Abstract
The ryanodine receptor (RyR), the largest calcium channel protein, has been studied because of its key roles in calcium signaling in cells. Insect RyRs are molecular targets for novel diamide insecticides. The target has been focused widely because of the diamides with high activity against lepidopterous pests and safety for nontarget organisms. To study our understanding of effects of diamides on RyR, we cloned the RyR gene from the oriental fruit moth, Grapholita molesta, which is the most serious pest of stone and pome tree fruits throughout the world, to investigate the modulation of diamide insecticides on RyR mRNA expression in G. molesta (GmRyR). The full-length cDNAs of GmRyR contain a unique 3′-UTR with 625 bp and an open reading frame of 15,402 bp with a predicted protein consisting of 5,133 amino acids. GmRyR possessed a high level of overall amino acid homology with insect and vertebrate isoforms, with 77–92% and 45–47% identity, respectively. Furthermore, five alternative splice sites were identified in GmRyR. Diagnostic PCR showed that the inclusion frequency of one optional exon (f) differed between developmental stages, a finding only found in GmRyR. The lowest expression level of GmRyR mRNA was in larvae, the highest was in male pupae, and the relative expression level in male pupae was 25.67 times higher than that of in larvae. The expression level of GmRyR in the male pupae was 8.70 times higher than in female pupae, and that in male adults was 5.70 times higher than female adults.
Keywords: ryanodine receptor, Grapholita molesta, alternative splice site, expression
Diamide insecticides, a novel class of insecticides, were developed in the 1990s (Lahm et al. 2005, Teixeira and Andaloro 2012). Currently, the commercialized diamide insecticides include one phthalic diamide (flubendiamide) and two anthranilic diamides (chlorantraniliprole and cyantraniliprole). The former was discovered by Nihon Nohyaku and codeveloped with Bayer; the later two were synthesized by DuPont and cocommercialized by DuPont and Syngenta. These insecticides have notably high activity against Lepidopteran species and are relatively safe to these insects’ natural enemies and mammals (Tohnishi 2005, Gopal and Mishra 2008, Dinter et al. 2009, Ioriatti et al. 2009, Tiwari and Stelinski 2012). The selectivity of diamide insecticides toward insects over mammals was determined to be due to the different isoforms of ryanodine receptors (RyRs), the target of diamides, in insects and mammals (Sattelle et al. 2008).
RyRs, the largest known calcium channel protein, are named after ryanodine, a plant alkaloid. The receptors are homomeric tetramers with a large hydrophilic N-terminal domain and a small membrane-spanning C-terminal domain. Each monomer of ∼550–580 kDa consists of ∼5,000 amino acids. This channel is located on the endoplasmic reticulum of muscles, neurons, and many other cell types (Ogawa and Murayama 1998, Fill and Copella 2002, Hamilton 2005, Sattelle et al. 2008). RyRs mediate many cellular and physiological activities by regulating the release of Ca2+ from the lumen of the sarcoplasmic and endoplasmic reticulum to the cytosol of muscle and nonmuscle cells, causing such effects as neurotransmitter release, hormone secretion, gene expression, and muscle contraction (Ogawa and Murayama 1998). Three RyR isoforms have been indentified in mammals. RyR1 is primarily observed in skeletal muscle, RyR2 is expressed with high levels in cardiac muscle, and RyR3 is found in many tissues with low abundance but is relatively abundant in the brain and diaphragm. Two isoforms (RyRα and RyRβ) were found in fish, amphibians, and birds (Ottini et al. 1996, Ogawa et al. 1999). However, only one RyR isoform exists in such insects as Drosophila melanogaster, Plutella xylostella, Sogatella furcifera, Leptinotarsa decemlineata, Ostrinia furnacalis, Cnaphalocrocis medinalis, Carposina sasakii, etc. (Xu et al. 2000; Sun et al. 2012; Wang et al. 2012a, 2013b; Yang et al. 2014; Sun et al. 2015a,b). These insects RyRs have ∼45–47% amino acid sequence identity with the three mammalian RyR isoforms (Sattelle et al. 2008).
Diamides are mainly used to control insects that attack food crops and vegetables (Ioriatti et al. 2009; Han et al. 2012; Roditakis et al. 2013; Campos et al. 2015). P. xylostella has shown high resistance to chlorantraniliprole and flubendiamide because of the high frequency of diamide use in southern China and certain countries in Southeast Asia (Troczka et al. 2012; Wang et al. 2013a; Lin et al. 2013; Guo et al. 2014a,b; Yan et al. 2014). However, these insecticides have not been widely used in controlling the pests of tree fruits. Chlorantraniliprole was reported to have shown excellent pest control activity on the oriental fruit moth (Jones et al. 2010), Grapholita molesta, a widely distributed pest that attacks stone and pome tree fruits. G. molesta was thought to originate from northwestern China, this pest is currently distributed throughout most peach, pear, and apple orchards in China (Zheng et al. 2013). To date, there have been no published reports of diamides with cross-resistance to the current insecticides. However, with the application of these chemicals for G. molesta, the problem of resistance may occur in the future. To clear the characterization of the receptors, the full-length RyR cDNA from G. molesta (GmRyR) was isolated and characterized, and the GmRyR mRNA expression pattern was investigated.
Materials and Methods
A laboratory colony of G. molesta was originally collected in 2012 from Xingcheng in Liaoning Province, China. Larvae were reared on immature apples without pesticides and an agar-free semiartificial diet in a laboratory for several generations under a photoperiod of 15:9 (L:D) h at 25 ± 1°C and 70–80% RH.
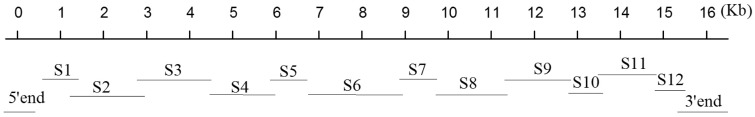
Total RNA was extracted from larvae, pupa, and adults according to the instructions in the RNAprep pure Tissue Kit (TIANGEN, China). The RNA pellet was dissolved in ddH2O. First strand cDNA was synthesized from 1 μl total RNA (650 μg/ml) using the Takara cDNA Synthesis Kit (Takara, China) following the manufacturer’s instructions. Fourteen fragments were amplified to obtain the full-length cDNA of GmRyR (Fig. 1).
Fig. 1.
PCR amplification and cloning of GmRyR cDNA.
Degenerate primers were designed by the method of Sun et al (2012, 2015a,b); specific primers were designed based on the obtained sequences (Table 1). The 5′and 3′ ends were amplified with nested PCR, with adaptor primers provided by the SMARTer RACE cDNA Amplification Kit (Clontech, Japan). Amplification of each fragment was performed with the following steps: an initial denaturing step at 94°C for 1 min followed by 30 cycles of 98°C for 10 s, 48–65°C (determined by the Tm of primers) for 30 s, and 72°C for 1–3 min (depending on the length of the amplified fragments), ending with an additional polymerization step at 72°C for 5 min. The PCR products of all fragments were purified and subcloned into the pMD19-T Simple Vector (Takara). JM109 (E. coli) competent cells transformed by the recombinant plasmids produced earlier were inoculated. The positive recombinant clones were then sequenced by BGI (Beijing, China).
Table 1.
Primers used in cloning G. molesta RyR cDNA
| Name | Primer namen | Primer sequence (5′–3′) | Description (length bp) |
|---|---|---|---|
| S1 | F1 | TTYCAYGTRACNCAYTGGTC | RT-PCR product S1 (824) |
| R1 | TGYTTYTCYTCGTGYTCCAT | ||
| S2 | F2 | TTCCGTGAACCTTGGCGAGA | RT-PCR product S2 (1,431) |
| R2 | GCAGGGATGTATCTTGTGGA | ||
| S3 | F3 | TGAGTTGCCGCTTCCTTCTT | RT-PCR product S3 (1,604) |
| R3 | CATCTGTTGGCGTTCCTTGA | ||
| S4 | F4 | GTAYACVAARGAYCARCCCAT | RT-PCR product S4 (1,482) |
| R4 | TCACCATCTCGCCAAGGTTCAC | ||
| S5 | F5 | MRDCCRCAYCARTGGGCTAG | RT-PCR product S5 (842) |
| R5 | GCRCCYTCVGCCATYTTCAT | ||
| S6 | F6 | CGAAAACTTGTTCCTGCCTC | RT-PCR product S6 (2,383) |
| R6 | ATACTCCGACAGCCGTGACA | ||
| S7 | F7 | CGHGARGCKGTBTCMGACTT | RT-PCR product S7 (854) |
| R7 | CKYTCVGCCATRTTYTGCAT | ||
| S8 | F8 | ACGGATTCAGCGACCCCATT | RT-PCR product S8 (1,394) |
| R8 | TGGTTGCCTTGAGTGGGAGT | ||
| S9 | F9 | TCWTAYTTRCCGTTCTGGTG | RT-PCR product S9 (1,586) |
| R9 | ATGTGGAGCAGCACCATCTC | ||
| S10 | F10 | ATMCAYGARCAAGARATGGA | RT-PCR product S10 (824) |
| R10 | CCTTCNARCATNGAHARCATCAT | ||
| S11 | F11 | AGTTGTCCAAGCACTCCTCG | RT-PCR product S11 (2,142) |
| R11 | CTCTTCGTGAGCCGCAAATG | ||
| S12 | F12 | TGGGACAARTTYGYRAAGAA | RT-PCR product S12 (716) |
| R12 | ATRAARCARTTGGAYTCCATGT | ||
| 3′end | 3′OF | CTGTGACGCATAATGGGAAGCA | RT-PCR product 3′RACE (1,143) |
| 3′IF | AAGAGGACGACGAGGTCAACAG | ||
| 5′end | 5′OR | AGAATCGCAGCACATCCCCACCG | RT-PCR product 5′RACE (977) |
| 5′IR | GGCTTGCGACATTACTGACCCTCC | ||
| RyR | RyR-F | GCTTCACCCGACGAGGCAGTGGAA | qRT-PCR (97) |
| RyR-R | CTTGTGCTTGCTTCTTCGCTTGTTCTC | ||
| GAPDH | GF | GCCAGCTACGACGCCATCAAGCA | qRT-PCR (109) |
| GR | CGCCGATGAAGTCAGAGGACACG | ||
| ASP-a | PaF | GAGCGAGCAGGATGATGTTT | diagnostic PCR for the presence of exon a |
| PaR | AATTTTCTTTGCCGGTCTCG | ||
| ASA-a | AaF | TCCGAGACCGGTAAAGGCA | diagnostic PCR for the absence of exon a |
| AaR | CCGTCGTGATGTGTCGTATG | ||
| AS-b | bF | TACAGCGGTAGTACAGAGTCG | diagnostic PCR for exon b |
| bR | TCGTATCTGTGGGTTAGGAC | ||
| AS-c | cF | CACCGCGGGTCGACGGAAAGT | diagnostic PCR for exon c |
| cR | TCGTATCTGTGGGTTAGGAC | ||
| ASP-d | GCCCAGTACAGCAGGTCAAG | diagnostic PCR for the presence of exon d | |
| PdR | AAGGCGTGGACTTGTAGCGA | ||
| ASA-d | AdF | AGTGTCACAG ACGAACCTCA | diagnostic PCR for the absence of exon d |
| AdR | AAGGCGTGGACTTGTAGCGA | ||
| ASP-e | PeF | CAGATGTCGTGACGGATTCA | diagnostic PCR for the presence of exon e |
| PeR | GGTGAGGAGGTCGTATGGGA | ||
| ASA-e | AeF | GTCTGGTGGC ACGGATTCAG | diagnostic PCR for the absence of exon e |
| AeR | GGTGAGGAGGTCGTATGGGA | ||
| ASP-f | PfF | TACTCGTTCTATCCGCTGCT | diagnostic PCR for the presence of exon f |
| PfR | AGCTCCGATTTTATGAGCCG | ||
| ASA-f | AfF | TCTTTACAGCAAACTGGGTT | diagnostic PCR for the absence of exon f |
| AfR | CCTCTTGTCCGATGTTCTCT |
Nucleotide sequences of full-length GmRyR cDNA were assembled by overlapping the 14 amplified fragments. GmRyR characterization was conducted using the same methods as performed in P. xylostella, Spodoptera exigua, and Ca. sasakii (Sun et al. 2012, 2015a,b).
The relative expression abundances of five GmRyR samples during three developmental stages (larvae, pupae, and adults) were measured using quantitative real-time PCR. Total RNA was extracted from whole bodies of 10 insects of each sample using the same method as earlier. First-strand cDNA was synthesized from 1,000 ng RNA using the High Capacity cDNA Reverse Transcription Kit (ABI, USA). Real-time qPCR for the RyR gene and GAPDH gene as an endogenous control from G. molesta were carried out in 20 µl reaction volumes containing 1 µl cDNA (200 ng/µl), 10 µl SYBR Premix Ex Taq (KAPA, USA), 0.4 µl each of forward and reverse primers (10 mM) (Table 1), and 8.2 µl ddH2O using the ABI 7500. Real-time PCR System (Applied Biosystems) in the same amplification condition: 95°C for 3 min followed by 40 cycles at 95°C for 3 s and 60°C for 20 s. Three biological replicates per sample were examined. The amplification efficiency of RyR and GAPDH were estimated by using E = (10−1/slope) − 1, where the slope was derived from the plot of the cycle threshold (Ct) value versus the log of the serially diluted template concentration, and computed to be 0.9584 and 0.9742, respectively. The data analysis model for quantification of the transcript level of GmRyR was computed according the 2−ΔΔCt method (Schmittgen and Livak 2008). The GmRyR expression data were shown as means ± SD. A statistical analysis was performed by Duncan’s multiple range test for significance (P < 0.05) using SPSS 20 (SPSS, Inc., Chicago, IL).
Diagnostic PCR analyses for the detection of alternative exons were used to detect the presence of each putative alternative exon in individual cDNA clones. Diagnostic PCR was used to determine the usage of each putative alternative exon for GmRyR mRNA from mature larvae, 4-d-old pupae (female and male), and 4-d-old adults (female and male). Data were collected from sets of 30 positive clones for each fragment and developmental stage. Table 1 lists the names and nucleotide sequences of primers used in diagnostic PCR reactions.
Results
The full-length cDNA of GmRyR (GenBank KM034750) is 16,299 bp long, as analyzed by overlaying all of the amplified sequenced fragments. The cDNA contains a 272 bp 5′-UTR, a unique 625 bp 3′-UTR with a 29-bp polyA tail, and an open reading frame (ORF) of 15,402 bp. From the ORF, a protein of 5,133 amino acid residues with molecular weight of 580.00 kDa is encoded with a predicted isoelectric point of 5.40.
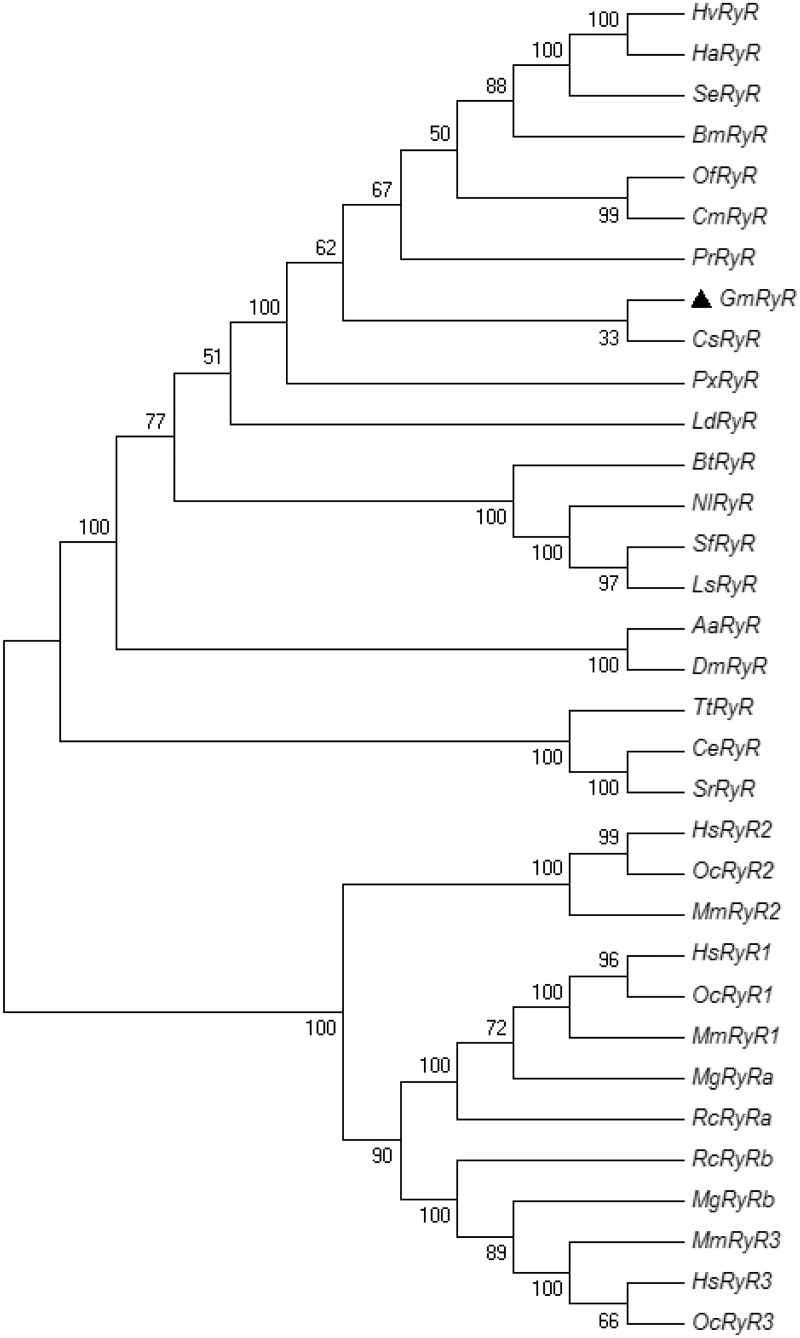
GmRyR showed amino acid identities with CsRyR, SeRyR, BmRyR, OfRyR, PxRyR, SfRyR, and DmRyR of 92, 93, 91, 93, 91, 79, and 79%, respectively. However, the amino acid identities of GmRyR with Oryctolagus cuniculus RyR1–3 was only 44–46%, and the amino acid identity of GmRyR with CeRyR was only 46% as well. To investigate the evolutionary relationships between GmRyR and 32 other RyR isoforms from 25 species, a phylogenetic analysis was performed using ClustalW and MEGA 6.0 based on the ORF amino acid sequences with high bootstrapping support in 1,000 replications (Fig. 2). The phylogenetic tree showed that insect RyRs are well segregated from invertebrate and vertebrate RyRs. GmRyR was clustered with CsRyR from the peach fruit moth, suggesting these two genes are closely related. A number of nucleotide differences were observed between these two overlapping clones. A total of 29 nucleotide differences resulted in 26 amino acid polymorphisms (Table 2). These polymorphisms were located both in the N- and C-terminal regions of GmRyR and may represent different alleles or few errors committed during the PCR procedure.
Fig. 2.
Phylogenetic tree of GmRyR and 32 other RyR isoforms. The GmRyR amino acid sequence was aligned with 32 representative RyR isoforms. The corresponding GenBank accession numbers are as follows: Heliothis virescens (HvRyR) ADE98118.1; Helicoverpa armigera (HaRyR) AIA23855.1; Sp. exigua (SeRyR) AFC36359.1; B. mori (BmRyR) DJ085056.1; O. furnacalis (OfRyR) AGH68757.1; C. medinalis (CmRyR) AFI80904.1; Pieris rapae (PrRyR) AGI62938.1; G. molesta (GmRyR) KM034750; Ca. sasakii (CsRyR) AHN16453.1; P. xylostella (PxRyR) AEI91094.1; L. decemlineata (LdRyR) AHW99830.1; Bemisia tabaci (BtRyR) AFK84957.1; Nilaparvata lugens (NlRyR) AIA23857.1; S. furcifera (SfRyR) AIA23859.1; Laodelphax striatella (LsRyR) AIA23858.1; Aedes aegypti (AaRyR) XP_001657320.1; D. melanogaster (DmRyR) AAM71083.1; Trichuris trichiura (TtRyR) CDW52896.1; Caenorhabditis elegans (CeRyR) BAA08309.1; Strongyloides ratti (SrRyR) CEF62113.1; Homo sapiens (HsRyR2) NP_001026.2; Or. cuniculus (OcRyR2) NP_001076226.1; Mus musculus (MmRyR2) NP_076357.2; H. sapiens (HsRyR1) NP_000531.2; Or. cuniculus (OcRyR1) NP_001095188.1; M. musculus (MmRyR1) NP_033135.2; Meleagris gallopavo (MgRyRa) ABY50125.1; Rana catesbeiana (RcRyRa) BAA04646.1; R. catesbeiana (RcRyRb) BAA04647.2; Me. gallopavo (MgRyRb) ABY50126.1; M. musculus (MmRyR3) NP_808320.2; H. sapiens (HsRyR3) CAA04798.1; Or. cuniculus (OcRyR3) NP_001076231.1. The neighbor-joining tree was generated in MEGA 6.0 with 1,000 bootstrap replicates.
Table 2.
Nucleotide and amino acid polymorphisms of GmRyR
| Nucleotide position* | Nucleotide exchange | Amino acid exchange# |
|---|---|---|
| 10 | G→A | A4→R |
| 11 | C→G | A4→R |
| 158 | A→G | G53→D |
| 190 | G→A | V64→M |
| 520 | T→C | S174→P |
| 614 | C→T | L205→P |
| 659 | T→C | F220→G |
| 660 | C→G | F220→G |
| 662 | G→T | G221→V |
| 663 | T→G | G221→V |
| 1,115 | G→A | R372→K |
| 2,047 | A→G | K683→E |
| 2,186 | A→G | N729→S |
| 2,300 | G→A | S767→N |
| 2,798 | C→T | P933→L |
| 4,050 | C→T | T1350→M |
| 5,193 | C→T | K1726→R |
| 6,262 | G→T | E2154→D |
| 6,686 | G→A | G2229→D |
| 6,700 | G→T | G2234→W |
| 6,965 | C→T | T2322→M |
| 7,048 | C→T | L2350→F |
| 8,192 | T→C | V2731→A |
| 8,705 | G→A | C2902→Y |
| 12,119 | A→G | Y4040→C |
| 13,544 | A→C | Q4515→P |
| 13,545 | G→A | L4516→S |
| 14,024 | T→C | V4675→A |
| 15,061 | A→G | I5021→V |
*The number of A in the initial methionine codon represents 1.
#The number of amino acid M in the initial ORF represents 1.
Conserved domains of GmRyR were predicted with four types of domains in the N-terminal region. The MIR (Mannosyltransferase, IP3R and RyR) domain was found at positions 217–398. Two RIH (RyR and IP3R Homology) domains (445–654, 2,235–2,462), three SPRY (SPla and RyR) domains (653–805, 1,085–1,217, 1,540–1,692), and four repeated RyR domains (859–953, 972–1,066, 2,836–2,929, 2,962–3,050) were also predicted. Six essential hydrophobic transmembrane domains (TM1–TM6) were predicted in the C-terminal region between amino acids 4,471 and 5,032, specifically at positions 4,471–4,493, 4,463–4,685, 4,743–4,765, 4,885–4,907, 4,933–4,955, and 5,013–5,032. Additionally, a RIH-associated domain (residues 4,009–4,134) and an EF-hand pair (Ca2+-binding sites, residues 4,222–4,270) were predicted at the C-terminal region of GmRyR.
Five putative alternative splice sites in GmRyR were revealed by the alignment of multiple cDNA clone sequences and the sequences of PCR products. They were found between nucleotides 265 and 279 (a), 3,421 and 3,519 (b/c), 4,459 and 4,521 (d), 8,845 and 8,862 (e), and 11,095 and 11,118 (f), respectively. Alternative splicing of nucleotides 3,421 and 3,519 forms one pair of mutually exclusive exons (b/c). Figure 3 shows the nucleotide and inferred amino acid sequences of the six alternative exons identified in this study.
Fig.3.
Nucleotide and putative amino acid sequences of alternatively spliced exons in the GmRyR gene.
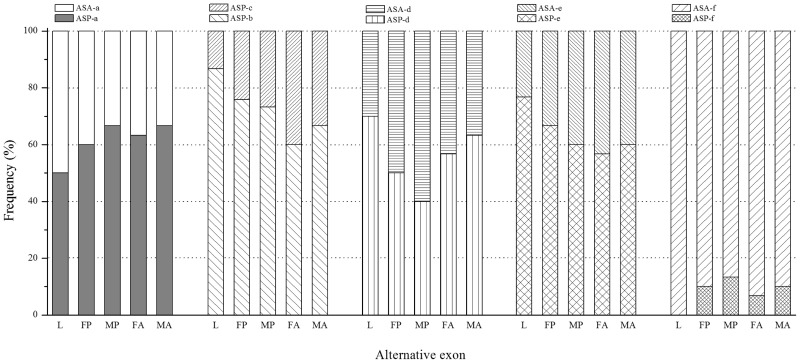
Diagnostic PCR analyses for the usage frequencies of each alternative exon in GmRyR mRNA are shown in Figure 4. The results show that the usage of exon f exhibited marked developmental regulation. Exon f was not present in all cDNA clones examined from larvae cDNA pools and was only present at low frequencies (6.67–13.33%) in the pupal and adult body cDNA pools.
Fig. 4.
Relative frequencies of individual GmRyR alternatively spliced exon usages. Alternative splicing exons were detected in fourth instar larvae (L), female pupae (FP), male pupae (MP), female adult (FA), and male adult (MA). ASA-a means that alternative splicing exon a is absent, ASP-a means that alternative splicing exon a is present. And other alternative exons (ASP-b, ASP-c, ASA-d, ASP-d, ASA-e, ASP-e, ASA-f, and ASP-f) were also showed in the same method.
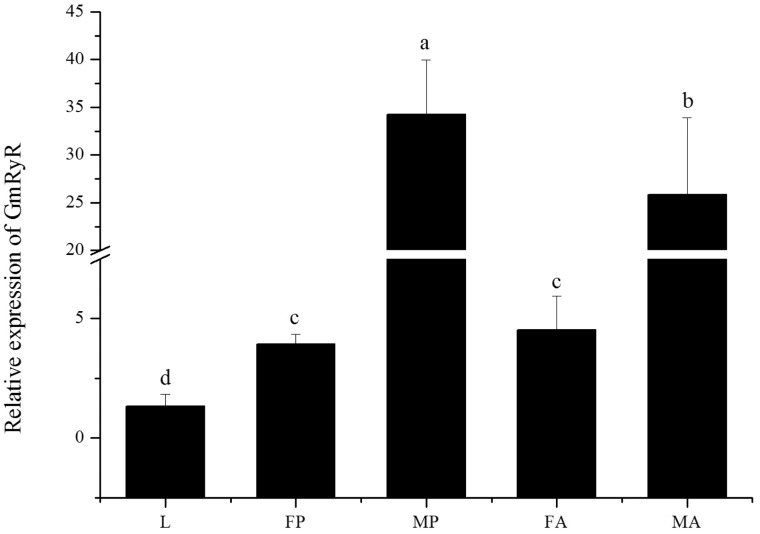
The relative expression level of GmRyR showed significant variation between different developmental stages and sexes (Fig. 5). The results showed that GmRyR had the lowest expression level in larvae and the highest in male pupae, and the relative expression level in male pupae was 25.67 (P < 0.001) times higher than that of in larvae. In addition, the expression level of GmRyR in male pupae was 8.70 (P = 0.001) times higher than in female pupae, and the expression level of GmRyR in male adults was 5.70 (P = 0.011) times higher when compared with that of female adults.
Fig. 5.
GmRyR relative expression level in fourth instar larvae (L), female pupae (FP), male pupae (MP), female adult (FA), and male adult (MA). The lowercases on the top of each bar means the statistic differences (P < 0.05).
Discussion
Ryanodine was isolated from extracts of the stem and root of Ryania Speciosa Vahl and exhibited high insecticidal activity to insects and mammals in 1948 (Rogers et al. 1948). A series of experiments found that ryanodine led to muscle contraction by regulating Ca2+ release. Thus, RyR was named after this alkaloid (Pessah et al. 1985). However, ryanodine was banned from use in pest control because of its toxicity to mammals. Although insect RyRs have been used as targets of insecticides since ryanodine was isolated, little research about RyRs was reported until the development of diamide insecticides. The diamide insecticides activate the calcium release channel that sensitize to ryanodine in insect and show high selectivity toward Lepidopteran insect pests over mammalian (Cordova et al. 2006). The diamides could evok typical symptoms for insect including poisoning, body contraction, feeding cessation, paralysis, and subsequent mortality. Because diamides are nonreactive in mammals, RyR isoforms from insects have been popular diamide targets, with increasing interest from the pesticide field since the 2000s. Recently, full-length RyRs from insects were cloned, namely PxRyR, CmRyR, OfRyR, BdRyR SfRyR, LdRyR, and SeRyR (Sun et al. 2012, 2015a; Wang et al. 2012a, 2013b, 2014; Yuan et al. 2014; Yang et al. 2014). The unique 625 bp 3′-UTR found in these species was different from other RyRs that reported, as it is the longest 3′-UTR found to date. The deduced 5,133 amino acid sequence of GmRyR showed higher amino acid identity (91–93%) with reported Lepidopteran insect RyRs compared other insect RyRs (78–79%). Furthermore, Lepidopteran insect RyRs share only 44–47% amino acid identities with mammalian isoforms. Therefore, the selectivity of diamide insecticides toward insect RyRs was suggested to be due to differences in RyR types between insects and mammals (Wang et al. 2012b). The spectrum of activity for diamides is wide, with flubendiamide targeting mostly Lepidoptera, while chlirantraniliprole and cyantraniliprole are effective in controlling Homoptera, Coleoptera, Diptera, and Thysanoptera species in addition to Lepidoptera (Teixeira and Andaloro 2013).
A multiple alignment of the C-terminal amino acid sequence from GmRyR with other reported RyRs, including BmRyR, PxRyR, OfRyR, HvRyR, DmRyR, and OcRyR1 (Or. cuniculus RyR), showed that the TM regions had high identities among the insect RyR isoforms aligned. The fragment motif, GVRAGGGIGD, sitting at residues 4,985–4,994 between TM5 and TM6 is known to form part of the pore-forming segments of the RyR Ca2+ release channel (Zhao et al. 1999). This motif from GmRyR shared complete identity with other all RyR isoforms. Moreover, the residue that enables Ca2+ sensitivity in the C-terminal domain of rabbit RyR1 (E4032) was also detected in GmRyR (E4170). Residues corresponding to I4897, R4913, and D4917 in OcRyR1, which were shown to play an important role in the activity and conductance of the RyR Ca2+ release channel, were also conserved in GmRyR (I4992, R5008, and D5012) (Zorzato et al. 1990).
Three binding regions have been predicted to be critical to diamide insecticide sensitivity. One region lies near the N-terminus (residues 183–290 from Bombyx mori) and two are located in the C-terminal TM domain (residues 4,610–4,655 from Drosophila and residue 4,946 from P. xylostella) (Kato et al. 2009, Troczka et al. 2012, Tao et al. 2013). Flubendiamide was identified to incorporate into the TM domain (amino acids 4,111–5,084) of BmRyR (Kato et al. 2009). Residues 188–295 of GmRyR share 95% identity and are equivalent with the 183–290 region of BmRyR; they both share 47–50% identity with mammalian RyRs. However, there were three sites in two regions with significant differences among species for the equivalent residues 188–295 of GmRyR (Table 3). AaRyR and DmRyR have a different residue in region I, three nematode species have three different residues in region II, and HsRyR1, MmRyR1, OcRyR1, and MgRyRα were different from other vertebrate RyRs. Table 3 indicated that region I residues were EV in Lepidoptera insects except for the peach fruit moth CsRyR, where they were EI. Region I residues were highly variable in other species. Region II residues contain eight amino acids, which have significant differences in all insects, except Noctuidae. Moreover, seven residues (N4922, N4924, N4935, L4950, L4981, N5013, and T5064) are unique to RyR isoforms from Lepidopteran insects (Wang et al. 2012a). Wang et al (2012a) and Cui et al. (2013) speculated that these residues might contribute to the differences in Ca2+ release channel properties between Lepidopteran and non-Lepidopteran insects RyRs (Wang et al. 2012a, Cui et al. 2013). However, we have different thoughts from Wang and Cui due to the insecticidal spectrum of diamides. Four special residues (D4452, P4504, V4647, and I4758) in residues 4,146–5,133 of GmRyR corresponding to residues 4,111–5,084 of BmRyR are shown in Table 2. Each residue is different among insects RyRs, nematode RyRs, and invertebrate RyRs. We think that these four residues might be involved in Ca2+ release from channels among Lepidoptera, non-Lepidoptera, nematode, and invertebrate RyRs. Isolation of microsomal membranes from insect muscles suggest that flubendiamide and chlorantraniliprole target a site localized in the pore of the insect RyR complex distinct from the ryanodine-binding site, suggesting that diamides can bind to several sites in RyR at the same time.
Table 3.
Differences of RyRs in the N-terminal and TM domains
| Animo acid position# |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 114 | 129 | 151 | 188–189 | 240–247 | 4,452 | 4,504 | 4,647 | 4,758 | ||
| Lepidoptera | GmRyR | L | Q | L | EV | TWSTEGGQ | D | P | V | I |
| CsRyR | L | Q | L | EI | TWSKEGGQ | D | P | V | I | |
| SeRyR | L | Q | L | EV | TWTKDGGQ | D | P | V | I | |
| HvRyR | L | Q | L | EV | TWTKDGGQ | D | P | V | I | |
| HaRyR | L | Q | L | EV | TWTKDGGQ | D | P | V | I | |
| BmRyR | L | Q | L | EV | TWTRDGGQ | D | P | V | I | |
| OfRyR | L | Q | L | EV | AWAKETGQ | D | P | V | I | |
| CmRyR | L | Q | L | EV | TWTKDGGL | D | P | V | I | |
| PxRyR | L | Q | L | EV | SWSNEGQH | D | P | V | I | |
| PrRyR | L | Q | L | EV | TWNKDGGL | D | P | V | I | |
| Coleoptera | LdRyR | Q | N | V | EI | TWDMEPGH | E | V | I | M |
| Homoptera | NlRyR | Q | N | V | DL | TWSEAPGQ | E | V | I | M |
| SfRyR | Q | N | V | DL | TWSEAPGQ | E | V | I | M | |
| LsRyR | Q | N | V | DL | TWSEAPGQ | E | V | I | M | |
| BtRyR | Q | N | V | DL | NWTETTGQ | E | V | I | M | |
| Diptera | AaRyR | N | N | V | DL | TWGQEPGQ | V | E | I | M |
| DmRyR | H | N | V | EQ | TWGREAGQ | V | E | I | M | |
| Nematode | TtRyR | V | T | I | MT | WSDHSQQN | S | V | V | L |
| CeRyR | V | N | I | YM | NWSEHPQH | S | A | S | L | |
| SrRyR | V | S | I | DQ | WSENQIHN | K | I | I | L | |
| Vertebrate | HsRyR1 | A | T | M | GE | ADS-DDQR | R | T | G | C |
| MmRyR1 | A | T | M | GE | SDS-DDQR | R | T | G | C | |
| OcRyR1 | A | T | M | GE | ADS-DDQR | R | T | G | C | |
| MgRyRα | S | T | L | GE | PEQGDERS | K | A | T | C | |
| RcRyRα | C | T | I | GD | TDQGEEQR | K | T | N | C | |
| HsRyR2 | S | T | I | GS | GEHGEEQR | K | A | K | C | |
| MmRyR2 | S | T | I | SS | GEHGEEQR | K | A | K | C | |
| OcRyR2 | S | T | I | GS | GEHGEEQR | K | A | K | C | |
| HsRyR3 | S | T | I | GN | TDQNDSQH | K | T | K | C | |
| MmRyR3 | S | T | I | GS | TDQNDSQH | K | T | K | C | |
| OcRyR3 | S | T | I | GN | TDQNDSQH | K | T | K | C | |
| MgRyRβ | S | T | I | GS | TDQGEEQR | K | T | K | C | |
| RcRyRβ | S | T | I | GN | TDQGEEQR | K | T | A | C | |
#The number of amino acid M in the initial ORF from GmRyR represents 1.
The putative adenine ring binding domain Y[GAST][VG] [KTQSN] was found at two locations (residues 1,145–1,148 [YSGS] and 5,098–5,101 [YTGQ]) in GmRyR. Two possible nucleotide-binding sites, identified on the basis of the consensus GXGXXG motif (Wierenga and Hol 1983), were located at positions 3,999–4,004 (GVGLEG) and 4,717–4,722 (GSGESG) in the GmRyR sequence. A potential calmodulin-binding site in GmRyR was recognized in residues 3,745–3,774, corresponding to residues 3,614–3,643 in OcRyR1 (Xiong et al. 1998). In addition, the binding sites of adenine nucleotide have no particularities compared with other Lepidopteran RyRs, such as O. furnacalis, C. medinalis, and P. xylostella (Wang et al. 2012a, Sun et al. 2012, Cui et al. 2013).
The presence of each putative alternative splice variant and expression of GmRyR mRNA in five discrete mRNA pools (larva, female pupa, male pupa, female adult, and male adult) were determined (Fig. 4). The alternative splice segments were different from CmRyR, OfRyR, BdRyR, and NlRyR (Wang et al. 2012, 2013; Cui et al. 2013; Yuan et al. 2014). Five alternative splice segments were located between amino acid residues 89–93 (a), 1,141–1,173 (b/c), 1,487–1,507 (d), 2,949–2,954 (e), and 3,699–3,706 (f). Interestingly, only the mutually exclusive exons (b/c) were also found in CmRyR, OfRyR, BdRyR, and NlRyR and were developmentally regulated (Wang et al. 2012a, 2014; Cui et al. 2013; Yuan et al. 2014). Furthermore, alternative splicing of residues (Ala3481-Gln3485) in RyR1 was thought to be developmentally regulated (Kimura et al. 2009). The BLAST results showed that residues 89–93 were present in CsRyR, PxRyR, HaRyR, HvRyR, and OfRyR. The analysis of alternatively spliced regions revealed that region b/c and d were located in the central part of the predicted SPRY domain and region e was located between the third and fourth RyR domains. The alternatively spliced region f was only in pupae and adults from GmRyR. Splice variants of RyR2 have been recognized for over a decade but their functions are as undefined to date. Moreover, Knight and Flexner (2007) confirmed that males were more sensitive to chlorantraniliprole-related disruption of mating than females. Thus, further studies are needed to elucidate the characterization and functions of each GmRyR splice variant.
There are some differences in RyR expression between invertebrates and vertebrates. RNA from eggs of G. molesta was not extracted because the eggs were not successfully collected. Real-time quantitative PCR showed that the expression level of GmRyR mRNA was quite different in larvae, pupae, and adults, particularly in pupae and adults of different sexes. The results showed the lowest expression level in larvae and the highest in male pupae. Wang et al. (2014) reported that NlRyR mRNA expression levels in macropterous female adults were significantly higher than that in brachypterous female adults as well as macropterous and brachypterous male adults. Guo et al. (2012) reported that the expression abundance of the RyR in the second-instar larvae and adults were considerably higher than those of the prepupae and pupae in P. xylostella. Wang et al. (2012b) showed that PxRyR expression level in larvae was higher than in pupae. The expression levels of OfRyR were gradually upregulated with increasing age (Cui et al. 2013), and different expression levels of RyR were shown in BdRyR (Yuan et al. 2014). In summary, although we cloned and characterized the expression levels of GmRyR, its properties and functions remain to be studied.
Acknowledgments
This research was conducted under the support of the Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP), the Fruiter Industry Technology System-Innovation Teams in Liaoning Province (LNGSCYTX-13/14), and the PhD Start-up Fund of the Natural Science Foundation of Liaoning Province (20141157).
References Cited
- Campos M. R., Silva T. B. M., Silva W. M., Silva J. E., Siqueira H. A. A. 2015. Susceptibility of Tuta absoluta (Lepidoptera: Gelechiidae) Brazilian populations to ryanodine receptor modulators. Pest Manag. Sci. 71: 537–544. [DOI] [PubMed] [Google Scholar]
- Cordova D., Benner E. A., Sacher M. D., Rauh J. J., Sopa J. S., Lahm G. P., Selby T. P., Stevenson T. M., Flexner L., Gutteridge S., et al. 2006. Anthranilic diamides: a new class of insecticides with a novel mode of action, ryanodine receptor activation. Pest. Biochem. Physiol. 84: 196–214. [Google Scholar]
- Cui L., Yang D. B., Yan X. J., Rui C. H., Wang Z. Y., Yuan H. Z. 2013. Molecular cloning, characterization and expression profiling of a ryanodine receptor gene in Asian Corn Borer, Ostrinia furnacalis (Guene´ e). PLoS One 8: e75825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinter A., Brugger K. E., Frost N. M., Woodward M. D. 2009. Chlorantraniliprole (Rynaxypyr): a novel DuPont™ insecticide with low toxicity and low risk for honey bees (Apis mellifera) and bumble bees (Bombus terrestris) providing excellent tools for uses in integrated pest management, pp. 984-996. In Proceedings, Hazards of Pesticides to Bees-10th International Symposium of the ICP-Bee Protection Group, 8–10 October 2008, Bucharest. [Google Scholar]
- Fill M., Copella J. A. 2002. Ryanodine receptor calcium release channels. Physiol. Rev. 82: 893–922. [DOI] [PubMed] [Google Scholar]
- Guo L., Tang B. Z., Dong W., Liang P., Gao X. W. 2012. Cloning, characterisation and expression profiling of the cDNA encoding the ryanodine receptor in diamondback moth, Plutella xylostella (L.) (Lepidoptera: Plutellidae). Pest Manag. Sci. 68: 1605–1614. [DOI] [PubMed] [Google Scholar]
- Guo L., Liang P., Zhou X. G., Gao X. W. 2014a. Novel mutations and mutation combinations of ryanodine receptor in a chlorantraniliprole resistant population of Plutella xylostella (L.). Sci. Rep. 4: 6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Wang Y., Zhou X. G., Li Z. Y., Liu S. Z., Liang P., Gao X. W. 2014b. Functional analysis of a pointmutation in the ryanodine receptor of Plutella xylostella (L.) associated with resistance to chlorantraniliprole. Pest Manag. Sci. 70: 1083–1089. [DOI] [PubMed] [Google Scholar]
- Gopal M., Mishra E. 2008. Analytical method for estimation of a new insecticide flubendiamide and its safety evaluation for usage in rice crop. Bull. Environ. Contam. Toxicol. 81: 360–364. [DOI] [PubMed] [Google Scholar]
- Hamilton S. L. 2005. Ryanodine receptors. Cell Calcium 38: 253–260. [DOI] [PubMed] [Google Scholar]
- Han W. S., Zhang S. F., Shen F. Y., Liu M., Ren C. C., Gao X. W. 2012. Residual toxicity and sublethal effects of chlorantraniliprole on Plutella xylostella (Lepidoptera: Plutellidae). Pest Manag. Sci. 68: 1184–1190. [DOI] [PubMed] [Google Scholar]
- Ioriatti C., Anfora G., Angeli G., Mazzoni V., Trona F. 2009. Effects of chlorantraniliprole on eggs and larvae of Lobesia botrana (Denis & Schiffermüller) (Lepidoptera: Tortricidae). Pest Manag. Sci. 65: 717–722. [DOI] [PubMed] [Google Scholar]
- Jones M. M., Robertson J. R., Weinzierl R. A. 2010. Susceptibility of oriental fruit moth (Lepidoptera: Tortricidae) larvae to selected reduced-risk insecticides. J. Econ. Entomol. 103: 1815–1820. [DOI] [PubMed] [Google Scholar]
- Kato K., Kiyonaka S., Sawaguchi Y., Tohnishi M., Masaki T., Yasokawa N., Mizuno Y., Mori E., Inoue K., Hamachi I., et al. 2009. Molecular characterization of flubendiamide sensitivity in the lepidopterous ryanodine receptor Ca2+ release channel. Biochemistry 48: 10342–10352. [DOI] [PubMed] [Google Scholar]
- Kimura T., Lueck J. D., Harvey P. J., Pace S. M., Ikemoto N., Casarotto M. G. 2009. Alternative splicing of RyR1 alters the efficacy of skeletal EC coupling. Cell Calcium 45: 264–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight A. L., Flexner L. 2007. Disruption of mating in codling moth (Lepidoptera: Tortricidae) by chlorantranilipole, an anthranilic diamide insecticide. Pest Manag. Sci. 63: 180–189. [DOI] [PubMed] [Google Scholar]
- Lahm G. P., Selby T. P., Freudenberger J. H., Stevenson T. M., Myers B. J., Seburyamo G., Smith B. K., Flexner L., Clark C. E., Cordov D. 2005. Insecticidal anthranilic diamides: a new class of potent ryanodine receptor activators. Bioorg. Med. Chem. Lett. 15: 4898–4906. [DOI] [PubMed] [Google Scholar]
- Lin Q. S., Jin F. L., Hu Z. D., Chen H. Y., Yin F., Li Z. Y., Dong X. L., Zhang D. Y., Ren S. X., Feng X. 2013. Transcriptome analysis of chlorantraniliprole resistance development in the diamondback moth Plutella xylostella. PLoS One 8: e72314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y., Murayama T. 1998. The structure and function of ryanodine receptors, pp. 5–22. In Sitsapesan R., Williams A. J. (eds.). Characteristics of Ryanodine Receptor Type 3 Isoform (RyR3) and Its Homologues. Imperial College Press, London. [Google Scholar]
- Ogawa Y., Murayama T., Kurebayashi N. 1999. Comparison of properties of Ca2+ release channels between rabbit and frog skeletal muscles. Mol. Cell. Biochem. 190: 191–201. [PubMed] [Google Scholar]
- Ottini L., Marziali G., Conti A., Charlesworth A., Sorrentino V. 1996. Alpha and beta isoforms of ryanodine receptor from chicken skeletal muscle are the homologues of mammalian RyR1 and RyR3. Biochem. J. 315: 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessah I. N., Waterhouse A. L., Casida J. E. 1985. The calcium ryanodine receptor complex of skeletal and cardiac muscle. Biochem. Biophys. Res. Commun. 128: 449–456. [DOI] [PubMed] [Google Scholar]
- Roditakis E., Skarmoutsou C., Staurakaki M., Martínez-Aguirre M. R., García-Vidal L., Bielza P., Haddi K., Rapisarda C., Rison J. L., Bassi A., et al. 2013. Determination of baseline susceptibility of European populations of Tuta absoluta (Meyrick) to indoxacarb and chlorantraniliprole using a novel dip bioassay method. Pest Manag. Sci. 69: 217–227. [DOI] [PubMed] [Google Scholar]
- Rogers E. F., Koniuszy F. R., Shavel J., Folkers K. 1948. Plant insecticides: ryanodine, a new alkaloid from Ryania speciosa Vahl. J. Am. Chem. Soc. 70: 3086–3088. [DOI] [PubMed] [Google Scholar]
- Sattelle D. B., Cordova D., Cheek T. R. 2008. Insect ryanodine receptors: molecular targets for novel pest control chemicals. Invert. Neurosci. 8: 107–119. [DOI] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. 2008. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Sun L. N., Cui L., Rui C. H., Yan X. J., Yang D. B., Yuan H. Z. 2012. Modulation of the expression of ryanodine receptor from Plutella xylostella as a result of diamide insecticide application. Gene 511: 265–273. [DOI] [PubMed] [Google Scholar]
- Sun L. N., Qiu G. S., Cui L., Ma C. S., Yuan H. Z. 2015a. Molecular characterization of a ryanodine receptor gene from Spodoptera exigua and its upregulation by chlorantraniliprole. Pest. Biochem. Physiol. 123: 56–63. [DOI] [PubMed] [Google Scholar]
- Sun L. N., Zhang H. J., Yan W. T., Ma C. S., Qiu G. S. 2015b. Molecular cloning and expression profling of a ryanodine receptor gene in the peach fruit moth (Carposina ssakii). Scientia Agricultura Sinica 48: 1971–1981. [Google Scholar]
- Tao Y., Gutteridge S., Benner E. A., Wu L. H., Rhoades D. F., Sacher M. D., Rivera M. A., Desaeger J., Cordova D. 2013. Identification of a critical region in the Drosophila ryanodine receptor that confers sensitivity to diamide insecticides. Insect Biochem. Mol. Biol. 43: 820–828. [DOI] [PubMed] [Google Scholar]
- Teixeira L. A., Andaloro G. T. 2012. Diamide insecticides: global efforts to address insect resistance stewardship challenges. Pest. Biochem. Physiol. 106: 76–78. [Google Scholar]
- Tiwari S., Stelinski L. L. 2013. Effects of cyantraniliprole, a novel anthranilic diamide insecticide, against Asian citrus psyllid under laboratory and field conditions. Pest Manag. Sci. 69: 1066–1072. [DOI] [PubMed] [Google Scholar]
- Tohnishi M., Nakao H., Furuya T., Seo A., Kodama H., Tsubata K., Fujioka S., Kodama H., Hirooka T., Nishimatsu T. 2005. Flubendiamide, a novel insecticide highly active against Lepidopterous insect pests. J. Pest. Sci. 30: 354–360. [Google Scholar]
- Troczka B., Zimmer C. T., Elias J., Schorn C., Bass C., Davies T. G. E., Field L. M., Williamson M. S., Slater R., Nauen R. 2012. Resistance to diamide insecticides in diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae) is associated with a mutation in the membrane-spanning domain of the ryanodine receptor. Insect Biochem. Mol. Biol. 42: 873–880. [DOI] [PubMed] [Google Scholar]
- Wang J., Liu Y. P., Gao J. K., Xie Z., Huang L., Wang W., Wang J. 2013a. Molecular cloning and mRNA expression of a ryanodine receptor gene in the cotton bollworm, Helicoverpa armigera. Pest. Biochem. Physiol. 107: 327–333. [DOI] [PubMed] [Google Scholar]
- Wang J., Xie Z., Gao J. K., Liu Y. P., Wang W. L., Huang L., Wang J. J. 2014. Molecular cloning and characterization of a ryanodine receptor gene in brown planthopper (BPH), Nilaparvata lugens (Stål). Pest Manag. Sci. 70: 790–797. [DOI] [PubMed] [Google Scholar]
- Wang J. J., Li Y. Q., Han Z. J., Zhu Y. L., Xie Z. J., Wang J., Liu Y. P., Li X. C. 2012a. Molecular characterization of a ryanodine receptor gene in the rice leaffolder, Cnaphalocrocis medinalis (Guenée). PLoS One 7: e36623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. L., Wu S. W., Yang Y. H., Wu Y. D. 2012b. Molecular cloning, characterization and mRNA expression of a ryanodine receptor gene from diamondback moth, Plutella xylostella. Pest. Biochem. Physiol. 102: 204–212. [Google Scholar]
- Wang X. X., Khakame S. K., Ye C., Yang Y. H., Wu Y. D. 2013b. Characterisation of field-evolved resistance to chlorantraniliprole in the diamondbackmoth, Plutella xylostella, from China. Pest Manag. Sci. 69: 661–665. [DOI] [PubMed] [Google Scholar]
- Wierenga R. H., Hol W. G. J. 1983. Predicted nucleotide-binding properties of p21 protein and its cancer-associated variant. Nature 302: 842–844. [DOI] [PubMed] [Google Scholar]
- Xiong H., Feng X. Y., Gao L., Xu L., Pasek D. A., Seok J. H., Meissner G. 1998. Identification of a two EF-hand Ca2+ binding domain in lobster skeletal muscle ryanodine receptor/Ca2+ release channel. Biochemistry 37: 4804–4814. [DOI] [PubMed] [Google Scholar]
- Xu X., Bhat M. B., Nishi M., Takeshima H., Ma J. 2000. Molecular cloning of cDNA encoding a Drosophila ryanodine receptor and functional studies of the carboxyl-terminal calcium release channel. Biophys. J. 78: 1270–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H. H., Xue C. B., Li G. Y., Zhao X. L., Che X. Z., Wang L. L. 2014. Flubendiamide resistance and Bi-PASA detection of ryanodine receptor G4946E mutation in the diamondback moth (Plutella xylostella L.). Pest. Biochem. Physiol. 115: 73–77. [DOI] [PubMed] [Google Scholar]
- Yang Y., Wan P. J., Hu X. X., Li G. Q. 2014. RNAi mediated knockdown of the ryanodine receptor gene decreases chlorantraniliprole susceptibility in Sogatella furcifera. Pest. Biochem. Physiol. 108: 58–65. [DOI] [PubMed] [Google Scholar]
- Yuan G. R., Shi W. Z., Yang W. J., Jiang X. Z., Dou W., Wang J. J. 2014. Molecular characteristics, mRNA expression, and alternative splicing of a ryanodine receptor gene in the oriental fruit fly, Bactrocera dorsalis (Hendel). PLoS One 9: e95199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M. C., Li P., Li X. L., Zhang L., Winkfein R. J., Chen S. W. 1999. Molecular identification of the ryanodine receptor pore-forming segment. J. Biol. Chem. 274: 25971–25974. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Peng X., Liu G., Pan H., Dorn S., Chen M. 2013. High genetic diversity and structured populations of the oriental fruit moth in its range of origin. PLoS One 8: e78476 doi:10.1371/journal.pone.0078476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorzato F., Fujii J., Otso K., Phillips M., Green N. M., Lai F. A., Meissner G., MacLennan D. H. 1990. Molecular cloning of cDNA encoding human and rabbit forms of the Ca2+ release channel (ryanodine receptor) of skeletal muscle sarcoplasmic reticulum. J. Biol. Chem. 265: 2244–2256. [PubMed] [Google Scholar]