Abstract
Inherited mutations in the rod visual pigment, rhodopsin, cause the degenerative blinding condition, retinitis pigmentosa (RP). Over 150 different mutations in rhodopsin have been identified and, collectively, they are the most common cause of autosomal dominant RP (adRP). Mutations in rhodopsin are also associated with dominant congenital stationary night blindness (adCSNB) and, less frequently, recessive RP (arRP). Recessive RP is usually associated with loss of rhodopsin function, whereas the dominant conditions are a consequence of gain of function and/or dominant negative activity. The in-depth characterisation of many rhodopsin mutations has revealed that there are distinct consequences on the protein structure and function associated with different mutations. Here we categorise rhodopsin mutations into seven discrete classes; with defects ranging from misfolding and disruption of proteostasis, through mislocalisation and disrupted intracellular traffic to instability and altered function. Rhodopsin adRP offers a unique paradigm to understand how disturbances in photoreceptor homeostasis can lead to neuronal cell death. Furthermore, a wide range of therapies have been tested in rhodopsin RP, from gene therapy and gene editing to pharmacological interventions. The understanding of the disease mechanisms associated with rhodopsin RP and the development of targeted therapies offer the potential of treatment for this currently untreatable neurodegeneration.
Keywords: rhodopsin, GPCR, retinal dystrophy, neurodegeneration, mutation, protein misfolding, proteostasis, therapy, protein traffic, endocytosis, CRISPR
1. Introduction
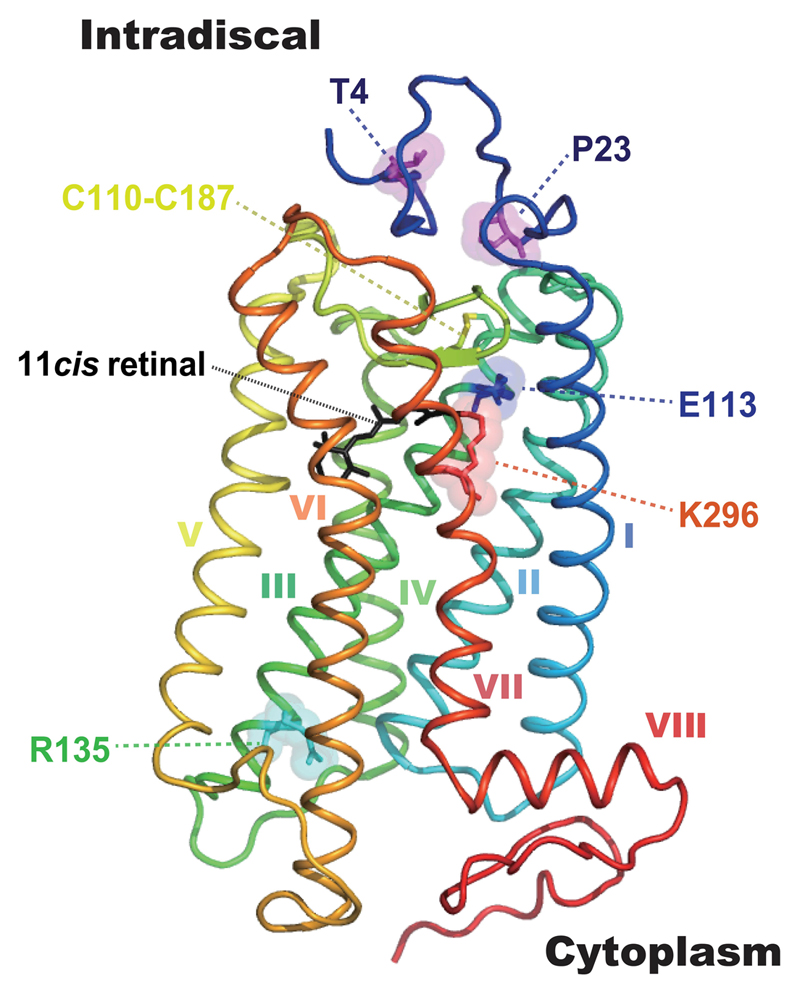
The outer retina of the mammalian eye possesses two types of ciliary photoreceptor, the rods and cones. Rods, with a peak absorption (λmax) of ~500 nm, contain the rod-specific rod opsin and are responsible for dim-light (scotopic) photoreception. Cones, with a λmax for visible light ranging from ~350-560 nm, contain one of up to three different cone opsins (ultraviolet/violet, middle- and long-wave sensitive). Image-forming photoreception is initiated by the absorption of a photon by the vitamin A derived chromophore 11-cis-retinal within rhodopsin, causing it to isomerise to the all-trans conformation. Whilst any opsin + 11-cis-retinal moiety is correctly termed a ‘rhodopsin’, the term rhodopsin is most commonly applied to the rod opsin + 11-cis-retinaldehyde photopigment complex (Lythgoe, 1979), and this convention will be followed in this review. Rhodopsin is the archetypal G protein-coupled receptor (GPCR) with seven transmembrane (TM) α-helices (Palczewski et al., 2000). The 11-cis-retinal acts an inverse agonist suppressing constitutive activity of the receptor, is located within the pocket formed by the α-helices and is coordinated through a protonated Schiff-base (PSB) linkage to a lysine residue in the 7th TM domain (K296) (Smith, 2010) (Figure 1).
Figure 1. The structure of rhodopsin.
The tertiary structure of bovine rhodopsin from the intradiscal N-terminus (dark blue) to the cytoplasmic C-terminus (red) containing 7 transmembrane motifs (α-helix I-VII) and cytosplamic helix VIII shown in a 3D model. 11-cis retinal (black), T4, P23, E113, R135, K296 and the disulphide bond site C110-C187 are highlighted. The 3D image was made using PyMol (PDB 1U19).
1.1. Identification of mutations in rhodopsin (RHO)
In the mammalian genome, the ciliary opsin family is composed of the rhodopsin gene (RHO); an ultraviolet/violet sensitive opsin gene (OPN1SW); middle/long-wave sensitive opsin genes (OPN1MW/LW); and the less well characterised opsin 3 (encephalopsin) gene (OPN3). These genes share at least three perfectly conserved intronic sites (Bellingham et al., 2003b) and appear to have arisen through duplications early in vertebrate evolution (Nördstrom et al., 2004). Subsequent to the cloning and characterisation of the bovine rhodopsin gene (Nathans and Hogness, 1983), the same authors published the structure and sequence of the human orthologue, RHO (Nathans and Hogness, 1984). With the exception of the intronless rod opsin gene of the Actinopterygii (ray-finned fish) (Bellingham et al., 2003a; Fitzgibbon et al., 1995), the RHO gene like all other currently characterised vertebrate rod opsin genes is composed of 5 exons, and the open reading frame predicts a 348 amino acid protein with a molecular weight of ~39 kDa.
The RHO gene was initially mapped to the long arm of chromosome 3 at 3q21-qter (Nathans et al., 1986), and is currently designated at 3q22.1 at coordinates 3:129,528,640-129,535,169 (build GRCh38.p10; http://www.ensembl.org/). Methodologies used to identify inherited retinal dystrophy (IRD) genes have recently been reviewed (Broadgate et al., 2017) and causative mutations in the RHO gene were identified by Dryja et al in 1990 (Dryja et al., 1990b), as the first genetic cause of retinitis pigmentosa (RP). The identified nucleotide substitution, c.68C>A, results in the following codon change CCC>CAC such that the normally observed proline (CCC) at position 23 of the rod opsin protein is substituted by a histidine (CAC) residue (p.P23H). Since this discovery, many different RHO mutations have been identified and many other genes for RP and other IRDs have been discovered, but P23H remains one of the most intensively studied causes of IRD. Further information regarding the genetic basis of IRDs can be accessed at the RetNet™ Retinal Information Network (https://sph.uth.edu/retnet/).
1.2. Phenotypes of rhodopsin associated retinal dystrophies
Mutations in RHO are associated with a classical form of RP, which is a rod-cone dystrophy caused by the dysfunction and death of rod cells, followed by the dysfunction and death of cone cells. A detailed clinical description of RP is beyond the scope of this review and readers are directed to this book chapter for more information (Gregory-Evans et al., 2013). Briefly, classical RP is characterised by a progressive loss of peripheral vision leading to so-called “tunnel vision”. The initial symptom is typically impaired dark adaptation and the development of “night blindness” (nyctalopia) through the loss of rod function starting in the 1st or 2nd decade of life (Hartong et al., 2006). As photoreceptor loss proceeds, there is a loss of pigmentation from the retinal pigment epithelium (RPE), and a build-up of intraretinal melanin deposits that can assume a “bone spicule” conformation. Central visual acuity is often preserved until the end stages of RP. Studies on the prevalence of non-syndromic RP in different populations indicate a frequency of between 1:3000 to 1:5000 individuals (Berson, 1993; Hamel, 2006; Hartong et al., 2006; Sharon et al., 2016), with between 15 and 35% of cases being reported as being of an autosomal dominant inheritance, depending on the population studied (Bravo-Gil et al., 2017; Hartong et al., 2006). In excess of 150 missense/nonsense RHO mutations are catalogued by the professional 2017.1 release of the Human Gene Mutation Database (Stenson et al., 2014) (http://www.hgmd.cf.ac.uk/).
Retinal pathologies resulting from mutations in the RHO gene can be inherited in either an autosomal dominant (ad) or autosomal recessive (ar) manner. Two disease states are associated with RHO mutations, RP and congenital stationary night blindness (CSNB). Whilst CSNB associated with a RHO mutation is inherited in an autosomal dominant manner (adCSNB), RP associated with a RHO mutation can be inherited in either an autosomal dominant (adRP) or autosomal recessive (arRP) form. Inherited variants in RHO are most often associated with adRP, while the recessive mode of inheritance is relatively uncommon.
1.2.1. CSNB
RHO associated adCSNB (MIM#610445; CSNBAD1; https://www.omim.org/entry/610445) is characterised by a lack of scotopic vision typified by an absence of detectable rod function by electroretinogram (ERG). Cone function typically appears normal. There are currently five RHO missense mutations associated with adCSNB: p.G90D (Rao et al., 1994; Sieving et al., 1995); p.T94I (al-Jandal et al., 1999); p.E113K (Reiff et al., 2016); p.A292E (Dryja et al., 1993) and p.A295V (Zeitz et al., 2008) (Table 1). Whilst normally non-progressive in nature, peripheral pigmentary changes and retinal degeneration have been noted as a feature in older patients (Sieving et al., 1995), suggesting potential overlap with an adRP phenotype later in life. The E113K mutation is striking in that both CSNB and adRP are present in the same pedigree with phenotype seemingly not linked to age of patient (Reiff et al., 2016). With the exception of the E113K mutation, constitutive activation of rhodopsin due to these mutations is believed to be the mechanism behind this form of adCSNB (Gross et al., 2003), see section 2 and classification (Table 1).
Table 1. Classification of adRP and adCSNB mutations.
Classification of rhodopsin mutations described in HGMD (http://www.hgmd.cf.ac.uk/) based on their experimentally studied biochemical and cellular characteristics. Italicised substitutions are observed in the gnomAD dataset at a frequency of ~ 1:80,000 or more and might be non-pathogenic substitutions. § might behave as class 4 after 11-cis-retinal rescue. † might behave as class 2 on overexpression, but class 4 in vivo.
| Classification | Key features | Mutations |
|---|---|---|
| 1 | Post Golgi trafficking and OS targeting | L328P, T342M, Q344R/P/ter, V345L/M, A346P, P347A/R/Q/L/S/T, ter349/Q/E |
| 2 | Misfolding, ER retention and instability | N15S§, T17M†, V20G§, P23A/H/L§, Q28H§, G51R/V, P53R, T58R/M, V87D, G89D, G106R/W, C110F/R/S/Y, E113K, L125R, W161R, A164E/V, C167R/W, P171Q/L/S, Y178N/D/C, E181K, G182S/V, C185R, C187G/Y, G188R/E, D190N/G/Y, H211R/P, C222R, P267R/L, S270R, K296N/E/M |
| 3 | Disrupted vesicular traffic and endocytosis | R135G/L/P/W |
| 4 | Altered post-translational modifications and reduced stability | T4K†, T17M†, M39R, N55K, G90V |
| 5 | Altered transducin activation | M44T, V137M |
| 6 | Constitutive activation | G90D, T94I, A292E, A295V |
| 7 | Dimerization deficiency | F45L, V209M, F220C |
| Unclassified | No observed biochemical or cellular defect or not studied in detail | P12R, R21C, Q28H, L40R, L46R, L47R, F52Y, F56Y, L57R, Y60ter, Q64ter, R69H, N78I, L79P, V87L, L88P, T92I, T97I, V104F, G109R, G114D/V, E122G, W126L/ter, S127F, L131P, Y136ter, C140S, T160T, M163T, A169P, P170H/R, S176F, P180A/S, Q184P, S186P/W, Y191C, T193M, M207R/K, V210F, I214N, P215L/T, M216R/L/K, R252P, T289P, S297R, A298D, K311E, N315ter, E341K, S343C |
1.2.2. arRP
Two missense RHO mutations, p.E150K (Azam et al., 2009; Kumaramanickavel et al., 1994; Saqib et al., 2015; Van Schil et al., 2016), and M253I (Collin et al., 2011); and two nonsense (premature stop codon) mutations, p.W161ter (Kartasasmita et al., 2011) and p.E249ter (Rosenfeld et al., 1992), are associated with arRP (Table 2). The observed homozygous recessive inheritance of loss-of-function RHO mutations (compound heterozygous RHO mutations have yet to be identified), suggests that scope for innocuous loss-of-function mutations in the RHO molecule is very restricted. This may account for the high number of adRP mutations. Interestingly, research on a knock-in (KI) mouse model of E150K suggests that this is actually a mild dominant mutation, which progresses more quickly in the homozygous state (Zhang et al., 2013). M253I was identified by homozygosity mapping and has not been studied in detail biochemically, but was also suggested to potentially be a mild mutation that is only pathogenic if present on both alleles (Collin et al., 2011). In contrast, it would appear that the p.W161ter and p.E249ter mutations might potentially cause a true null phenotype since the locations of the nucleotide substitutions, c.482G>A (TGG>TAG) and c.745G>T (GAG>TAG) in exon 2 and exon 4 respectively, suggest that nonsense mediated decay is likely to clear the premature stop codon containing mRNA transcript to an unknown degree (Hernan et al., 2011; Roman-Sanchez et al., 2016). Rhodopsin is essential for rod cell function and rod cell survival, as homozygous rhodopsin (Rho) knock-out (KO) mice do not develop outer segments (OS) and show a progressive retinal degeneration (Humphries et al., 1997; Lem et al., 1999). By contrast, the heterozygous Rho KO mice do not display overt retinal degeneration, but show alterations in their photoresponses, OS morphology and have smaller disk membranes within their OS (Makino et al., 2012; Rakshit and Park, 2015).
Table 2. RHO mutations associated with an arRP phenotype.
| Mutation | Notes | References |
|---|---|---|
| E150K | Found in consanguineous Indian, Pakistani and Turkish families1-4. Heterozygous carriers appear normal2. Knock-in mouse model suggests mild dominant mutation5. |
1Kumaramanickavel et al., (1994) 2Azam et al., (2009) 3Saqib et al., (2015) 4Van Schil et al., (2016) 5Zhang et al. (2013) |
| W161ter | Found in non-consanguineous Indonesian families - heterozygous carriers appear normal6. Mutant transcript subject clearance by nonsense-mediated decay (NMD)7. |
6Kartasasmita et al., (2010) 7Roman-Sanchez et al., (2016) |
| E249ter | Found in consanguineous French-Canadian family8. Asymptomatic heterozygous carriers show slightly depressed ERGs9. Mutant transcript subject to clearance by nonsense-mediated decay (NMD)7,10. |
8Rosenfeld et al., (1992) 9Rosenfeld et al., (1995) 10Hernan et al., (2011) |
| M253I | Found in patient of consanguineous Turkish descent - heterozygous parents were asymptomatic11. M253I was found in the heterozygous state in 5 out of 35 LCA patients in Canadian study12. |
11Collin et al., (2011) 12Mezer et al., (2006) |
1.2.3. adRP
Currently more than 150 documented missense/nonsense RHO mutations are associated with an adRP phenotype (Table 1). Collectively, they are the most common cause of adRP, accounting for 20-30% of all cases (Sullivan et al., 2006). Whilst the P23H rod opsin mutation is arguably the most studied, it also likely represents a founder effect (Farrar et al., 1990), appearing to be confined to North America and seemingly accounting for ~10% of adRP cases in the USA in patients with a western European origin (Sullivan et al., 2006). The null alleles for arRP and lack of aggressive retinal degeneration in heterozygous Rho KO mouse models (Humphries et al., 1997; Lem et al., 1999), suggest that haploinsufficiency is not the disease mechanism and that dominant mutations are typically gain-of-function or dominant-negative mutations. Several attempts have been made to classify RHO mutations on the basis of (a) their clinical manifestation e.g. (Cideciyan et al., 1998; Gal et al., 1997; Krebs et al., 2010), and (b) their biochemical and cellular behaviour e.g. (Kaushal and Khorana, 1994; Krebs et al., 2010; Mendes et al., 2005; Rakoczy et al., 2011; Sung et al., 1991). Cideciyan et al. (1998) developed a broad phenotype-genotype correlation of adRP RHO mutations consisting of two classes (Table 3). Clinical classifications typically have two major classes: class A that has early onset severe rod dysfunction; whilst class B is normally associated with a later onset, less severe phenotype with a slower progression (Table 3). The difference in rates of presentation are likely related to the underlying biochemical and cellular defects associated with the mutation, but other factors such as genetic modifiers and environment can also affect the presentation of a condition and/or lead to interfamilial variability (Iannaccone et al., 2006). Examples of known modifiers include different alleles of Rpe65 and light exposure, which are discussed in depth later. Here we have collated the available biochemical and cellular information to suggest potential classes for each dominant mutation (Table 1, Figure 2); however, given that RHO mutations cause RP through multiple potential mechanisms, this classification is not mutually exclusive, with some mutants potentially leading to consequences that are applicable to more than one class.
Table 3. Clinical classification of rhodopsin RP mutations.
Class A phenotype refers to a severe early-onset loss of rod function. Class B refers to a milder phenotype, with retained rod function at least in certain retinal regions.
| Phenotype | Mutation | Reference |
|---|---|---|
| Class A | L88P1, L131P1, R135L/G/W2, E181K2, Q344P1, V345L2, P347L2 |
1Audo et al., (2010) 2Cideciyan et al., (1998) 3Iannaccone et al., (2006) |
| Class B | N15S1, T17M2, P23H2, T58R2, Q64ter2, V87D2, G89D2, G106R2, P180A3, G188R3, D190G/Y2, T193M2, M207K2, Q312ter2, T342M2, Q344ter2 |
1Audo et al., (2010) 2Cideciyan et al., (1998) 3Iannaccone et al., (2006) |
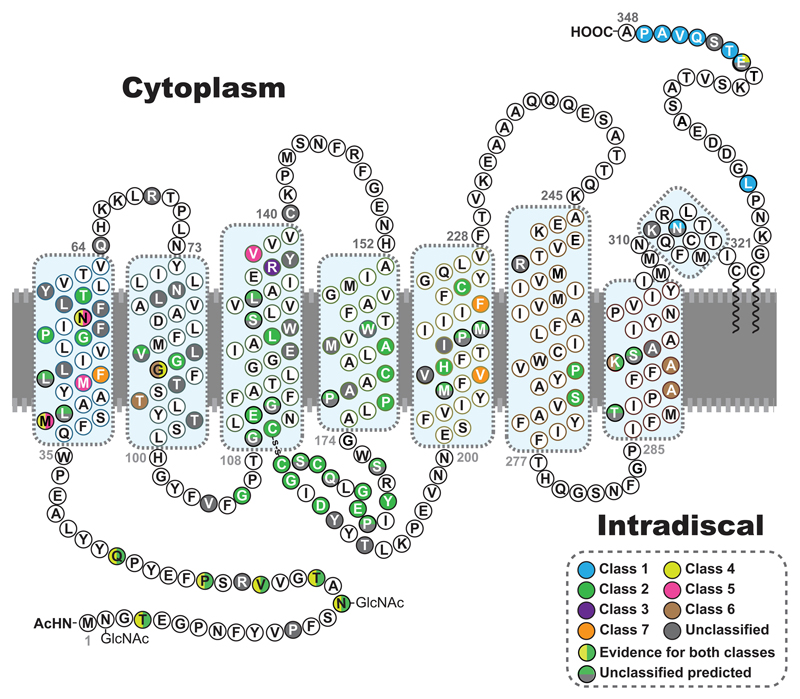
Figure 2. The position of rhodopsin mutations.
The primary amino acid sequence of human rhodopsin is shown as a secondary structure schematic to show the position of different classes of mutation. Class 1, post-Golgi trafficking and outer segment targeting (blue). Class 2 misfolding, ER retention and instability (green). Class 3, disrupted vesicular trafficking and endocytosis (violet). Class 4, altered post-translational modifications and reduced stability (yellow). Class 5 altered transducin activation (pink). Class 6 constitutive activation (brown). Class 7 dimerization deficiency (orange). Where there is evidence for more than one type of class it is shown with a vertical colour split. Uncategorised mutations, or those with no biochemical or cellular defects are shown in grey. Those with predicted effects on folding or binding from FoldX (Rakoczy et al., 2011) are shown with a horizontal colour split. The 8 alpha helix motifs of rhodopsin are highlighted by blue boxes.
1.3. Variation in the RHO gene, when is a variant a mutation?
Given that RHO was the first gene in which RP causing mutations were identified (Dryja et al., 1990a; Dryja et al., 1990b), it received intensive screening in RP cohorts across the globe, and it was thus tempting to assume that all variants in RHO identified in individuals with RP were likely pathogenic. Caution needs to be used, however, as some changes could be benign, especially where evidence of familial transmission is absent, such as in isolated patients, where there is no family history, or familial cases where other individuals in a pedigree have not been assessed and genotyped. The collection of data on human genetic variation has enabled researchers to investigate the frequency of nucleotide changes on an as yet unprecedented global scale (Lek et al., 2016). We used the Genome Aggregation Database (gnomAD; http://gnomad.broadinstitute.org/), which contains 123,136 exome sequences and 15,496 whole-genome sequences from unrelated individuals and excludes patients with severe paediatric disease, to determine the allele frequencies for potential missense/nonsense RHO mutations and compared against the currently catalogued RHO mutations. Whilst presence in the gnomAD dataset does not exclude the possibility of late onset disease causing alleles being present, based on the prevalence of adRP associated with RHO mutations in the range between 1:20,000 and 1:150,000, we estimate that a fully penetrant adRP causing RHO variant cannot be more common than 1:20,000 (0.0005) and would more likely have a minor allele frequency of <1:80,000 (<0.0000125), otherwise they would be very common disease associated alleles. These analyses revealed that missense changes that had previously been reported as potential RHO ‘mutations’ such as G51A (Cideciyan et al., 1998; Macke et al., 1993), V104I (Macke et al., 1993), A333V (Eisenberger et al., 2013), and T340M (Clark et al., 2010; Stone, 2003), are almost certainly benign single nucleotide polymorphisms (SNPs) with allele frequencies of between 1:1000 (G51A) and 1:10,000 (A333V). As a control, the T320N substitution that has been shown not to segregate with adRP (Mandal et al., 2005) exhibits a gnomAD frequency of 1:10,000. Therefore, we have excluded these five substitutions from our classifications. Whilst the presence of rare RHO variants in the general population (italicised variants in Table 1) questions their potential to cause fully penetrant aggressive adRP, they could still be associated with very mild late onset disease, arRP, or potentially act as disease modifiers. Nevertheless, these data reinforce the need for thorough genetics, such as segregation analyses, and in depth functional analyses to confirm pathogenicity.
2. Structural and biochemical basis of rhodopsin RP mutants
Rhodopsin is a member of the class A family of GPCRs. Upon activation by light, rhodopsin couples with the G protein transducin to initiate the first step in vision. Rhodopsin is synthesised in the rod cell inner segments (IS) and transported to rod OS, where it is densely packed into stacks of membranous discs. Fully folded rhodopsin adopts a seven TM helical bundle configuration in these disc membranes (Figure 1). The N-terminus resides in the interior of the rod discs, while the C-terminus is in the cytoplasm of the OS. Rod opsin combines with its 11-cis-retinal ligand to form the dark-state rhodopsin pigment (visible λmax 500 nm).
Biochemical examination of rhodopsin RP mutants have largely been undertaken by expressing them in cell culture model systems, such as COS-1 and HEK293 cells. Such studies led to the initial identification of two major classes of mutant, class I and class II (Kaushal and Khorana, 1994; Sung et al., 1991). Upon recombinant expression, class I mutants formed rhodopsin pigment and reached the cell surface; whereas, class II mutants misfolded, formed rhodopsin pigment poorly, if at all, and were retained fully or partly within the endoplasmic reticulum (ER). Upon further examination using more relevant animal model systems (Table 4), class I mutants were found to be defective in trafficking to the OS, caused by a change to the C-terminal OS trafficking motif. This classification system was expanded to six groups based on additional criteria, such as thermal stability, transducin activation capability, constitutive activation and receptor endocytosis (Mendes et al., 2005). Here we have updated this classification and expanded to include a new class of rhodopsin mutation (Table 1).
Table 4.
Commonly used mammalian models of rhodopsin RP (KO = knock-out; KI = knock-in; Tg = transgenic).
| Mutation | Species | Methodology | Reference(s) |
|---|---|---|---|
| Rho KO | Mouse | Homologous recombination | Humphries et al., (1997) Lem et al., (1999) |
| T4R | Dog | Spontaneous | Kijas et al (2002) |
| T17M (Tg) | Mouse | Human Transgene | Li et al (1998) |
| V20G-P23H-P27L (VPP) (Tg) |
Mouse | Mouse Transgene | Naash et al., (1993) |
| P23H (Tg) | Mouse | Human Transgene | (Olsson et al., 1992) |
| P23H (KI) | Mouse | Homologous recombination | Sakami et al., (2011) |
| P23H-GFP (KI) | Mouse | RHO-GFP Homologous Recombination |
Price et al., (2011) |
| P23H (Tg) | Rat P23H Line 1 (P23H-1) P23H Line 2 (P23H-2) P23H Line 3 (P23H-3) |
Mouse Transgene (1 high copy=fast) (3=slow) |
Pennesi et al., (2008) |
| P23H (Tg) | Pig | Human Transgene | Ross et al., (2011) |
| G90D (Tg) | Mouse | Mouse Transgene | Sieving et al., (2001) |
| E150K (KI) | Mouse | Homologous recombination | Zhang et al (2013) |
| S334ter (Tg) | Rat S334ter Line 3 (S334ter-3) S334ter Line 4 (S334ter-4) S334ter Line 5 (S334ter-5) S334ter Line 7 (S334ter-7) S334ter Line 9 (S334ter-9) |
Mouse Transgene | Pennesi et al., (2008) |
| Q344ter (Tg) | Mouse | Mouse Transgene | Sung et al., (1994) |
| P347L (Tg) | Rabbit | Rabbit Transgene | Kondo et al., (2009) |
| P347L (Tg) | Pig | Pig Transgene | Petters et al., (1997) |
| P347S (Tg) | Mouse | Human Transgene | Li et al., (1996) |
| P347S (Tg) | Pig | Pig Transgene | Kraft et al., (2005) |
2.1. Structural and biochemical insight into rhodopsin RP mutants
It is unclear why so many, often single point (missense), mutations in rhodopsin have such a large impact on folding of rhodopsin (class 2) and ultimately trigger such deleterious outcomes for rod cell structure and viability. Wild-type (WT) rhodopsin is an exquisitely evolved photon-capturing device that has very low background noise. As such, the intricate nature of the interactions that keep the TM helical bundle intact, yet permit full receptor activation upon absorption of a single photon by the retinal chromophore moiety, imply a delicately balanced two state machine. As vision, and especially scotopic vision, is adapted to operation in low light, it might be that rhodopsin folding and the photoreceptor ER quality control system also evolved to allow only correctly folded receptors to reach the light sensing OS destination. In the case of adRP, the very high expression levels and biosynthesis rates of rhodopsin could lead to the production of considerable amounts of even mildly defective protein (25% of total rod cell protein). The requirement for this scale of enhanced folding or degradation might be too demanding for the rod cell to manage, ultimately leading to a breakdown of proteostasis. For these reasons, it is important to understand the differences in severity of RP (age of onset, extent and rate of degeneration) of different point mutations and correlate these with their biochemical and cellular consequences. It is also important to understand the precise molecular basis of the structural defects brought about by the different mutants. It is challenging to collect structural data for mutants that are, by definition, misfolded; however, such insight could be necessary for the development of therapeutic approaches that target the misfolded mutant receptors directly.
2.2. The distribution of RP mutants in rhodopsin
RP mutations are located throughout the rod opsin apoprotein (Figure 2). The molecular mechanisms of disease can be linked to different types of receptor malfunction.
2.2.1. RP mutations located in the N-terminus of rhodopsin: the role of the N-terminal cap
In addition to P23H, other mutations located in the N-terminus of rhodopsin (residues M1 to Q35) include T4K, P23A/L, N15K, T17M, V20G and Q28H. The mutations in this N-terminal segment are sometimes associated with a relatively mild disease, with RP developing in later life and with slowly advancing symptoms (Table 3). Interestingly these mutations are often associated with sector RP, whereby retinal damage is more prevalent at light-exposed regions (sectors) of the retina (Jacobson et al., 1991; Kemp et al., 1992; Krill et al., 1970; Ramon et al., 2014; Van Woerkom and Ferrucci, 2005). The pattern of dystrophy in these sector mutants suggests a role of light exposure that is supported by biochemical data for non-N-terminal mutants e.g. N55K (Ramon et al., 2014). Therefore, these N-terminal mutants might not exert their deleterious effects entirely though protein misfolding in the ER, as indicated by early cell culture based experiments; however, light could also affect their biogenesis through altering the availability of 11-cis-retinal, which could otherwise improve mutant receptor biogenesis (Tam and Moritz, 2007).
The crystal structure of rhodopsin revealed how the N-terminal segment of rhodopsin folds into a cap-like structure that covers the second extracellular loop (EL2) ‘plug’ and is in contact with EL1 and EL3 (Figure 1) (Li et al., 2004). Unlike most GPCRs that bind their ligands via an ‘open’ extracellular domain, rhodopsin and other visual receptors acquire the retinal ligand via distinct transient pores that open between the TM helices. This has allowed the extracellular (intradiscal) domain to evolve such that it contributes to the thermal stability of rhodopsin as well as the receptor activation process (Ahuja et al., 2009). This idea is supported by the observation that further stabilising the interaction between the N-terminal cap and the EL loops by engineering in an additional disulphide bond between residues N2(C) and D282(C) increases the thermal stability of the pigment and makes possible the isolation of the rod opsin apoprotein in detergent (Xie et al., 2003).
Early experiments relied upon expression of rod opsin mutants in cell culture models (COS-1, HEK293) for preparation of mutant pigments (Kaushal and Khorana, 1994; Sung et al., 1991). In these experiments transfected cells were treated with the 11-cis-retinal ligand during purification, and prior to assessment of pigment formation capability. These results suggested that the N-terminal mutants were members of class 2 (misfolded, retained in the ER). Subsequent experiments using transfected cells treated with 11-cis or 9-cis retinal during expression revealed that N-terminal mutants could in fact form pigments, if protein misfolding was rescued by the ligand functioning as a pharmacological chaperone (Krebs et al., 2010; Li et al., 1998a; Mendes and Cheetham, 2008; Noorwez et al., 2003; Saliba et al., 2002). We later showed that all the RP mutants in the N-terminus could be rescued by this procedure; however, all the pigments produced were defective and displayed much lower levels of thermal stability and had altered photobleaching and signal transduction behaviour (Opefi et al., 2013). Furthermore, a subset of these mutants could be repaired in the N2C/D282C rhodopsin background. That is, they could form pigment after completion of expression and by post-harvest treatment of the transfected cells with 11-cis-retinal. Additionally, these repaired mutant pigments had close to WT-like properties, but still showed decreased stability. Taken together, these results suggest that the N-terminal cap is needed to stabilise the rod opsin protein and the pigment, but it also plays a role in the rhodopsin photocycle and signalling through transducin. One explanation for the functional role of the N-terminal cap lies in its close proximity to the extracellular loops and extracellular ends of the TM helices. Solid state magic angle spinning (MAS) nuclear magnetic resonance (NMR) experiments have revealed changes in hydrogen bond interactions between TM6 and EL2 take place during light activation of rhodopsin (Kimata et al., 2016). The N-terminal cap positioned over the EL2 could be required for the correct orientation of EL2 during the photoactivation process.
In a separate study of rod opsin expression in transgenic C. elegans nervous system, the P23H protein was expressed at low levels, due to degradation and aggregation (Chen et al., 2014). The P23H could be partially rescued with 9-cis-retinal and form a purified pigment that had WT-like photocycle and signal transduction properties; however, the P23H mutant pigment formed slowly and was unstable (Chen et al., 2014). The capacity for pharmacological chaperone rescue with retinal suggests that in photoreceptors, in the presence of 11-cis-retinal, it might be stabilised and sufficiently folded to leave the ER and reach the rod OS. It is not clear from animal model studies, however, when 11-cis-retinal is inserted during rod opsin biogenesis. The possibility exists that if P23H pigment can reach the rod OS, even if in small amounts, it could disrupt the integrity of discs though its inherent instability (see sections 3.4 and 5.1.1). This scenario is reminiscent of that described for class 4 mutants where receptor instability in the OS is the most likely explanation for cytotoxity (Mendes et al., 2005). Thus, we can consider class 2 mutant receptor sub populations where folding is rescued by pharmacological chaperone 11-cis-retinal to gain a class 4 mutant phenotype.
2.2.2. RP mutations in the transmembrane helices
Rhodopsin RP mutants have been described in each of the seven TM segments (Figure 2). Many of these are point mutations that bring about the introduction of a charged amino acid into the TM segment, for example, L40R, L46R, G51R, P53R and T58R in TM helix 1 (Table 1). In most cases, it is likely that the presence of the basic (or acidic) residue is preventing insertion of the domain into the ER membrane and thus resulting in the type 2 misfolding phenotype often associated with such mutations.
More recent investigation into mutations located in TM1 focused on M39R and N55K associated with sector RP (Ramon et al., 2014). These mutants were expressed at lower levels in COS-1 cells and their pigment profiles indicated some degree of misfolding and decreased thermal stability. These pigments behaved differently in response to light, properties that could explain the sector RP pathology.
RP mutations have also been found in the 11-cis-retinal attachment site K296 (reviewed in (Park, 2014), as well as the K296 PSB counterion E113. Substitutions at K296 have been proposed to cause RP due to the close association of its constitutively phosphorylated form with arrestin, similar to class 3 mutants, and not due to activation of transducin (Park, 2014). The K296E mutant, however, also misfolds and is retained in the ER on heterologous expression (Saliba et al., 2002), suggesting a partial class 2 phenotype. The recently identified E113K mutation has been shown to result in moderately progressive RP, as well as CSNB, in the same family (Reiff et al., 2016). The relatively mild phenotypes brought about by the E113K mutation are difficult to reconcile with previous cell culture and biochemical studies. When the E113K mutant was expressed using COS-1 cells it was unable to form rhodopsin pigment and had abnormal mobility on SDS-PAGE, both hallmarks of a misfolded receptor of the class 2 mutants (Sakmar et al., 1989). One possible explanation for this paradox is that if 11-cis-retinal is present during folding and functions as a pharmacological chaperone to rescue E113K folding (as described in section 2.2.1) then there could be a complete or near complete rescue. The E113K pigment that is formed might be constitutively active and lead to rod dysfunction similar to class 6 CSNB mutants.
RP mutants in the TM helices could also give rise to the loss of side chains required for structure and function (e.g. H211P) or introduce bulky side chains into regions of tight helix packing. Crystal structures reveal the 7TM bundle of rhodopsin contains both tightly packed and more loosely packed helices (Sanchez-Reyes et al., 2017). Two separate interior packing clusters (1&2) were identified that mediate interactions between TM helices H1 and H2 or H3 and H4 respectively. These packing clusters are conserved in GPCRs and are composed of amino acids with small side chains. The residue A164 is a component of packing cluster 2 and presumably its mutation to a more bulky valine (A164V) could disturb rhodopsin folding and stability by disrupting formation of this packing core leading to misfolding (Sanchez-Reyes et al., 2017; Stojanovic et al., 2003).
2.3. Structural studies of CSNB mutants
Class 2 rhodopsin RP mutants have not been amenable to structural biology methods, such as X-ray crystallography, or NMR, because they are misfolded; however, insight into the molecular defects of rhodopsin mutations can be obtained by the study of mutant pigments where folding is not affected. For example, Singhal et al. (2016), collected X-ray diffraction data from crystals of the rod opsin and light-activated forms of CSNB mutants, G90D and T94I. They found that a new salt bridge was formed between the aspartate residue in G90D and the K296 PSB retinal attachment site. This new salt bridge was proposed to be the reason for the increased basal activation in the dark of this mutant. By contrast, in T94I the introduced hydrophobic side chain prolonged the lifetime of the metarhodopsin II (meta II) state by establishing a contact with K296 (Singhal et al., 2016). Therefore, both mutations alter the dark state by weakening the interaction between the PSB and its counterion E113, a concept that could also explain increased basal receptor activity for other CSNB mutants. It is important to understand why such mutations do not go on to cause the degenerative RP phenotype, or do so very slowly. One possibility is that signalling defects of this nature give rise to mild changes in rod cell physiology. Another possibility is that they are so mild that rod cell degeneration is very slow and is affected by other genetic and environmental modifiers.
2.4. Dimerization deficiency in adRP mutants
The ability for rhodopsin to form higher order oligomers in animal tissue was demonstrated by atomic force microscopy (Fotiadis et al., 2003). These observations have subsequently been supported by various biochemical and biophysical means (Fotiadis et al., 2003; Jastrzebska et al., 2015; Kota et al., 2006). Furthermore, rows of dimers have been observed by cryo-electron tomography in dark adapted mouse photoreceptors (Gunkel et al., 2015). The role of these oligomeric rhodopsin structures in photoreceptor function remains controversial, but there is evidence that dimeric and multimeric interactions might begin during folding in the ER, as the biogenesis of mutant protein can affect the WT (Mendes and Cheetham, 2008; Rajan and Kopito, 2005; Saliba et al., 2002). Recently, three previously reported RHO mutations that did not have a documented biochemical phenotype were examined for their ability to perform phospholipid scrambling and form dimers (Ploier et al., 2016). These amino acid positions (F45, V209 and F220) are all located at the surface of TM1 or TM5 (Figure 2). The authors report that the variants (F45L, V209M and F220C) behaved as monomers, unlike the WT protein which formed dimers or multimers, suggesting that these mutations potentially lead to RP through an inability to dimerise (Ploier et al., 2016). We propose this defect could be classified as class 7 (Table 1). Variants at these three residues, however, are present in the general population in gnomAD at relatively high frequencies (F45L, 0.000022; V209M, 0.000012; F220C 0.00002 and F220L 0.000049). Furthermore, genetic studies have shown that F45L, V209M and F220L did not segregate with disease in families (Davies et al., 2012; Dryja et al., 2000; Lim et al., 2009), which suggests that they are not disease causing mutations. Therefore, at present, the significance of these residues and their effect on rhodopsin oligomerisation and lipid homeostasis in rhodopsin RP remains uncertain.
3. Rhodopsin biogenesis, post-translational modifications and trafficking
Rhodopsin is co-translationally translocated into the rod photoreceptor ER and trafficked to the OS, via the Golgi apparatus on membranous vesicle carriers (Figure 3). Early in its biogenesis, the N-terminus of rhodopsin becomes glycosylated at two asparagine residues: N2 and N15 (Figure 1 and 2). For many proteins, glycosylation is necessary for the binding of ER chaperones (typically lectins) to assist protein folding and targeting misfolded species for ER-associated degradation (ERAD) (Helenius and Aebi, 2001). Although glycosylation is not required for WT rod opsin to fold sufficiently to exit the ER, glycosylation is essential for mutant rhodopsin ERAD (Saliba et al., 2002). Furthermore, a study of transgenic frogs found that non-glycosylated rhodopsin (N2S/N15S) did not accumulate in the ER and trafficked to the OS, similar to T4K, T4N and T17M (Tam and Moritz, 2009). The expression of these mutants caused retinal degeneration in a light dependent manner, highlighting the importance of glycosylation at N15 for photoreceptor survival (Tam and Moritz, 2009). In addition, monoglycosylated rhodopsin is expressed at high levels and trafficked to the OS in the T4R rhodopsin dog retina, and also causes a light dependent degeneration (Zhu et al., 2004). Glycosylation is important for rhodopsin function, non-glycosylated mutants activate transducin poorly (Kaushal et al., 1994), and expression of a non-glycosylated mutant rod opsin in mouse rods led to very low levels of expression and OS that did not form (Murray et al., 2015a).
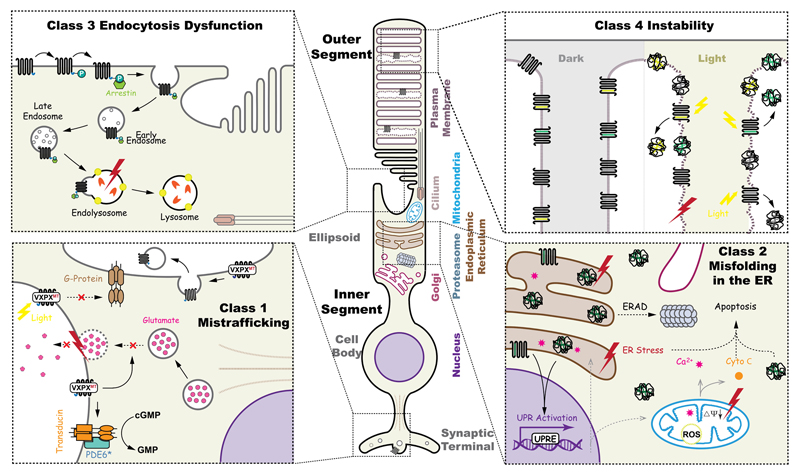
Figure 3. Schematic illustration of the potential pathogenic consequences caused by rhodopsin mutations.
A rod photoreceptor has distinct regions; including, the outer segments (OS) and inner segments (IS), cell body and synaptic terminals. Pathogenic mutations in rhodopsin disturb several cellular pathways, including endocytosis dysfunction, structural instability of the OS, mistrafficking, and misfolding. Top left, class 3, hyperphosyphorylated rhodopsin is bound by arrestin (green), the Rho-arrestin complex is endocytosed and disrupts vesicular traffic. Top right, upon illumination, unstable rhodopsin mutants (class 2, 4 and 7) could aggregate in the disk of OS, thus causing damage of the plasma membrane homeostasis. Bottom left, class 1 mutations that affect the ciliary targeting signal, VXPX, are mislocalised, including at the synapse, where they could inhibit synaptic vesicle fusion or be abnormally activate transducin. Misfolding class 2 rhodopsin mutants in the ER might induce ER stress, degraded by ERAD and potentially aggregate, leading to the activation of pro-apoptotic pathways.
In addition to glycosylation, rhodopsin is palmitoylated at the cysteine residues C322 and C323 (Ovchinnikov et al., 1988). This modification anchors the C-terminal tail to the disc membrane and loss of palmitoylation alters transducin activation (Wang et al., 2005). The formation of a disulphide bond between C110 (at the base of TM3) and C187 (in EL2) connects TM3 and TM5 and is structurally essential for rhodopsin (Hwa et al., 1999; Karnik et al., 1988; McKibbin et al., 2007). This modification helps maintain the structure of rhodopsin in its correct conformation throughout its synthesis and transport to the OS (Murray et al., 2009), and is important for the stability of the meta II structure and its coupling to transducin activation (Davidson et al., 1994). Misfolded rhodopsin due to mutations in the intradiscal domain are characterized by the formation of an incorrect disulphide bond between C185 and C187, as opposed to the correct and highly conserved C110–C187 disulphide bond (Athanasiou et al., 2014; Hwa et al., 1999; Liu et al., 1996). Rhodopsin has an OS targeting signal located within its cytoplasmic C-terminal tail, the VXPX motif (Sung and Chuang, 2010; Tam et al., 2000). Deficiencies in this motif lead to mislocalisation of rhodopsin via mistrafficking and rod degeneration (Sung et al., 1994; Tam et al., 2000).
3.1. Misfolding, ER retention and instability
As described earlier, studies from cell culture models have shown that misfolded P23H rhodopsin is retained in the ER, suggesting the existence of an ER mechanism that does not permit further traffic of misfolded rhodopsin (Athanasiou et al., 2014; Athanasiou et al., 2012; Kaushal and Khorana, 1994; Kosmaoglou et al., 2009; Sung et al., 1991). Animal models of class 2 mutants (Table 1 and Table 4), including P23H, also show ER retention and degradation of mutant rhodopsin accompanied by photoreceptor cell death; including Drosophila melanogaster (Wang and Montell, 2007), Xenopus laevis (Tam and Moritz, 2006, 2009), mice (Frederick et al., 2001; Olsson et al., 1992), rats (Lee et al., 2003) and pigs (Ross et al., 2012). More recently, P23H KI mice were generated with the same gene dosage as in adRP, in which at least 90% of the P23H was degraded and the remainder that exited the ER was found in structurally disorganised OS (Chiang et al., 2015; Sakami et al., 2011).
Rhodopsin misfolding and ER retention has been shown to cause ER stress (Lin et al., 2007). Accumulation of unfolded proteins in the ER causes ER stress and cells respond to ER stress by activating the unfolded protein response (UPR), a signalling network initiated by the ER-anchored receptors IRE1α, PERK and ATF6. The UPR aims to restore ER protein-folding homeostasis, but can turn into a lethal signal when the stress is too severe or prolonged, leading to cell death (Cao and Kaufman, 2012; Remondelli and Renna, 2017).
Increases in BiP (binding immunoglobulin protein; HSPA5) expression are a hallmark of UPR activation. In Drosophila, the levels of heat shock cognate protein-3 (Hsc3), which is the homologue of the mammalian ER Hsp70, BiP, were increased in flies expressing the class 2 mutants G93D, and P37H (the P23H equivalent) in rhodopsin (Rh1) and in flies that lack NinaA, a peptidyl-prolyl cis-trans isomerase chaperone that is required for Rh1 maturation (Griciuc et al., 2010b; Mendes et al., 2009). BiP mRNA levels were also highly elevated in HEK293 cells overexpressing P23H rod opsin, but not in cells expressing WT rod opsin (Lin et al., 2007). There was increased activation of the IRE1/XBP-1 UPR pathway, which could mediate the changes in BiP levels together with other branches of the UPR (Griciuc et al., 2010b; Lin et al., 2007; Mendes et al., 2009). Additionally, in P23H rats, BiP mRNA levels were elevated after photoreceptor differentiation, but declined when degeneration progressed and this change was accompanied by the induction of pro-apoptotic factors, such as CHOP, suggesting that the ER stress could not be overcome (Lin et al., 2007). Studies of the P23H KI mouse, however, showed lower levels of ER stress and efficient ERAD of the mutant protein (Chiang et al., 2015), suggesting that overexpression of rhodopsin might be a factor in the ER stress observed in these transgenic models.
P23H rhodopsin is targeted for ERAD and proteasomal degradation by the ubiquitin proteasome system (UPS), but if degradation fails then rhodopsin can aggregate to form inclusions that are ubiquitylated and recruit Hsp70 molecular chaperones (Illing et al., 2002; Saliba et al., 2002). Ubiquitylated inclusions, however, are rarely seen in photoreceptors in animal models in vivo, suggesting that aggregation is either infrequent or could lead to cell death before the formation of microscopically visible inclusions. Further evidence for a potential role of the proteasome in the pathogenesis of rhodopsin RP is that aggregation of P23H leads to a global impairment of proteasomal function, which could ultimately compromise the degradation of other misfolded protein substrates of the UPS (Bennett et al., 2005). Indeed, an accumulation of a ubiquitin, Ub(G76V)–GFP, proteasomal substrate reporter was observed in P23H mouse photoreceptors, whereas other mouse models of retinal degeneration that do not involve protein misfolding did not show any proteasomal overload, suggesting proteasomal insufficiency might contribute to pathogenesis in the P23H transgenic mouse model (Lobanova et al., 2013).
A potential type of dominant-negative effect of class 2 mutants at the level of the ER has also been observed, whereby the presence of the mutant protein interferes with the biogenesis and traffic of the WT protein. For example, co-expression of WT and P23H rhodopsin in vitro resulted in the formation of inclusions containing the WT protein (Mendes and Cheetham, 2008; Saliba et al., 2002), and co-aggregation of the mutant and WT protein with enhanced proteasome-mediated degradation of the WT protein (Rajan and Kopito, 2005). This property might not be shared by all rod opsin mutants, however, as the G188R mutant also aggregates, but has only small effects on the WT protein and they do not co-aggregate (Gragg et al., 2016). Therefore, mutations in rhodopsin that cause misfolding can lead to enhanced mutant, and WT, rhodopsin degradation; potential ER stress; aggregation and alter photoreceptor homeostasis (Figure 3).
3.2. Post Golgi trafficking and OS targeting
Point mutations at the C-terminus of rod opsin (e.g. V345M, P347S) and C-terminal truncations (e.g. Q334ter) have been reported to affect OS transport (Sung et al., 1994; Tam et al., 2000), and are regarded as class 1 (Table 1, Figure 2). As discussed earlier, class 1 mutants have no obvious defects in heterologous cell culture expression models; however, expression in photoreceptors revealed their striking effect on post ER trafficking. The majority of class 1 mutants are clustered at the rhodopsin C-terminus (Sung et al., 1993; Sung et al., 1994; Tam et al., 2000). The integrity of the rhodopsin C-terminal OS targeting signal VXPX-COOH is important for photoreceptor survival, as mutations in this highly conserved motif cause severe forms of RP (Table 3) (Berson et al., 2002). The expression of C-terminal truncations of bovine rhodopsin in HEK293 cells showed that the proximal region (311-316) of the carboxyl terminus is critical for the proper folding and stability of the rhodopsin molecule, but that amino acids 316 to 348 were not necessary for the activation of transducin (Weiss et al., 1994). Different studies in transgenic mice, rats, pigs and frogs carrying mutations in this C-terminal sorting signal resulted in mistrafficking, leading to the accumulation of rhodopsin at the IS, plasma membrane and synapses (Concepcion and Chen, 2010; Green et al., 2000; Li et al., 1998b; Sung et al., 1994; Tam et al., 2000). In frogs, rhodopsin without the VXPX motif cannot bind to ADP Ribosylation Factor 4 (Arf4), a small G protein important for packaging the rhodopsin transport carrier (RTC) vesicles in the IS (Deretic and Wang, 2012). Consequently, RTC packaging and trafficking becomes compromised, leading to accumulation or mistrafficking to IS structures, including endosomes, lysosomes, and the Golgi apparatus. Arf4 recruits other trafficking components, such as ASAP1 (ArfGAP with SH3 Domain Ankyrin Repeat and PH Domain 1), Rab11, Rabin8, and Rab8, to the RTCs. Defects in the coordination of these trafficking components can contribute to rhodopsin mislocalisation (Wang et al., 2012). The role of Arf4 in rhodopsin transport has been challenged, however, by a recent study of Arf4 deletion in mouse photoreceptors, which showed no protein mislocalisation or retinal degeneration (Pearring et al., 2017). Instead, it was suggested that the IFT20 subunit of the intraflagellar transport (IFT) particle was important for rhodopsin sorting, or transport from the Golgi to the OS in mammals, as inducible knockout of IFT20 in mouse photoreceptors caused rhodopsin accumulation in the Golgi membranes (Keady et al., 2011).
The presence of a large quantity of mutant rhodopsin is likely to significantly alter the physical properties, the composition, and the function of the affected membranes in different photoreceptor compartments (Figure 3) (Deretic, 2006). It has been suggested that accumulation of truncated rhodopsin in various ectopic membrane locations can be toxic and contribute to photoreceptor cell death by disrupting synaptic transmission, overloading the degradation machinery or decreasing the availability of functional proteins important for the phototransduction process in the regions where the aberrant rhodopsin is concentrated (Alfinito and Townes-Anderson, 2002). For example, mislocalised Q344ter can ectopically initiate signalling in the IS and outer nuclear compartments (Concepcion and Chen, 2010), and prolonged photoreceptor responses due to loss of phosphorylation sites results in an S334ter mutant results in abnormal rhodopsin deactivation and might lead to OS shortening and photoreceptor cell death (Lee and Flannery, 2007). In contrast, expression of a K296R substitution, which blocks the ability of rod opsin to bind chromophore and activate transducin, in combination with Q344ter in Xenopus laevis did not alter the rate of degeneration, suggesting that the effects of mislocalised rhodopsin are not mediated by the activated form of rhodopsin (Tam et al., 2006). Therefore, the mechanisms of mislocalised rhodopsin mediated cell death are currently unclear.
3.3. Disrupted vesicular traffic and endocytosis
In addition to the effect of C-terminal mutations on rhodopsin traffic, other mutations can affect vesicle traffic. In particular, inherited amino acid substitutions at R135 in rhodopsin cause a severe and fast progressing form of RP (Table 1 and 3) (Iannaccone et al., 2006). This residue is conserved in all rhodopsin (class A) family GPCRs, and it has been suggested to play an important role in the regulation of G protein signalling. Mutations at this residue form their own class (class 3), as they have distinct biochemical and cellular defects compared to the other characterised rhodopsin mutants (Aguila et al., 2014; Mendes et al., 2005; Chuang et al., 2004). In particular, the R135L mutant appears to fold normally and exit the ER, but similar to some of the constitutively active mutants, it is constitutively phosphorylated and binds with a high affinity to visual arrestin in the absence of chromophore (Aguila et al., 2014; Chuang et al., 2004). The R135L rhodopsin recruits cytosolic arrestin to the plasma membrane, and the rhodopsin-arrestin complex is internalized into the endocytic pathway, leading to an accumulation in the photoreceptor IS and preventing traffic to the OS (Figure 3). Rhodopsin class 3 mutants might cause disease by disrupting the dynamic interplay between different endocytic compartments and rendering them dysfunctional. The resulting imbalances, for example in iron and nutrient levels, might cause subsequent photoreceptor degeneration (Song and Dunaief, 2013); however, as yet there are no good animal models of class 3 mutants, so the precise mechanisms remain to be elucidated.
3.4. Altered post-translational modifications and reduced stability
The class 4 group of mutations (Table 1) do not appear to directly affect rhodopsin folding at physiological expression levels, but instead affect rhodopsin stability and posttranslational modifications. A naturally occurring dog model of RP caused by a T4R substitution (Table 4) disrupts glycosylation at N2 (Kijas et al., 2002). These dogs are extremely sensitive to light-mediated retinal damage and exhibit high levels of the AP-1 apoptosis marker after acute exposure to light (Cideciyan et al., 2005; Zhu et al., 2004). No evidence of UPR activation was observed in the canine T4R rhodopsin model, but Hsp70 transcription levels were upregulated in the light-exposed retinas, suggesting induction of the heat shock response (HSR); however, no differences in the protein levels of Hsp70 (or Hsp90) were observed (Marsili et al., 2015). This could reflect a relatively small increase in protein level, or that the stress led to cell death before the mRNA could be translated into protein.
Similarly, the T17M rhodopsin mutation abolishes glycosylation at the N15 site of rhodopsin and mouse models of T17M are also very susceptible to light damage (White et al., 2007). Studies in transgenic frogs of T4K and T17M mutations supported light activation as the cause of pathology, as abolishing photoexposure significantly reduced rod cell death (Tam et al., 2014). This highlights how environmental light exposure could influence disease progression in adRP patients. Overexpression models of T17M, however, share some of the characteristics of class 2 mutants with ER retention and induction of the UPR (Jiang et al., 2014b; Kunte et al., 2012). Interestingly, studies of P23H KI mice, P23H-GFP KI mice and in Xenopus have demonstrated that, similar to class 4 mutants, some P23H rhodopsin can escape the ER and be transported to the OS (Price et al., 2011; Sakami et al., 2011; Tam and Moritz, 2007; Tam et al., 2014). In the OS the mutant rhodopsin can undergo time dependent aggregation leading to vesiculo-tubular structures and destabilisation of rod photoreceptor disk membranes (Haeri and Knox, 2012; Haeri et al., 2013) that are exacerbated by light (Bogea et al., 2015) (Figure 3). Therefore, inhibition of rhodopsin glycosylation by T4R and T17M mutations, or a failure of the ER quality control machinery to retain class 2 mutant proteins, results in protein reaching the OS that is unstable and affected by exposure to light, leading to photoreceptor cell death. Mutations at these residues cause retinal degeneration predominantly in the lower hemisphere, consistent with light-induced damage from overhead sources (Jacobson et al., 1991; Kemp et al., 1992; White et al., 2007). Some sector RP mutants, such as N55K, could also fall into this class of mutations (Table 1), as they display apparently, normal folding but have inherent structural instability (Ramon et al., 2014). The effect of the oligomerisation mutants on OS stability is not currently clear, but it is possible that they could disrupt the higher order organisation of rhodopsin within discs and have similar effects on photoreceptor survival.
3.5. Altered transducin activation
The M44T and V137M RP associated variants share spectral and structural features with WT rhodopsin (Table 1), but have significantly increased transducin initial activation rates (Andrés et al., 2003), such that they are class 5 (Figure 2). It was suggested that these mutants have altered stoichiometric balance of the different proteins involved in the phototransduction biochemical reactions. These mutants, however, have no activity in the dark or absence of chromophore, suggesting that constitutive activity is not the basis of their pathogenesis (Park, 2014).
3.6. Constitutive activation
Class 6 mutants (Table 1, Figure 2) show constitutive activation of rod opsin in the absence of chromophore and in the dark. The constitutive activation of the phototransduction cascade has been suggested as a potential mechanism of cell death in RP. Intriguingly, the majority of rhodopsin mutations clustered around the chromophore binding pocket lead to constitutive activation, and are associated with rod dysfunction through CSNB, not rod cell death with secondary cone cell death in RP (Gross et al., 2003). For example, the G90D mutation has been described as causing CSNB (Sieving et al., 1995). Studies in vitro showed that the G90D mutant activates the visual cascade in the dark or without chromophore (Rao et al., 1994). Constitutive activity was also observed in the G90D transgenic mice and frogs, acting as if in the presence of continuous background light (Dizhoor et al., 2008). Interestingly, another mutation at the same G90 position, G90V, was reported to be associated with RP not CSNB (Toledo et al., 2011). Furthermore, the K296E mutation that causes RP can also activate transducin constitutively. This mutant was proposed to cause RP due to its ability to sequester cellular components needed for the shut-off process, such as arrestin, similar to class 3 mutants, rather than its constitutive activity (Chen et al., 2006); however, other reports suggest that K296E is also prone to misfolding and be retained in the ER, similar to class 2 mutants (Saliba et al., 2002; Tam and Moritz, 2006). G90V also affects the thermal stability of the protein (Toledo et al., 2011), suggesting some overlap with class 4 mutants.
Two other CSNB mutations leading to constitutive transducin activation are A292E and T94I (al-Jandal et al., 1999; Dryja et al., 1993). A292E and T94I constitutively activate transducin in vitro in the absence of chromophore, by disrupting the interaction between E113 and K296 that contributes to stabilisation of the protein in the inactive state. The T94I mutant has a long-lived meta II intermediate in comparison to the WT and the other CSNB mutants (Gross et al., 2003; Ramon et al., 2003). Aberrant activation of rhodopsin leads to the inability of the rod cell to respond to dim light, which manifests as night blindness; however, it appears that rod dysfunction alone is not sufficient to lead to rod cell death and RP.
4. Photoreceptor cell death
Rhodopsin RP mutations lead to photoreceptor cell death by a two-stage process. Firstly, the rod photoreceptors degenerate and this leads to the second stage, which is death of cone photoreceptors. Despite the availability of various animal models, the mechanisms underlying the eventual loss of cone photoreceptors and the loss of daylight vision remain largely unknown.
Classical apoptosis depends on activity of caspase-type proteases, with caspase-3 as the prototypic mediator and executioner of apoptotic cell death (Porter and Janicke, 1999). Caspase-independent apoptosis includes calpain and PARP (poly(ADP-ribose) polymerase) activity. Calpains are a group of calcium-activated proteases that have been implicated in neuronal degeneration in a number of different neuronal tissues, including the retina. Calpain activity may result in an increased production of reactive oxygen species (ROS) and accumulation of oxidative DNA damage by abolishing AIF (apoptosis-inducing factor) oxidoreductase activity and OGG1 (8-Oxoguanine glycosylase)-dependent DNA repair activity. In conjunction with ROS production, calpain activity has been suggested as a cause of lysosomal membrane disruption with the resultant leakage and activation of various hydrolytic enzymes, in particular cathepsins (Wang et al., 2009).
Early studies on RP proposed apoptosis as the final common pathway in retinal degeneration (Chang et al., 1993). While a number of publications suggested an involvement of apoptosis, a more recent study of P23H and S334ter rats identified the presence of non-apoptotic markers of cell death in these two rhodopsin models (Kaur et al., 2011). Both P23H and S334ter rats showed activation of calpain and PARP, associated with calpastatin down-regulation, increased oxidative DNA damage and accumulation of PARpolymers. Interestingly, activation of caspases-3 and -9 and cytochrome c leakage were observed only in the S334ter mutant, which also showed increased expression of PARP-1. These results suggested that downstream cell death mechanisms triggered by different genetic mutations, in different species, share a number of key components (Kaur et al., 2011). The involvement of calpains in retinal degeneration was also demonstrated after light damage in the T4R dog (Marsili et al., 2015).
Disruption of ER function affects other cellular pathways including oxidative stress (Bhandary et al., 2012), cytosolic Ca2+-release (Figure 3) (Kaufman and Malhotra, 2014) and inflammation (Garg et al., 2012). For instance, studies on the rhodopsin mutants T17M and E349X have shown significant up-regulation of pro-inflammatory markers associated with an ER-stress response (Rana et al., 2014).
Recent studies have shown that cellular necrosis can also contribute to cone and rod degeneration in animal models of retina degeneration (Murakami et al., 2012; Trichonas et al., 2010). Necrosis, which was traditionally thought to be an uncontrolled process of cell death, is now known to also have a regulated component (Holler et al., 2000; Matsumura et al., 2000). The receptor-interacting protein kinases (RIP), RIP1 and RIP3, have been identified as critical mediators of programmed necrosis (Vandenabeele et al., 2010). It has been suggested that the ER-stress response in the P23H-1 rat results in necroptosis in rod photoreceptors due to up-regulation of RIP1/RIP3 complexes and increased mitochondrial fission factor DRP1 and mitochondrial protein phosphatase PGAM5 activity, that subsequently leads to an influx of calcium into mitochondria to trigger necroptosis (Viringipurampeer et al., 2016). Rod photoreceptors undergoing necroptotic cell death could release their cellular contents acting as damage-associated molecular patterns (DAMPS), which leads to inflammasome activation in cones (Murakami et al., 2015). Studies of the P23H-1 rat model suggest that the secondary cone cell death occurs via pro-inflammatory mechanisms and it was proposed that activation of the NLRP3-dependent inflammasome could drive cone photoreceptor cell death in rhodopsin RP (Viringipurampeer et al., 2016).
In contrast, a study using four different mouse models of RP, including rhodopsin RP, suggested that cone cell death could also be the result of prolonged starvation, caused by activation of autophagy. Downregulation of the insulin/mTOR pathway, a key pathway that regulates cellular metabolism, was observed during the period of cone degeneration leading to the suggestion that glucose uptake and/or the intracellular levels of glucose may be compromised in cones in RP (Punzo et al., 2009). A recent study in mice suggested that loss of mTOR does not affect cone survival; however, deregulation of this pathway in diabetes decreased cone function (Ma et al., 2015). Therefore, although the mechanisms of rod and cone cell death in rhodopsin RP remain to be fully defined and might vary for different mutations, the opportunity exists to intervene and preserve vision.
5. Therapeutic approaches to rhodopsin RP
Photoreceptors require a stringent regulation of proteostasis for their function and viability. Therefore, their vulnerability to the toxic gain-of-function effects of mutant rhodopsin could reflect their long-term inability to maintain proteostasis in the presence of this additional stress. Proteostasis mechanisms have been a therapeutic target in several studies which aimed to correct misfolding, reduce rhodopsin aggregation, enhance the degradation of mutant rhodopsin, stimulate autophagy or enhance cell survival. Manipulation of these mechanisms can be achieved by either the use of drugs that restore proteostasis (i.e. pharmacological chaperones, kosmotropes, molecular chaperone- or autophagy-inducers) or by targeting key components in the photoreceptor proteostasis network (e.g. ER quality control, ERAD, UPR) or with cell death inhibitors and neurotrophic factors. Some of these strategies have been shown to prolong cell survival both in cell culture systems and in animal models of disease, indicating the potential importance of proteostasis for controlling rhodopsin RP.
5.1. Approaches for restoring proteostasis
5.1.1. Pharmacological chaperones and methods to improve folding
Pharmacological chaperones are substrate-specific, small molecules that are designed to target directly the protein structure and shift the protein folding equilibrium towards the native state. For example, retinoids (9-cis and 11-cis-retinal) or non-isomerisable retinoid analogues (11-cis-7-ring retinal) have been shown to promote P23H rod opsin trafficking and folding in vitro (Krebs et al., 2010; Mendes and Cheetham, 2008; Noorwez et al., 2003; Noorwez et al., 2009; Opefi et al., 2013; Saliba et al., 2002), and protect cells from the toxic effects of mutant rod opsin (Mendes and Cheetham, 2008). While improving rod opsin folding in vitro opened the possibility for pharmacological rescue of P23H, and other class 2 rod opsin mutants, to treat this form of retinal degeneration, promoting P23H folding and stability with such compounds in vivo has been proved to be challenging (see section 5.3).
More recently, the AMPK activator metformin was used as an alternative approach to promote rhodopsin folding, via regulation of protein translation (Athanasiou et al., 2017a). Metformin improved P23H folding and traffic and reduced cell death in cell models, but did not appear to act as a pharmacological chaperone, as it did not bind directly to rhodopsin. Instead it improved folding through altering the cellular milieu and the rate of translation. In contrast to the effect in cell culture, metformin accelerated retinal degeneration in the P23H-1 rat model and P23H KI mouse, as the metformin-rescued P23H that trafficked to the OS in larger amounts was still intrinsically unstable and destabilised the OS (Athanasiou et al., 2017a). This would agree with other studies of P23H stability (Chen et al., 2014; Opefi et al., 2013) and the potential of mutant P23H that escapes ER quality control to disrupt and destabilise OS structure (Bogea et al., 2015; Haeri and Knox, 2012; Haeri et al., 2013; Sakami et al., 2011). Therefore, these studies suggest that correction of mutant rhodopsin folding, without improving rhodopsin stability, could further compromise the OS and exacerbate rod cell death. Thereby, emphasising the need to focus on other treatment strategies such as promoting mutant rhodopsin degradation, reducing rhodopsin aggregation, and/or promote mutant opsin stability by other means, such as the use of allosteric ligands.
5.1.2. Kosmotropes
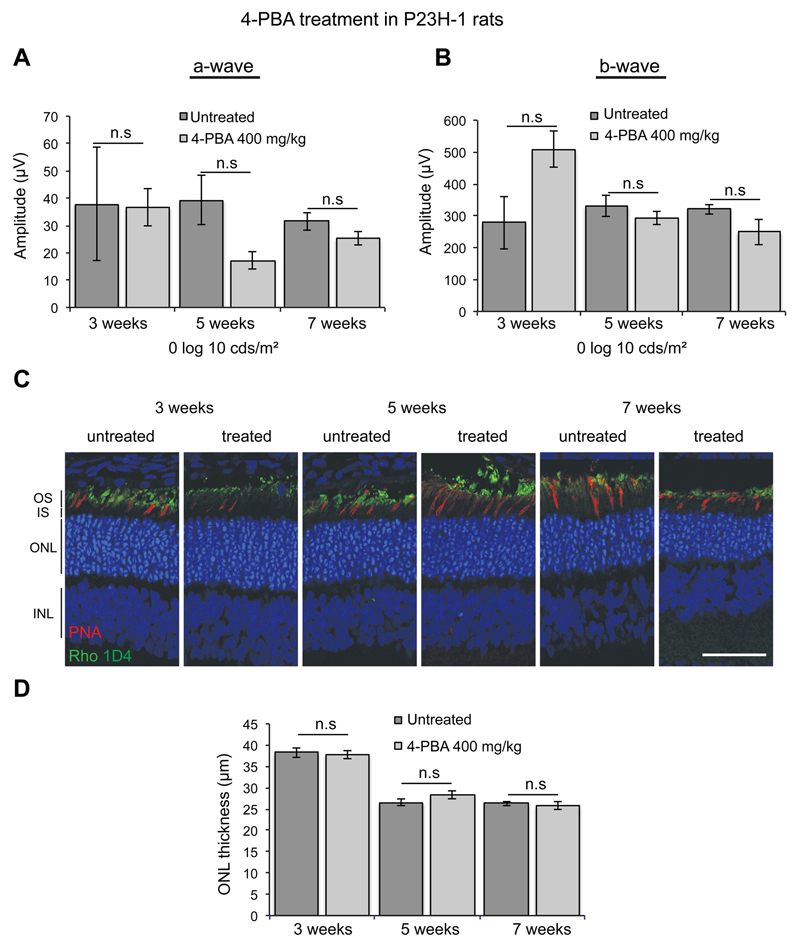
Kosmotropes, also known as chemical chaperones, are small, low molecular weight compounds that can improve the stability of proteins in their native conformation and reduce aggregation. Kosmotropes bind to proteins non-specifically and, therefore, have a potential to be used to a variety of protein misfolding diseases. The kosmotrope sodium 4-phenylbutyrate (4-PBA) is clinically approved for use in children with urea cycle disorders and for the treatment of sickle cell disease, thalassemia, and cystic fibrosis (Perlmutter, 2002). In vitro studies have shown that 4-PBA could decrease P23H rod opsin aggregation (Mendes and Cheetham, 2008) and reduce UPR signalling and ER stress associated with the T17M rod opsin mutation by promoting its degradation (Jiang et al., 2014a). Additionally, 4-PBA prevented the induction of the UPR and reduced apoptosis in RP17 associated with carbonic anhydrase mutations (Bonapace et al., 2004). In glaucoma, 4-PBA has been shown to decrease misfolded myocillin aggregation and apoptosis both in cells, and in mice (Yam et al., 2007; Zode et al., 2011). Importantly, administration of 4-PBA in the Rpe65 (R91W) mouse model of Leber congenital amarausis (LCA), which has mislocalisation and misfolding of cone opsin due a deficiency of 11-cis-retinal, led to improvement of cone opsin traffic accompanied by improvement in cone survival and vision (Li et al., 2016). Despite these data and the protection observed in the P23H cell model (Mendes and Cheetham, 2008), we did not observe any improvement of photoreceptor function, or amelioration of degeneration, in the P23H-1 rat model (Table 4) after systemic treatment with 4-PBA for up to 7 weeks as assessed by ERG and retinal histology (Figure 4). This suggests that, in contrast to misfolded cone opsins, 4-PBA does not act as a chemical chaperone for misfolded rhodopsin in vivo and therefore it does not protect against photoreceptor degeneration in the P23H rat model of RP. By contrast, the kosmotrope curcumin was reported to improve photoreceptor function and morphology in P23H-1 rats by reducing P23H aggregation and ER stress and by enhancing rhodopsin localisation to the rod OS (Vasireddy et al., 2011).
Figure 4. 4-PBA does not affect photoreceptor function and survival in P23H-1 rats.
P23H-1 rats were either left untreated or treated from P21 to P49 with 400 mg/kg 4-PBA (n = 4 for each condition) via IP injection. (A) Scotopic ERG’s were performed as previously described (Aguila et al., 2014). No significant differences were observed in P23H-1 rats that had received the treatment at any time point and at any intensity (-6 to 2.7 log cds/m2) (data not shown). Average at 0 log cds/m2 for a-wave and b-wave of P23H-1 rats at 3, 5 and 7 weeks of age. Values are means ± SEM. n.s non-significant values, 2 way ANOVA. (B) Retinal histology for measuring the outer nuclear layer (ONL) thickness was made on digital images of stained cryosections, every 500 microns from the optic nerve outwards for both the inferior and superior hemisphere. The ONL thickness across the whole from 4 animals at each time point was averaged and analysed using Graphpad Prism (Sigmastat). (C) Representative images of the ONL in treated and untreated P23H-1 rats at 3. 5 and 7 weeks stained with DAPI (blue), anti-rhodopsin antibody 1D4 (green) and cone marker peanut agglutinin (PNA) (red). Scale bar = 50 microns.
The chemical chaperone tauroursodeoxycholic acid (TUDCA) is a component of bear bile that inhibits apoptosis by preventing Bax from being transported to the mitochondria to initiate caspase release (Boatright et al., 2009). TUDCA has excited a lot of interest in eye research, as it has been shown to be beneficial in several retinal degeneration models. For instance, TUDCA preserved photoreceptor structure and function in an LCA (Lrat-/-) model (Fu and Zhang, 2014; Zhang et al., 2012), in the Rd1 (Lawson et al., 2016) and Rd10 mouse models of phosphodiesterase (PDE) RP (Drack et al., 2012; Oveson et al., 2011; Phillips et al., 2008) and prevented obesity in Bardet-Biedl syndrome Type I mice (Drack et al., 2012). Importantly, TUDCA slowed retinal degeneration in the lower copy number and slower degenerating P23H-3 rat (Table 4) model of rhodopsin RP, preserving photoreception function and survival and improving their connectivity with horizontal and bipolar cells (Fernandez-Sanchez et al., 2011). TUDCA also preserved photoreceptor function and survival in the fast degenerative P23H-1 rat model, since we observed significant increase in the a-wave, which corresponds to photoreceptor function, (Figure 5A) and outer nuclear layer (ONL) thickness (Figure 5B) in treated rats. We did not observe the same protection, however, after a longer treatment with TUDCA in the P23H KI mouse model (Figure 5C-D). TUDCA’s main suggested mechanism of action is to reduce ER stress. While the UPR is induced in the P23H-1 rats (Lin et al., 2007; Parfitt et al., 2014), the P23H KI model shows only activation of the IRE1 branch leading to a robust degradation of P23H and no PERK activation (Chiang et al., 2015), which might explain why TUDCA did not have a similar effect in these two models. Despite the effectiveness of TUDCA to protect photoreceptors in a range of animal models, its use in man could be limited by the required efficacious dose (500 mg/kg) and its unpleasant taste. In a recent study, however, Fernandez-Sanchez et al. (2017) demonstrated that biodegradable microspheres containing traces of TUDCA could be administered intravitreally in P23H-3 rats and prevent retinal degeneration and vision loss. This method provided a safe and long-term intraocular delivery of TUDCA with a neuroprotective effect, hence opening up the possibility for future clinical trials with this drug, if the effect can be validated in other models with the right gene dosage.
Figure 5. TUDCA improves photoreceptor function and survival in P23H-1 rats but has no effect in P23H KI mice.
P23H-1 rats were received 500 mg/kg TUDCA (n=9) or vehicle-PBS (n=7) from P21-P35 and P23H KI mice were received 500 mg/kg TUDCA (n=8) or vehicle-PBS (n=7) from P21-P43. (A, C) Scotopic ERG’s were performed as previously described (Athanasiou et al., 2017) at -5 to 1-log intensities (data not shown). (A) Average at 0 log cds/m2 for a-wave and b-wave of P23H-1 rats showing significant improvement of a-wave in TUDCA-treated compared to vehicle-treated P23H-1 rats (*p<0.05). The b-wave is not significantly affected (n.s) by TUDCA treatment. (B, D) OCT analysis was performed as previously described across the nasal-temporal meridian (Athanasiou et al., 2017a). The mean ONL thickness across the whole retina is significantly enhanced in TUDCA-treated P23H-1 rats (**p<0.01). (C) Average at 0 log cds/m2 for a-wave and b-wave of P23H-KI mice showing no significant changes in both the a-wave and b-wave in TUDCA-treated compared to vehicle-treated P23H-KI mice (n.s). (D) The mean ONL thickness across the whole retina does not alter in TUDCA-treated mice (n.s). (A-D) Values are mean ± SEM. Unpaired two-sided Student's t test.
5.1.3. Molecular chaperone inducers and autophagy inducers
An alternative way to restore proteostasis is through manipulation of the cellular milieu and, in particular, the adaptive machinery that maintains proteostasis: the molecular chaperone machinery; the HSR; the UPR; and autophagy. The induction of the HSR is inhibited by the binding of molecular chaperones to heat shock factor (HSF1), which keep it as a monomer. Following cell stress the chaperones are recruited to misfolded proteins and allow HSF1 to trimerise and signalling cascades complete the activation. Therefore, inhibition of molecular chaperones can induce the HSR by releasing HSF1 and enhancing cell stress, which stimulates the production of molecular chaperones. The Hsp90 inhibitors and HSR inducers geldanamycin, radicicol and 17-AAG reduced P23H aggregation and cell death in cell culture (Mendes and Cheetham, 2008). Furthermore, 17-AAG, in combination with temporary permeabilisation of the blood retinal barrier, supressed retinal degeneration in an IMPDH1 mouse model (Tam et al., 2010b). Moreover, a single low systemic dose of the blood retinal barrier permeable Hsp90 inhibitor, HSP990, stimulated the HSR in the retina, reduced P23H aggregation, improved ERG responses and protected against photoreceptor cell death in P23H-1 rats (Aguila et al., 2014). Hsp90 inhibition also protected against the adverse effects of R135L rod opsin on endocytosis in cells and in the retina. This was not mediated through the HSR, however, but was a direct consequence of inhibiting Hsp90 function. Hsp90 functions as an essential chaperone in the maturation of many proteins, in particular protein kinases that can function as oncogenes, making it an attractive target for cancer therapy. Hsp90 inhibition reduced rhodopsin R135L hyperphosphorylation and arrestin binding, because of post-translation loss of rhodopsin kinase (GRK1), which mediates mutant rhodopsin hyperphosphorylation and is an Hsp90 client kinase (Aguila et al., 2014). Sustained Hsp90 inhibition also affected the biogenesis of PDE, and affected ERG responses (Aguila et al., 2014). A range of Hsp90 inhibitors have been developed and used in clinical trials for cancer (Butler et al., 2015); therefore, targeting Hsp90 in oncology, or as a way to induce the HSR, could also affect normal vision and other means of inducing the HSR might be more appropriate. Interestingly, the heat shock protein co-inducer arimoclomol might be a good alternative as it stimulated both the HSR and the UPR pathways in P23H retina, reduced P23H aggregation and ameliorated photoreceptor degeneration in both P23H-1 and P23H-3 rat models (Parfitt et al., 2014).
Another major cellular pathway for dealing with misfolded proteins and their removal is autophagy. Treatment with the autophagy-inducer rapamycin in cells expressing P23H reduced P23H aggregation (Mendes and Cheetham, 2008), and enhanced P23H degradation and colocalisation with autophagic markers (Kaushal, 2006). Systemic administration of rapamycin to P23H-3 rats slowed rod cell degeneration, but did not affect cones suggesting that inhibition of the mTOR pathway, and activation of autophagy is particularly protective for rod cells (Sizova et al., 2014).
5.1.4. Targeting individual proteins to restore proteostasis
Targeting key components of the photoreceptor proteostasis network could also enable photoreceptors to deal with misfolded rhodopsin. For instance, the chaperone network between ER degradation-enhancing α-mannosidase-like 1 (EDEM1), ERdj5 (DNAJC10) and BiP has been shown to interact with mutant rhodopsin and individual overexpression of these proteins reduced P23H aggregation and promoted rhodopsin degradation in cell models (Athanasiou et al., 2014; Athanasiou et al., 2012; Kosmaoglou et al., 2009). BiP overexpression also reduced ER stress and photoreceptor cell death in P23H rats (Gorbatyuk et al., 2010). Moreover, studies in Drosophila have shown that valosin containing protein, VCP/p97, can also enhance mutant Rh1 degradation (Griciuc et al., 2010a); however, genetic inactivation of VCP protected flies from retinal degeneration, suggesting that excessive rhodopsin dislocation from the ER and/or degradation can be detrimental to photoreceptors (Griciuc et al., 2010b).
Several studies have reported the induction of UPR in retinal degeneration; however, it is still uncertain whether activation of UPR is an adaptive, protective mechanism, part of the disease pathology, or a secondary effect due to the death of photoreceptors. The data from different rhodopsin models support each of these potential outcomes, in particular, for the PERK effectors ATF4 and CHOP, which is proposed to be a pro-apoptotic factor. For instance, reduction of CHOP levels in the T17M mouse model of RP led to reduction of photoreceptor function and survival. This was accompanied by a reduction of rhodopsin mRNA levels and down-regulation of important photoreceptor transcriptional factors, such as neural retinal leucine zipper (Nrl) and cone-rod homeobox (Crx), suggesting that CHOP plays a protective role in T17M-related RP (Nashine et al., 2013). By contrast, reduction of CHOP levels in the VPP (P23H) transgenic mouse model did not affect photoreceptor function. In later stages of disease, however, CHOP depleted mice showed less degeneration in the central retina suggesting that CHOP reduction might influence late stage responses of surrounding cells and/or other ongoing cellular responses in the retina to dying photoreceptors and thereby prolong cell survival in older animals (Adekeye et al., 2014). Moreover, depletion of CHOP in the P23H KI mouse model did not affect photoreceptor numbers, indicating that photoreceptor cell death in this model occurs independently of CHOP (Chiang et al., 2015). Interestingly, there was still an induction of IRE1 in the P23H KI retina in this study after many of the photoreceptors had died, suggesting that the cell death might be independent of the UPR and could arise from other retinal cells (Chiang et al., 2015).
On the other hand, reduction of ATF4 protected T17M rhodopsin mice from retinal degeneration by reducing apoptosis and promoting autophagy, whereas overexpression of ATF4 in the retina of T17M mice via AAV delivery accelerated retinal degeneration (Bhootada et al., 2016). In addition, intravitreal administration of KIRA6, which was designed to inhibit regulated IRE1-dependent mRNA decay (RIDD), a process that cleaves RNA to either preserve ER homeostasis or induce cell death, appeared to preserve photoreceptor numbers in P23H-1 rats (Ghosh et al., 2014), suggesting that activation of the UPR is part of the pathology. In contrast, studies of the P23H KI mouse revealed no c-Jun N-terminal kinase activation or RIDD, suggesting an adaptive response of the UPR and not chronic cell death activation (Chiang et al., 2015; Hiramatsu et al., 2015).
Furthermore, arimoclomol treatment, which potentiated all the three UPR branches in P23H rats correlated with reduced retinal degeneration, indicating that UPR activation per se is not detrimental (Parfitt et al., 2014). More recently, inhibition of the PERK branch of the UPR with the specific inhibitor GSK2606414 accelerated retinal degeneration and enhanced rhodopsin aggregation in P23H-1 rats, whereas inhibition of eIF2a phosphatase with salubrinal improved photoreceptor survival (Athanasiou et al 2017b), further suggesting that activation of PERK in this adRP model is part of a protective mechanism. In addition, enhanced ERAD and the induction of ER stress in Drosophila protects against Rh1 mutants (Kang and Ryoo, 2009; Mendes et al., 2009), suggesting a protective role for ERAD and the UPR. Considering all the above, further studies need to be conducted in different models of rhodopsin RP with the right gene dosage to elucidate the exact role of each of the UPR branches in photoreceptor degeneration.
5.2. Histone deacetylase inhibitors
Valpropic acid (VPA) has been used clinically as an anticonvulsant (epilepsy), mood stabiliser (bipolar disorder) and for migraine prevention. VPA is a pleiotropic drug, but is thought to act mainly as a histone deacetylase (HDAC) inhibitor. HDAC inhibitors can alter gene expression non-specifically and can have protective or negative effect in different cell and animal models of neurodegeneration (Chuang et al., 2009). In animal models, VPA provided neuroprotection and regeneration of retinal ganglion cells after optic nerve crush in Sprague Dawley (SD) rats (Biermann et al., 2011) and after retinal ischemia in Wistar rats (Zhang et al., 2011). Moreover, VPA alone or in combination with guanabenz and a caspase-12 inhibitor increased the ONL thickness and preserved photoreceptor function in a Bardet Biedl syndrome mouse model (BBS12-/-) (Mockel et al., 2012).
Additionally, VPA was reported to be protective in the P23H Xenopus model (Vent-Schmidt et al., 2017); however, there was no improvement in mutant rhodopsin trafficking, something that had been previously suggested as being VPA’s mode of action (Clemson et al., 2011). Instead, there was a reduction in global levels of rhodopsin, suggesting enhanced rhodopsin degradation, as previously seen after dark rearing in the same model (Tam and Moritz, 2007), and the effect was mediated by HDAC inhibition (Vent-Schmidt et al., 2017). Interestingly, the same doses that were protective in the P23H model exacerbated retinal degeneration in the T17M model of RP, where light, as opposed to misfolding, is more important for cell death. Although the effect was mediated by HDAC inhibition, the mechanism of T17M exacerbation remains unclear as VPA did not interfere with rhodopsin degradation or trafficking. VPA also exacerbated retinal degeneration in Xenopus S344ter and T4K models (Vent-Schmidt et al., 2017). Therefore, the effect of VPA was dependent by both genotype and environmental conditions. The study of Vent-Schmidt et al. (2017) was not the only one that showed opposing effects of VPA in different retinal degeneration models. VPA treatment significantly reduced photoreceptor loss in the fast-degenerative rd1 mouse model by P20, but accelerated degeneration in the slow-degenerative rd10 model, even though both are caused by mutations in PDE (PDE6) (Mitton et al., 2014). The authors also reported that VPA can cause photoreceptor loss in the immature retina, since they observed reduced ONL thickness in WT mice when administered at P9 but not at P17 (Mitton et al., 2014).
We tested VPA in the P23H-1 rat model by providing it in drinking water from postnatal (P) day P21 to P35. Interestingly, we observed that VPA treatment decreased photoreceptor function and survival, as the VPA-treated rats showed significantly lower a-waves and thinner ONL compared to the vehicle-treated rats (Figure 6), suggesting that VPA is detrimental for this model of RP, which is the opposite of what was observed in the P23H Xenopus model (Vent-Schmidt et al., 2017).
Figure 6. Valproic acid worsens photoreceptor function and survival in P23H-1 rats.
P23H-1 rats were received VPA (7.1 gm/Liter) in drinking water (n=9) or water (n=8) from P21-P35. (A) Scotopic ERG’s were performed as previously described (Athanasiou et al., 2017a) at -5 to 1-log intensities (data not shown). Average at 0 log cds/m2 for a-wave and b-wave of P23H-1 rats showing significant reduction of a-wave in VPA-treated animals (**p<0.01). The b-wave is not significantly affected (n.s) by VPA treatment. Values are mean ± SEM. Unpaired two-sided Student's t test. (B) OCT analysis was performed as previously described (Athanasiou et al., 2017a) across the nasal-temporal meridian. The mean ONL thickness across the whole retina is significantly reduced in VPA-treated animals (**p<0.001). Values are mean ± SEM. Unpaired two-sided Student's t test.
A published report of off-label VPA use for the treatment of RP (Clemson et al., 2011) was used as the basis for establishing a clinical trial in the USA, but this report has attracted a lot of controversy. This was due to the use of non-genotyped patients, low numbers, the short duration, the study design, statistical analysis and lack of reporting any side effects (Sandberg et al., 2011; Sisk, 2012; van Schooneveld et al., 2011). In another study, short-term treatment with oral VPA for three months in 10 patients with established RP showed improvement in visual acuity and vision (Shanmugam et al., 2012). Similarly, other studies followed which tested the efficacy of VPA in RP patients for up to 6 or 12 months and reported some positive effect to visual function of patients (Iraha et al., 2016; Kumar et al., 2014); however, all the aforementioned studies had similar limitations with the Clemson et al. study. A longer follow up of patients (up to 4 years) with several retinal dystrophies, including a total of 31 patients (21 RP patients and some from the Clemson et al., study) showed no benefit of VPA treatment and secession of treatment in 30% of the patients due to adverse effects of the drug (Bhalla et al., 2013). Additionally, epileptic patients can develop colour vision defects after VPA treatment (Verrotti et al., 2004), questioning further the clinical utility of VPA for visual problems. Collectively, these studies raise several concerns about the efficacy of VPA, highlight the importance of taking into account the specific genetic defects and potential differences between mutations before conducting a clinical trial and suggest that VPA cannot be used indiscriminately for the treatment of RP.
5.3. Nutritional supplements - Vitamin A palmitate
Vitamin A palmitate alone, or in combination with docosahexaenoic acid (DHA) and lutein, are the only prescribed treatments for RP; however, there is an ongoing debate in the scientific community for their efficacy to slow down the course of the disease. In 1993, Berson et al. investigated the effect of 15,000 IU/d supplementation of vitamin A and/or 400 IU/d of vitamin E and trace amounts of both vitamins, in 601 randomised patients with RP for up to 6 years and the results were assessed by performing 30 Hz cone flicker ERG. They reported a protective effect of vitamin A at high doses and a detrimental effect of vitamin E (Berson et al., 1993). Later, in 2004, the same group showed that the addition of DHA capsules to high dose (15,000 IU/d) vitamin A supplementation did not slow the course of the disease (Berson et al., 2004a); however, in a parallel study, DHA was protective in the first two out of four years in patients who did not take any vitamin A before the study (Berson et al., 2004b). In a third trial, Berson et al. (2010) tested the addition of 12 mg/day lutein to high dose of vitamin A (15,000 IU/d) and concluded that this combination with a high fish oil diet, where DHA is a major component, could benefit RP patients (Berson et al., 2010). Despite their positive outcomes, all the aforementioned studies have been criticised: for the methodology and statistical analysis; the robustness of the data, as there were no significant benefits in any measure of visual function; and that conclusions were drawn based on secondary outcomes and not primary outcome measures (Marmor, 1993; Massof and Finkelstein, 1993; Massof and Fishman, 2010; Norton, 1993).
Perhaps more importantly, these studies were not conducted in genotyped patients and there is growing evidence that vitamin A should be avoided in some IRD patients. For example, patients that have mutations in the Stargardt’s disease gene ABCA4, which can also cause arRP, are vulnerable to high levels of vitamin A (Charbel Issa et al., 2015). High doses of vitamin A could also cause liver damage and could cause severe birth defects if taken during pregnancy (Massof and Fishman, 2010). Interestingly, vitamin A deprivation prevented light-induced retinal degeneration in transgenic X. laevis expressing T4K opsin, suggesting that vitamin A supplementation could be detrimental to patients that carry the T4K rhodopsin mutation and patients that have sector RP (Tam et al., 2014).
In contrast, supplementation of vitamin A decreased the rate of retinal degeneration in the mouse T17M RP model, however, it had no effect in the P347S model (Li et al., 1998a). These data could support the hypothesis that increased levels of vitamin A could act as a pharmacological chaperone to promote mutant rhodopsin folding. Vitamin A deprivation also induced retinal degeneration in a P23H Xenopus model, reiterating the importance of vitamin A for retinal homeostasis (Tam et al., 2010a); however, evidence from Rpe65 polymorphisms in mice suggests that excess 11-cis-retinal may exacerbate P23H mutant opsin disease progression (Samardzija et al., 2006a). The Rpe65 polymorphism that has increased RPE65 activity, and presumably higher 11-cis-retinal levels, were associated with faster degeneration in the P23H mice, thereby illustrating how genetic modifiers could influence the progression of rhodopsin adRP. One of the limitations for the use of vitamin A is that 11-cis-retinal is produced efficiently via the visual cycle in the retina and therefore is more than adequate in most young individuals with a healthy diet. Increasing the amount of retinoid availability combined with light exposure could result in unnaturally high levels of all-trans retinal, which is toxic to photoreceptors (Chen et al., 2012). Their use as pharmacological chaperones is also limited as cis-retinoids change conformation when bleached, and because locked forms do not discriminate between opsins, they could potentially affect cone opsin function. Therefore, similar to VPA, vitamin A supplementation needs to be carefully considered and cannot be given broadly to RP patients, regardless of genetic diagnosis.
5.4. Prolonging cell survival with cell death inhibitors or neurotrophic factors
Cell death pathways can also be targeted for the treatment of rhodopsin RP. For instance, cone survival, together with preservation of ONL thickness and improvement of rod function was observed after treatment of the P23H-1 rat model with the cell death inhibitors necrostatin-1s (RIP1 inhibitor) and N-acetylcysteine (an inhibitor of NLRP3), while calpeptin (calpain inhibitor) improved only rod function and survival, but did not affect cones (Viringipurampeer et al., 2016).
Ablation of caspase-1 preserved retinal morphology and increased rhodopsin expression in the VPP/GHL (P23H) mouse (Table 4), but not in rd1 and light-induced retinal degeneration models, where caspase-1 is also abnormally overexpressed. The protection in this rhodopsin mouse model was only transient, however, providing more evidence that additional pathways, independent of caspase-1 are also involved in photoreceptor death process (Samardzija et al., 2006b). In addition, ablation of caspase-7 in T17M rhodopsin mice preserved retinal structure and function even from light-induced damage and it was suggested this was mediated by reducing ATF6 and PERK signalling and reducing JNK-induced apoptosis (Choudhury et al., 2013). Finally, viral delivery of the X-linked inhibitor of apoptosis (XIAP), which inhibits the action of caspases-3, -7 and -9 preserved both photoreceptor structure and function in the P23H-1 model, but protected only photoreceptor structure in the S334ter rhodopsin model highlighting that the different nature of rhodopsin mutations can affect the protection offered by targeting common apoptotic pathways (Leonard et al., 2007).
Another way to protect the degenerating retina in rhodopsin RP is to provide neuroprotection through supplementation with neurotrophic factors. The advantage of neurotrophic factors is that they can potentially improve vision for a range of retinal disorders, as they act on neuronal survival irrespective of the genetic cause. Ciliary neurotrophic factor (CNTF) is the most extensively studied factor both preclinically in different models of retinal degeneration, and in the clinic. Viral-mediated delivery of CNTF in the Rho KO mouse model led to a significant increase in photoreceptor survival up to 3 months (Liang et al., 2001), while recombinant intravitreal delivery in the S334ter rat model led to increased cone survival through the preservation of cone OS (Li et al., 2010). CNTF supplementation in the peripherin 2 mouse model (Prph2rd2/rd2), however, led to rod photoreceptor dysfunction (Schlichtenbrede et al., 2003). A recent clinical report on a long-term follow up of RP patients that received implant-mediated CNTF supplementation found no serious adverse effects, but also no evidence of efficacy or improved retinal structure and visual function (Birch et al., 2016). Transgenic mediated overexpression of brain derived neurotrophic factor (BDNF) provided structural and functional rescue in the presence of Q334ter rhodopsin (Okoye et al., 2003). Overexpression of glial-derived neurotrophic factor (GDNF) using adeno-associated virus (AAV) vectors was also shown to provide structural and functional rescue in both the S334ter rat (Dalkara et al., 2011) and in other models of retinal degeneration such as the Prph2rd2/rd2 and in the Royal College of Surgeons (RCS) rat (Buch et al., 2006). More recently, viral-mediated erythropoietin supplementation in the P23H transgenic mouse led to a structural and functional rescue that persisted for 7 months (Rex et al., 2016).
Furthermore, subretinal injections of the Rod-derived Cone Viability Factor (RdCVF) preserved cone photoreceptor numbers and photopic function in P23H-1 rats (Yang et al., 2009), while systemic viral delivery to neonates and intravitreal viral administration of RdCVF rescued both the structure and function of cone photoreceptors in the rd10 and P23H KI mouse models (Byrne et al., 2015). Interestingly, AAV-mediated delivery of the longer counterpart RdCVFL early in the disease did not rescue cone cells, but improved rod function, increased levels of rhodopsin, and decreased the by-products of cellular oxidative stress in dark reared rd10 animals (Byrne et al., 2015). It remains to be seen if clinical application of any of these neurotrophic factors will replicate their preclinical efficacy without deleterious effects.
5.4. Genetic modification strategies
5.4.1. RNA interference, ribozymes and antisense oligonucleotides
Initial gene therapy approaches to treat dominant RP in animal models have involved knockdown of the mutant allele or complete suppression of all rhodopsin followed by supplementation with a rhodopsin cDNA resistant to the silencing agent. The first proof of concept involved AAV-mediated delivery of a ribozyme targeting the P23H allele which provided functional and structural rescue in the P23H transgenic rat for up to 8 months (LaVail et al., 2000). Ribozymes are difficult to design, however, and not suitable for all variants. Indeed, the P23H allele in this rat model was adapted to be optimal for ribozyme cleavage (Orhan et al., 2015).
In contrast, allele specific targeting by antisense oligonucleotides targeting the P23H allele in the P23H-1 rat model showed some functional rescue a month following intravitreal administration (Murray et al., 2015b). Furthermore, allele independent RNA interference applications have demonstrated rhodopsin reductions of up to 90% with subsequent supplementation of a resistant WT allele that can rescue the P23H mouse for up to 9 months, and the P347S mouse for up to 10 weeks (Chadderton et al., 2009; O'Reilly et al., 2007).
More recently, a different AAV-based approach to silence mutant rhodopsin expression utilised engineered proteins that bind cis-acting elements upstream of the rhodopsin gene, reducing its expression (Botta et al., 2016). Their efficiency to target rhodopsin mutations was shown in the P347S mouse where a functional improvement was seen two weeks post-treatment and also in the porcine retina, where total rhodopsin knockdown was followed by supplementation with human rhodopsin (Botta et al., 2016).
5.4.2. Gene therapy
Gene supplementation is an option to treat recessive disorders where the target gene product is lacking, but is less certain in dominant disease with a clear gain of function effect, as opposed to haploinsufficiency. Surprisingly, gene supplementation with WT rhodopsin was shown to provide protection from cell death in the P23H mouse (Mao et al., 2011). In this study, WT rhodopsin overexpression in the mutant mice improved retinal structure and function up to 6 months post-treatment, indicating that overexpression could provide benefit. These data provide further evidence for a dominant negative effect of class 2 mutants and the protective potential of the WT allele. In the same study, however, it was noted that rhodopsin overexpression in WT mice caused retinal degeneration, indicating that the rhodopsin level is critical and needs to be carefully controlled as it has a protective effect only in the presence of a dominant mutant and across a small window of rhodopsin expression.
Additionally, gene augmentation with protective factors can also provide protection against cell death. For example, P23H rats that received AAV-mediated delivery of proinsulin, which has been characterised as a neuroprotective molecule, had functional rescue for up to 4 months (Fernandez-Sanchez et al., 2012), indicating that certain types of gene augmentation might be applicable to ameliorate adRP phenotype.
5.4.3. CRISPR/Cas9
CRISPR/Cas9 technology is at the forefront of gene editing research attracting much attention because of its efficiency and stringency in genomic targeting and due to its simplicity to design a guiding molecule directing the nuclease at the locus of interest. CRISPR/Cas9 applications rely on either the inactivation of the cleaved genomic locus following non-homologous end joining (NHEJ) or its precise correction following homology directed repair (HDR). In the post-mitotic retina HDR is very inefficient; therefore, most applications focusing on targeting mutant alleles have aimed to render them inactive using NHEJ. As CRISPR/Cas9 is still at early stages for in vivo applications, there are only a few studies investigating its use to treat rhodopsin-mediated RP. For instance, an innovative study by Yu et al., (2017) used AAV-mediated CRISPR/Cas9 inactivation of Nrl, a rod-specific transcription factor, to alter rod cell fate determination in the rd10, Rho KO and P347S rhodopsin mouse models and reported improvement of rod survival and preservation of cone function.
In a rhodopsin targeted approach, electroporation of a plasmid encoding for a Cas9 nuclease and a guide RNA (gRNA) targeting the P23H allele into mouse retina showed a marked reduction in expression of P23H on a rhodopsin null background (Latella et al., 2016). In this study, in addition to the reduced levels of P23H, a high proportion of genomic editing was reported (up to 33%). It is important to note that as electroporation efficiency in the retina is low, the analyses were performed in the GFP selected cells that had received the CRISPR/Cas9 plasmid (as visualised by reporter gene expression) and not in whole retinal lysates. More recently, a similar approach was used to target the P23H allele as well as an HDR-mediated correction in vitro in iPS cells, followed by in vivo NHEJ-mediated targeting in the pig retina (Burnight et al., 2017).
Moreover, Bakondi et al. (2016) used a similar approach to target the S334ter allele in transgenic rats and they reported an improvement in function following electroporation of the targeting plasmid. This exciting technology, however, is still in its infancy and faces significant technical challenges and more broadly applicable approaches. For example, in the case of rhodopsin adRP, any CRISPR approach must be allele specific or it would convert an adRP phenotype to a more severe arRP one, and these studies have not been done on human rhodopsin genomic sequences in the context of one mutant and one WT allele. Furthermore, a recent study of CRISPR/Cas9 targeting in fertilised Xenopus eggs showed that in-frame deletions frequently caused dominant retinal degeneration associated with rhodopsin biosynthesis defects, while frameshift phenotypes were consistent with knockout (Feehan et al., 2017). Given that NHEJ is random, there is almost a 1:3 chance that CRISPR gene editing could replace one dominant rhodopsin mutation with another dominant mutation within individual rods, even if the editing were allele specific.
Furthermore, while all the above studies do not report any off-target effects and several studies suggested a low incidence of off-target mutations using CRISPR/Cas9 (Iyer et al., 2015; Smith et al., 2014; Veres et al., 2014), a recent study reported multiple unexpected off-target effects after using CRISPR/Cas9 in vivo (Schaefer et al., 2017). However, several concerns have been raised in the scientific community regarding the interpretation of the data, the low numbers of animals and the overall experimental design of this study and support the hypothesis that these off target effects are due to genetic variation between the animals studied and not due to CRISPR/Cas9 treatment (Kim et al., 2017; Lareau et al., 2017; Wilson et al., 2017). Nevertheless, improving CRISPR specificity and efficacy is important because even a low frequency of unexpected mutations can be harmful and any potential off target effects should be considered and tested in more detail before any clinical application (Tasan and Zhao, 2017).
6. Concluding remarks
Immense progress has been made in the understanding of molecular pathways that lead to retinal degeneration associated with rhodopsin mutations. Conflicting therapeutic outcomes from different animal models reflect the complexity of this type of retinal degeneration. They also highlight the importance of understanding the molecular consequences of the many different types of mutation and developing mutation mechanism-dependent therapeutic strategies, regardless of whether these are dealing with the cause of the disease or the consequences of the disease. These approaches are getting closer to the clinic and the enhanced understanding of rhodopsin function informs, not only GPCR biology, but also more efficient patient-tailored therapies.
Glossary.
CRISPR/Cas9: a method for specific gene editing that has been developed from studying bacterial immunity. The Cas9 nuclease cleaves specific DNA sequences based on a guide RNA.
ERG: the electroretinogram measures the electrical responses of various cell types within the retina. The a-wave arises from the photoreceptors and the b-wave corresponds to the propagation through the retina. A scotopic ERG is done in dim light after dark adaptation to measure rod responses, whereas a photopic ERG measures cone responses.
Heat shock response: the cellular response to imbalanced proteostasis in the cytoplasm and nucleus.
Mutation: a change in genomic DNA, and potentially protein, that is associated with disease.
Off-label: the prescription of an approved drug for an indication it has not been approved for.
Outer nuclear layer (ONL): the layer of nuclei outermost in the neural retina, corresponding to photoreceptor nuclei.
Proteostasis: the balance of protein synthesis, modification, traffic and degradation that enables function of the proteome and maintenance of protein homeostasis.
Retinitis pigmentosa: an inherited retinal dystrophy, usually characterised by rod cell dysfunction and degeneration followed by cone cell dysfunction and degeneration.
Rhodopsin: any opsin + 11-cis-retinal moiety is correctly termed a ‘rhodopsin’, but here we use the term rhodopsin to refer to the rod opsin + 11-cis-retinaldehyde photopigment complex of rod cells.
Schiff Base: the product of the chemical association of an aldehyde with a primary amine. 11-cis-retinal forms a Schiff base with rhodopsin via lysine 296.
Unfolded protein response: the cellular response to ER stress and disturbed proteostasis in the ER.
Variant: a sequence change in the DNA of unknown significance.
Acknowledgements
We are grateful to the reviewers for their considered and constructive comments that have improved the depth and content of this review. We are grateful to Professor Molday (Department of Biochemistry and Molecular Biology, University of British Columbia, Canada) for the gift of the Rho-1D4 antibody, Professor Matt LaVail (University of California San Francisco, USA) for providing P23H-1 transgenic rats and to Dalila Bevilacqua for assisting with the drug treatments. This work was supported by RP Fighting Blindness (GR580, GR572 and GR563), The BIG Lottery Fund, Fight for Sight (1439, 1791 and 1489/1490), and The Wellcome Trust (092621 and 205041 to M.E.C and multiuser equipment grant 099173).
Abbreviations
- 4-PBA
sodium 4-phenylbutyrate
- AAV
Adeno-associated virus
- ad
autosomal dominant
- adRP
autosomal dominant retinitis pigmentosa
- adCSNB
autosomal dominant congenital stationary night blindness
- AIF
apoptosis inducing factor
- ar
autosomal recessive
- Arf4
ADP Ribosylation Factor 4
- arRP
autosomal recessive retinitis pigmentosa
- ASAP1
ArfGAP with SH3 Domain Ankyrin Repeat and PH Domain 1
- ATF6
activating transcription factor 6
- BDNF
brain derived neurotrophic factor
- BiP
binding immunoglobulin protein
- Cas9
CRISPR associated protein 9
- CHOP
C/EBP homologous protein
- CNTF
Ciliary neurotrophic factor
- CRISPR
Clustered Regularly Interspaced Short Palindromic Repeats
- CRX
cone-rod homeobox
- CSNB
congenital stationary night blindness
- DAMPS
damage-associated molecular patterns
- DHA
docosahexaenoic acid
- Drp1
dynamin-related protein 1
- EDEM1
ER degradation-enhancing α-mannosidase-like 1
- EL
extracellular loop
- ER
endoplasmic reticulum
- ERAD
ER associated degradation
- ERG
electroretinogram
- GDNF
glial-derived neurotrophic factor
- GPCR
G-protein coupled receptor
- GRK1
rhodopsin kinase
- gRNA
guide RNA
- HDAC
histone deacetylase
- HDR
homology directed repair
- HSF1
heat shock factor
- HSR
heat shock response
- IFT
intraflagellar transport
- IRD
inherited retinal dystrophy
- IRE1α
inositol-requiring enzyme 1
- IS
inner segment
- KI
knock-in
- KO
knock-out
- LCA
Leber congenital amaurosis
- MAS
magic angle spinning
- Meta II
metarhodopsin II
- NHEJ
non-homologous end joining
- NMR
nuclear magnetic resonance
- NRL
neural retinal leucine zipper
- OGG1
8-Oxoguanine glycosylase
- ONL
outer nuclear layer
- OS
outer segment
- P
post-natal
- PARP
poly(ADP-ribose) polymerase
- PDE
phosphodiesterase
- PERK
protein kinase RNA-like endoplasmic reticulum kinase
- PSB
protonated Schiff base
- RCS
Royal College of Surgeons
- RdCVF
Rod-derived Cone Viability Factor
- RIDD
regulated IRE1-dependent mRNA decay
- RIP
receptor-interacting protein kinases
- ROS
reactive oxygen species
- RP
retinitis pigmentosa
- RPE
retinal pigment epithelium
- RTC
rhodopsin transport carrier
- SD
Sprague Dawley
- TM
transmembrane
- TUDCA
tauroursodeoxycholic acid
- UPR
unfolded protein response
- VPA
valproic acid
- WT
wild-type
- VCP
valosin containing protein
- XBP-1
X-box binding protein 1
- XIAP
X-linked inhibitor of apoptosis
References
- Adekeye A, Haeri M, Solessio E, Knox BE. Ablation of the proapoptotic genes CHOP or Ask1 does not prevent or delay loss of visual function in a P23H transgenic mouse model of retinitis pigmentosa. PLoS One. 2014;9:e83871. doi: 10.1371/journal.pone.0083871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguila M, Bevilacqua D, McCulley C, Schwarz N, Athanasiou D, Kanuga N, Novoselov SS, Lange CA, Ali RR, Bainbridge JW, Gias C, et al. Hsp90 inhibition protects against inherited retinal degeneration. Hum Mol Genet. 2014;23:2164–2175. doi: 10.1093/hmg/ddt613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja S, Hornak V, Yan EC, Syrett N, Goncalves JA, Hirshfeld A, Ziliox M, Sakmar TP, Sheves M, Reeves PJ, Smith SO, et al. Helix movement is coupled to displacement of the second extracellular loop in rhodopsin activation. Nature structural & molecular biology. 2009;16:168–175. doi: 10.1038/nsmb.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- al-Jandal N, Farrar GJ, Kiang AS, Humphries MM, Bannon N, Findlay JB, Humphries P, Kenna PF. A novel mutation within the rhodopsin gene (Thr-94-Ile) causing autosomal dominant congenital stationary night blindness. Human mutation. 1999;13:75–81. doi: 10.1002/(SICI)1098-1004(1999)13:1<75::AID-HUMU9>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Alfinito PD, Townes-Anderson E. Activation of mislocalized opsin kills rod cells: a novel mechanism for rod cell death in retinal disease. Proc Natl Acad Sci U S A. 2002;99:5655–5660. doi: 10.1073/pnas.072557799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés A, Garriga P, Manyosa J. Altered functionality in rhodopsin point mutants associated with retinitis pigmentosa. Biochemical and Biophysical Research Communications. 2003;303:294–301. doi: 10.1016/s0006-291x(03)00328-0. [DOI] [PubMed] [Google Scholar]
- Athanasiou D, Aguila M, Opefi CA, South K, Bellingham J, Bevilacqua D, Munro PM, Kanuga N, Mackenzie FE, Dubis AM, Georgiadis A, et al. Rescue of mutant rhodopsin traffic by metformin-induced AMPK activation accelerates photoreceptor degeneration. Hum Mol Genet. 2017a;26:305–319. doi: 10.1093/hmg/ddw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou D, Aguila D, Bellingham J, Kanuga N, Adamson P, Cheetham ME. The role of the ER stress response protein PERK in rhodopsin retinitis pigmentosa. Hum Mol Genet. 2017b doi: 10.1093/hmg/ddx370. in press ddx370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou D, Bevilacqua D, Aguila M, McCulley C, Kanuga N, Iwawaki T, Chapple JP, Cheetham ME. The co-chaperone and reductase ERdj5 facilitates rod opsin biogenesis and quality control. Hum Mol Genet. 2014;23:6594–6606. doi: 10.1093/hmg/ddu385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou D, Kosmaoglou M, Kanuga N, Novoselov SS, Paton AW, Paton JC, Chapple JP, Cheetham ME. BiP prevents rod opsin aggregation. Molecular biology of the cell. 2012;23:3522–3531. doi: 10.1091/mbc.E12-02-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audo I, Manes G, Mohand-Said S, Friedrich A, Lancelot ME, Antonio A, Moskova-Doumanova V, Poch O, Zanlonghi X, Hamel CP, Sahel JA, et al. Spectrum of rhodopsin mutations in French autosomal dominant rod-cone dystrophy patients. Invest Ophthalmol Vis Sci. 2010;51:3687–3700. doi: 10.1167/iovs.09-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam M, Khan MI, Gal A, Hussain A, Shah ST, Khan MS, Sadeque A, Bokhari H, Collin RW, Orth U, van Genderen MM, et al. A homozygous p.Glu150Lys mutation in the opsin gene of two Pakistani families with autosomal recessive retinitis pigmentosa. Mol Vis. 2009;15:2526–2534. [PMC free article] [PubMed] [Google Scholar]
- Bakondi B, Lv W, Lu B, Jones MK, Tsai Y, Kim KJ, Levy R, Akhtar AA, Breunig JJ, Svendsen CN, Wang S. In Vivo CRISPR/Cas9 Gene Editing Corrects Retinal Dystrophy in the S334ter-3 Rat Model of Autosomal Dominant Retinitis Pigmentosa. Mol Ther. 2016;24:556–563. doi: 10.1038/mt.2015.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellingham J, Tarttelin EE, Foster RG, Wells DJ. Structure and evolution of the teleost extraretinal rod-like opsin (errlo) and ocular rod opsin (rho) genes: is teleost rho a retrogene? J Exp Zool B Mol Dev Evol. 2003a;297:1–10. doi: 10.1002/jez.b.18. [DOI] [PubMed] [Google Scholar]
- Bellingham J, Wells DJ, Foster RG. In silico characterisation and chromosomal localisation of human RRH (peropsin)--implications for opsin evolution. BMC Genomics. 2003b;4:3. doi: 10.1186/1471-2164-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EJ, Bence NF, Jayakumar R, Kopito RR. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Molecular cell. 2005;17:351–365. doi: 10.1016/j.molcel.2004.12.021. [DOI] [PubMed] [Google Scholar]
- Berson EL. Retinitis pigmentosa. The Friedenwald Lecture. Invest Ophthalmol Vis Sci. 1993;34:1659–1676. [PubMed] [Google Scholar]
- Berson EL, Rosner B, Sandberg MA, Hayes KC, Nicholson BW, Weigel-DiFranco C, Willett W. A randomized trial of vitamin A and vitamin E supplementation for retinitis pigmentosa. Arch Ophthalmol. 1993;111:761–772. doi: 10.1001/archopht.1993.01090060049022. [DOI] [PubMed] [Google Scholar]
- Berson EL, Rosner B, Sandberg MA, Weigel-DiFranco C, Brockhurst RJ, Hayes KC, Johnson EJ, Anderson EJ, Johnson CA, Gaudio AR, Willett WC, et al. Clinical trial of lutein in patients with retinitis pigmentosa receiving vitamin A. Arch Ophthalmol. 2010;128:403–411. doi: 10.1001/archophthalmol.2010.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson EL, Rosner B, Sandberg MA, Weigel-DiFranco C, Moser A, Brockhurst RJ, Hayes KC, Johnson CA, Anderson EJ, Gaudio AR, Willett WC, et al. Clinical trial of docosahexaenoic acid in patients with retinitis pigmentosa receiving vitamin A treatment. Arch Ophthalmol. 2004a;122:1297–1305. doi: 10.1001/archopht.122.9.1297. [DOI] [PubMed] [Google Scholar]
- Berson EL, Rosner B, Sandberg MA, Weigel-DiFranco C, Moser A, Brockhurst RJ, Hayes KC, Johnson CA, Anderson EJ, Gaudio AR, Willett WC, et al. Further evaluation of docosahexaenoic acid in patients with retinitis pigmentosa receiving vitamin A treatment: subgroup analyses. Arch Ophthalmol. 2004b;122:1306–1314. doi: 10.1001/archopht.122.9.1306. [DOI] [PubMed] [Google Scholar]
- Berson EL, Rosner B, Weigel-DiFranco C, Dryja TP, Sandberg MA. Disease progression in patients with dominant retinitis pigmentosa and rhodopsin mutations. Invest Ophthalmol Vis Sci. 2002;43:3027–3036. [PubMed] [Google Scholar]
- Bhalla S, Joshi D, Bhullar S, Kasuga D, Park Y, Kay CN. Long-term follow-up for efficacy and safety of treatment of retinitis pigmentosa with valproic acid. Br J Ophthalmol. 2013;97:895–899. doi: 10.1136/bjophthalmol-2013-303084. [DOI] [PubMed] [Google Scholar]
- Bhandary B, Marahatta A, Kim H-R, Chae H-J. Mitochondria in relation to cancer metastasis. Journal of Bioenergetics and Biomembranes. 2012;44:623–627. doi: 10.1007/s10863-012-9464-x. [DOI] [PubMed] [Google Scholar]
- Bhootada Y, Kotla P, Zolotukhin S, Gorbatyuk O, Bebok Z, Athar M, Gorbatyuk M. Limited ATF4 Expression in Degenerating Retinas with Ongoing ER Stress Promotes Photoreceptor Survival in a Mouse Model of Autosomal Dominant Retinitis Pigmentosa. PLoS One. 2016;11:e0154779. doi: 10.1371/journal.pone.0154779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biermann J, Boyle J, Pielen A, Lagreze WA. Histone deacetylase inhibitors sodium butyrate and valproic acid delay spontaneous cell death in purified rat retinal ganglion cells. Mol Vis. 2011;17:395–403. [PMC free article] [PubMed] [Google Scholar]
- Birch DG, Bennett LD, Duncan JL, Weleber RG, Pennesi ME. Long-term Follow-up of Patients With Retinitis Pigmentosa Receiving Intraocular Ciliary Neurotrophic Factor Implants. Am J Ophthalmol. 2016;170:10–14. doi: 10.1016/j.ajo.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatright JH, Nickerson JM, Moring AG, Pardue MT. Bile acids in treatment of ocular disease. J Ocul Biol Dis Infor. 2009;2:149–159. doi: 10.1007/s12177-009-9030-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogea TH, Wen RH, Moritz OL. Light Induces Ultrastructural Changes in Rod Outer and Inner Segments, Including Autophagy, in a Transgenic Xenopus laevis P23H Rhodopsin Model of Retinitis Pigmentosa. Invest Ophthalmol Vis Sci. 2015;56:7947–7955. doi: 10.1167/iovs.15-16799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonapace G, Waheed A, Shah GN, Sly WS. Chemical chaperones protect from effects of apoptosis-inducing mutation in carbonic anhydrase IV identified in retinitis pigmentosa 17. Proc Natl Acad Sci U S A. 2004;101:12300–12305. doi: 10.1073/pnas.0404764101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botta S, Marrocco E, de Prisco N, Curion F, Renda M, Sofia M, Lupo M, Carissimo A, Bacci ML, Gesualdo C, Rossi S, et al. Rhodopsin targeted transcriptional silencing by DNA-binding. Elife. 2016;5:e12242. doi: 10.7554/eLife.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Gil N, Gonzalez-Del Pozo M, Martin-Sanchez M, Mendez-Vidal C, Rodriguez-de la Rua E, Borrego S, Antinolo G. Unravelling the genetic basis of simplex Retinitis Pigmentosa cases. Sci Rep. 2017;7:41937. doi: 10.1038/srep41937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadgate S, Yu J, Downes SM, Halford S. Unravelling the genetics of inherited retinal dystrophies: Past, present and future. Prog Retin Eye Res. 2017;59:53–96. doi: 10.1016/j.preteyeres.2017.03.003. [DOI] [PubMed] [Google Scholar]
- Buch PK, MacLaren RE, Duran Y, Balaggan KS, MacNeil A, Schlichtenbrede FC, Smith AJ, Ali RR. In contrast to AAV-mediated Cntf expression, AAV-mediated Gdnf expression enhances gene replacement therapy in rodent models of retinal degeneration. Mol Ther. 2006;14:700–709. doi: 10.1016/j.ymthe.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Burnight ER, Gupta M, Wiley LA, Anfinson KR, Tran A, Triboulet R, Hoffmann JM, Klaahsen DL, Andorf JL, Jiao C, Sohn EH, et al. Using CRISPR-Cas9 to Generate Gene-Corrected Autologous iPSCs for the Treatment of Inherited Retinal Degeneration. Mol Ther. 2017 doi: 10.1016/j.ymthe.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler LM, Ferraldeschi R, Armstrong HK, Centenera MM, Workman P. Maximizing the Therapeutic Potential of HSP90 Inhibitors. Mol Cancer Res. 2015;13:1445–1451. doi: 10.1158/1541-7786.MCR-15-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne LC, Dalkara D, Luna G, Fisher SK, Clerin E, Sahel JA, Leveillard T, Flannery JG. Viral-mediated RdCVF and RdCVFL expression protects cone and rod photoreceptors in retinal degeneration. J Clin Invest. 2015;125:105–116. doi: 10.1172/JCI65654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao SS, Kaufman RJ. Unfolded protein response. Current biology : CB. 2012;22:R622–626. doi: 10.1016/j.cub.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Chadderton N, Millington-Ward S, Palfi A, O'Reilly M, Tuohy G, Humphries MM, Li T, Humphries P, Kenna PF, Farrar GJ. Improved retinal function in a mouse model of dominant retinitis pigmentosa following AAV-delivered gene therapy. Mol Ther. 2009;17:593–599. doi: 10.1038/mt.2008.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang GQ, Hao Y, Wong F. Apoptosis: final common pathway of photoreceptor death in rd, rds, and rhodopsin mutant mice. Neuron. 1993;11:595–605. doi: 10.1016/0896-6273(93)90072-y. [DOI] [PubMed] [Google Scholar]
- Charbel Issa P, Barnard AR, Herrmann P, Washington I, MacLaren RE. Rescue of the Stargardt phenotype in Abca4 knockout mice through inhibition of vitamin A dimerization. Proc Natl Acad Sci U S A. 2015;112:8415–8420. doi: 10.1073/pnas.1506960112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Shi G, Concepcion FA, Xie G, Oprian D, Chen J. Stable rhodopsin/arrestin complex leads to retinal degeneration in a transgenic mouse model of autosomal dominant retinitis pigmentosa. J Neurosci. 2006;26:11929–11937. doi: 10.1523/JNEUROSCI.3212-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Jastrzebska B, Cao P, Zhang J, Wang B, Sun W, Yuan Y, Feng Z, Palczewski K. Inherent instability of the retinitis pigmentosa P23H mutant opsin. J Biol Chem. 2014;289:9288–9303. doi: 10.1074/jbc.M114.551713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Okano K, Maeda T, Chauhan V, Golczak M, Maeda A, Palczewski K. Mechanism of all-trans-retinal toxicity with implications for stargardt disease and age-related macular degeneration. J Biol Chem. 2012;287:5059–5069. doi: 10.1074/jbc.M111.315432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang WC, Kroeger H, Sakami S, Messah C, Yasumura D, Matthes MT, Coppinger JA, Palczewski K, LaVail MM, Lin JH. Robust Endoplasmic Reticulum-Associated Degradation of Rhodopsin Precedes Retinal Degeneration. Molecular neurobiology. 2015;52:679–695. doi: 10.1007/s12035-014-8881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury S, Bhootada Y, Gorbatyuk O, Gorbatyuk M. Caspase-7 ablation modulates UPR, reprograms TRAF2-JNK apoptosis and protects T17M rhodopsin mice from severe retinal degeneration. Cell Death Dis. 2013;4:e528. doi: 10.1038/cddis.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang DM, Leng Y, Marinova Z, Kim HJ, Chiu CT. Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends Neurosci. 2009;32:591–601. doi: 10.1016/j.tins.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JZ, Vega C, Jun W, Sung CH. Structural and functional impairment of endocytic pathways by retinitis pigmentosa mutant rhodopsin-arrestin complexes. J Clin Invest. 2004;114:131–140. doi: 10.1172/JCI21136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cideciyan AV, Hood DC, Huang Y, Banin E, Li ZY, Stone EM, Milam AH, Jacobson SG. Disease sequence from mutant rhodopsin allele to rod and cone photoreceptor degeneration in man. Proc Natl Acad Sci U S A. 1998;95:7103–7108. doi: 10.1073/pnas.95.12.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cideciyan AV, Jacobson SG, Aleman TS, Gu D, Pearce-Kelling SE, Sumaroka A, Acland GM, Aguirre GD. In vivo dynamics of retinal injury and repair in the rhodopsin mutant dog model of human retinitis pigmentosa. Proc Natl Acad Sci U S A. 2005;102:5233–5238. doi: 10.1073/pnas.0408892102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark GR, Crowe P, Muszynska D, O'Prey D, O'Neill J, Alexander S, Willoughby CE, McKay GJ, Silvestri G, Simpson DA. Development of a diagnostic genetic test for simplex and autosomal recessive retinitis pigmentosa. Ophthalmology. 2010;117:2169–2177 e2163. doi: 10.1016/j.ophtha.2010.02.029. [DOI] [PubMed] [Google Scholar]
- Clemson CM, Tzekov R, Krebs M, Checchi JM, Bigelow C, Kaushal S. Therapeutic potential of valproic acid for retinitis pigmentosa. Br J Ophthalmol. 2011;95:89–93. doi: 10.1136/bjo.2009.175356. [DOI] [PubMed] [Google Scholar]
- Collin RW, van den Born LI, Klevering BJ, de Castro-Miro M, Littink KW, Arimadyo K, Azam M, Yazar V, Zonneveld MN, Paun CC, Siemiatkowska AM, et al. High-resolution homozygosity mapping is a powerful tool to detect novel mutations causative of autosomal recessive RP in the Dutch population. Invest Ophthalmol Vis Sci. 2011;52:2227–2239. doi: 10.1167/iovs.10-6185. [DOI] [PubMed] [Google Scholar]
- Concepcion F, Chen J. Q344ter Mutation Causes Mislocalization of Rhodopsin Molecules That Are Catalytically Active: A Mouse Model of Q344ter-Induced Retinal Degeneration. PLOS ONE. 2010;5:e10904. doi: 10.1371/journal.pone.0010904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalkara D, Kolstad KD, Guerin KI, Hoffmann NV, Visel M, Klimczak RR, Schaffer DV, Flannery JG. AAV mediated GDNF secretion from retinal glia slows down retinal degeneration in a rat model of retinitis pigmentosa. Mol Ther. 2011;19:1602–1608. doi: 10.1038/mt.2011.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson FF, Loewen PC, Khorana HG. Structure and function in rhodopsin: replacement by alanine of cysteine residues 110 and 187, components of a conserved disulfide bond in rhodopsin, affects the light-activated metarhodopsin II state. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:4029–4033. doi: 10.1073/pnas.91.9.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies WI, Downes SM, Fu JK, Shanks ME, Copley RR, Lise S, Ramsden SC, Black GC, Gibson K, Foster RG, Hankins MW, et al. Next-generation sequencing in health-care delivery: lessons from the functional analysis of rhodopsin. Genet Med. 2012;14:891–899. doi: 10.1038/gim.2012.73. [DOI] [PubMed] [Google Scholar]
- Deretic D. A role for rhodopsin in a signal transduction cascade that regulates membrane trafficking and photoreceptor polarity. Vision Res. 2006;46:4427–4433. doi: 10.1016/j.visres.2006.07.028. [DOI] [PubMed] [Google Scholar]
- Deretic D, Wang J. Molecular assemblies that control rhodopsin transport to the cilia. Vision Res. 2012;75:5–10. doi: 10.1016/j.visres.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizhoor AM, Woodruff ML, Olshevskaya EV, Cilluffo MC, Cornwall MC, Sieving PA, Fain GL. Night Blindness and the Mechanism of Constitutive Signaling of Mutant G90D Rhodopsin. The Journal of Neuroscience. 2008;28:11662. doi: 10.1523/JNEUROSCI.4006-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drack AV, Dumitrescu AV, Bhattarai S, Gratie D, Stone EM, Mullins R, Sheffield VC. TUDCA slows retinal degeneration in two different mouse models of retinitis pigmentosa and prevents obesity in Bardet-Biedl syndrome type 1 mice. Invest Ophthalmol Vis Sci. 2012;53:100–106. doi: 10.1167/iovs.11-8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryja TP, Berson EL, Rao VR, Oprian DD. Heterozygous missense mutation in the rhodopsin gene as a cause of congenital stationary night blindness. Nat Genet. 1993;4:280–283. doi: 10.1038/ng0793-280. [DOI] [PubMed] [Google Scholar]
- Dryja TP, McEvoy JA, McGee TL, Berson EL. Novel rhodopsin mutations Gly114Val and Gln184Pro in dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2000;41:3124–3127. [PubMed] [Google Scholar]
- Dryja TP, McGee TL, Hahn LB, Cowley GS, Olsson JE, Reichel E, Sandberg MA, Berson EL. Mutations within the rhodopsin gene in patients with autosomal dominant retinitis pigmentosa. The New England journal of medicine. 1990a;323:1302–1307. doi: 10.1056/NEJM199011083231903. [DOI] [PubMed] [Google Scholar]
- Dryja TP, McGee TL, Reichel E, Hahn LB, Cowley GS, Yandell DW, Sandberg MA, Berson EL. A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature. 1990b;343:364–366. doi: 10.1038/343364a0. [DOI] [PubMed] [Google Scholar]
- Eisenberger T, Neuhaus C, Khan AO, Decker C, Preising MN, Friedburg C, Bieg A, Gliem M, Charbel Issa P, Holz FG, Baig SM, et al. Increasing the yield in targeted next-generation sequencing by implicating CNV analysis, non-coding exons and the overall variant load: the example of retinal dystrophies. PLoS One. 2013;8:e78496. doi: 10.1371/journal.pone.0078496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar GJ, Kenna P, Redmond R, McWilliam P, Bradley DG, Humphries MM, Sharp EM, Inglehearn CF, Bashir R, Jay M, et al. Autosomal dominant retinitis pigmentosa: absence of the rhodopsin proline----histidine substitution (codon 23) in pedigrees from Europe. Am J Hum Genet. 1990;47:941–945. [PMC free article] [PubMed] [Google Scholar]
- Feehan JM, Chiu CN, Stanar P, Tam BM, Ahmed SN, Moritz OL. Modeling Dominant and Recessive Forms of Retinitis Pigmentosa by Editing Three Rhodopsin-Encoding Genes in Xenopus Laevis Using Crispr/Cas9. Sci Rep. 2017;7:6920. doi: 10.1038/s41598-017-07153-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Sanchez L, Bravo-Osuna I, Lax P, Arranz-Romera A, Maneu V, Esteban-Perez S, Pinilla I, Puebla-Gonzalez MDM, Herrero-Vanrell R, Cuenca N. Controlled delivery of tauroursodeoxycholic acid from biodegradable microspheres slows retinal degeneration and vision loss in P23H rats. PLoS One. 2017;12:e0177998. doi: 10.1371/journal.pone.0177998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Sanchez L, Lax P, Isiegas C, Ayuso E, Ruiz JM, de la Villa P, Bosch F, de la Rosa EJ, Cuenca N. Proinsulin slows retinal degeneration and vision loss in the P23H rat model of retinitis pigmentosa. Hum Gene Ther. 2012;23:1290–1300. doi: 10.1089/hum.2012.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Sanchez L, Lax P, Pinilla I, Martin-Nieto J, Cuenca N. Tauroursodeoxycholic acid prevents retinal degeneration in transgenic P23H rats. Invest Ophthalmol Vis Sci. 2011;52:4998–5008. doi: 10.1167/iovs.11-7496. [DOI] [PubMed] [Google Scholar]
- Fitzgibbon J, Hope A, Slobodyanyuk SJ, Bellingham J, Bowmaker JK, Hunt DM. The rhodopsin-encoding gene of bony fish lacks introns. Gene. 1995;164:273–277. doi: 10.1016/0378-1119(95)00458-i. [DOI] [PubMed] [Google Scholar]
- Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Atomic-force microscopy: Rhodopsin dimers in native disc membranes. Nature. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- Frederick JM, Krasnoperova NV, Hoffmann K, Church-Kopish J, Ruther K, Howes K, Lem J, Baehr W. Mutant rhodopsin transgene expression on a null background. Invest Ophthalmol Vis Sci. 2001;42:826–833. [PubMed] [Google Scholar]
- Fu Y, Zhang T. Pathophysilogical mechanism and treatment strategies for Leber congenital amaurosis. Adv Exp Med Biol. 2014;801:791–796. doi: 10.1007/978-1-4614-3209-8_99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal A, Apfelstedt-Sylla E, Janecke AR, Zrennert E. Rhodopsin mutations in inherited retinal dystrophies and dysfunctions. Progress in Retinal and Eye Research. 1997;16:51–79. [Google Scholar]
- Garg AD, Krysko DV, Verfaillie T, Kaczmarek A, Ferreira GB, Marysael T, Rubio N, Firczuk M, Mathieu C, Roebroek AJ, Annaert W, et al. A novel pathway combining calreticulin exposure and ATP secretion in immunogenic cancer cell death. The EMBO journal. 2012;31:1062–1079. doi: 10.1038/emboj.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R, Wang L, Wang ES, Perera BG, Igbaria A, Morita S, Prado K, Thamsen M, Caswell D, Macias H, Weiberth KF, et al. Allosteric inhibition of the IRE1alpha RNase preserves cell viability and function during endoplasmic reticulum stress. Cell. 2014;158:534–548. doi: 10.1016/j.cell.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbatyuk MS, Knox T, LaVail MM, Gorbatyuk OS, Noorwez SM, Hauswirth WW, Lin JH, Muzyczka N, Lewin AS. Restoration of visual function in P23H rhodopsin transgenic rats by gene delivery of BiP/Grp78. Proc Natl Acad Sci U S A. 2010;107:5961–5966. doi: 10.1073/pnas.0911991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gragg M, Kim TG, Howell S, Park PS. Wild-type opsin does not aggregate with a misfolded opsin mutant. Biochim Biophys Acta. 2016;1858:1850–1859. doi: 10.1016/j.bbamem.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green ES, Menz MD, LaVail MM, Flannery JG. Characterization of rhodopsin mis-sorting and constitutive activation in a transgenic rat model of retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2000;41:1546–1553. [PubMed] [Google Scholar]
- Gregory-Evans K, Pennesi ME, Weleber RG. Retinitis Pigmentosa and Allied Disorders. In: Sadda SR, Hinton DR, Schachat AP, Sadda SR, Wilkinson CP, Wiedemann P, Schachat AP, editors. Retina. 5th ed. W.B. Saunders; London: 2013. pp. 761–835. [Google Scholar]
- Griciuc A, Aron L, Piccoli G, Ueffing M. Clearance of Rhodopsin(P23H) aggregates requires the ERAD effector VCP. Biochim Biophys Acta. 2010a;1803:424–434. doi: 10.1016/j.bbamcr.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Griciuc A, Aron L, Roux MJ, Klein R, Giangrande A, Ueffing M. Inactivation of VCP/ter94 suppresses retinal pathology caused by misfolded rhodopsin in Drosophila. PLoS Genet. 2010b:6. doi: 10.1371/journal.pgen.1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross AK, Rao VR, Oprian DD. Characterization of rhodopsin congenital night blindness mutant T94I. Biochemistry. 2003;42:2009–2015. doi: 10.1021/bi020613j. [DOI] [PubMed] [Google Scholar]
- Gunkel M, Schoneberg J, Alkhaldi W, Irsen S, Noe F, Kaupp UB, Al-Amoudi A. Higher-order architecture of rhodopsin in intact photoreceptors and its implication for phototransduction kinetics. Structure. 2015;23:628–638. doi: 10.1016/j.str.2015.01.015. [DOI] [PubMed] [Google Scholar]
- Haeri M, Knox BE. Rhodopsin mutant P23H destabilizes rod photoreceptor disk membranes. PLoS One. 2012;7:e30101. doi: 10.1371/journal.pone.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeri M, Knox BE, Ahmadi A. Modeling the flexural rigidity of rod photoreceptors. Biophys J. 2013;104:300–312. doi: 10.1016/j.bpj.2012.11.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel C. Retinitis pigmentosa. Orphanet J Rare Dis. 2006;1:40. doi: 10.1186/1750-1172-1-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368:1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- Hernan I, Gamundi MJ, Planas E, Borras E, Maseras M, Carballo M. Cellular expression and siRNA-mediated interference of rhodopsin cis-acting splicing mutants associated with autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2011;52:3723–3729. doi: 10.1167/iovs.10-6933. [DOI] [PubMed] [Google Scholar]
- Hiramatsu N, Chiang WC, Kurt TD, Sigurdson CJ, Lin JH. Multiple Mechanisms of Unfolded Protein Response-Induced Cell Death. Am J Pathol. 2015;185:1800–1808. doi: 10.1016/j.ajpath.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler N, Zaru R, Micheau O, Thome M, Attinger A, Valitutti S, Bodmer JL, Schneider P, Seed B, Tschopp J. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nature immunology. 2000;1:489–495. doi: 10.1038/82732. [DOI] [PubMed] [Google Scholar]
- Humphries MM, Rancourt D, Farrar GJ, Kenna P, Hazel M, Bush RA, Sieving PA, Sheils DM, McNally N, Creighton P, Erven A, et al. Retinopathy induced in mice by targeted disruption of the rhodopsin gene. Nat Genet. 1997;15:216–219. doi: 10.1038/ng0297-216. [DOI] [PubMed] [Google Scholar]
- Hwa J, Reeves PJ, Klein-Seetharaman J, Davidson F, Khorana HG. Structure and function in rhodopsin: further elucidation of the role of the intradiscal cysteines, Cys-110, -185, and -187, in rhodopsin folding and function. Proc Natl Acad Sci U S A. 1999;96:1932–1935. doi: 10.1073/pnas.96.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannaccone A, Man D, Waseem N, Jennings BJ, Ganapathiraju M, Gallaher K, Reese E, Bhattacharya SS, Klein-Seetharaman J. Retinitis pigmentosa associated with rhodopsin mutations: Correlation between phenotypic variability and molecular effects. Vision Res. 2006;46:4556–4567. doi: 10.1016/j.visres.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Illing ME, Rajan RS, Bence NF, Kopito RR. A rhodopsin mutant linked to autosomal dominant retinitis pigmentosa is prone to aggregate and interacts with the ubiquitin proteasome system. J Biol Chem. 2002;277:34150–34160. doi: 10.1074/jbc.M204955200. [DOI] [PubMed] [Google Scholar]
- Iraha S, Hirami Y, Ota S, Sunagawa GA, Mandai M, Tanihara H, Takahashi M, Kurimoto Y. Efficacy of valproic acid for retinitis pigmentosa patients: a pilot study. Clin Ophthalmol. 2016;10:1375–1384. doi: 10.2147/OPTH.S109995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V, Shen B, Zhang W, Hodgkins A, Keane T, Huang X, Skarnes WC. Off-target mutations are rare in Cas9-modified mice. Nat Methods. 2015;12:479. doi: 10.1038/nmeth.3408. [DOI] [PubMed] [Google Scholar]
- Jacobson SG, Kemp CM, Sung CH, Nathans J. Retinal function and rhodopsin levels in autosomal dominant retinitis pigmentosa with rhodopsin mutations. Am J Ophthalmol. 1991;112:256–271. doi: 10.1016/s0002-9394(14)76726-1. [DOI] [PubMed] [Google Scholar]
- Jastrzebska B, Chen Y, Orban T, Jin H, Hofmann L, Palczewski K. Disruption of Rhodopsin Dimerization with Synthetic Peptides Targeting an Interaction Interface. J Biol Chem. 2015;290:25728–25744. doi: 10.1074/jbc.M115.662684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Xiong S, Xia X. Chemical chaperone 4-phenylbutyrate prevents endoplasmic reticulum stress induced by T17M rhodopsin. Cell Biosci. 2014a;4:75. doi: 10.1186/2045-3701-4-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Xiong S, Xia X. Retinitis pigmentosaassociated rhodopsin mutant T17M induces endoplasmic reticulum (ER) stress and sensitizes cells to ER stress-induced cell death. Molecular medicine reports. 2014b;9:1737–1742. doi: 10.3892/mmr.2014.1987. [DOI] [PubMed] [Google Scholar]
- Kang MJ, Ryoo HD. Suppression of retinal degeneration in Drosophila by stimulation of ER-associated degradation. Proc Natl Acad Sci U S A. 2009;106:17043–17048. doi: 10.1073/pnas.0905566106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnik SS, Sakmar TP, Chen HB, Khorana HG. Cysteine residues 110 and 187 are essential for the formation of correct structure in bovine rhodopsin. Proc Natl Acad Sci U S A. 1988;85:8459–8463. doi: 10.1073/pnas.85.22.8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartasasmita A, Fujiki K, Iskandar E, Sovani I, Fujimaki T, Murakami A. A novel nonsense mutation in rhodopsin gene in two Indonesian families with autosomal recessive retinitis pigmentosa. Ophthalmic Genet. 2011;32:57–63. doi: 10.3109/13816810.2010.535892. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ, Malhotra JD. Calcium trafficking integrates endoplasmic reticulum function with mitochondrial bioenergetics. Biochim Biophys Acta. 2014;1843:2233–2239. doi: 10.1016/j.bbamcr.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J, Mencl S, Sahaboglu A, Farinelli P, van Veen T, Zrenner E, Ekstrom P, Paquet-Durand F, Arango-Gonzalez B. Calpain and PARP activation during photoreceptor cell death in P23H and S334ter rhodopsin mutant rats. PLoS One. 2011;6:e22181. doi: 10.1371/journal.pone.0022181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal S. Effect of rapamycin on the fate of P23H opsin associated with retinitis pigmentosa (an American Ophthalmological Society thesis) Trans Am Ophthalmol Soc. 2006;104:517–529. [PMC free article] [PubMed] [Google Scholar]
- Kaushal S, Khorana HG. Structure and function in rhodopsin. 7. Point mutations associated with autosomal dominant retinitis pigmentosa. Biochemistry. 1994;33:6121–6128. doi: 10.1021/bi00186a011. [DOI] [PubMed] [Google Scholar]
- Kaushal S, Ridge KD, Khorana HG. Structure and function in rhodopsin: the role of asparagine-linked glycosylation. Proc Natl Acad Sci U S A. 1994;91:4024–4028. doi: 10.1073/pnas.91.9.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keady BT, Le YZ, Pazour GJ. IFT20 is required for opsin trafficking and photoreceptor outer segment development. Molecular biology of the cell. 2011;22:921–930. doi: 10.1091/mbc.E10-09-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp CM, Jacobson SG, Roman AJ, Sung CH, Nathans J. Abnormal rod dark adaptation in autosomal dominant retinitis pigmentosa with proline-23-histidine rhodopsin mutation. Am J Ophthalmol. 1992;113:165–174. doi: 10.1016/s0002-9394(14)71529-6. [DOI] [PubMed] [Google Scholar]
- Kijas JW, Cideciyan AV, Aleman TS, Pianta MJ, Pearce-Kelling SE, Miller BJ, Jacobson SG, Aguirre GD, Acland GM. Naturally occurring rhodopsin mutation in the dog causes retinal dysfunction and degeneration mimicking human dominant retinitis pigmentosa. Proceedings of the National Academy of Sciences. 2002;99:6328–6333. doi: 10.1073/pnas.082714499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-T, Park J, Kim D, Kim K, Bae S, Schlesner M, Kim J-S. Questioning unexpected CRISPR off-target mutations in vivo. bioRxiv. 2017 [Google Scholar]
- Kimata N, Pope A, Eilers M, Opefi CA, Ziliox M, Hirshfeld A, Zaitseva E, Vogel R, Sheves M, Reeves PJ, Smith SO. Retinal orientation and interactions in rhodopsin reveal a two-stage trigger mechanism for activation. Nature communications. 2016;7:12683. doi: 10.1038/ncomms12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Sakai T, Komeima K, Kurimoto Y, Ueno S, Nishizawa Y, Usukura J, Fujikado T, Tano Y, Terasaki H. Generation of a transgenic rabbit model of retinal degeneration. Invest Ophthalmol Vis Sci. 2009;50:1371–1377. doi: 10.1167/iovs.08-2863. [DOI] [PubMed] [Google Scholar]
- Kosmaoglou M, Kanuga N, Aguila M, Garriga P, Cheetham ME. A dual role for EDEM1 in the processing of rod opsin. Journal of cell science. 2009;122:4465–4472. doi: 10.1242/jcs.055228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota P, Reeves PJ, Rajbhandary UL, Khorana HG. Opsin is present as dimers in COS1 cells: identification of amino acids at the dimeric interface. Proc Natl Acad Sci U S A. 2006;103:3054–3059. doi: 10.1073/pnas.0510982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft TW, Allen D, Petters RM, Hao Y, Peng YW, Wong F. Altered light responses of single rod photoreceptors in transgenic pigs expressing P347L or P347S rhodopsin. Mol Vis. 2005;11:1246–1256. [PubMed] [Google Scholar]
- Krebs MP, Holden DC, Joshi P, Clark CL, 3rd, Lee AH, Kaushal S. Molecular mechanisms of rhodopsin retinitis pigmentosa and the efficacy of pharmacological rescue. J Mol Biol. 2010;395:1063–1078. doi: 10.1016/j.jmb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- Krill AE, Archer D, Martin D. Sector retinitis pigmentosa. Am J Ophthalmol. 1970;69:977–987. doi: 10.1016/0002-9394(70)91042-1. [DOI] [PubMed] [Google Scholar]
- Kumar A, Midha N, Gogia V, Gupta S, Sehra S, Chohan A. Efficacy of oral valproic acid in patients with retinitis pigmentosa. J Ocul Pharmacol Ther. 2014;30:580–586. doi: 10.1089/jop.2013.0166. [DOI] [PubMed] [Google Scholar]
- Kumaramanickavel G, Maw M, Denton MJ, John S, Srikumari CR, Orth U, Oehlmann R, Gal A. Missense rhodopsin mutation in a family with recessive RP. Nat Genet. 1994;8:10–11. doi: 10.1038/ng0994-10. [DOI] [PubMed] [Google Scholar]
- Kunte MM, Choudhury S, Manheim JF, Shinde VM, Miura M, Chiodo VA, Hauswirth WW, Gorbatyuk OS, Gorbatyuk MS. ER Stress Is Involved in T17M Rhodopsin-Induced Retinal Degeneration. Investigative Ophthalmology & Visual Science. 2012;53:3792–3800. doi: 10.1167/iovs.11-9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau C, Clement K, Hsu JY, Pattanayak V, Joung JK, Aryee MJ, Pinello L. “Unexpected mutations after CRISPR-Cas9 editing in vivo” are most likely pre-existing sequence variants and not nuclease-induced mutations. bioRxiv. 2017 [Google Scholar]
- Latella MC, Di Salvo MT, Cocchiarella F, Benati D, Grisendi G, Comitato A, Marigo V, Recchia A. In vivo Editing of the Human Mutant Rhodopsin Gene by Electroporation of Plasmid-based CRISPR/Cas9 in the Mouse Retina. Mol Ther Nucleic Acids. 2016;5:e389. doi: 10.1038/mtna.2016.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVail MM, Yasumura D, Matthes MT, Drenser KA, Flannery JG, Lewin AS, Hauswirth WW. Ribozyme rescue of photoreceptor cells in P23H transgenic rats: long-term survival and late-stage therapy. Proc Natl Acad Sci U S A. 2000;97:11488–11493. doi: 10.1073/pnas.210319397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson EC, Bhatia SK, Han MK, Aung MH, Ciavatta V, Boatright JH, Pardue MT. Tauroursodeoxycholic Acid Protects Retinal Function and Structure in rd1 Mice. Adv Exp Med Biol. 2016;854:431–436. doi: 10.1007/978-3-319-17121-0_57. [DOI] [PubMed] [Google Scholar]
- Lee D, Geller S, Walsh N, Valter K, Yasumura D, Matthes M, LaVail M, Stone J. Photoreceptor Degeneration in Pro23His and S334ter Transgenic Rats. In: LaVail MM, Hollyfield JG, Anderson RE, editors. Retinal Degenerations: Mechanisms and Experimental Therapy. Springer; US Boston, MA: 2003. pp. 297–302. [DOI] [PubMed] [Google Scholar]
- Lee ES, Flannery JG. Transport of Truncated Rhodopsin and Its Effects on Rod Function and Degeneration. Investigative ophthalmology & visual science. 2007;48:2868–2876. doi: 10.1167/iovs.06-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lem J, Krasnoperova NV, Calvert PD, Kosaras B, Cameron DA, Nicolo M, Makino CL, Sidman RL. Morphological, physiological, and biochemical changes in rhodopsin knockout mice. Proc Natl Acad Sci U S A. 1999;96:736–741. doi: 10.1073/pnas.96.2.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard KC, Petrin D, Coupland SG, Baker AN, Leonard BC, LaCasse EC, Hauswirth WW, Korneluk RG, Tsilfidis C. XIAP protection of photoreceptors in animal models of retinitis pigmentosa. PLoS One. 2007;2:e314. doi: 10.1371/journal.pone.0000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- Li S, Samardzija M, Yang Z, Grimm C, Jin M. Pharmacological Amelioration of Cone Survival and Vision in a Mouse Model for Leber Congenital Amaurosis. J Neurosci. 2016;36:5808–5819. doi: 10.1523/JNEUROSCI.3857-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Sandberg MA, Pawlyk BS, Rosner B, Hayes KC, Dryja TP, Berson EL. Effect of vitamin A supplementation on rhodopsin mutants threonine-17 --> methionine and proline-347 --> serine in transgenic mice and in cell cultures. Proc Natl Acad Sci U S A. 1998a;95:11933–11938. doi: 10.1073/pnas.95.20.11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Snyder WK, Olsson JE, Dryja TP. Transgenic mice carrying the dominant rhodopsin mutation P347S: evidence for defective vectorial transport of rhodopsin to the outer segments. Proc Natl Acad Sci U S A. 1996;93:14176–14181. doi: 10.1073/pnas.93.24.14176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tao W, Luo L, Huang D, Kauper K, Stabila P, Lavail MM, Laties AM, Wen R. CNTF induces regeneration of cone outer segments in a rat model of retinal degeneration. PLoS One. 2010;5:e9495. doi: 10.1371/journal.pone.0009495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZY, Wong F, Chang JH, Possin DE, Hao Y, Petters RM, Milam AH. Rhodopsin transgenic pigs as a model for human retinitis pigmentosa. Invest Ophthalmol Vis Sci. 1998b;39:808–819. [PubMed] [Google Scholar]
- Liang FQ, Aleman TS, Dejneka NS, Dudus L, Fisher KJ, Maguire AM, Jacobson SG, Bennett J. Long-term protection of retinal structure but not function using RAAV.CNTF in animal models of retinitis pigmentosa. Mol Ther. 2001;4:461–472. doi: 10.1006/mthe.2001.0473. [DOI] [PubMed] [Google Scholar]
- Lim KP, Yip SP, Cheung SC, Leung KW, Lam ST, To CH. Novel PRPF31 and PRPH2 mutations and co-occurrence of PRPF31 and RHO mutations in Chinese patients with retinitis pigmentosa. Arch Ophthalmol. 2009;127:784–790. doi: 10.1001/archophthalmol.2009.112. [DOI] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Garriga P, Khorana HG. Structure and function in rhodopsin: correct folding and misfolding in two point mutants in the intradiscal domain of rhodopsin identified in retinitis pigmentosa. Proc Natl Acad Sci U S A. 1996;93:4554–4559. doi: 10.1073/pnas.93.10.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanova ES, Finkelstein S, Skiba NP, Arshavsky VY. Proteasome overload is a common stress factor in multiple forms of inherited retinal degeneration. Proc Natl Acad Sci U S A. 2013;110:9986–9991. doi: 10.1073/pnas.1305521110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lythgoe JN. The Ecology of Vision. Clarendon Press; Oxford: 1979. [Google Scholar]
- Ma S, Venkatesh A, Langellotto F, Le YZ, Hall MN, Ruegg MA, Punzo C. Loss of mTOR signaling affects cone function, cone structure and expression of cone specific proteins without affecting cone survival. Exp Eye Res. 2015;135:1–13. doi: 10.1016/j.exer.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macke JP, Davenport CM, Jacobson SG, Hennessey JC, Gonzalez-Fernandez F, Conway BP, Heckenlively J, Palmer R, Maumenee IH, Sieving P, et al. Identification of novel rhodopsin mutations responsible for retinitis pigmentosa: implications for the structure and function of rhodopsin. Am J Hum Genet. 1993;53:80–89. [PMC free article] [PubMed] [Google Scholar]
- Makino CL, Wen XH, Michaud NA, Covington HI, DiBenedetto E, Hamm HE, Lem J, Caruso G. Rhodopsin expression level affects rod outer segment morphology and photoresponse kinetics. PLoS One. 2012;7:e37832. doi: 10.1371/journal.pone.0037832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal MN, Heckenlively JR, Burch T, Chen L, Vasireddy V, Koenekoop RK, Sieving PA, Ayyagari R. Sequencing arrays for screening multiple genes associated with early-onset human retinal degenerations on a high-throughput platform. Invest Ophthalmol Vis Sci. 2005;46:3355–3362. doi: 10.1167/iovs.05-0007. [DOI] [PubMed] [Google Scholar]
- Mao H, James T, Jr, Schwein A, Shabashvili AE, Hauswirth WW, Gorbatyuk MS, Lewin AS. AAV delivery of wild-type rhodopsin preserves retinal function in a mouse model of autosomal dominant retinitis pigmentosa. Hum Gene Ther. 2011;22:567–575. doi: 10.1089/hum.2010.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmor MF. A randomized trial of vitamin A and vitamin E supplementation for retinitis pigmentosa. Arch Ophthalmol. 1993;111:1460–1461. doi: 10.1001/archopht.1993.01090110018004. author reply 1463-1465. [DOI] [PubMed] [Google Scholar]
- Marsili S, Genini S, Sudharsan R, Gingrich J, Aguirre GD, Beltran WA. Exclusion of the unfolded protein response in light-induced retinal degeneration in the canine T4R RHO model of autosomal dominant retinitis pigmentosa. PLoS One. 2015;10:e0115723. doi: 10.1371/journal.pone.0115723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massof RW, Finkelstein D. Supplemental vitamin A retards loss of ERG amplitude in retinitis pigmentosa. Arch Ophthalmol. 1993;111:751–754. doi: 10.1001/archopht.1993.01090060039019. [DOI] [PubMed] [Google Scholar]
- Massof RW, Fishman GA. How strong is the evidence that nutritional supplements slow the progression of retinitis pigmentosa? Arch Ophthalmol. 2010;128:493–495. doi: 10.1001/archophthalmol.2010.46. [DOI] [PubMed] [Google Scholar]
- Matsumura H, Shimizu Y, Ohsawa Y, Kawahara A, Uchiyama Y, Nagata S. Necrotic death pathway in Fas receptor signaling. The Journal of cell biology. 2000;151:1247–1256. doi: 10.1083/jcb.151.6.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKibbin C, Toye AM, Reeves PJ, Khorana HG, Edwards PC, Villa C, Booth PJ. Opsin stability and folding: the role of Cys185 and abnormal disulfide bond formation in the intradiscal domain. J Mol Biol. 2007;374:1309–1318. doi: 10.1016/j.jmb.2007.10.013. [DOI] [PubMed] [Google Scholar]
- Mendes CS, Levet C, Chatelain G, Dourlen P, Fouillet A, Dichtel-Danjoy ML, Gambis A, Ryoo HD, Steller H, Mollereau B. ER stress protects from retinal degeneration. The EMBO journal. 2009;28:1296–1307. doi: 10.1038/emboj.2009.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes HF, Cheetham ME. Pharmacological manipulation of gain-of-function and dominant-negative mechanisms in rhodopsin retinitis pigmentosa. Hum Mol Genet. 2008;17:3043–3054. doi: 10.1093/hmg/ddn202. [DOI] [PubMed] [Google Scholar]
- Mendes HF, van der Spuy J, Chapple JP, Cheetham ME. Mechanisms of cell death in rhodopsin retinitis pigmentosa: implications for therapy. Trends in molecular medicine. 2005;11:177–185. doi: 10.1016/j.molmed.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Mezer E, Sutherland J, Goei SL, Heon E, Levin AV. Utility of molecular testing for related retinal dystrophies. Can J Ophthalmol. 2006;41:190–196. doi: 10.1139/I06-007. [DOI] [PubMed] [Google Scholar]
- Mitton KP, Guzman AE, Deshpande M, Byrd D, DeLooff C, Mkoyan K, Zlojutro P, Wallace A, Metcalf B, Laux K, Sotzen J, et al. Different effects of valproic acid on photoreceptor loss in Rd1 and Rd10 retinal degeneration mice. Mol Vis. 2014;20:1527–1544. [PMC free article] [PubMed] [Google Scholar]
- Mockel A, Obringer C, Hakvoort TB, Seeliger M, Lamers WH, Stoetzel C, Dollfus H, Marion V. Pharmacological modulation of the retinal unfolded protein response in Bardet-Biedl syndrome reduces apoptosis and preserves light detection ability. J Biol Chem. 2012;287:37483–37494. doi: 10.1074/jbc.M112.386821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Ikeda Y, Nakatake S, Miller JW, Vavvas DG, Sonoda KH, Ishibashi T. Necrotic cone photoreceptor cell death in retinitis pigmentosa. Cell Death & Disease. 2015;6:e2038. doi: 10.1038/cddis.2015.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Matsumoto H, Roh M, Suzuki J, Hisatomi T, Ikeda Y, Miller JW, Vavvas DG. Receptor interacting protein kinase mediates necrotic cone but not rod cell death in a mouse model of inherited degeneration. Proc Natl Acad Sci U S A. 2012;109:14598–14603. doi: 10.1073/pnas.1206937109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AR, Fliesler SJ, Al-Ubaidi MR. Rhodopsin: the functional significance of asn-linked glycosylation and other post-translational modifications. Ophthalmic Genet. 2009;30:109–120. doi: 10.1080/13816810902962405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AR, Vuong L, Brobst D, Fliesler SJ, Peachey NS, Gorbatyuk MS, Naash MI, Al-Ubaidi MR. Glycosylation of rhodopsin is necessary for its stability and incorporation into photoreceptor outer segment discs. Human Molecular Genetics. 2015a;24:2709–2723. doi: 10.1093/hmg/ddv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray SF, Jazayeri A, Matthes MT, Yasumura D, Yang H, Peralta R, Watt A, Freier S, Hung G, Adamson PS, Guo S, et al. Allele-Specific Inhibition of Rhodopsin With an Antisense Oligonucleotide Slows Photoreceptor Cell Degeneration. Invest Ophthalmol Vis Sci. 2015b;56:6362–6375. doi: 10.1167/iovs.15-16400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naash MI, Hollyfield JG, al-Ubaidi MR, Baehr W. Simulation of human autosomal dominant retinitis pigmentosa in transgenic mice expressing a mutated murine opsin gene. Proc Natl Acad Sci U S A. 1993;90:5499–5503. doi: 10.1073/pnas.90.12.5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashine S, Bhootada Y, Lewin AS, Gorbatyuk M. Ablation of C/EBP homologous protein does not protect T17M RHO mice from retinal degeneration. PLoS One. 2013;8:e63205. doi: 10.1371/journal.pone.0063205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans J, Hogness DS. Isolation, sequence analysis, and intron-exon arrangement of the gene encoding bovine rhodopsin. Cell. 1983;34:807–814. doi: 10.1016/0092-8674(83)90537-8. [DOI] [PubMed] [Google Scholar]
- Nathans J, Hogness DS. Isolation and nucleotide sequence of the gene encoding human rhodopsin. Proc Natl Acad Sci U S A. 1984;81:4851–4855. doi: 10.1073/pnas.81.15.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans J, Piantanida TP, Eddy RL, Shows TB, Hogness DS. Molecular genetics of inherited variation in human color vision. Science. 1986;232:203–210. doi: 10.1126/science.3485310. [DOI] [PubMed] [Google Scholar]
- Noorwez SM, Kuksa V, Imanishi Y, Zhu L, Filipek S, Palczewski K, Kaushal S. Pharmacological chaperone-mediated in vivo folding and stabilization of the P23H-opsin mutant associated with autosomal dominant retinitis pigmentosa. J Biol Chem. 2003;278:14442–14450. doi: 10.1074/jbc.M300087200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noorwez SM, Sama RR, Kaushal S. Calnexin improves the folding efficiency of mutant rhodopsin in the presence of pharmacological chaperone 11-cis-retinal. J Biol Chem. 2009;284:33333–33342. doi: 10.1074/jbc.M109.043364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nördstrom K, Larsson TA, Larhammar D. Extensive duplications of phototransduction genes in early vertebrate evolution correlate with block (chromosome) duplications. Genomics. 2004;83:852–872. doi: 10.1016/j.ygeno.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Norton EW. A randomized trial of vitamin A and vitamin E supplementation for retinitis pigmentosa. Arch Ophthalmol. 1993;111:1460. doi: 10.1001/archopht.1993.01090110018003. author reply 1463-1465. [DOI] [PubMed] [Google Scholar]
- O'Reilly M, Palfi A, Chadderton N, Millington-Ward S, Ader M, Cronin T, Tuohy T, Auricchio A, Hildinger M, Tivnan A, McNally N, et al. RNA interference-mediated suppression and replacement of human rhodopsin in vivo. Am J Hum Genet. 2007;81:127–135. doi: 10.1086/519025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoye G, Zimmer J, Sung J, Gehlbach P, Deering T, Nambu H, Hackett S, Melia M, Esumi N, Zack DJ, Campochiaro PA. Increased expression of brain-derived neurotrophic factor preserves retinal function and slows cell death from rhodopsin mutation or oxidative damage. J Neurosci. 2003;23:4164–4172. doi: 10.1523/JNEUROSCI.23-10-04164.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson JE, Gordon JW, Pawlyk BS, Roof D, Hayes A, Molday RS, Mukai S, Cowley GS, Berson EL, Dryja TP. Transgenic mice with a rhodopsin mutation (Pro23His): a mouse model of autosomal dominant retinitis pigmentosa. Neuron. 1992;9:815–830. doi: 10.1016/0896-6273(92)90236-7. [DOI] [PubMed] [Google Scholar]
- Opefi CA, South K, Reynolds CA, Smith SO, Reeves PJ. Retinitis pigmentosa mutants provide insight into the role of the N-terminal cap in rhodopsin folding, structure, and function. J Biol Chem. 2013;288:33912–33926. doi: 10.1074/jbc.M113.483032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orhan E, Dalkara D, Neuille M, Lechauve C, Michiels C, Picaud S, Leveillard T, Sahel JA, Naash MI, Lavail MM, Zeitz C, et al. Genotypic and phenotypic characterization of P23H line 1 rat model. PLoS One. 2015;10:e0127319. doi: 10.1371/journal.pone.0127319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov Yu A, Abdulaev NG, Bogachuk AS. Two adjacent cysteine residues in the C-terminal cytoplasmic fragment of bovine rhodopsin are palmitylated. FEBS Lett. 1988;230:1–5. doi: 10.1016/0014-5793(88)80628-8. [DOI] [PubMed] [Google Scholar]
- Oveson BC, Iwase T, Hackett SF, Lee SY, Usui S, Sedlak TW, Snyder SH, Campochiaro PA, Sung JU. Constituents of bile, bilirubin and TUDCA, protect against oxidative stress-induced retinal degeneration. J Neurochem. 2011;116:144–153. doi: 10.1111/j.1471-4159.2010.07092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Parfitt DA, Aguila M, McCulley CH, Bevilacqua D, Mendes HF, Athanasiou D, Novoselov SS, Kanuga N, Munro PM, Coffey PJ, Kalmar B, et al. The heat-shock response co-inducer arimoclomol protects against retinal degeneration in rhodopsin retinitis pigmentosa. Cell Death Dis. 2014;5:e1236. doi: 10.1038/cddis.2014.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PS. Constitutively active rhodopsin and retinal disease. Advances in pharmacology. 2014;70:1–36. doi: 10.1016/B978-0-12-417197-8.00001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearring JN, San Agustin JT, Lobanova ES, Gabriel CJ, Lieu EC, Monis WJ, Stuck MW, Strittmatter L, Jaber SM, Arshavsky VY, Pazour GJ. Loss of Arf4 causes severe degeneration of the exocrine pancreas but not cystic kidney disease or retinal degeneration. PLoS Genet. 2017;13:e1006740. doi: 10.1371/journal.pgen.1006740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennesi ME, Nishikawa S, Matthes MT, Yasumura D, LaVail MM. The relationship of photoreceptor degeneration to retinal vascular development and loss in mutant rhodopsin transgenic and RCS rats. Exp Eye Res. 2008;87:561–570. doi: 10.1016/j.exer.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmutter DH. Chemical chaperones: a pharmacological strategy for disorders of protein folding and trafficking. Pediatr Res. 2002;52:832–836. doi: 10.1203/00006450-200212000-00004. [DOI] [PubMed] [Google Scholar]
- Petters RM, Alexander CA, Wells KD, Collins EB, Sommer JR, Blanton MR, Rojas G, Hao Y, Flowers WL, Banin E, Cideciyan AV, et al. Genetically engineered large animal model for studying cone photoreceptor survival and degeneration in retinitis pigmentosa. Nat Biotechnol. 1997;15:965–970. doi: 10.1038/nbt1097-965. [DOI] [PubMed] [Google Scholar]
- Phillips MJ, Walker TA, Choi HY, Faulkner AE, Kim MK, Sidney SS, Boyd AP, Nickerson JM, Boatright JH, Pardue MT. Tauroursodeoxycholic acid preservation of photoreceptor structure and function in the rd10 mouse through postnatal day 30. Invest Ophthalmol Vis Sci. 2008;49:2148–2155. doi: 10.1167/iovs.07-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploier B, Caro LN, Morizumi T, Pandey K, Pearring JN, Goren MA, Finnemann SC, Graumann J, Arshavsky VY, Dittman JS, Ernst OP, et al. Dimerization deficiency of enigmatic retinitis pigmentosa-linked rhodopsin mutants. Nature communications. 2016;7:12832. doi: 10.1038/ncomms12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter AG, Janicke RU. Emerging roles of caspase-3 in apoptosis. Cell death and differentiation. 1999;6:99–104. doi: 10.1038/sj.cdd.4400476. [DOI] [PubMed] [Google Scholar]
- Price BA, Sandoval IM, Chan F, Simons DL, Wu SM, Wensel TG, Wilson JH. Mislocalization and degradation of human P23H-rhodopsin-GFP in a knockin mouse model of retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2011;52:9728–9736. doi: 10.1167/iovs.11-8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punzo C, Kornacker K, Cepko CL. Stimulation of the insulin/mTOR pathway delays cone death in a mouse model of retinitis pigmentosa. Nature neuroscience. 2009;12:44–52. doi: 10.1038/nn.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan RS, Kopito RR. Suppression of wild-type rhodopsin maturation by mutants linked to autosomal dominant retinitis pigmentosa. J Biol Chem. 2005;280:1284–1291. doi: 10.1074/jbc.M406448200. [DOI] [PubMed] [Google Scholar]
- Rakoczy EP, Kiel C, McKeone R, Stricher F, Serrano L. Analysis of disease-linked rhodopsin mutations based on structure, function, and protein stability calculations. J Mol Biol. 2011;405:584–606. doi: 10.1016/j.jmb.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Rakshit T, Park PS. Impact of reduced rhodopsin expression on the structure of rod outer segment disc membranes. Biochemistry. 2015;54:2885–2894. doi: 10.1021/acs.biochem.5b00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon E, Cordomi A, Aguila M, Srinivasan S, Dong X, Moore AT, Webster AR, Cheetham ME, Garriga P. Differential light-induced responses in sectorial inherited retinal degeneration. J Biol Chem. 2014;289:35918–35928. doi: 10.1074/jbc.M114.609958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon E, del Valle LJ, Garriga P. Unusual thermal and conformational properties of the rhodopsin congenital night blindness mutant Thr-94 --> Ile. J Biol Chem. 2003;278:6427–6432. doi: 10.1074/jbc.M210929200. [DOI] [PubMed] [Google Scholar]
- Rana T, Shinde VM, Starr CR, Kruglov AA, Boitet ER, Kotla P, Zolotukhin S, Gross AK, Gorbatyuk MS. An activated unfolded protein response promotes retinal degeneration and triggers an inflammatory response in the mouse retina. Cell Death Dis. 2014;5:e1578. doi: 10.1038/cddis.2014.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VR, Cohen GB, Oprian DD. Rhodopsin mutation G90D and a molecular mechanism for congenital night blindness. Nature. 1994;367:639–642. doi: 10.1038/367639a0. [DOI] [PubMed] [Google Scholar]
- Reiff C, Owczarek-Lipska M, Spital G, Roger C, Hinz H, Juschke C, Thiele H, Altmuller J, Nurnberg P, Da Costa R, Neidhardt J. The mutation p.E113K in the Schiff base counterion of rhodopsin is associated with two distinct retinal phenotypes within the same family. Sci Rep. 2016;6:36208. doi: 10.1038/srep36208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remondelli P, Renna M. The Endoplasmic Reticulum Unfolded Protein Response in Neurodegenerative Disorders and Its Potential Therapeutic Significance. Frontiers in Molecular Neuroscience. 2017;10:187. doi: 10.3389/fnmol.2017.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex TS, Kasmala L, Bond WS, de Lucas Cerrillo AM, Wynn K, Lewin AS. Erythropoietin Slows Photoreceptor Cell Death in a Mouse Model of Autosomal Dominant Retinitis Pigmentosa. PLoS One. 2016;11:e0157411. doi: 10.1371/journal.pone.0157411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman-Sanchez R, Wensel TG, Wilson JH. Nonsense mutations in the rhodopsin gene that give rise to mild phenotypes trigger mRNA degradation in human cells by nonsense-mediated decay. Exp Eye Res. 2016;145:444–449. doi: 10.1016/j.exer.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld PJ, Cowley GS, McGee TL, Sandberg MA, Berson EL, Dryja TP. A null mutation in the rhodopsin gene causes rod photoreceptor dysfunction and autosomal recessive retinitis pigmentosa. Nat Genet. 1992;1:209–213. doi: 10.1038/ng0692-209. [DOI] [PubMed] [Google Scholar]
- Rosenfeld PJ, Hahn LB, Sandberg MA, Dryja TP, Berson EL. Low incidence of retinitis pigmentosa among heterozygous carriers of a specific rhodopsin splice site mutation. Invest Ophthalmol Vis Sci. 1995;36:2186–2192. [PubMed] [Google Scholar]
- Ross JW, Fernandez de Castro JP, Zhao J, Samuel M, Walters E, Rios C, Bray-Ward P, Jones BW, Marc RE, Wang W, Zhou L, et al. Generation of an inbred miniature pig model of retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2012;53:501–507. doi: 10.1167/iovs.11-8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakami S, Maeda T, Bereta G, Okano K, Golczak M, Sumaroka A, Roman AJ, Cideciyan AV, Jacobson SG, Palczewski K. Probing mechanisms of photoreceptor degeneration in a new mouse model of the common form of autosomal dominant retinitis pigmentosa due to P23H opsin mutations. J Biol Chem. 2011;286:10551–10567. doi: 10.1074/jbc.M110.209759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakmar TP, Franke RR, Khorana HG. Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc Natl Acad Sci U S A. 1989;86:8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba RS, Munro PM, Luthert PJ, Cheetham ME. The cellular fate of mutant rhodopsin: quality control, degradation and aggresome formation. Journal of cell science. 2002;115:2907–2918. doi: 10.1242/jcs.115.14.2907. [DOI] [PubMed] [Google Scholar]
- Samardzija M, Wenzel A, Naash M, Reme CE, Grimm C. Rpe65 as a modifier gene for inherited retinal degeneration. Eur J Neurosci. 2006a;23:1028–1034. doi: 10.1111/j.1460-9568.2006.04639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samardzija M, Wenzel A, Thiersch M, Frigg R, Reme C, Grimm C. Caspase-1 ablation protects photoreceptors in a model of autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2006b;47:5181–5190. doi: 10.1167/iovs.06-0556. [DOI] [PubMed] [Google Scholar]
- Sanchez-Reyes OB, Cooke ALG, Tranter DB, Rashid D, Eilers M, Reeves PJ, Smith SO. G Protein-Coupled Receptors Contain Two Conserved Packing Clusters. Biophys J. 2017;112:2315–2326. doi: 10.1016/j.bpj.2017.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg MA, Rosner B, Weigel-DiFranco C, Berson EL. Lack of scientific rationale for use of valproic acid for retinitis pigmentosa. Br J Ophthalmol. 2011;95:744. doi: 10.1136/bjo.2010.198176. [DOI] [PubMed] [Google Scholar]
- Saqib MA, Nikopoulos K, Ullah E, Sher Khan F, Iqbal J, Bibi R, Jarral A, Sajid S, Nishiguchi KM, Venturini G, Ansar M, et al. Homozygosity mapping reveals novel and known mutations in Pakistani families with inherited retinal dystrophies. Sci Rep. 2015;5:9965. doi: 10.1038/srep09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer KA, Wu WH, Colgan DF, Tsang SH, Bassuk AG, Mahajan VB. Unexpected mutations after CRISPR-Cas9 editing in vivo. Nat Methods. 2017;14:547–548. doi: 10.1038/nmeth.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichtenbrede FC, MacNeil A, Bainbridge JW, Tschernutter M, Thrasher AJ, Smith AJ, Ali RR. Intraocular gene delivery of ciliary neurotrophic factor results in significant loss of retinal function in normal mice and in the Prph2Rd2/Rd2 model of retinal degeneration. Gene Ther. 2003;10:523–527. doi: 10.1038/sj.gt.3301929. [DOI] [PubMed] [Google Scholar]
- Shanmugam PM, Minija CK, Ramanjulu R, Tekwani P, Saxena M. Effect of short-term oral valproic Acid on vision and visual field in retinitis pigmentosa. Ophthalmol Ther. 2012;1:6. doi: 10.1007/s40123-012-0006-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon D, Kimchi A, Rivolta C. OR2W3 sequence variants are unlikely to cause inherited retinal diseases. Ophthalmic Genet. 2016;37:366–368. doi: 10.3109/13816810.2015.1081252. [DOI] [PubMed] [Google Scholar]
- Sieving PA, Fowler ML, Bush RA, Machida S, Calvert PD, Green DG, Makino CL, McHenry CL. Constitutive "light" adaptation in rods from G90D rhodopsin: a mechanism for human congenital nightblindness without rod cell loss. J Neurosci. 2001;21:5449–5460. doi: 10.1523/JNEUROSCI.21-15-05449.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieving PA, Richards JE, Naarendorp F, Bingham EL, Scott K, Alpern M. Dark-light: model for nightblindness from the human rhodopsin Gly-90-->Asp mutation. Proc Natl Acad Sci U S A. 1995;92:880–884. doi: 10.1073/pnas.92.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal A, Guo Y, Matkovic M, Schertler G, Deupi X, Yan EC, Standfuss J. Structural role of the T94I rhodopsin mutation in congenital stationary night blindness. EMBO reports. 2016;17:1431–1440. doi: 10.15252/embr.201642671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisk RA. Valproic acid treatment may be harmful in non-dominant forms of retinitis pigmentosa. Br J Ophthalmol. 2012;96:1154–1155. doi: 10.1136/bjophthalmol-2012-301950. [DOI] [PubMed] [Google Scholar]
- Sizova OS, Shinde VM, Lenox AR, Gorbatyuk MS. Modulation of cellular signaling pathways in P23H rhodopsin photoreceptors. Cell Signal. 2014;26:665–672. doi: 10.1016/j.cellsig.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Gore A, Yan W, Abalde-Atristain L, Li Z, He C, Wang Y, Brodsky RA, Zhang K, Cheng L, Ye Z. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell. 2014;15:12–13. doi: 10.1016/j.stem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SO. Structure and activation of the visual pigment rhodopsin. Annu Rev Biophys. 2010;39:309–328. doi: 10.1146/annurev-biophys-101209-104901. [DOI] [PubMed] [Google Scholar]
- Song D, Dunaief JL. Retinal iron homeostasis in health and disease. Frontiers in Aging Neuroscience. 2013;5:24. doi: 10.3389/fnagi.2013.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovic A, Hwang I, Khorana HG, Hwa J. Retinitis pigmentosa rhodopsin mutations L125R and A164V perturb critical interhelical interactions: new insights through compensatory mutations and crystal structure analysis. J Biol Chem. 2003;278:39020–39028. doi: 10.1074/jbc.M303625200. [DOI] [PubMed] [Google Scholar]
- Stone EM. Finding and interpreting genetic variations that are important to ophthalmologists. Trans Am Ophthalmol Soc. 2003;101:437–484. [PMC free article] [PubMed] [Google Scholar]
- Sullivan LS, Bowne SJ, Birch DG, Hughbanks-Wheaton D, Heckenlively JR, Lewis RA, Garcia CA, Ruiz RS, Blanton SH, Northrup H, Gire AI, et al. Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa: a screen of known genes in 200 families. Invest Ophthalmol Vis Sci. 2006;47:3052–3064. doi: 10.1167/iovs.05-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung CH, Chuang JZ. The cell biology of vision. The Journal of cell biology. 2010;190:953–963. doi: 10.1083/jcb.201006020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung CH, Davenport CM, Hennessey JC, Maumenee IH, Jacobson SG, Heckenlively JR, Nowakowski R, Fishman G, Gouras P, Nathans J. Rhodopsin mutations in autosomal dominant retinitis pigmentosa. Proc Natl Acad Sci U S A. 1991;88:6481–6485. doi: 10.1073/pnas.88.15.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung CH, Davenport CM, Nathans J. Rhodopsin mutations responsible for autosomal dominant retinitis pigmentosa. Clustering of functional classes along the polypeptide chain. J Biol Chem. 1993;268:26645–26649. [PubMed] [Google Scholar]
- Sung CH, Makino C, Baylor D, Nathans J. A rhodopsin gene mutation responsible for autosomal dominant retinitis pigmentosa results in a protein that is defective in localization to the photoreceptor outer segment. J Neurosci. 1994;14:5818–5833. doi: 10.1523/JNEUROSCI.14-10-05818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam BM, Moritz OL. Characterization of rhodopsin P23H-induced retinal degeneration in a Xenopus laevis model of retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2006;47:3234–3241. doi: 10.1167/iovs.06-0213. [DOI] [PubMed] [Google Scholar]
- Tam BM, Moritz OL. Dark rearing rescues P23H rhodopsin-induced retinal degeneration in a transgenic Xenopus laevis model of retinitis pigmentosa: a chromophore-dependent mechanism characterized by production of N-terminally truncated mutant rhodopsin. J Neurosci. 2007;27:9043–9053. doi: 10.1523/JNEUROSCI.2245-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam BM, Moritz OL. The role of rhodopsin glycosylation in protein folding, trafficking, and light-sensitive retinal degeneration. J Neurosci. 2009;29:15145–15154. doi: 10.1523/JNEUROSCI.4259-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam BM, Moritz OL, Hurd LB, Papermaster DS. Identification of an outer segment targeting signal in the COOH terminus of rhodopsin using transgenic Xenopus laevis. The Journal of cell biology. 2000;151:1369–1380. doi: 10.1083/jcb.151.7.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam BM, Noorwez SM, Kaushal S, Kono M, Moritz OL. Photoactivation-induced instability of rhodopsin mutants T4K and T17M in rod outer segments underlies retinal degeneration in X. laevis transgenic models of retinitis pigmentosa. J Neurosci. 2014;34:13336–13348. doi: 10.1523/JNEUROSCI.1655-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam BM, Qazalbash A, Lee HC, Moritz OL. The dependence of retinal degeneration caused by the rhodopsin P23H mutation on light exposure and vitamin a deprivation. Invest Ophthalmol Vis Sci. 2010a;51:1327–1334. doi: 10.1167/iovs.09-4123. [DOI] [PubMed] [Google Scholar]
- Tam BM, Xie G, Oprian DD, Moritz OL. Mislocalized rhodopsin does not require activation to cause retinal degeneration and neurite outgrowth in Xenopus laevis. J Neurosci. 2006;26:203–209. doi: 10.1523/JNEUROSCI.3849-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam LC, Kiang AS, Campbell M, Keaney J, Farrar GJ, Humphries MM, Kenna PF, Humphries P. Prevention of autosomal dominant retinitis pigmentosa by systemic drug therapy targeting heat shock protein 90 (Hsp90) Hum Mol Genet. 2010b;19:4421–4436. doi: 10.1093/hmg/ddq369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasan I, Zhao H. Targeting Specificity of the CRISPR/Cas9 System. ACS Synth Biol. 2017;6:1609–1613. doi: 10.1021/acssynbio.7b00270. [DOI] [PubMed] [Google Scholar]
- Toledo D, Ramon E, Aguila M, Cordomi A, Perez JJ, Mendes HF, Cheetham ME, Garriga P. Molecular mechanisms of disease for mutations at Gly-90 in rhodopsin. J Biol Chem. 2011;286:39993–40001. doi: 10.1074/jbc.M110.201517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trichonas G, Murakami Y, Thanos A, Morizane Y, Kayama M, Debouck CM, Hisatomi T, Miller JW, Vavvas DG. Receptor interacting protein kinases mediate retinal detachment-induced photoreceptor necrosis and compensate for inhibition of apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21695–21700. doi: 10.1073/pnas.1009179107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Schil K, Karlstetter M, Aslanidis A, Dannhausen K, Azam M, Qamar R, Leroy BP, Depasse F, Langmann T, De Baere E. Autosomal recessive retinitis pigmentosa with homozygous rhodopsin mutation E150K and non-coding cis-regulatory variants in CRX-binding regions of SAMD7. Sci Rep. 2016;6:21307. doi: 10.1038/srep21307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schooneveld MJ, van den Born LI, van Genderen M, Bollemeijer JG. The conclusions of Clemson et al concerning valproic acid are premature. Br J Ophthalmol. 2011;95:153. doi: 10.1136/bjo.2010.194373. author reply 153-154. [DOI] [PubMed] [Google Scholar]
- Van Woerkom C, Ferrucci S. Sector retinitis pigmentosa. Optometry. 2005;76:309–317. doi: 10.1016/s1529-1839(05)70314-6. [DOI] [PubMed] [Google Scholar]
- Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nature reviews. Molecular cell biology. 2010;11:700–714. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- Vasireddy V, Chavali VR, Joseph VT, Kadam R, Lin JH, Jamison JA, Kompella UB, Reddy GB, Ayyagari R. Rescue of photoreceptor degeneration by curcumin in transgenic rats with P23H rhodopsin mutation. PLoS One. 2011;6:e21193. doi: 10.1371/journal.pone.0021193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vent-Schmidt RYJ, Wen RH, Zong Z, Chiu CN, Tam BM, May CG, Moritz OL. Opposing Effects of Valproic Acid Treatment Mediated by Histone Deacetylase Inhibitor Activity in Four Transgenic X. laevis Models of Retinitis Pigmentosa. J Neurosci. 2017;37:1039–1054. doi: 10.1523/JNEUROSCI.1647-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veres A, Gosis BS, Ding Q, Collins R, Ragavendran A, Brand H, Erdin S, Cowan CA, Talkowski ME, Musunuru K. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrotti A, Lobefalo L, Priolo T, Rapinese M, Trotta D, Morgese G, Gallenga PE, Chiarelli F. Color vision in epileptic adolescents treated with valproate and carbamazepine. Seizure. 2004;13:411–417. doi: 10.1016/j.seizure.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Viringipurampeer IA, Metcalfe AL, Bashar AE, Sivak O, Yanai A, Mohammadi Z, Moritz OL, Gregory-Evans CY, Gregory-Evans K. NLRP3 inflammasome activation drives bystander cone photoreceptor cell death in a P23H rhodopsin model of retinal degeneration. Hum Mol Genet. 2016;25:1501–1516. doi: 10.1093/hmg/ddw029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Morita Y, Mazelova J, Deretic D. The Arf GAP ASAP1 provides a platform to regulate Arf4- and Rab11-Rab8-mediated ciliary receptor targeting. The EMBO journal. 2012;31:4057–4071. doi: 10.1038/emboj.2012.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Montell C. Phototransduction and retinal degeneration in Drosophila. Pflugers Archiv : European journal of physiology. 2007;454:821–847. doi: 10.1007/s00424-007-0251-1. [DOI] [PubMed] [Google Scholar]
- Wang Y, Dawson VL, Dawson TM. Poly(ADP-ribose) signals to mitochondrial AIF: A key event in parthanatos. Experimental Neurology. 2009;218:193–202. doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wen XH, Ablonczy Z, Crouch RK, Makino CL, Lem J. Enhanced shutoff of phototransduction in transgenic mice expressing palmitoylation-deficient rhodopsin. J Biol Chem. 2005;280:24293–24300. doi: 10.1074/jbc.M502588200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss ER, Osawa S, Shi W, Dickerson CD. Effects of carboxyl-terminal truncation on the stability and G protein-coupling activity of bovine rhodopsin. Biochemistry. 1994;33:7587–7593. doi: 10.1021/bi00190a011. [DOI] [PubMed] [Google Scholar]
- White DA, Fritz JJ, Hauswirth WW, Kaushal S, Lewin AS. Increased sensitivity to light-induced damage in a mouse model of autosomal dominant retinal disease. Invest Ophthalmol Vis Sci. 2007;48:1942–1951. doi: 10.1167/iovs.06-1131. [DOI] [PubMed] [Google Scholar]
- Wilson CJ, Fennell T, Bothmer A, Maeder ML, Reyon D, Cotta-Ramusino C, Fernandez CA, Marco E, Barrera LA, Jayaram H, Albright CF, et al. The experimental design and data interpretation in ‘Unexpected mutations after CRISPR Cas9 editing in vivo’ by Schaefer et al. are insufficient to support the conclusions drawn by the authors. bioRxiv. 2017 [Google Scholar]
- Xie G, Gross AK, Oprian DD. An opsin mutant with increased thermal stability. Biochemistry. 2003;42:1995–2001. doi: 10.1021/bi020611z. [DOI] [PubMed] [Google Scholar]
- Yam GH, Gaplovska-Kysela K, Zuber C, Roth J. Sodium 4-phenylbutyrate acts as a chemical chaperone on misfolded myocilin to rescue cells from endoplasmic reticulum stress and apoptosis. Invest Ophthalmol Vis Sci. 2007;48:1683–1690. doi: 10.1167/iovs.06-0943. [DOI] [PubMed] [Google Scholar]
- Yang Y, Mohand-Said S, Danan A, Simonutti M, Fontaine V, Clerin E, Picaud S, Leveillard T, Sahel JA. Functional cone rescue by RdCVF protein in a dominant model of retinitis pigmentosa. Mol Ther. 2009;17:787–795. doi: 10.1038/mt.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Mookherjee S, Chaitankar V, Hiriyanna S, Kim JW, Brooks M, Ataeijannati Y, Sun X, Dong L, Li T, Swaroop A, Wu Z. Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nature communications. 2017;8:14716. doi: 10.1038/ncomms14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitz C, Gross AK, Leifert D, Kloeckener-Gruissem B, McAlear SD, Lemke J, Neidhardt J, Berger W. Identification and functional characterization of a novel rhodopsin mutation associated with autosomal dominant CSNB. Invest Ophthalmol Vis Sci. 2008;49:4105–4114. doi: 10.1167/iovs.08-1717. [DOI] [PubMed] [Google Scholar]
- Zhang N, Kolesnikov AV, Jastrzebska B, Mustafi D, Sawada O, Maeda T, Genoud C, Engel A, Kefalov VJ, Palczewski K. Autosomal recessive retinitis pigmentosa E150K opsin mice exhibit photoreceptor disorganization. J Clin Invest. 2013;123:121–137. doi: 10.1172/JCI66176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Baehr W, Fu Y. Chemical chaperone TUDCA preserves cone photoreceptors in a mouse model of Leber congenital amaurosis. Invest Ophthalmol Vis Sci. 2012;53:3349–3356. doi: 10.1167/iovs.12-9851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Tong N, Gong Y, Qiu Q, Yin L, Lv X, Wu X. Valproate protects the retina from endoplasmic reticulum stress-induced apoptosis after ischemia-reperfusion injury. Neurosci Lett. 2011;504:88–92. doi: 10.1016/j.neulet.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Zhu L, Jang GF, Jastrzebska B, Filipek S, Pearce-Kelling SE, Aguirre GD, Stenkamp RE, Acland GM, Palczewski K. A naturally occurring mutation of the opsin gene (T4R) in dogs affects glycosylation and stability of the G protein-coupled receptor. J Biol Chem. 2004;279:53828–53839. doi: 10.1074/jbc.M408472200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zode GS, Kuehn MH, Nishimura DY, Searby CC, Mohan K, Grozdanic SD, Bugge K, Anderson MG, Clark AF, Stone EM, Sheffield VC. Reduction of ER stress via a chemical chaperone prevents disease phenotypes in a mouse model of primary open angle glaucoma. J Clin Invest. 2011;121:3542–3553. doi: 10.1172/JCI58183. [DOI] [PMC free article] [PubMed] [Google Scholar]