Abstract
Whether nucleic acids that circulate in blood have any patho-physiological functions in the host have not been explored. We report here that far from being inert molecules, circulating nucleic acids have significant biological activities of their own that are deleterious to healthy cells of the body. Fragmented DNA and chromatin (DNAfs and Cfs) isolated from blood of cancer patients and healthy volunteers are readily taken up by a variety of cells in culture to be localized in their nuclei within a few minutes. The intra-nuclear DNAfs and Cfs associate themselves with host cell chromosomes to evoke a cellular DNA-damage-repair-response (DDR) followed by their incorporation into the host cell genomes. Whole genome sequencing detected the presence of tens of thousands of human sequence reads in the recipient mouse cells. Genomic incorporation of DNAfs and Cfs leads to dsDNA breaks and activation of apoptotic pathways in the treated cells. When injected intravenously into Balb/C mice, DNAfs and Cfs undergo genomic integration into cells of their vital organs resulting in activation of DDR and apoptotic proteins in the recipient cells. Cfs have significantly greater activity than DNAfs with respect to all parameters examined, while both DNAfs and Cfs isolated from cancer patients are more active than those from normal volunteers. All the above pathological actions of DNAfs and Cfs described above can be abrogated by concurrent treatment with DNase I and/or anti-histone antibody complexed nanoparticles both in vitro and in vivo. Taken together, our results that circulating DNAfs and Cfs are physiological, continuously arising, endogenous DNA damaging agents with implications to ageing and a multitude of human pathologies including initiation of cancer.
Keywords: Ageing, apoptosis, cancer, circulating chromatin, circulating DNA, circulating nucleic acids, circulating nucleosomes, DNA damage, DNA damage response, DNA double-strand breaks, DNA repair
1. Introduction
Apoptosis is a natural biological process that leads to cellular fragmentation and release of nuclear material in the form of mono- and oligo-nucleosomes (van Nieuwenhuijze et al. 2003). It has been estimated that several hundred billion to a trillion cells die in the adult human body daily due to normal physiology to be replaced by a similar number generated through mitosis (Fliedner et al. 2002). The existence of an efficient scavenging system notwithstanding, considerable amount of apoptotic genetic material enters the circulation in normal individuals (Zhong et al. 2007), and in elevated levels in a multitude of acute and chronic human pathologies including cancer (Holdenrieder et al. 2001; Chang et al. 2003; Lam et al. 2003; Lui et al. 2003; Trejo-Becerril et al. 2003; Zeerleder et al. 2003; Gal et al. 2004; Holdenrieder and Stieber 2004; Kremer et al. 2005; Butt et al. 2006; Chiu et al. 2006; Rhodes et al. 2006; Umetani et al. 2006; Pisetsky and Ullal 2010; Tsai et al. 2011; Mittra et al. 2012). The levels of circulating nucleic acids increase with advancing age (Jylhävä et al. 2011; Mittra et al. 2012), and foetal DNA that circulates in maternal plasma has been used for pre-natal diagnosis of genetic abnormalities (Kitzman et al. 2012). However, whether nucleic acids that circulate in blood have any patho-physiological role to play in the host is only beginning to be explored (Mittra et al. 2006; Mittra et al. 2010; Mittra et al. 2012; Rekha et al. 2013). We report here results of the first systematic investigation into the biological properties of fragmented DNA (DNAfs) and chromatin (Cfs) isolated from the blood of cancer patients and healthy individuals. The objective of the present study was to determine whether circulating nucleic acids have any biological functions of their own. We show by a series of experiments conducted in vitro in cultured cells as well as in vivo in mice that DNAfs and Cfs are not inert molecules but have significant patho-physiological activities that are deleterious to healthy cells of the body. They freely enter healthy cells and damage their DNA by integrating into their genomes, thereby acting as a physiological, continuously arising, endogenous DNA damaging agents.
2. Materials and methods
2.1. Blood collection
Informed written consent was obtained from human subjects recruited for the study as approved by the Institutional Review Board of Advanced Centre for Treatment, Research and Education in Cancer, Tata Memorial Centre, Navi Mumbai, India. Six mL of blood was collected from patients suffering from advanced cancers of various organs (stages III–IV). The collected blood was processed either for separation of plasma or for serum for DNAfs or Cfs isolation respectively. Alternatively, of the 6 mL of collected blood, 3 mL was used for separation of plasma for DNAfs isolation, and the remaining 3 mL was used for separation of serum for isolation of Cfs. Blood was also collected from age- and sex-matched healthy volunteers and processed as above. The details of cancer patients and healthy volunteers from whom blood was collected for this study are given in supplementary table 1. Blood was allowed to stand at room temperature for 2 h prior to collecting plasma or serum. Since the quantity of tumour-derived DNA in circulation is highly variable (Leary et al. 2012), pooled plasma/serum (typically from ~5 patients) was used to isolate DNAfs and Cfs in order to maintain inter-experimental consistency.
2.2. Isolation, characterization and quantification of DNAfs and Cfs
DNAfs: Circulating DNAfs were isolated from plasma using NucleoSpin® Plasma XS kit (Macherey-Nagel, Germany), which is specifically designed for this purpose. The DNA isolated was quantified using Quanti-iT™ PicoGreen® dsDNA Assay Kit (Invitrogen, USA. Catalogue No. P7589) and the amount of DNA was expressed as ng/mL. The nature and integrity of DNA was determined by agarose gel electrophoresis. Because of the presence of DNase in circulating blood, although at low concentrations, it is possible that at least a portion of ‘free’ DNA remains within apoptotic bodies that originate during apoptosis of cancer cells (Halicka et al. 2000; Takeshita et al. 2004). However, since the DNA purification protocol includes use of Proteinase K and binding of DNA to silica filter, it is highly unlikely that the purified DNAfs contained apoptotic bodies.
Cfs: Circulating Cfs were isolated from serum by a method developed in our laboratory as described herein. Serum samples were centrifuged at 700,000g for 16 h at 4°C and the pellet obtained was lysed with lysis buffer provided with the Cell Death Detection ELISAplus kit described below (Roche Diagnostics GmbH, Germany). The resulting solution was centrifuged for 16 h at 700,000g, and the pellet obtained was suspended in PBS and passed through an affinity column containing a mixture of biotinylated antihistone antibodies (20 μg each of H1, H2A, H2B, H3, H4 in a volume of 1 mL) bound to Pierce® Streptavidin Plus Ultralink® Resin (Thermo Scientific, USA) (1 mL). The washing step was omitted, and the column was directly eluted with low salt (0.25 M NaCl), and Cfs were recovered by ultra-centrifugation as described above. The final pellet was re-suspended in PBS in a volume corresponding to the initial serum volume from which the pellet had been obtained (1mL). The presence of chromatin in the final suspension was confirmed by a sandwich ELISA assay, which is meant specifically for the detection of nucleosomes/chromatin (Cell Death Detection ELISAplus kit, Roche Diagnostics GmbH, Germany). This ELISA assay was also used for quantification of the amount of Cfs present in the final suspension by measuring the absorbance kinetics at 405 nm. The amount of chromatin present was expressed as Arbitrary Units/mL. Cfs were further characterized by electron microscopy as described by Vollenweider, Sogo and Koller (Vollenweider et al. 1975). Grids were examined in a FEI Tecnai 12 BioTwin transmission electron microscope fitted with a SIS Megaview III CCD camera. To detect the presence of DNA in the isolated Cfs, the latter were suspended in buffer containing 0.5% SDS and 0.5 mg/mL proteinase K, incubated at 50°C for 1 h and analysed by 1% agarose gel electrophoresis (Schnitzler 2001). Western blot analysis of Cfs was performed to confirm the presence of histones using polyclonal antibodies against the respective histone proteins (Santa Cruz Biotechnology INC., USA). The DNA content of Cfs was estimated using Quanti-iT™ PicoGreen® dsDNA Assay Kit (Invitrogen™, USA) and the amount of DNA was expressed as ng/mL.
2.3. Isolation, quantification and fluorescent labelling of RNA
RNA from pooled plasma from cancer patients was isolated using Plasma/Serum Circulating RNA Purification Kit (Norgen Biotek Corp, Canada. Catalogue No. 30000) according to the manufacturer’s instructions. The kit uses a proprietary resin which ensures the isolation of all sizes of circulating and exosomal RNA, including microRNA. RNA was quantified using Nanodrop 1000 spectrophotometer (Thermo Scientific, USA) and labelled using ULS™ microRNA Labeling Kit (With Cy3) (Kreatech Diagnostics, The Netherlands. Catalogue No. EA-037).
2.4. Reconstitution of chromatin
Chromatin was reconstituted with DNA purified from pooled plasma from cancer patients using the In Vitro Chromatin Assembly Kit (Diagenode, Belgium. Catalogue No. ca-vitro-001) as per the manufacturer’s instructions. The standard assembly reaction contained 2 μg of DNAfs and purified core histones at 1:1 histone:DNA ratio, 15 μL ATP, 3.5 μL remodeling spacing factor and nucleosome assembly protein-1 that catalyses the deposition of histones into extended periodic nucleosomes. Efficiency of assembly formation was evaluated by gel electrophoresis. The DNA content of reconstituted Cfs (RCfs) was estimated using Quanti-It PicoGreen kit and amount of DNA was expressed as ng/100 μL.
2.5. Synthesis of pullulan-histone antibody nanoconjugates
Synthesis of pullulan-histone antibody nanoconjugates was performed as described in our recent publication using H4 IgG (Rekha et al. 2013). The process involved three steps, namely, (1) activation of pullulan: typically Pullulan (Sigma Aldrich™, USA) was dissolved in 20 mM borax buffer, mixed with Traut’s reagent (2-Iminothiolane, Sigma Aldrich™, USA) under continuous stirring (final pH 7.0) and dialysed against 0.1 M sodium phosphate (pH 7.4) containing 0.15 M NaCl and 1 mM EDTA; (2) activation of H4 IgG: H4 IgG (200 μg/mL) was mixed with 3-maleimido benzoyl NHS (Sigma Aldrich™, USA) and kept for half an hour at 25°C for the reaction to be completed; (3) conjugation of activated pullulan with activated H4 IgG: activated H4 IgG was conjugated by vigorous stirring with activated pullulan to form monodispersed nanoconjugates.
2.6. Treatment of cells in culture
Cell lines used in our study were: NIH3T3 (mouse fibroblast), B/CMBA.OV (mouse ovary), MM55.K (mouse kidney), 3T3-L1 (mouse adipocyte), HeLa (human cervical cancer). All cell lines were obtained from the American Type Culture Collection, USA. Cells were seeded at a density of 6×104 cells/35 mm culture dishes in 1.5 mL of Dulbecco’s Modified Eagle Medium (DMEM) (Gibco® Life Technologies, USA. Catalogue No. 12800-017) and maintained at 37°C in the humidified atmosphere of 5% CO2 in air. After 16 h (cell number ~100,000 / dish), Cfs suspended in PBS and DNAfs suspended in elution buffer diluted in PBS (total volume=100 μL for both) were added to the cells and treatment was continued for periods as mentioned in the text. No transfection agent(s) was used in any of our experiments. Control cells were treated with 100 μL PBS alone. The medium was changed every 72 h when required. Equal concentrations of DNAfs (5 ng) and Cfs (5 ng equivalent of DNA) from the same patient pool were usually used for treatment of cells, unless mentioned otherwise in the text.
2.7. Development of single-cell clones from DNAfs- and Cfs-treated cells
NIH3T3 cells (10×104) were treated with DNAfs and Cfs (5ng DNA each) and cells were allowed to grow. After 5 passages, several single-cell clones were developed from DNAfs- and Cfs-treated cells by serial dilution. Two randomly selected clones from DNAfs-treated cells (E10 and E12) and Cfs-treated cells (B2 and D5) were used for further experimentation.
2.8. Fluorescent labelling of DNAfs, Cfs and RCfs and detection of their intracellular fate by laser confocal microscopy
DNAfs were non-enzymatically labelled in a 50 μL reaction with Platinum Bright™ 550 Red/Orange Nucleic Acid Labeling Kit (Kreatech Diagnostics, The Netherlands. Catalogue No. GLK-004) as per the manufacturer’s protocol. Cfs were dually labelled; the DNA component was labelled using Platinum Bright™ 550 Red/Orange Nucleic Acid Labeling Kit, while the protein component was labelled with ATTO 488 NHS-ester (ATTO-TEC GmbH, Germany. Catalogue No. AD 488-35) in accordance with the manufacturer’s protocol (www.atto-tec.com). RCfs were similarly dually labelled with Platinum Bright™ and ATTO 488 NHS-ester. NIH3T3 cells grown overnight on coverslips were treated with 100 μL of fluorescently labelled DNAfs, Cfs or RCfs (10 ng DNA each) for varying periods. After treatment, cells were fixed in 4% paraformaldehyde for 20 min, washed in PBS, mounted onto clean glass slides with Vecta-shield and intracellular uptake of labelled DNAfs, Cfs and RCfs and their localization in DAPI-stained nuclei was visualized using Zeiss differential laser scanning confocal microscopy (LSCM) platform. Optical sections were captured at depths that passed through the nuclei. Fifty nuclei were analysed in each case, and the number of nuclei showing positive fluorescent signals as well as the average number of signals per nucleus were recorded.
2.9. Detection of chromosomal association of labelled DNAfs, Cfs and RCfs
NIH3T3 cells were treated with labelled DNAfs, Cfs and RCfs (10 ng DNA each) and metaphase spreads were prepared after 6 h and observed under fluorescent microscope. Fifty metaphases were analysed in each case and the numbers of metaphases showing positive fluorescent signals as well as the average number of signals per metaphase were recorded.
2.10. Detection of human DNA sequences in mouse cells in vitro and in vivo by FISH
Several single-cell clones were established from NIH3T3 cells treated with DNAfs and Cfs, and four randomly selected clones were analysed by FISH for presence of human DNA on metaphase spreads (DNAfs-derived clones: E10, E12; Cfs-derived clones: B2, D5). A mixture of Texas Red-labelled human whole-genomic (1 μL) and biotinylated human pan-centrometric probes (1 μL) (Chrom-Bios GmbH, Germany. Catalogue No. HGDNAOR10 and HPANCBI10) was used. For the detection of pan-centrometric signals, hybridized slides were treated with 100 μL of Avidin-FITC conjugated secondary antibody (1:200 dilution in 4× SSC/0.1 % Tween 20). Image acquisition and analysis were performed using the Spectral Bio-Imaging System (Applied Spectral Imaging, Israel). Fifty metaphases were examined for each clone for presence of human DNA signals. The human specific FISH probes were unreactive to mouse cells.
For in vivo experiments, Balb/C mice (6–8 weeks old, weighing ~20 g) obtained from the Institute Animal House were used for the study. They were housed in the Animal House Facility of the Institute. Mice were injected intravenously with DNAfs (100 ng) and Cfs (100 ng DNA equivalent) and animals were sacrificed on day 7 by cervical dislocation and their vital organs removed, fixed in formalin and processed for FISH. Control animals were injected with 100 μl of saline. The human FISH probes used here were unreactive to mouse DNA. The protocol for in vivo experiments in mice was approved by the Institutional Animal Ethics Committee (IAEC) of Advanced Centre for Treatment, Research and Education in Cancer, Tata Memorial Centre, Navi Mumbai, India.
2.11. Detection of human DNA in mouse cells by whole genome sequencing
Whole genome sequencing of two clones each derived from DNAfs-treated (E10 and E12) and Cfs-treated (B2 and D5) cells was undertaken to detect the presence of human-specific sequences in mouse cells. Intact high quality genomic DNA was isolated to generate whole genome libraries for sequencing on the Illumina GA IIX (Genotypic Technology (P) Ltd, India). For whole genome libraries, 3 μg of genomic DNA was made up to 100 μL with nuclease-free water (Ambion® Life Technologies, USA) and sonicated using a Bioruptor (Diagenode, Belgium) (30 pulses on high at 30 s ON and 30 s OFF) to obtain desired fragment lengths ranging between 150 and 600 bp. Libraries for whole genome sequencing were constructed according to a modification of the TruSeq DNA library protocol outlined in ‘TruSeq DNA Sample preparation guide’ (Part # 15005180; Rev. A; Nov 2010). The libraries were quantified using Nanodrop and validated for quality. The latter was performed by running the library on a High Sensitivity Bioanalyzer Chip (Agilent) to check for quality and size distribution. DNA library fragments were diluted, denatured and hybridized to a lawn of oligonucleotides immobilized on the flow-cell surface. Hybridized DNA template was amplified using immobilized oligonucleotides as primers. Each hybridized template, using the process of isothermal bridge amplification, resulted in the formation of clusters comprised of roughly 1000 clonal copies. Paired end-sequencing was performed on the Illumina GA IIX to generate 54-bases-long reads for Cfs-derived clones and 100-bases-long reads for DNAfs-derived clones respectively with an average of 1.2× coverage genome-wide across samples (supplementary table 2).
Bioinformatics analysis: hg19 and mm9 reference sequences were used as human and mouse references respectively. BWA v 0.6.1.software was used for aligning the reads to the reference genomes. The sequence reads from study accession ERP000354, submitted by the Sanger center to the NCBI Sequence read archive, were used as a mouse control in our experiments. Default parameters (best hit) were used for alignment of sequence reads to a reference genome sequence. The reads were aligned to a reference genome using a short read aligner, and matched to a reference genome, after subtracting the aligned reads (supplementary figure 1).
To identify presence of human Alu elements in the mouse cell clones, the DNA reads were compared to a database of Alu elements specific to the hg19 human genome downloaded from the UCSC genome browser database (Karolchik et al. 2014) employing the UCSC table browser (Karolchik et al. 2004) using offline BLAST (Altschul et al. 1990). An e-value of 0.05 was used as a cut-off for all alignments generated using threshold of 80% and above sequence identity without gaps.
PCR amplification of human Alu elements: Genomic DNA was PCR amplified using primers for the Alu elements (HSU14570 and HSAL002744): 5′GAATGGCGTGAACC CGGG3′ and 5′TTTTGAGACGGAGTCTCGCTC3′ for HSU14570; 5′CACCTTGTCCTCCCAAAGTG3′ and 5′ TGCTCAGAAATCATTTCATG3′ for HSAL002744. The PCR products were purified using column (NucleoSpin Gel and PCR Clean-up kit), sanger sequencing performed and sequencing traces were aligned using NCBI blast as well as Mutation Surveyor V 4.0.9.
2.12. Detection of activated DDR and apoptosis-related proteins by immuno-fluorescence in vitro and in vivo
In vitro studies: NIH3T3 cells were seeded on cover slips at a density of 6 ×104 cells and allowed to grow overnight (16 h) and subjected to treatment with DNAfs, Cfs and RCfs (5 ng DNA each) for 6 h in duplicate experiments. Control cells were treated with 100 μL PBS alone. Cells were fixed with 4% paraformaldehyde for 20 min at room temperature, permeabilized in 0.2% Triton for 30 min, blocked in 3% BSA (Bovine Serum Albumin) for 1 h and immuno-stained overnight with various specific antibodies against DDR proteins. Cells were immediately mounted on slides with Vecta-shield and images were acquired through Spectral Bio-Imaging System (Applied Spectral Imaging, Israel). In general, 50 DAPI-stained nuclei were examined in duplicate in each experiment; the number of nuclei showing at least two foci were considered as positive. The antibodies used for detection of the various activated DDR proteins and their sources are given in supplementary table 3. Anti-rabbit/anti-mouse secondary antibodies used were labelled with FITC (Abcam®, UK. Catalogue No. ab6717/ab6785), Texas Red (Abcam® UK. Catalogue No. Ab6883) or Rhodamine (Merck Millipore, USA. Catalogue No. AP160R) as appropriate. Onset of apoptosis was determined by assessing the status of mitochondrial membrane potential by labelling with BD™ MitoScreen (JC-1) kit using 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazol-carbocyanine iodide (BD Biosciences, USA. Catalogue No. 551302) and cell-associated fluorescence was detected after addition of JC-1 dye. The number of cells showing green fluorescence was counted. Activation of Cytochrome-C (Merck-Millipore, Germany. Catalogue No. 05-479) and Caspase-3 were quantified by immuno-fluorescence using the respective antibodies and the number of cells showing positive fluorescence was recorded. All experiments were done in duplicate; 50 cells were analysed in each case and nuclei showing at least two foci were considered as positive. The average number of positive cells was recorded.
In vivo studies: Balb/C mice (6–8 weeks old, weighing ~20 g) were injected intravenously with DNAfs (100 ng) and Cfs (containing equivalent of 100 ng DNA) in 100 μL of buffer. Control mice were injected with 100 μL of saline. Two mice were used for treatment in each case. After 24 h, animals were anesthetized with CO2 and blood was collected from orbital plexus. Animals were then sacrificed by cervical dislocation and the following organs were removed and snap frozen in liquid nitrogen: lung, liver, brain, heart, kidney, spleen, pulmonary artery, skin and muscle and processed for cryo-sectioning. The collected blood was processed for isolation of peripheral blood mononuclear cells (PBMCs) by Ficoll gradient centrifugation. Cryo-sections of tissues and PBMCs smeared on slides were processed for immunofluorescence staining against γ-H2AX and active Caspase-3 markers. At least 1000 DAPI-stained nuclei per animal were examined from 10 randomly chosen areas of various tissues, and in case of PBMCs, 100 nuclei per animal were examined. In both cases, the number of nuclei showing positive foci (γ-H2AX) and number of nuclei showing positive fluorescence (Caspase-3) were recorded.
2.13. Experiments using specific inhibitors (DNase I and CNPs)
Several experiments were performed both in vitro and in vivo to examine whether cellular/nuclear entry of DNAfs and Cfs and the multiple biological activities that they affect could be inhibited by specific inhibitors, namely, DNase I and CNPs. In case of treatment with DNAfs, inhibition experiments were done using DNase I while in case of Cfs, they were done using both DNase I and CNPs. For in vitro experiments, cells grown at a density of 10×104 were treated with DNAfs and Cfs (5 ng DNA each) for 6 h in the presence or absence of DNase I (0.05 U/mL) and/or CNPs (5 μg H4 IgG/mL) and the treated cells were analysed for the various specified parameters. For in vivo experiments, mice were given a single intravenous injection of DNAfs and Cfs (100 ng DNA each) through tail vein with and without additional administration of DNase I (15 mg/kg i.p.) and/or CNPs (50 μg H4 IgG/mouse i.p.). DNase I and CNPs treatment was started 4 h prior to DNAfs and Cfs administration and continued for 24 h or 7 days as specified. DNase I was injected at 12 hourly intervals while CNPs were administered every 24 h. Animals were sacrificed and their vital organs, namely, heart, lung, liver and brain were removed for analysis of the specified parameters.
2.14. Statistical analysis
Statistical analysis was performed using GraphPad Prism 5 (GraphPad Software, Inc., USA. Version 5.0). Data were compared using Chi-square analysis and Student’s t-test as appropriate. The tests used have been indicated in appropriate places in legends to figures.
3. Results
3.1. Characterization of DNAfs and Cfs
When run on 1% agarose gels, DNAfs isolated from plasma generated a smear pattern indicating their extensive size heterogeneity (supplementary figure 2A). The presence of chromatin in the isolated Cfs fractions was confirmed by the Cell Death Detection ELISAplus designed exclusively for the detection of nucleosomes/chromatin. When isolated Cfs were characterized by electron microscopy (EM), they were found to retain the typical beads-on-a-string appearance of nucleosomes (supplementary figure 2B upper left-hand panel). Under EM, Cfs were heterogeneous in size ranging from 10 nm to several hundred nm. Size heterogeneity of DNA fragments isolated from Cfs was further evident on agarose gel electrophoresis wherein a smear pattern, similar to that seen with DNAfs, was obtained. Two enriched zones of DNA fragments were present corresponding to sizes of ~2.5 kb and <250 base pairs (supplementary figure 2B upper right-hand panel). While the latter may represent DNA derived from mono- and dinucleosomes, the former is likely derived from significantly larger number of nucleosome units (>15). Western blot analysis of Cfs using polyclonal antibodies against histone confirmed the presence of histone proteins H2A, H2B, H3 and H4 (supplementary figure 2B lower panel).
3.2. Cellular entry, nuclear localization and chromosomal association of DNAfs and Cfs
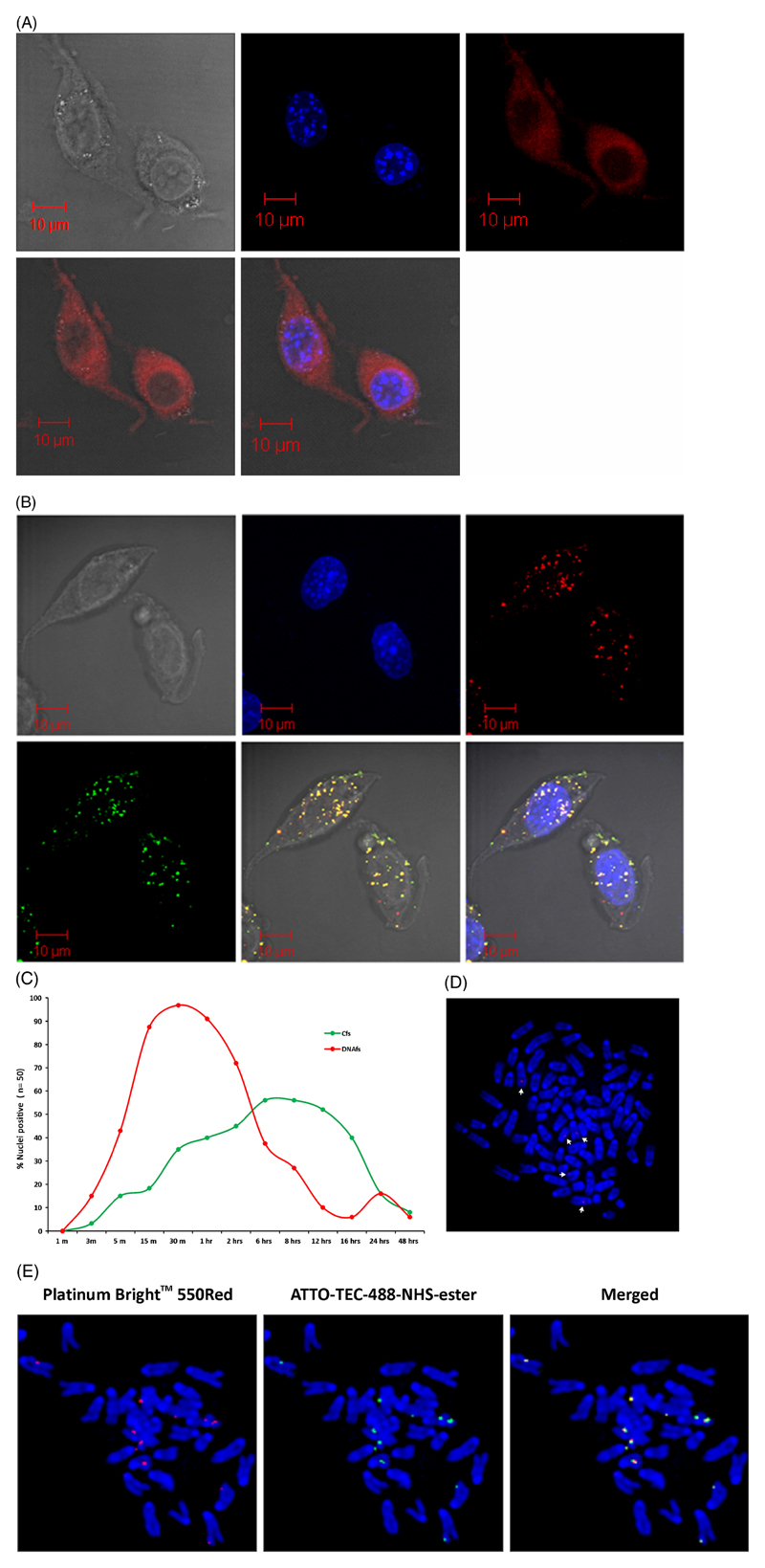
We treated NIH3T3 cells with fluorescently labelled DNAfs and Cfs and examined the recipient cells by LSCM at various time points. Both DNAfs and Cfs were readily taken up by the recipient cells within minutes, and numerous fluorescent signals of varying sizes could be detected in the cytoplasm and nuclei of the treated cells (figure 1A and B). Intracellular DNAfs appeared as fine particles, while Cfs were larger in size and were more discrete. Temporal studies to monitor the rate of nuclear uptake of fluorescently labelled nucleic acids revealed that the uptake of DNAfs was rapid and that fluorescent signals were detectable in nuclei of treated cells as early as at 3 min (figure 1C). DNAfs uptake reached a maximum by 30 min by which time almost 100% of the nuclei (49/50) contained fluorescent signals. Flow cytometric analysis revealed that DNAfs internalized at 30 min corresponded to 3.88% of the genomic DNA. The proportion of cells showing intracellular signals declined steadily thereafter to reach a near baseline level by 16 h, indicating that nuclear DNAfs were being actively degraded. The uptake of Cfs was more gradual reaching a peak accumulation at 6–8 h when 29/50 nuclei (58%) showed fluorescent signals. A statistical comparison of peak nuclear uptake of DNAfs (30 min) and Cfs (6 h) showed a significant greater uptake of DNAfs than Cfs (p<0.0001). These experiments were repeated several times.
Figure 1.
Cellular entry, nuclear uptake and chromosomal association of fluorescently labelled DNAfs and Cfs. NIH3T3 cells (10×104) were treated with DNAfs labelled with ULS (red) and Cfs dual-labelled with ULS (red) and ATTO-TEC (green) (10 ng DNA in all experiments). (A) Intracellular fate of DNAfs at 30 min as analysed by LSCM. Numerous fine fluorescent particles are seen in the cytoplasm and in the nucleus. DIC, DAPI and ULS pictures are represented in different panels. (B) Intracellular fate of Cfs at 6 h as analysed by LSCM. Presence of dual-labelled Cfs in the cytoplasm and nuclei are clearly seen. The red and green signals appear yellow in colour when the images are overlapped. (C) Kinetics of nuclear uptake of fluorescently labelled DNAfs and Cfs analysed by LSCM. Fifty nuclei were analysed at each time-point and the percentage of positive nuclei was recorded. Nuclei containing at least two fluorescent spots were considered as positive. (D and E) Association of fluorescently labelled DNAfs (D) and Cfs (E) with chromosomes of treated cells. NIH3T3 cells were treated with labelled DNAfs and Cfs and metaphase spreads were prepared 6 h after treatment and analysed by fluorescence microscopy. Note that the labelled DNA particles are considerably smaller in size than Cfs particles.
To further examine the intra-nuclear fate of DNAfs and Cfs, we prepared metaphase spreads from cells previously treated for 6 h with fluorescently labelled DNAfs and Cfs and examined them by fluorescent microscopy. Both DNAfs and Cfs had the ability to associate themselves with the chromosomes of treated cells (figure 1D and E). The association of fluorescent DNAfs with condensed chromosomes was infrequent and the signals appeared considerably smaller in size compared to those of Cfs. A statistical comparison of the extent of chromosomal association of DNAfs (9/50 metaphases) and Cfs (20/50 metaphases) showed a significant greater degree of chromosomal association in favour of Cfs (p=0.0275). Thus, it is noteworthy that although DNAfs were taken up more avidly by nuclei of treated cells compared to Cfs (98% vs 58%; p<0.0001), the latter associated themselves with chromosomes with greater efficiency that did DNAfs (40% vs 18%; p=0.0275). These experiments were repeated at least once.
Negative control experiments: We conducted several negative control experiments to validate the above findings and found the following: (1) RNA purified from plasma of cancer patients labelled with Cy3 was taken up sparingly by the cells. Although we occasionally saw a few low-intensity fluorescent signals in the cytoplasm, we never detected any fluorescent RNA signals in the nuclei of the treated cells (supplementary figure 3A); (2) We next examined whether fluorescently labelled proteins (α-lactalbumin, MW~14kDa) had the ability to enter into cells, especially into their nuclei. We found no fluorescent signals in any of the cells examined (supplementary figure 3B); (3) We tested for the ability of the fluorescent labels themselves to enter into cells and their nuclei. We observed that while both ATTO label and Platinum Bright Dye had the ability to enter into the cytoplasm of the recipient cells, neither entered the nuclei (supplementary figure 3C and D).
In order to examine whether nuclear uptake of DNAfs and Cfs are dependent on the metabolic activity of the cell, we investigated the intracellular fate of DNAfs and Cfs when added to cells in the presence of a metabolic poison (Actinomycin D, 0.0005 μg/mL) and at low temperature (31°C). These conditions have been widely reported to inhibit metabolic activity of cultured cells (Gawel-Thompson and Greene 1989; Fulda et al. 2000; Sakurai et al. 2005; Marchant et al. 2008; Pliss et al. 2013). There was a significant reduction in nuclear uptake of DNAfs and Cfs under both these conditions (supplementary figure 3E, F, G and H).
Experiments using specific inhibitors (DNase I and CNPs): We next investigated if nuclear uptake of DNAfs and Cfs could be prevented by treatment with DNase I (0.05 U) and/or CNPs (5 μg of H4 IgG). Bovine pancreatic DNase I was obtained from Sigma Aldrich (Catalogue No. DN25-1G). DNase I significantly inhibited nuclear uptake of DNAfs (supplementary figure 3I); and with respect to Cfs, this was equally inhibited by both DNase I and CNPs (supplementary figure 3J).
3.3. Experiments with reconstituted Cfs
When DNAfs purified from plasma from cancer patients were reconstituted in vitro into RCfs, fluorescently dual-labelled, and applied to cultured cells, RCfs exhibited cellular and nuclear uptake properties that were similar to those seen with serum-derived Cfs with 26/50 nuclei (52%) showing presence of fluorescent signals compared to 29/50 (58%) seen in case of native Cfs (supplementary figure 4A). Notably, the kinetics of cellular uptake of RCfs, unlike that of DNAfs from which they had been prepared which peaks at 30 min, was retarded to reach a maximum at 6 h similar to that seen with native Cfs (supplementary figure 4B). Further, the association of RCfs with mitotic condensed chromosomes could be clearly seen in 10/50 metaphases after 6 h as compared to 20/50 seen in case with native Cfs (supplementary figure 4C). These results confirmed that reconstitution of DNAfs into RCfs in vitro imparted properties to the latter that were similar to those of serum-derived Cfs. This experiment was repeated on at least 3 occasions.
3.4. Genomic integration of DNAfs and Cfs
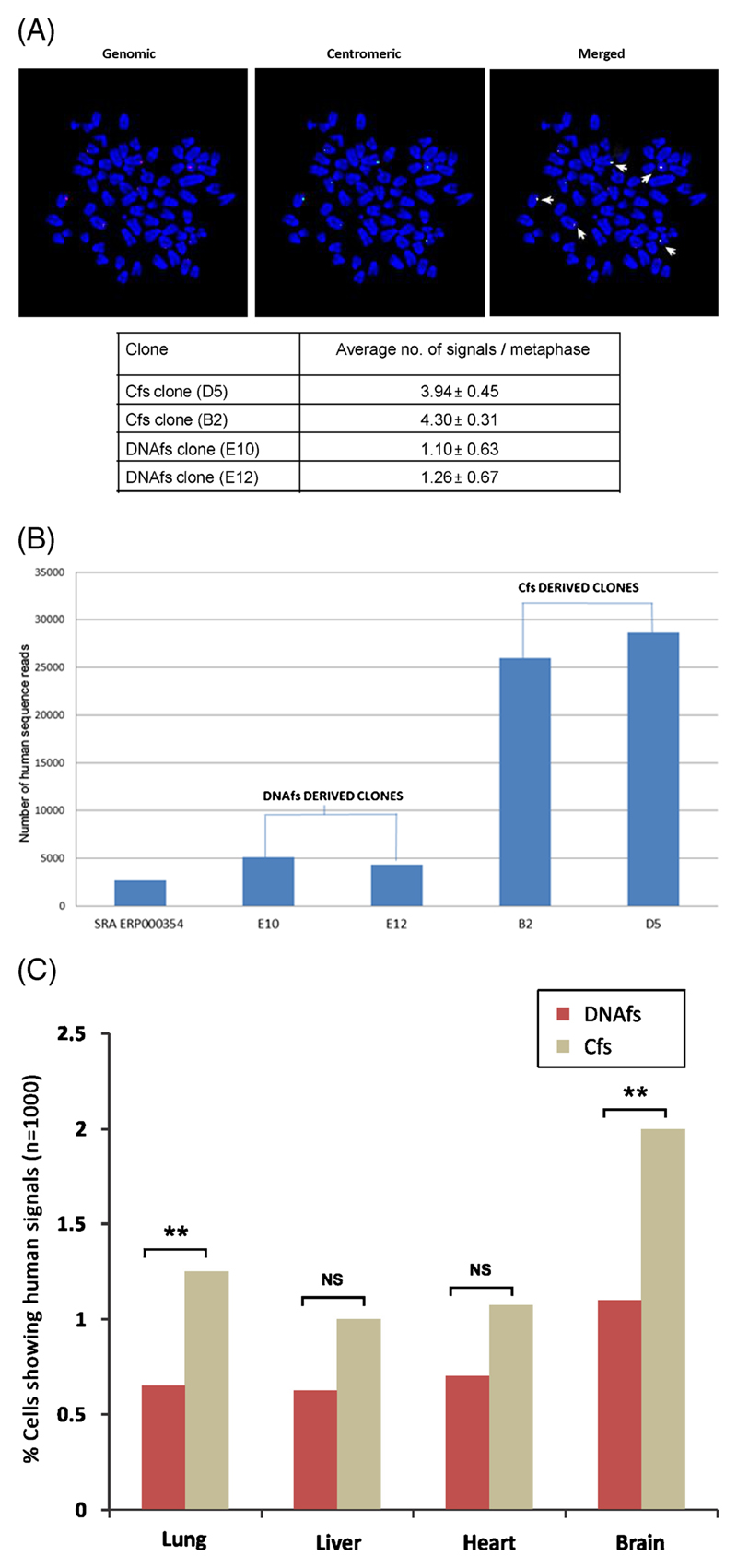
In vitro experiments (using FISH): We analysed single-cell clones derived from cells treated with DNAfs (E10 and E12) and Cfs (B2 and D5) by FISH using human whole genomic and human pan-centromeric probes. Control experiments confirmed that these probes did not cross-hybridize with mouse DNA. In all four clones examined we could clearly detect positive signals indicating the presence of human DNA in these mouse cell clones (figure 2A upper panel). However, many more (~4-fold) human signals were detectable in Cfs-derived clones B2 and D5 than were detectable in DNA-derived clones E10 and E12; these differences were highly statistically significant (p<0.01 to p<0.0001) (figure 2A lower panel). Interestingly, we frequently found human genomic and pan-centromeric signals to co-localize on chromosomal arms indicating common sites of integration (discussed later). These experiments were repeated at least on two occasions.
Figure 2.
Integration of DNAfs and Cfs into host cell genomes. (A) Representative FISH images showing presence of human genomic (red) and pan-centromeric (green) signals in mouse cell clone (B2) derived from NIH3T3 cells treated with Cfs. Upon merging the images, red and green signals frequently co-localize giving yellow fluorescence (marked by arrows). Summary results of FISH analysis depicted in a tabular form shows significantly higher frequency of human-specific signals (genomic + centromeric) in Cfs compared to DNAfs-derived clones. Fifty metaphases were counted in each case and the mean number of signals per metaphase was recorded and analysed by Student’s t-test. D5 vs E10 (p=0.0004); D5 vs E12 (p=0.0013); B2 vs E10 (p=0.0001); B2 vs E12 (p=0.0001). (B) Whole genome sequencing to detect presence of human sequence reads in DNAfs-derived and Cfs-derived clones. The histogram shows number of reads that are strictly human in nature in single-cell clones generated from DNAfs (E10, E12) and Cfs (B2, D5) treated NIH3T3 cells as well as in mouse reference genome (SRA ERP000354). The 2720 human sequence reads detected in the mouse reference genome are either an artifact of inefficiency of our alignment algorithm or an artifact of sequencing in the mouse reference genome. Control vs E10 (Chi-sq. 727.30; p<10–10); Control vs E12 (Chi-sq. 468.19; p<10–10); Control vs B2 (Chi-sq. 7502.97; p<10–10); Control vs D5 (Chi-sq. 4654.724; p<10–10); E10 vs B2 (Chi-sq. 4124.458; p<10–10); E10 vs D5 (Chi-sq. 1896.15; p<10–10); E12 vs B2 (Chi-sq. 4542.31; p<10–10); E12 vs D5 (Chi-sq. 2270.34; p<10–10). (C) In vivo FISH detection of human DNA in nuclei of vital organs of mice following intravenous injection of DNAfs and Cfs. Mice were intravenously injected with DNAfs and Cfs (100 ng DNA each) through tail vein and sacrificed on day 7. Control animals were injected with 100 μl of saline. Vital organs were removed and processed for FISH. The experiments were done in duplicate i.e., with two animals in each group. Two thousand cells were counted for each tissue per animal and the percentage of nuclei showing human-specific signals (genomic and/or centromeric) was calculated and analysed by Chi-square test. **p<0.01. The control animals did not show any human signals.
In vitro experiments (using next-generation sequencing): To further confirm our FISH findings, we undertook whole genome sequencing of the above DNAfs- and Cfs-derived clones. In the DNAfs-derived clones E10 and E12, we obtained 100-bases-long paired-end reads on Illumina GA II X with 42 million and 40 million total paired-end reads in the two clones, respectively (supplementary table 2). In Cfs-derived clones B2 and D5 we obtained 54-bases-long paired-end reads with 39 million and 58 million total paired-end reads in the two clones, respectively (supplementary table 2). Of these, 145364 and 144886 reads from DNAfs-derived clones aligned to the human genome using BWA default parameters (best hit), while significantly higher numbers, viz. 598644 and 889932 from Cfs-derived clones aligned to the human genome with perfect match (supplementary figure 5). Next, to eliminate the fraction of reads conserved between mouse and human among these aligned reads, we performed computational subtraction by back-aligning the presumptive human reads to mouse reference sequences using identical criterion of BWA default parameters. The set of reads that remained unmapped after the subtractive phases were considered to be purely human in origin. The number of strictly human reads contained in the DNAfs-derived E10 and E12 clones were several fold lower than those contained in Cfs-derived B2 and D5 clones. In clones E10 and E12, 5106 and 4354 reads respectively were strictly human in nature, compared to 25979 and 28694 reads in clones B2 and D5 respectively (figure 2B; supplementary figure 5). This indicated a significantly higher (5- to 6-fold) efficiency of genomic integration of human DNA sequences in Cfs-derived compared to DNAfs-derived clones. These findings confirmed our results with FISH performed on the same clones which had demonstrated a ~4-fold higher human signals in the Cfs-derived compared to the DNAfs-derived clones (figure 2A lower panel). It needs to be mentioned that in the mouse reference genome used as controls downloaded from NCBI, perfect match to the human reference genome was found to be of the order of 7088 reads, of which 2720 reads were found to be of human in nature (supplementary figure 5). These 2720 sequence reads in mouse reference genome are either an artifact of inefficiency of our alignment algorithm, or an artifact of sequencing in the mouse reference genome.
We were curious to find out whether human repetitive Alu elements could be detected in the mouse cell clones. After adjusting for Alu reads found in the control mouse reference genome, we identified 47 unique Alu elements that belonged to 8 different Alu families in the Cfs-derived B2 clone and 35 unique Alu elements that belonged to 8 different Alu families in the Cfs-derived D5 clone. In the DNAfs-derived clones, we found 19 unique Alu elements representing 5 Alu families in the E10 clone, and 23 unique Alu elements representing 5 Alu families in the E12 clone (supplementary table 4; supplementary figure 6). We validated the predicted Alu elements by PCR in 9 randomly chosen reads found in the Cfs- and DNAfs-derived clones with primers designed ~100 bases apart. Seven out of 9 PCR reactions amplified a ~100 bp fragment, while in human genomic DNA extracted from DOK cells as template (positive control), all reactions amplified a ~100 bp fragment. None of the PCR reactions amplified human Alu from mouse genomic DNA extracted from NIH3T3 cells (negative control). Four randomly selected PCR products amplified from Cfs- and DNAfs-derived clones were confirmed by Sanger sequencing (supplementary figure 7A, B and C). This data further confirm the presence of human DNA in the mouse cell clones.
In vivo experiments (using FISH): Since DNAfs and Cfs could access genomic DNA of cells in culture for integration, the possibility existed that such a phenomenon occurred naturally within the body involving circulating DNAfs and Cfs. To examine this, mice were injected intravenously with DNAfs and Cfs (100 ng DNA) and were sacrificed 7 days later and their vital organs were removed and processed for FISH. We clearly detected human-specific FISH signals, both whole-genomic and pan-centromeric, in heart, lung, liver and brain, of mice (supplementary figure 8B). Animals injected with vehicle alone did not show any FISH signals (supplementary figure 8A). We once again found that Cfs integrated with greater efficiency than did DNAfs and this difference was statistically significant with respect to lung and brain (p<0.01) (figure 2C). Interestingly, brain showed the maximum number of human DNA signals in comparison to other vital organs. Since brain cells seldom divide, these data suggested that integration of DNAfs and Cfs can occur in interphase (G0) cells. They also suggest that DNAfs and Cfs can cross the blood–brain barrier and access cells within the brain for their integration. The incoming human DNA apparently remains permanently integrated into the mouse genome since human FISH signals were detectable in vital organs even when animals were sacrificed 3 months after Cfs injection. These experiments were repeated once.
In vivo experiments using specific inhibitors (DNase I and CNPs): We administered DNAfs and Cfs (100 ng DNA each) intravenously into mice and investigated whether the simultaneous administration of DNase I and/or CNPs could inhibit their genomic integration into cells of vital organs. The animals were injected intravenously with DNAfs and Cfs as described above and were treated i.p. daily with DNase I and/or CNPs. Animals were sacrificed on day 7 and their vital organs, namely, heart, lung, liver and brain, were analysed by FISH for the presence of human DNA signals. We clearly found that genomic integration of DNAfs was significantly inhibited by treatment with DNase I in all organs except the heart, while that of Cfs integration was significantly inhibited in all organs by treatment with CNPs and in the liver by DNase I (supplementary figure 8C and D).
3.5. Intracellular DNAfs and Cfs activate DDR and apoptotic pathways
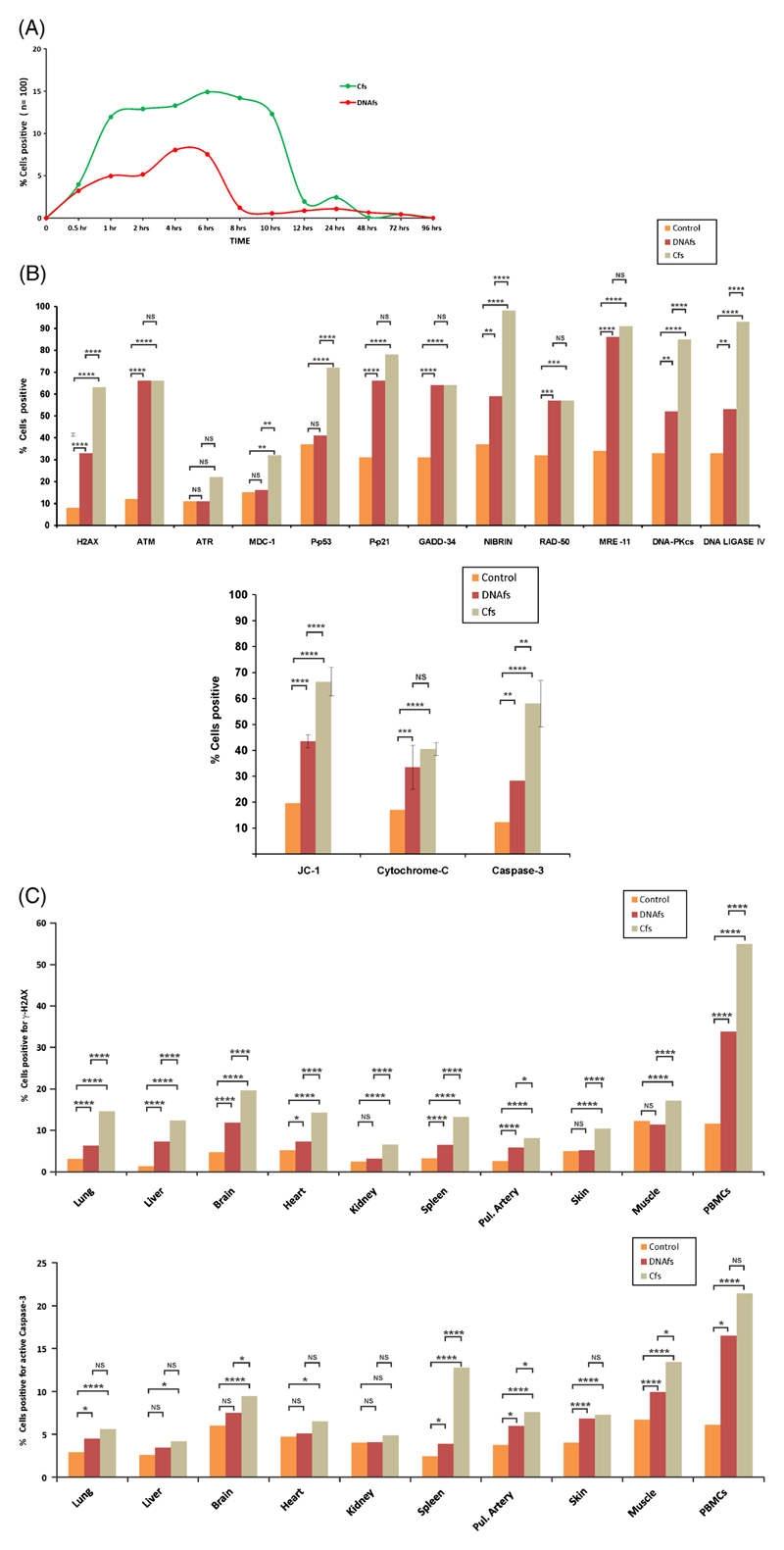
In vitro experiments: To investigate if genomic integration of DNAfs and Cfs led to damage to DNA of the host cells, we undertook indirect immuno-fluorescence analysis of H2AX activation, an indicator of dsDNA breaks, at various time points following treatment of cells with DNAfs and Cfs. We observed that both DNAfs and Cfs activated H2AX (figure 3A). Kinetic studies revealed that γH2AX-positive cells increased gradually following treatment, and phosphorylation reached a peak at around 6 h in both cases (figure 3A). This was followed by a decline in number of γH2AX-positive cells; and in case of Cfs, the decline was more gradual with relatively high levels of phosphorylation persisting until about 12 h as opposed to 8 h for DNAfs. It should be noted that the degree of H2AX phosphorylation was greater in response to Cfs than to DNAfs at all time points. When RCfs were reconstituted from DNAfs and used in similar experiments, it was seen that RCfs were also capable of activating H2AX with a kinetics that was similar to that of native Cfs (supplementary figure 9; figure 3A). The above experiments were repeated once.
Figure 3.
Activation of DNA damage and apoptotic pathways in response to DNAfs and Cfs derived from cancer patients. (A) Kinetics of H2AX phosphorylation in nucleic of NIH3T3 cells treated with DNAfs and Cfs. DNAfs and Cfs were isolated from pooled samples from cancer patients. NIH3T3 cells (10×104) were treated with DNAfs and Cfs (5 ng DNA each) and cells were processed for immuno-fluorescence to detect H2AX activation at various time points. Fifty nuclei in duplicate were analysed at each time-point and the percentage of nuclei showing positive signals was calculated. Nuclei with two or more fluorescent foci were considered as positive. (B) Analysis of various proteins involved in the DDR (upper panel) and apoptotic (lower panel) pathways induced by DNAfs and Cfs. NIH3T3 cells (10×104) were treated as described above and cells were processed for immuno-fluorescence at 6 h following treatment for detection of DDR proteins. Nuclei with two or more fluorescent foci were considered as positive. Experiments were done in duplicate; 50 cells were counted in each case and the percentage of positive cells was calculated and analysed by Chi-square test. For analysis of activation of apoptotic pathways, cells were treated as described above for 24 h and processed for detection of JC-1, Cytochrome-C and Caspase-3 by immuno-fluorescence. For JC-1, cell-associated fluorescence was detected after addition of JC-1 dye and the percentage of cells showing green fluorescence was calculated. For Cytochrome-C and Caspase-3, fluorescence was detected following antibody treatment and the percentage of positive cells was calculated. Experiments were done in duplicate, and 50 cells were examined in each case and analysed by Chi-square test. **p<0.01, ***p<0.001, ****p<0.0001. (C) In vivo activation of H2AX (upper panel) and active Caspase-3 (lower panel) in various organs and in PBMCs following intravenous injection of DNAfs and Cfs (100ng DNA each). Animals were sacrificed after 24 h, and various organs and PBMCs were processed for immuno-fluorescence. Control animals were injected with 100 μl of saline. The experiments were done in duplicate i.e., with two animals in each group. At least 1000 DAPI-stained nuclei per animal were examined from 10 randomly chosen areas of various tissues, and in case of PBMCs, 100 nuclei per animal were examined. In both cases, the number of nuclei showing positive foci (γ-H2AX) and number of cells showing positive fluorescence (Caspase-3) were recorded. The percentage of nuclei showing fluorescent foci (γ-H2AX) and percentage of cells showing positive nuclear fluorescence (active Caspase-3) were calculated and analysed by Chi-square test. *p<0.05, ****p<0.0001.
We next undertook a detailed analysis of activation of proteins of repair pathways to DNA damage following treatment of cells for 6 h with DNAfs and Cfs. The DDR proteins examined, in addition to γ-H2AX, included ATM, ATR, MDC1, p-p53, p-p21, GADD34, Nibrin, Rad50, Mre11, as well as those involved in DNA repair by non-homologous-end-joining (NHEJ), namely, DNA-PKcs and DNA ligase IV (figure 3B upper panel). It is seen from figure 3B that while Cfs significantly activated all DDR proteins except ATR, DNAfs did not activate ATR, MDC-1 and P-p53. In general, Cfs were more active in inducing DDR than were DNAfs.
DDR induction is not restricted to NIH3T3 cells: We tested the effect of DNAfs and Cfs treatment in three other cell lines of mouse origin, and in a transformed human cell line (HeLa) (supplementary figure 10). We found that, as in case of NIH3T3 cells, the other cell lines also responded to Cfs by activating H2AX (range p<0.01 to p<0.0001). DNAfs also activated H2AX in these cells; however, they were significant only with respect to B/CMBA.Ov (mouse ovarian epithelial) and 3T3-L1 (mouse adipocytes) cells (p<0.01 for both), but not with respect to HeLa (human cervical cancer) and MM55.K (mouse kidney epithelial) cells.
We next investigated whether DNAfs and Cfs activated the apoptotic pathway (figure 3B lower panel). The following apoptotic markers were examined (at 24 h): (1) JC-1 (5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazol-carbocyanine iodide), an indicator of status of mitochondrial membrane potential, (2) Cytochrome-c and (3) active Caspase-3. The latter two are involved in mitochondria-mediated apoptosis. DNAfs and Cfs significantly increased the induction of all the three markers of apoptosis when compared to untreated control cells (range p<0.01 to p<0.0001), and with respect to JC-1 and Caspase-3, Cfs was significantly more active than DNAfs (p<0.0001 and p<0.01 respectively). These data indicate that DNAfs and Cfs, particularly the latter, perturb the balance of nuclear-mitochondrial cross-talk and, thereby, activate the induction of apoptosis.
In vitro experiments using specific inhibitors (DNase I and CNPs): We clearly observed that both H2AX and Caspase-3 activation by DNAfs was significantly inhibited by treatment with DNase I (supplementary figure 11A and C), while that by Cfs was significantly inhibited by treatment with both DNase I and CNPs (supplementary figure 11B and D).
In vivo experiments: Intravenous injection of DNAfs and Cfs (100 ng DNA each) showed γ-H2AX signals in all tissues examined, as well as in peripheral blood mononuclear cells (PBMCs) at 24 h (supplementary figure 12A and B; figure 3C upper panel). Cfs treatment resulted in a significantly greater number of cells showing evidence of DNA double-strand breaks compared to those in control animals in all organs and in PBMCs (p<0.0001). H2AX activation was most pronounced in PBMCs with over 50% of cells showing evidence of dsDNA breaks. DNAfs was also able to activate H2AX in most organs (p<0.05 or p<0.0001) except kidney, skin and muscle. However, in every instance, Cfs was significantly the more potent inducer of γ-H2AX than were DNAfs (p<0.05 or p<0.0001). The DNA repair process is apparently completed within days since γ-H2AX levels reached baseline values when animals were sacrificed and their vital organs analysed on day 7 after i.v., injection of DNAfs and Cfs. The activation of active Caspase-3 in various organs and PBMCs are given in figure 3C (lower panel). Once again, Cfs significantly increased Caspase-3 in all organs (except kidney), as well as in PBMCs (p<0.05 or p<0.0001). DNAfs, too, activated Caspase-3, but only with respect to lung, spleen, pulmonary artery, skin, muscle and PBMCs (p<0.05 or p<0.0001). In general, Cfs were more potent inducers of active Caspase-3 than were DNAfs.
In vivo experiments using specific inhibitors (DNase and CNPs): We then investigated whether phosphorylation of H2AX and activation of Caspase-3 in response to DNAfs and Cfs in mice could be inhibited by the simultaneous administration of DNase I and/or CNPs. We administered DNAfs and Cfs (100 ng DNA each) intravenously into mice which were concurrently treated intraperitoneally with DNase I and/or CNPs. The animals were sacrificed 24 h after treatment and vital organs, namely, heart, lung, liver and brain as well as PBMCs, were examined by immunofluorescence for activation of H2AX and Caspase-3. We clearly observed that both H2AX and Caspase-3 activation by DNAfs was significantly inhibited by treatment with DNase I in several organs (supplementary figure 12C and D). Cfs-induced H2AX activation was significantly inhibited in all organs and in PBMCs by both CNPs and DNase I, while that of Caspase-3 was significantly inhibited by both CNPs and DNase I in lung, liver and brain (supplementary figure 12E and F).
3.6. DNAfs and Cfs derived from healthy individuals activate DDR and apoptotic pathways
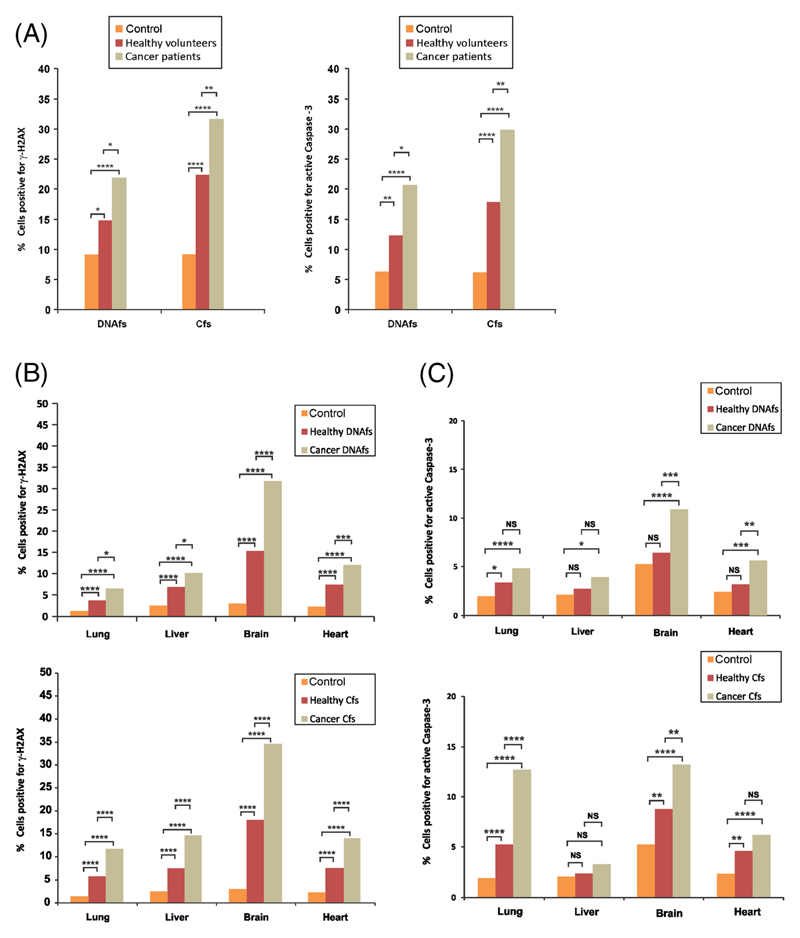
In vitro experiments: Since DNAfs and Cfs also circulate in healthy individuals, we decided to investigate whether DNAfs and Cfs isolated from healthy volunteers were also active in inducing DDR and apoptosis. For the sake the of comparison, we also assessed these parameters in response to DNAfs and Cfs isolated from cancer patients matched for age and sex in the same experiments. Both DNAfs and Cfs isolated from healthy volunteers were indeed capable of significantly increasing the induction of γ-H2AX in NIH3T3 cells (p<0.05 and p<0.0001 respectively) (figure 4A left-hand panel) as well as active Caspase-3 (p<0.01 and p<0.0001 respectively) (figure 4A right-hand panel). Interestingly, in spite of an equal amount of DNA having been applied to cells, DNAfs and Cfs isolated from cancer patients were significantly more active than those from healthy individuals with respect to both γ-H2AX and active Caspase-3 (p<0.05 and p<0.01 for DNAfs and Cfs respectively). It is possible that greater activity of cancer-derived DNAfs and Cfs is due to tumour-related changes in DNA sequences making integration into host cell genomes problematic.
Figure 4.
Induction of γH2AX and active Caspase-3 by DNAfs and Cfs derived from healthy volunteers. Samples from cancer patients were also analysed for comparison. DNAfs and Cfs were isolated from pooled plasma/serum of healthy volunteers and age- and sex-matched cancer patients. (A) In vitro analysis of γ-H2AX and active Caspase-3. NIH3T3 cells (10×104) were treated with DNAfs and Cfs (5 ng DNA each) for 6 h for detection of γ-H2AX (left) and for 24 h for detection of active Caspase -3 (right) by immuno-fluorescence. For γ-H2AX (left-hand panel), 300 nuclei were counted and the percentage of nuclei showing positive foci were calculated and analysed by Chi-square test. For active Caspase-3 (right-hand panel), 200 cells were counted and the percentage of cells showing positive fluorescent signals were calculated and analysed by Chi-square test. *p<0.05, **p<0.01, ****p<0.0001. (B) In vivo detection of γ-H2AX activation by DNAfs (upper panel) and Cfs (lower panel). Mice were injected intravenously with DNAfs and Cfs (100 ng DNA each) and vital organs were removed after 24 h and processed for γH2AX by immuno-fluorescence as described earlier. Control animals were injected with 100 μl of saline. The experiments were done in duplicate i.e., with two animals in each group. One thousand cells from each animal were analysed and the percentage of nuclei with positive fluorescent foci was calculated and analysed by Chi-square test. *p<0.05, **p<0.01, ****p<0.0001. (C) In vivo analysis of Caspase-3 activation by DNAfs (upper panel) and Cfs (lower panel). Mice were injected intravenously with DNAfs and Cfs (100 ng DNA each) and vital organs were removed after 24 h and processed for active Caspase-3 by immuno-fluorescence as described earlier. Control animals were injected with 100 μl of saline. The experiments were done in duplicate i.e., with two animals in each group; one thousand cells from each animal were analysed and the percentage of cells with positive fluorescence was calculated and analysed by Chi-square test. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
In vivo experiments: Assessment of γ-H2AX response following intravenous injection of DNAfs and Cfs (100 ng DNA) from healthy individuals reinforced the in vitro findings in that both were able to activate H2AX in vital organs of mice (p<0.01 and p<0.0001) (figure 4B upper and lower). Of the four vital organs, brain was the most responsive with respect to H2AX activation (p<0.0001). Strikingly, although the same amount of DNA (100 ng) was used for treatment, the H2AX activation was significantly greater in response to both DNAfs- and Cfs-derived from cancer patients, than those derived from healthy individuals (range p<0.05 to p<0.0001) (figure 4B upper and lower). This difference between cancer patients and healthy individuals was particularly pronounced with respect to Cfs. Caspase-3 was also activated by both DNAfs and Cfs from healthy individuals (figure 4C upper and lower). In case of DNAfs, this difference was significant only with respect to lung (p<0.05), while in the case of Cfs, the activation of Caspase-3 was evident in all organs (p<0.0001) except liver. Again, although the same amount of DNA (100 ng) was used for treatment, the Caspase-3 activation was greater in response to DNAfs and Cfs derived from cancer patients, than those derived from healthy individuals. This was especially so in case of brain and heart with respect to DNAfs (p<0.001 and p<0.01) and in the case of lung and brain with respect to Cfs (p<0.0001 and p<0.01).
3.7. Genomic integration of DNAfs and Cfs involve dsDNA break repair
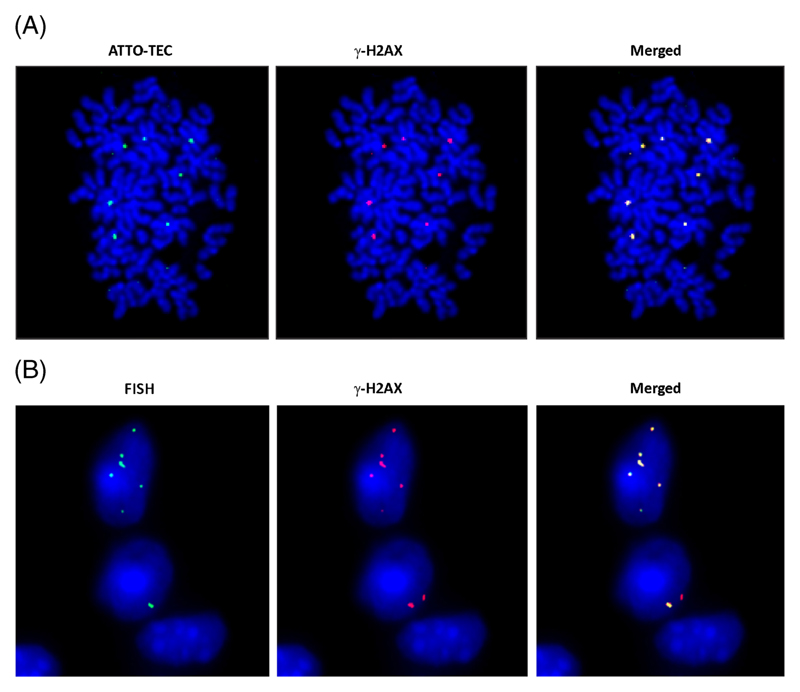
In order to investigate whether integration of human DNA sequences into mouse-cell genomes that we had detected by FISH and NGS (figure 2A and B) involves DNA double-strand break repair, we treated NIH3T3 cells with Cfs that were pre-labelled in their histones with ATTO-TEC (green) and looked for co-localization of these green signals on chromosomal arms on metaphase spreads with red signals generated by Texas Red-labelled secondary antibodies that bound to primary antibodies against γ-H2AX (figure 5A). Many co-localized spots were clearly seen, indicating that integration of Cfs was accompanied by a repair response to dsDNA breaks. We undertook similar co-localization experiments in vivo in animals injected with Cfs (100 ng DNA) and sacrificed them at 24 h. Immuno-FISH experiments using human genomic probes and γ-H2AX antibodies clearly revealed co-localized signals in nuclei of cells of vital organs, indicating that H2AX is also activated in vivo at sites of integration of Cfs (figure 5B).
Figure 5.
Genomic integration of Cfs involves DNA double-strand break repair. (A) In vitro demonstration of γ-H2AX foci at sites of Cfs integration. NIH3T3 cells (10×104) were treated for 6 h with Cfs (5 ng DNA) labelled in their protein with ATTO-TEC (green) and metaphase spreads were prepared. Immuno-fluorescence images were developed using antibody to γ-H2AX (red). Co-localization of green and red signals were clearly visible. (B) In vivo demonstration of γ-H2AX foci at the sites of Cfs integration. Mice were injected i.v. with Cfs (100 ng DNA) and sacrificed 24 h later. Sections of brain were processed for immuno-FISH using human-specific genomic probe (green) and antibody against γ-H2AX (red). Co-localization of green and red signals are clearly visible.
4. Discussion
DNAfs and Cfs derived from apoptotic cells are known to circulate in the blood of healthy individuals and in elevated levels in a host of acute and chronic disease conditions including ageing and cancer (Holdenrieder et al. 2001; Chang et al. 2003; Lam et al. 2003; Lui et al. 2003; Trejo-Becerril et al. 2003; Zeerleder et al. 2003; Gal et al. 2004; Holdenrieder and Stieber 2004; Kremer et al. 2005; Butt et al. 2006; Chiu et al. 2006; Rhodes et al. 2006; Umetani et al. 2006; Swarup and Rajeswari 2007; Zhong et al. 2007; Holdenrieder and Stieber 2009; Pisetsky and Ullal 2010; Rykova et al. 2010; Jylhävä et al. 2011; Tsai et al. 2011; Mittra et al. 2012). Our study shows that far from being inert molecules, DNAfs and Cfs have significant biological activities of their own that are deleterious to healthy cells of the body. Our systematic investigation of the biological properties of circulating DNAfs and Cfs isolated from blood of cancer patients and healthy volunteers has led to the following novel observations: (1) Fluorescently labelled DNAfs and Cfs are readily taken up by cells in culture to be localized in their nuclei within minutes; (2) the intra-nuclear DNAfs and Cfs rapidly associate themselves with host cell chromosomes, evoking a DDR; (3) the activated DNA repair pathways facilitate the incorporation of DNAfs and Cfs into the host cell genomes; (4) FISH detected the presence of human DNA sequences in single-cell clones derived from mouse cells treated with DNAfs and Cfs; (5) whole genome sequencing of these clones confirmed the presence of tens of thousands of human sequence reads in recipient mouse cell genomes as well as in the presence of several human Alu elements; (6) genomic integration of DNAfs and Cfs leads to dsDNA breaks and activation of apoptotic pathways in the treated cells; (7) when injected intravenously, DNAfs and Cfs undergo genomic integration resulting in activation of DDR and apoptosis of cells of vital organs of mice; (8) Cfs were significantly more active than DNAfs with respect to all parameters examined; (9) DNAfs and Cfs from cancer patients were more active than those from normal volunteers; and (10) finally, treatment with DNase I and CNPs led to the abrogation of all the above pathological effects of DNAfs and Cfs both in vitro and in vivo. Taken together these results suggest that DNAfs and Cfs act as physiological, continuously arising, endogenous DNA damaging agents that induce dsDNA breaks and apoptosis in healthy cells by integrating into their genomes.
We made the striking observation that DNAfs and Cfs could freely enter into cells, especially their nuclei, without assistance and activate proteins of DDR and apoptotic pathways. This observation finds support from several lines of evidence which indicate that, while high-molecular-weight DNA is incapable of entering into cells (Shih et al. 1979), fragmented DNA and chromatin, such as those arising from apoptotic cells, can access healthy cells and enter their nuclei to affect biological actions. For example, horizontal transfer of oncogenes has been reported following co-culture of apoptotic bodies with mouse fibroblast cells leading to malignant transformation of the latter (Bergsmedh et al. 2001). DNA released from leukemic cells in the form of nucleosomes can integrate themselves into the chromosomes of surrounding stromal cells and induce DNA damage and apoptosis (Dvořáková et al. 2013). Mutated K-ras oncogene carried in plasma from colon cancer patients can be taken up by mouse fibroblast cells, leading to their oncogenic transformation, and K-ras sequences could be detected in these cells by PCR and FISH (García-Olmo et al. 2010, Trejo-Becerril et al. 2012). Genes reconstituted into chromatin in vitro are readily taken up by cells in culture to localize in the nuclei of recipient cells, leading to the suggestion that chromatinization of DNA maybe an efficient means for gene delivery (Wagstaff et al. 2008). Circulating DNA can cross the blood–brain barrier, and male foetal DNA have been found in maternal brain cells (Chan et al. 2012). Fragmented DNA released by bacteria in the peritoneal cavity of frogs can enter brain cells and synthesize RNA (Anker and Stroun 1972). Taken together, the above studies clearly show that fragmented DNA or chromatin from sources other than human blood or plasma can also freely enter into cells, especially their nuclei, and activate biological effects in the target cells.
We did not attempt, in this study, to investigate the mechanism(s) by which DNAfs and Cfs are taken up by recipient cells. Nonetheless, our experiments with a metabolic poison (Actinomycin D) and at low temperature (31°C) clearly demonstrated that cellular/nuclear uptake of DNAfs and Cfs are energy dependent and require an active metabolic machinery of the recipient cells (supplementary figure 3E, F, G and H). DNAfs were internalized into the nuclei with greater efficiency than were Cfs; while the peak DNAfs uptake occurred at 30 min with 98% of the nuclei showing presence of fluorescent signals, nuclear uptake of Cfs peaked at 6–8 h when 58% of the nuclei showed presence of fluorescent Cfs signals (p<0.0001) (figure 1C). However, the reverse was true with respect to chromosomal association: Cfs associated with chromosomes with greater efficiency (40%) than did DNAfs (18%) (p=0.0275). In any event, following their nuclear uptake, both DNAfs and Cfs were rapidly degraded with those that had associated with chromosomes apparently escaping degradation (figure 1C).
Throughout our studies we consistently observed that, when equated for amount of DNA applied, Cfs were significantly more active in every respect compared to DNAfs, both in vitro and in vivo. For example, Cfs were more efficient (1) in associating themselves with chromosomes (figure 1D and E); (2) in their ability to integrate into host cell genomes (figure 2A, B and C); and (3) in inducing DDR and apoptosis (figures 3B and 3C). These findings lead us to propose that, although DNAfs have the ability to rapidly enter into cells, naked DNA fragments remain biologically inert until they are able to convert themselves into Cfs-like structures by complexing with newly synthesized intracellular histones. The DNA-histone assemblies are, however, not as efficient as incoming native Cfs in their ability either to induce DDR or to integrate themselves into host cell genomes. This interpretation may be relevant in another context wherein we observed a discordance between the kinetics of nuclear uptake and that of H2AX activation by DNAfs and Cfs. Although DNAfs were rapidly taken up by recipient cell nuclei peaking at 30 min, as opposed to 6–8 h for Cfs (figure 1C), the kinetics of H2AX activation was similar, both reaching a maximum at ~6 h (figure 3A). This suggests that while Cfs can activate H2AX soon after entry, for DNAfs, there is a lag period before H2AX can be activated. The lag period may represent the time required for DNAfs to reconstitute themselves into Cfs-like structures by complexing with intracellular histones.
We clearly detected the presence of human DNA signals in single-cell clones derived from DNAfs- and Cfs-treated mouse cells using human-specific whole-genomic and pancentromeric FISH probes (figure 2A). Since the threshold value for detection of FISH signals has been estimated to be of the order of 30–50 kb (Frengen et al. 1997), our detection of human signals in mouse cells by FISH suggested that multiple small fragments of DNAfs and Cfs might have linked up by non-homologous end-joining (NHEJ) to produce long concatamers of discontinuous DNA segments before integrating into host cell genomes (discussed later). NHEJ of discontinuous DNAfs and Cfs could have been facilitated by DNA-ligase IV and DNA-PKcs that were found to be activated during DDR (figure 3B). We clearly demonstrated that genomic integration of these concatamers involved dsDNA break repair by detecting co-localization of ATTO-TEC or FISH signals with those of γ-H2AX on chromosomal arms or in nuclei of treated cells (figure 5A and B). The heterogeneously ligated concatamers apparently elicit DNA repair pathways involving homologous and non-homologous recombinations for their integration and repair in the host cell genomes (discussed later).
Massively parallel next-generation sequencing of single-cell clones derived from DNAfs- and Cfs-treated mouse cells provided additional line of evidence for the presence of human DNA in mouse-cell genomes at single base resolution. We found 5- to 6-fold higher abundance of human DNA in Cfs-derived clones than DNAfs-derived ones (figure 2B; supplementary figure 5). This finding is consistent with our FISH experiments, wherein the numbers of human signals were approximately ×4 higher in the same Cfs-derived clones compared to the DNAfs-derived clones (figure 2A). Computational analysis also detected a higher abundance of human Alu repeats in Cfs-derived clones compared to DNAfs-derived clones (supplementary table 4; supplementary figure 6). Given that the computational subtraction pipeline adopted in this study (supplementary figure 1) was conservative and maximally stringent allowing zero mismatch of human reads that might be present in the mouse cell clones when aligned to human reference sequence, we anticipated a much higher underlying abundance of human sequences in these transformed mouse cell clones than what we uncovered. It is likely that human reads harbouring SNPs, or those that are tumour-derived and harbour somatic alterations, have remained unaccounted. Taken together, these observations give additional insights into the panoply of apparently random integration of human DNA that occurs in mouse cells and provide the groundwork for using new sequencing technologies in future studies to investigate preferential integration sites, if any.
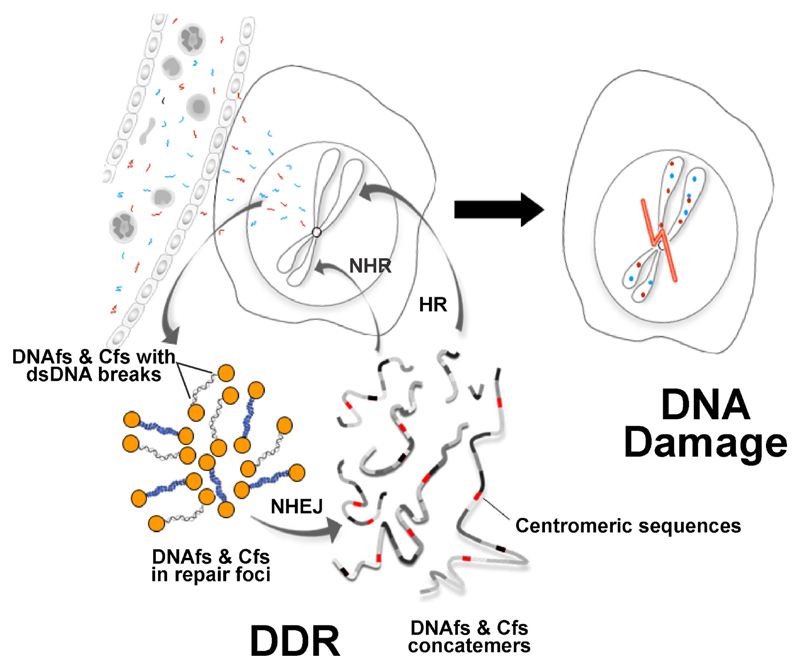
Based on these observations, we propose a new working model for DNA damage and mutagenesis that are induced by circulating DNAfs and Cfs (figure 6). It is well established from classical experiments that when genomic DNA is damaged by ionizing and UV-radiation and chemicals, the damage to DNA is followed by activation of DDR pathways which attempt to repair the damage. The new model, on the other hand, proposes that when DNAfs and Cfs enter the recipient cell, the latter perceives the dsDNA breaks present in their two ends as damaged ‘self’ DNA and, in an attempt to repair the ‘perceived damage’, activate proteins of the DDR pathway. The activated DNA repair proteins facilitate the integration of DNAfs and Cfs into host cell genomes by homologous and/or non-homologous recombination; and it is this event of DNA integration into host cell genomes that brings about the damage to host cell DNA (figure 5A and B). Thus, paradoxically, it is the activation of DDR that brings about damage to DNA rather than maintenance of DNA integrity. The model further proposes that the activation of DNA-PKcs and DNA ligase-IV, proteins that are involved in NHEJ, leads to linking up of many of the internalized DNAfs and Cfs to form long concatamers of discontinuous DNA segments which form new substrates for genomic integration (Burma et al. 2006). The proposal of DNA concatamerization is supported by the finding that genomic and centromeric signals frequently co-localize on chromosomal arms during FISH analysis (figure 2A), suggesting that the concatamers often harbour centromeric sequences. The above model of DNA damage and repair as proposed by us is represented schematically in figure 6. It must be pointed out, however, that more experimental work is needed to substantiate the model.
Figure 6.
Schematic representation of proposed model of DNA damage. NHEJ = non-homologous end-joining; HR = homologous recombination; NHR = non-homologous recombination.
It becomes clear from the present work that circulating nucleic acids, far from being biologically inert particles, have significant deleterious functions in the host. They freely enter healthy cells and damage their DNA by integrating into their genomes. While the genome is exposed to a number of xenobiotics and DNA damaging events, they are usually transient and do not cause permanent damage. However, circulating nucleic acids are ubiquitous and continuously arising, inflicting repeated damage to the somatic DNA. This automatically suggests that the somatic genome may not be stable, but rather remains in a state of turmoil characterized by dsDNA breaks, genomic instability and apoptosis affected by integration of circulating DNAfs and Cfs. These events may lead to deletions, duplications and rearrangements causing DNA mosaicism, which is being increasingly uncovered in somatic cells (Jacobs et al. 2012; Laurie et al. 2012; McConnell et al. 2013). The uptake and genomic integration of circulating nucleic acids and the concomitant DNA damage that they induce may contribute to ageing as well as to initiation and progression of cancer by triggering genetic events and consequent genomic rearrangements that underlie both these interrelated processes (Vijg and Dollé 2002; Hoeijmakers 2009; Stephens et al. 2009; Trejo-Becerril et al. 2012). DNAfs and Cfs may also be implicated in a multitude of other acute and chronic human disorders that are associated with elevated levels of circulating nucleic acids thereby making them one of the key players in maintenance of human health and disease. (Holdenrieder et al. 2001; Chang et al. 2003; Lam et al. 2003; Lui et al. 2003; Trejo-Becerril et al. 2003; Zeerleder et al. 2003; Gal et al. 2004; Holdenrieder and Stieber 2004; Kremer et al. 2005; Butt et al. 2006; Chiu et al. 2006; Rhodes et al. 2006; Umetani et al. 2006; Pisetsky and Ullal 2010; Tsai et al. 2011; Mittra et al. 2012).
Supplementary Material
Supplementary materials pertaining to this article are available on the Journal of Biosciences Website at http://www.ias.ac.in/jbiosci/mar2015/supp/Mittra.pdf
Acknowledgements
We sincerely thank Dr LC Padhy for most generously offering his time during numerous scientific discussions, for his invaluable suggestions and for his critical inputs into the manuscript. We thank Dr AN Ghosh for taking the EM images. We acknowledge the contribution made by the many lab members who have contributed to this project over the years. Special thanks are due to Nagnath Swamy, Shazia Mansoor, Nabila Akhtar, Mansoor Ali, Rashmi Malvee, Arpit Bhargava, Preetam Bala, Mihir Shetty, Ajay Choudhary and Mihir Parmar. This study was supported by the Department of Atomic Energy, Govt. of India, through its grant CTCTMC to Tata Memorial Centre awarded to IM. AD is supported by an Intermediate fellowship from the Wellcome Trust/DBT India Alliance (IA/I/11/2500278), by a grant from Department of Biotechnology, Govt. of India (BT/PR2372/AGR/36/696/2011), and intramural grants (Seed-In-Air 2897, TMH Plan Project 2712 and IRB 92). PU is supported by Senior Research Fellowship from Council of Scientific & Industrial Research, Govt. of India.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anker P, Stroun M. Bacterial ribonucleic acid in the frog brain after a bacterial peritoneal infection. Science. 1972;178:621–623. doi: 10.1126/science.178.4061.621. [DOI] [PubMed] [Google Scholar]
- Bergsmedh A, Szeles A, Henriksson M, Bratt A, Folkman MJ, Spetz AL, Holmgren L. Horizontal transfer of oncogenes by uptake of apoptotic bodies. Proc Natl Acad Sci USA. 2001;98:6407–6411. doi: 10.1073/pnas.101129998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burma S, Chen BP, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst) 2006;5:1042–1048. doi: 10.1016/j.dnarep.2006.05.026. [DOI] [PubMed] [Google Scholar]
- Butt AN, Shalchi Z, Hamaoui K, Samadhan A, Powrie J, Smith S, Janikoun S, Swaminathan R. Circulating nucleic acids and diabetic complications. Ann NY Acad Sci. 2006;1075:258–270. doi: 10.1196/annals.1368.034. [DOI] [PubMed] [Google Scholar]
- Chan WF, Gurnot C, Montine TJ, Sonnen JA, Guthrie KA, Nelson JL. Male microchimerism in the human female brain. PLoS One. 2012;7:e45592. doi: 10.1371/journal.pone.0045592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CP, Chia RH, Wu TL, Tsao KC, Sun CF, Wu JT. Elevated cell-free serum DNA detected in patients with myocardial infarction. Clin Chim Acta. 2003;327:95–101. doi: 10.1016/s0009-8981(02)00337-6. [DOI] [PubMed] [Google Scholar]
- Chiu TW, Young R, Chan LY, Burd A, Lo DY. Plasma cell-free DNA as an indicator of severity of injury in burn patients. Clin Chem Lab Med. 2006;44:13–17. doi: 10.1515/CCLM.2006.003. [DOI] [PubMed] [Google Scholar]
- Dvořáková M, Karafiát V, Pajer P, Kluzáková E, Jarkovská K, Peková S, Krutílková L, Dvořák M. DNA released by leukemic cells contributes to the disruption of the bone marrow microenvironment. Oncogene. 2013;32:5201–5209. doi: 10.1038/onc.2012.553. [DOI] [PubMed] [Google Scholar]
- Fliedner TM, Graessle D, Paulsen C, Reimers K. Structure and function of bone marrow hemopoiesis: mechanisms of response to ionizing radiation exposure. Cancer Biother Radiopharm. 2002;17:405–426. doi: 10.1089/108497802760363204. [DOI] [PubMed] [Google Scholar]
- Frengen E, Thomsen PD, Brede G, Solheim J, de Jong PJ, Prydz H. The gene cluster containing the LCAT gene is conserved between human and pig. Cytogenet Cell Genet. 1997;76:53–57. doi: 10.1159/000134515. [DOI] [PubMed] [Google Scholar]
- Fulda S, Meyer E, Debatin KM. Metabolic inhibitors sensitize for CD95 (APO-1/Fas)-induced apoptosis by down- regulating Fas-associated death domain-like interleukin 1- converting enzyme inhibitory protein expression. Cancer Res. 2000;60:3947–3956. [PubMed] [Google Scholar]
- Gal S, Fidler C, Lo YM, Taylor M, Han C, Moore J, Harris AL, Wainscoat JS. Quantitation of circulating DNA in the serum of breast cancer patients by real-time PCR. Br J Cancer. 2004;90:1211–1215. doi: 10.1038/sj.bjc.6601609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Olmo DC, Domínguez C, García-Arranz M, Anker P, Stroun M, García-Verdugo JM, García-Olmo D. Cell free nucleic acids circulating in the plasma of colorectal cancer patients induce the oncogenic transformation of susceptible cultured cells. Cancer Res. 2010;70:560–567. doi: 10.1158/0008-5472.CAN-09-3513. [DOI] [PubMed] [Google Scholar]
- Gawel-Thompson KJ, Greene RM. Epidermal growth factor: modulator of murine embryonic palate mesenchymal cell proliferation, polyamine biosynthesis, and polyamine transport. J Cell Physiol. 1989;140:359–370. doi: 10.1002/jcp.1041400222. [DOI] [PubMed] [Google Scholar]
- Halicka HD, Bedner E, Darzynkiewicz Z. Segregation of RNA and separate packaging of DNA and RNA in apoptotic bodies during apoptosis. Exp Cell Res. 2000;260:248–256. doi: 10.1006/excr.2000.5027. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- Holdenrieder S, Stieber P, Bodenmüller H, Busch M, Fertig G, Fürst H, Schalhorn A, Schmeller N, et al. Nucleosomes in serum of patients with benign and malignant diseases. Int J Cancer. 2001;95:114–120. doi: 10.1002/1097-0215(20010320)95:2<114::aid-ijc1020>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Holdenrieder S, Stieber P. Apoptotic markers in cancer. Clin Biochem. 2004;37:605–617. doi: 10.1016/j.clinbiochem.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Holdenrieder S, Stieber P. Clinical use of circulating nucleosomes. Crit Rev Clin Lab Sci. 2009;46:1–24. doi: 10.1080/10408360802485875. [DOI] [PubMed] [Google Scholar]
- Jacobs KB, Yeager M, Zhou W, Wacholder S, Wang Z, Rodriguez-Santiago B, Hutchinson A, Deng X, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44:651–658. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jylhävä J, Kotipelto T, Raitala A, Jylhä M, Hervonen A, Hurme M. Aging is associated with quantitative and qualitative changes in circulating cell-free DNA: the vitality 90+ study. Mech Ageing Dev. 2011;132:20–26. doi: 10.1016/j.mad.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42:D764–D770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzman JO, Snyder MW, Ventura M, Lewis AP, Qiu R, Simmons LE, Gammill HS, Rubens CE, et al. Noninvasive whole-genome sequencing of a human fetus. Sci Transl Med. 2012;4:137ra76. doi: 10.1126/scitranslmed.3004323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer A, Wilkowski R, Holdenrieder S, Nagel D, Stieber P, Seidel D. Nucleosomes in pancreatic cancer patients during radiochemotherapy. Tumour Biol. 2005;26:44–49. doi: 10.1159/000084339. [DOI] [PubMed] [Google Scholar]
- Lam NY, Rainer TH, Chan LY, Joynt GM, Lo YM. Time course of early and late changes in plasma DNA in trauma patients. Clin Chem. 2003;49:1286–1291. doi: 10.1373/49.8.1286. [DOI] [PubMed] [Google Scholar]
- Laurie CC, Laurie CA, Rice K, Doheny KF, Zelnick LR, McHugh CP, Ling H, Hetrick KN, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet. 2012;44:642–650. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, O’Shaughnessy J, Kinzler KW, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012;4:162ra154. doi: 10.1126/scitranslmed.3004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui YY, Woo KS, Wang AY, Yeung CK, Li PK, Chau E, Ruygrok P, Lo YM. Origin of plasma cell-free DNA after solid organ transplantation. Clin Chem. 2003;49:495–496. doi: 10.1373/49.3.495. [DOI] [PubMed] [Google Scholar]
- Marchant RJ, Al-Fageeh MB, Underhill MF, Racher AJ, Smales CM. Metabolic rates, growth phase, and mRNA levels influence cell-specific antibody production levels from in vitro-cultured mammalian cells at sub-physiological temperatures. Mol Biotechnol. 2008;39:69–77. doi: 10.1007/s12033-008-9032-0. [DOI] [PubMed] [Google Scholar]
- McConnell MJ, Lindberg MR, Brennand KJ, Piper JC, Voet T, Cowing-Zitron C, Shumilina S, Lasken RS, et al. Mosaic copy number variation in human neurons. Science. 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittra I, Mishra PK, Mansoor S, Samant U, Patkar V, Sharma S, Ali M, Padhy LC. Cancer Res. 8 Suppl. Vol. 70. Washington, DC. Philadelphia (PA): AACR: 2010. Apr 17–21, Circulating chromatin is a novel DNA damaging agent that induces genomic instability and malignant transformation. (abstract); in Proceedings of the 101st Annual Meeting of the American Association for Cancer Research. Abstract number LB-103. [Google Scholar]
- Mittra I, Nair NK, Mishra PK. Nucleic acids in circulation: Are they harmful to the host? J Biosci. 2012;37:301–312. doi: 10.1007/s12038-012-9192-8. [DOI] [PubMed] [Google Scholar]
- Mittra I, Samant U, Modi GK, Mishra PK, Bhuvaneswar GS. A method for ex-vivo separation of apoptotic chromatin fragments from blood or plasma for prevention and treatment of diverse human diseases. US patent application no. FPAA819PCT dated 27.10.2006. 2006
- Pisetsky DS, Ullal AJ. The blood nucleome in the pathogenesis of SLE. Autoimmun Rev. 2010;10:35–37. doi: 10.1016/j.autrev.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliss A, Malyavantham KS, Bhattacharya S, Berezney R. Chromatin dynamics in living cells: identification of oscillatory motion. J Cell Physiol. 2013;228:609–616. doi: 10.1002/jcp.24169. [DOI] [PubMed] [Google Scholar]
- Rekha MR, Pal K, Bala P, Shetty M, Mittra I, Bhuvaneshwar GS, Sharma CP. Pullulan-histone antibody nanoconjugates for the removal of chromatin fragments from systemic circulation. Bio materials. 2013;34:6328–6338. doi: 10.1016/j.biomaterials.2013.05.019. [DOI] [PubMed] [Google Scholar]
- Rhodes A, Wort SJ, Thomas H, Collinson P, Bennett ED. Plasma DNA concentration as a predictor of mortality and sepsis in critically ill patients. Crit Care. 2006;10:R60. doi: 10.1186/cc4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rykova EY, Laktionov PP, Vlassov VV. Circulating nucleic acids in health and disease. In: Kikuchi Y, Rykova E, editors. Extracellular Nucleic Acids: Nucleic Acids And Molecular Biology. Vol. 25. Berlin Heidelberg; Springer-Verlag: 2010. pp. 93–128. [Google Scholar]
- Sakurai T, Itoh K, Liu Y, Higashitsuji H, Sumitomo Y, Sakamaki K, Fujita J. Low temperature protects mammalian cells from apoptosis initiated by various stimuli in vitro. Exp Cell Res. 2005;309:264–272. doi: 10.1016/j.yexcr.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Schnitzler GR. Isolation of histones and nucleosome coresfrom mammalian cells. Curr Protoc Mol Biol. 2001 doi: 10.1002/0471142727.mb2105s50. Chapter 21: Supplement 50, Unit 21.5.1–21.5.12. [DOI] [PubMed] [Google Scholar]
- Shih C, Shilo BZ, Goldfarb MP, Dannenberg A, Weinberg RA. Passage of phenotypes of chemically transformed cells via transfection of DNA and chromatin. Proc Natl Acad Sci USA. 1979;76:5714–5718. doi: 10.1073/pnas.76.11.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup V, Rajeswari MR. Circulating (cell-free) nucleic acids–a promising, non-invasive tool for early detection of several human diseases. FEBS Lett. 2007;581:795–799. doi: 10.1016/j.febslet.2007.01.051. [DOI] [PubMed] [Google Scholar]
- Takeshita H, Nakajima T, Mogi K, Kaneko Y, Yasuda T, Iida R, Kishi K. Rapid quantification of DNase I activity in one-microliter serum samples. Clin Chem. 2004;50:446–448. doi: 10.1373/clinchem.2003.025304. [DOI] [PubMed] [Google Scholar]
- Trejo-Becerril C, Pérez-Cárdenas E, Taja-Chayeb L, Anker P, Herrera-Goepfert R, Medina-Velázquez LA, Hidalgo-Miranda A, Pérez-Montiel D, et al. Cancer progression mediated by horizontal gene transfer in an in vivo model. PLoS One. 2012;7:e52754. doi: 10.1371/journal.pone.0052754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trejo-Becerril C, Pérez-Cárdenas E, Treviño-Cuevas H, Taja-Chayeb L, García-López P, Segura-Pacheco B, Chávez-Blanco A, Lizano-Soberon M, et al. Circulating nucleosomes and response to chemotherapy: an in vitro, in vivo and clinical study on cervical cancer patients. Int J Cancer. 2003;104:663–668. doi: 10.1002/ijc.11003. [DOI] [PubMed] [Google Scholar]
- Tsai NW, Lin TK, Chen SD, Chang WN, Wang HC, Yang TM, Lin YJ, Jan CR, et al. The value of serial plasma nuclear and mitochondrial DNA levels in patients with acute ischemic stroke. Clin Chim Acta. 2011;412:476–479. doi: 10.1016/j.cca.2010.11.036. [DOI] [PubMed] [Google Scholar]
- Umetani N, Giuliano AE, Hiramatsu SH, Amersi F, Nakagawa T, Martino S, Hoon DS. Prediction of breast tumor progression by integrity of free circulating DNA in serum. J Clin Oncol. 2006;24:4270–4276. doi: 10.1200/JCO.2006.05.9493. [DOI] [PubMed] [Google Scholar]
- van Nieuwenhuijze AE, van Lopik T, Smeenk RJ, Aarden LA. Time between onset of apoptosis and release of nucleosomes from apoptotic cells: putative implications for systemic lupus erythematosus. Ann Rheum Dis. 2003;62:10–14. doi: 10.1136/ard.62.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijg J, Dollé ME. Large genome rearrangements as a primary cause of aging. Mech Ageing Dev. 2002;123:907–915. doi: 10.1016/s0047-6374(02)00028-3. [DOI] [PubMed] [Google Scholar]
- Vollenweider HJ, Sogo JM, Koller T. A routine method for protein-free spreading of double- and single-stranded nucleic acid molecules. Proc Natl Acad Sci USA. 1975;72:83–87. doi: 10.1073/pnas.72.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff KM, Fan JY, De Jesus MA, Tremethick DJ, Jans DA. Efficient gene delivery using reconstituted chromatin enhanced for nuclear targeting. FASEB J. 2008;22:2232–2242. doi: 10.1096/fj.07-099911. [DOI] [PubMed] [Google Scholar]
- Zeerleder S, Zwart B, Wuillemin WA, Aarden LA, Groeneveld AB, Caliezi C, van Nieuwenhuijze AE, van Mierlo GJ, et al. Elevated nucleosome levels in systemic inflammation and sepsis. Crit Care Med. 2003;31:1947–1951. doi: 10.1097/01.CCM.0000074719.40109.95. [DOI] [PubMed] [Google Scholar]
- Zhong XY, Hahn S, Kiefer V, Holzgreve W. Is the quantity of circulatory cell-free DNA in human plasma and serum samples associated with gender, age and frequency of blood donations? Ann Hematol. 2007;86:139–143. doi: 10.1007/s00277-006-0182-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.