Abstract
The regulation of gene expression by transcription factors serves a critical function in cell proliferation. Zinc-finger protein 224 (ZNF224), a Krüppel-associated-box-containing zinc finger protein, is known to serve a crucial function in integrating the transcriptional co-factors that activate transcriptional regulation pathways in the cell. A previous study demonstrated that ZNF224 enhances cell proliferation by downregulating the expression of p21 and p53. The present study identified mediator complex subunit 28 (MED28) as a potential binding partner for ZNF224; this was confirmed by co-immunoprecipitation and a surface plasmon resonance assay. Additionally, the KRAB domain at the N-terminal of ZNF224 interacts with the MED domain of MED28. Bimolecular fluorescence complementation analysis revealed that ZNF224 associates with MED28 in the nucleus. In addition, ZNF224 was rapidly degraded upon treatment with the DNA-damaging agent camptothecin (CPT). Transient overexpression of MED28 inhibited the CPT-mediated degradation of ZNF224, resulting in increased colony formation by MCF-7 cells. The molecular mechanisms that underlie the biological outcomes of MED28 expression have not yet been fully elucidated. The present study provides molecular evidence for the function of ZNF224 and MED28 in the DNA-damage response.
Keywords: breast cancer, cancer cell proliferation, DNA damage, mediator complex subunit 28, zinc-finger protein 224
Introduction
Maintaining genetic stability is an important process in all organisms; failure to protect cells from genotoxic stress caused by DNA-damaging agents, including ionizing radiation, ultraviolet radiation and reactive oxygen species, may cause tumour formation (1). The DNA damage response (DDR) signaling pathway is the primary response mechanism against these stresses (1,2). DDR serves a crucial function in DNA repair since it protects the integrity of the genome from genotoxic agents by controlling cell-cycle checkpoints, resulting in cell-cycle arrest or apoptosis in eukaryotic cells (3). The fate of the cell, survival or death, upon DNA damage is determined following DNA repair, cell-cycle checks and the expression of apoptotic proteins (4). In arrested cells, polymerases are inactivated to inhibit DNA replication and transcription and are reactivated once DNA repair is complete (5). In a non-repaired or checkpoint-bypassed cell, however, cell death, via mechanisms including apoptosis, necrosis or mitotic catastrophe, depends on the balance between pro-survival and pro-apoptotic factors (5,6). Proteins that induce cell death via checkpoint mechanisms are known as tumor suppressors; however, overactive proliferative proteins known as oncogenes may induce cancer formation (7).
Tumor protein p53 (hereafter p53) is a tumor suppressor protein that is occasionally referred to as ‘the guardian of the genome’ (8,9). Previous research has shown that p53 is a primary component of the intracellular signaling response against genotoxic stress and that multiple types of human cancer exhibit inactivated p53 owing to inherited mutations (9). Under normal conditions, p53 is commonly inactivated and degraded in a ubiquitin-dependent manner; however, it is upregulated and stabilized under genetically unstable conditions, such as DNA damage, to suppress the progression of tumorigenesis (10). The primary function of p53 upon DNA damage is to regulate the expression of target genes associated with cell fate (11). A previous study revealed that zinc finger protein 224 (ZNF224) functions as an oncogene in breast cancer cells and downregulates expression of p53 and cyclin dependent kinase 1A (CDK1A, also known as p21) upon DNA damage (12). Although the mRNA expression levels of ZNF224 were unaffected by the DDR, ZNF224 protein levels showed a gradual decrease (12). These results indicated that post-translational modification to ZNF224 or the formation of a complex with another regulatory protein may mediate its inhibition upon DNA damage.
The human genome encodes ~30 mediator complex subunit (MED) proteins that control intracellular signaling to the nucleus (13). The major function of these mediators is to transduce signals from transcription factors to RNA polymerase II to aid the regulation of transcription; it has recently been determined that mediators serve a function as co-activators or co-repressors, depending on their interacting proteins (14). MED28 has been demonstrated to promote the migration and proliferation of breast cancer cells through interactions with the mitogen-activated protein kinase kinase-1 (MEK-1) signaling pathway (15–17). However, the target ligands of MED28 have not been validated, either in vitro or in vivo. To improve understanding of the roles of MED28 in proliferating cancer cells, the present study identified its interacting protein as ZNF224 using proteomic and structural approaches. In addition, a cell-based fluorescence assay revealed their functional co-localization in breast cancer cells. The present study also investigated the role of MED28 targeting ZNF224, and the overexpression of MED28 demonstrated a significant difference in terms of differentiation compared to the control sample by colony forming experiment in breast cancer cells.
Materials and methods
Cell culture
Cells from the 293 and human adenoma breast cancer MCF-7 cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM; Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone; GE Healthcare Life Sciences) and 1% penicillin/streptomycin (Hyclone; GE Healthcare Life Sciences).
Plasmids
Constructs expressing Flag-ZNF224 (12), Myc-MED28 (18), and 6× His-MED28 (18) were previously verified. HA-tagged N-terminus of ZNF224 construct was kindly provided by Dr. Constanzo (University of Naples Federico II, Naples, Italy). Flag-ZNF224 and Myc-MED28 constructs were subcloned into pE green fluorescence protein (GFP) and pmCherry vectors, respectively. The following primers were used to synthesize the glutathione S-transferase (GST)-MED28 deletion mutants: Full-length forward, 5′-CGCGGATCCATGGCGGCTC-3′ and reverse, 5′-CCGCTCGAGTCACGTTGGCTTCAG-3′; amino acid (aa) 1–71 forward, 5′-CGCGGATCCATGGCGGCTC-3′ and reverse, 5′-CCGCTCGAGTCATTCCTGATCGGTGCC-3′; aa 1–150 forward, 5′-CGCGGATCCATGGCGGCTC-3′ and reverse, 5′-CCGCTCGAGTCAGTGCTGCACGTTGAT-3′; aa 43–176 forward, 5′-GGAATTCCATATGACTTTGGTGGACGAG-3′ and reverse, 5′-CCGCTCGAGTCACTTCAGAGGTGC-3′; aa 61–176 forward, 5′-GGAATTCCATATGAGTCAGGACTATGTCAATGG-3′ and reverse, 5′-CCGCTCGAGTCACTTCAGAGGTGC-3′; aa 72–176 (MED domain) forward, 5′-CGCGGATCCAAGAAATTCGAACCG-3′ and reverse, 5′-CCGCTCGAGTCACTTCAGAGGTGCA-3′. The above sequences were amplified by PCR using AccuPower PCR premix (Bioneer Corporation, Daejeon, Korea) and inserted into the pGEX-4T3 vector using the BamHI and XhoI restriction sites (GE Healthcare Life Sciences, Little Chalfont, UK) according to the manufacturer's protocol. The thermocycler conditions were as follows: Initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C, annealing at melting temperature (Tm) 55–58°C and extension at 72°C for 1 min each. ZNF224 and MED28 cDNA clones were subcloned into pBiCF-VN173-Flag and pBiFC-VC155-HA vectors, respectively (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany).
Mammalian cell transfection
Plasmid DNA constructs 1 µg DNA were transfected into cells using a mixture of 150 mM NaCl and polyethylenimine (Polysciences, Inc., Warrington, PA, USA) according to the manufacturer's protocol, and cells were plated in a humidified chamber atmosphere comprising 95% air and 5% CO2 at 37°C for 24 h prior to transfection. All subsequent experiments were performed 24 h after transfection.
Colony-forming assay
MCF-7 cells (3×103 cell/well) were transfected with Myc-MED28 and/or Flag-ZNF224, and seeded on 6-well plates and maintained in a humidified chamber atmosphere comprising 95% air and 5% CO2 at 37°C for 24 h. After transfection, 10 µM CPT (Camptothecin; Sigma-Aldrich; Merck KGaA) which dissolved in dimethyl sulfoxide (DMSO) was treated for 2 weeks. Following CPT treatment, cells were fixed with 4% paraformaldehyde at 25°C for 10 min and stained using crystal violet solution (0.05% crystal violet, 1% formaldehyde, 1% methanol and 1X PBS) at 25°C for 30 min. Stained cells were washed with water by dropping gently, and air dried at room temperature. The number of colonies were quantified using the Nikon digital photo camera COOLPIX P310 and ImageJ program (version 1.6.0; National Institutes of Health, Bethesda, MD, USA).
Western blotting and immunoprecipitation
The 293 cells were co-transfected with Flag-ZNFF224, HA-ZNF224-N, Flag-ZNF255 and Myc-MED28 vectors and maintained in a humidified chamber atmosphere comprising 95% air and 5% CO2 at 37°C for 24 h. For immunoprecipitation, cells were harvested and lysed with lysis buffer (50 mM HEPES, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 10 mM NaF, 1 mM NaOV3, 1 mM PMSF, 1 mM EDTA) containing a protease inhibitor cocktail (Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 4°C for 30 min, and centrifuged at 25,000 × g for 10 min at 4°C. Purified 1 µg anti-Flag (cat. no. F3165; Sigma-Aldrich; Merck KGaA) and 1 µg anti-Myc antibodies (cat no. sc-40; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) were incubated with 500 µg of protein extracts at 4°C for 4 h. Then, protein A/G PLUS agarose beads (cat. no. sc-2003; Santa Cruz Biotechnology, Inc.) were added and rotated at 0.04 × g at 4°C for 2 h on Rotator (FINEPCR, Gunpo-si, Korea). The beads were collected by centrifugation at 3,000 × g for 1 min, and washed three times in lysis buffer and resuspended in sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer [120 mM Tris-HCl (pH 6.8), 20% glycerol, 4% SDS, 28.8 mM 2-mercaptoethanol, 0.01% bromophenol blue]. The samples were boiled in SDS-PAGE sample buffer [120 mM Tris-HCl (pH 6.8), 20% glycerol, 4% SDS, 28.8 mM 2-mercaptoethanol, 0.01% bromophenol blue] at 100°C for 10 min and separated by SDS-PAGE on a 12.5% gel. For western blotting, the whole cell lysates were quantified using a Bradford protein assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA) through SpectroStar (BMG Labtech GmbH, Ortenberg, Germany). The whole cell lysates (30 µg) and immunoprecipitation sample (30 µl) were subjected to SDS-PAGE, and transferred to a polyvinyldene fluoride membrane (EMD Millipore, Billerica, MA, USA). Each transferred membrane was blocked with TBS with Tween-20 (TBS-T) buffer [20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.2% Tween-20] containing 5% skim milk at 25°C for 1 h, and then incubated with the appropriate primary antibodies diluted in 1% bovine serum albumin (BSA; Bovogen, Victoria, Australia) in TBS-T buffer at 4°C for overnight. The following primary antibodies were used: M2 anti-Flag (cat. no. F3165; 1:3,000; Sigma-Aldrich; Merck KGaA), anti-Myc (cat. no. sc-40; 1:1,000; Santa Cruz Biotechnology, Inc.), anti-HA (cat. no. sc-57592; 1:1,000; Santa Cruz Biotechnology, Inc.), anti-ZNF224 (cat. no. ab168669; 1:1,000; Abcam, Cambridge, UK), anti-MED28 (1:10,000; laboratory-made), and anti-alpha-tubulin (1:20,000; laboratory-made). The membranes were washed three times in TBS-T buffer, and subjected to incubation with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G as a secondary antibody (cat. no. 31430; 1:25,000; Thermo Fisher Scientific, Inc.) at 25°C for 1 h. An enhanced chemiluminescence detection system (Santa Cruz Biotechnology, Inc.) was used for detection according to the manufacturer's protocol, and the membranes were exposed to the X-ray film (Fujifilm, Tokyo, Japan) in a dark room, and then developed by Vivid X-ray developer and Fixer solution (Duksan, Seoul, Korea) according to the manufacturer's protocol.
GST fusion protein purification and pull-down assays
BL-21 cells expressing the recombinant protein of GST-tagged MED28 full length and deletion mutants were lysed in a lysis buffer [10 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 200 mM PMSF, 5 mM DTT, 1% Triton X-100, 100 µg/ml lysozyme and PIC] at 4°C for 15 min. Cell lysates expressing GST-tagged proteins were purified using Glutathione Separose™ 4B (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) at 4°C for 2 h according to the manufacturer's protocol. Purified 10 µg GST-tagged proteins were incubated with Flag-ZNF224 transfected 293 cells lysate at 4°C for 1 h. The beads were collected by centrifugation at 3,000 × g for 1 min, and extensively washed three times with cell lysis buffer (50 mM HEPES, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 10 mM NaF, 1 mM NaOV3, 1 mM PMSF, 1 mM EDTA) containing a protease inhibitor cocktail (Thermo Fisher Scientific, Inc.). The bound proteins were eluted by boiling in the SDS-PAGE sample buffer [120 mM Tris-HCl (pH 6.8), 20% glycerol, 4% SDS, 28.8 mM 2-mercaptoethanol, 0.01% bromophenol blue] at 100°C for 10 min, and 30 µg of whole cell lysate and 30 µl of samples were subjected to SDS-PAGE and western blotting analysis as described above with an M2 anti-Flag (cat. no. F3165; 1:3,000; Sigma-Aldrich; Merck KGaA) antibody.
Matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) and liquid chromatography-mass spectrometry (LC-MS/MS) analysis
MED28 overexpressed cells were lysed with lysis buffer (50 mM HEPES, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 10 mM NaF, 1 mM NaOV3, 1 mM PMSF, 1 mM EDTA) at 4°C for 30 min, and precipitated with an anti-MED28 antibody and protein A/G PLUS-agarose (Santa Cruz Biotechnology, Inc.). The precipitates were then boiled in SDS-PAGE sample buffer at 100°C for 10 min and subjected to SDS-PAGE. Bands of interest on the SDS-PAGE gel were in-gel digested with trypsin (Promega Corporation, Madison, WI, USA) for 4°C for 30 min. The peptides were loaded on an Agilent 1100 Series nano-LC and LTQ-mass spectrometer (Thermo Electron; Thermo Fisher Scientific, Inc.) for LC-MS/MS analysis. For LC separation, 0.1% formic acid in deionized water and 0.1% formic acid in acetonitrile were used for mobile phase A and B, respectively. Mass spectra were acquired on a full mass scan (400–1,800 m/z) by MS/MS and LTQ. The database search criteria were as follows: taxonomy, Homo sapiens; fixed modification; carboxyamidomethylated (+57) at cysteine residues; variable modification, oxidized (+16) at methionine residues; maximum allowed missed cleavage, 1; and MS tolerance, 100 ppm. Common contaminants including trypsin and keratin were excluded.
Analysis of ZNF224 protein stability
MCF-7 cells were transfected with the 1 µg Myc-empty vector and 1 µg Myc-MED28 maintained in a humidified chamber atmosphere comprising 95% air and 5% CO2 at 37°C for 24 h. Cells are treated with 100 µg cycloheximide for 24 h post-transfection. Cells were harvested at 0, 1, 2, 4 and 8 h following cycloheximide treatment, and the levels of ZNF224, MED28 and α-tubulin were determined by western blotting of total cell lysates as described above.
Fluorescence detection
For the fluorescence assay, MCF-7 cells were co-transfected with pEGFP-ZNF224 (0.5 µg) and pmCherry-MED28 (0.5 µg), and seeded (2×105 cells/well) onto sterile coverslips in 12-well plates the day prior to transfection. MCF-7 cells were washed three times with 1× PBS and fixed with 4% paraformaldehyde (cat. no. CNP015-1000; CellNest, Gyeonggi-do, Korea) at 25°C for 10 min. The coverslips were mounted on glass slides using mounting solution (Biomeda Corp., Foster City, CA, USA) and images were captured under a fluorescence confocal microscope under a ×60 magnification (Carl Zeiss AG, Oberkochen, Germany).
Bimolecular fluorescence complementation (BiFC) assay
MCF-7 cells were co-transfected with 1 µg pBiFC-VN173-ZNF224-Flagand and 1 µg pBiFC-VC155-MED28-HA plasmids, and seeded (3×105 cells/well) onto 6-well plates. Then, the cells were maintained in a humidified chamber atmosphere comprising 95% air and 5% CO2 at 37°C for 24 h. BiFC-induced GFP images were captured under a fluorescence microscope at a ×20 magnification (Nikon Corporation, Tokyo, Japan).
Surface plasmon resonance (SPR) assay
SPR experiments were performed using the SR7500DC Dual Channel system (Reichert, Inc., Depew, NY, USA) according to the manufacturer's protocol. Flag-ZNF224 proteins (50 µg) were immobilized on CMDH chips (Reichert Technologies) according to the manufacturer's protocol. For the Flag-ZNF224-6xHis-MED28 binding analysis, each concentration (0, 1.25, 2.5 and 5 µM) of 6X His-MED28 protein in 1X PBS buffer was injected as an analyte. BSA was used as a control. The scrubber2 program (version 2.0.0004; BioLogic Software Pty., Ltd., Campbell, ACT, Australia) was used to evaluate the dissociation constant (KD).
Statistical analysis
The bands from all western blot analysis were adjusted by ImageJ program (version 1.6.0; National Institutes of Health) for statistical analysis. All experiments were performed at least three times and analyses were performed with Microsoft Excel (Microsoft Corporation, Redmond, WA, USA) and GraphPad Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA). When two groups were compared, statistical differences were assessed with an unpaired two-tailed Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
MED28 is a binding partner of ZNF224
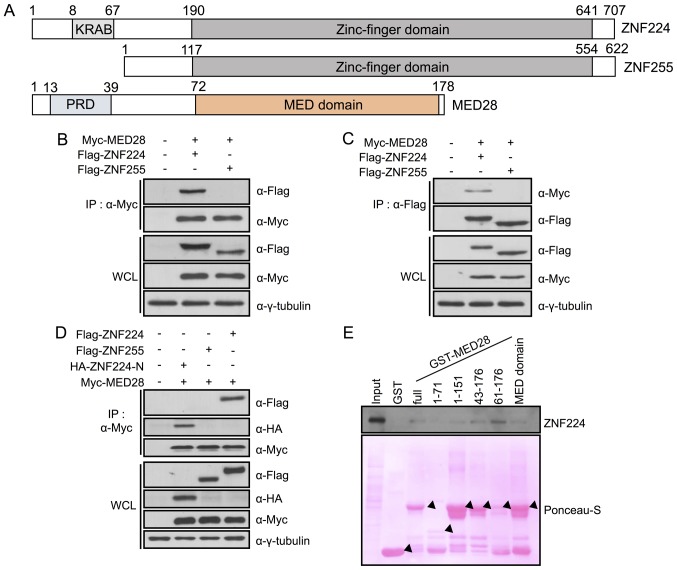
It was previously revealed that ZNF224 regulates the transcription of TP53 and p21 upon DNA damage (12); however, the exact molecular mechanism of this interaction is, to the best of our knowledge, unknown. It was hypothesized that the physical interaction of ZNF224 with its binding partner protein in the nucleus affected the expression of downstream genes. Therefore, a pulldown assay was performed using 293 cell lysates in the presence of purified GST or GST-MED28. Mass analysis using MALDI-TOF identified ZNF224 as a binding candidate of MED28 (data not shown). Sequence analysis revealed that ZNF224 has a canonical Krüppel-associated box (KRAB) domain and 19 tandem-repeat C2H2 zinc finger domains; ZNF255, its alternative transcription product, lacks the KRAB domain (19). In addition, MED28 possesses a proline-rich (aa 13–39) and MED domain (Fig. 1A) (20). To determine whether MED28 is an essential binding partner of ZNF224, a co-immunoprecipitation assay was performed following transfection of Myc-MED28 with Flag-ZNF224 or Flag-ZNF255. A GST pull-down assay using anti-Myc or anti-Flag antibodies revealed that ZNF224, but not ZNF255, specifically associated with MED28, indicating that the KRAB domain of ZNF224 may bind to MED28 (Fig. 1B and C). To investigate this further, co-immunoprecipitation was performed following transfection of Myc-MED28 with the HA-ZNF224 N-terminal KRAB domain (aa 1–67), Flag-ZNF224 or Flag-ZNF255. This result confirmed that the KRAB domain, which is found at the N-terminal of ZNF224, specifically interacts with MED28 (Fig. 1D). To identify the ZNF224-interacting domain of MED28, GST-fused MED28-deletion mutants were designed, designated aa 1–71, aa 1–151, aa 43–176, aa 61–176, and aa 72–176 (identified as the MED domain), and examined their interactions with ZNF224 using a GST pull-down assay (Fig. 1E). The results revealed that the MED domain of MED28 is critical for association with ZNF224 (Fig. 1E). Next, an SPR assay was performed to evaluate the binding affinity of ZNF224 with MED28. ZNF224 bound to MED28 with affinity (KD=3.4×10−6 M), but not to BSA used as negative control. Collectively, these observations revealed that MED28 is a ZNF224-interacting protein through specific binding regions, and that ZNF224/MED28 interactions may be associated with the regulation of downstream genes.
Figure 1.
MED28 is a ZNF224-binding protein. (A) Protein structure of ZNF224/ZNF255 and mediator complex subunit 28 (MED28). ZNF224 has conserved KRAB (aa 8–67) and zinc finger (aa 190–641) domains. MED28 has a PRD (aa 13–39) and MED domain (aa 72–178), which is shared between the paralogous mediator complex subunit proteins. Myc-MED28 was transfected into 293 cells in the presence of Flag-ZNF224 or Flag-ZNF255 for 24 h. Cell extracts were subjected to an immunoprecipitation assay using (B) anti-Myc or (C) anti-Flag antibodies; samples were separated by SDS-PAGE with WCL. Immunoblotting was performed with the indicated antibodies, and γ-tubulin was used as a loading control. (D) Myc-MED28 was transfected into 293 cells in the presence of Flag-ZNF224, Flag-ZNF255, or HA-ZNF224-N (aa 1–67). Cell lysates were precipitated using an anti-Myc antibody and immunoblotted with the indicated antibodies. (E) Ponceau-S stain showing purified GST (lane 2), GST-MED28 full length (lane 3), aa 1–71 (lane 4), aa 1–151 (lane 5), aa 43–176 (lane 6), aa 61–176 (lane 7), and aa 72–176 (MED domain, lane 8). Cell lysates from Flag-ZNF224-transfected 293 cells were incubated with GST fusion proteins for 4 h, and samples were analyzed using SDS-PAGE and immunoblotted with an anti-Flag antibody. MED28, mediator complex subunit 28; ZNF224, zinc finger protein 224; aa, amino acid; KRAB, Krüppel-associated box; PRD, proline rich domain; HA, hemagglutinin; Myc, Myc proto-oncogene protein; WCL, whole cell lysates; GST, glutathione S-transferase.
Nucleus-specific interactions of ZNF224 with MED28
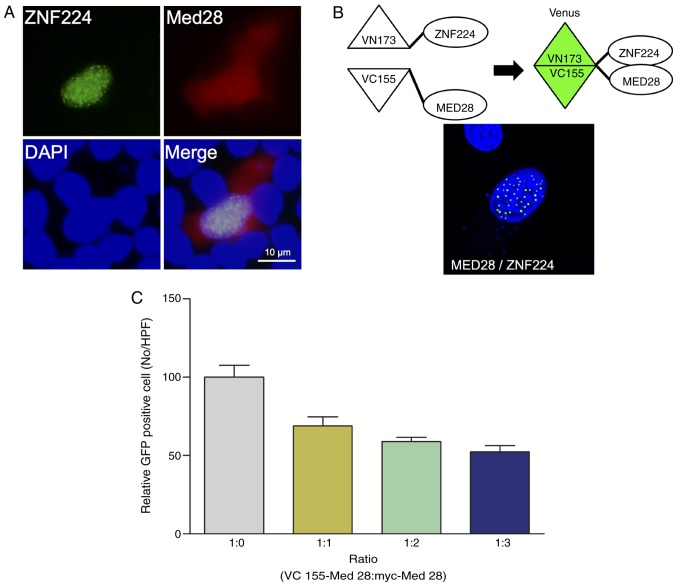
Previously, it was shown that ZNF224 downregulates the transcription of p53 and p21, and that MED28 is localized to the cytosol and nucleus (12,18). In addition, it is known that ZNF224 primarily localizes to the nucleus and forms a punctate pattern (12). Therefore, we hypothesized that the interaction between ZNF224 and MED28 may occur in the nucleus. To verify this hypothesis, the cellular co-localization of ZNF224 and MED28 was examined using fluorescence microscopy. ZNF224 co-localized with MED28 in the nucleus (Fig. 2A). To confirm the nuclear interaction of ZNF224 with MED28 further, a BiFC assay. ZNF224 was annealed to the N-terminus of the Venus fluorescent protein (VN173-ZNF224) and MED28 to the C-terminus of Venus (VC155-MED28) and co-transfected them into HeLa cells. As shown in Fig. 2B, the BiFC signal was detected only in the nucleus of the cells, confirming that the interaction of ZNF224 with MED28 primarily occurs in nucleus (Fig. 2A). To confirm the nuclear-specific interaction further, the expression level of Myc-MED28 was increased in a dose-dependent manner to act as a competitor for VC155-MED28, the number of GFP positive cells was evaluated. The results showed that the number of GFP-positive cells decreased as the concentration of Myc-MED28 increased (Fig. 2C), indicating that the nuclear interaction between ZNF224 and MED28 is specific.
Figure 2.
Intracellular co-localization of ZNF224 and MED28. (A) MCF-7 cells transfected with pEGFP-ZNF224 (green) and pmCherry-MED28 (red) were scanned with a confocal microscope (magnification, ×60). The nucleus was stained with DAPI (blue). (B) Schematic diagram of the BiFC assay and monitoring of the interaction between ZNF224 and MED28. MCF-7 cells were plated onto coverslips and co-transfected with BiFC constructs. After 24 h, the coverslips were fixed and processed for the fluorescence assay. (C) MCF-7 cells were transiently co-transfected with BiFC constructs and Myc-MED28 at different doses, and GFP-positive cells were counted and evaluated. BiFC, bimolecular fluorescence complementation; GFP, green fluorescent protein.
MED28 prevents the degradation of ZNF224 upon DNA damage
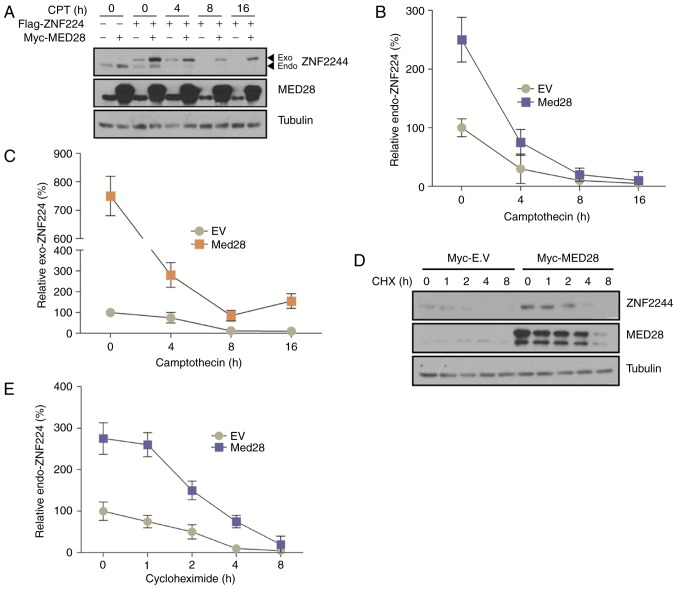
Multiple KRAB-ZFPs function as transcriptional repressors during DDR in cancer cells (21). In a previous study, it was found that ZNF224, a KRAP-ZFP, served a function as a transcriptional repressor upon DNA damage and that the overexpression of ZNF224 results in tumor formation (12). In addition, it was observed that ZNF224 was degraded in a time-dependent manner upon DNA damage (12). Previous studies have revealed that MEDs possess functions as co-repressors or co-activators of transcription in eukaryotic cells (14,22). However, it is not clear whether MED28 functions as a co-repressor or co-activator of ZNF224, or how MED28 affects ZNF224 in DDR. To address this question, the degradation of ZNF224 was examined in response to DNA damage in the presence or absence of MED28. Notably, MED28 expression increased the stability of endogenous and exogenous ZNF224 upon DNA damage (Fig. 3). Because the level of ZNF224 translation could affect protein stability, ZNF224 protein stability in the presence or absence of overexpressed MED28 was examined following treatment with cycloheximide. The vector inducing overexpression of MED28 maintained higher levels of ZNF224 expression than the empty vector (Fig. 3B). These results indicated that ZNF224 is degraded by DDR (although the enzyme that regulates the stability of ZNF224 has not been identified), and that MED28 may inhibit the turnover of ZNF224 by forming a MED28/ZNF224 complex or inhibiting the binding of DDR-mediated proteases.
Figure 3.
ZNF224 is stabilized by MED28 upon DNA damage. (A-C) Myc-MED28 and/or Flag-ZNF224 co-transfected MCF-7 cells were incubated with 100 µM CPT for the indicated time periods prior to harvesting. Endogenous and exogenous ZNF224 and MED28 were detected using an anti-ZNF224 and anti-MED28 antibodies, respectively; α-tubulin was used as the loading control (n=3, P<0.03 vs. EV control). (D and E) MCF-7 cells were transfected with EV or Myc-MED28. After 24 h, cells were treated with cycloheximide and harvested at the indicated times. Cell extracts were examined by immunoblotting with the indicated antibodies (n=3, P<0.001). MED28, mediator complex subunit 28; ZNF224, zinc finger protein 224; EV, empty vector; CPT, camptothecin; CHX, cycloheximide; Exo, exogenous; Endo, endogenous.
MED28 increases cell proliferation upon DNA damage
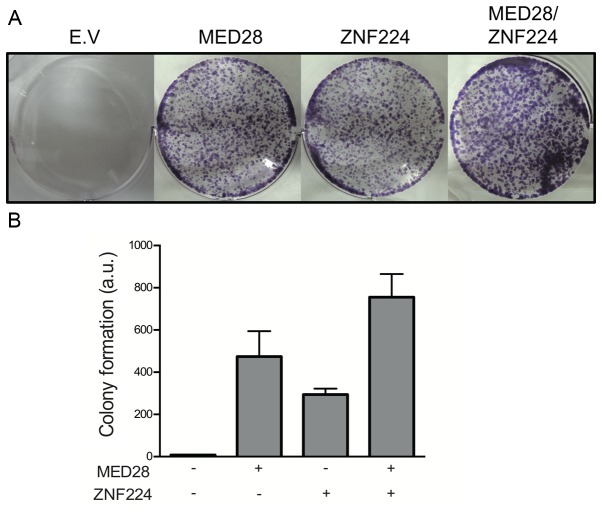
Cell-cycle progression is accompanied by checkpoint phases (23). This phenomenon generally occurs in proliferating cells through the activation of checkpoint proteins to prevent genome instability; thereafter, the cell will typically resume normal proliferation (24). Since p53 and p21 are activated during DNA repair (25), it was hypothesized that MED28 may disrupt DNA repair through ZNF224-mediated repression of p53 and p21, leading to increased cell proliferation. To quantify the function of MED28 in the recovery of cell proliferation following DNA damage, the colony-forming ability of MCF-7 cells was assessed following treatment with camptothecin (CPT), a DNA-damaging agent known to be an inhibitor of topoisomerase I (26). The colony-forming assay revealed that co-transfection with MED28 and ZNF224 considerably increased the proliferation rate of MCF-7 cells (Fig. 4). This result indicated that the interaction of MED28 with ZNF224 induced abnormal cell proliferation following DNA damage.
Figure 4.
ZNF224 and MED28 synergistically promote colony formation in MCF-7 cells. (A) MCF-7 cells transfected with the indicated expression vectors were seeded on 6-well plates and treated with 10 µM camptothecin to induce DNA damage response and apoptosis. After 2 weeks, the colonies were stained with crystal violet and the number of colonies was counted. (B) This experiment was statistically analyzed and normalized across 3 separate experiments. EV, empty vector; MED28, mediator complex subunit 28; ZNF224, zinc finger protein 224.
Discussion
ZNF224 serves a function as a transcriptional repressor in the regulation of gene expression through interactions with multiple co-factors, including KRAB-associated protein-1, protein arginine methyltransferase 5, DEP domain-containing 1 and Wilms' tumor gene 1 (19,27–31). To the best of our knowledge, previous studies investigating the function of MED28 have focused only on phenotypes or relative intracellular signaling in mammalian cells (17,20). In the present study, MED28 was screened as a binding partner of ZNF224. In addition, the present study revealed that MED28 was associated with intracellular signals through a physical interaction with ZNF224 upon DNA damage.
The KRAB domain is a core transcriptional repressor domain in KRAB-ZFPs and mediates interactions with co-repressors (21). Interaction-domain mapping analysis using multiple deletion mutants revealed that the KRAB domain of ZNF224 is critical for its association with MED28 (Fig. 1). A previous study demonstrated that the zinc-finger domains of KRAB-ZFPs are critical for binding to DNA at the promoter of the target gene, whereas the remaining zinc-finger domains are required for protein-protein interactions (32). However, the present study indicated that the KRAB domain may also be required for substrate binding.
Interaction analysis using multiple MED28-deletion mutants derived from Escherichia coli also revealed interactions between MED28 and ZNF224. In addition, the deletion mutant containing the MED domain of MED28 exhibited an increased binding affinity to ZNF224 compared with that of any other deletion mutant. This result indicated that the post-translational modification of MED28 is not required for interaction with ZNF224 and that the C-terminal MED domain of MED28 is essential for mediating the association with ZNF224. In addition, co-localization of ZNF224 with MED28 was observed in the nucleus. Therefore, the results of the present study indicated that ZNF224 interacts with MED28 in the nucleus to repress the transcription of target genes.
In vitro colony-forming assays can be used to predict the outcome of cancer therapies, and are often used to test anticancer drugs (33). Therefore, the present study assessed the association between ZNF224 and MED28 under chemo-sensitive conditions using a DNA-damaging agent in cancer cells; their expression increased the incidence of colony formation compared with the non-expressing group. This result indicated that the interaction between ZNF224 and MED28 has synergetic effects for cancer cell survival and proliferation, against the cell-cycle checkpoint system upon DNA damage. ZNF224 is highly expressed in breast and bladder cancer (12,30,34). In addition, MED28 is abundantly expressed in breast, colon, and prostate cancer (35). It was determined that the interaction between MED28 and ZNF224 downregulates tumour suppressors, including p53 and p21, resulting in increased cancer cell proliferation compared with normal conditions. Further study is required to identify how the interaction between ZNF224 and MED28 is regulated in cancer and how the development of an inhibitor to suppress the interaction between ZNF224 and MED28 may contribute to effective cancer treatments. Collectively, the data suggests that the novel protein MED28 increases the proliferative ability of breast cancer cells by the regulation of the ZNF224 protein against DNA damage responses.
Acknowledgements
The present study was supported by the Basic Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (grant no. NRF-2012R1A1A2040602).
References
- 1.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 2.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinert T. DNA damage and checkpoint pathways: Molecular anatomy and interactions with repair. Cell. 1998;94:555–558. doi: 10.1016/S0092-8674(00)81597-4. [DOI] [PubMed] [Google Scholar]
- 4.Zhou BB, Elledge SJ. The DNA damage response: Putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 5.Roos WP, Thomas AD, Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer. 2016;16:20–33. doi: 10.1038/nrc.2015.2. [DOI] [PubMed] [Google Scholar]
- 6.Vitale I, Galluzzi L, Castedo M, Kroemer G. Mitotic catastrophe: A mechanism for avoiding genomic instability. Nat Rev Mol Cell Biol. 2011;12:385–392. doi: 10.1038/nrm3115. [DOI] [PubMed] [Google Scholar]
- 7.Freed-Pastor W, Prives C. Targeting mutant p53 through the mevalonate pathway. Nat Cell Biol. 2016;18:1122–1124. doi: 10.1038/ncb3435. [DOI] [PubMed] [Google Scholar]
- 8.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 9.Stracquadanio G, Wang X, Wallace MD, Grawenda AM, Zhang P, Hewitt J, Zeron-Medina J, Castro-Giner F, Tomlinson IP, Goding CR, et al. The importance of p53 pathway genetics in inherited and somatic cancer genomes. Nat Rev Cancer. 2016;16:251–265. doi: 10.1038/nrc.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muller PA, Vousden KH. Mutant p53 in cancer: New functions and therapeutic opportunities. Cancer Cell. 2014;25:304–317. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menendez D, Inga A, Resnick MA. The expanding universe of p53 targets. Nat Rev Cancer. 2009;9:724–737. doi: 10.1038/nrc2730. [DOI] [PubMed] [Google Scholar]
- 12.Cho JG, Park S, Lim CH, Kim HS, Song SY, Roh TY, Sung JH, Suh W, Ham SJ, Lim KH, Park SG. ZNF224, Krüppel like zinc finger protein, induces cell growth and apoptosis-resistance by down-regulation of p21 and p53 via miR-663a. Oncotarget. 2016;7:31177–31190. doi: 10.18632/oncotarget.8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malik S, Roeder RG. The metazoan mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allen BL, Taatjes DJ. The mediator complex: A central integrator of transcription. Nat Rev Mol Cell Biol. 2015;16:155–166. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu M, Sartippour MR, Zhang L, Norris AJ, Brooks MN. Targeted inhibition of EG-1 blocks breast tumor growth. Cancer Biol Ther. 2007;6:936–941. doi: 10.4161/cbt.6.6.4189. [DOI] [PubMed] [Google Scholar]
- 16.Lee MF, Pan MH, Chiou YS, Cheng AC, Huang H. Resveratrol modulates MED28 (Magicin/EG-1) expression and inhibits epidermal growth factor (EGF)-induced migration in MDA-MB-231 human breast cancer cells. J Agric Food Chem. 2011;59:11853–11861. doi: 10.1021/jf202426k. [DOI] [PubMed] [Google Scholar]
- 17.Huang CY, Chou YH, Hsieh NT, Chen HH, Lee MF. MED28 regulates MEK1-dependent cellular migration in human breast cancer cells. J Cell Physiol. 2012;227:3820–3827. doi: 10.1002/jcp.24093. [DOI] [PubMed] [Google Scholar]
- 18.Yu MA, Cho JG, Kim KI, Jo YJ, Sung JH, Yang HB, Park SG. Generation of med28 specific monoclonal antibodies. Monoclon Antib Immunodiagn Immunother. 2015;34:30–35. doi: 10.1089/mab.2014.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Florio F, Cesaro E, Montano G, Izzo P, Miles C, Costanzo P. Biochemical and functional interaction between ZNF224 and ZNF255, two members of the Kruppel-like zinc-finger protein family and WT1 protein isoforms. Hum Mol Genet. 2010;19:3544–3556. doi: 10.1093/hmg/ddq270. [DOI] [PubMed] [Google Scholar]
- 20.Lu M, Zhang L, Maul RS, Sartippour MR, Norris A, Whitelegge J, Rao JY, Brooks MN. The novel gene EG-1 stimulates cellular proliferation. Cancer Res. 2005;65:6159–6166. doi: 10.1158/0008-5472.CAN-04-4016. [DOI] [PubMed] [Google Scholar]
- 21.Lupo A, Cesaro E, Montano G, Zurlo D, Izzo P, Costanzo P. KRAB-Zinc finger proteins: A repressor family displaying multiple biological functions. Curr Genomics. 2013;14:268–278. doi: 10.2174/13892029113149990002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myers LC, Kornberg RD. Mediator of transcriptional regulation. Annu Rev Biochem. 2000;69:729–749. doi: 10.1146/annurev.biochem.69.1.729. [DOI] [PubMed] [Google Scholar]
- 23.Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17:93–115. doi: 10.1038/nrc.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 25.Sperka T, Wang J, Rudolph KL. DNA damage checkpoints in stem cells, ageing and cancer. Nat Rev Mol Cell Biol. 2012;13:579–590. doi: 10.1038/nrm3420. [DOI] [PubMed] [Google Scholar]
- 26.Gokduman K. Strategies targeting DNA topoisomerase i in cancer chemotherapy: Camptothecins, nanocarriers for camptothecins, organic non-camptothecin compounds and metal complexes. Curr Drug Targets. 2016;17:1928–1939. doi: 10.2174/1389450117666160502151707. [DOI] [PubMed] [Google Scholar]
- 27.Medugno L, Florio F, De Cegli R, Grosso M, Lupo A, Costanzo P, Izzo P. The Krüppel-like zinc-finger protein ZNF224 represses aldolase A gene transcription by interacting with the KAP-1 co-repressor protein. Gene. 2005;359:35–43. doi: 10.1016/j.gene.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 28.Cesaro E, De Cegli R, Medugno L, Florio F, Grosso M, Lupo A, Izzo P, Costanzo P. The Kruppel-like zinc finger protein ZNF224 recruits the arginine methyltransferase PRMT5 on the transcriptional repressor complex of the aldolase A gene. J Biol Chem. 2009;284:32321–32330. doi: 10.1074/jbc.M109.043349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montano G, Ullmark T, Jernmark-Nilsson H, Sodaro G, Drott K, Costanzo P, Vidovic K, Gullberg U. The hematopoietic tumor suppressor interferon regulatory factor 8 (IRF8) is upregulated by the antimetabolite cytarabine in leukemic cells involving the zinc finger protein ZNF224, acting as a cofactor of the Wilms' tumor gene 1 (WT1) protein. Leuk Res. 2016;40:60–67. doi: 10.1016/j.leukres.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 30.Harada Y, Kanehira M, Fujisawa Y, Takata R, Shuin T, Miki T, Fujioka T, Nakamura Y, Katagiri T. Cell-permeable peptide DEPDC1-ZNF224 interferes with transcriptional repression and oncogenicity in bladder cancer cells. Cancer Res. 2010;70:5829–5839. doi: 10.1158/0008-5472.CAN-10-0255. [DOI] [PubMed] [Google Scholar]
- 31.Lee TH, Lwu S, Kim J, Pelletier J. Inhibition of Wilms tumor 1 transactivation by bone marrow zinc finger 2, a novel transcriptional repressor. J Biol Chem. 2002;277:44826–44837. doi: 10.1074/jbc.M205667200. [DOI] [PubMed] [Google Scholar]
- 32.Moore M, Klug A, Choo Y. Improved DNA binding specificity from polyzinc finger peptides by using strings of two-finger units; Proc Natl Acad Sci USA; 2001; pp. 1437–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salmon SE. Human tumor colony assay and chemosensitivity testing. Cancer Treat Rep. 1984;68:117–125. [PubMed] [Google Scholar]
- 34.Gur-Dedeoglu B, Konu O, Bozkurt B, Ergul G, Seckin S, Yulug IG. Identification of endogenous reference genes for qRT-PCR analysis in normal matched breast tumor tissues. Oncol Res. 2009;17:353–365. doi: 10.3727/096504009788428460. [DOI] [PubMed] [Google Scholar]
- 35.Zhang L, Maul RS, Rao J, Apple S, Seligson D, Sartippour M, Rubio R, Brooks MN. Expression pattern of the novel gene EG-1 in cancer. Clin Cancer Res. 2004;10:3504–3508. doi: 10.1158/1078-0432.CCR-03-0467. [DOI] [PubMed] [Google Scholar]