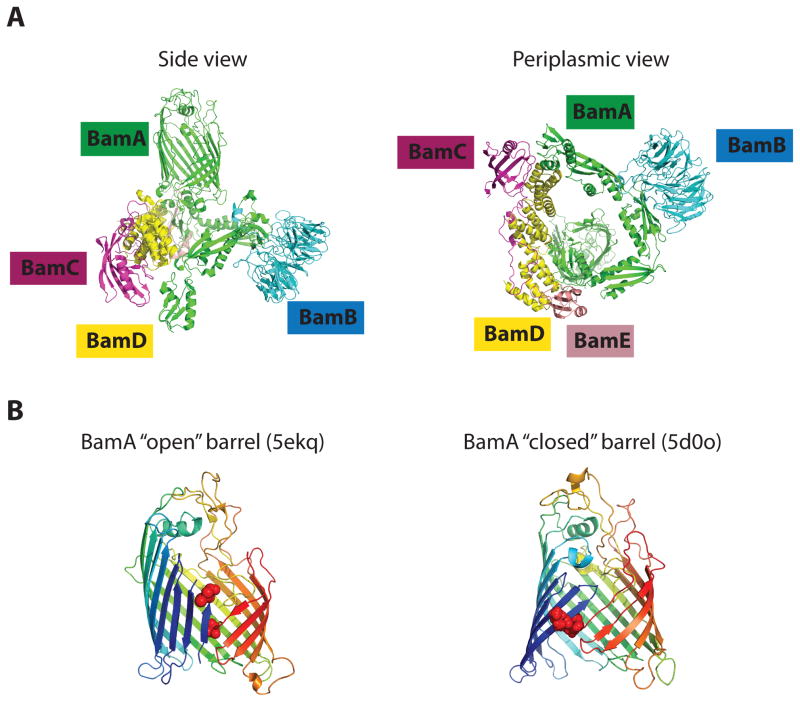
Figure 2. The structure of the Bam complex.
(A) The structure of the Bam complex by cryo-EM (5LJO) (37). The central component, BamA, contains a β-barrel and a periplasmic domain, consisting of five POTRA domains that scaffold lipoproteins BamB-E. POTRA domains together with lipoproteins form a ring-like structure with the cavity underneath the BamA barrel. Note, that the position of the ring relative to BamA barrel varies substantially in different structures of the Bam complex.
(B) The architecture of the BamA barrel and the seam in two different crystal structures of BamA (2; 31). The residues of the seam used for a disulfide crosslinking are highlighted by the red spheres. These structures have been proposed to represent BamA in its “open” and “closed” conformations.
Because BamA contains a transmembrane β-barrel domain, BamA is believed to play a central role in the assembly and insertion of the incoming β-barrel substrate into the OM. The barrel domain of BamA consists of 16-stranded β-sheet enclosing a large water filled lumen (1; 65; 68) (Fig. 2A and B). Extracellular loops of BamA form a dome on top of BamA, enclosing the barrel from the top and preventing solute passage through the BamA pore. Extracellular loop (eL) 6 partially inserts inside the lumen and interacts with β-strands 14–16. The barrel itself displays two intriguing features (Fig. 2A and B). One is that the height of the BamA barrel, dictated by the membrane-spanning hydrophobic belt, is significantly shorter proximal to the seam at only 9–12 Å compared to 20 Å on the opposite side of the barrel. The second is that the seam displays poor hydrogen bonding between the first and the last barrel β-strands. Both of these features will be discussed below in light of proposed mechanisms of BamA function.
The periplasmic domain of BamA consists of five POTRA domains, P1-5 (26; 27; 45) (Fig. 2A). Although the POTRA domains do not share high degree sequence similarity, they display a conserved structural fold, comprising of a pair of antiparallel helices covering a three-stranded β sheet. Despite having highly similar structures, the POTRA domains are functionally distinct. P5 provides an interaction interface between BamA and BamCDE subcomplex (45), while P3 is important for BamA interaction with BamB and P1 is important for BamA interaction with a chaperone SurA (93). Although P1 and P2 are important for BamA function, deletion of P1-2 is not lethal; the remaining POTRA domains are essential (45).
The structure of the Bam complex revealed that the POTRA domains of BamA do not simply hang in the periplasm, but together with Bam lipoproteins form a ring-like structure which is predicted to lay parallel to the membrane plane (2; 31; 35; 37) (Fig. 2A). Molecular dynamic (MD) simulations of the BamA in the membrane releveled that all five POTRA domains have high affinity for the membrane, and establish multiple hydrogen bonds with phospholipid head groups and partition directly into the membrane via highly conserved tryptophan residues (25). Partitioning of these tryptophans into the detergent micelle was later observed in the cryo-EM structure of the Bam complex (37). The same structure also revealed that the Bam lipoproteins also intercalated into the micelle via N-terminal residues in the case of BamB and E and the hydrophobic loop of BamD (37). Despite overall similarity in the periplasmic ring architecture, all of the structures vary significantly in the orientation of the periplasmic ring relatively to BamA barrel. It is not clear, which one of these orientations, if any, represent in vivo state of the Bam complex. It is possible that interaction of the ring with the membrane and/or a substrate would limit its flexibility towards a particular orientation. Alternatively, the ring may sample several alternative conformations during the assembly process.
The periplasmic ring occludes the cavity, and the POTRA domains and BamD make up its inner surface (2; 31; 35; 37) (Fig. 2A). BamA interaction with BamD is mediated primarily through P5. Although some interaction between BamD and P2 was also observed, it is unlikely to have functional significance, because deletion of P5 or a disruption of a single salt bridge in P5/BamD interface is sufficient to fully separate BamCDE and BamAB subcomplexes (45; 75).
BamD is a central and essential component of the Bam complex (59). One of the functions of BamD is a substrate recognition (34). BamD binds unfolded OMP substrates, and the binding site lays within the C-terminal OMP sequences which closely resembles the mitochondrial β signal (33; 34). The β signal targets mitochondrial OMPs to a homologous Sam complex, and the BamD analog, Sam35, is involved in this recognition (55). Ability to recognize the substrates is an essential function of BamD, and the peptides containing the β signal, which bind BamD, are toxic because they inhibit β barrel assembly (34). However, the function of BamD is not limited to the initial substrate recognition, because BamD remains closely associated with the substrate throughout entire OMP assembly and must play an active role during this process (40; 56). One of these roles is to regulate BamA conformational cycling through the direct interaction with P5 (see BamA conformational mobility section for more details) (75; 76). This direct interaction allows both proteins coordinate their activities during the OMP assembly. However, the requirement for this direct and stable interaction can be bypassed by introducing activating mutations in both bamA and bamD, which allows both proteins work in concert but independently of each other (75).
The function of non-essential lipoproteins is even more elusive. BamB interacts with BamA directly via P3, while BamE and BamC interact primary with BamD and likely modulate BamD interactions with the substrate and BamA. Loss of bamE or bamC does not confer general OMP assembly defect, but bamE alters BamA conformation in a manner similar to an activating mutation in bamD (76; 77; 87). Several lines of evidence suggest that although non-essential, these lipoproteins may be required for assembly of specific substrates. For example, assembly of large, difficult substrates (such as LptD, TolC, autotransporters) is not affected by single mutations in non-essential components, (10; 58; 78). In contrast, deletion of bamB strongly affects high volume trimeric substrates, such as porins and the maltodextrin transporter LamB (10). Deletion of bamE does not affect assembly of any single OMP but is required for assembly of RcsF/OMP complexes (50). Because these Bam substrates are so structurally diverse, their assembly may require specialized activities of the Bam complex. Bam lipoproteins may either directly perform these activities or enhance the efficiency of BamAD core components towards specific substrates.
3. Models for the β-barrel assembly
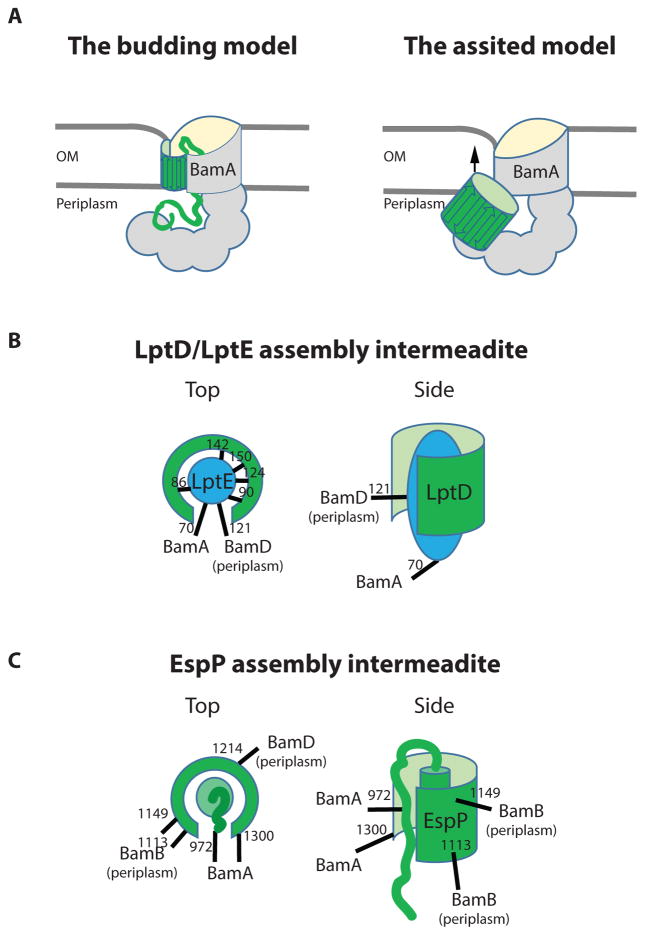
Because a β-sheet is amphipathic, having a hydrophobic interface on one side and hydrophilic on the other, β-barrels cannot be fully membrane integrated prior to seam closure forming the cylinder. Hence, two models were proposed for how Bam functions (Figure 3A). In the budding model, β-strands of an incoming OMP are templated at the open BamA seam forming a hybrid BamA/OMP barrel, and once all β-strands are formed, the new barrel closes and buds off the BamA into the OM. In the BamA assisted model, β-barrel folding begins on the periplasmic side of the OM and is integrated as one unit into the OM by the Bam complex. While several lines of in vitro and in vivo evidence exist to support the BamA assisted model, there is no direct exprimental evidence for the budding model, but it has recently been revitalized after crystallographic observation of a BamA seam configuration.
Figure 3. The proposed models for BamA function and experimental evidence for the folding in the periplasm.
(A) The BamA budding model suggests that BamA opens laterally and templates the β-strands of an incoming OMP. As a result, a hybrid BamA/OMP barrel is formed. Once all OMP β-strand are templated, the OMP barrel buds off laterally into the OM (67). The BamA assisted model suggests that OMP folding is driven by their intrinsic thermodynamic properties and occurs largely at the periplasmic side of the OM. The function of BamA is to reduce the kinetic barrier for membrane integration by creating a membrane defect (illustrated by OM thinning) in the proximity to the BamA seam (24).
(B) LptD/LptE assembly intermediate represented by the lptD4213 assembly-defective mutant (56). The LptD barrel (green) is largely formed because it established contacts LptE (blue), but not fully closed because LptE is still crosslinked to BamD. Sites of LptE crosslinking to the indicated proteins are shown. This LptD/E complex remains in the periplasm because it crosslinks to the periplasmic lipoprotein BamD. The N-terminal domains of LptD and LptE lipid moieties are not shown for clarity.
(C) EspP assembly intermediate represented by espP_G1066A or G1081A assembly-defective mutants or WT under the conditions of slow growth (40; 71). EspP barrel largely formed because the α-helix of the passenger domain is buried inside the barrel. The barrel remains open to facilitate translocation of the passenger domain, which can still be crosslinked to BamA. Sites of EspP crosslinking to the indicated proteins are shown. This partially folded EspP barrel remains in the periplasm because it crosslinks to the periplasmic lipoproteins BamB and BamD, and cannot be crosslinked to LPS. Only the C-terminal part of the passenger domain is shown for clarity.
3.1 The budding model
Initial structures of the BamA barrel domain revealed poor hydrogen bond formation between first and last β-strands (Fig. 2B) (68). This configuration of the seam was not surprising because it has been known for a long time that the BamA barrel has the lowest thermodynamic stability in comparison to other β-barrels (9). Barrel stability depends in large part on the number of hydrogen bonds formed between β-strands, and this low BamA stability with a melting temperature (Tm) around 37° C is intriguing, because it suggests that the BamA barrel may be highly dynamic at this physiological temperature. Moreover, several labs identified mutations in bamA, which destabilize the BamA barrel even further (19; 90). This destabilization is evident by the change in BamA migration in semi-denatured SDS-PAGE gels. The barrel of BamA WT like the barrels of other OMPs is resistant to SDS denaturing unless heated and the folded form migrates faster than the unfolded protein in an SDS-PAGE gel. Several mutations within the barrel domain result in the production of the BamA protein that migrates like the unfolded protein in SDS-gels indicating that the BamA barrel is SDS-sensitive without heat denaturing, demonstrating that the mutations alter the conformation of the barrel domain. Although these mutant BamA proteins migrate as unfolded proteins on SDS-PAGE, we will refer to this alternative conformation as “SDS-labile” instead of “unfolded” to avoid confusion, as such mutant BamA proteins are fully functional (19; 90).
Observation of a weakened BamA seam configuration led to the suggestion that BamA barrel can open laterally exposing unpaired β strands in BamA (67) (Fig. 3A). This opening would create sites for β-strand templating of an incoming OMP, which would get access to the lateral gate by first being threaded through the lumen of BamA barrel. To demonstrate the ability of BamA to open laterally MD simulation of BamA were performed in synthetic lipids under conditions of elevated temperatures (65° C) (67; 68). This choice of temperature is particularly surprising knowing the low thermostability of the BamA barrel. When MD simulations were repeated with BamA incorporated into the asymmetric lipid bilayer resembling the E. coli OM and under physiological temperatures, no lateral opening was observed (25).
Later structures of the Bam complex confirmed the poor interaction between the strands of the seam; however, the exact configuration of the seam varied, particularly in the conformation and tilt of β16 (Fig. 2B) (2; 31; 35; 37). Gu et al, compared the structure of the Bam complex with and without BamB and suggested that the BamA seam, instead of opening/closing laterally, undergoes a scissor-like movement to change barrel conformation from an inward (periplasmic)- to an outward open state and that BamB regulates this confirmation change (31). Later a cryo-EM structure of the Bam complex showed a partially open BamA barrel, and that this was the only state of BamA in the pentameric complex (37).
It is important to note, that all of the structures solved are of a detergent-solubilized BamA and therefore, may not represent BamA in its native membrane environment. Since an opened BamA barrel was observed in the absence of an OMP substrate, it is difficult to imagine how such a BamA conformation would interact with the outer membrane. BamA contains a large, water-filled cavity, and lateral opening of the barrel would expose it to the hydrophobic core of the lipid bilayer. More explanation is needed for how such a thermodynamically unfavorable conformation is achieved without an external energy source.
Perhaps the main argument used to support the budding model is the observation that strains expressing BamA with the seam crosslinked by a disulfide bond are not viable (67) (Fig. 2B). While the result is interesting, this whole-cell based assay does not provide direct evidence for a β-stand templating at the BamA seam, and it does not differentiate between alternative explanations to as why Cys crosslinked BamA is not functional. For example, this assay cannot differentiate between reduced kinetics and loss of function of BamA. Indeed, in vitro studies with purified proteins show that Cys crosslinked BamA shows reduced kinetics and not loss of function (see below). Moreover, such crosslinking may result in overall conformational changes of BamA barrel (compare the BamA structure in “open” and “closed” conformation (Fig. 2B), or it may limit the mobility of BamA barrel domain, including extracellular loops, preventing conformational changes of BamA during the assembly process.
Reduced kinetics versus a loss of function
If BamA indeed opens laterally to template β-strands, this function of BamA is expected to be essential. One the other hands, even if the function of BamA is only partially compromised (e.g. due to changes in conformational dynamics), it can also result in loss of viability because OMPs exist in large excess (over one-hundred fold) to the Bam complexes and accumulation of unfolded OMPs in the periplasm is toxic. This phenotype would be similar to the synthetic lethal phenotype caused by loss of the non-essential components. For example, the presence of bamE or bamB is not required for viability, however, double deletion is lethal unless suppressor mutations in bamA are present (90) or the σE envelope stress response is pre-activated or kinetically enhanced to minimize periplasmic damage (52). Under these suppressed conditions, the bamB bamE double mutant grows normally, suggesting that BamB and BamE do not perform an essential function but are rather required for the full efficiency of the Bam machine. Recent studies have shown using an in vitro system with OmpT as a substrate that the Cys crosslinked BamA variant is still able to assemble OmpT into proteoliposome but with reduced kinetics (37). This result is inconsistent with the budding model, in which opening of the seam is expected to be fully required for β-barrel folding.
BamA conformational mobility
Several experimental studies provided strong of evidence that BamA undergoes conformation changes during the assembly process that involve functional communication between different domains of BamA, including the POTRA domains, the barrel, and eL6, and that the Bam lipoproteins regulate such conformational changes by direct interaction with the POTRA domains.
bamD_R197L is a gain of function mutation in bamD, which allows BamD to function independently of its stable interaction with BamA (75). Interestingly, bamD_R197L biases BamA to a conformation in which BamA barrel becomes sensitive to extracellular protease and eL6 becomes accessible to a Mal-PEG labeling (76). The same phenotype, on the other hand, is also conferred by a loss of function mutation in bamE demonstrating the opposite role of BamE and BamD in regulating BamA conformation (76; 77). This result led to the model in which BamA cycles between two alternative conformational states. While single bamE or bamD_R197L mutations only bias BamA towards one of the conformations and do not confer an OMP assembly defect, combination of the two result in a synthetic lethal phenotype, suggesting that when BamA is arrested in one of the conformations, it is not functional (76). BamD interacts with BamA through the P5 domain, and the salt bridge between BamD_R197 and BamA_E373 residues is of particular importance (75). While the bamA_E373K mutation is lethal due to the disruption of the BamA/D interaction, bamA_E373A mutant is viable but displays the OM permeability indicative of a partial loss of function in BamA. Importantly, a suppressor mutation of this phenotype mapped to the eL6, supporting the functional communication between P5/BamD and eL6 (75). Since P5 and eL6 do not interact directly, it is likely that this communication is transduced through the conformation change in the barrel itself.
The same functional connection is also evident from a suppressor analysis of bamA mutations that disrupt the proper interaction of eL6 with the barrel (19). These mutations, bamA_R661G (in eL6) or bamA_D740G (in the barrel) result in an OM permeability phenotype, and BamA accumulates in these strains as an SDS-labile form suggesting altered conformation of BamA barrel domain. Several suppressor mutations in the barrel but also in P5 were isolated. Interestingly, these suppressors restored OM permeability but did not restore SDS-resistant conformation of the barrel, indicating that the SDS-labile form of BamA is fully functional (19).
In a separate study, several bamA mutations that alter eL6 or the barrel domain where isolated as suppressors of bamE bamB synthetic lethality (90). These mutations restored OMP assembly and allowed growth of the bamE bamB strain at non-permissive temperature. Several of these mutant BamAs also had different mobility on the SDS-PAGE suggesting altered barrel conformation (25). These barrel and eL6 altering mutations, essentially confer a gain of function phenotype bypassing the requirement for BamA communication with the periplasmic lipoproteins. One of the mutations, bamA_F494L, was later shown to also be a partial bypass suppressor of bamD (63). Although bamD is still essential, it can be almost completely depleted in bamA_F494L strain without affecting viability (63).
Restricting BamA flexibility by Cys crosslinking may affect different substrates differently. While none of the β-barrels, except for BamA itself and LptD, are essential, an assembly defect of a non-essential substrate may cause harmful effects if it accumulates at the Bam complex and inhibits Bam activity. Therefore, it is critical to resolve the underlying defects in the Cys-crosslinked BamA strains before drawing fundamental conclusions about the importance of the lateral gate.
3.2. BamA assisted model
One of the features of β-barrel proteins is the unusually high free energy of folding, and β-barrel proteins can spontaneously fold in vitro in the presence of a detergent or into thin lipid bilayers (24; 46). Biophysical studies of such unassisted folding revealed a multi-step process (36; 47; 48). First, an OMP binds to the membrane surface, and the secondary structures start to form at the membrane interface. The OMP then goes through a transition step, also referred as a molten globule, in which β-hairpins start to form and penetrate the membrane in a coordinated manner. This process continues until the barrel is fully formed and membrane integrated. Such spontaneous folding can only occur in thin membranes made up by the short acyl chain lipids and at a temperature at or above phase transition, demonstrating the requirement of a membrane defect for an insertion (15). Native E. coli phospholipids, such as phosphoethanolamine (PE) and or phosphoglycerol (PG), do not support spontaneous OMP folding and hence a model was proposed that the Bam complex functions to reduce the kinetic barrier of membrane insertion by either creating a membrane defect and/or stabilizing certain transition conformations of the OMP at the membrane (28). Consistent with this, incorporation of BamA into the PE-based liposomes accelerates OMP folding (28). Analysis of the rate of such BamA- assisted folding revealed that BamA does not establish significant thermodynamic interactions with the substrates (72), and this transient interaction is in contrast to what is expected from the budding model in which a BamA/OMP hybrid barrel is formed.
The crystal structure of BamA revealed that the barrel domain is only 9–12 Å thick next to its seam suggesting that it may cause a membrane defect either by membrane thinning or by a hydrophobic mismatch between the membrane and aromatic rings of the BamA barrel (68). The thinning of the phospholipid bilayer was observed in BamA containing proteoliposomes by electron microscopy (86). Such thinning can create a membrane defect similar to what was observed in in vitro studies with short-chain lipid containing vesicles. However, the exact nature of such a membrane defect in vivo is not understood. Recent MD simulations of the full-length BamA and the cryo-EM structure of the Bam complex in a detergent micelle suggested that Bam complex interactions with the OM are not limited to the BamA barrel domain, but may also involve the POTRA domains and the Bam lipoproteins (25; 37). Though the function of these interactions with the membrane is currently unknown, it is possible that they contribute to the alteration of membrane architecture around the Bam complex.
The BamA-assisted model is based on the intrinsic properties of the β-barrels protein which serve as a driving force for folding (thermodynamic pull) and the function of the Bam complex is not to actively fold proteins per se but to reduce the activation energy for a membrane insertion (kinetic push) (24) (Fig. 3A). Many aspects of this model have been worked out in vitro using biophysical methods, which allow tracking of the folding intermediates and their membrane insertion. However, in vitro studies are limited by inability to reconstitute the LPS-containing asymmetric bilayer to resemble the OM, and Bam-assisted folding has been reconstituted only for relatively simple monomeric substrates. In vivo, characterization of the β-barrel assembly pathway is limited because it is essential and cannot be significantly perturbed and assembly intermediates are short-lived, making them difficult to characterize. However, in vivo studies of complex, slow folding substrates, such as LptD and EspP provided evidence that the β-barrel folding is initiated and largely occurs in the periplasm/periplasmic side of the OM followed by the membrane insertion (Fig. 3B and C). Thus, the available in vivo evidence supports the BamA-assisted model rather than the budding model.
4. Experimental evidence for β-barrel folding at the periplasmic side of the OM
Studies with LptD
LptD is an essential OM component of the LPS translocon, which is directly responsible for translocation of LPS molecules into the outer leaflet of the OM (95) (Fig. 1). It is the largest β-barrel (26 strands) and it also contains an N-terminal periplasmic domain (5; 12; 18; 74). LptD exists in 1:1 complex with lipoprotein LptE. LptE largely resides inside the lumen of LptD forming an extensive network of site-specific contacts. LptE serves three distinct functions: it is required for LptD assembly; it plugs large LptD lumen so it cannot function as a pore and participates directly in LPS translocation (11; 29; 60). The LptD assembly pathway is very complex. Not only is LptD folded around its partner LptE, but it also contains two non-consecutive disulfide bonds, which are formed and rearranged during the LptD assembly process. There are two N-terminal cysteines, C31, and C173 and two C-terminal cysteines, C724 and C725. Upon secretion in the periplasm, the N-terminal Cys residues form a consecutive disulfide bond, resulting in [1,2] oxidized LptD, a reaction catalyzed by DsbA (13; 79). The rearrangement of [1,2] oxidized LptD into the mature form with non-consecutive disulfide bonds ([1,3][2,4]-LptD) happens after [1,2] LptD is folded around LptE, which brings C-terminal Cys residues in proximity to N-terminal Cys residues (11). Accordingly, conditions that limit LptE stall LptD folding and LptD accumulates in the [1,2] form (56). In addition, when LptE is limiting the [1,2] form of LptD does not accumulate on the Bam complex, demonstrating that LptE is recognized by the Bam complex first, and suggesting that LptE is required not only for LptD folding but also for a recognition of LptD by the Bam complex (56).
Because of the complexity of the process, assembly of LptD/E complex is relatively slow making it possible to track assembly intermediates using pulse-chase analysis (13). The LptD oxidation status and the formation of site-specific contacts with LptE can be monitored in this process, which allows not only the characterization of different assembly intermediates but also the establishment temporal order in the assembly process (11–13; 79). Studies of assembly-defective substrates can be even more informative, as these defective substrates are often slowed down or stalled at a particular assembly step.
One such study concerned an assembly-defective mutant of lptD, lptD4213 (56). lptD4213 is 23 amino acid deletion in eL4, and this mutant was instrumental to the discovery of the Bam complex (7; 20; 80; 81; 94). LptD4213 accumulates in [1,2] oxidized form and unlike LptD WT can be easily crosslinked to the Bam complex as an assembly intermediate. Interaction studies of LptD4213 revealed that it is stalled on the Bam complex together with LptE. Moreover, it exists as a partially folded barrel, because multiple site-specific contacts, which are characteristic of mature LptD/LptE complex, have been established between LptD4213 and LptE (Fig. 3B). Although largely folded, the barrel of LptD4213 is not entirely closed and LptE residues, which face the bulge of LptD barrel in the mature complex, can still be crosslinked to BamA and BamD. Because BamD is the periplasmic component of the Bam complex, this crosslinking clearly establishes that such partially folded LptD/LptE still remains exposed to the periplasm. This result is in striking contrast to the proposed budding model. It also worth noting that the BamA barrel is simply not big enough to accommodate LptE in its lumen to allow LptD folding around LptE at the BamA lateral gate. Although the precise sites of interaction between LptD4213 and BamA and BamD are unknown, the site-specific crosslinking between LptE and BamA and BamD suggests that BamA and BamD interact with the N-terminal and C-terminal parts of LptD barrel domain (Fig. 3B). Once the barrel is closed, it is released from the Bam complex into the OM.
One can argue that LptD may be unique in its assembly pathway as it is the only β-barrel, which requires another partner protein, LptE, for its assembly. However, many larger β-barrels feature the presence of the plug domains or large extracellular loops folded inside the lumen of the β-barrel, which may serve a similar function.
One of the groups of β-barrels that features plug domains is TonB-dependent transporters, including FhuA (23; 57). Their plug domains share large interaction interface with the lumen with multiple hydrogen bonds and salt bridges. Although the assembly of such barrels have not been studied in detail, it is striking that FhuA shares the same chaperone pathways as LptD, and these are the only two proteins known to absolutely require SurA but also dependent on Skp/FkpA (84; 91). The plug and the barrel domains of FhuA can be separated into two polypeptides, yet form a functional translocon (6). While at the time, it was taken as an indication that the plug is inserted into the lumen after the barrel is formed, we suggest it may function in the assembly as separate protein, similar to LptE. Consistent with this notion, deletion of the plug domain of FhuA results in ten-fold lower accumulation of the “barrel only” FhuA, which is indicative of an assembly defect and the levels of the “barrel only” FhuA can be restored by co-expressing the plug domain as a separate polypeptide (6).
Several larger OMPs contain extracellular loops folded inside the barrel, for example eL3 of trimeric porins, such as OmpC, F, LamB and PhoE (3; 14; 83; 98) as well as eL6 of BamA itself. Early studies of PhoE showed that when Cys residues were introduced in eL3 and the lumen, they formed a disulfide bond in the periplasm in a DsbA-dependent manner, and because this disulfide formation requires of at least some degree of tertiary structure formation it was suggested that barrel folding begins in the periplasm (22). Recent studies focused on BamA eL6 have shown that the formation of proper interaction network between eL6 and BamA lumen is required for BamA folding (96). The disruption of this interaction by introducing amino acid substitutions at the eL6 (e.g. BamA_V660A/R661A) result in a BamA assembly defect; this altered BamA accumulates in the periplasm on BamD and is the subject for a degradation by the periplasmic protease DegP. Moreover, proper eL6 folding inside the lumen is a prerequisite for membrane insertion because the assembly and membrane integration of BamA_ V660A/R661A can be rescued by introducing a disulfide bond to tether eL6 to the lumen (96). These results suggest that eL6 may play a role in BamA barrel folding similar to the role of LptE in LptD folding. Because eL6 interacts with several β-strands, the β-sheet has to be at least partially formed to establish this tertiary interaction, further supporting the model for periplasmic folding of β-barrels.
Studies with EspP
EspP is an autotransporter expressed by enterohemorrhagic E. coli (EHEC) (8). Like other members of the serine protease autotransporters of Enterobacteriaceae (SPATE) family, it consists of an N-terminal passenger domain, which is released by autoproteolytic cleavage from the C-terminal β-barrel domain. (16). Earlier models of autotransporter mechanism suggested that the β-barrel domain is sufficient for translocation of the passenger domain, hence the name autotransporter. However, later crystallographic analysis showed that the barrel is not big enough for a passenger translocation, and this prompted a number of mechanistic studies which revealed that the translocation event is coordinated with barrel folding at the Bam complex (38–40; 71). These studies were not only important for our current understanding of the mechanism of autotransporter secretion (reviewed in (4)) but also made several important implications for β-barrel assembly at the Bam complex.
These autotransporter studies also utilized a combination of pulse-chase experiments and site-specific crosslinking with several assembly/translocation intermediates that stall EspP at different stages. Folding of the EspP barrel begins in the periplasm, as evidenced by incorporation of the C-terminal part of the passenger domain into the partially folded EspP barrel in such a way that it becomes inaccessible to proteolysis and site-specific labeling even in permeabilized cells. This conformation resembles that of mature EspP (Fig. 3C). Assembly defective mutant proteins EspP_G1066A or G1081A are slowed down at the Bam complex in a pretranslocation state (71). At this stage, the C-terminal part of the passenger domain adopts a hairpin conformation but has not yet emerged on the cell surface. EspP_G1066A and G1081A remain periplasmic because they can be crosslinked not only to BamA but also to periplasmic lipoproteins, such as BamB and BamD, and unlike the EspP_WT cannot be crosslinked to LPS. The other variant, EspP_586TEV, stalls EspP during the translocation process, at a stage in which the first C-terminal passenger residues emerge at the cell surface, but the translocation event has not been completed because the passenger is not released and still can be crosslinked to BamA and SurA (38). Importantly, at this stage the barrel of EspP_586TEV is still not fully inserted into the OM because barrel does not establish site-specific crosslinked to LPS and remains its crosslinking with BamB and BamD (Fig. 3C). The intermediates described above can also be detected using the WT protein under conditions when EspP assembly is slowed down by growth at 20° C (40; 71).
Although EspP and LptD represent completely different BamA substrates, the similarity between their assembly pathways is remarkable (Fig. 3B and C), suggesting it may apply more broadly to the assembly of other β-barrel proteins. These experimental studies provide strong evidence that the barrel folding occurs in the periplasm/periplasmic side of the OM and that neither EspP nor LptD is templated at the BamA lateral gate. It is true that LptD and EspP are large and complex substrates and may require specialized activities of the Bam complex for their assembly; however, it is unlikely that other, more simple, β-barrel proteins would require β-strand templating while bigger and more complex substrates do not. In addition, results of site-specific crosslinking suggest that BamA and BamD interact with limited sites on the substrates, mainly limited to the N- and C-terminal parts of the barrel. This may be important for restricting the conformational freedom of an OMP, which will trigger β-barrel formation driven by the intrinsic thermodynamic properties of the OMPs, similar to what is observed in vitro.
5. Concluding remarks
Understanding the mechanism of β-barrel folding/insertion into the OM will require understanding how individual members of the complex functionally interact with incoming substrates and the conformational changes in both substrate and machine that occur during this process. Structural studies of the Bam complex in the absence of the substrate and away from its native environment cannot reveal all of the dynamic changes the Bam complex undergoes during the OMP assembly. As previously demonstrated with transcription (RNA polymerase) and translation (ribosome), isolation and characterization of assembly intermediates are critical for advancing the field. Structural information on kinetically stable intermediates can provide snapshots of the various dynamic events during the assembly reaction. Finally, because the OM may play a critical role in β-barrel folding, the question of how the Bam complex functions within this unique asymmetric bilayer requires the characterization of assembly intermediates in whole cells (or OM vesicles).
References
- 1.Albrecht R, Schutz M, Oberhettinger P, Faulstich M, Bermejo I, et al. Structure of BamA, an essential factor in outer membrane protein biogenesis. Acta Crystallogr D Biol Crystallogr. 2014;70:1779–89. doi: 10.1107/S1399004714007482. [DOI] [PubMed] [Google Scholar]
- 2.Bakelar J, Buchanan SK, Noinaj N. The structure of the beta-barrel assembly machinery complex. Science. 2016;351:180–6. doi: 10.1126/science.aad3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basle A, Rummel G, Storici P, Rosenbusch JP, Schirmer T. Crystal structure of osmoporin OmpC from E. coli at 2.0 A. J Mol Biol. 2006;362:933–42. doi: 10.1016/j.jmb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein HD. Looks can be deceiving: recent insights into the mechanism of protein secretion by the autotransporter pathway. Mol Microbiol. 2015;97:205–15. doi: 10.1111/mmi.13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Botos I, Majdalani N, Mayclin SJ, McCarthy JG, Lundquist K, et al. Structural and Functional Characterization of the LPS Transporter LptDE from Gram-Negative Pathogens. Structure. 2016;24:965–76. doi: 10.1016/j.str.2016.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun M, Endriss F, Killmann H, Braun V. In vivo reconstitution of the FhuA transport protein of Escherichia coli K-12. J Bacteriol. 2003;185:5508–18. doi: 10.1128/JB.185.18.5508-5518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun M, Silhavy TJ. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol Microbiol. 2002;45:1289–302. doi: 10.1046/j.1365-2958.2002.03091.x. [DOI] [PubMed] [Google Scholar]
- 8.Brunder W, Schmidt H, Karch H. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol Microbiol. 1997;24:767–78. doi: 10.1046/j.1365-2958.1997.3871751.x. [DOI] [PubMed] [Google Scholar]
- 9.Burgess NK, Dao TP, Stanley AM, Fleming KG. Beta-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. J Biol Chem. 2008;283:26748–58. doi: 10.1074/jbc.M802754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charlson ES, Werner JN, Misra R. Differential effects of yfgL mutation on Escherichia coli outer membrane proteins and lipopolysaccharide. J Bacteriol. 2006;188:7186–94. doi: 10.1128/JB.00571-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chimalakonda G, Ruiz N, Chng SS, Garner RA, Kahne D, Silhavy TJ. Lipoprotein LptE is required for the assembly of LptD by the beta-barrel assembly machine in the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A. 2011;108:2492–7. doi: 10.1073/pnas.1019089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chng SS, Ruiz N, Chimalakonda G, Silhavy TJ, Kahne D. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc Natl Acad Sci U S A. 2010;107:5363–8. doi: 10.1073/pnas.0912872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chng SS, Xue M, Garner RA, Kadokura H, Boyd D, et al. Disulfide rearrangement triggered by translocon assembly controls lipopolysaccharide export. Science. 2012;337:1665–8. doi: 10.1126/science.1227215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowan SW, Schirmer T, Rummel G, Steiert M, Ghosh R, et al. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–33. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 15.Danoff EJ, Fleming KG. Membrane defects accelerate outer membrane beta-barrel protein folding. Biochemistry. 2015;54:97–9. doi: 10.1021/bi501443p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dautin N. Serine protease autotransporters of enterobacteriaceae (SPATEs): biogenesis and function. Toxins (Basel) 2010;2:1179–206. doi: 10.3390/toxins2061179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong C, Yang X, Hou HF, Shen YQ, Dong YH. Structure of Escherichia coli BamB and its interaction with POTRA domains of BamA. Acta Crystallogr D Biol Crystallogr. 2012;68:1134–9. doi: 10.1107/S0907444912023141. [DOI] [PubMed] [Google Scholar]
- 18.Dong H, Xiang Q, Gu Y, Wang Z, Paterson NG, et al. Structural basis for outer membrane lipopolysaccharide insertion. Nature. 2014;511:52–6. doi: 10.1038/nature13464. [DOI] [PubMed] [Google Scholar]
- 19.Dwyer RS, Ricci DP, Colwell LJ, Silhavy TJ, Wingreen NS. Predicting functionally informative mutations in Escherichia coli BamA using evolutionary covariance analysis. Genetics. 2013;195:443–55. doi: 10.1534/genetics.113.155861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eggert US, Ruiz N, Falcone BV, Branstrom AA, Goldman RC, et al. Genetic basis for activity differences between vancomycin and glycolipid derivatives of vancomycin. Science. 2001;294:361–4. doi: 10.1126/science.1063611. [DOI] [PubMed] [Google Scholar]
- 21.Endo T, Kawano S, Yamano K. BamE structure: the assembly of beta-barrel proteins in the outer membranes of bacteria and mitochondria. EMBO Rep. 2011;12:94–5. doi: 10.1038/embor.2010.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eppens EF, Nouwen N, Tommassen J. Folding of a bacterial outer membrane protein during passage through the periplasm. EMBO J. 1997;16:4295–301. doi: 10.1093/emboj/16.14.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science. 1998;282:2215–20. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 24.Fleming KG. A combined kinetic push and thermodynamic pull as driving forces for outer membrane protein sorting and folding in bacteria. Philos Trans R Soc Lond B Biol Sci. 2015:370. doi: 10.1098/rstb.2015.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleming PJ, Patel DS, Wu EL, Qi Y, Yeom MS, et al. BamA POTRA Domain Interacts with a Native Lipid Membrane Surface. Biophys J. 2016;110:2698–709. doi: 10.1016/j.bpj.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gatzeva-Topalova PZ, Walton TA, Sousa MC. Crystal structure of YaeT: conformational flexibility and substrate recognition. Structure. 2008;16:1873–81. doi: 10.1016/j.str.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gatzeva-Topalova PZ, Warner LR, Pardi A, Sousa MC. Structure and flexibility of the complete periplasmic domain of BamA: the protein insertion machine of the outer membrane. Structure. 2010;18:1492–501. doi: 10.1016/j.str.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gessmann D, Chung YH, Danoff EJ, Plummer AM, Sandlin CW, et al. Outer membrane beta-barrel protein folding is physically controlled by periplasmic lipid head groups and BamA. Proc Natl Acad Sci U S A. 2014;111:5878–83. doi: 10.1073/pnas.1322473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grabowicz M, Andres D, Lebar MD, Malojcic G, Kahne D, Silhavy TJ. A mutant Escherichia coli that attaches peptidoglycan to lipopolysaccharide and displays cell wall on its surface. Elife. 2014;3:e05334. doi: 10.7554/eLife.05334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gromiha MM, Suwa M. Current developments on beta-barrel membrane proteins: sequence and structure analysis, discrimination and prediction. Curr Protein Pept Sci. 2007;8:580–99. doi: 10.2174/138920307783018712. [DOI] [PubMed] [Google Scholar]
- 31.Gu Y, Li H, Dong H, Zeng Y, Zhang Z, et al. Structural basis of outer membrane protein insertion by the BAM complex. Nature. 2016;531:64–9. doi: 10.1038/nature17199. [DOI] [PubMed] [Google Scholar]
- 32.Hagan CL, Kim S, Kahne D. Reconstitution of outer membrane protein assembly from purified components. Science. 2010;328:890–2. doi: 10.1126/science.1188919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hagan CL, Westwood DB, Kahne D. bam Lipoproteins Assemble BamA in vitro. Biochemistry. 2013;52:6108–13. doi: 10.1021/bi400865z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hagan CL, Wzorek JS, Kahne D. Inhibition of the beta-barrel assembly machine by a peptide that binds BamD. Proc Natl Acad Sci U S A. 2015;112:2011–6. doi: 10.1073/pnas.1415955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han L, Zheng J, Wang Y, Yang X, Liu Y, et al. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat Struct Mol Biol. 2016;23:192–6. doi: 10.1038/nsmb.3181. [DOI] [PubMed] [Google Scholar]
- 36.Huysmans GH, Baldwin SA, Brockwell DJ, Radford SE. The transition state for folding of an outer membrane protein. Proc Natl Acad Sci U S A. 2010;107:4099–104. doi: 10.1073/pnas.0911904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iadanza MG, Higgins AJ, Schiffrin B, Calabrese AN, Brockwell DJ, et al. Lateral opening in the intact beta-barrel assembly machinery captured by cryo-EM. Nat Commun. 2016;7:12865. doi: 10.1038/ncomms12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ieva R, Bernstein HD. Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc Natl Acad Sci U S A. 2009;106:19120–5. doi: 10.1073/pnas.0907912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ieva R, Skillman KM, Bernstein HD. Incorporation of a polypeptide segment into the beta-domain pore during the assembly of a bacterial autotransporter. Mol Microbiol. 2008;67:188–201. doi: 10.1111/j.1365-2958.2007.06048.x. [DOI] [PubMed] [Google Scholar]
- 40.Ieva R, Tian P, Peterson JH, Bernstein HD. Sequential and spatially restricted interactions of assembly factors with an autotransporter beta domain. Proc Natl Acad Sci U S A. 2011;108:E383–91. doi: 10.1073/pnas.1103827108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishida H, Garcia-Herrero A, Vogel HJ. The periplasmic domain of Escherichia coli outer membrane protein A can undergo a localized temperature dependent structural transition. Biochim Biophys Acta. 2014;1838:3014–24. doi: 10.1016/j.bbamem.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 42.Jansen KB, Baker SL, Sousa MC. Crystal structure of BamB bound to a periplasmic domain fragment of BamA, the central component of the beta-barrel assembly machine. J Biol Chem. 2015;290:2126–36. doi: 10.1074/jbc.M114.584524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim KH, Aulakh S, Paetzel M. Crystal structure of beta-barrel assembly machinery BamCD protein complex. J Biol Chem. 2011;286:39116–21. doi: 10.1074/jbc.M111.298166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim KH, Paetzel M. Crystal structure of Escherichia coli BamB, a lipoprotein component of the beta-barrel assembly machinery complex. J Mol Biol. 2011;406:667–78. doi: 10.1016/j.jmb.2010.12.020. [DOI] [PubMed] [Google Scholar]
- 45.Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–4. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 46.Kleinschmidt JH. Folding of beta-barrel membrane proteins in lipid bilayers - Unassisted and assisted folding and insertion. Biochim Biophys Acta. 2015;1848:1927–43. doi: 10.1016/j.bbamem.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 47.Kleinschmidt JH, Bulieris PV, Qu J, Dogterom M, den Blaauwen T. Association of neighboring beta-strands of outer membrane protein A in lipid bilayers revealed by site-directed fluorescence quenching. J Mol Biol. 2011;407:316–32. doi: 10.1016/j.jmb.2011.01.021. [DOI] [PubMed] [Google Scholar]
- 48.Kleinschmidt JH, Tamm LK. Time-resolved distance determination by tryptophan fluorescence quenching: probing intermediates in membrane protein folding. Biochemistry. 1999;38:4996–5005. doi: 10.1021/bi9824644. [DOI] [PubMed] [Google Scholar]
- 49.Knowles TJ, Browning DF, Jeeves M, Maderbocus R, Rajesh S, et al. Structure and function of BamE within the outer membrane and the beta-barrel assembly machine. EMBO Rep. 2011;12:123–8. doi: 10.1038/embor.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Konovalova A, Mitchell AM, Silhavy TJ. A lipoprotein/beta-barrel complex monitors lipopolysaccharide integrity transducing information across the outer membrane. Elife. 2016:5. doi: 10.7554/eLife.15276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konovalova A, Perlman DH, Cowles CE, Silhavy TJ. Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of beta-barrel proteins. Proc Natl Acad Sci U S A. 2014;111:E4350–8. doi: 10.1073/pnas.1417138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Konovalova A, Schwalm JA, Silhavy TJ. A Suppressor Mutation That Creates a Faster and More Robust sigmaE Envelope Stress Response. J Bacteriol. 2016;198:2345–51. doi: 10.1128/JB.00340-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Konovalova A, Silhavy TJ. Outer membrane lipoprotein biogenesis: Lol is not the end. Philos Trans R Soc Lond B Biol Sci. 2015:370. doi: 10.1098/rstb.2015.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–9. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 55.Kutik S, Stojanovski D, Becker L, Becker T, Meinecke M, et al. Dissecting membrane insertion of mitochondrial beta-barrel proteins. Cell. 2008;132:1011–24. doi: 10.1016/j.cell.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 56.Lee J, Xue M, Wzorek JS, Wu T, Grabowicz M, et al. Characterization of a stalled complex on the beta-barrel assembly machine. Proc Natl Acad Sci U S A. 2016;113:8717–22. doi: 10.1073/pnas.1604100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Locher KP, Rees B, Koebnik R, Mitschler A, Moulinier L, et al. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell. 1998;95:771–8. doi: 10.1016/s0092-8674(00)81700-6. [DOI] [PubMed] [Google Scholar]
- 58.Mahoney TF, Ricci DP, Silhavy TJ. Classifying beta-Barrel Assembly Substrates by Manipulating Essential Bam Complex Members. J Bacteriol. 2016;198:1984–92. doi: 10.1128/JB.00263-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, et al. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol. 2006;61:151–64. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- 60.Malojcic G, Andres D, Grabowicz M, George AH, Ruiz N, et al. LptE binds to and alters the physical state of LPS to catalyze its assembly at the cell surface. Proc Natl Acad Sci U S A. 2014;111:9467–72. doi: 10.1073/pnas.1402746111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matsuyama S, Yokota N, Tokuda H. A novel outer membrane lipoprotein, LolB (HemM), involved in the LolA (p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli. EMBO J. 1997;16:6947–55. doi: 10.1093/emboj/16.23.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.May JM, Sherman DJ, Simpson BW, Ruiz N, Kahne D. Lipopolysaccharide transport to the cell surface: periplasmic transport and assembly into the outer membrane. Philos Trans R Soc Lond B Biol Sci. 2015:370. doi: 10.1098/rstb.2015.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Misra R, Stikeleather R, Gabriele R. In vivo roles of BamA, BamB and BamD in the biogenesis of BamA, a core protein of the beta-barrel assembly machine of Escherichia coli. J Mol Biol. 2015;427:1061–74. doi: 10.1016/j.jmb.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Narita SI, Tokuda H. Bacterial lipoproteins; biogenesis, sorting and quality control. Biochim Biophys Acta. 2016 doi: 10.1016/j.bbalip.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 65.Ni D, Wang Y, Yang X, Zhou H, Hou X, et al. Structural and functional analysis of the beta-barrel domain of BamA from Escherichia coli. FASEB J. 2014;28:2677–85. doi: 10.1096/fj.13-248450. [DOI] [PubMed] [Google Scholar]
- 66.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Noinaj N, Kuszak AJ, Balusek C, Gumbart JC, Buchanan SK. Lateral opening and exit pore formation are required for BamA function. Structure. 2014;22:1055–62. doi: 10.1016/j.str.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Noinaj N, Kuszak AJ, Gumbart JC, Lukacik P, Chang H, et al. Structural insight into the biogenesis of beta-barrel membrane proteins. Nature. 2013;501:385–90. doi: 10.1038/nature12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, Kahne D. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat Rev Microbiol. 2016;14:337–45. doi: 10.1038/nrmicro.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pautsch A, Schulz GE. Structure of the outer membrane protein A transmembrane domain. Nat Struct Biol. 1998;5:1013–7. doi: 10.1038/2983. [DOI] [PubMed] [Google Scholar]
- 71.Pavlova O, Peterson JH, Ieva R, Bernstein HD. Mechanistic link between beta barrel assembly and the initiation of autotransporter secretion. Proc Natl Acad Sci U S A. 2013;110:E938–47. doi: 10.1073/pnas.1219076110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Plummer AM, Fleming KG. BamA Alone Accelerates Outer Membrane Protein Folding In Vitro through a Catalytic Mechanism. Biochemistry. 2015;54:6009–11. doi: 10.1021/acs.biochem.5b00950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Plummer AM, Fleming KG. From Chaperones to the Membrane with a BAM! Trends Biochem Sci. 2016;41:872–82. doi: 10.1016/j.tibs.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qiao S, Luo Q, Zhao Y, Zhang XC, Huang Y. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature. 2014;511:108–11. doi: 10.1038/nature13484. [DOI] [PubMed] [Google Scholar]
- 75.Ricci DP, Hagan CL, Kahne D, Silhavy TJ. Activation of the Escherichia coli beta-barrel assembly machine (Bam) is required for essential components to interact properly with substrate. Proc Natl Acad Sci U S A. 2012;109:3487–91. doi: 10.1073/pnas.1201362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rigel NW, Ricci DP, Silhavy TJ. Conformation-specific labeling of BamA and suppressor analysis suggest a cyclic mechanism for beta-barrel assembly in Escherichia coli. Proc Natl Acad Sci U S A. 2013;110:5151–6. doi: 10.1073/pnas.1302662110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rigel NW, Schwalm J, Ricci DP, Silhavy TJ. BamE modulates the Escherichia coli beta-barrel assembly machine component BamA. J Bacteriol. 2012;194:1002–8. doi: 10.1128/JB.06426-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rossiter AE, Leyton DL, Tveen-Jensen K, Browning DF, Sevastsyanovich Y, et al. The essential beta-barrel assembly machinery complex components BamD and BamA are required for autotransporter biogenesis. J Bacteriol. 2011;193:4250–3. doi: 10.1128/JB.00192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ruiz N, Chng SS, Hiniker A, Kahne D, Silhavy TJ. Nonconsecutive disulfide bond formation in an essential integral outer membrane protein. Proc Natl Acad Sci U S A. 2010;107:12245–50. doi: 10.1073/pnas.1007319107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ruiz N, Falcone B, Kahne D, Silhavy TJ. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell. 2005;121:307–17. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 81.Ruiz N, Wu T, Kahne D, Silhavy TJ. Probing the barrier function of the outer membrane with chemical conditionality. ACS Chem Biol. 2006;1:385–95. doi: 10.1021/cb600128v. [DOI] [PubMed] [Google Scholar]
- 82.Sandoval CM, Baker SL, Jansen K, Metzner SI, Sousa MC. Crystal structure of BamD: an essential component of the beta-Barrel assembly machinery of gram-negative bacteria. J Mol Biol. 2011;409:348–57. doi: 10.1016/j.jmb.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schirmer T, Keller TA, Wang YF, Rosenbusch JP. Structural basis for sugar translocation through maltoporin channels at 3.1 A resolution. Science. 1995;267:512–4. doi: 10.1126/science.7824948. [DOI] [PubMed] [Google Scholar]
- 84.Schwalm J, Mahoney TF, Soltes GR, Silhavy TJ. Role for Skp in LptD assembly in Escherichia coli. J Bacteriol. 2013;195:3734–42. doi: 10.1128/JB.00431-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Simpson BW, May JM, Sherman DJ, Kahne D, Ruiz N. Lipopolysaccharide transport to the cell surface: biosynthesis and extraction from the inner membrane. Philos Trans R Soc Lond B Biol Sci. 2015:370. doi: 10.1098/rstb.2015.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sinnige T, Weingarth M, Renault M, Baker L, Tommassen J, Baldus M. Solid-state NMR studies of full-length BamA in lipid bilayers suggest limited overall POTRA mobility. J Mol Biol. 2014;426:2009–21. doi: 10.1016/j.jmb.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 87.Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, Silhavy TJ. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci U S A. 2007;104:6400–5. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sklar JG, Wu T, Kahne D, Silhavy TJ. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007;21:2473–84. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Snijder HJ, Ubarretxena-Belandia I, Blaauw M, Kalk KH, Verheij HM, et al. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature. 1999;401:717–21. doi: 10.1038/44890. [DOI] [PubMed] [Google Scholar]
- 90.Tellez R, Jr, Misra R. Substitutions in the BamA beta-barrel domain overcome the conditional lethal phenotype of a DeltabamB DeltabamE strain of Escherichia coli. J Bacteriol. 2012;194:317–24. doi: 10.1128/JB.06192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vertommen D, Ruiz N, Leverrier P, Silhavy TJ, Collet JF. Characterization of the role of the Escherichia coli periplasmic chaperone SurA using differential proteomics. Proteomics. 2009;9:2432–43. doi: 10.1002/pmic.200800794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vogt J, Schulz GE. The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence. Structure. 1999;7:1301–9. doi: 10.1016/s0969-2126(00)80063-5. [DOI] [PubMed] [Google Scholar]
- 93.Vuong P, Bennion D, Mantei J, Frost D, Misra R. Analysis of YfgL and YaeT interactions through bioinformatics, mutagenesis, and biochemistry. J Bacteriol. 2008;190:1507–17. doi: 10.1128/JB.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–45. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 95.Wu T, McCandlish AC, Gronenberg LS, Chng SS, Silhavy TJ, Kahne D. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A. 2006;103:11754–9. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wzorek JSJL, Tomasek David, Hagan Christine L, Kahne Daniel E. Membrane integration of an essential β-barrel protein prerequires burial of an extracellular loop. 2017 doi: 10.1073/pnas.1616576114. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yakushi T, Yokota N, Matsuyama S, Tokuda H. LolA-dependent release of a lipid-modified protein from the inner membrane of Escherichia coli requires nucleoside triphosphate. J Biol Chem. 1998;273:32576–81. doi: 10.1074/jbc.273.49.32576. [DOI] [PubMed] [Google Scholar]
- 98.Yamashita E, Zhalnina MV, Zakharov SD, Sharma O, Cramer WA. Crystal structures of the OmpF porin: function in a colicin translocon. EMBO J. 2008;27:2171–80. doi: 10.1038/emboj.2008.137. [DOI] [PMC free article] [PubMed] [Google Scholar]