Abstract
Please cite this paper as: Vincent et al. (2010) Experimental inoculation of pigs with pandemic H1N1 2009 virus and HI cross‐reactivity with contemporary swine influenza virus antisera. Influenza and Other Respiratory Viruses 4(2), 53–60
Background A novel A/H1N1 was identified in the human population in North America in April 2009. The gene constellation of the virus was a combination from swine influenza A viruses (SIV) of North American and Eurasian lineages that had never before been identified in swine or other species.
Objectives The objectives were to (i) evaluate the clinical response of swine following experimental inoculation with pandemic H1N1 2009; (ii) assess serologic cross‐reactivity between H1N1 2009 and contemporary SIV antisera; and (iii) develop a molecular assay to differentiate North American‐lineage SIV from H1N1 2009.
Methods Experiment 1: Weaned pigs were experimentally infected with A/California/04/2009 (H1N1). Experiment 2: The cross‐reactivity of a panel of US SIV H1N1 or H1N2 antisera with three isolates of pandemic A/H1N1 was evaluated. Experiment 3: A polymerase chain reaction (PCR)‐based diagnostic test was developed and validated on samples from experimentally infected pigs.
Results and Conclusions In experiment 1, all inoculated pigs demonstrated clinical signs and lesions similar to those induced by endemic SIV. Viable virus and antigen were only detected in the respiratory tract. In experiment 2, serologic cross‐reactivity was limited against H1N1 2009 isolates, notably among virus antisera from the same HA phylogenetic cluster. The limited cross‐reactivity suggests North American pigs may not be fully protected against H1N1 2009 from previous exposure or vaccination and novel tests are needed to rapidly diagnose the introduction of H1N1 2009. In experiment 3, an RT–PCR test that discriminates between H1N1 2009 and endemic North American SIV was developed and validated on clinical samples.
Keywords: H1N1 2009, hemagglutination inhibition assay, influenza A virus, pathogenesis, swine
Introduction
In March–April 2009, a novel pandemic H1N1 emerged in the human population in North America. 1 The gene constellation of the emerging virus was demonstrated to be a combination of genes from swine influenza A viruses (SIV) of North American and Eurasian lineages that had never before been identified in swine or other species. The emergent H1N1 quickly spread in the human population and the outbreak reached pandemic level 6 as declared by the World Health Organization on 11 June 2009. Although the eight gene segments of the novel virus share lineage with available sequences of corresponding genes from SIV from North America and Eurasia, no closely related ancestral SIV with this gene combination has been identified in North America or elsewhere in the world. 2 , 3 Other than sporadic transmission to humans, 4 , 5 SIV of the H1N1 subtype historically have been distinct from avian and other mammalian H1N1 influenza viruses in characteristics of host specificity, serologic cross‐reactivity, and/or nucleotide sequence. Since 1997–1998 in North America, multiple subtypes of endemic SIV (H3N2, H1N1, and H1N2) co‐circulate in most major swine producing regions of the USA and Canada (reviewed in Ref. 6). The North American SIV have a triple reassortant internal gene (TRIG) constellation consisting of six genes excluding the surface glycoproteins, hemagglutinin (HA), and neuraminidase (NA), with the six TRIG genes being derived from swine, avian, and human influenza viruses. Additionally, introduction of H1N1 and H1N2 viruses with the HA and NA genes originating from contemporary human seasonal influenza A viruses (hu‐like H1) that are genetically and antigenically distinct from the classical swine H1 lineage were reported in pigs in Canada. 7 The viruses identified in Canadian pigs were human virus lineage in entirety or double (human‐swine) reassortants. Since 2005, hu‐like H1N1 and H1N2 viruses have emerged in swine herds across the USA as human‐swine reassortants possessing the TRIG. 8 Four phylogenetic clusters (α, β, γ, and δ) of H1 SIV are now endemic in US swine. 8 , 9 However, to date, Eurasian lineage SIVs have not been reported in the USA, thus the potential impact of transmission of the pandemic H1N1 virus to the US pig population is unknown.
In experiment 1, weaned pigs were experimentally inoculated with A/CA/04/2009 H1N1 to evaluate clinical signs and lesions during acute infection with the human pandemic virus in the swine host. As North American lineage H1 SIV are endemic in the USA, it is also important to understand the role that existing herd immunity against endemic swine H1 may play in protecting the swine population from the human pandemic H1N1 2009. The serologic cross‐reactivity of a panel of swine antisera generated against H1N1 or H1N2 SIV was investigated against three novel human A/H1N1 isolates in experiment 2 to predict the susceptibility of pigs with pre‐existing immunity against North American lineage H1 swine viruses against the pandemic H1N1 2009. To rapidly differentiate the novel H1N1 from endemic North American H1 SIV, in experiment 3 a restriction fragment length polymorphism (RFLP) developed for the matrix (M) gene was evaluated for use as a method of discriminating H1 genotypes in swine specimens.
Materials and methods
Virus isolates
A/California/04/2009 (CA/09) (cell passage 1), A/New York/18/2009 (NY/09) (egg passage 3), and A/Mexico/4108/2009 (MX/09) (egg passage 2) received from the Centers for Disease Control and Prevention (CDC) were propagated in Madin–Darby Canine Kidney (MDCK) cells for use in the studies described below.
In vivo study
In experiment 1, four 5‐week‐old cross‐bred pigs from a herd free of SIV and porcine reproductive and respiratory syndrome virus (PRRSV) were housed in ABSL3 isolation and cared for in compliance with the Institutional Animal Care and Use Committee of the National Animal Disease Center. Pigs were inoculated intra‐tracheally with 2 ml of 1 × 105 50% tissue culture infectious dose (TCID50) of CA/09 (H1N1)v as previously described. 9 Four additional age‐ and source‐matched pigs were maintained as negative controls. All pigs were screened for influenza A nucleoprotein antibody by ELISA (MultiS ELISA; IDEXX, Westbrook, ME, USA) before the start of the study to ensure absence of prior immunity. Pigs were observed twice daily for signs of clinical disease and fever. Nasal swabs (Fisherbrand Dacron swabs; Fisher Scientific, Pittsburg, PA, USA) were taken and placed into 2 ml minimal essential medium (MEM) on 0, 1, 2, 3, 4, and 5 days post‐infection (dpi) to evaluate nasal virus shedding and stored at −80°C until study completion. Pigs were humanely euthanized with a lethal dose of pentobarbital (Sleepaway; Fort Dodge Animal Health, Fort Dodge, IA, USA) on 5 dpi to evaluate lung lesions and viral load in the lung and other selected tissues. Fresh samples were taken from lung, tonsil, inguinal lymph node, liver, spleen, kidney, semitendinosus skeletal muscle (ham), and colon contents (feces) using individual sterile instruments for each tissue. Fresh necropsy samples were stored at −80°C until processed for downstream assays. Additional samples of the same tissues were fixed in 10% buffered formalin and processed by routine methods for histopathologic and immunohistopathologic examination. Immunohistochemical methods for the detection of influenza antigen in tissues using monoclonal antibody against type A nucleoprotein were employed as previously described. 10 Bronchoalveolar lavage fluid (BALF) samples from 5 dpi were screened for aerobic bacterial growth on blood agar and Casmin (Nicotinamide adenine dinucleotide (NAD) enriched) plates. Diagnostic polymerase chain reaction (PCR) for PCV2 11 and Mycoplasma hyopneumoniae, 12 or an in‐house reverse transcriptase PCR (RT–PCR) for PRRSV were conducted on nucleic acid extracts from BALF.
Virus isolation from tissues
Frozen necropsy samples were utilized to assess tissue viral load. Approximately 500 mg of tissue or colon contents (feces) were placed in a 1·5‐ml pestle tube and homogenized in 300 μl of PBS with antibiotics. Subsequently, 200 μl of the tissue homogenate, serum, or nasal swab sample was then placed on confluent MDCK cells in 24‐well plates to incubate for 1 h. After 1 h of incubation, the sample was removed and 400‐μl MEM w/TPCK trypsin was added. The plate was checked at 24 and 48 h for cytopathic effects. After 48 h, 200 μl of cell culture supernatant from each well of the 24‐well plate was subsequently passed onto a confluent 48‐well plate after a freeze and thaw cycle. After 48 h, evidence of cytopathic effects was evaluated and presence of virus antigen confirmed by immuno‐cytochemical staining. Two replicates of tissue virus isolation were conducted. Virus titers in BALF and nasal swabs were determined on MDCK cells in 96‐well plates.
RNA extraction from tissues and fluids
The MagMax Microarray (Ambion, Austin, TX, USA) protocol for RNA extraction from tissues was followed. Approximately 10 mg of each tissue was placed in a 1·5‐ml pestle tube and homogenized in 300 μl of PBS w/antibiotics. Tissue homogenate (100 μl) from above was added to bromo–chloro–propane (10 μl), incubated for 5 min at room temperature, centrifuged at 12 000 × g for 10 min at 4°C and 100 μl of the aqueous phase was transferred to the processing plate for MagMax RNA extraction as per manufacturer’s instructions. The MagMax Viral RNA Isolation (Ambion) kit protocol was used as per manufacturer’s instructions for serum, nasal swab, or BALF by adding 50 μl of serum, nasal swab, or BALF sample to the Mag Max plate for RNA extraction.
Real time RT–PCR
A TaqMan assay targeting the matrix gene was performed as previously described 13 with modification as per the USDA–APHIS National Veterinary Services Laboratory protocol to increase sensitivity for the pandemic virus matrix gene by adding a pandemic H1N1 2009 matched reverse primer.
Serologic assays
In experiment 2, 38 polyclonal antisera from pigs immunized with 19 H1 SIV isolated during 1999–2008 were tested against the pandemic viruses in a standard hemagglutination inhibition (HI) assay. 14 Each of the four phylogenetic clusters (α, β, γ, and δ) 8 , 9 of endemic North American H1 SIV were represented in the panel of sera as previously described 15 and with additional SIV isolates using the same methodology (A.L. Vincent, unpublished). Virus isolates used to generate antisera are listed in Table 3. Immunized pigs received intramuscular injections of 1 × 106 TCID50 per ml or approximately 64–128 HA units of UV‐inactivated influenza virus combined with a commercial adjuvant (Emulsigen D; MVP Laboratories, Inc., Ralston, NE, USA), followed by one or two booster doses 2–3 weeks apart until sufficient homologous HI titers were reached. For use in the HI assay, sera were heat inactivated at 56°C for 30 min, then treated to remove non‐specific HA inhibitors and natural serum agglutinins with receptor destroying enzyme followed by treatment with a 20% suspension of kaolin (Sigma Aldrich, St Louis, MO, USA) and adsorption with 0·5% turkey red blood cells (RBCs). The HI assays were then performed with the CA/09, NY/09, and MX/09 viruses as antigens and turkey RBC using standard techniques. 14 Additionally, 14 antisera from pigs immunized with five commercial vaccines or vaccines in the process of eventual licensure from the USDA–APHIS Center for Veterinary Biologics were tested against the pandemic H1N1 2009 viruses. Commercial vaccine antisera against either individual virus components or the combination of vaccines were supplied by the manufacturers for use in the study. Vaccine A (FluSure® XP; Pfizer Animal Health, New York, NY, USA) is a fully licensed trivalent commercial product containing cluster IV H3N2, γ‐cluster H1N1, and δ‐cluster H1N1 SIV as vaccine seed viruses. Vaccine B (MaxiVac Excell® 5.0; Intervet/Schering‐Plough, Boxmeer, the Netherlands) is a pentavalent product under review for full licensure containing clusters I and IV H3N2 and β‐, γ‐, and δ‐cluster H1 SIV. Vaccine C (Pneumostar® SIV; Novartis Animal Health, Basel, Switzerland) is a fully licensed bivalent commercial product containing H3N2 and α‐cluster H1N1, and a trivalent product that is under review for full licensure with the addition of a γ‐cluster H1 SIV to the H3N2 and α‐cluster H1N1 in the bivalent product. Vaccine D (Newport Labs, Worthington, MN, USA) is a bivalent autogenous vaccine containing β‐ and γ‐cluster H1 SIV. Vaccine E (Boehringer‐Ingelheim‐Vetmedica, St Joseph, MO, USA) is a subunit vaccine for H1 SIV in the research and development stage. The reported HI titer is the reciprocal of the highest dilution of serum where inhibition of viral agglutination of RBC was observed with a particular serum and virus pair.
Table 3.
Serologic cross‐reactivity between swine anti‐influenza sera and pandemic H1N1 2009
| Antiserum | Homologous | CA/09 | NY/09 | MX/09 |
|---|---|---|---|---|
| H1α phylogenetic cluster | ||||
| A/sw/MN/37866/99 | 1280 | 20 | 40 | 160 |
| 2560 | 80 | 40 | 320 | |
| A/sw/MN/02053/08 | 320 | <10 | <10 | 20 |
| 320 | <10 | <10 | <10 | |
| A/sw/MN/02093/08 | 320 | <10 | <10 | <10 |
| 320 | <10 | <10 | <10 | |
| H1β phylogenetic cluster | ||||
| A/sw/NC/36883/02 | 640 | 40 | 40 | 160 |
| 640 | 20 | 20 | 80 | |
| A/sw/IA/00239/04 | 1280 | 20 | 10 | 80 |
| 1280 | 20 | 20 | 40 | |
| A/sw/KY/02086/08 | 640 | <10 | <10 | <10 |
| 80 | <10 | <10 | <10 | |
| A/sw/NE/02013/08 | 640 | <10 | <10 | <10 |
| 160 | <10 | <10 | <10 | |
| A/sw/NC/03084/08 | 640 | <10 | <10 | <10 |
| 320 | <10 | <10 | <10 | |
| H1γ phylogenetic cluster | ||||
| A/sw/MN/1192/01 | 320 | <10 | 40 | 160 |
| 80 | <10 | <10 | 20 | |
| A/sw/MN/00194/03 | 1280 | <10 | 40 | 160 |
| 320 | <10 | 40 | 320 | |
| A/sw/KS/00246/04 | 1280 | <10 | 40 | 160 |
| 1280 | <10 | 40 | 160 | |
| A/sw/OH/511445/07 | 640 | 20 | 20 | 160 |
| 2560 | 80 | 40 | 640 | |
| A/sw/MO/02060/08 | 1280 | 40 | 20 | 160 |
| 640 | 80 | 40 | 320 | |
| A/sw/OH/02026/08 | 640 | 40 | <10 | 80 |
| 160 | <10 | <10 | 10 | |
| A/sw/NC/02023/08 | 320 | <10 | <10 | 40 |
| 320 | <10 | 20 | 80 | |
| A/sw/IA/02096/08 | 160 | <10 | <10 | <10 |
| 80 | <10 | <10 | <10 | |
| H1δ phylogenetic cluster | ||||
| A/sw/TX/01976/08 | 320 | <10 | <10 | <10 |
| 160 | <10 | <10 | <10 | |
| A/sw/IA/02039/08 | 320 | <10 | <10 | <10 |
| 160 | <10 | <10 | <10 | |
| A/sw/MN/02011/08 | 2560 | <10 | <10 | <10 |
| 640 | <10 | <10 | <10 | |
| Commercial vaccine strains | ||||
| Vaccine A | 1280 | 10 | <10 | 20 |
| 1280 | <10 | <10 | <10 | |
| 640 | <10 | <10 | <10 | |
| 640 | <10 | <10 | <10 | |
| Vaccine B | 320 | <10 | 10 | 40 |
| 640 | <10 | <10 | <10 | |
| 640 | 40 | 10 | 40 | |
| Vaccine C | – | <10 | <10 | <10 |
| – | 10 | <10 | 10 | |
| Vaccine D | 1280 | 20 | 10 | 40 |
| 1280 | 40 | 20 | 80 | |
| 1280 | 20 | 10 | 40 | |
| Vaccine E | – | <10 | <10 | <10 |
| – | <10 | <10 | <10 | |
CA/09, A/California/04/2009; NY/09, A/New York/18/2009; MX/09, A/Mexico/4108/2009.
RFLP analysis
To identify the Eurasian lineage matrix gene found in pandemic H1N1 2009 by RFLP analysis, isolated RNA was amplified by RT–PCR using forward primer 5′‐CATCCCGTCAGGCCCCCTCA and reverse primer 5′‐CATGGCCTCCGCTGCCTGTT in experiment 3. Specifically, 4 μl RNA was subjected to amplification by the One‐Step RT–PCR Kit (Qiagen, Valencia, CA, USA) using a final concentration of each primer at 0.6 μm with 5 units of RNase inhibitor (Invitrogen, Carlsbad, CA, USA) included in a total volume of 25 μl. Cycling conditions were as follows: 1 cycle of 50°C for 30 min followed by 95°C for 15 min; 35 cycles of 94°C for 30 s, 58°C for 40 s, and 72°C for 1 min; 1 cycle of 72°C for 10 min followed by a 4°C hold. After amplification, 17 μl was digested for 2 h with restriction endonuclease AlwNI under manufacturer’s suggested conditions (New England BioLabs, Ipswich, MA, USA). Digested samples were analyzed using a 1·2% agarose gel. Products migrated as an uncleaved 568‐bp fragment for endemic North American SIV matrix genes or as cleaved (40, 185, and 343 bp) fragments for the Eurasian SIV‐lineage matrix gene in the pandemic H1N1 2009.
Results
A/CA/04/2009 induced clinical disease in pigs
All pigs were free of influenza A virus and influenza A virus antibodies prior to the start of the experiment. All pigs were negative for extraneous viral and M. hyopneumoniae in the BALF at 5 dpi and were negative for significant aerobic bacterial growth from BALF. Negative control pigs remained negative for influenza A virus. All pigs infected with CA/09 developed clinical disease and elevated rectal temperatures beginning as early as 24 h post‐infection and extending to 4–5 dpi. Mean rectal temperatures peaked on 2 dpi at 40·4°C with a maximum individual temperature of 41·4°C. Clinical signs were typical of acute respiratory illness with SIV, including lethargy, inappetence, and increased respiration rate and respiratory effort. Nasal virus shedding was detected as early as 1 dpi by RT–PCR and 2 dpi by virus isolation on MDCK cells (Table 1). Virus isolation identified no biologically viable virus outside the respiratory tract whereas viral nucleic acid was identified at low levels in serum and lymph node (Table 2).
Table 1.
Nasal shedding and viral load in the lung by RT–PCR and virus titration*
| Pig | Nasal swab | Bronchoalveolar lavage fluids | ||||
|---|---|---|---|---|---|---|
| 1 dpi | 2 dpi | 3 dpi | 4 dpi | 5 dpi | 5 dpi | |
| 446 | +/0 | +/0·5 | +/3·5 | +/4·5 | +/4·5 | +/3·7 |
| 447 | −/0 | −/0 | +/0·5 | +/1·5 | +/2·3 | +/4·3 |
| 448 | +/0 | −/0 | +/1·7 | +/2·5 | +/3·5 | +/4·3 |
| 449 | −/0 | +/2·5 | +/4·5 | +/4·5 | +/3·5 | +/4·7 |
*RT–PCR result/log10 TCID50 titer/mL.
Table 2.
Detection of virus by RT–PCR and virus isolation in blood and tissues at 5 dpi*
| Pig | Lung | Tonsil | Lymph node | Serum | Liver | Kidney | Spleen | Muscle | Feces |
|---|---|---|---|---|---|---|---|---|---|
| 446 | +/− | +/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− |
| 447 | +/+ | +/+ | +/− | +/− | −/− | −/− | −/− | −/− | −/− |
| 448 | +/+ | +/+ | −/− | −/− | −/− | −/− | −/− | −/− | −/− |
| 449 | +/− | +/− | −/− | +/− | −/− | −/− | −/− | −/− | −/− |
*RT–PCR result/virus isolation result.
All pigs had cranioventral lung consolidation with the percentage of lung involved ranging from 10% to 22%. Microscopic lesions in lungs were typical of influenza virus infection in pigs and were characterized by multifocal to widespread necrotizing bronchitis and bronchiolitis, light peribronchiolar lymphocytic cuffing, and mild multifocal interstitial pneumonia. Airway damage involved all levels of the intrapulmonary respiratory tract and varied from acute epithelial necrosis and sloughing or bronchioles lined by attenuated epithelium and filled with necrotic cellular debris to subacute lesions characterized by disorganized proliferative epithelium lining recovering bronchi and bronchioles. Occasional alveoli contained necrotic cellular debris, but most were clear of debris or inflammatory infiltrates. Tracheal epithelium was focally attenuated in two pigs. No microscopic lesions were observed in other tissues. Lung tissues from all pigs were positive for immunohistochemical staining of influenza A viral antigen. Virus antigen was detected in attached and sloughing necrotic epithelial cells in large to small bronchioles and occasionally in cells or necrotic debris in alveolar lumens (Figure 1B). Only an occasional tracheal epithelial cell contained virus antigen. No viral antigen was detected in liver, kidney, spleen, lymph nodes, tonsil, or skeletal muscle in any of the pigs (data not shown).
Figure 1.
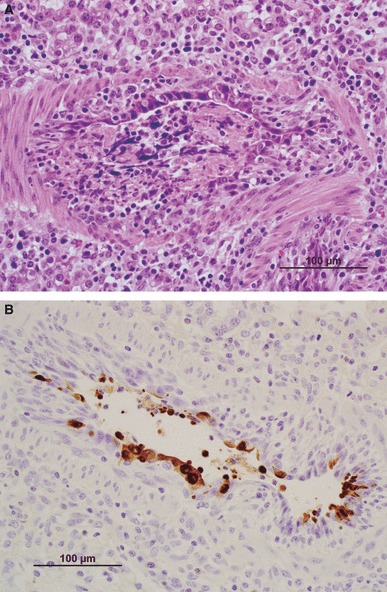
(A) Large bronchiole with severe acute necrosis and sloughing of epithelial cells. Remaining epithelial layer of attenuated cells only partially lines the lumen that is filled with necrotic debris. Light mixed leukocytic infiltrate is evident in the lamina propria and surrounding tissue. (B) Necrotizing bronchiolitis with influenza virus‐infected epithelial cells sloughing into the lumen identified by immunohistochemistry.
Serologic cross‐reactivity
The cross‐reactivity of the panel of swine H1 antisera against the three pandemic H1N1 2009 isolates is summarized in Table 3. A reciprocal HI titer for individual pig antiserum is reported for each virus isolate. Cross‐reactivity ranged from minimal to moderate among the three viral antigens based on reduction between the homologous and heterologous reciprocal HI titers. The greatest cross‐reactivity was demonstrated between A/Mexico/4108/2009 and antisera generated against SIV from the H1γ phylogenetic cluster. Cross‐reactivity with antisera from vaccinated pigs was limited against all three 2009 pandemic viral antigens.
Discrimination of pandemic H1N1 2009 by RFLP analysis of the matrix gene
An alignment of endemic swine influenza matrix gene nucleotide sequences with CA/09 (FJ966085, 972 bp) revealed two unique sites (nt 72 and 415) for restriction endonuclease AlwNI. To assess whether this site could be used to differentiate the novel H1N1 isolate from the present circulating SIV based on the matrix gene, a panel of 24 SIV isolated from cases of respiratory disease from US swine and three pandemic isolates were analyzed using the RFLP test. As shown in Figure 2, the M genes of none of the 24 viruses isolated from US swine were susceptible to digestion with AlwNI (lanes 1–24), whereas the M genes of all three human isolates were cleaved into 40, 185, and 343 bp fragments (lanes 25, 30, 31). RNA extracted directly from BALF from animals infected with CA/09 were also examined by this method (lanes 26–29) and produced identical results as the human isolates.
Figure 2.
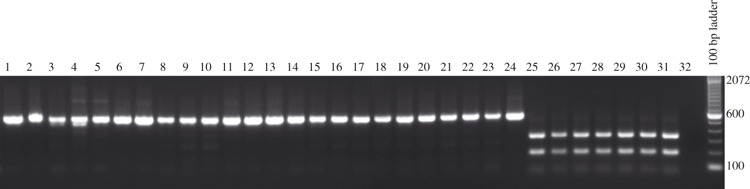
Restriction fragment length polymorphism analysis of different swine influenza isolates. 1: A/Sw/MN/002083/2007 (HuH1N1), 2: A/Sw/MN/00194/2003 (H1N2), 3: A/Sw/IA/00239/2004 (H1N1), 4: A/Sw/MN/37866/1999 (H1N1), 5: A/Sw/NC/36883/2002 (rH1N1), 6: 2007 H1N1 field isolate (H1N1), 7: A/Sw/OH/51445/1/2007 (H1N1), 8: A/Sw/NC/02084/2008 (H1N1), 9: A/Sw/MN/02093/2008 (H1N1), 10: A/Sw/IA/02096/2008 (H1N1), 11: 2005 H1N1 field isolate (H1N1), 12: 2005 H1N1 field isolate (H1N1), 13: A/Sw/IL/3743/2007 (H1N2), 14: A/Sw/IL/00685/2005 (H1N1), 15: A/Sw/TX/01976/2008 (H1N2), 16: A/Sw/MN/02011/2008 (H1N1), 17: A/Sw/NE/02013/2008 (H1N1), 18: A/Sw/NC/02023/2008 (H1N1), 19: A/Sw/IA/02039/2008 (H1N2), 20: A/Sw/OH/02026/2008 (H1N1), 21: A/Sw/MN/02053/2008 (H1N1), 22: A/Sw/MO/02060/2008 (H1N1), 23: A/Sw/NC/00573/2005 (H1N1), 24: A/Sw/MN/1192/2001 (H1N2), 25: A/CA/04/2009 (H1N1)v, 26: A/CA/04/09 (H1N1)v BALF 446, 27: A/CA/04/09 (H1N1)v BALF 447, 28: A/CA/04/09 (H1N1)v BALF 448, 29: A/CA/04/09 (H1N1)v BALF 449, 30: A/NY/18/09 (H1N1)v, 31: A/Mexico/168/09 (H1N1)v, 32: RT–PCR Negative Control.
Discussion
Although the pandemic H1N1 2009 viruses are genetically related to SIV of North American and Eurasian lineages, the constellation of the eight gene segments in the pandemic H1N1 2009 is not known to circulate widely in pigs. Additionally, the HA from the pandemic H1N1 viruses circulating in the human population are phylogenetically distinct from the nearest swine virus sequences. As the virulence of the pandemic H1N1 2009 in pigs was not known, we experimentally infected weaned pigs with one isolate of pandemic H1N1 2009, CA/09. We demonstrated that weaned pigs are susceptible to infection and clinical disease induced by CA/09. Importantly, the infection was characteristic of acute influenza illness in swine. Clinical signs, pathologic changes, and virus replication were restricted to the respiratory tract. The results reported here are consistent with recent reports of experimental infections with other strains of pandemic H1N1 virus, where pigs developed pyrexia, anorexia, and dyspnea within several days following challenge. 16 , 17 Likewise, there have been reports of swine becoming infected in the field with the pandemic H1N1 virus in which the pigs displayed mild respiratory disease (http://www.oie.int/eng/en_index.htm), similar to outbreaks with endemic SIV.
In the study reported here, tissues outside the respiratory tract were found to be negative by virus isolation at 5 dpi. Only respiratory tract samples were positive by both real‐time RT–PCR and virus isolation. The inguinal lymph node from one pig and serum from two pigs were positive for viral RNA, but lymph node and serum samples from all pigs were negative by virus isolation. This demonstrates the sensitivity of the RT–PCR assay in that viral nucleic acid in lymph tissue and blood can be detected by RT–PCR when infectious virus cannot. In contrast, all day 5 post‐infection BALF, nasal swabs, and lung tissue homogenates from two out of four pigs were positive by virus isolation and RT–PCR. Lung samples from two pigs were negative in both replicates of virus isolation but positive by RT–PCR. The negative virus isolation results from lung tissues were likely due to sampling and/or the presence of a severe inflammatory response in the lungs of these two pigs leading to virus inactivation. A future study is planned to evaluate influenza levels in tissues by VI and real‐time RT–PCR at additional time points.
Limited serologic cross‐reactivity with the pandemic H1N1 2009 isolates was demonstrated in HI tests with sera from pigs infected or vaccinated with contemporary H1 SIV, although variation in cross‐reactivity between viral antigens was apparent. The reduction in reactivity was demonstrated by the eightfold or greater decrease in titer when compared to homologous HI titer. An eightfold or greater loss in reactivity is considered a significant reduction that may be predictive of decreased protection from challenge. NY/09 was used as antigen against the swine anti‐SIV panel with results similar to those obtained with CA/09. However, MX/09 demonstrated broader cross‐reactivity with the NADC H1 serum reference panel. This was especially apparent for sera from pigs immunized with viruses from the H1γ HA phylogenetic cluster. The cross‐reactivity with antisera from viruses of this cluster is important as this is the HA clade in which the HA from the 2009 pandemic H1N1 is most closely related. 1 Thus, it is possible to speculate that pre‐existing immunity to certain currently circulating H1 SIV strains may provide some level of protection in pigs against the pandemic virus. However, the differences between the pandemic H1N1 2009 isolates suggest a possibility of biologic variation in host and/or virus properties responsible for the variation in serologic cross‐reactivity. The CA/09 HA gene has 99·1% identity at the amino acid level with the NY/09 and MX/09 viruses, with four to five amino acid changes over the entire HA molecule depending on the reference sequences compared. However, none of these changes occurs at the receptor binding site or at putative antigenic sites, making the mechanism behind the differences in antigenicity unclear. The effect this variation would have on preventing transmission of the pandemic H1N1 2009 from the human population to pig populations with prior immunity to SIV remains unknown.
Serologic cross‐reactivity with antisera from pigs vaccinated with five US commercial vaccines was additionally assessed by HI with the three pandemic H1N1 isolates. Cross‐reactivity was consistently low between the vaccine antisera and all pandemic H1N1 2009 novel viruses tested, although titers were slightly higher with the isolate from Mexico. This suggests that currently available vaccines may provide only limited protection against infection with the novel human H1N1. Future studies will investigate a subset of the vaccines from this in vitro study in a vaccination efficacy challenge model to validate the serum cross‐reactivity.
To discriminate between the endemic North American SIV isolates and the pandemic H1N1 2009, an RT–PCR–RFLP assay was developed and evaluated on a panel of virus isolates as well as clinical samples from the pigs experimentally infected with CA/09. None of the matrix gene amplicons from the panel of endemic SIV were cut by AlwNI. The matrix genes from the three examined pandemic H1N1 2009 isolates as well as the BALF samples from pigs infected with CA/09 were cleaved by AlwNI, demonstrating the usefulness of the diagnostic test to discriminate North American SIV lineage matrix genes from Eurasian SIV lineage matrix genes present in the pandemic H1N1 2009. In addition to monitoring for the emergence of the pandemic H1N1 2009 in the swine population in toto, tests that identify specific gene segments such as the one described here will be useful for monitoring changes in the gene constellation of viruses should reassortment between the pandemic virus and endemic SIV occur.
The pig has been suggested to be a ‘mixing vessel’ for emergence of influenza viruses with pandemic potential due to the presence of both avian and mammalian receptors expressed by respiratory tract epithelial cells. 18 However, both receptors linkage types have been additionally detected in the respiratory tract of humans 19 and in the trachea and intestine of quail. 20 These findings suggest the potential hosts for reassortment events between avian and mammalian influenza A viruses are not as limited as once thought. The pandemic H1N1 2009 demonstrates the potential for viruses with genes from swine lineages to emerge and spread in the human population. Although the novel reassortant H1N1 has not been identified circulating endemically in swine, reassortants between the North American and Eurasian lineage swine viruses have been identified from pigs in China. 2 It is apparent that pigs may be infected at least transiently with wholly avian and/or human viruses, allowing reassortment with swine viruses to acquire avian and/or human virus gene segments. 7 , 21 , 22 , 23 , 24 , 25 , 26 The 2009 pandemic H1N1 underscores the potential risk to human and animal populations of other influenza virus subtypes and genotypes that may evolve with the SIV TRIG backbone. Increased surveillance and monitoring for the pandemic A/H1N1 as well as other SIV in both the swine and human populations are critical to understand the dynamic ecology of influenza A viruses in susceptible host populations.
Acknowledgements
The authors thank David Michael and Hillary Horst for technical assistance and Dr Becky Jepsen, Brian Pottebaum, and Jason Huegel for assistance with animal studies. We thank Dr Crystal Loving for critical review of the manuscript and Dr Marie Gramer (University of Minnesota) for providing SIV isolates. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. Funding was provided by USDA–ARS and DHHS–CDC.
References
- 1. Garten RJ, Davis CT, Russell CA et al. Antigenic and genetic characteristics of swine‐origin 2009 A(H1N1) influenza viruses circulating in humans. Science 2009; 325:197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smith GJD, Vijaykrishna D, Bahl J et al. Origins and evolutionary genomics of the 2009 swine‐origin H1N1 influenza A epidemic. Nature 2009; 459:1122–1125. [DOI] [PubMed] [Google Scholar]
- 3. Trifonov V, Khiabanian H, Rabadan R. Geographic dependence, surveillance, and origins of the 2009 influenza A (H1N1) virus. N Eng J Med 2009; 361:115–119. [DOI] [PubMed] [Google Scholar]
- 4. Myers KP, Olsen CW, Gray GC. Cases of swine influenza in humans: a review of the literature. Clin Infect Dis 2007; 44:1084–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shinde V, Bridges CB, Uyeki TM, Shu B, Balish A, Xu X et al. Triple‐reassortant swine influenza A (H1) in humans in the United States, 2005–2009. N Eng J Med 2009; 360:2616–2625. [DOI] [PubMed] [Google Scholar]
- 6. Vincent AL, Ma W, Lager KM, Janke BH, Richt JA. Swine influenza viruses a North American perspective. Adv Virus Res 2008; 72:127–154. [DOI] [PubMed] [Google Scholar]
- 7. Karasin AI, Carman S, Olsen CW. Identification of human H1N2 and human‐swine reassortant H1N2 and H1N1 influenza A viruses among pigs in Ontario, Canada (2003 to 2005). J Clin Microbiol 2006; 44:1123–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vincent AL, Ma W, Lager KM, Gramer MR, Richt JA, Janke BH. Characterization of a newly emerged genetic cluster of H1N1 and H1N2 swine influenza virus in the United States. Virus Genes 2009; 39:176–185. [DOI] [PubMed] [Google Scholar]
- 9. Vincent AL, Swenson SL, Lager KM, Gauger PC, Loiacono C, Zhang Y. Characterization of an influenza A virus isolated from pigs during an outbreak of respiratory disease in swine and people during a county fair in the United States. Vet Microbiol 2009; 137:51–59. 19203846 [Google Scholar]
- 10. Vincent LL, Janke BH, Paul PS, Halbur PG. A monoclonal‐antibody‐based immunohistochemical method for the detection of swine influenza virus in formalin‐fixed, paraffin‐embedded tissues. J Vet Diagn Invest 1997; 9:191–195. [DOI] [PubMed] [Google Scholar]
- 11. Opriessnig T, Yu S, Gallup JM et al. Effect of vaccination with selective bacterins on conventional pigs infected with type 2 porcine circovirus. Vet Pathol 2003; 40:521–529. [DOI] [PubMed] [Google Scholar]
- 12. Strait EL, Madsen ML, Minion FC et al. Real‐time PCR assays to address genetic diversity among strains of Mycoplasma hyopneumoniae . J Clin Microbiol 2008; 46:2491–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Spackman E, Suarez DL. Type A influenza virus detection and quantitation by real‐time RT‐PCR. Methods Mol Biol Clifton, NJ 2008; 436:19–26. [DOI] [PubMed] [Google Scholar]
- 14. WHO . WHO Manual on Animal Infuenza Diagnosis and Surveillance, 2nd edn, 2002. Available at http://www.wpro.who.int/NR/rdonlyres/EFD2B9A7‐2265‐4AD0‐BC98‐97937B4FA83C/0/manualonanimalaidiagnosisandsurveillance.pdf (Accessed 25 May 2009).
- 15. Vincent AL, Lager KM, Ma W et al. Evaluation of hemagglutinin subtype 1 swine influenza viruses from the United States. Vet Microbiol 2006; 118:212–222. [DOI] [PubMed] [Google Scholar]
- 16. Lange E, Kalthoff D, Blohm U, Teifke JP, Breithaupt A, Maresch C et al. Pathogenesis and transmission of the novel swine‐origin influenza virus A/H1N1 after experimental infection of pigs. J Gen Virol 2009;90(Pt 9):2119–2123. [DOI] [PubMed] [Google Scholar]
- 17. Brookes SM, Irvine RM, Nunez A et al. Influenza A (H1N1) infection in pigs. Vet Rec 2009; 164:760–761. [DOI] [PubMed] [Google Scholar]
- 18. Ito T, Couceiro JN, Kelm S et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol 1998; 72:7367–7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature 2006; 440:435–436. [DOI] [PubMed] [Google Scholar]
- 20. Wan H, Perez DR. Quail carry sialic acid receptors compatible with binding of avian and human influenza viruses. Virology 2006; 346:278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karasin AI, West K, Carman S, Olsen CW. Characterization of avian H3N3 and H1N1 influenza A viruses isolated from pigs in Canada. J Clin Microbiol 2004; 42:4349–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Olsen CW, Karasin A, Erickson G. Characterization of a swine‐like reassortant H1N2 influenza virus isolated from a wild duck in the United States. Virus Res 2003; 93:115–121. [DOI] [PubMed] [Google Scholar]
- 23. Olsen CW, Karasin AI, Carman S et al. Triple reassortant H3N2 influenza A viruses, Canada, 2005. Emerg Infect Dis 2006; 12:1132–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu H, Zhang GH, Hua RH et al. Isolation and genetic analysis of human origin H1N1 and H3N2 influenza viruses from pigs in China. Biochem Biophys Res Commun 2007; 356:91–96. [DOI] [PubMed] [Google Scholar]
- 25. Zhou NN, Senne DA, Landgraf JS et al. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol 1999; 73:8851–8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ma W, Vincent AL, Gramer MR et al. Identification of H2N3 influenza A viruses from swine in the United States. Proc Natl Acad Sci U S A 2007; 104:20949–20954. [DOI] [PMC free article] [PubMed] [Google Scholar]


