Abstract
Glycerol is an abundant by-product during biodiesel production and additionally has several assets compared to sugars when used as a carbon source for growing microorganisms in the context of biotechnological applications. However, most strains of the platform production organism Saccharomyces cerevisiae grow poorly in synthetic glycerol medium. It has been hypothesized that the uptake of glycerol could be a major bottleneck for the utilization of glycerol in S. cerevisiae. This species exclusively relies on an active transport system for glycerol uptake. This work demonstrates that the expression of predicted glycerol facilitators (Fps1 homologues) from superior glycerol-utilizing yeast species such as Pachysolen tannophilus, Komagataella pastoris, Yarrowia lipolytica and Cyberlindnera jadinii significantly improves the growth performance on glycerol of the previously selected glycerol-consuming S. cerevisiae wild-type strain (CBS 6412-13A). The maximum specific growth rate increased from 0.13 up to 0.18 h−1 and a biomass yield coefficient of 0.56 gDW/gglycerol was observed. These results pave the way for exploiting the assets of glycerol in the production of fuels, chemicals and pharmaceuticals based on baker's yeast.
Keywords: Yeast, Saccharomyces cerevisiae, Glycerol, Transport, Glycerol facilitator, Fps1, Stl1
Graphical abstract
Highlights
-
•
S. cerevisiae strains growing in synthetic glycerol medium with μmax up to 0.18 h−1.
-
•
The sole genetic modification was the expression of a predicted glycerol facilitator.
-
•
Facilitated diffusion can completely replace native glycerol/H+ symport.
-
•
Biomass yield coefficients of engineered S. cerevisiae comparable to K. pastoris.
-
•
Exploiting the assets of glycerol as feedstock in S. cerevisiae bioprocesses visible.
1. Introduction
Throughout the last decades the yeast Saccharomyces cerevisiae has become a favorite production organism in industrial biotechnology and a suitable chassis in synthetic biology. Apart from being used for bioethanol production, various genetically engineered strains have been generated to produce a large number of valuable compounds, such as recombinant proteins, lactic and succinic acid, butanols, isoprenoids and polyketides (Borodina and Nielsen, 2014, Hong and Nielsen, 2012, Nevoigt, 2008).
Glycerol is accumulated as a waste product in the biodiesel, oleo-chemical, and bioethanol industries (Aldiguier et al., 2004, Thompson and He, 2006). Apart from its abundance, a main advantage of glycerol as a carbon source for biotechnological applications is that it does not exert the so-called Crabtree effect (Crabtree, 1929). This means that high biomass yields (as well as yields for biomass-associated products such as heterologous proteins) could be achieved without strenuous substrate monitoring strategies which are inevitable in aerated, glucose-based cultures. Controlling the described parameters becomes particularly challenging when the production is carried out in large-scale bioreactors where microorganisms are subjected to fluctuations in concentrations of e.g. substrate and oxygen (Delvigne et al., 2006, Enfors et al., 2001, Fowler and Dunlop, 1989). In addition, the liquid nature of glycerol may reduce dilution effects of the culture broth when used as a carbon source in bioprocesses run in fed-batch mode. In principle, the high degree of reduction can also become an asset of glycerol in the future provided that fermentative production of a small molecule compound is envisaged and glycerol is catabolized via a fermentative rather than the respiratory route. Notably, certain bacteria are able to ferment glycerol under anaerobic conditions (Yazdani and Gonzalez, 2007) and certain wild-type non-Saccharomyces yeast species (Liu et al., 2012) as well as engineered strains (Hong et al., 2010, Kata et al., 2016) have been demonstrated to produce at least some ethanol from glycerol.
In spite of its assets glycerol has hitherto been neglected as a carbon source for industrial bioprocesses employing S. cerevisiae as a production host. This is due to the fact that wild-type S. cerevisiae strains do not exhibit acceptable growth rates in synthetic glycerol medium if no growth-supporting medium supplements (such as amino acids and nucleic bases) are added (Swinnen et al., 2013). Recently, a wild-type S. cerevisiae isolate (CBS 6412) was identified that grows with a maximum specific growth rate (µmax) of ~0.1 h−1 in synthetic glycerol medium (Swinnen et al., 2013). A haploid meiotic segregant of this diploid strain (referred to as CBS 6412-13A) was isolated that shows a µmax of ~0.13 h−1.
Several non-conventional yeast species have been reported to show growth rates on glycerol significantly higher than described for S. cerevisiae. For example, Lages et al. (1999) studied growth on glycerol of 42 different yeast species. Among them Cyberlindnera jadinii (syn.: Candida utilis, Pichia jadinii) showed the highest µmax of 0.32 h−1. Other yeast species, such as Yarrowia lipolytica, Komagataella pastoris (syn.: Pichia pastoris) and Pachysolen tannophilus, have been reported to reach growth rates in the range of 0.26–0.30 h−1 (Liu et al., 2012, Mattanovich et al., 2009, Workman et al., 2013).
The molecular basis for the more efficient glycerol utilization in the above-mentioned yeast species as compared to S. cerevisiae is not clear, but first indications towards glycerol uptake being a major limitation in S. cerevisiae have already been provided by Gancedo et al. (1968). The study demonstrated that glycerol uptake (described as membrane permeability) in the studied S. cerevisiae strain was ~105 fold lower as compared to C. jadinii (syn.: C. utilis).
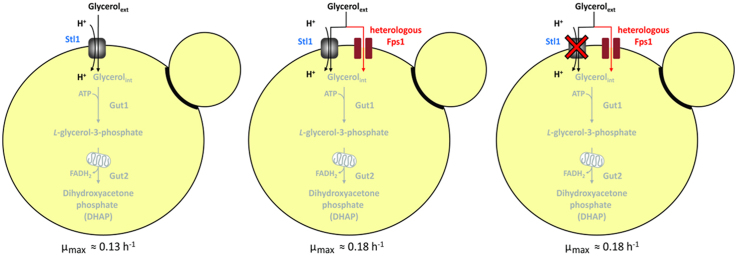
In S. cerevisiae, glycerol is transported into the cell via a glycerol/H+-symporter encoded by STL1 (Ferreira et al., 2005). In addition, S. cerevisiae expresses a glycerol facilitator encoded by FPS1 (Luyten et al., 1995). However, the same authors showed that in contrast to the deletion of STL1, the deletion of FPS1 does neither significantly impair growth on glycerol nor glycerol uptake. Obviously, the glycerol facilitator of S. cerevisiae, if at all, only marginally contributes to glycerol uptake by mediating passive diffusion (Oliveira et al., 2003). Further studies proved that the main function of S. cerevisiae Fps1 is to control the release of glycerol during osmoregulation when cells are growing on glucose (Tamas et al., 1999).
Using a S. cerevisiae strain barely growing on glycerol (µmax=0.02 h−1), it has been previously demonstrated that the overexpression of an Fps1 homologue (encoded by FPS2 and predicted to be a glycerol facilitator) from the P. tannophilus strain CBS 4044 can restore growth on glycerol of the respective S. cerevisiae stl1 deletion mutant (Liu et al., 2013). In contrast, overexpression of the endogenous S. cerevisiae Fps1 from the same strong TEF1 promoter was not able to overcome this growth defect suggesting that the Fps1 homologues from P. tannophilus and S. cerevisiae possess different functions with respect to glycerol import (Liu et al., 2013). As the results indicated that P. tannophilus Fps2 can function as a glycerol uptake system via facilitated diffusion in S. cerevisiae, it was of great interest to test whether this and other glycerol facilitators from superior glycerol-utilizing yeast species are able to further improve the growth on glycerol of the above-mentioned, previously isolated S. cerevisiae wild-type strain CBS 6412-13A. This strain is, in contrast to many laboratory and industrial yeast strains, naturally able to grow in synthetic glycerol medium without any supplements (µmax=0.13 h−1).
2. Materials and methods
2.1. Strains, plasmids, medium composition and culture conditions
All yeast strains and plasmids used in this study are listed in Table S1 (Supplementary data). Yeast cells were cultivated on solid YPD medium containing 10 g L−1 yeast extract, 20 g L−1 peptone, 20 g L−1 glucose, and 15 g L−1 agar. For selection of engineered cells after transformation, the solid medium was supplemented either with G418 (200 mg L−1) or phleomycin (20 mg L−1). All experiments assessing growth in liquid medium containing glycerol in the Growth Profiler 1152 were performed in synthetic medium according to Verduyn et al. (1992) containing 5 g L−1 (NH4)2SO4, 3 g L−1 KH2PO4, 0.5 g L−1 MgSO4·7H2O, 15 mg L−1 EDTA, 4.5 mg L−1 ZnSO4·7H2O, 0.84 mg L−1 MnCl2·2H2O, 0.3 mg L−1 CoCl2·6H2O, 0.3 mg L−1 CuSO4·5H2O, 0.4 mg L−1 NaMoO4·2H2O, 4.5 mg L−1 CaCl2·2H2O, 3 mg L−1 FeSO4·7H2O, 1 mg L−1 H3BO3, and 0.1 mg L−1 KI. After heat sterilization of the medium, filter-sterilized vitamins were added. Final vitamin concentrations were 0.05 mg L−1 d-(+)-biotin, 1 mg L−1 D-pantothenic acid hemicalcium salt, 1 mg L−1 nicotinic acid, 25 mg L−1 myo-inositol, 1 mg L−1 thiamine chloride hydrochloride, 1 mg L−1 pyridoxine hydrochloride, and 0.2 mg L−1 4-aminobenzoic acid. The carbon source added to the medium was either 20 g L−1 glucose or 60 mL L−1 glycerol. The pH of the synthetic glucose and glycerol medium was adjusted to 6.5 with 2 M KOH or 4.0 with 2 M H3PO4, respectively. For growth characterizations in bioreactors, synthetic medium contained 10 g L−1 glycerol as a carbon source and was adjusted to a pH of 5.0. Medium used for pre-cultivations of cells for bioreactor experiments contained 100 mM potassium hydrogen phthalate buffer.
Escherichia coli DH5α was used for plasmid construction and isolation, and cells were routinely cultivated in lysogeny broth (LB) supplemented with 100 mg L−1 ampicillin. Plasmids were isolated by using the QIAprep Spin Miniprep Kit (Qiagen, Hilden, Germany).
2.2. General molecular biology techniques
All primers used within this study are listed in Table S2 (Supplementary data). All PCRs for cloning were performed using Phusion® High-Fidelity DNA Polymerase (New England BioLabs, Frankfurt am Main, Germany) and PCR conditions were adapted to the guidelines of the manufacturer. Restriction enzymes and T4 DNA ligase were obtained from Thermo Fisher Scientific (Waltham, MA, USA) and used according to the manufacturer's instructions. All PCR products for cloning and transformation were purified by using the QIAquick PCR Purification Kit (Qiagen).
2.3. Construction of cassettes for expression of Fps1 homologues in S. cerevisiae
The sequences encoding for the glycerol facilitator homologues from C. jadinii DSM 2361, K. pastoris X-33, P. tannophilus CBS 4044 and Y. lipolytica IBT 446 (referred to as CjFPS1, KpFPS1, PtFPS2 and YlFPS1, respectively) were first amplified by PCR from genomic DNA using the respective primers that included restriction sites for subsequent cloning (Table S2). The sequence of CjFPS1 was derived from the whole genome sequence deposited under the GenBank accession nos. DG000065–DG000077 (Tomita et al., 2012) by identification of conserved regions from various known FPS1 coding sequences. Primers for the amplification of KpFPS1, PtFPS2 and YlFPS1 were designed according to the GenBank accession numbers XM_002494184, JQ481632 and XM_503595, respectively. After purification, the fragments were cloned into the multiple cloning site of the S. cerevisiae expression vector p416TEF (TEF1 promoter and CYC1 terminator) by restriction, ligation and subsequent transformation of E. coli DH5α yielding plasmids p416TEFCjFPS1, p416TEFKpFPS1, p416TEFPtFPS2 and p416TEFYlFPS1.
2.4. Genome modifications in S. cerevisiae
Transformations of expression and deletion cassettes into S. cerevisiae were performed according to the lithium acetate method described by Gietz et al. (1995). Expression cassettes encoding the different glycerol facilitator homologues were PCR-amplified from the respective constructed plasmids (2.3) and integrated into chromosome VII of CBS 6412-13A using the long terminal repeat (LTR) YGLCτ3 as a target site (Flagfeldt et al., 2009). Integration was achieved in two steps. First, two PCR fragments containing (i) a counter selectable, galactose-inducible growth inhibitory sequence (referred to as GALp-GIN11M86) and (ii) the kanMX4 selectable marker were co-transformed and integrated by homologous recombination at the target locus. The GALp-GIN11M86 cassette was PCR-amplified from plasmid pGG119 using primers 155 and 175, while the kanMX4 expression cassette was amplified from plasmid pUG6 using primers 171 and 176. After its integration at the target locus, the entire GALp-GIN11M86/kanMX4 cassette was replaced by the respective expression cassette encoding one of the FPS1 homologues. Templates for amplifying the FPS expression cassettes (including promoter and terminator) were the above-described p416TEF vectors (see Section 2.3). PCR amplification for each cassette was conducted by employing the same primer pair (257 and 258) generating flanking sequences of 60 bp homologous to chromosomal regions upstream and downstream of the previously integrated GALp-GIN11M86/kanMX4 cassette at the ends of the PCR product. Cells bearing the GALp-GIN11M86/kanMX4 cassette were transformed with the resulting PCR products and transformants were selected on solid synthetic medium containing 2% galactose as the sole carbon source in order to induce the growth inhibitory sequence GIN11M86.
The deletion of STL1 was achieved by integration of a disruption cassette carrying the phleomycin resistance gene (ble). The disruption cassette was PCR-amplified from pUG66 using primers 16 and 17. Primers contained 5′ terminal sequences resulting in a PCR product with 40-bp flanking sequences homologous to regions in the promoter and the open reading frame of the STL1 gene, respectively. After purification, the disruption cassette was used to transform the strains CBS 6412-13A CjFPS1 and CBS 6412-13A PtFPS2.
Correct integrations of all expression cassettes integrated at the YGLCτ3 locus as well as STL1 disruption were verified by diagnostic PCRs as described by Swinnen et al. (2013) using TaKaRa Ex Taq Polymerase and buffer according to the manufacturer's guidelines (Merck KGaA, Darmstadt, Germany).
2.5. Quantitative analysis of growth on glycerol in the Growth Profiler 1152
The Growth Profiler 1152 (Enzyscreen, Haarlem, The Netherlands) was used for quantitative analysis of growth on glycerol in white Krystal 24-well clear bottom microplates (Porvair Sciences, Leatherhead, UK) as described by Swinnen et al. (2013). It allowed the determination of culture densities (expressed as green value or G-value) in each single well of the microplate at intervals of 40 min. The obtained G-values were afterwards converted into OD600 values (referred to as OD600 equivalents) using a calibration curve for cell culture ODs between 0.1 and 2.5 with the equation of the best fit line for S. cerevisiae: OD600 equivalent=6.1761×10−8×G-value3.4784, for C. jadinii: 1.6667×10−7×G-value3.3259, for P. tannophilus: 2.4481×10−7×G-value3.2911, for K. pastoris: 2.6407×10−7×G-value3.1582, and for Y. lipolytica: 3.5056×10−7×G-value3.1333. According to these standard curves, the upper limit for a reliable determination of culture densities is approximately at an OD600 value of 2.5.
2.6. Growth characterization in bioreactors
All reactor-based cultivations were carried out in triplicates using Sartorius 1 L bioreactors (Sartorius, Stedim Biotech, Göttingen, Germany) with equivalent working volumes and equipped with two Rushton six-blade disc turbines. The pH electrode (Mettler Toledo, OH, USA) was calibrated according to the manufacturer's standard procedures. The bioreactor was sparged with sterile atmospheric air and off-gas concentrations of oxygen and carbon dioxide were measured with a Prima Pro Process Mass Spectrometer (Thermo Fischer Scientific), calibrated monthly with gas mixtures containing 5% (v/v) CO2, 0.04% (v/v) ethanol and methanol, 1% (v/v) argon, 5% (v/v) and 15% (v/v) oxygen all with nitrogen as carrier gas (Linde Gas, AGA, Enköping, Sweden). The temperature was maintained at 30 °C throughout the cultivation and pH controlled by automatic addition of 2 M NaOH or 2 M H2SO4. Culture conditions were pH 5.0, stirring rate of 800 rpm and air flow of 1 vvm. Inocula were prepared in two steps by sub-culturing each strain in shake flasks with 100 mL of synthetic glycerol medium (2.1) with orbital shaking at 150 rpm and 30 °C. The cells were first cultivated for 5–7 days, after which they were used to inoculate a second set of shake flasks to an OD600 of ~0.1. At mid-exponential phase, ~2–3 days later, these cultures were harvested, washed twice in 0.9% saline and used to inoculate the bioreactors to an OD600 of ~0.1. The strains were cultivated until carbon depletion and the culture had entered stationary phase as monitored by off-gas CO2 concentration (~30 to 45 h). HPLC, OD600 and dry weight (DW) samples were taken during the exponential growth phases for metabolite profiling. HPLC analyses were performed as described by Liu et al. (2013). Standard yeast biomass composition on C-mol basis was assumed to be CH1.8O0.5N0.2 (24.6g DW C-mol−1) for all strains (Villadsen et al., 2011).
3. Results and discussion
3.1. Growth performance of four selected non-conventional yeast species in synthetic glycerol medium
As the growth rates on glycerol reported for the non-Saccharomyces yeast species mentioned in the introduction were obtained from different studies, they might not directly be comparable. For a direct comparison, representative strains of the four above-mentioned yeast species (C. jadinii DSM 2361, K. pastoris X-33, P. tannophilus CBS 4044 and Y. lipolytica IBT 446) and the previously selected S. cerevisiae isolate CBS 6412-13A were tested in synthetic glycerol medium without the addition of any supplements. Growth performances on glycerol were recorded using the Growth Profiler 1152. As expected, the non-conventional yeast species generally showed much better growth on glycerol as compared to the S. cerevisiae strain CBS 6412-13A (Fig. 1) even though the latter has been one of the best S. cerevisiae isolates with regard to the glycerol growth phenotype among 52 isolates tested under the same conditions (Swinnen et al., 2013). The highest µmax of ~0.42 h−1 was observed for C. jadinii DSM 2361 (Table S3, Supplementary data). This growth rate was even remarkably higher than the one previously reported for this species. Y. lipolytica IBT 446 and P. tannophilus CBS 4044 exhibited maximum specific growth rates about twice that of S. cerevisiae CBS 6412-13A (~0.27 and ~0.26 h−1, respectively). The µmax of the K. pastoris strain X-33 (~0.18 h−1) was the lowest among the non-conventional yeast species employed in this study, but still higher than that of S. cerevisiae CBS 6412-13A (~0.13 h−1). Interestingly, the rate for K. pastoris was also found to be lower than reported in the literature (see Section 1). The observed differences may be explained by the different strains and/or conditions such as medium composition and pH used in the various studies. For example, we used a pH of 4 for comparative growth measurements in synthetic glycerol medium since the S. cerevisiae strains tested in our laboratory showed a significantly better growth performance as compared to pH 6, where exponential growth was only observed after a prolonged phase of linearly increasing optical density (unpublished data). Growth analyses of the different species in synthetic glycerol medium at pH 6 revealed that except S. cerevisiae CBS 6412-13A only the strain Y. lipolytica IBT 446 was significantly affected in its µmax by the pH. The latter strain grew even better at pH 6 (Table S3, Supplementary data).
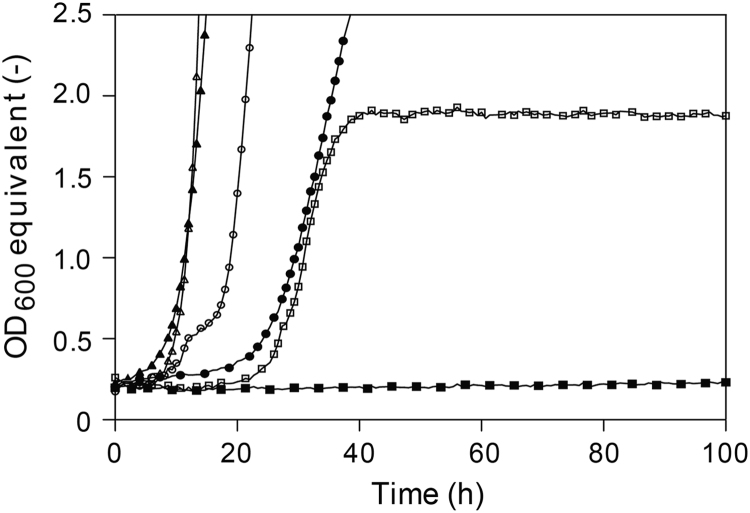
Fig. 1.
Growth performance of four selected yeast species, known for superior glycerol utilization, in synthetic glycerol medium in comparison to the S. cerevisiae strain CBS 6412-13 A. Growth was recorded in synthetic medium containing 6% (v/v) glycerol as the sole carbon source using the Growth Profiler 1152. Pre- and intermediate cultures were prepared in synthetic medium containing 2% (w/v) glucose as the sole carbon source. Growth curves are shown for C. jadinii DSM 2361 (open triangles), P. tannophilus CBS 4044 (closed triangles), Y. lipolytica IBT 446 (open circles), K. pastoris X-33 (open squares), and CBS 6412-13A (closed circles). The laboratory S. cerevisiae strain CEN.PK113-1A not growing on glycerol at all was used as a negative control (closed squares).
It must be generally mentioned that the extensive lag phases shown in Fig. 1, Fig. 2, Fig. 3 and in Table S3 were caused by the fact that cultures were pre-grown in glucose-containing medium. This was necessary for the wild-type S. cerevisiae strain in order to achieve a sufficiently high cell density within a reasonable time period. As expected all lag phases almost completely disappeared if the main culture was directly inoculated with glycerol-pre-grown cells (Fig. S1, Supplementary data).
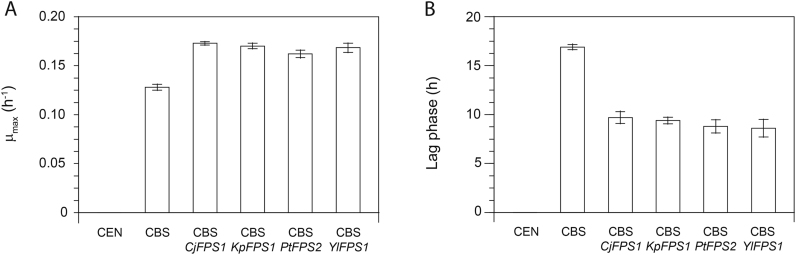
Fig. 2.
Growth performance of the S. cerevisiae wild-type strain CBS 6412-13A (CBS) and engineered derivatives expressing a glycerol facilitator gene from C. jadinii (CBS CjFPS1), K. pastoris (CBS KpFPS1), P. tannophilus (CBS PtFPS2) or Y. lipolytica (CBS YlFPS1) in synthetic medium containing glycerol as the sole carbon source. The expression of all glycerol facilitators was controlled by the S. cerevisiae TEF1 promoter and the CYC1 terminator, and one copy of the respective expression cassette was integrated into the genome. The laboratory S. cerevisiae strain CEN.PK113-1A (CEN) not growing on glycerol at all was used as a negative control. The cells were grown in synthetic medium containing 6% (v/v) glycerol as the sole source of carbon and growth was recorded using the Growth Profiler 1152. For pre- and intermediate cultures synthetic medium with 2% (w/v) glucose was used. After glucose depletion the intermediate cultures were used to inoculate synthetic glycerol medium resulting in a lag phase due to the change of the carbon source. Maximum specific growth rates (µmax) (A) and duration of lag phases after inoculation into synthetic glycerol medium (B) are shown. All mean values and standard deviations for error bar preparation were obtained from at least three biological replicates.
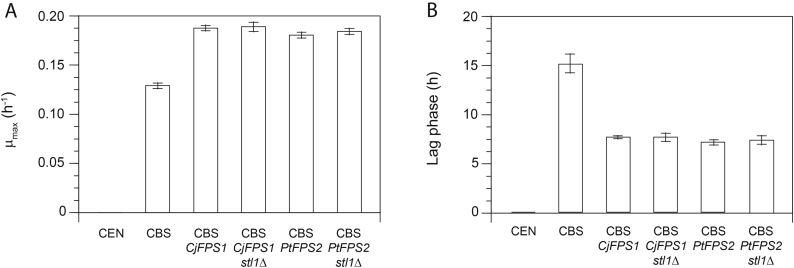
Fig. 3.
Growth performance of stl1 deletion mutants of the S. cerevisiae strain CBS 6412-13A expressing a glycerol facilitator gene from C. jadinii (CBS CjFPS1) or P. tannophilus (CBS PtFPS2) in synthetic glycerol medium. Growth on glycerol was analysed in synthetic medium with 6% (v/v) glycerol using the Growth Profiler 1152. The non-engineered CBS 6412-13A (CBS) and the laboratory strain CEN.PK113-1A (CEN) were used as controls. Pre- and intermediate cultures were performed in synthetic glucose (2% (w/v)) medium and after glucose depletion the intermediate cultures were used to inoculate the synthetic glycerol medium, which led to the observation of lag phases after the switch of the carbon source. Maximum specific growth rates (µmax) (A) and duration of lag phases after inoculation into the synthetic glycerol medium (B) are shown. All mean values as well as standard deviations for the preparation of error bars were obtained from at least three biological replicates.
3.2. Heterologous expression of Fps1 homologues improves glycerol utilization of S. cerevisiae strain CBS 6412-13A
The cassettes for the expression of Fps1 homologues from the above-mentioned four non-Saccharomyces yeast species were constructed and integrated into the genome of the S. cerevisiae strain CBS 6412-13A (see 2.3, 2.4). Notably, all strains expressing a heterologous Fps1 homologue grew significantly better in synthetic glycerol medium compared to the reference strain CBS 6412-13A (Fig. 2). Interestingly, no substantial differences with respect to the origin of the glycerol facilitator were observed. Maximum growth rates of all engineered strains increased by ~30–40% to values between 0.16 and 0.18 h−1 while the duration of the lag phases after cell transfer from synthetic glucose medium to synthetic glycerol medium decreased by almost 50% for all tested strains (Fig. 2). As expected, these lag phases also almost completely abolished when the glucose in the intermediate culture was replaced by glycerol and exponentially growing cells were used for inoculation of the main culture (Fig. S1, Supplementary data).
The µmax achieved here for S. cerevisiae strains on glycerol were impressive even though the mere fact that Fps2 from P. tannophilus can restore growth of a stl1 deletion mutant was known (Liu et al., 2013). It must be noted that the previous results were obtained in a derivative of CEN.PK which is a laboratory strain that is only able to grow in glycerol medium after adding medium supplements such as amino acids and nucleic bases. The expression of the P. tannophilus FPS2 in this strain only improved the growth rate from ~0.02 to ~0.07 h−1 although medium supplements were present. In contrast, the growth rates achieved in the current study by expressing a heterologous glycerol facilitator in the strain CBS 6412-13A were comparable to the one recorded for the K. pastoris strain X-33 (see Section 3.1), a strain that is grown in glycerol medium even for commercial purposes. We therefore believe that our S. cerevisiae strains exhibit growth rates that are already in a range acceptable for industrial applications exploiting glycerol as a carbon source and can also be used as a starting point for further improvements.
In the context of a separate study conducted by our laboratory (Swinnen et al., 2016), the FPS1 from C. jadinii DSM 2361 was overexpressed in the wild-type S. cerevisiae laboratory strain CEN.PK113-1A. This did not result in any quantifiable growth on glycerol. Notably, the same synthetic glycerol medium and conditions were used. The result implies that other factors limit the growth on glycerol in this wild-type strain. Indeed, after transferring the major genetic determinants for the superior growth phenotype of CBS 6412-13A to CEN.PK113-1A by reverse engineering, the overexpression of CjFPS1 exerted its beneficial effect to growth on glycerol and resulted in an increase of µmax from 0.08 to ~0.11 h−1.
3.3. Deletion of STL1 does not impair the superior glycerol growth performance of strains expressing Fps1 homologues
All constructed strains still carry a functional (native) STL1 gene and therefore an active glycerol transport system in addition to the respective heterologous glycerol facilitator. STL1 expression is even known to be strongly induced at the transcriptional level after shifting cells from glucose to glycerol as the carbon source (Ferreira et al., 2005, Roberts and Hudson, 2006). In order to evaluate the contribution of Stl1 to the glycerol uptake of the strains expressing a heterologous glycerol facilitator, the endogenous STL1 gene was exemplarily deleted in strains CBS 6412-13A CjFPS1 and CBS 6412-13A PtFPS2. Surprisingly, deletion of the glycerol/H+-symporter did not exert any effect on the glycerol growth phenotype of the respective strains and the aforementioned high growth rates were retained (Fig. 3). This result implies that each of the heterologous glycerol facilitators was able to fully replace the endogenous active glycerol uptake of S. cerevisiae and even exceeds the glycerol transport capacities of the native Stl1. Notably, the exclusive presence of facilitated diffusion rendered glycerol uptake by the engineered cells energy independent.
Plasmid-based expression of a second copy of the CjFPS1 did not further improve the growth on glycerol of CBS 6412-13A CjFPS1 (data not shown). One can conclude that another metabolic step than substrate uptake became rate-controlling for glycerol utilization capacity in strains expressing a heterologous glycerol facilitator. This information might be useful for subsequent strain improvement approaches aiming at even higher µmax on glycerol.
It should be mentioned that, after achieving the relatively high growth rates on glycerol in our constructed strains, we briefly checked once more the impact of the overexpression of the endogenous FPS1 from S. cerevisiae under the control of the same strong TEF1 promoter (on a CEN/ARS plasmid) in both the CBS 6412-13A wild-type and stl1∆ background. As expected, there was no improvement of growth on glycerol in the first and no growth at all in the second strain (data not shown). This finding is in agreement to previous studies demonstrating that the function of S. cerevisiae Fps1 is to control the release of glycerol after osmotic shock rather than glycerol uptake (Tamas et al., 1999). It has been already discussed by Liu et al. (2013) that the S. cerevisiae Fps1 protein shows a different protein length compared to the Fps2 from P. tannophilus. Our current data confirmed that it is indeed the type of Fps (and not just the simple overexpression) which is crucial for its function as a glycerol uptake system. In fact, the other three selected Fps homologues show a much higher degree of homology to Fps2 from P. tannophilus than to Fps1 from S. cerevisiae.
3.4. Strain characterization in controlled batch cultivations in bioreactors
All aforementioned quantitative data on growth performance in synthetic glycerol medium using the Growth Profiler were obtained from non-controlled batch cultures grown in 24-well plates. While these conditions allow for convenient and precise determination of initial maximal growth rates, they are not suitable for calculating product (biomass and CO2) yields and carbon balances. In fact, the pH rapidly dropped (mainly due to the utilization of (NH4)2SO4 as a nitrogen source) and cell growth ceased long before glycerol was depleted.
To better evaluate product yields, pH-controlled bioreactor experiments with the strains CBS 6412-13A CjFPS1 and CBS 6412-13A PtFPS2 as well as the corresponding stl1 deletion strains were conducted in synthetic glycerol medium. In order to reduce cultivation times, a substrate concentration of only 10 g L−1 of glycerol was used (as compared to 60 mL L−1 used in the Growth Profiler experiments). After carbon source depletion, yield coefficients for both biomass and CO2 were determined (Table 1). The wild-type strain CBS 6412-13A was not able to grow at all under these conditions which confirmed a previous observation that no growth could be observed in synthetic medium with this strain as long as glycerol concentrations ≤20 g L−1 were used (unpublished results). The strains bearing a glycerol facilitator showed yield coefficients for biomass that lay between 0.67 and 0.69 C-mol C-mol−1 (up to 0.56 gDW/gglycerol), which was comparable to yield coefficients previously obtained using K. pastoris (Ghosalkar et al., 2008). The respective yield coefficients for CO2 were between 0.29 and 0.31 C-mol C-mol−1. No additional (fermentative) products could be detected implying that glycerol had been solely converted into biomass and CO2. The maximum specific growth rates of the strains determined based on both optical density and dry weight measurements (between ~0.15 and 0.17 h−1 as shown in Table 1) were slightly lower as compared to our above-mentioned results obtained with the Growth Profiler 1152 in 24-well plates. A very likely explanation is the lower concentration of glycerol in the growth medium used for the bioreactor experiments.
Table 1.
pH-controlled bioreactor experiments for characterizing the growth on glycerol of the S. cerevisiae strain CBS 6412-13A expressing FPS1 homologues from either C. jadinii DSM 2361 or P. tannophilus CBS 4044 in both wild-type and stl1 deletion background. All strains were cultivated in synthetic medium containing 10 g L−1 glycerol as the sole carbon source and pH was controlled at 5.0. Mean values and standard deviations for growth rates and yield coefficients (Ysx: biomass on substrate and Ysc: CO2 on substrate) were obtained from three biological replicates.
| Strain |
Growth rate (h−1) |
Yield coefficients (C-mol C-mol−1) |
||
|---|---|---|---|---|
| OD600 | Dry weight | Ysx | Ysc | |
| CBS 6412-13A CjFPS1 | 0.17±0.01 | 0.17±0.00 | 0.68±0.01 | 0.31±0.03 |
| CBS 6412-13A PtFPS2 | 0.15±0.01 | 0.15±0.01 | 0.69±0.01 | 0.30±0.00 |
| CBS 6412-13A CjFPS1 stl1Δ | 0.15±0.00 | 0.16±0.00 | 0.68±0.02 | 0.29±0.01 |
| CBS 6412-13A PtFPS2 stl1Δ | 0.15±0.01 | 0.16±0.00 | 0.68±0.02 | 0.29±0.01 |
4. Conclusions
This study shows that the sole expression of a single copy of a heterologous glycerol facilitator from yeast species with superior glycerol utilization significantly improves the maximum specific growth rate on glycerol of the moderately growing S. cerevisiae wild-type strain CBS 6412-13A. The fact that the biomass yield coefficient reached levels comparable to the yeast species K. pastoris is very promising with regard to the development of S. cerevisiae-based processes for the production of biomass-related compounds using the respiratory carbon source glycerol and therefore avoiding the Crabtree effect that prevents high biomass yields in sugar-based media. The results obtained in this work might also be a first step towards the use of glycerol's reducing power for the fermentative production of small molecules.
Acknowledgements
This work was funded through the ERA-NET scheme of the 7th EU Framework Program (IPCRES; German Federal Ministry of Education and Research, Project No. 031A344). We thank Johan M. Thevelein (KU Leuven, Belgium) for kindly providing us with the plasmid pGG119, Sebastian Springer (Jacobs University Bremen gGmbH) for providing us with K. pastoris X-33 as well as Solvejg Sevecke, Vedya Daron, Nikola Gyurchev, Joseph Zimba, and Shorai Sandra Jaffet for technical support. We thank Tina Johansen for technical assistance at the Bioreactor Platform, Technical University of Denmark.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.meteno.2016.09.001.
Appendix A. Supplementary material
Supplementary material
.
References
- Aldiguier A.S., Alfenore S., Cameleyre X., Goma G., Uribelarrea J.L., Guillouet S.E., Molina-Jouve C. Synergistic temperature and ethanol effect on Saccharomyces cerevisiae dynamic behaviour in ethanol bio-fuel production. Bioprocess Biosyst. Eng. 2004;26:217–222. doi: 10.1007/s00449-004-0352-6. [DOI] [PubMed] [Google Scholar]
- Borodina I., Nielsen J. Advances in metabolic engineering of yeast Saccharomyces cerevisiae for production of chemicals. Biotechnol. J. 2014;9:609–620. doi: 10.1002/biot.201300445. [DOI] [PubMed] [Google Scholar]
- Crabtree H.G. Observations on the carbohydrate metabolism of tumours. Biochem. J. 1929;23:536–545. doi: 10.1042/bj0230536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvigne F., Lejeune A., Destain J., Thonart P. Stochastic models to study the impact of mixing on a fed-batch culture of Saccharomyces cerevisiae. Biotechnol. Prog. 2006;22:259–269. doi: 10.1021/bp050255m. [DOI] [PubMed] [Google Scholar]
- Enfors S.O., Jahic M., Rozkov A., Xu B., Hecker M., Jurgen B., Kruger E., Schweder T., Hamer G., O’Beirne D., Noisommit-Rizzi N., Reuss M., Boone L., Hewitt C., McFarlane C., Nienow A., Kovacs T., Tragardh C., Fuchs L., Revstedt J., Friberg P.C., Hjertager B., Blomsten G., Skogman H., Hjort S., Hoeks F., Lin H.Y., Neubauer P., van der Lans R., Luyben K., Vrabel P., Manelius A. Physiological responses to mixing in large scale bioreactors. J. Biotechnol. 2001;85:175–185. doi: 10.1016/s0168-1656(00)00365-5. [DOI] [PubMed] [Google Scholar]
- Ferreira C., van Voorst F., Martins A., Neves L., Oliveira R., Kielland-Brandt M.C., Lucas C., Brandt A. A member of the sugar transporter family, Stl1p is the glycerol/H+ symporter in Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16:2068–2076. doi: 10.1091/mbc.E04-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagfeldt D.B., Siewers V., Huang L., Nielsen J. Characterization of chromosomal integration sites for heterologous gene expression in Saccharomyces cerevisiae. Yeast. 2009;26:545–551. doi: 10.1002/yea.1705. [DOI] [PubMed] [Google Scholar]
- Fowler J.D., Dunlop E.H. Effects of reactant heterogeneity and mixing on catabolite repression in cultures of Saccharomyces cerevisiae. Biotechnol. Bioeng. 1989;33:1039–1046. doi: 10.1002/bit.260330813. [DOI] [PubMed] [Google Scholar]
- Gancedo C., Gancedo J.M., Sols A. Glycerol metabolism in yeasts. Pathways of utilization and production. Eur. J. Biochem. 1968;5:165–172. doi: 10.1111/j.1432-1033.1968.tb00353.x. [DOI] [PubMed] [Google Scholar]
- Ghosalkar A., Sahai V., Srivastava A. Optimization of chemically defined medium for recombinant Pichia pastoris for biomass production. Bioresour. Technol. 2008;99:7906–7910. doi: 10.1016/j.biortech.2008.01.059. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl R.H., Willems A.R., Woods R.A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Hong K.K., Nielsen J. Metabolic engineering of Saccharomyces cerevisiae: a key cell factory platform for future biorefineries. Cell. Mol. Life Sci. 2012;69:2671–2690. doi: 10.1007/s00018-012-0945-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W.K., Kim C.H., Heo S.Y., Luo L.H., Oh B.R., Seo J.W. Enhanced production of ethanol from glycerol by engineered Hansenula polymorpha expressing pyruvate decarboxylase and aldehyde dehydrogenase genes from Zymomonas mobilis. Biotechnol. Lett. 2010;32:1077–1082. doi: 10.1007/s10529-010-0259-z. [DOI] [PubMed] [Google Scholar]
- Kata I., Semkiv M.V., Ruchala J., Dmytruk K.V., Sibirny A.A. Overexpression of the genes PDC1 and ADH1 activates glycerol conversion to ethanol in the thermotolerant yeast Ogataea (Hansenula) polymorpha. Yeast. 2016 doi: 10.1002/yea.3175. [DOI] [PubMed] [Google Scholar]
- Lages F., Silva-Graca M., Lucas C. Active glycerol uptake is a mechanism underlying halotolerance in yeasts: a study of 42 species. Microbiology. 1999;145:2577–2585. doi: 10.1099/00221287-145-9-2577. [DOI] [PubMed] [Google Scholar]
- Liu X., Jensen P.R., Workman M. Bioconversion of crude glycerol feedstocks into ethanol by Pachysolen tannophilus. Bioresour. Technol. 2012;104:579–586. doi: 10.1016/j.biortech.2011.10.065. [DOI] [PubMed] [Google Scholar]
- Liu X., Mortensen U.H., Workman M. Expression and functional studies of genes involved in transport and metabolism of glycerol in Pachysolen tannophilus. Micro. Cell Fact. 2013;12:27–36. doi: 10.1186/1475-2859-12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luyten K., Albertyn J., Skibbe W.F., Prior B.A., Ramos J., Thevelein J.M., Hohmann S. Fps1, a yeast member of the MIP family of channel proteins, is a facilitator for glycerol uptake and efflux and is inactive under osmotic stress. EMBO J. 1995;14:1360–1371. doi: 10.1002/j.1460-2075.1995.tb07122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattanovich D., Graf A., Stadlmann J., Dragosits M., Redl A., Maurer M., Kleinheinz M., Sauer M., Altmann F., Gasser B. Genome, secretome and glucose transport highlight unique features of the protein production host Pichia pastoris. Micro. Cell Fact. 2009;8:29–41. doi: 10.1186/1475-2859-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevoigt E. Progress in metabolic engineering of Saccharomyces cerevisiae. Microbiol Mol. Biol. Rev. 2008;72:379–412. doi: 10.1128/MMBR.00025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira R., Lages F., Silva-Graca M., Lucas C. Fps1p channel is the mediator of the major part of glycerol passive diffusion in Saccharomyces cerevisiae: artefacts and re-definitions. Biochim. Biophys. Acta. 2003;1613:57–71. doi: 10.1016/s0005-2736(03)00138-x. [DOI] [PubMed] [Google Scholar]
- Roberts G.G., Hudson A.P. Transcriptome profiling of Saccharomyces cerevisiae during a transition from fermentative to glycerol-based respiratory growth reveals extensive metabolic and structural remodeling. Mol. Genet. Genom. 2006;276:170–186. doi: 10.1007/s00438-006-0133-9. [DOI] [PubMed] [Google Scholar]
- Swinnen S., Ho P.W., Klein M., Nevoigt E. Genetic determinants for enhanced glycerol growth of Saccharomyces cerevisiae. Metab. Eng. 2016;36:68–79. doi: 10.1016/j.ymben.2016.03.003. [DOI] [PubMed] [Google Scholar]
- Swinnen S., Klein M., Carrillo M., McInnes J., Nguyen H.T., Nevoigt E. Re-evaluation of glycerol utilization in Saccharomyces cerevisiae: characterization of an isolate that grows on glycerol without supporting supplements. Biotechnol. Biofuels. 2013;6:157–168. doi: 10.1186/1754-6834-6-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamas M.J., Luyten K., Sutherland F.C., Hernandez A., Albertyn J., Valadi H., Li H., Prior B.A., Kilian S.G., Ramos J., Gustafsson L., Thevelein J.M., Hohmann S. Fps1p controls the accumulation and release of the compatible solute glycerol in yeast osmoregulation. Mol. Microbiol. 1999;31:1087–1104. doi: 10.1046/j.1365-2958.1999.01248.x. [DOI] [PubMed] [Google Scholar]
- Thompson J.C., He B.B. Characterization of crude glycerol from biodiesel production from multiple feedstocks. Appl. Eng. Agric. 2006;22:261–265. [Google Scholar]
- Tomita Y., Ikeo K., Tamakawa H., Gojobori T., Ikushima S. Genome and transcriptome analysis of the food-yeast Candida utilis. PLoS One. 2012;7:e37226. doi: 10.1371/journal.pone.0037226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verduyn C., Postma E., Scheffers W.A., Van Dijken J.P. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- Villadsen J., Nielsen J., Liden G. Third edition. Bioreaction Engineering Principles; 2011. Bioreaction Engineering Principles; pp. 1–561. [Google Scholar]
- Workman M., Holt P., Thykaer J. Comparing cellular performance of Yarrowia lipolytica during growth on glucose and glycerol in submerged cultivations. AMB Express. 2013;3:58–66. doi: 10.1186/2191-0855-3-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdani S.S., Gonzalez R. Anaerobic fermentation of glycerol: a path to economic viability for the biofuels industry. Curr. Opin. Biotechnol. 2007;18:213–219. doi: 10.1016/j.copbio.2007.05.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material