Abstract
In this study, a red mutant was obtained through in vitro regeneration of a wild purple potato. High-performance liquid chromatography and Mass spectrometry analysis revealed that pelargonidin-3-O-glucoside and petunidin-3-O-glucoside were main anthocyanins in the mutant and wild type tubers, respectively. In order to thoroughly understand the mechanism of anthocyanin transformation in two materials, a comparative transcriptome analysis of the mutant and wild type was carried out through high-throughput RNA sequencing, and 295 differentially expressed genes (DEGs) were obtained. Real-time qRT-PCR validation of DEGs was consistent with the transcriptome date. The DEGs mainly influenced biological and metabolic pathways, including phenylpropanoid biosynthesis and translation, and biosynthesis of flavone and flavonol. In anthocyanin biosynthetic pathway, the analysis of structural genes expressions showed that three genes, one encoding phenylalanine ammonia-lyase, one encoding 4-coumarate-CoA ligase and one encoding flavonoid 3′,5′-hydroxylasem were significantly down-regulated in the mutant; one gene encoding phenylalanine ammonia-lyase was significantly up-regulated. Moreover, the transcription factors, such as bZIP family, MYB family, LOB family, MADS family, zf-HD family and C2H2 family, were significantly regulated in anthocyanin transformation. Response proteins of hormone, such as gibberellin, abscisic acid and brassinosteroid, were also significantly regulated in anthocyanin transformation. The information contributes to discovering the candidate genes in anthocyanin transformation, which can serve as a comprehensive resource for molecular mechanism research of anthocyanin transformation in potatoes.
Introduction
Potato (Solanum tuberosum L.) has been the fifth largest crop in the world. It is nutritious with a reputation of "underground apple". Red and purple-fleshed potatoes additionally contain 2 times higher anthocyanin content in comparison with white-flesh potatoes [1]. Anthocyanin, a family of water-soluble bioactive flavonoids, is in charge of producing various colors such as red, blue, and purple in plants. It can be classified into six main types: pelargonidin, cyanidin, delphinidin, peonidin, petunidin, and malvidin [2]. Potatoes with anthocyanins have benefits on health. The studies with rats demonstrate anthocyanins extracted from potatoes can significantly increase SOD activity and antioxidant capacity, and protect from injury [3, 4]. Anthocyanins in potatoes exert an anti-bacterial activity against different bacterial strains and a slight activity against three fungal strains [5].
Currently, owing to the potential advantages of anthocyanin to human health, the researches of anthocyanin biosynthesis in potato have been widely investigated. These researches are mainly concentrated on structural genes and regulation genes encoding transcription factor. In the aspect of structural genes, some structural genes have also been cloned and their functions are identified. Earlier researches show that R locus and P locus are involved in the coloration of potato tuber skin [6, 7]. P is epistatic to R [8]. P encodes flavonoid 3′,5′-hydroxylase (F3′5′H) [9]. F3′5′H is responsible for producing blue/purple petunidin-based anthocyanins and its function is verified by an immobilized transposon dTstu1 in F3′5′H [10]. R encodes dihydroflavonol 4-reductase (DFR) [11, 12]. DFR causes not only an increase in red pelargonidin, but also an increase in purple petunidin in potato tubers [13, 14]. Other structural genes involved in anthocyanin biosynthesis, such as CHS (CHSJ and CHSG), CHI, F3H and 3GT, are cloned from potato roots, stems, leaves, flowers and tubers [15]. CHS and CHI can cause a significant increase in pelargonidin and petunidin [16]. 3GT can deepen tuber color and increase anthocyanin content in potato tubers [17].
In the aspect of transcription factors, there are lots of transcription factors regulating anthocyanin biosynthesis discovered in potato. The transcription factor I is required as the third allele locus in anthocyanin biosynthesis in the tuber skin of diploid potatoes [8, 9]. When the potato lacks the functional allele I, the tuber exhibits white regardless of the presence of R and P. I is recessively epistatic to P and R [9]. D is another transcription factor required for the red and purple anthocyanin in the tuber skin of tetraploid potatoes, and is allelic to I in diploid potatoes [7]. D encodes the transcription factor R2R3MYB, and co-segregates with purple and red tubers in tetraploid potatoes [18]. R2R3MYB transcription factor AN1 has an effect on promoting anthocyanin biosynthesis in tubers and leaves of potatoes. The effect can be enhanced in the presence of the co-expression factors [19]. The co-expression factors mainly include bHLH and WD40-repeat proteins. A bHLH transcription regulator is co-localized with QTL on chromosome 9 [17, 20]. With the co-expression factor StbHLH1, the effect of AN1 on promoting anthocyanin biosynthesis is significantly improved [19, 21]. WD40-repeat gene StAN11 is cloned and significantly increases the accumulation of anthocyanin in potato tubers through its overexpression [22]. MYB, bHLH and WD40 can form the MYB-bHLH-WD anthocyanin regulatory complex to improve anthocyanin biosynthesis in potato [23].
Recently, with the development of the next-generation sequencing, transcriptome is used to comprehensively analyze the anthocyanin biosynthesis in potato. Liu carried out a comparative transcriptome analysis of white potato cultivar ‘Xin Daping’ and purple potato cultivar ‘Hei Meiren’. New versions of the pathway genes and potential transcription factors in potato anthocyanin biosynthesis were discovered [24]. Kyoungwon performed a comparative transcriptome analysis of three colored potato cultivars: light-red ‘Hongyoung’, dark-purple ‘Jayoung’ and white ‘Atlantic’. Regulatory networks of red anthocyanins and purple anthocyanin in potato were established [25].
Although some articles have been reported on the anthocyanin biosynthesis of potato, insight into the regulation of anthocyanin transformation have remained future objectives. In addition, the used materials in previous researches have different genetic backgrounds, and it possibly influence the screening of differential genes and decrease the accuracy of comparative transcriptome analysis. It is urgent to use the homologous materials to study anthocyanin transformation mechanism for enriching the metabolic pathway of anthocyanin.
In this study, we fortunately obtained a red mutant SD140 from a purple wild potato SD92. Both materials were homologous, which facilitated our comparative transcriptome analysis. In order to study the mechanism of anthocyanin transformation, we identified the main types of anthocyanins and carried out a comparative transcriptome analysis of SD140 and SD92.
Materials and methods
Plant sources
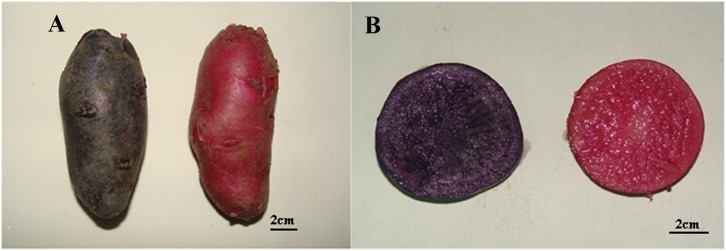
SD92, commonly known as Hei Jingang, is tetraploid with purple skin and purple flesh tuber. A red tuber mutant SD140 with red skin and flesh was obtained through in vitro regeneration of SD92 tuber slices (Fig 1). Two materials were planted in a greenhouse at 20 ± 2°C under a 16 h light/8 h dark cycle at the Institute of Vegetables and Flowers, Shandong Academy of Agricultural Sciences at the normal sowing time (August 10, 2016) in Jinan. The fresh tubers (diameter 4-5cm) were harvested, and cleaned with sterilized water. The tubers were immediately used for pigment extraction and two biological replicates were used. The tubers for mRNA extraction were frozen in liquid nitrogen and stored at -80°C, three biological replicates were used.
Fig 1. Phenotypic differences in SD92 and SD140.
(A) whole tuber, (B) sectional drawing.
Pigment extraction and purification
Potato tubers (250 g) were washed with distilled water and smashed in 500 mL of a mixture containing 50% alcohol and 1.6% citric acid. The resulting mixture was filtrated twice and centrifuged at 4000 rpm. The supernatant was transferred into a column filled with the pretreated macroporous resin AB-8 for 6 h, and the pigments absorbed in AB-8 were eluted with 95% alcohol. The eluate was concentrated with rotary evaporator at 30°C and dried in vacuo for 12 h. The laboratory protocol was deposited in protocols.io website (http://dx.doi.org/10.17504/protocols.io.ma4c2gw).
Analytical high-performance liquid chromatography
The pigment extractions were analyzed by the ultra-performance liquid chromatography (UPLC) system (Waters, Milford, Massachusetts, USA) equipped with Waters acquity PDA detector and a BEH C18 column (2.1×100mm, 1.7μm particle size). Acetonitrile (mobile phase A) and 2% methanoic acid (mobile phase B) were used as the eluent at a flow rate of 0.3 mL/min. The gradient conditions for sample elution were as follows: 5% A 95% B, 18 min; 20% A 80% B, 20 min; 100% A, 22 min; 100% A, 22.1 min; 5% A 95% B, 25 min; 5% A 95% B. The anthocyanins were detected at 530 nm using PAD. The column temperature was kept at 45°C. The volume for each injection was 1 uL.
Mass spectrometry
Mass spectrometry (MS) analysis was performed on a quadrupole time-of-flight (Q-TOF) mass spectrometry (WATERS MALDI SYNAPT) system. The anthocyanins were analyzed in Positive-ion (PI) mode. The optimized conditions were: the capillary and sampling cone voltage of 3.5kV and 30 V, respectively; the source block temperature of 100°C; desolvation temperature, 400°C; desolvation gas flow, 500 L/h; cone gas flow, 50 L/h; detector voltage, 1800 V. Data were gathered in the v mode, with a scan accumulation time of 1 s and the TOF data being collected between m/z 50 and 1,500. The MS-MS experiments were performed using a collision energy of 6 eV, which was optimized for each compound.
cDNA library construction and high-throughput sequencing
According to the manufacturer’s protocol, total RNA of the sample was extracted using modified Trizol reagent (Ambion, USA). And then RNA was digested with 10U DNaseI (Thermo, USA) for 1 h at 37°C. RNA quality and purity were detected by Agilent 2100 and NanoDrop (USA). The poly (A) mRNA was enriched using Oligo (dT) magnetic beads from digested RNA and was interrupted into short fragments. Using the short mRNA fragments as templates, First-strand cDNA were synthesized using random hexamer primers. The second strand was synthesized by buffer, dNTPs, Rnase H, and DNA polymerase I. The double-stranded cDNA was obtained by PCR with illumina primers. The two-stranded cDNA was purified with magnetic beads. The cohesive ends were repaired and the base "A" was added to the 3′ end. The fragments with suitable size were selected and were enriched by PCR amplication. The cDNA library was qualified and quantified with the Agilent 2100 Bioanalyzer (USA) and ABI StepOnePlus Real-Time PCR System (USA). The qualified library was sequenced by use of Illumina HiSeqX-Ten (USA). In total, six cDNA libraries were constructed in this study. All the raw reads were acquired by RNA-seq. In order to obtain clean reads, we filtered low quality reads, adaptor sequences and the reads in which unknown base N content was too high.
We deposited all the sequence data of the six cDNA libraries into NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/), and the accession number is SRP125987.
Differentially expressed genes identification
All clean reads were mapped to the reference transcripts of Solanum tuberosum L. (PGSC_DM_v3.4_transcript-update, http://solanaceae.plantbiology.msu.edu/data) using the Bowtie2 (v2.2.5). Statistics and bioinformatics analyses were carried out on the number of clean reads mapped to reference genes and genome, sequencing saturation, and random sequencing distribution. And then gene expression levels were calculated based on the number of reads mapped to the reference sequences. To identify the differentially expressed genes (DEGs) in the two Samples, DEseq2 algorithm was used. For each gene, the average expression level in the SD92 group (Control-avg) and in the SD140 group (Treat-avg) were calculated, respectively. DEGs were screened according to the following criteria: |log2(Treatment-avg /Control-avg)| ≥ 1.00 and adjusted p-value ≤ 0.05.
Gene Ontology (GO) enrichment and KEGG pathway enrichment analysis of DEGs
The DEGs were mapped to the corresponding GO terms and KEGG Orthology (KO) pathways. We used the R software phyper perform GO functional enrichment and KEGG functional enrichment. The p-value formula (https://en.wikipedia.org/wiki/Hypergeometric_distribution) in hypergeometric test is:
Then false discovery rate (FDR) for each p-value was calculated, and the terms which FDR is not larger than 0.01 were defined as a significant enrichment.
Transcription factor (TF) coding ability prediction in DEGs
We used getorf (EMBOSS: 6.5.7.0) to detect the ORF of each DEG, then used hmmsearch (v3.0) to align the ORF to the TF protein domain (data from PlantfDB), and finally identified TF according to the regulation based on the TF family characteristics.
Real-time qRT-PCR validation of the genes expression difference
To validate the transcriptome sequencing results, the transcription level of DEGs in the transcriptome was verified by real-time qRT-PCR. RNA samples used for qRT-PCR were same as those used in high-throughput sequencing experiments. Gene specific primers were listed in S1 Table. 25 μL of 2× UltraSYBR Mixture (CWBIO, CHINA), 1 μL cDNA, and 50 μL of total reaction volume were applied to all the reactions following the manufacturer’s method. The qPCR reaction was performed using an iCycler iQ (Bio-Rad, Hercules, CA, USA). The qPCR conditions were 37 cycles of 15 s at 95°C and 1 min at 60°C, followed by 65°C to 95°C melting curve detection. Three biological replicates were performed and 18S RNA was used as a reference gene.
Results
Pigment analysis
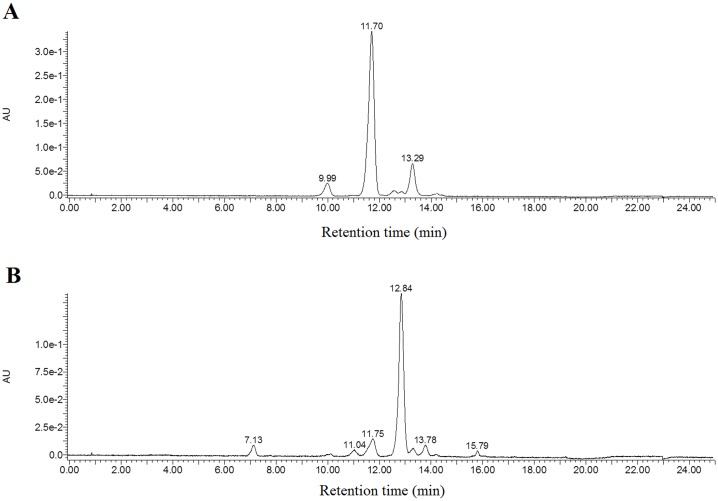
Fig 1 showed the potato tubers appearance (A) and the cross section (B) of SD140 and SD92, respectively. SD140 was obtained by the regeneration of tuber slices of SD92. After regeneration, both skin and flesh of SD92 changed from purple to red. Pure anthocyanins were obtained through pigment extraction, concentration and purification from the fresh tubers. In order to isolate the pure anthocyanins, HPLC was performed. For the anthocyanins extracted from SD140, three major independent peaks were obtained, and their retention times were located at 9.99, 11.70 and 13.29 minutes, respectively (Fig 2A). Similarly, six major peaks were acquired from SD92 extractions, and their retention times were located at 7.13, 11.04, 11.75, 12.84, 13.78, 15.79 minutes, respectively (Fig 2B). All these peaks were further identified by the quadrupole time-of-flight (Q-TOF) mass spectrometry, and the results were presented in Table 1. As for SD92, the MS analysis of the largest peak at the retention time of 11.70 minutes displayed an [M]+ ion at m/z 933.3 and a major fragmentation in MS2 at m/z 317.1. The peaks at 9.99 and 11.70 min revealed a major fragmentation in MS2 at m/z 317.1 (S1 Fig). The results were confirmed on the basis of HPLC retention time, elution order, spectroscopic data, MS fragmentation pattern, MS2, and previous findings [26–30]. The MS2 fragmentation of 317.1 at m/z corresponded to petunidin (Table 1). The results confirmed that the main anthocyanin in SD92 was petunidin. As for SD140, the MS analysis results of the largest peak at 12.84 min showed an [M]+ ion at m/z 887.3 and a major fragmentation in MS2 at m/z 271.1 (S2 Fig). The peaks at 7.13, 11.04, and 15.79 min exhibited a major fragmentation in MS2 at m/z 271.1. The MS2 fragmentation of 271.1 at m/z corresponded to pelargonidin. The results confirmed that the main anthocyanin in SD140 was pelargonidin (Table 1).
Fig 2. High-performance liquid chromatogram profile of anthocyanin extraction in tubers.
(A) SD92, (B) SD140.
Table 1. Peak assignments of anthocyanins extracted from the tubers of SD92 and SD140.
| Sample | Retention time (min) | Molecular ion [M]+ (m/z) | Molecular weight (Da) | MS2 (m/z) | Tentativel identification |
|---|---|---|---|---|---|
| SD92 | 9.99 | 949.3 | 950.3 | 317.1 | Petunidin |
| 919.3 | 920.3 | 303.1 | Delphinidin | ||
| 11.70 | 933.3 | 934.3 | 317.1 | Petunidin | |
| 13.29 | 917.3 | 918.3 | 301.1 | Peonidin | |
| SD140 | 7.13 | 579.2 | 580.2 | 271.1 | Pelargonidin |
| 11.04 | 903.4 | 904.4 | 271.1 | Pelargonidin | |
| 11.75 | 933.4 | 934.4 | 317.1 | Petunidin | |
| 12.84 | 887.3 | 888.3 | 271.1 | Pelargonidin | |
| 13.78 | 887.4 | 888.4 | 271.1 | Pelargonidin | |
| 917.4 | 918.4 | 271.1 | Pelargonidin | ||
| 15.79 | 725.3 | 725.3 | 271.1 | Pelargonidin |
Transcriptome analysis
RNA sequencing and mapping of the sequence reads
Illumina sequencing technology was used to perform high-throughput RNA sequencing of SD92 and SD140, respectively. After removing the adaptor sequences and low-quality reads, 44.15–44.95 Mb clean reads were acquired from each sample. The clean reads were mapped to the genome. The mapped ratio of each sample reached 55.63%-61.49% (S2 Table). After transcripts were reconstructed, we acquired 37460 transcripts. 15232 transcripts were novel, of which 3641 were novel genes (Table 2).
Table 2. Summary of gene expression.
| Sample | Replication | Clean reads (Mb) | Total mapping ratio (%) | Uniquely mapping ratio (%) | Total transcriptnumber | Total gene number | Novel gene number |
|---|---|---|---|---|---|---|---|
| SD92 | 1 | 44.42 | 72.92 | 36.24 | 37460 | 26193 | 3082 |
| 2 | 44.15 | 73.58 | 35.88 | 37117 | 25943 | 3085 | |
| 3 | 44.46 | 72.90 | 31.30 | 36126 | 25445 | 3041 | |
| SD140 | 1 | 44.95 | 72.52 | 33.15 | 36835 | 25880 | 3068 |
| 2 | 44.43 | 75.42 | 35.20 | 36747 | 26117 | 3109 | |
| 3 | 44.81 | 72.29 | 32.89 | 36530 | 25730 | 3050 |
To identify each gene expression in SD92 and SD140 tubers, the clean reads were mapped to the complete reference sequences, which included the reference gene sequence and novel gene. The results demonstrated that 32.39–39.51Mb clean reads were mapped in each sample. These values accounted for 72.29%-75.42% of each library (Table 2). Based on the mapped results, we counted the read coverage and the reads distribution of each transcript. The random reads distribution on the transcripts indicated that the read positions were evenly distributed in each gene (S3 Fig). Reads coverage on transcripts analysis showed that the average percentage of each gene covered by reads was approximately 50% (S4 Fig). Therefore, the sequencing data should accurately reflect gene expression and can be used for differential gene expression analysis.
Global gene expression pattern analysis
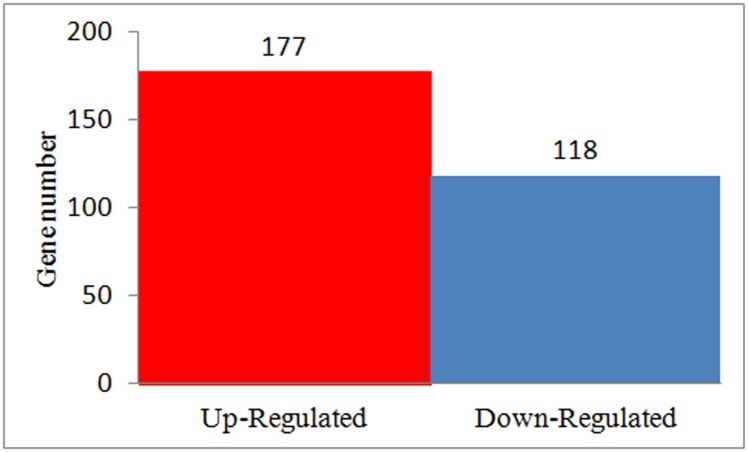
In SD92 and SD140, there were 22679 and 22667 genes expressed in all three biological replicates, respectively (S5 Fig). To explore the molecular mechanism involved in anthocyanin transformation, we analyzed the DEGs in three biological replicates. 267 DEGs were obtained between SD92 and SD140. Among these DEGs, 177 (60.00%) were up-regulated and 118 (40.00%) were down-regulated (Fig 3) in SD140 compared to SD92.
Fig 3. Statistics of differently expressed genes.
To facilitate the global analysis of gene expression, GO analysis of DEGs between SD92 and SD140 was performed. The results revealed 98 DEGs were classified. The DEGs were classified into the three main GO categories: biological process, cellular component and molecular function (S6 Fig). For further review of the GO classification of the DEGs, we also categorized each GO term into its sub-categories. The results indicated the sub-categories significantly enriched in the biological process category primarily contained biological regulation, localization, and response to stimulus. The DEGs in this category were associated with the regulation of catalytic activity, transcription, and macromolecule metabolic process. Cell part, cell, organelle, organelle part, membrane and membrane part were the main sub-categories belonging to the cellular component category. For the molecular function category, the main sub-categories were binding, catalytic activity, transporter activity and molecular function regulator. Catalytic activity was involved in putative RING-H2 finger protein, disulfide-isomerase protein and ras-related RABA1f protein. Proteins in the binding sub-category included beta-galactosidase, calreticulin, luminal-binding protein, kinesin-related protein, probably inactive leucine-rich repeat receptor and heat shock 70 kDa protein.
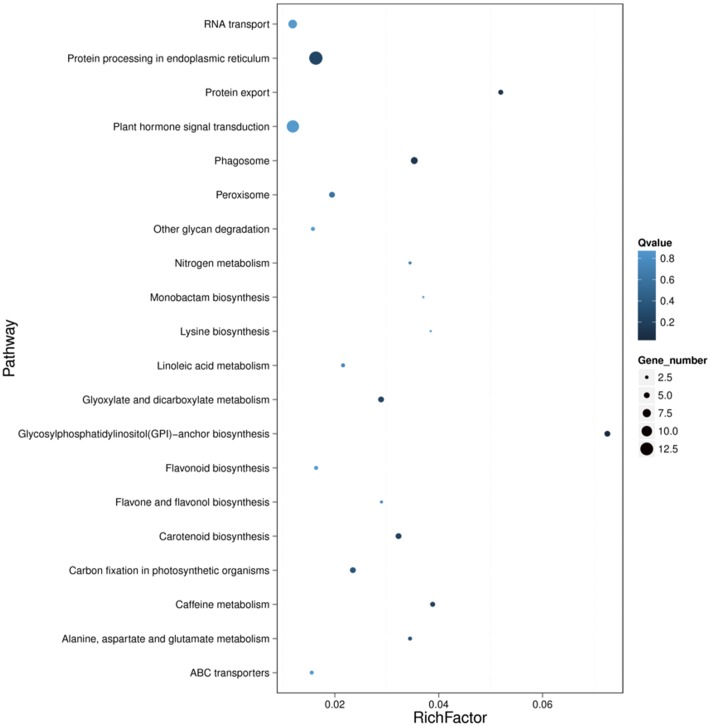
KEGG analysis was performed to obtain the difference in the metabolic pathway in SD92 and SD140 (Fig 4). Among the DEGs, 220 genes corresponded to KO codes, including 126 significantly up-regulated genes and 94 significantly down-regulated genes in SD140. To review the function of the DEGs, we further clustered each KO term of the DEGs. The results showed that the DEGs were mostly enriched in transport and catabolism, translation, biosynthesis of other secondary metabolites and transcription. These clusters included the following pathways: protein processing in endoplasmic reticulum (13 genes), plant hormone signal transduction (12 genes), RNA transport (8 genes), flavonoid biosynthesis (3 genes), flavone and flavonol biosynthesis (2 genes), phenylpropanoid biosynthesis (7 genes) and spliceosome (7 gens).
Fig 4. KEGG enrichment analysis of DEGs.
Gene expression changes of anthocyanin biosynthesis in potato
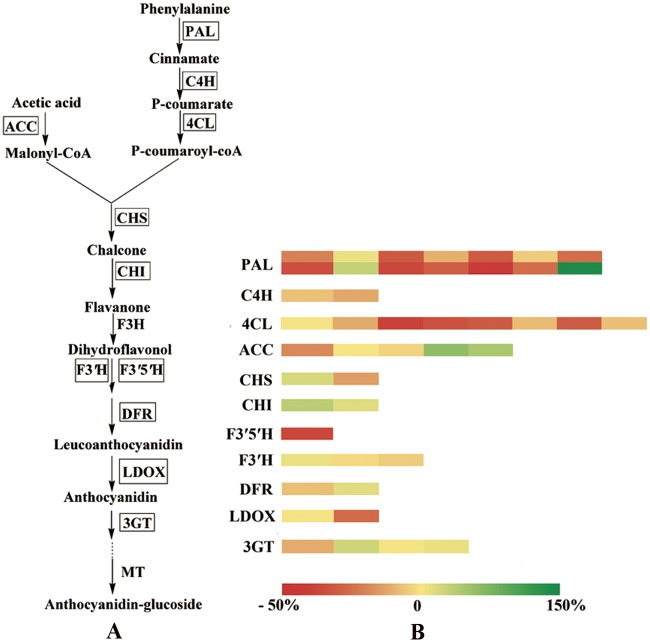
Considering that the main type of anthocyanins in SD92 was different from SD140, and that the results of KEGG enrichment of DEGs showed differences in flavonoid metabolic pathways between SD92 and SD140, we selected 41 genes to investigate the pathway of anthocyanin biosynthesis. Of the selected genes, 8 were new genes. The criterion for selection was that the gene expression level was not less than 20 reads at least in one material (Fig 5 and S3 Table). The 41 genes mainly encoded the following enzymes: phenylalanine ammonia-lyase (PAL), 4-coumarate-CoA ligase (4CL), chalcone synthase (CHS), flavonoid 3′,5′-hydroxylase (F3′5′H), flavonoid 3′-hydroxylase (F3′H), dihydroflavonol 4-reductase (DFR), leucoanthocyanidin dioxygenase (LDOX), anthocyanidin 3-O-glucosyltransferase (3GT). These enzymes had two characteristics. One was that these enzymes possessed isozymes. Phenylalanine ammonia-lyase included two isozymes: PAL and PAL1. Chalcone synthase mainly included two isozymes: CHS1B, CHS2. 4-Coumarate-CoA ligase included 4CL2 and 4CL. The other was that these enzymes belonged to a multi-gene family. PAL and 4CL families comprised 14 and 9 genes, respectively. The differences of 41 gene expression levels in anthocyanin biosynthetic pathway were investigated according to p-value. If p-value ≤ 0.05, the gene was significantly regulated. The result showed the expression of four genes significantly changed, and other genes almost didn’t significantly changed. Four changed genes included two genes encoding PAL, one gene encoding 4CL, one gene encoding F3′5′H. To investigate the change range of these significantly changed genes, we calculated the rate of change by using the following formula: (Treatment-avg—Control-avg)/ Control-avg ×100% (S3 Table). The results indicated that in SD140 one gene encoding PAL was up-regulated by 144%, another gene encoding PAL was down-regulated by 42.58%; one gene encoding 4CL was down-regulated by 39.83%; one gene encoding F3′5′H was down-regulated by 37.66%.
Fig 5. Gene expression levels of anthocyanin biosynthesis in potato tubers.
(A) The metabolic pathway of anthocyanin biosynthesis; the genes encoding enzymes enclosed in boxes were detected in this transcriptome, (B) heatmap of the rate of change of gene expression.
Analysis of transcription factors in DEGs
Transcription factor is a kind of protein binding the specific nucleotide sequence of a gene and regulates the transcription of the related gene. The GO analysis indicated that there existed differences in transcription and DNA binding between SD92 and SD140. To determine the molecular mechanism of anthocyanin transformation, we examined the differential expression of transcription factors between SD92 and SD140. The results revealed the transcription factors were encoded by 18 DEGs (Table 3). These transcription factors belonged to 12 families. Alfin family, bZIP family, C3H family, FHA family, GRAS family, LOB family, MADS family were down-regulated in SD140 and were mainly involved in histone acetyltransferase, transcription factor HBP-1, zinc finger protein ZAT11, zinc finger CCCH and uncharacterized protein. S1Fa family, Tify family, zf-HD family were up-regulated in SD140 and were mainly involved in vacuolar amino acid transporter YPQ2, plastidic glucose transporter, zinc-finger homeodomain protein 9 and protein RADIALIS. The AP2-EREBP family contained AP2 ethylene-responsive transcription factor ANT and ethylene-responsive transcription factor 4-like. ANT was up-regulated and 4-like was down-regulated. The MYB family comprised myb-related protein MYBAS1 and protein RADIALIS 3. MYBAS1 was up-regulated, and RADIALIS 3 was down-regulated in SD140.
Table 3. Transcription factors analysis in DEGs.
| TF | #Gene | Function | Up/Down- Regulation |
|---|---|---|---|
| Alfin-like | BGI_novel_G000417 | histone acetyltransferase HAC1-like isoform X1 | Down |
| AP2-EREBP | PGSC0003DMG400004921 | AP2-like ethylene-responsive transcription factor ANT | Up |
| AP2-EREBP | PGSC0003DMG400022823 | ethylene-responsive transcription factor 4-like | Down |
| bZIP | PGSC0003DMG400026364 | transcription factor HBP-1b(c38)-like isoform X1, X3, X6, X2 | Down |
| C2H2 | PGSC0003DMG400030803 | protein TRANSPARENT TESTA 1-like, RNA-binding protein 24-like | Up |
| C2H2 | PGSC0003DMG400015534 | zinc finger protein ZAT11-like | Down |
| C3H | BGI_novel_G003371 | zinc finger CCCH domain-containing protein 24 | Down |
| FHA | BGI_novel_G001032 | uncharacterized protein LOC102601992 | Down |
| GRAS | PGSC0003DMG400029921 | scarecrow-like protein 8 | Down |
| LOB | PGSC0003DMG400008649 | LOB domain-containing protein 4 | Down |
| MADS | PGSC0003DMG400022867 | uncharacterized protein LOC102601973 | Down |
| MYB | PGSC0003DMG400028282 | myb-related protein MYBAS1 | Up |
| MYB | PGSC0003DMG400015461 | protein RADIALIS | Down |
| S1Fa | PGSC0003DMG400010043 | probable vacuolar amino acid transporter YPQ2 | Up |
| S1Fa | PGSC0003DMG400016530 | probable plastidic glucose transporter 3 | Up |
| Tify | PGSC0003DMG400001178 | protein TIFY 5A | Up |
| Tify | BGI_novel_G002514 | protein TIFY 5A | Up |
| zf-HD | PGSC0003DMG400033694 | zinc-finger homeodomain protein 9 | Up |
Analysis of plant hormone signal transduction in DEGs
Hormones are growth regulators, which affect plant germination, rooting, flowering, fruiting, sex determination, dormancy and shedding. The KEGG analysis indicated that there were differences in plant hormone signal transduction between SD92 and SD140. In order to deeply analyze the molecular mechanism of anthocyanin transformation, the genes expression differences in hormone signal transduction between SD92 and SD140 were investigated. The results indicated that 12 genes expression in hormone signal transduction exhibited significant change (Table 4). The hormone signal transduction included GA, ABA, IAA, BR, Ethylene and Salicylic acid signal transduction. As for SD140, in GA signal transduction, one gene encoding scarecrow-like protein 8 was significantly down-regulated; in ABA signal transduction, one gene encoding abscisic acid receptor PYL4 was down-regulated; in BR signal transduction, one gene encoding D-type cyclin family 3 subgroup 3 and one gene encoding inactive leucine-rich repeat receptor-like protein kinase IMK2 were up-regulated, one gene encoding BRI1 kinase inhibitor 1 and one gene encoding LRR receptor-like serine/ threonine-protein kinase At2g24230 were down-regulated.
Table 4. Plant hormone signal transduction analysis in DEGs.
| Hormone | GeneID | Kegg Orthology | Nr Description | Up/Down- Regulation |
|---|---|---|---|---|
| Auxin | PGSC0003DMG400008504 | K13946 | auxin transporter-like protein 2 | Up |
| Auxin | PGSC0003DMG402002635 | K14484 | IAA15 | Down |
| Auxin | PGSC0003DMG400005338 | K14484 | auxin-responsive protein IAA | Down |
| Auxin | BGI_novel_G000882 | K14486 | Auxin response factor 8 | Down |
| Brassinosteroid | PGSC0003DMG400008001 | K14505 | D-type cyclin family 3 subgroup 3 | Up |
| Brassinosteroid | PGSC0003DMG400013198 | K04733,K20359,K04730,K13415 | inactive leucine-rich repeat receptor-like protein kinase IMK2 | Up |
| Brassinosteroid | PGSC0003DMG400008020 | K14499 | BRI1 kinase inhibitor 1 | Down |
| Brassinosteroid | PGSC0003DMG400028478 | K04730, K13415 | LRR receptor-like serine/ threonine—protein kinase At2g24230 | Down |
| Ethylene | PGSC0003DMG400021341 | K14514 | sucrose-phosphatase 1 isoform X2 lycopersicum] | Down |
| Abscisic acid | PGSC0003DMG400023949 | K14496 | abscisic acid receptor PYL4—like | Down |
| Salicylic acid | PGSC0003DMG400026364 | K14431 | transcription factor HBP-1b (c38)—like isoform X6,X2,X1, X3 | Down |
| Gibberellin | PGSC0003DMG400029921 | K14777, K14494 | scarecrow-like protein 8 | Down |
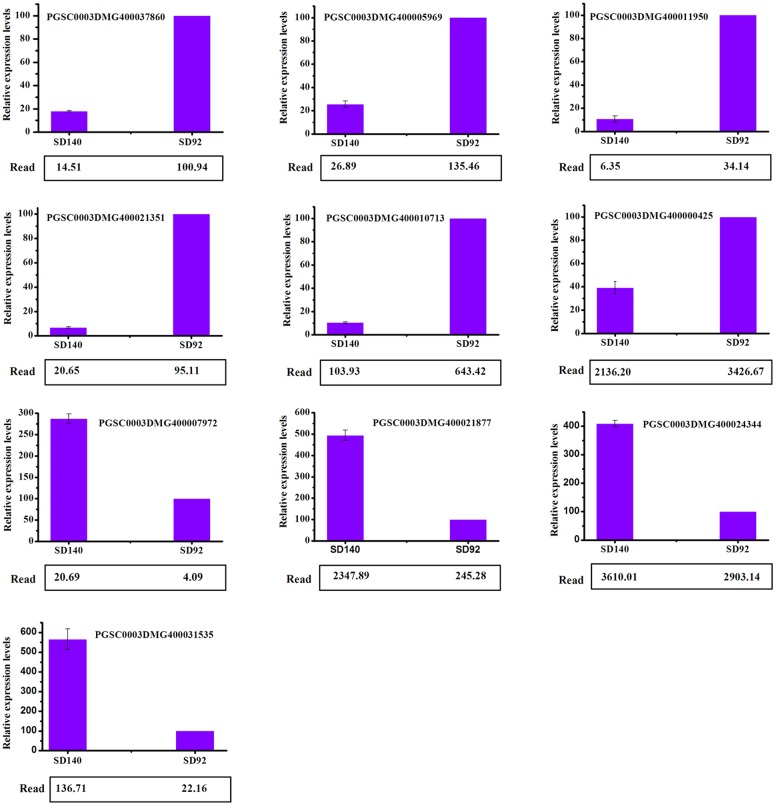
Validation of RNA-Seq-based gene expression by qRT-PCR
To validate the RNA-Seq results, a qRT-PCR analysis was performed on 10 genes. These genes included eight randomly selected DEGs and two genes in the anthocyanin metabolic pathway. Eight randomly selected DEGs were involved in DNA-directed RNA polymerase, ubiquitin-protein ligase, ferredoxin, translation initiation factor, xyloglucosyl transferase and ABA 8′-hydroxylase. Two genes in the anthocyanin biosynthetic pathway encoded F3′5′H and LDOX. The qRT-PCR result indicated that all 10 genes expression pattern was accordant with the RNA-Seq date (Fig 6).
Fig 6. qRT-PCR verification of expression of selected genes.
Discussion
Effect of structural genes expression differences on anthocyanin transformation in anthocyanin biosynthesis pathway
Anthocyanin biosynthesis pathway has been elucidated in many plant species. Different species have different structural genes. Many structural genes have been cloned from model species, such as maize and petunia [17]. In this study, the structural genes PAL, ACC, 4CL, C4H, CHS, CHI, F3′H, F3′5′H, ANS, DFR and 3GT were detected by analyzing the transcriptome of SD92 and SD140. The results indicated that the expressions levels of F3′H were both low in two materials. F3′H is responsible for biosynthesis of peonidin [14]. So the predicated peonidin levels were low in the two materials, which were consistent with our HPLC-MS analysis. These results further confirmed that F3′H was responsible for the biosynthesis of peonidin in potato.
Additionally, the differential expression analysis of structural genes in anthocyanin biosynthesis revealed that one PAL, one 4CL and one F3′5′H were significantly down-regulated in SD140; one PAL was significantly up-regulated. PAL and 4CL belong to anthocyanin biosynthetic upstream genes, and they are involved in multiple metabolic pathways [31, 32]. These genes expression changes may not cause changes in anthocyanin biosynthesis. We paid more attention to the gene change in downstream. F3′5′H belongs to anthocyanin biosynthetic downstream gene. F3′5′H plays a key role in regulating different types of anthocyanin biosynthesis [14]. We found that it was significantly down-regulated in anthocyanin transformation.
Effect of F3′5′H on anthocyanin transformation
In this work, petunidin-3-O-glucoside and pelairgonidin-3-O-glucoside were found to be responsible for purple tuber and red tuber, respectively, and these findings were consistent with previous results [9, 15]. F3′H and F3′5′H, belonging to the cytochrome P-450 total branch, are key enzymes controlling different branches of anthocyanin biosynthesis in some species [14]. When both F3′H and F3′5′H are present, F3′5′H is predominant. In other words, F3′5′H not only promotes anthocyanins synthesis pathway to 3′ 5′-hydroxylated anthocyanins but also stops the biosynthetic pathway to 3′-hydroxylated anthocyanins [33, 34]. Besides, F3′5′H expression was much higher and F3′H expression was much lower, so F3′5′H had a main effect on the color of potato tuber. F3′5′H controls the biosynthesis of petunidin-based anthocyanins [17]. Silenced F3′5′H, inactivation and degeneration of F3′5′H can decrease petunidin-based anthocyanins [35, 36]. The decreased petunidin in SD140 was caused by the decreased expression of F3′5′H, which was verified by qRT-PCR. Moreover, the main type of anthocyanin in the purple tubers and the red tubers were petunidin and pelargonidin, respectively. Therefore, the transformation from petunidin to pelargonidin in tuber was related to the decrease of F3′5′H expression level and dihydrokaempferol directly synthesize pelargonidin [14]. So it was possible that the transcript of F3′5′H was influenced.
Effect of transcription factors on anthocyanin transformation
Transcription factors are adaptor molecules. It can be used to detect regulatory sequences in the DNA and target the assembly of protein complexes controlling gene expression [37]. Transcription factors play important roles on controlling plant development and differentiation. In this study, the transcription factor families, including AP2-EREBP family, C2H2 family, MYB family, S1Fa family, Tify family, GRAS family, Alfin family, C3H family, FHA family, LOB family, MADS family and zf-HD family, significantly changed during anthocyanin transformation.
In plants, R2R3MYB family, a Myb-type transcription factor, is closely related to flavonoid metabolic pathways. The R2R3MYB family includes a lot of subfamilies in different species. The subfamilies, such as Stan2, VvMYBPA1, VvMYB5b and VvMYB5a, can regulate anthocyanin biosynthesis by activating structural genes. All the activated genes include F3′5′H [11, 38, 39]. There are also inhibitory factors in the MYB family, such as SlTRY (CPC) and MYB182. They inhibit the anthocyanin biosynthesis [40, 41]. In our study, the expressions of the genes encoding the MYB family members were significantly different in potatoes with different types of anthocyanins. It was possible that these MYB family members regulated anthocyanin transformation from petunidin to pelargonidin by regulating F3′5′H, which switched from 3′-hydroxylation to 3′5′-hydroxylation anthocyanin synthesis pathway. In MYB family, MYBAS1 was up-regulated, which suggested it promoted the transformation from petunidin to pelargonidin by repressing F3′5′H expression. RADIALIS 3 was down-regulated, which suggested it inhibited the transformation from petunidin to pelargonidin.
Besides, bZIP family also played an important role in anthocyanin transformation. bZIP, an alkaline leucine zipper transcription factor, comprised a basic part and a leucine zipper part [42]. bZIP transcription factors G/HBF-1, LONG HYPOCOTYL 5 (HY5) and HY5 HOMOLOG (HYH) can increase the structural genes expression of CHS, CHI, F3H and DFR and promote anthocyanin biosynthesis by binding the promoter of the structural genes [43, 44, 45]. bZIP transcription factors also improve anthocyanin biosynthesis by binding the R2R3MYB transcription factor PAP1 promoter region [46, 47]. R2R3MYB can regulate the expression of F3′5′H, which regulates transformation from petunidin to pelargonidin [36]. Therefore, bZIP can regulate anthocyanin transformation from petunidin to pelargonidin by F3′5′H. In this study, a gene encoding bZIP transcription factor HBP-1b (c38) -like isoform X1, X3, X6, X2 was down-regulated. It suggested that the HBP-1b (c38) inhibited the transformation from petunidin to pelargonidin by MYB.
MADS, a transcription factor, consists of the MADS-box, the I, the K and the C regions [48]. MADS family is closely related to anthocyanin biosynthesis [49–51]. MADS transcription factors increase anthocyanin biosynthesis by activating the genes encoding CHS, CHI, F3H, DFR, ANS and UFTG [49, 52]. MADS transcription factor VmTDR4 plays an important role in accumulation of anthocyanins through the up-regulation of R2R3MYB family [53]. R2R3MYB can regulate anthocyanin transformation through F3′5′H [36]. So MADS can regulate anthocyanin transformation through F3′5′H. In this study, a gene encoding MADS was significantly down-regulated in SD140. It was possible that the MADS suppressed the transformation from petunidin to pelargonidin through regulating F3′5′H.
In this study, the transcription factors were significantly regulated, such as AP2-EREBP family, C2H2 family, S1Fa family, Tify family, GRAS family, Alfin family, C3H family, FHA family, LOB family, and zf-HD family. The functions of these transcription factors were worth further research.
Effect of plant hormone on anthocyanin transformation
Plant hormone can influence anthocyanin biosynthesis. GA signal is involved in anthocyanin biosynthesis pathway. GA can decrease anthocyanin accumulation under low temperature or phosphate starvation [54, 55]. The depression effect under low temperature was determined by two bZIP transcription factors (HY5 and HYH) [54]. In this study, one DEG encoding scarecrow-like protein 8 in GA signal pathway, and one DEG encoding bZIP transcription factor HBP-1b(c38) were both down-regulated. The results suggested that scarecrow-like protein 8 inhibited the transformation through HBP-1b(c38). ABA can independently improve anthocyanin accumulation and also has a synergic effect with sugar [56, 57]. ABA regulates anthocyanin biosynthesis via MYB, bHLH and bZIP [58–60]. In this work, one DEG with the function of ABA receptor PYL4 in ABA signal pathway was down-regulated. Additonallly, the aforementioned analysis indicated that MYB and bZIP regulated anthocyanin transformation. Therefore, it could be concluded that ABA receptor PYL4 inhibited the transformation from petunidin to pelargonidin by MYB and bZIP. BR also affects JA-induced anthocyanin accumulation by the WD-repeat/MYB/bHLH transcriptional complexes [61]. In current work, D-type cyclin family 3 and inactive leucine-rich repeat receptor- like protein kinase IMK2 were up-regulated, BRI1 kinase inhibitor 1 and LRR receptor serine/threonine-protein kinase At2g24230 were down-regulated. Besides, the above-mentioned findings confirmed that MYB and bZIP regulated anthocyanin transformation. As a result, it could be infered that with the aid of the MYB and bZIP, the two up-regulated BR response proteins inhibited anthocyanin transformation and the two down-regulated BR response proteins promoted anthocyanin transformation.
Moreover, the response proteins in signal transduction of IAA and ethylene were significantly regulated in anthocyanin transformation in this work, The functions of these hormone response proteins were worth further research.
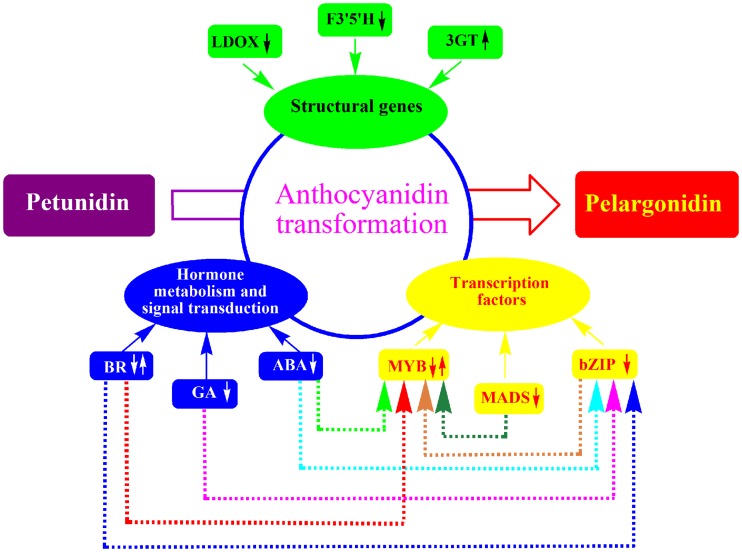
Conclusion
In this study, both pigments and the comparative transcriptome analysis of purple potato tuber and its red mutant were carried out. It revealed the decreased expression level of F3′5′H was mainly responsible for anthocyanin transformation from petunidin to pelargonidin in potato tubers. Transcription factors bZIP family, MSD family regulated the transformation through MYB family, which regulated F3′5′H expression. Response proteins of ABA, GA, BR also affected the transformation through transcription factors.The structural genes, transcription factors and their related factors formed a complex that affected the transformation from petunidin to pelargonidin in potato tuber (Fig 7). The transcriptome analysis provided valuable information for anthocyanin transformation.
Fig 7. A complex that regulates the anthocyanin transformation in potato tuber.
Supporting information
(TIF)
(TIF)
SD140:a,b,c. SD92: d,e,f.
(TIF)
SD140:a,b,c. SD92: d,e,f.
(TIF)
A: SD92, B: SD140.
(TIF)
A threshold of corrected p value ≤ 0.001 was used to judge the significantly enriched GO terms in DEGs.
(TIF)
(DOC)
(DOC)
(DOC)
Acknowledgments
We thank Associate Professor Yumeng Huo for providing advice on manuscript preparation.
Data Availability
All the RNA sequencing data are available from the NCBI Sequence Read Archive using SRA accession number SRP125987.
Funding Statement
This work was supported by two fundings. One was the “Youth Funding of Shandong Academy of Agricultural Sciences (Grant No. 2014QNM13)” sponsored by Shandong Academy of Agricultural Sciences. The other funding is “Modern Agriculture Industrial Technology System Funding of Shandong Province (Grant No. SDAIT-10-022-05)” sponsored by Shandong Provincial Department of Agriculture.
References
- 1.Brown CR, Culley D, Bonierbale M, Amorós W. Anthocyanin, carotenoid content, and antioxidant values in native South American potato cultivars. HortScience, 2007; 42 (7): 1733–1736. [Google Scholar]
- 2.Martin C, Gerats T. Control of pigment biosynthesis genes during petal development. Plant Cell. 1993;5: 1253–1264. doi: 10.1105/tpc.5.10.1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han KH, Sekikawa M, Shimada K, Hashimoto M, Hashimoto N, Noda T, et al. Anthocyanin-rich purple potato flake extract has antioxidant capacity and improves antioxidant potential in rats. Br J Nutr. 2006; 96: 1125–1133. [DOI] [PubMed] [Google Scholar]
- 4.Han KH, Hashimoto N, Hashimoto M, Noda T, Shimada KI, Lee CH, et al. Red potato extract protect from D-Galactosamine-induced liver injury rats. Biosci Biotechnol Biochem. 2006;70: 2285–2288. doi: 10.1271/bbb.60097 [DOI] [PubMed] [Google Scholar]
- 5.Bontempo P, Carafa V, Grassi R, Basile A, Tenore GC, Formisano C, et al. Antioxidant, antimicrobial and anti-proliferative activities of Solanum tuberosum L. var. Vitelotte. Food Chem Toxicol. 2013; 55: 304–312. doi: 10.1016/j.fct.2012.12.048 [DOI] [PubMed] [Google Scholar]
- 6.Salaman RN. The inheritance of colour and other characters in the potato. Mol Gen Genet 1911; 5:192–193. [Google Scholar]
- 7.Dodds KS, Long DH. The inheritance of colour in diploid potatoes. II. A three-factor linkage group. J Genet. 1956; 54: 27–41. [Google Scholar]
- 8.Dodds KS, Long DH. The inheritance of colour in diploid potatoes. I. Types of anthocyanidins and their genetic loci. J Genet. 1955; 53: 136–149. [Google Scholar]
- 9.Jung CS, Griffiths HM, De Jong MD, Cheng SP, Bodis M, De Jong WS. The potato P locus codes for flavonoid 3′,5′-hydroxylase. Theor Appl Genet. 2005; 110: 269–275. doi: 10.1007/s00122-004-1829-z [DOI] [PubMed] [Google Scholar]
- 10.Momose M, Abe Y, Ozeki Y. Miniature inverted-repeat transposable elements of stowaway are active in potato. Genetics Soc America. 2010; 186:59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Jong WS, De Jong DM, De Jong H, Kalazich J, Bodis M. An allele of dihydrofavonol 4-reductase associated with the ability to produce red anthocyanin pigments in potato (Solanum tuberosumL.). Theor Appl Genet. 2003; 107: 1375–1383. doi: 10.1007/s00122-003-1395-9 [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Cheng S, De Jong D, GriYths H, Halitschke R, De Jong W. The potato R locus codes for dihydroflavonol 4-reductase. Theor Appl Genet. 2009; 119: 931–937. doi: 10.1007/s00122-009-1100-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stobieckia M, MatysiakKatab I, Franski R, Skala J, Szopa J. Monitoring changes in anthocyanin and steroid alkaloid glycoside content in lines of transgenic potato plants using liquid chromatography/mass spectrometry. Phytochemistry. 2003; 62: 959–969. [DOI] [PubMed] [Google Scholar]
- 14.Holton TA, Cornish EC. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell. 1995; 7: 1071–1083. doi: 10.1105/tpc.7.7.1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei Q, Wang Q, Feng Z. Increased accumulation of anthocyanins in transgenic potato tubers by overexpressing the 3GT gene. Plant Biotechnol Rep. 2012; 6: 69–75. [Google Scholar]
- 16.Lukaszewicz M, Matysiak-kata I, Skala J, Fecka I, Cisowski W, Szopa J. Antioxidant capacity; 52: 1526–1533. [DOI] [PubMed] [Google Scholar]
- 17.Lu Q, Yang Q. cDNA cloning and expression of anthocyanin biosynthetic genes in wild potato (Solanum pinnatisectum). Afr J Biotechnol. 2006; 5: 811–818. [Google Scholar]
- 18.Jung CS, GriYths HM, De Jong DM, Cheng SP, Bodis M, Kim TS, et al. The potato developer (D) locus encodes an R2R3 MYB transcription factor that regulates expression of multiple anthocyanin structural genes in tuber skin. Theor Appl Genet. 2009; 120: 45–57. doi: 10.1007/s00122-009-1158-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Damelia V, Aversano R, Batelli G, Caruso I, Castellano Moreno M, Castro-Sanz AB, et al. High AN1 variability and interaction with basic helix-loop-helix co-factors related to anthocyanin biosynthesis in potato leaves. Plant J. 2014; 80:527–540. doi: 10.1111/tpj.12653 [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Jung CS, De Jong WS. Genetic analysis of pigmented tuber flesh in potato. Theor Appl Genet. 2009; 119:143–150. doi: 10.1007/s00122-009-1024-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Wang K, Espley RV, Wang L, Yang H, Yu B, et al. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J Exp Bot. 2016; 67: 2159–2176. doi: 10.1093/jxb/erw014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Wang B, Wang M. Cloning and characterization of a potato StAN11 gene involved in anthocyanin biosynthesis regulation. J Integr Plant Biol. 2014; 56: 364–372. doi: 10.1111/jipb.12136 [DOI] [PubMed] [Google Scholar]
- 23.Fogelman E, Tanami S, Ginzberg I. Anthocyanin synthesis in native and wound periderms of potato. Physiol Plant. 2015; 153: 616–626. doi: 10.1111/ppl.12265 [DOI] [PubMed] [Google Scholar]
- 24.Liu YH, Wang KL, Deng C, Warran B, Li W, Yu B, et al. Comparative transcriptome analysis of white and purple potato to identify genes involved in anthocyanin biosynthesis. PLoS ONE. 2015. June 8 doi: 10.1371/journal.pone.0129148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kyoungwon C, Kwang-Soo C, Hwang-Bae S, In JH, Hong Su-Young, Lee Hyerim, et al. Network analysis of the metabolome and transcriptome reveals novel regulation of potato pigmentation. J Exp Bot. 2016; January 4 doi: 10.1093/jxb/erv549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garciaa-Beneytezeva E, Cabello F, Revilla E. Analysis of grape and wine anthocyanins by HPLC-MS. J. Agric. Food Chem. 2003; 51: 5622–5629. doi: 10.1021/jf0302207 [DOI] [PubMed] [Google Scholar]
- 27.Ramirez JE, Zambrano R, Sepúlveda B, Kennelly EJ, Simirgiotis MJ. Anthocyanins and antioxidant capacities of six Chilean berries by HPLC-HR-ESI-ToF-MS. Food Chem. 2015; 176: 106–114. doi: 10.1016/j.foodchem.2014.12.039 [DOI] [PubMed] [Google Scholar]
- 28.Pertuzatti PB, Becker P, Barcia MT, Gomes Rebello LP, Gómez-Alonso S, Teixeira Duarte RM, et al. Antimicrobial activity and differentiation of anthocyanin profiles of rabbiteye and highbush blueberries using HPLC-DAD-ESI-MSn and multivariate analysis. J Fuct Foods. 2016; 26: 506–516. [Google Scholar]
- 29.Lee JH, Kang NS, Shin SO, Shin SH, Lim SG, Suh DY, Baek IY, Park KY, Ha TJ. Characterisation of anthocyanins in the black soybean (Glycine max L.) by HPLC-DAD-ESI/MS analysis. Food Chemistry 112 (2009) 226–231. [Google Scholar]
- 30.Lee MJ, Park JS, Choi DS, Jung MY. Characterization and quantitation of anthocyanins in purple-fleshed sweet potatoes cultivated in Korea by HPLC-DAD and HPLC-ESIQTOF-MS/MS. Agric. Food Chem. 2013, 61, 3148–3158. [DOI] [PubMed] [Google Scholar]
- 31.Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu Rev Plant Biol, 2003; 54: 519–546. doi: 10.1146/annurev.arplant.54.031902.134938 [DOI] [PubMed] [Google Scholar]
- 32.Yan J, Yu L, Xu S, Zhu W, Gu WH. Apigenin accumulation and expression analysis of apigenin biosynthesis relative genes in celery. Sci. Hortic. 2014; 165: 218–224. [Google Scholar]
- 33.Castellarin SD, Gaspero GD. Transcriptional control of anthocyanin biosynthetic genes in extreme phenotypes for berry pigmentation of naturally occurring grapevines. BMC Plant Biol. 2007; 7: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimada Y, Ohbayashi M, Nakano-Shimada R, Okinaka Y, Kiyokawa S, Kikuchi Y. Genetic engineering of the anthocyanin biosynthetic pathway with flavonoid-3′5′-hydroxylase: specific switching of the pathway in petunia. Plant Cell rep. 2001; 20: 456–462. [Google Scholar]
- 35.Rommens CM, Richael CM, Yan CM, Yan H, Navarre DA, Ye J, et al. Engineered native pathways for high kaempferol and caffeoylquinate production in potato. Plant Biotechnol J. 2008; 6:870–886. doi: 10.1111/j.1467-7652.2008.00362.x [DOI] [PubMed] [Google Scholar]
- 36.Wessinger CA, Rausher MD. Ecological transition predictably associated with gene degeneration. Mol Biol Evol. 2015; 32:347–354. doi: 10.1093/molbev/msu298 [DOI] [PubMed] [Google Scholar]
- 37.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011; 25: 2227–2241. doi: 10.1101/gad.176826.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bogs J, Jaffé FW, Takos AM, Walker AR, Robinson SP. The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol. 2007; 143: 1347–1361. doi: 10.1104/pp.106.093203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Azuma A, Kobayashi S, Goto-Yamamoto N, Shiraishi M, Mitani N, Yakushiji H, et al. Color recovery in berries of grape (Vitis vinifera L.) ‘Benitaka’, a bud sport of ‘Italia’, is caused by a novel allele at the VvmybA1 locus. Plant Sci. 2009; 176: 470–478. doi: 10.1016/j.plantsci.2008.12.015 [DOI] [PubMed] [Google Scholar]
- 40.Wada T, Kunihiro A, Tominaga-Wada R. Arabidopsis CAPRICE (MYB) and GLABRA3 (bHLH) control tomato (Solanum lycopersicum) anthocyanin biosynthesis. PLoS One. 2014; 9, e109093 doi: 10.1371/journal.pone.0109093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshida K, Ma D, Constabel CP. The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes. Plant Physiol. 2015; 3: 693–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hurst HC. Transcription factors. 1: bZIP proteins. Protein Profile. 1994; 2:123–168. [PubMed] [Google Scholar]
- 43.Zhang Y, Zheng S, Liu Z, Wang L, Bi Y. Both HY5 and HYH are necessary regulators for low temperature—induced anthocyanin accumulation in Arabidopsis seedlings. J Plant Physiol. 2011; 4: 367–374. [DOI] [PubMed] [Google Scholar]
- 44.Zhang HY, He H, Wang X, Wang XF, Yang X, Li L, et al. Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 2011; 65: 346–358. doi: 10.1111/j.1365-313X.2010.04426.x [DOI] [PubMed] [Google Scholar]
- 45.Droge-Laser W, Kaiser A, Lindsay WP, Halkier BA, Loake GJ, Doerner P, et al. Rapid stimulation of a soybean protein—serine kinase that phosphorylates a novel bZIP DNA-binding protein, G /HBF-1, during the induction of early transcription-dependent defenses. EMBO J. 1997; 4: 726–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shin DH, Choi MG, Kim K, Bang G, Cho M, Choi SB, et al. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013; 587: 1543–1547. doi: 10.1016/j.febslet.2013.03.037 [DOI] [PubMed] [Google Scholar]
- 47.Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 2005; 57: 155–171. doi: 10.1007/s11103-004-6910-0 [DOI] [PubMed] [Google Scholar]
- 48.Shore P, Sharrocks AD. The MADS-box family of transcription factors. Eur. J. Biochem. 1995; 229: 1–13. [DOI] [PubMed] [Google Scholar]
- 49.Lalusin AG, Nishita K, Kim SH, Ohta M, Fujimura T. A new MADS-box gene (IbMADS10) from sweet potato (Ipomoea batatas (L.) Lam) is involved in the accumulation of anthocyanin. Mol Genet Genomics. 2006; 275: 44–54. doi: 10.1007/s00438-005-0080-x [DOI] [PubMed] [Google Scholar]
- 50.Pan ZJ, Chen YY, Du JS, Chen YY, Chung MC, Tsai WC, et al. Flower development of Phalaenopsis orchid involves functionally divergent SEPALLATA-like genes. New Phytol. 2014; 202: 1024–1042. doi: 10.1111/nph.12723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun Y, Qiu Y, Duan M, Wang JL, Zhang XH, Wang HP, et al. Identification of anthocyanin biosynthesis related microRNAs in a distinctive Chinese radish (Raphanus sativus L.) by high-throughput sequencing. Mol Genet Genom. 2017; 292: 215–229. [DOI] [PubMed] [Google Scholar]
- 52.Sasaki K, Yamaguchi H, Nakayama M, Aida R, Ohtsubo N. Comodification of class B genes TfDEF and TfGLO in Torenia fournieri Lind. alters both flower morphology and inflorescence architecture. Plant Mol Biol. 2014; 86: 319–334. doi: 10.1007/s11103-014-0231-8 [DOI] [PubMed] [Google Scholar]
- 53.Jaakola L, Poole M, Jones MO, Kamarainen-Karppinen T, Koskimaki JJ, Hohtola A, et al. A SQUAMOSA MADS Box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol. 2010; 153: 1619–1629. doi: 10.1104/pp.110.158279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang YQ, Liu ZJ, Liu RZ, Hao HL, Bi YR. Gibberellins negatively regulate low temperature-induced anthocyanin accumulation in a HY5/HYH-dependent manner. Plant Signaling & Behavior. 2011; 6(5): 632–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang CF, Gao XH, Liao LL, Harberd NP, Fu XD. Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in arabidopsis. Plant Physiology. 2007; 145: 1460–1470. doi: 10.1104/pp.107.103788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen X, Zhao K, Liu L, Zhang K, Yuan H, Liao X, et al. A role for PacMYBA in ABA-regulated anthocyanin biosynthesis in red-colored sweet cherry cv. Hong Deng (Prunus avium L.). Plant cell physiol. 2014; 55: 862–880. doi: 10.1093/pcp/pcu013 [DOI] [PubMed] [Google Scholar]
- 57.Tonelli C, Cominelli E, Allegra D, Galbiati M. Plant tolerance to drought and salinity: modulation of transcription factors. Proceedings of the 18th International Conference on Arabidopsis Research, 2007; 176.
- 58.Koyama K, Sadamatsu K, Goto-Yamamoto N. Abscisic acid stimulated ripening and gene expression in berry skins of the Cabernet Sauvignon grape. Functional & Integrative Genomics, 2009; 10(3): 367–381. [DOI] [PubMed] [Google Scholar]
- 59.Qin YH, Zhang XH, Feng L, Li WW, Xu ZS, Li LC, et al. Response of millet transcription factor gene SibZIP42 to high salt and ABA treatment in transgenic Arabidopsis. Scientia Agricultura Sinica. 2016; 17: 3276–3286. [Google Scholar]
- 60.Palma-Silva C, Ferro M, Bacc M, Turchetto-Zolet AC. De novo assembly and characterization of leaf and floral transcriptomes of the hybridizing bromeliad species (Pitcairnia spp.) adapted to Neotropical Inselbergs. Mol Ecol Resour. 2016; doi: 10.1111/1755-0998.12504 [DOI] [PubMed] [Google Scholar]
- 61.Peng ZH, Han CY, Yuan LB, Zhang K, Huang HM, Ren CM. Brassinosteroid enhances jasmonate-induced anthocyanin accumulation in Arabidopsis seedlings. J Integr Plant Biol, 2011; 53(8): 632–640 doi: 10.1111/j.1744-7909.2011.01042.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
SD140:a,b,c. SD92: d,e,f.
(TIF)
SD140:a,b,c. SD92: d,e,f.
(TIF)
A: SD92, B: SD140.
(TIF)
A threshold of corrected p value ≤ 0.001 was used to judge the significantly enriched GO terms in DEGs.
(TIF)
(DOC)
(DOC)
(DOC)
Data Availability Statement
All the RNA sequencing data are available from the NCBI Sequence Read Archive using SRA accession number SRP125987.