Abstract
To obtain accurate and reliable results from quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) analysis, it is necessary to select suitable reference genes as standards for normalizing target gene expression data. QRT-PCR is a popular analytical methodology for studying gene expression and it has been used widely in studies of Aphis gossypii Glover in recent years. However, there is absence of study on the stability of the expression of reference genes in A. gossypii. In this study, eight commonly used candidate reference genes, including 18S, 28S, β-ACT, GAPDH, EF1α, RPL7, α-TUB, and TBP, were evaluated under various experimental conditions to assess their suitability for use in the normalization of qRT-PCR data. The optimal number of reference genes was determined using the geNorm program, and the suitability of particular reference genes was empirically validated by performing normalizations of expression data for the HSP70 gene. The results showed the most suitable combinations of reference genes for the different experimental conditions. For experiments based on divergent developmental stages, EF1α, β-ACT, and RPL7 are the optimal reference gene combination, both EF1α and β-ACT are the optimal combination used in the experiments of different geographical populations, whereas for experiments of the temperature changes, the combination of GAPDH and RPL7 is optimal, both 18S and β-ACT are an optimal combination for feeding assay experiments. These research results should be useful for the selection of the suitable reference genes to obtain reliable qRT-PCR data in the gene expression study of A. gossypii.
Keywords: Aphis gossypii, gene expression, normalization, qRT-PCR, reference gene
Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) is one of the most effective, sensitive, and economical methods for gene expression analysis (Bustin 2000, VanGuilder et al. 2008, Derveaux et al. 2010, Lu et al. 2013, Liang et al. 2014). Although qRT-PCR is widely used to analyze gene expression in biological research, there remain a number of problems that limit its use. One of the biggest challenges in qRT-PCR analysis is normalization of the variations caused by some errors in RNA extraction and purification, reverse-transcription, efficiency of PCR amplification, etc. (Huggett et al. 2005, Udvardi et al. 2008, Bustin et al. 2009). Several strategies have been applied in attempts to normalize these variations in qRT-PCR analysis, these include normalization of sample size, ensuring the quality and quantity of RNA, and removing DNA contamination, among others (Vandesompele et al. 2002, Huggett et al. 2005). Of such strategies, the most widely used is the selection of appropriate reference gene(s) to normalize nonspecific variation or errors (Huggett et al. 2005, Liang et al. 2014). In the last two decades, several genes were used as reference genes, as their expression levels remained relatively constant in different experimental conditions (Bustin 2002). The expression of an ideal reference gene used in quantitative gene expression studies should not be influenced by various conditions of the experiment (Schmittgen and Zakraisek 2000). However, no reference genes are expressed stably across all possible experimental conditions (Vandesompele et al. 2002, Derveaux et al. 2010). In addition, the expression of several conventional reference genes, including 18S RNA, β-ACT, and GAPDH has been shown to vary extensively under particular experimental conditions or in response to external stimuli (Zhu et al. 2001, Glare et al. 2002, Radonić et al. 2004, Zhu and Altmann 2005, Shen et al. 2010, Yuan et al. 2014). Clearly, for given set of experimental biological samples, it is necessary to select suitable reference genes for use in the normalization of qRT-PCR analytical results. To date, several studies have systematically assessed insect reference genes across different experimental conditions. These insects include the honey bee Apis mellifera L. (Lourenco et al. 2008), the silkworm Bombyx mori L. (Wang et al. 2008), the fruit fly Drosophila melanogaster Meigen (Ponton et al. 2011), the diamondback moth Plutella xylostella l. (Fu et al. 2013), and the tobacco whitefly Bemisia tabaci Gennadius (Li et al. 2013, Liang et al. 2014).
The cotton aphid, Aphis gossypii Glover (Hemiptera: Aphididae), is an important insect pest in cotton and cucurbit fields that causes economic damage both through direct feeding and through virus transmission (Cao et al. 2008). In recent years, qRT-PCR has been widely used to quantify gene expression levels in diverse studies of A. gossypii, such as, studies of insecticide resistance, ecological adaption, and olfaction mechanisms (Cao et al. 2008, 2014; Pan et al. 2010; Li et al. 2013). Some studies have showed that at least two or three reference genes should be used to achieve accurate normalization (Thellin et al. 1999, Vandesompele et al. 2002). However, in the aforementioned studies, the researchers only used one reference gene (18S, GAPDG, or β-ACT) to normalize the variation of mRNA levels of genes of interest for all of the diverse experimental conditions. These less than ideal experimental practices likely resulted from the lack of empirical data about which reference genes in A. gossypii are most appropriate for qRT-PCR gene expression analysis.
We conducted the present study to ameliorate this situation and to enable the empirically informed selection of suitable reference genes for future studies with A. gossypii. Eight commonly used normalization genes (18S, 28S, β-ACT, GAPDH, EF1α, RPL7, α-TUB, and TBP) were selected for analyzing their performance under several different experimental conditions in A. gossypii. Whereafter, a target gene (HSP70) was selected and used as the validation on the performance of the reference genes. Our results should be useful for the selection of suitable, reliable reference genes in modern molecular genetic analyses in A. gossypii.
Materials and Methods
Insects
The laboratory strain of A. gossypii used in this study was collected in 1999 from cotton fields in the Xinjiang Uygur Autonomous Region, China, and has been maintained in our laboratory for more than 15 years. The aphids were reared on cotton seedlings in controlled conditions of 20–23°C, 60% RH, and a photoperiod of 16:8 (L:D) h, as described previously (Cao et al. 2008, Pan et al. 2009). Other populations used in this experiment were collected from cotton fields in Shawan (Xinjiang Uygur Autonomous Region), Xiangyang (Hubei province), Binzhou (Shandong province), and Zhengzhou (Henan province) in 2014.
Biotic Factors
Developmental Stages
Two hundred first-day nymphs, 50 alate and 50 apterous A. gossypii adults were collected in RNase-free tubes for each replication, respectively. The samples were in triplicate and then snap frozen in the liquid nitrogen before stored at −80°C for RNA extraction.
Field Populations
Four field collected populations from Shawan (Xinjiang Uygur Autonomous Region), Xiangyang (Hubei Province), Binzhou (Shandong Province), and Zhengzhou (Henan Province) were used in this study.
Abiotic Factors
Temperature Treatment
Apterous adults were exposed to various temperatures (15, 20, 25, 30, and 35°C, respectively) for 1 h. In total, 50 adults at each temperature point were collected for RNA extraction.
Feeding Assays
Artificial diet: Sterilized glass tubes that open at both ends (3 cm in length, 2 cm in diameter) were used for the in vitro feeding assay. One end of the tube was covered by two layers of parafilm, 200 µl of a 0.5 M sterile sucrose solution was sandwiched between the two parafilm layers (artificial diet). Fifty healthy apterous adults were gently placed into the tube with a brush, and the tube was sealed with a piece of Chinese art paper by solid glue. Aphids were allowed to feed on this artificial diet for 24 h. After that, live aphids were collected using as RNA extraction. For the cotton leaf feeding assays, 50 healthy apterous adults were gently brushed from cotton leaves and collected for RNA extraction.
Reference Gene Selection and Primer Design
Eight commonly used reference genes were selected, including 18S ribosomal RNA (rRNA) (18S), 28S rRNA (28S), beta actin (β-ACT), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), elongation factor 1 alpha (EF1α), ribosomal protein L7 (RPL7), beta-tubulin (β-TUB), and TATA box binding protein-associated factor (TBP). Primer Premier 5.0 software was used to design the primers. Details for the primers used in this study are listed in Table 1.
Table 1.
Primer pairs used for qRT-PCR analysis of candidate reference genes and HSP70, a target gene
| Gene symbol | Gene name | Accession number | Sequence (5′–3′) | Product length (bp) | Efficiency (%) | Tm (°C) | R2 |
|---|---|---|---|---|---|---|---|
| β-ACT | Beta-Actin | KF018928.1 | F: TCTTGGGAATGGAATCTTGC | 254 | 98.10 | 60 | 0.994 |
| R: GGACAGAGAAGCCAAGATGG | |||||||
| 18S | 18S ribosomal | KF018922.1 | F: ATTGACGGAAGGGCACC | 157 | 106.09 | 60 | 0.998 |
| R: CGCTCCACCAACTAAGAACG | |||||||
| 28S | 28S ribosomal | KC796354.1 | F: GAGGTCCGTAGCGATTCTGA | 105 | 99.25 | 60 | 0.996 |
| R: GAGGGAAACTTCGGAGGGA | |||||||
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | KP676380 | F: ACTACTGTTCATGCAACCACCG | 272 | 94.47 | 60 | 0.998 |
| R: GCTGCTTCCTTAACCTTATCCT | |||||||
| EF1α | Elongation factor 1 alpha | EU019874.1 | F: GAAGCCTGGTATGGTTGTCGT | 187 | 107.53 | 60 | 0.998 |
| R: GGGTGGGTTGTTCTTTGTG | |||||||
| TBP | TATA box binding protein | AGT79997.1 | F: TGCTCCGAGTGAAGAAAAGG | 171 | 103.78 | 60 | 0.994 |
| R: ACGGGCAAATGACTAGTGGA | |||||||
| RPL7 | Ribosomal protein L7 | KP676382 | F: TGCCGGAGTCTGTACTCAA | 255 | 94.70 | 60 | 0.999 |
| R: TCACACCACGAATACGCA | |||||||
| α-TUB | Alpha-Tubulin | KP676379 | F: CCGTCAATTGTTCCACCCTG | 195 | 91.33 | 60 | 0.998 |
| R: CCAGATCCAGTACCACCTCC | |||||||
| HSP70 | Heat shock protein 70 gene | KP676381 | F: TCGCCTGTCTCAAGCCGAAAT | 98 | 102.99 | 60 | 0.991 |
| R: GGTTCTTTGCCGCGATCTTG |
F, forward primer; R, reverse primer; Tm, melting temperature; R2, coefficient of correlation.
RNA Extraction and cDNA Synthesis
Total RNA was isolated with TRIzol reagent (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. The purity and concentration of total RNA were assessed with a NAS-99 spectrophotometer (ACTGene). RNA samples with an A260/A280 ratio ranging from 1.8 to 2.0 and an A260/A230 ratio >2.0 were used for cDNA synthesis. First-strand complementary DNA was synthesized from 1 μg of total RNA with a PrimeScript RT reagent kit with gDNA Eraser (Takara, Dalian, China) following the manufacturer’s instructions and stored at −20°C until use.
Quantitative Real-Time PCR
Quantitative real-time PCR was performed using ROX’s Platinum SYBR Green qPCR SuperMix-UDG kit (Invitrogen) on an Applied Biosystems 7500 Real-Time PCR system (Applied Biosystems, Foster city, CA). The reactions were performed in a 25 μl volume of a mixture containing 1 μl cDNA template, 12.5 μl SYBR Green qPCR SuperMix-UDG, 0.5 μl of each primer, and 10.5 μl of nuclease-free water. The thermocycling program was as follows: 50°C for 2 min, 95°C for 2 min, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. At the end of each PCR run, a melting curve analysis from 65 to 95°C was applied to all reactions to ensure the specificity of the amplified product. Standard curves were created based on a fivefold dilution series of cDNA (1:5, 1:25, 1:125, 1:625, 1:3125, and 1:15625). The corresponding qRT-PCR efficiencies (E) were calculated according to the equation: (Pfaffl 2001). Each sample was prepared as three biological replicates, and each reaction was analyzed with three technical replications.
Analysis of the Stability of Reference Gene Expression
The expression stability of the eight selected reference genes was evaluated with the delta cycle threshold (Ct) method and three commonly used software tools, geNorm version 3.5 (Vandesompele et al. 2002), Normfinder version 0.953 (Andersen et al. 2004), and BestKeeper (Pfaffl et al. 2004).
The geNorm software initially calculates the value of gene expression stability (M) and generates a stability ranking; genes with the lowest M value have the most stable expression. geNorm also calculates pair-wise variation Vn/Vn + 1, which represents the variation between two sequential normalization factors and determines the optimal number of reference genes required for accurate normalization. Vn/Vn + 1 ratio below 0.15 suggest that the use of an additional reference gene would not significantly improve normalization. NormFinder software is a model-based approach to identifying suitable reference genes for use in normalization (Andersen et al. 2004). The candidate gene with the lowest value is considered to be the most stable reference gene. The MS Excel-based software BestKeeper and the comparative delta Ct method were also used to select optimal reference genes. We also used a user-friendly web-based comprehensive tool, RefFinder (http://www.leonxie.com/referencegene.php) to evaluate and select reference genes. RefFinder integrates the aforementioned major computational programs (geNorm, Normfinder, BestKeeper, and the comparative delta Ct method) to compare and rank the tested candidate reference genes. RefFinder assigns an appropriate weight to an individual gene and calculates the geometric mean of these weights for the overall final ranking.
Validation of Reference Gene Selection
To evaluate the validity of the optimized selection of reference genes, the expression levels of the heat shock protein 70 gene (HSP70) were analyzed in different experimental conditions (temperature treatment, population, and developmental stages). For each experimental condition, the expression profiles of the gene HSP70 were normalized using only one reference gene (the most stable reference gene [NF1] and the least stable reference gene [NF8]) and several stable reference genes (NF(1 − n)) recommended by RefFinder. The relative expression levels of HSP70 in different samples were calculated according to the 2−ΔΔCt method (Livak and Schmittgen 2001). The target gene expression normalized by the least stable reference gene, and the recommended combination of reference genes were calculated using a Student’s t-test, implemented in SPSS 17.0 software with a significance level set at P = 0.05.
Results
PCR Amplification Efficiencies and Expression Levels of Candidate Reference Genes
Traditional PCR was used to evaluate the primer specificity of the eight reference genes and the one target gene of interest used. This analysis showed that each primer pair produced a single product, and sequencing of these products showed that they had 100% identity with the fragment sequences used to design the primers (data not shown). Analysis of melting curves showed that there were single peaks for each primer pairs further demonstrated that each of the primer pairs amplified a unique product. A standard curve was generated for each gene using fivefold serial dilutions of cDNA. The amplification efficiencies of all the primer pairs were between 91.33 and 107.53%, and the correlation coefficients (R2) ranged from 0.991 to 0.999 (Table 1).
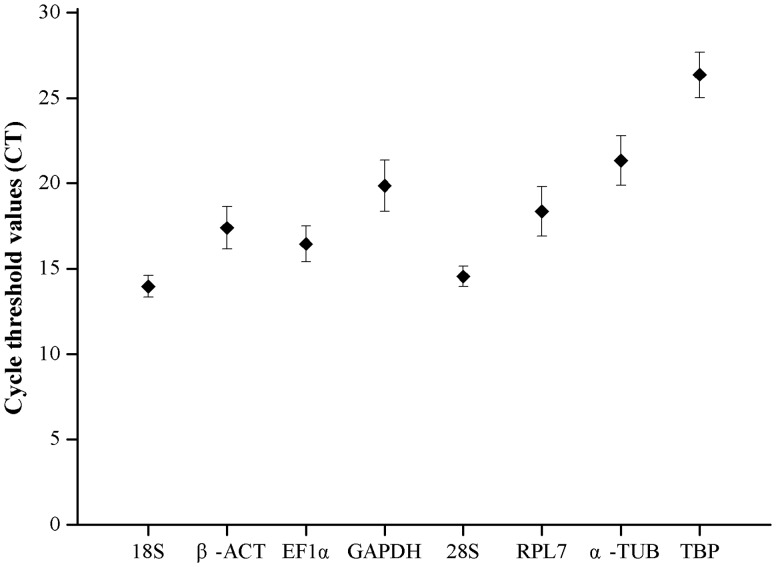
The Ct values were adopted to compare the transcript abundance of the selected genes in different samples. The mean Ct values of the eight reference genes varied significantly. The means of the Ct values ranged from 13.96 to 26.35, with the lowest and highest Ct values obtained from 18S (12.73) and TBP (28.55). 18S had the highest mean expression levels, followed by 28S (14.55), EF1α (16.45), β-ACT (17.39), RPL7 (18.35), GAPDH (19.85), α-TUB (21.33), and TBP (26.35) (Fig. 1).
Fig. 1.
Expression levels of candidate reference genes of A. gossypii. The expression levels of candidate reference genes in samples are shown in terms of the Ct values. The ‘black boxes’ indicate the mean value of replicated samples, and the whiskers indicate the standard deviation of the mean.
Expression Stability of the Candidate Reference Genes
Developmental Stage Experiments
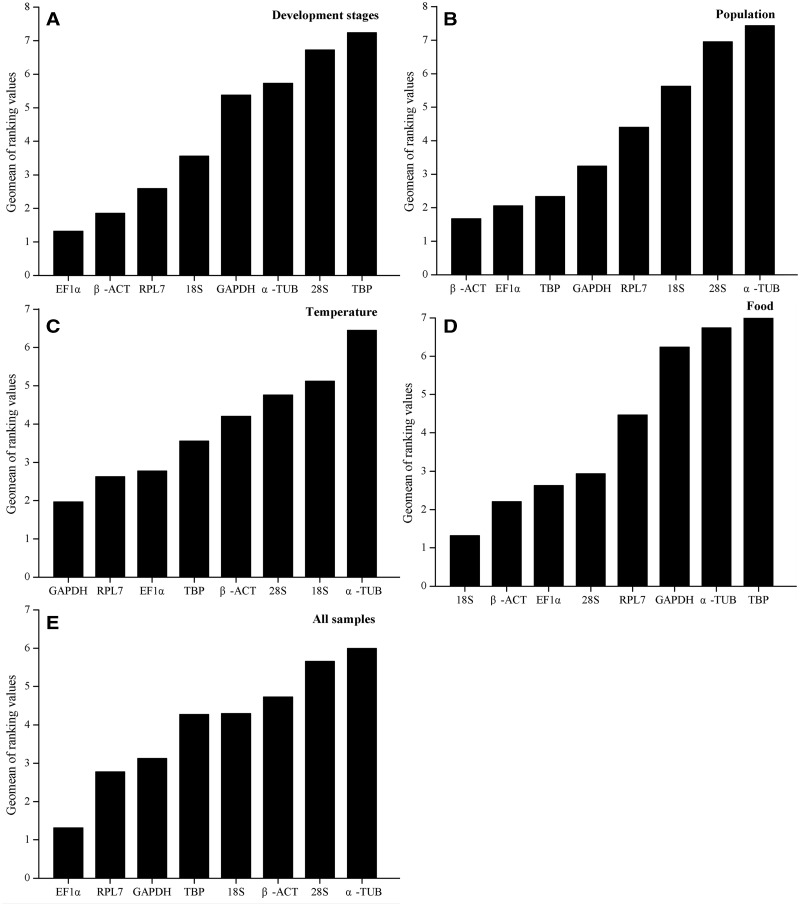
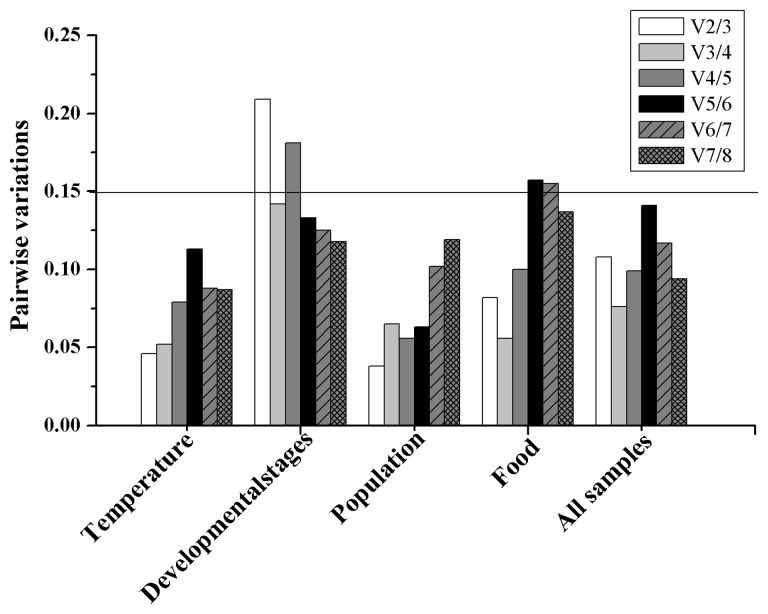
The overall expression stability rankings generated by three methods (delta Ct method, NormFinder, and geNorm) were almost identical, the top three stable reference genes were EF1α, β-ACT, and RPL7. The BestKeeper method ranked β-ACT, 18S, and EF1α as the top three stable reference genes (Table 2). According to the RefFinder results, the stability rankings from the most stable to the least stable across different developmental stages were as follows: EF1α, β-ACT, RPL7, 18S, GAPDH, α-TUB, 28S, and TBP (Fig. 2A). For geNorm analysis, the V3/4 was below the threshold of V = 0.15 (Fig. 3), indicating that three most stable genes are needed for a reliable normalization. Therefore, for the developmental stage experiments, the three most stable reference genes (EF1α, β-ACT, and RPL7) were appropriate to normalization (Table 3).
Table 2.
Expression stability of the candidate reference genes under different experimental conditions
| Conditions | Reference gene | ΔCT |
BestKeeper |
Normfinder |
geNorm |
||||
|---|---|---|---|---|---|---|---|---|---|
| Stability | Rank | Stability | Rank | Stability | Rank | Stability | Rank | ||
| Developmental stages | 18S | 0.92 | 4 | 0.474 | 2 | 0.711 | 5 | 0.614 | 4 |
| 28S | 1.06 | 7 | 0.603 | 4 | 0.917 | 8 | 0.915 | 8 | |
| β-ACT | 0.77 | 2 | 0.32 | 1 | 0.349 | 2 | 0.571 | 3 | |
| GAPDH | 0.94 | 5 | 0.852 | 7 | 0.707 | 4 | 0.809 | 6 | |
| EF1α | 0.74 | 1 | 0.559 | 3 | 0.272 | 1 | 0.504 | 1 | |
| RPL7 | 0.82 | 3 | 0.716 | 5 | 0.484 | 3 | 0.528 | 2 | |
| α-TUB | 1.00 | 6 | 0.77 | 6 | 0.795 | 6 | 0.754 | 5 | |
| TBP | 1.06 | 7 | 1.003 | 8 | 0.872 | 7 | 0.865 | 7 | |
| Population | 18S | 0.61 | 6 | 0.365 | 4 | 0.483 | 6 | 0.485 | 7 |
| 28S | 0.62 | 7 | 0.458 | 8 | 0.488 | 7 | 0.413 | 6 | |
| β-ACT | 0.40 | 1 | 0.255 | 2 | 0.096 | 1 | 0.18 | 4 | |
| GAPDH | 0.49 | 4 | 0.426 | 7 | 0.403 | 4 | 0.145 | 3 | |
| EF1α | 0.42 | 2 | 0.338 | 3 | 0.258 | 3 | 0.141 | 2 | |
| RPL7 | 0.50 | 5 | 0.418 | 5 | 0.410 | 5 | 0.14 | 1 | |
| α-TUB | 0.76 | 8 | 0.420 | 6 | 0.68 | 8 | 0.554 | 8 | |
| TBP | 0.46 | 3 | 0.086 | 1 | 0.187 | 2 | 0.272 | 5 | |
| Temperature | 18S | 0.69 | 5 | 0.222 | 2 | 0.555 | 7 | 0.408 | 7 |
| 28S | 0.97 | 6 | 0.219 | 1 | 0.945 | 8 | 0.550 | 8 | |
| β-ACT | 0.44 | 3 | 0.820 | 7 | 0.284 | 5 | 0.097 | 3 | |
| GAPDH | 0.4 | 1 | 0.779 | 5 | 0.204 | 3 | 0.085 | 1 | |
| EF1α | 0.44 | 3 | 0.562 | 3 | 0.103 | 1 | 0.228 | 5 | |
| RPL7 | 0.42 | 2 | 0.810 | 6 | 0.27 | 4 | 0.087 | 2 | |
| α-TUB | 0.59 | 4 | 0.905 | 8 | 0.523 | 6 | 0.281 | 6 | |
| TBP | 0.44 | 3 | 0.659 | 4 | 0.152 | 2 | 0.181 | 4 | |
| Food | 18S | 0.64 | 1 | 0.542 | 3 | 0.306 | 1 | 0.214 | 1 |
| 28S | 0.74 | 4 | 0.435 | 1 | 0.385 | 3 | 0.359 | 5 | |
| β-ACT | 0.71 | 3 | 0.524 | 2 | 0.499 | 4 | 0.232 | 2 | |
| GAPDH | 0.92 | 5 | 1.108 | 7 | 0.672 | 6 | 0.563 | 6 | |
| EF1α | 0.65 | 2 | 0.607 | 4 | 0.320 | 2 | 0.242 | 3 | |
| RPL7 | 0.74 | 4 | 0.666 | 5 | 0.551 | 5 | 0.253 | 4 | |
| α-TUB | 1.17 | 7 | 0.924 | 6 | 1.016 | 7 | 0.732 | 7 | |
| TBP | 1.18 | 8 | 1.436 | 8 | 1.073 | 8 | 0.843 | 8 | |
Fig. 2.
Expression stability of the candidate reference genes under different experimental conditions. The average expression stability of the reference genes was calculated using the Geomean method by RefFinder. A lower Geomean of ranking value indicates more stable expression. (A) Different developmental stages, (B) different populations, (C) apterous adult A. gossypii treated with varying temperatures, (D) apterous adult A. gossypii fed different foods, (E) pooled samples.
Fig. 3.
Optimal number of reference genes for normalization in A. gossypii. The pair-wise variation (Vn/Vn + 1) was analyzed by geNorm software between the normalization factors NFn and NFn + 1 to determine the optimal number of reference genes required for accurate normalization in a given class of experiment. A value lower than 0.15 indicates that the use of additional reference genes would not significantly improve normalization.
Table 3.
Best performing reference genes in A. gossypii for the different experimental conditions
| Experimental conditions | Preferable reference genes | |||
|---|---|---|---|---|
| Biotic factors | Developmental stages | EF1α | RPL7 | β-ACT |
| Population | EF1α | β-ACT | ||
| Abiotic factors | Temperature treatment | RPL7 | GAPDH | |
| Food | β-ACT | 18S | ||
| Pooled samples | EF1α | RPL7 | ||
Population Experiments
The stability rankings generated by the delta Ct method, BestKeeper, and NormFinder were similar, β-ACT, EF1α, and TBP were identified as the three most stably expressed reference genes. The geNorm analysis, however, indicated that RPL7, EF1α, and GAPDH were the most stably expressed reference genes. The RefFinder ranking, from the most stable to the least stable gene in the different populations, was as follows: β-ACT, EF1α, TBP, GAPDH, RPL7, 18S, 28S, and α-TUB (Fig. 2B). The geNorm results showed that all of the Vn/n + 1 values were below the proposed 0.15 cut-off (Fig. 3). Thus, two reference genes were enough to normalize the gene expression levels in qRT-PCR analyses. Therefore, the combination of β-ACT and EF1α as reference gene was the most suitable for normalizing qRT-PCR data in the population experiments (Table 3).
Temperature Treatments
The geNorm and delta Ct method identified GAPDH, RPL7, and β-ACT as the most stably expressed reference genes. 28S and 18S were identified as the top two most stably expressed reference genes by BestKeeper. However, these two genes were identified as the least stably expressed reference genes by NormFinder and geNorm (Table 2). The RefFinder stability rankings from the most stable to the least stable gene under the varying temperature treatments were as follows: GAPDH, RPL7, EF1α, TBP, β-ACT, 28S, 18S, and α-TUB (Fig. 2C). The geNorm analysis showed that all of the Vn/n + 1 values were below the 0.15 cut-off value (Fig. 3). The combination of GAPDH and RPL7 was recommended for normalizing the qRT-PCR data in the temperature treatment experiments (Table 3).
Feeding Assays
The stability rankings generated by the delta Ct method, geNorm, and NormFinder identified 18S, β-ACT, and EF1α as the three most stably expressed reference genes. BestKeeper analysis indicated that 18S, 28S, and β-ACT were the three most stably expressed reference genes. The RefFinder stability rankings, from the most stable to the least stable gene, under the different food assays were as follows: 18S, β-ACT, EF1α, 28S, RPL7, GAPDH, α-TUB, and TBP (Fig. 2D). The geNorm analysis revealed that the V2/3 value was below the 0.15 cut-off (Fig. 3), and recommended 18S and β-ACT used as normalizing the analytical qRT-PCR data for the feeding assays (Table 3).
Pooled Data of Various Experiments
The expression stability of candidate reference genes was analyzed in pooled data across all of the various experiments. According to RefFinder, the stability ranking, from the most stable to the least stable across all samples, was as follows: EF1α, RPL7, GAPDH, TBP, 18S, β-ACT, 28S, and α-TUB (Fig. 2E). geNorm analysis indicated that all of the pair-wise variation values were below the proposed 0.15 cut-off value (Fig. 3). EF1α and RPL7 were identified as the most stably expressed reference genes for qRT-PCR normalization (Table 3). Usually, two reference genes were enough to normalize the qRT-PCR analytical results.
Validation of Reference Genes Selection
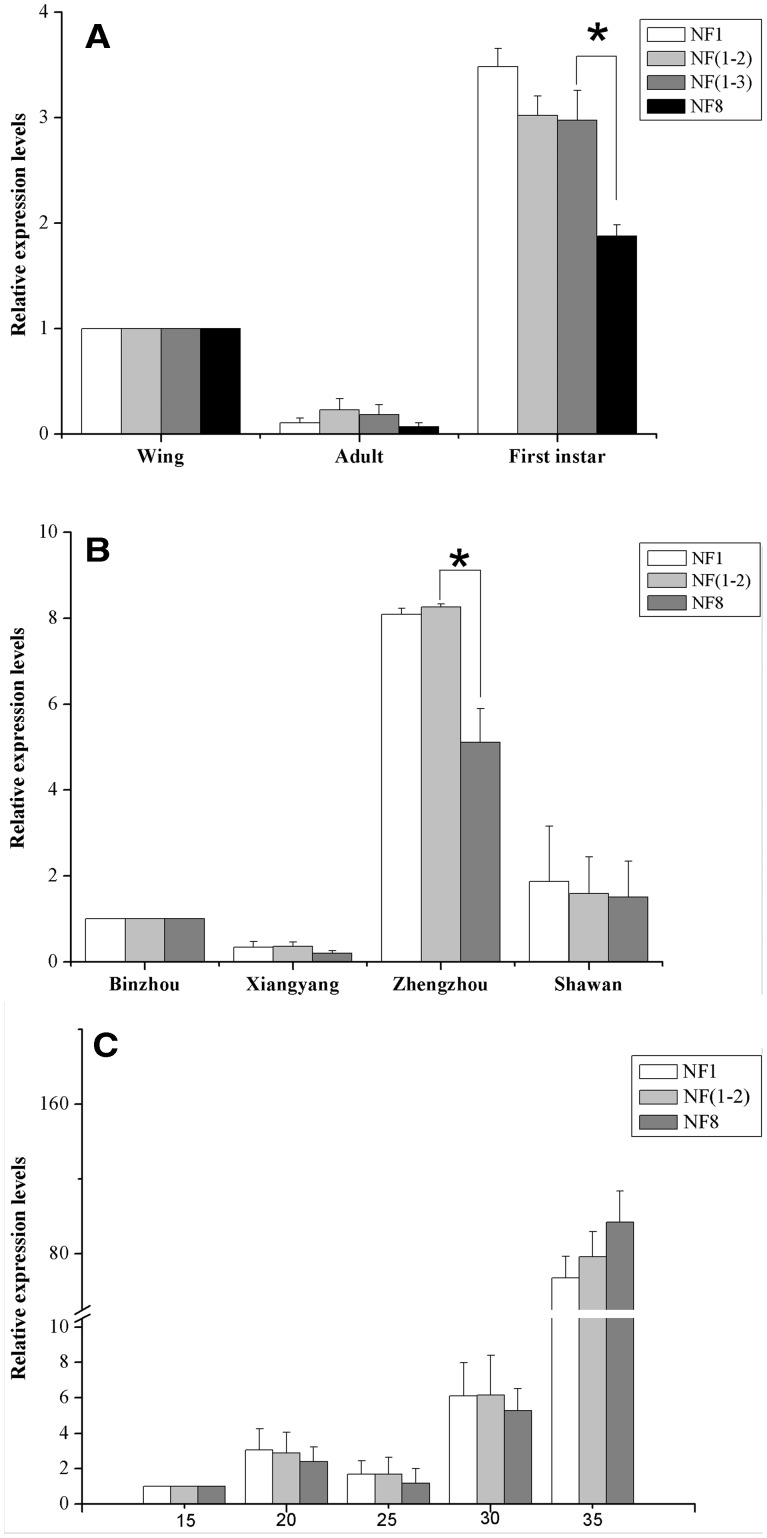
To assess the performance of selected reference genes, the expression level of HSP70 was analyzed in the same experimental conditions used for the comparisons of the expression stability of the reference genes. The similar expression levels were obtained in the developmental stage experiments when normalized using the most stable reference gene (EF1α), the combination of the two most stable reference genes (EF1α and β-ACT), and the combination of the three most stable reference genes (EF1α, β-ACT, and RPL7), and HSP70 transcript levels were higher in first-instar nymphs compared with both alate and apterous adults (Fig. 4A). However, when normalized with the least stable reference gene (TBP), no evident difference was observed between first-instar nymphs and either of the adult forms. The expression level of HSP70 normalized by the combination of the three best reference genes was significantly different from the expression level calculated using the least suitable reference gene (TBP) in the first-instar nymphs (P < 0.05) (Fig. 4A). For the experiments with the different geographical populations, the HSP70 transcript level was higher in the Zhengzhou population than in the other three populations, no matter whether it was normalized by the most stable reference gene (β-ACT), the combination of the two most stable reference genes (β-ACT and EF1α), or the least stable reference gene (α-TUB) (Fig. 4B). The expression levels of HSP70 normalized using the most stable reference gene were not different from using the combination of recommended reference genes in all four populations. However, when normalized using the least suitable reference gene, the expression levels of HSP70 were found to be significantly different in Zhengzhou population (P < 0.05) (Fig. 4B). The expression profiles of HSP70 were significantly different at the different temperatures, no matter whether the most stable reference gene (GAPDH), the combination of the two most stable reference genes (GAPDH, RPL7) or the least stable reference gene (α-TUB) was used for the normalization. The HSP70 expression levels were lower in the treatment groups by 15–30°C than by the 35°C (Fig. 4C).
Fig. 4.
Relative expression levels of a target gene of interest (HSP70) were calculated using different sets of reference genes. (A) Different expression levels in three developmental stages, (B) different expression levels in four different populations, (C) different expression levels in varying temperature treatments. The data represent the mean values ± SD. Bars represent the means and standard deviations of three biological replicates.
Discussion
Gene expression analyses are extremely important and popular in modern biological research and could lead to a better understanding of the molecular mechanisms underlying many biological processes. qRT-PCR has become one of the most commonly utilized technologies for the analysis of gene expression in recent years, because of its particular advantages, such as sensitive, accurate, and effective. However, the limitations of qRT-PCR method could result in the changes of gene expression results dependent on biotic and abiotic factors. The most serious concern is probably the inaccurate results caused by inappropriate methods that rely on the expression of a less than fully suitable reference gene. Previous studies have demonstrated that two or three reference genes should be used to ensure more accurate results (Thellin et al. 1999, Vandesompele et al. 2002), and the different combination of reference genes should be used in different experimental conditions (Lu et al. 2013, Yang et al. 2014, Zhang et al. 2014). Compounding this situation, most publications only use a single reference gene for all qRT-PCR data normalization, and these genes are often selected based on popularity rather than on their suitability for a particular experimental context. In this light, there can be no doubt that the selection of suitable A. gossypii reference genes for qRT-PCR data normalization is very important for improving research focused on this economically devastating pest.
In this study, we identified eight candidate reference genes and evaluated the performance of these genes for normalization in different experimental conditions. The results showed that the expression stability of the four most commonly used reference genes (18S, β-ACT, GAPDH, and EF1α) in A. gossypii was varied. Of these four reference genes, EF1α was found to have the least changes in expression level in various experimental conditions. EF1α plays an important role in protein synthesis and has been used as a stably expressed reference gene for normalization in qRT-PCR studies. In the present study, EF1α was the most stable reference gene across developmental stages. The similar results were obtained in A. glycines (Bansal et al. 2012) and Helicoverpa armigera (Zhang et al. 2014). Furthermore, EF1α also exhibited the stable expression in population samples, food assays, and temperature treatments (Table 2; Fig. 2). In other species, such as Bombus lucorum, Chortoicetes terminifera, and B. tabaci, EF1α has been identified as a stably expressed reference gene (Horňáková et al. 2010, Chapuis et al. 2011, Liang et al. 2014).
rRNAs, including 18S rRNA and 28S rRNA, are commonly used reference genes, as the levels of rRNA are thought to be less likely to vary dependent on different conditions (Zhong and Simons 1999, Bustin 2000). Some studies have suggested that 18S or 28S is an ideal reference gene for normalization of qRT-PCR data (Goidin et al. 2001, Bagnall and Kotze 2010). However, we found that 28S was not expressed at constant levels in different experimental conditions, and it was ranked as one of the two least suitable reference genes under the majority of the experimental conditions (Fig. 2). Although 18S is one of the most commonly used reference genes in A. gossypii, the present results indicated that 18S was also not a perfect reference gene in A. gossypii. 18S was as the most stable reference gene only present in food treatments, whereas the expression of 18S was varied significantly under other experimental conditions (Fig. 2). This unstable phenomenon of 18S as reference gene was also found in other insect species including Bactrocera dorsalis (Shen et al. 2010), B. tabaci (Li et al. 2013, Liang et al. 2014), Spodoptera exigua (Hubner) (Zhu et al. 2014), and Nilaparvata lugens Stål (Yuan et al. 2014).
In previous gene expression studies of A. gossypii, 18S, β-ACT, and GAPDH were the most frequently used reference genes for normalizing qRT-PCR data (Cao et al. 2008, 2014; Pan et al. 2009; Gu et al. 2013; Gong et al. 2014). However, our results showed that GAPDH is also not an ideal reference gene for all the studies of A. gossypii, it is only suitable under certain experimental conditions and needed to cooperate with other reference genes to obtain a reliable result. For instance, in the temperature stress experiments, the combination of GAPDH and RPL7 could be suitable for normalizing the qRT-PCR data. Similarly, when 18S and β-ACT were used for normalizing the analytical qRT-PCR results of feeding assays, an accurate result could be obtained. It suggests that the commonly used reference genes are often not well-suited for normalization of all qRT-PCR data, empirically demonstrates that researchers should pay more attention than they typically do when selected genes to use as internal controls for normalization.
As there is no universally accepted method for the characterization of the suitability of reference genes used in the normalization of qRT-PCR data, the stability of the expression of eight candidate reference genes was evaluated via four commonly used programs (geNorm, NormFinder, BestKeeper, and the comparative ΔCt method) for data generated in different experimental conditions. Notably, the suitability rankings of the reference genes were different with the different programs. This difference can be attributed to the fact of the distinct algorithms for each program. Perhaps not surprisingly, we found that no single reference gene was ranked as the most stably expressed gene in all of the various experimental conditions. This emphasizes the need to identify the most reliable reference genes for each experimental condition (Vandesompele et al. 2002). According to the algorithms of RefFinder, EF1α, β-ACT, GAPDH, and 18S were the most stably expressed reference genes in the developmental stage, population, temperature treatment, and feeding assay experiments. GeNorm is a widely used software that not only ranks the reference genes by calculating an M value (the average expression stability) but also analyzes pair-wise variation (V) to determine the optimal number of reference genes required for accurate normalization. According to the geNorm algorithms, several sets of reference genes were identified for suitable normalizing qRT-PCR data in A. gossypii under different conditions. To validate these selected reference genes, the expression levels of HSP70, an important stress-inducible heat shock protein gene (Bettencourt et al. 2007), were analyzed in different developmental stages, temperature treatments, and populations. The results demonstrated that using unsuitable reference gene(s) for normalization might lead to deviated results. Therefore, using appropriate reference genes for normalization is one of the important preconditions for accurate estimation of target gene expression.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31330064).
References Cited
- Andersen C. L., Jensen J. L., Ørntoft T. F. 2004. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64: 5245–5250. [DOI] [PubMed] [Google Scholar]
- Bagnall N. H., Kotze A. C. 2010. Evaluation of reference genes for real-time PCR quantification of gene expression in the Australian sheep blowfly, Lucilia cuprina. Med. Vet. Entomol. 24: 176–181. [DOI] [PubMed] [Google Scholar]
- Bansal R., Mamidala P., Mian M. A., Mittapalli O., Michel A. P. 2012. Validation of reference genes for gene expression studies in Aphis glycines (Hemiptera: Aphididae). J. Econ. Entomol. 105: 1432–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt B. R., Catherine C. H., Nimali M. 2007. Polyglutamine expansion in Drosophila: thermal stress and Hsp70 as selective agents. J. Biosci. 32: 537–547. [DOI] [PubMed] [Google Scholar]
- Bustin S. A. 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25: 169–193. [DOI] [PubMed] [Google Scholar]
- Bustin S. A. 2002. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 29: 23–39. [DOI] [PubMed] [Google Scholar]
- Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M. W., Shipley G. L., et al. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55: 611–622. [DOI] [PubMed] [Google Scholar]
- Cao C. W., Zhang J., Gao X. W., Liang P., Guo H. L. 2008. Overexpression of carboxylesterase gene associated with organophosphorous insecticide resistance in cotton aphids, Aphis gossypii (Glover). Pestic. Biochem. Phys. 90: 175–180. [Google Scholar]
- Cao D. P., Liu Y., Walker W. B., Li J. H., Wang G. R. 2014. Molecular characterization of the Aphis gossypii olfactory receptor gene families. PLoS One 9: e101187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuis M. P., Tohidi-Esfahani D., Dodgson T., Blondin L., Ponton F., Cullen D., Simpson S. J., Sword G. A. 2011. Assessment and validation of a suite of reverse transcription-quantitative PCR reference genes for analyses of density-dependent behavioural plasticity in the Australian plague locust. BMC Mol. Biol. 12: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derveaux S., Vandesompele J., Hellemans J. 2010. How to do successful gene expression analysis using real-time PCR. Methods 50: 227–230. [DOI] [PubMed] [Google Scholar]
- Fu W., Xie W., Zhang Z., Wang S. L., Wu Q. J., Liu Y., Zhou X. M., Zhou X. G., Zhang Y. J. 2013. Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: Plutellidae). Int. J. Biol. Sci. 9: 792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glare E. M., Divjak M., Bailey M. J., Walters E. H. 2002. β-Actin and GAPDH housekeeping gene expression in asthmatic airways is variable and not suitable for normalising mRNA levels. Thorax 57: 765–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goidin D., Mamessier A., Staquet M. J., Schmitt D., Berthier-Vergnes O. 2001. Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and beta-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal. Biochem. 295: 17–21. [DOI] [PubMed] [Google Scholar]
- Gong Y. H., Yu X. R., Shang Q. L., Shi X. Y., Gao X. W. 2014. Oral delivery mediated RNA interference of a carboxylesterase gene results in reduced resistance to organophosphorus insecticides in the cotton aphid, Aphis gossypii Glover. PLoS One 9: e102823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S. H., Wu K. M., Guo Y. Y., Field L. M., Pickett J. A., Zhang Y. J., Zhou J. J. 2013. Identification and expression profiling of odorant binding proteins and chemosensory proteins between two wingless morphs and a winged morph of the cotton aphid Aphis gossypii glover. PLoS One 8: e73524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horňáková D., Matoušková P., Kindl J., Valterová I., Pichová I. 2010. Selection of reference genes for real-time polymerase chain reaction analysis in tissues from Bombus terrestris and Bombus lucorum of different ages. Anal. Biochem. 397: 118–120. [DOI] [PubMed] [Google Scholar]
- Huggett J., Dheda K., Bustin S., Zumla A. 2005. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 6: 279–284. [DOI] [PubMed] [Google Scholar]
- Li R. M., Xie W., Wang S. L., Wu Q. J., Yang N. N., Yang X., Pan H. P., Zhou X. M., Bai L. Y., Xu B. Y., et al. 2013. Reference gene selection for qRT-PCR analysis in the sweetpotato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). PLoS One 8: e53006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. Q., Zhang S., Luo J. Y., Wang C. Y., Lv L. M., Dong S. L., Cui J. J. 2013. Ecological adaption analysis of the cotton aphid (Aphis gossypii) in different phenotypes by transcriptome comparison. PLoS One 8: e83180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P., Guo Y. J., Zhou X. G., Gao X. W. 2014. Expression profiling in Bemisia tabaci under insecticide treatment: indicating the necessity for custom reference gene selection. PLoS One 9: e87514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lourenco A. P., Mackert A., Cristino A. S., Simoes P. Z. L. 2008. Validation of reference genes for gene expression studies in the honey bee, Apis mellifera, by quantitative real-time RT-PCR. Apidologie 39: 372–385. [Google Scholar]
- Lu Y. H., Yuan M., Gao X. W., Kang T. H., Zhan S., Wan H., Li J. H. 2013. Identification and validation of reference genes for gene expression analysis using quantitative PCR in Spodoptera litura (Lepidoptera: Noctuidae). PLoS One 8: e68059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y. O., Guo H. L., Gao X. W. 2009. Carboxylesterase activity, cDNA sequence, and gene expression in malathion susceptible and resistant strains of the cotton aphid, Aphis gossypii. Comput. Biochem. Phys. B 152: 266–270. [DOI] [PubMed] [Google Scholar]
- Pan Y. O., Shang Q. L., Fang K., Zhang J., Xi J. H. 2010. Down-regulated transcriptional level of Ace1 combined with mutations in Ace1 and Ace2 of Aphis gossypii are related with omethoate resistance. Chem. Biol. Interact. 188: 553–557. [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., Tichopad A., Prgomet C., Neuvians T. P. 2004. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26: 509–515. [DOI] [PubMed] [Google Scholar]
- Ponton F., Chapuis M. P., Pernice M., Sword G. A., Simpson S. J. 2011. Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J. Insect Physiol. 57: 840–850. [DOI] [PubMed] [Google Scholar]
- Radonić A., Thulke S., Mackay I. M., Landt O., Siegert W., Nitsche A. 2004. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 313: 856–862. [DOI] [PubMed] [Google Scholar]
- Schmittgen T. D., Zakraisek B. A. 2000. Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J. Biochem. Biophys. Methods 46: 69–81. [DOI] [PubMed] [Google Scholar]
- Shen G. M., Jiang H. B., Wang X. N., Wang J. J. 2010. Evaluation of endogenous references for gene expression profiling in different tissues of the oriental fruit fly Bactrocera dorsalis (Diptera: Tephritidae). BMC Mol. Biol. 11: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thellin O., Zorzi W., Lakaye B., De Borman B., Coumans B., Hennen G., Grisar T., Igout A., Heinen E. 1999. Housekeeping genes as internal standards: use and limits. J. Biotechnol. 75: 291–295. [DOI] [PubMed] [Google Scholar]
- Udvardi M. K., Czechowski T., Scheible W. R. 2008. Eleven golden rules of quantitative RT-PCR. Plant Cell 20: 1736–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Roy N. Van, De Paepe A., Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: research0034.1‐0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanGuilder H. D., Vrana K. E., Freeman W. M. 2008. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques 44: 619–626. [DOI] [PubMed] [Google Scholar]
- Wang G. H., Xia Q. Y., Cheng D. J., Duan J., Zhao P., Chen J., Zhu L. 2008. Reference genes identified in the silkworm Bombyx mori during metamorphism based on oligonucleotide microarray and confirmed by qRT-PCR. Insect Sci. 15: 405–413. [Google Scholar]
- Yang Q. B., Li Z., Cao J. J., Zhang S. D., Zhang H. J., Wu X. Y., Zhang Q. W., Liu X. X. 2014. Selection and assessment of reference genes for quantitative PCR normalization in migratory locust Locusta migratoria (Orthoptera: Acrididae). PLoS One 9: e98164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Lu Y. H., Zhu X., Wan H., Shakeel M., Zhan S., Jin B., Li J. H. 2014. Selection and evaluation of potential reference genes for gene expression analysis in the brown planthopper Nilaparvata lugens (Hemiptera: Delphacidae) using reverse-transcription quantitative PCR. PLoS One 9: e86503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. D., An S. H., Li Z., Wu F. M., Yang Q. B., Liu Y. C., Cao J. J., Zhang H. J., Zhang Q. W., Liu X. X. 2014. Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene 555: 393–402. [DOI] [PubMed] [Google Scholar]
- Zhong H., Simons J. W. 1999. Direct comparison of GAPDH, β-Actin, cyclophilin, and 28S rRNA as internal standards for quantifying RNA levels under hypoxia. Biochem. Biophys. Res. Commun. 259: 523–526. [DOI] [PubMed] [Google Scholar]
- Zhu G. Z., Chang Y. S., Zuo J., Dong X. Y., Zhang M., Hu G. X., Fang F. D. 2001. Fudenine, a C-terminal truncated rat homologue of mouse prominin, is blood glucose-regulated and can up-regulate the expression of GAPDH. Biochem. Biophys. Res. Commun. 281: 951–956. [DOI] [PubMed] [Google Scholar]
- Zhu L. J., Altmann S. W. 2005. mRNA and 18S-RNA coapplication-reverse transcription for quantitative gene expression analysis. Anal. Biochem. 345: 102–109. [DOI] [PubMed] [Google Scholar]
- Zhu X., Yuan M., Shakeel M., Zhang Y. J., Wang S. L., Wang X., Zhan S., Kang T. H., Li J. H. 2014. Selection and evaluation of reference genes for expression analysis using qRT-PCR in the beet armyworm Spodoptera exigua (Hubner) (Lepidoptera: Noctuidae). PLoS One 9: e84730. [DOI] [PMC free article] [PubMed] [Google Scholar]