Abstract
Pyrolysis offers a straightforward approach for the deconstruction of plant cell wall polymers into bio-oil. Recently, there has been substantial interest in bio-oil fractionation and subsequent use of biological approaches to selectively upgrade some of the resulting fractions. A fraction of particular interest for biological upgrading consists of polysaccharide-derived substrates including sugars and sugar dehydration products such as levoglucosan and cellobiosan, which are two of the most abundant pyrolysis products of cellulose. Levoglucosan can be converted to glucose-6-phosphate through the use of a levoglucosan kinase (LGK), but to date, the mechanism for cellobiosan utilization has not been demonstrated. Here, we engineer the microbe Pseudomonas putida KT2440 to use levoglucosan as a sole carbon and energy source through LGK integration. Moreover, we demonstrate that cellobiosan can be enzymatically converted to levoglucosan and glucose with β-glucosidase enzymes from both Glycoside Hydrolase Family 1 and Family 3. β-glucosidases are commonly used in both natural and industrial cellulase cocktails to convert cellobiose to glucose to relieve cellulase product inhibition and to facilitate microbial uptake of glucose. Using an exogenous β-glucosidase, we demonstrate that the engineered strain of P. putida can grow on levoglucosan up to 60 g/L and can also utilize cellobiosan. Overall, this study elucidates the biological pathway to co-utilize levoglucosan and cellobiosan, which will be a key transformation for the biological upgrading of pyrolysis-derived substrates.
Keywords: Pseudomonas putida KT2440, Levoglucosan kinase, B-glucosidase, Cellobiosan, Pyrolysis, Biofuels
Highlights
-
•
Levoglucosan kinase is engineered into Pseudomonas putida KT2440.
-
•
Cellobiosan can be cleaved to levoglucosan and glucose by β-glucosidases.
-
•
This provides a path forward to co-utilize levoglucosan and cellobiosan.
-
•
These transformations will be important for hybrid processing applications.
1. Introduction
Thermal processes for biomass deconstruction, such as pyrolysis and liquefaction, offer rapid, effective methods for the depolymerization of plant cell wall components (Czernik and Bridgwater, 2004, Mohan et al., 2006, Yang et al., 2007, Laird et al., 2009). These processes typically produce heterogeneous slates of compounds derived from polysaccharides and lignin that can potentially be upgraded simultaneously over chemical catalysts (Zhang et al., 2007, Mortensen et al., 2011, Xiu and Shahbazi, 2012, Wang et al., 2013), integrated into petroleum refinery streams (Talmadge et al., 2014), or fractionated through a wide variety of approaches and subsequently upgraded in a more selective manner to a broader slate of fuels and chemicals (Brown, 2005, Brown, 2007, Lian et al., 2010, Jarboe et al., 2011, Lian et al., 2012). Recently, there has been substantial emphasis placed on selective fractionation of pyrolysis-derived substrates (Lian et al., 2010, Pollard et al., 2012, Rover et al., 2014, Liang et al., 2013, Rover et al., 2014, Gooty et al., 2014, Gooty et al., 2014) and the use of biological approaches to selectively upgrade at least some of the resulting fractions. Biological approaches are particularly attractive, as metabolic engineering enables both the broadening of substrate specificity as well as the targeted production of single products of interest (Brown, 2005, Brown, 2007, Jarboe et al., 2011). This general concept of combining thermal deconstruction with subsequent biological upgrading has been dubbed “hybrid processing” by Brown and co-workers (Brown, 2005, Brown, 2007, Jarboe et al., 2011).
One particular pyrolysis fraction of interest for biological upgrading consists of polysaccharide-derived substrates. In typical fast pyrolysis schemes, levoglucosan and cellobiosan are the most abundantly produced dehydration products of cellulose (Patwardhan et al., 2011, Patwardhan et al., 2009, Bai and Brown, 2014, Bai et al., 2013). Multiple tandem catalytic-biological schemes have been developed to fractionate levoglucosan-rich streams from bio-oil, hydrolyze it to glucose, and upgrade it to ethanol, for example (Lian et al., 2010, Shafizadeh and Stevenson, 1982, Helle et al., 2007, Bennett et al., 2009, Yu and Zhang, 2003, Yu and Zhang, 2003, Chan and Duff, 2010). Lian et al. developed a process that used solvent fractionation to separate phenolics from pyrolytic sugars, hydrolyze the levoglucosan to glucose, and then use a biological step to either produce ethanol or fatty acids (Lian et al., 2010). Bennett et al. examined the optimal conditions to produce glucose from a levoglucosan-rich fraction of bio-oil, and noted a 216% yield of glucose, attributing the high yield to cellobiosan and other oligomeric forms of cellulose present in the bio-oil fraction (Bennett et al., 2009).
These types of processes require an intermediate catalytic step to produce glucose, but another approach has been developed that enables biological upgrading directly from levoglucosan (Prosen et al., 1993, Lian et al., 2013, Layton et al., 2011). Namely, levoglucosan can be converted to glucose-6-phosphate (G6P) through use of a levoglucosan kinase (LGK) (Layton et al., 2011, Zhuang and Zhang, 2002, Kitamura et al., 1991, Dai et al., 2009), the structure of which was recently reported (Bacik et al., 2015). Prosen et al. screened multiple fungi and yeasts on levoglucosan-enriched substrates and demonstrated substantial growth but only after the removal of lignin-derived aromatics, suggesting that tolerance to lignin-derived aromatic compounds is a major issue in these streams (Prosen et al., 1993). The observed growth was likely due to the presence of endogenous LGK enzymes (Kitamura et al., 1991). The LGK enzyme present in Lipomyces starkeyi has been shown to be quite active for conversion of levoglucosan to G6P (Dai et al., 2009). Jarboe et al. subsequently engineered the L. starkeyi LGK gene into an ethanol-producing strain of Escherichia coli (KO11), and were able to obtain near-complete conversion of levoglucosan and produce ethanol. Similar to observations from Prosen et al. (1993) and other studies (Chan and Duff, 2010), the authors note that substrate toxicity is likely to be a major factor for the feasibility of these biological approaches to upgrading pyrolytic sugar streams. Subsequently, Jarboe et al. conducted a detoxification study of pyrolytic sugars using an overliming method. The authors were able to demonstrate a nearly 10-fold improvement in ethanol production relative to no cleanup using the same engineered E. coli strain (Chi et al., 2013).
Along with levoglucosan, significant amounts of cellobiosan often form during fast pyrolysis (Rover et al., 2014, Bai and Brown, 2014, Helle et al., 2007, Chi et al., 2013, Choi et al., 2014, Radlein et al., 1987, Scott et al., 1997, Johnston and Brown, 2014, Tessini et al., 2011). Radlein identified cellobiosan in 1987 as a major component of anhydrosugars after pyrolysis of Avicel (between 6% and 15% of the liquid product). To our knowledge, direct cellobiosan utilization has not been previously reported. However, to move towards a consolidated biological process for production of fuels or chemicals from pyrolytic sugar streams, it will be necessary to enable the biological utilization of these highly abundant molecules.
Here, we engineer a solvent-tolerant microbe, Pseudomonas putida KT2440, to utilize levoglucosan through the heterologous expression of the levoglucosan kinase from L. starkeyi. Furthermore, we demonstrate biological cellobiosan utilization using β-glucosidase-mediated hydrolysis of cellobiosan. Accordingly, our results provide a trajectory towards more complete biological utilization of pyrolytic sugar streams.
2. Materials and methods
2.1. Plasmid and strain construction
The lgk gene from L. starkeyi was codon optimized using Gene Designer software from DNA 2.0 and synthesized as a gBlock by Integrated DNA Technologies. The sequence of this codon optimized gene has been deposited at GenBank under the accession number KU377145. This fragment was cloned into plasmid pMFL76 which is derived from the commercial PCR-Blunt II Topo vector (ThermoFisher), with the addition of two 1-kb genomic regions located in proximity of the rpoS region of the genome from P. putida KT2440 to enable homologous recombination mediated genomic integration into strain KT2440. The Ptac-lgk was inserted in between these 1 kb homology regions and transformed into KT2440. A single kanamycin-resistant transformant was isolated and growth in M9 levoglucosan was confirmed, to generate strain FJPO3.
2.2. Media and growth conditions
Most growths were in a modified formulation of M9 minimal salts as described previously (Linger et al., 2014). Briefly, 6.78 g Na2PO4 (anhydrous), 3.0 g KH2PO4, and 0.50 g NaCl were dissolved in 750 mL deionized H2O. The pH was adjusted to 7.4 with 10 N NaOH and brought to 900 mL with H2O. The solution was autoclaved for 15 min at 121 °C and allowed to cool. One hundred µL of 1 M CaCl2 (100 µM final) and 2 mL of 1 M MgSO4-7H2O (2 mM final), 1 mL of 100 mM FeSO4, (100 µM final) and 10 mL 1 M (NH4)2SO4 (10 mM final) was added. In the case of Fig. 1C, mM (NH4)2SO4, was used to induce mcl-PHA production. All growths were performed at 30 °C and in shake-flasks except during growth curve analysis (Section 2.3).
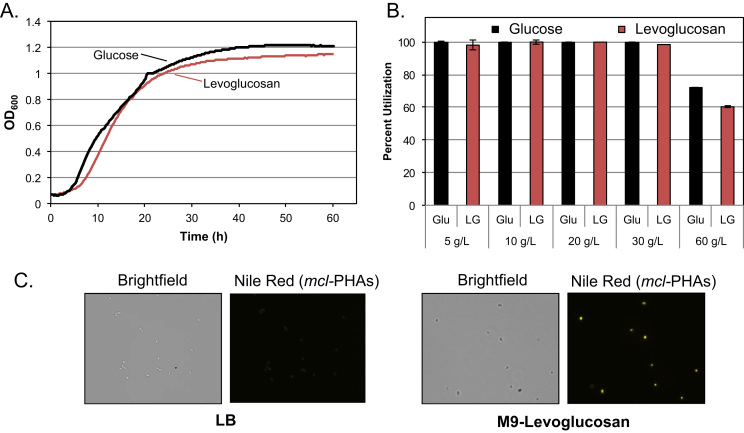
Fig. 1.
The engineered P. putida strain is capable of growth and polyhydroxyalkanoate production using levoglucosan as the sole carbon source. (A) Growth curve analysis of levoglucosan in M9 minimal medium supplemented with either glucose or levoglucosan using a Bioscreen-C Automated Growth Curves Analysis System. (B) HPLC analysis shows the percent utilization of either glucose or levoglucosan from cultures of FJPO3 grown in shake-flasks. (C) Brightfield and fluorescence microscopy of strain FJPO3 grown on LB and M9-levoglucosan, with Nile Red staining prior to fluorescence microscopy to stain mcl-PHAs.
2.3. Growth curves and growth rate analysis
Growth curves were generated using a Bioscreen C automated microbiology growth curve analysis system from Growth Curves USA. Overnight cultures were diluted to 0.02 OD600 in a total volume of 300 µL per well. Incubations were performed at 30 °C with continuous shaking at maximum intensity and turbidity measurements (OD420-580) were collected every 15 min for the duration of the experiments. Spectrophotometric blanks were subtracted from the measured values and each growth curve represents the average of three independent cultures. The maximum specific growth rate (µmax) was calculated as the maximum slope of the log phase of the growth curve over any given 4 hour period during the course of the experiment (Franden et al., 2009). In the case of the growths containing β-glucosidases, relevant cultures were spiked with twenty micrograms of ag1 (Agrobacterium sp. β-glucosidase, Megazyme) prior to inoculation in Bioscreen C Honeycomb plates. Plates were held at 40 °C with shaking for 1 h immediately prior to inoculation.
2.4. Microscopic visualization of mcl-PHA production
One mL of saturated cultures were centrifuged at 5000xg for 5 min. The growth medium was removed via aspiration and the cell pellet was washed twice in phosphate-buffered saline (PBS), pH 7.4. Cells were suspended in 100 µL of PBS with 10 µg/mL Nile Red (Molecular Probes, ThermoFisher) and incubated for 15 min at room temperature in the dark. Cells were centrifuged, washed once in PBS and immobilized on microscope slides by mixing with 1% low-melting-temperature agarose in a 1:1 ratio. Images were acquired using a Nikon Eclipse 80i microscope. Nile Red fluorescence was detected between 560 and 590 nm using band-pass filtering.
2.5. High-performance liquid chromatography (HPLC)
Following the β-glucosidase reaction period, the tubes were placed on ice, filtered through a 0.2 µm filter and analyzed via High-Performance Liquid Chromatography (HPLC; Agilent 1100 series system (Agilent USA, Santa Clara, CA)) using a Shodex SP0810 carbohydrate column with de-ashing guard cartridges (BioRad Laboratories, Hercules, CA) run at 85 °C with ultra-pure water as the isocratic mobile phase at a flow rate of 0.6 mL/min. A refractive index detector was used for compound detection. By-products were identified by co-elution at the same retention time with pure compounds. Standard curves for substrate (cellobiosan) and products (glucose and levoglucosan) were also generated in order to quantify results. Additionally, enzymes were analyzed via HPLC in buffer alone (without cellobiosan) to ensure no carry-over products inherent to the enzyme preparations.
2.6. β-glucosidase enzymes used in this study
(1) Aspergillus niger bgl1 (Glycoside Hydrolase Family 3) obtained from Megazyme (Cat number E-BGLUC, GenBank Accession number: AJ132386), (2) Phanaerochate chrysosporium bgl1A (Glycoside Hydrolase Family 3) obtained from Megazyme (Cat number E-BGOSPC, GenBank Accession number:AAC26489), (3) Thermotoga maritima bglA (Glycoside Hydrolase Family 1) obtained from Megazyme (Cat number E-BGOSTM, GenBank Accession number: CAA52276.1), (4) Agrobacterium sp. abg (Glycoside Hydrolase Family 1) obtained from Megazyme (Cat number E-BGOSAG, GenBank Accession number: AAA22085.1).
2.7. 7-glucosidase mediated hydrolysis of cellobiosan
Each reaction was set up in 1.5 mL microcentrifuge tubes using cellobiosan at a concentration of 2 mg/mL in the manufacturers recommended buffers: 50 mM sodium maleate, pH 6.5 (T. maritima and Agrobacterium sp.), and 100 mM sodium acetate, pH 5.0 (P. chrysosporium and A. niger). In each reaction, 2.5–20 µg of enzyme was loaded and total reaction volume was 400 µL. A no-enzyme reaction was run simultaneously to ensure cellobiosan cleavage was enzyme-dependent. Reactions were run at 40 °C using a dry-block for 90 min. In a separate experiment evaluating the substrate concentration, 0.125 μM of purified A. niger β-glucosidase was incubated at 40 °C in a 96-well microtiter plate with 0, 0.5, 1, 2, 4, 8, 16 or 32 mM cellobiose or cellobiosan. Reactions were initiated by the addition of enzyme and quenched by boiling in a thin walled PCR tube at 0.5, 1, 5 and 10 min. Following the reaction periods, the tubes were placed on ice, filtered through a 0.2 µm filter and analyzed via HPLC using a carbohydrate column. Standard curves were generated for the substrate (cellobiosan) and products (glucose and levoglucosan) to quantify results. Additionally, the enzymes were analyzed via HPLC in buffer alone (without the addition of cellobiosan) to ensure no carry-over products inherent to the enzyme preparations.
3. Results
3.1. Engineering levoglucosan utilization in P. putida KT2440
Levoglucosan kinase (LGK) from L. starkeyi was codon optimized and synthesized for expression using the strong and constitutively active hybrid Ptac promoter, shown previously to function in P. putida KT2440 (Bagdasarian et al., 1983, Johnson and Beckham, 2015). P. putida KT2440 was transformed with a Ptac-lgk construct to generate strain FJPO3. Fig. 1A shows growth curves of FJPO3 in M9 medium supplemented with either 7.5 g/L glucose or 7.5 g/L levoglucosan. Growth profiles in these carbon sources at this concentration are quite similar, suggesting efficient utilization of levoglucosan in strain FJPO3. In addition, total levoglucosan utilization is similar to glucose utilization and FJPO3 has the capacity to grow at concentrations as high as 60 g/L (Fig. 1B).
P. putida produces medium chain-length polyhydroxy-alkanoates (mcl-PHAs), which are high-value polymers that can serve as plastics, adhesive precursors, and as precursors to chemical building blocks or hydrocarbons (Linger et al., 2014, Chen, 2009). As mcl-PHAs represent a potential target to generate from pyrolytic sugars, we wanted to ensure that P. putida would generate mcl-PHAs using levoglucosan as its sole carbon source. Using Nile Red fluorescence, it is qualitatively clear that FJPO3 produces mcl-PHAs in N-limited medium [M9 1 mM (NH4)2SO4] containing levoglucosan as a sole carbon source, but not in the N- and C-rich LB medium (Fig. 1C).
3.2. Cellobiosan is cleaved by β-glucosidases to levoglucosan and glucose
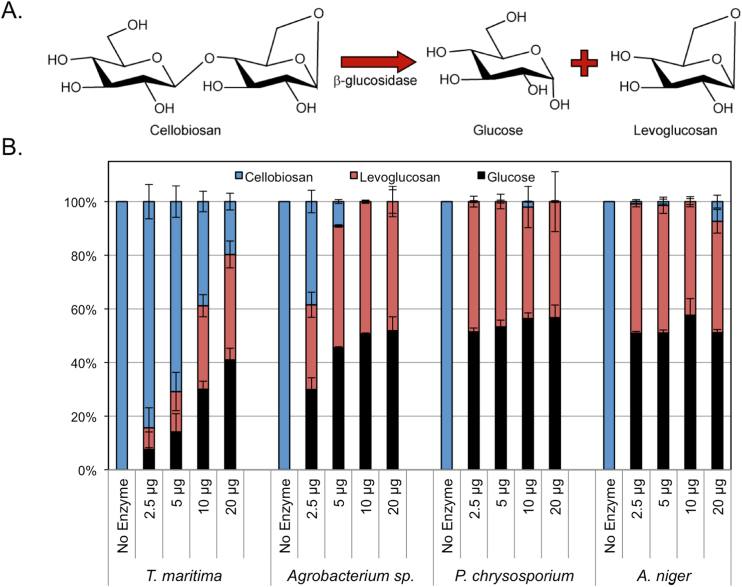
The mechanism of biological turnover of cellobiosan has not been reported previously to our knowledge. A potential mechanism for catabolism of cellobiosan is cleavage of the β-1,4-glycosidic bond connecting the levoglucosan and glucose moieties. In nature, β-glucosidases (from Glycoside Hydrolase Families 1 and 3, GH1 and GH3, respectively) cleave cellobiose to liberate two glucose molecules, and indeed, this mechanism is of paramount importance for overcoming product inhibition during enzymatic depolymerization of cellulose by cellulases (Chundawat et al., 2011, Payne et al., 2015). Given the similarity of cellobiose and cellobiosan, we hypothesized that β-glucosidases may be able to cleave the glycosidic bond in cellobiosan in a similar fashion (Fig. 2A). To test this hypothesis, we first tested the activity of four β-glucosidases against cellobiosan. The chosen enzymes comprise two representatives each from GH Families 1 and 3 as well as two fungal and two bacterial enzymes: Aspergillus niger bgl1 (GH Family 3), Phanaerochate chrysosporium bgl1A (GH Family 3), Thermotoga maritima bglA (GH Family 1), and Agrobacterium sp. abg (GH Family 1). We obtained highly purified forms of these four β-glucosidases from Megazyme and performed in vitro assays using HPLC to monitor cellobiosan disappearance and to identify the products. As shown in Fig. 2B, all four β-glucosidases are able to cleave cellobiosan to generate glucose and levoglucosan. Four different enzyme loadings are shown for each enzyme.
Fig. 2.
Cellobiosan is cleaved to glucose and levoglucosan by all tested β-glucosidases. (A) The reaction of cellobiosan to glucose and levoglucosan. (B) Conversion as a function of enzyme loading (no enzyme, 2.5, 5, 10, and 20 µg of enzyme loadings) in 400 µL reaction vessels with 2 mg/mL of cellobiosan for 90 min reaction times at 40 °C.
HPLC analysis shows the disappearance of cellobiosan and the appearance of glucose and levoglucosan with the addition of beta-glucosidase enzymes to the reaction.
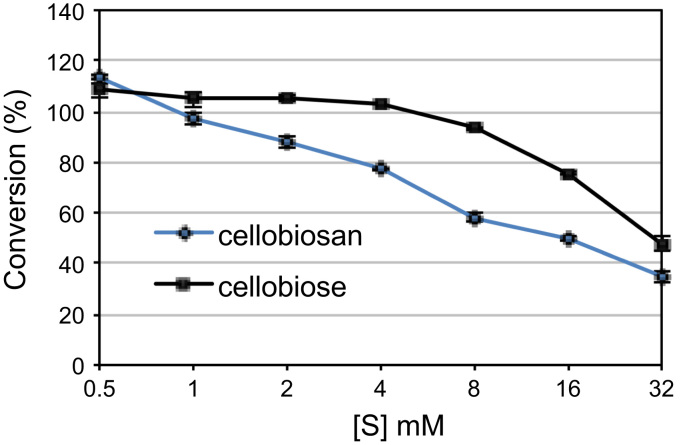
The results of this assay show unequivocally that β-glucosidases are capable of hydrolyzing the β-1,4-glycosidic bond in cellobiosan to liberate glucose and levoglucosan as products. Additionally, it suggests that this activity is conserved from β-glucosidases derived from multiple GH families (GH1 and GH3), and from both prokaryotic (Agrobacterium sp. and T. maritima) and eukaryotic organisms (P. chrysosporium and A. niger), and from mesophilic (A. niger and Agrobacterium sp.), thermophilic (P chrysosporium), and hyperthermophilic organisms (T. maritima). Importantly, the data in Fig. 2 were generated at a single temperature (40 °C); we note that this is not the optimum temperature reported by Megazyme for all enzymes tested. Accordingly, conclusions about the relative differences β-glucosidase efficacy on cellobiosan are only relevant at the conditions tested. When we evaluated the ability of A. niger β-glucosidase to hydrolyze increasing concentrations of cellobiosan or cellobiose, in both cases we found that the percent conversion decreased as substrate concentration increased. This is expected from cellobiose, which is known to cause substrate and product inhibition (Teugjas and Valjamae, 2013); however, the inhibition was more dramatic with cellobiosan (Fig. 3). Both reactions showed no further conversion after the first minute (data not shown), ruling out the possibility that slower reaction kinetics cause this decrease in conversion after 10 min. It is not clear whether this inhibition is due to the increase in levoglucosan as cellobiosan is cleaved or substrate inhibition caused by cellobiosan itself.
Fig. 3.
Conversion of substrates to products as a function of substrate concentration. Conversion of cellobiose or cellobiosan to glucose or glucose and levoglucosan, respectively by A. niger β-glucosidase (abg) after a 10 min of incubation.
3.3. Addition of a β-glucosidase enables growth of P. putida-LGK on cellobiosan
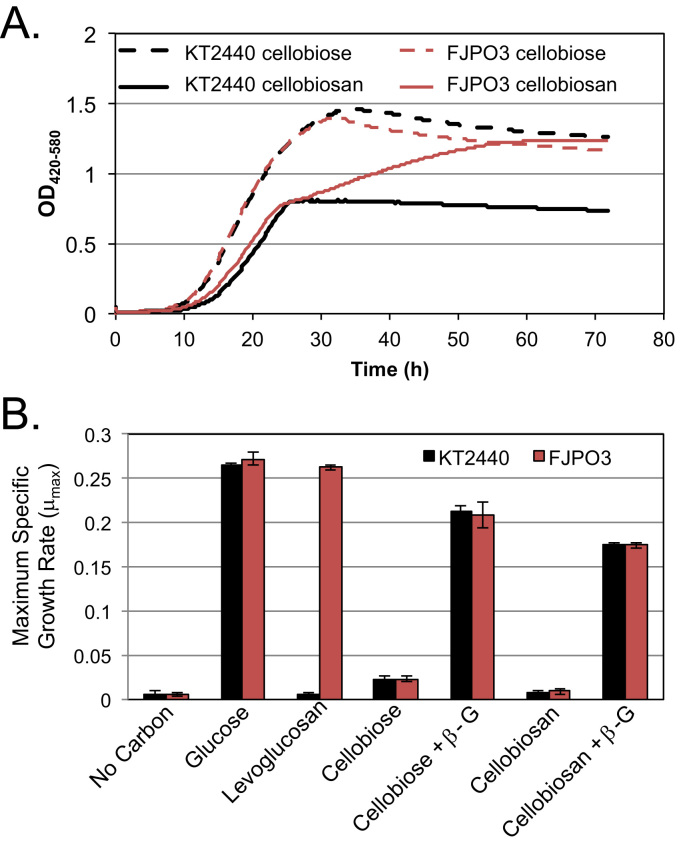
To demonstrate the ability of P. putida strain FJPO3 to grow using cellobiosan as its sole carbon source, we performed growths using various carbon sources with and without the addition of the Agrobacterium sp. abg β-glucosidase (Fig. 4). Growth profiles of KT2440 and FJPO3 are virtually indistinguishable in M9 medium containing cellobiose with the addition of abg suggesting equivalent glucose usage between the strains (Fig. 4A). Conversely, in the cellobiosan plus abg conditions, it is clear that while both FJPO3 and KT2440 are capable of utilizing the glucose component of cellobiosan, FJPO3 continues growth after KT2440's growth ceases, albeit at a reduced rate. Importantly, we observed no growth of either strain in either cellobiose or cellobiosan without the addition of abg. For clarity, these data are not depicted on the growth curves of Fig. 4A. The maximum specific growth rates (μmax) for all conditions are shown in Fig. 4B. Fig. 4B additionally depicts the max growth rate in M9 media containing levoglucosan and glucose as the sole carbon source.
Fig. 4.
Growth of P. putida on cellobiose and cellobiosan with the addition of exogenous β-glucosidase. (A) Growth curve of strain FJPO3 and parent strain KT2440 on β-glucosidase hydrolyzed cellobiose or cellobiosan (both at 10 g/L), using OD420-580 to track growth (B) Maximum specific growth rates of both KT2440 and FJPO3 in M9 medium with the annotated sole carbon sources.
4. Discussion and conclusions
Developing biocatalysts that span the breadth of molecules present in thermochemical-derived substrates and in parallel developing detoxification strategies is of critical importance to the viability of hybrid processing (Brown, 2005, Brown, 2007, Lian et al., 2010, Jarboe et al., 2011, Lian et al., 2012, Liang et al., 2013, Lian et al., 2013, Layton et al., 2011, Chi et al., 2013). To that end, we demonstrated that the addition of β-glucosidase in an LGK strain of P. putida KT2440 enables complete utilization of cellobiosan via hydrolytic cleavage of the glycosidic linkage, liberating glucose and levoglucosan. Given the range of enzymes tested, we anticipate that many β-glucosidases will exhibit the same activity on cellobiosan. This finding will enable understanding, implementing, and manipulating cellobiosan utilization in any number of microbial organisms. β-glucosidases can be added exogenously or could be expressed and secreted directly from the microbial biocatalyst.
Going forward, P. putida KT2440 is a promising strain for hybrid processing applications given its inherent ability to tolerate toxic environments, the ability to rapidly engineer the microbe, and its ability to utilize a large number of lignin-derived aromatic compounds (Nikel et al., 2014, Jiménez et al., 2002, Poblete-Castro et al., 2012). Strains of P. putida have been engineered to catabolize a broad range of substrates including xylose (Meijnen et al., 2008), phenol (Vardon et al., 2015), and now levoglucosan, among others. The hybrid processing concept from thermochemical deconstruction of biomass is closely mirrored by work from our group in biological funneling of lignin-derived streams using P. putida KT2440 and other microbes (Linger et al., 2014, Johnson and Beckham, 2015, Vardon et al., 2015, Salvachúa et al., 2015). Lignin-derived streams also contain a significant amount of heterogeneous substrates and are also quite toxic. P. putida, alongside other microbes such as Rhodococcus jostii RHA1, Acinetobacter sp. ADP1, and Amycolatopsis sp., are quite tolerant to lignin-derived streams and are able to catabolize a broad range of substrates, thus potentially offering good starting candidates for the biological component of hybrid processing.
Acknowledgments
We thank the US Department of Energy's Bioenergy Technologies Office (DOE-BETO) for funding this work via Contract No. DE-AC36-08GO28308 with the National Renewable Energy Laboratory. We thank Mary Biddy, Michael Guarnieri, Christopher Johnson, and Brandon Knott of NREL and Robert Brown, Kirsten Davis, Laura Jarboe, Marjorie Rover, Ryan Smith, and Zhiyou Wen of Iowa State University for helpful discussions.
References
- Bacik J.-P. Producing Glucose 6-Phosphate from Cellulosic Biomass: structural insights into levoglucosan bioconversion. J. Biol. Chem. 2015;290(44):26638–26648. doi: 10.1074/jbc.M115.674614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdasarian M.M., Amann E., Lurz R., Rückert B., Bagdasarian M. Activity of the hybrid trp-lac (tac) promoter of Escherichia coli in Pseudomonas putida. Construction of broad-host-range, controlled-expression vectors. Gene. 1983;26(2):273–282. doi: 10.1016/0378-1119(83)90197-x. [DOI] [PubMed] [Google Scholar]
- Bai X., Brown R.C. Modeling the physiochemistry of levoglucosan during cellulose pyrolysis. J. Anal. Appl. Pyrol. 2014;105:363–368. [Google Scholar]
- Bai X., Johnston P., Sadula S., Brown R.C. Role of levoglucosan physiochemistry in cellulose pyrolysis. J. Anal. Appl. Pyrol. 2013;99:58–65. [Google Scholar]
- Bennett N.M., Helle S.S., Duff S.J. Extraction and hydrolysis of levoglucosan from pyrolysis oil. Bioresour. Technol. 2009;100(23):6059–6063. doi: 10.1016/j.biortech.2009.06.067. [DOI] [PubMed] [Google Scholar]
- Brown, R.C. 2005. Biomass refineries based on hybrid thermochemical‐biological processing‐an overview Biorefineries-Industrial Processes and Products: Status Quo and Future Directions 2005 227 252.
- Brown R.C. Hybrid thermochemical/biological processing. Appl. Biochem. Biotechnol. 2007;137(1–12):947–956. doi: 10.1007/s12010-007-9110-y. [DOI] [PubMed] [Google Scholar]
- Chan J.K., Duff S.J. Methods for mitigation of bio-oil extract toxicity. Bioresour. Technol. 2010;101(10):3755–3759. doi: 10.1016/j.biortech.2009.12.054. [DOI] [PubMed] [Google Scholar]
- Chen G.-Q. A microbial polyhydroxyalkanoates (PHA) based bio-and materials industry. Chem. Soc. Rev. 2009;38(8):2434–2446. doi: 10.1039/b812677c. [DOI] [PubMed] [Google Scholar]
- Chi Z. Overliming detoxification of pyrolytic sugar syrup for direct fermentation of levoglucosan to ethanol. Bioresour. Technol. 2013;150:220–227. doi: 10.1016/j.biortech.2013.09.138. [DOI] [PubMed] [Google Scholar]
- Choi Y.S., Johnston P.A., Brown R.C., Shanks B.H., Lee K.-H. Detailed characterization of red oak-derived pyrolysis oil: Integrated use of GC, HPLC, IC, GPC and Karl-Fischer. J. Anal. Appl. Pyrol. 2014;110:147–154. [Google Scholar]
- Chundawat S.P.S., Beckham G.T., Himmel M.E., Dale B.E. Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu. Rev. Chem. Biomol. Eng. 2011;2:6.1–6.25. doi: 10.1146/annurev-chembioeng-061010-114205. [DOI] [PubMed] [Google Scholar]
- Czernik S., Bridgwater A. Overview of applications of biomass fast pyrolysis oil. Energy Fuels. 2004;18(2):590–598. [Google Scholar]
- Dai J. Cloning of a novel levoglucosan kinase gene from Lipomyces starkeyi and its expression in Escherichia coli. World J. Microbiol. Biotechnol. 2009;25(9):1589–1595. [Google Scholar]
- Franden M.A., Pienkos P.T., Zhang M. Development of a high-throughput method to evaluate the impact of inhibitory compounds from lignocellulosic hydrolysates on the growth of Zymomonas mobilis. J. Biotechnol. 2009;144(4):259–267. doi: 10.1016/j.jbiotec.2009.08.006. [DOI] [PubMed] [Google Scholar]
- Gooty A.T., Li D., Berruti F., Briens C. Kraft-lignin pyrolysis and fractional condensation of its bio-oil vapors. J. Anal. Appl. Pyrol. 2014;106:33–40. [Google Scholar]
- Gooty A.T., Li D., Briens C., Berruti F. Fractional condensation of bio-oil vapors produced from birch bark pyrolysis. Sep. Purif. Technol. 2014;124:81–88. [Google Scholar]
- Helle S., Bennett N.M., Lau K., Matsui J.H., Duff S.J. A kinetic model for production of glucose by hydrolysis of levoglucosan and cellobiosan from pyrolysis oil. Carbohydr. Res. 2007;342(16):2365–2370. doi: 10.1016/j.carres.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Jarboe L.R., Wen Z., Choi D., Brown R.C. Hybrid thermochemical processing: fermentation of pyrolysis-derived bio-oil. Appl. Microbiol. Biotechnol. 2011;91(6):1519–1523. doi: 10.1007/s00253-011-3495-9. [DOI] [PubMed] [Google Scholar]
- Jiménez J.I., Miñambres B., Garcia J.L., Díaz E. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ. Microbiol. 2002;4(12):824–841. doi: 10.1046/j.1462-2920.2002.00370.x. [DOI] [PubMed] [Google Scholar]
- Johnson C.W., Beckham G.T. Aromatic catabolic pathway selection for optimal production of pyruvate and lactate from lignin. Metab. Eng. 2015;28:240–247. doi: 10.1016/j.ymben.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Johnston P.A., Brown R.C. Quantitation of sugar content in pyrolysis liquids after acid hydrolysis using high-performance liquid chromatography without neutralization. J. Agric. Food Chem. 2014;62(32):8129–8133. doi: 10.1021/jf502250n. [DOI] [PubMed] [Google Scholar]
- Kitamura Y., Abe Y., Yasui T. Metabolism of Levoglucosan (1, 6-Anhydro-α-d-glucopyranose) in Microorganisms. Agric. Biol. Chem. 1991;55(2):515–521. [Google Scholar]
- Laird D.A., Brown R.C., Amonette J.E., Lehmann J. Review of the pyrolysis platform for coproducing bio‐oil and biochar. Biofuels Bioprod. Biorefin. 2009;3(5):547–562. [Google Scholar]
- Layton D.S., Ajjarapu A., Choi D.W., Jarboe L.R. Engineering ethanologenic Escherichia coli for levoglucosan utilization. Bioresour. Technol. 2011;102(17):8318–8322. doi: 10.1016/j.biortech.2011.06.011. [DOI] [PubMed] [Google Scholar]
- Lian J. Separation, hydrolysis and fermentation of pyrolytic sugars to produce ethanol and lipids. Bioresour. Technol. 2010;101(24):9688–9699. doi: 10.1016/j.biortech.2010.07.071. [DOI] [PubMed] [Google Scholar]
- Lian J., Garcia-Perez M., Chen S. Fermentation of levoglucosan with oleaginous yeasts for lipid production. Bioresour. Technol. 2013;133:183–189. doi: 10.1016/j.biortech.2013.01.031. [DOI] [PubMed] [Google Scholar]
- Lian J., Garcia-Perez M., Coates R., Wu H., Chen S. Yeast fermentation of carboxylic acids obtained from pyrolytic aqueous phases for lipid production. Bioresour. Technol. 2012;118:177–186. doi: 10.1016/j.biortech.2012.05.010. [DOI] [PubMed] [Google Scholar]
- Liang Y. Utilization of acetic acid-rich pyrolytic bio-oil by microalga Chlamydomonas reinhardtii: Reducing bio-oil toxicity and enhancing algal toxicity tolerance. Bioresour. Technol. 2013;133:500–506. doi: 10.1016/j.biortech.2013.01.134. [DOI] [PubMed] [Google Scholar]
- Linger J.G. Lignin valorization through integrated biological funneling and chemical catalysis. Proc. Nat. Acad. Sci. 2014;111(33):12013–12018. doi: 10.1073/pnas.1410657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijnen J.-P., de Winde J.H., Ruijssenaars H.J. Engineering Pseudomonas putida S12 for efficient utilization of d-xylose and l-arabinose. Appl. Environ. Microbiol. 2008;74(16):5031–5037. doi: 10.1128/AEM.00924-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan D., Pittman C.U., Steele P.H. Pyrolysis of wood/biomass for bio-oil: a critical review. Energy Fuels. 2006;20(3):848–889. [Google Scholar]
- Mortensen P.M., Grunwaldt J.-D., Jensen P.A., Knudsen K., Jensen A.D. A review of catalytic upgrading of bio-oil to engine fuels. Appl. Catal. A: Gen. 2011;407(1):1–19. [Google Scholar]
- Nikel P.I., Martínez-García E., de Lorenzo V. Biotechnological domestication of pseudomonads using synthetic biology. Nat. Rev. Microbiol. 2014;12(5):368–379. doi: 10.1038/nrmicro3253. [DOI] [PubMed] [Google Scholar]
- Patwardhan P.R., Dalluge D.L., Shanks B.H., Brown R.C. Distinguishing primary and secondary reactions of cellulose pyrolysis. Bioresour. Technol. 2011;102(8):5265–5269. doi: 10.1016/j.biortech.2011.02.018. [DOI] [PubMed] [Google Scholar]
- Patwardhan P.R., Satrio J.A., Brown R.C., Shanks B.H. Product distribution from fast pyrolysis of glucose-based carbohydrates. J. Anal. Appl. Pyrol. 2009;86(2):323–330. [Google Scholar]
- Payne C.M. Fungal cellulases. Chem. Rev. 2015;115(3):1308–1448. doi: 10.1021/cr500351c. [DOI] [PubMed] [Google Scholar]
- Poblete-Castro I., Becker J., Dohnt K., Dos Santos V.M., Wittmann C. Industrial biotechnology of Pseudomonas putida and related species. Appl. Microbiol. Biotechnol. 2012;93(6):2279–2290. doi: 10.1007/s00253-012-3928-0. [DOI] [PubMed] [Google Scholar]
- Pollard A., Rover M., Brown R. Characterization of bio-oil recovered as stage fractions with unique chemical and physical properties. J. Anal. Appl. Pyrol. 2012;93:129–138. [Google Scholar]
- Prosen E.M., Radlein D., Piskorz J., Scott D.S., Legge R.L. Microbial utilization of levoglucosan in wood pyrolysate as a carbon and energy source. Biotechnol. Bioeng. 1993;42(4):538–541. doi: 10.1002/bit.260420419. [DOI] [PubMed] [Google Scholar]
- Radlein D.S., Grinshpun A., Piskorz J., Scott D.S. On the presence of anhydro-oligosaccharides in the sirups from the fast pyrolysis of cellulose. J. Anal. Appl. Pyrol. 1987;12(1):39–49. [Google Scholar]
- Rover M.R. Production of clean pyrolytic sugars for fermentation. ChemSusChem. 2014;7(6):1662–1668. doi: 10.1002/cssc.201301259. [DOI] [PubMed] [Google Scholar]
- Rover M.R., Johnston P.A., Whitmer L.E., Smith R.G., Brown R.C. The effect of pyrolysis temperature on recovery of bio-oil as distinctive stage fractions. J. Anal. Appl. Pyrol. 2014;105:262–268. [Google Scholar]
- Salvachúa D., Karp E.M., Nimlos C.T., Vardon D.R., Beckham G.T. Towards lignin consolidated bioprocessing: simultaneous lignin depolymerization and product generation by bacteria. Green Chem. 2015;17(11):4951–4967. [Google Scholar]
- Scott, D. Legge, R. Piskorz, J. Majerski, P. Radlein, D. 1997. Fast pyrolysis of biomass for recovery of specialty chemicals. Developments in Thermochemical Biomass Conversion. Springer, pp. 523–535.
- Shafizadeh F., Stevenson T.T. Saccharification of douglas‐fir wood by a combination of prehydrolysis and pyrolysis. J. Appl. Poly. Sci. 1982;27(12):4577–4585. [Google Scholar]
- Talmadge M.S. A perspective on oxygenated species in the refinery integration of pyrolysis oil. Green Chem. 2014;16(2):407–453. [Google Scholar]
- Tessini C. High performance thin layer chromatography determination of cellobiosan and levoglucosan in bio-oil obtained by fast pyrolysis of sawdust. J. Chromatogr. A. 2011;1218(24):3811–3815. doi: 10.1016/j.chroma.2011.04.037. [DOI] [PubMed] [Google Scholar]
- Teugjas H., Valjamae P. Selecting beta-glucosidases to support cellulases in cellulose saccharification. Biotechnol. Biofuels. 2013;6(1):105. doi: 10.1186/1754-6834-6-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardon D.R. Adipic acid production from lignin. Energy Environ. Sci. 2015;8:617–628. [Google Scholar]
- Wang H., Male J., Wang Y. Recent advances in hydrotreating of pyrolysis bio-oil and its oxygen-containing model compounds. ACS Cataly. 2013;3(5):1047–1070. [Google Scholar]
- Xiu S., Shahbazi A. Bio-oil production and upgrading research: a review. Renew. Sustain. Energy Rev. 2012;16(7):4406–4414. [Google Scholar]
- Yang H., Yan R., Chen H., Lee D.H., Zheng C. Characteristics of hemicellulose, cellulose and lignin pyrolysis. Fuel. 2007;86(12):1781–1788. [Google Scholar]
- Yu Z., Zhang H. Ethanol fermentation of acid-hydrolyzed cellulosic pyrolysate with Saccharomyces cerevisiae. Bioresour. Technol. 2003;90(1):95–100. doi: 10.1016/s0960-8524(03)00093-2. [DOI] [PubMed] [Google Scholar]
- Yu Z., Zhang H. Pretreatments of cellulose pyrolysate for ethanol production by Saccharomyces cerevisiae, Pichia sp. YZ-1 and Zymomonas mobilis. Biomass and Bioenergy. 2003;24(3):257–262. [Google Scholar]
- Zhang Q., Chang J., Wang T., Xu Y. Review of biomass pyrolysis oil properties and upgrading research. Energy Convers. Manag. 2007;48(1):87–92. [Google Scholar]
- Zhuang X., Zhang H. Identification, characterization of levoglucosan kinase, and cloning and expression of levoglucosan kinase cDNA from Aspergillus niger CBX-209 in Escherichia coli. Protein Express. Purif. 2002;26(1):71–81. doi: 10.1016/s1046-5928(02)00501-6. [DOI] [PubMed] [Google Scholar]